Abstract
Simultaneous visualization of the teeth and periodontium is of significant clinical interest for image-based monitoring of periodontal health. We recently reported the application of a dual-modality photoacoustic-ultrasound (PA-US) imaging system for resolving periodontal anatomy and periodontal pocket depths in humans. This work utilized a linear array transducer attached to a stepper motor to generate 3D images via maximum intensity projection. This prior work also used a medical head immobilizer to reduce artifacts during volume rendering caused by motion from the subject (e.g., breathing, minor head movements). However, this solution does not completely eliminate motion artifacts while also complicating the imaging procedure and causing patient discomfort. To address this issue, we report the implementation of an image registration technique to correctly align B-mode PA-US images and generate artifact-free 2D cross-sections. Application of the deshaking technique to PA phantoms revealed 80% similarity to the ground truth when shaking was intentionally applied during stepper motor scans. Images from handheld sweeps could also be deshaken using an LED PA-US scanner. In ex vivo porcine mandibles, pigmentation of the enamel was well-estimated within 0.1 mm error. The pocket depth measured in a healthy human subject was also in good agreement with our prior study. This report demonstrates that a modality-independent registration technique can be applied to clinically relevant PA-US scans of the periodontium to reduce operator burden of skill and subject discomfort while showing potential for handheld clinical periodontal imaging.
1. Introduction
Periodontitis affects nearly 50% of Americans exerting systemic effects on the body [1]. Chronic inflammation from periodontitis has been implicated as a risk factor for cardiovascular diseases [2–4], cancer [5], and dementia [6]. Thus, tools to diagnose periodontal disease at an early stage and the development of new measurement techniques are urgently needed [7].
Ultrasound (US) imaging systems with frequencies ≤20 MHz have previously been used to image facial crestal bone or the cementoenamel junction but lack spatial resolution and contrast [8,9]. The integration of optical excitation via photoacoustic imaging significantly increases the potential applications in oral health [10]. We recently utilized a dual-modality photoacoustic-ultrasound (PA-US) system to non-invasively image the pocket depth and geometry [11,12]. This approach offered 0.01 mm precision and provided a full contour of the pocket. Even though promising results were obtained, larger-scale clinical translation of this approach is limited by shaking artifacts caused by motion from the subject and operator. In prior work, a medical head immobilizer was used to stabilize the subject, but this failed to completely eliminate motion artifacts while also causing some discomfort to subjects [11,12].
To address these issues, we implement here an image registration technique for deshaking periodontal PA-US images. Image registration methods try to align two or more images of a specific scene. In general, there are two methods for registration: interactive and automated [13]. In the interactive method, a set of landmarks is manually selected and used to estimate the transformation models between the two images. This method requires an experienced and accurate operator. The work can be tedious, repetitive, and time-consuming. Consequently, automated registration methods are essential, and various methods have been proposed in this field [14] including the Harris-Laplacian, scale invariant feature transform (SIFT), speeded up robust features (SURF), maximally stable extremal regions (MSER), and modality independent neighborhood descriptor (MIND).
Automated methods typically have at least three steps: feature detection, feature matching, and model estimation [15]. Most of the differences in image registration algorithms are related to feature detection. The Harris-Laplacian algorithm looks for corners as features [16]. It is invariant to scale changes and rotation. The SIFT algorithm extracts key points through a pyramid structure that is invariant to scale, rotation, and brightness [17]. However, the number of these points is undesirable, and it takes a long time to create the vector describing the features. SURF is also invariant to scale and rotation, and its performance is acceptable in terms of speed [16]. MSER uses regional features and is independent of geometric and radiometric changes [18]. MIND [19] is based on a concept called self-similarity. It can be used for linear and deformable registration processes. This algorithm uses edges, corner points, and texture for features. Of these algorithms, we applied MIND for deshaking because it provides more sensitivity to the structural information of images [19].
In this paper, we report on the results obtained with a stepper-motor and handheld PA-US for noninvasive profiling of dental and periodontal anatomy. Our approach uses an image registration method based on MIND to correct for shaking artifacts produced by irregular hand movements during handheld scanning and subject motion during motor-based scanning. Phantom, ex vivo, and human data were collected and compared for qualitative and quantitative evaluation. Our results demonstrate that shaking artifacts from 3D PA-US images of the periodontal anatomy, enamel pigmentation, and pocket depth can be algorithmically removed to allow for accurate measurement and visualization of periodontal features in clinically relevant imaging scenarios.
2. Materials and methods
2.1. Experimental study
2.1.1. Equipment and imaging setup
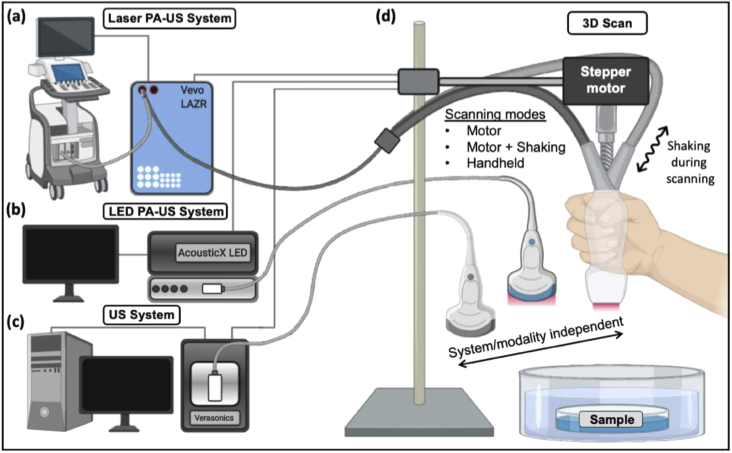
This study used three imaging systems. Two commercial photoacoustic-ultrasound (PA-US) systems both capable of simultaneous ultrasound and photoacoustic data acquisition were employed: a laser-integrated, high-frequency system [Fig. 1(a), Vevo LAZR, Visualsonics] and an LED-based system [Fig. 1(b), AcousticX, Cyberdyne Inc.]. The Vevo system was used with a linear array transducer: LZ-550 (Fc = 40 MHz, FOV = 10 × 14 mm (d × w)) and a Q-switched Nd:YAG laser (4-6 ns pulsewidth) followed by an optical parametric oscillator (OPO) laser operating from 680–970 nm. The LZ-550 was used for swine and human mandible imaging. The framerate was 5 Hz. The position of the transducer was controlled with a stepper motor (scan distance = 17 mm, step size = 0.054 mm). The AcousticX system utilized a 128-element linear array transducer (Fc = 10 MHz, FOV = 4 cm × 3.5 cm (d × w)) coupled with two 690-nm LED linear arrays. The LED repetition rate was 4K Hz and the framerate was 30 Hz. The scan distance for phantoms was 25 mm and 40 mm for the forearm epidermis imaging (step size = 0.081 mm). We also used a commercial ultrasound data acquisition system named Vantage (Verasonics, Inc.) for ultrasound imaging of the depth phantom and swine jaw [Fig. 1(c)]. This system can collect signal from 256 channels simultaneously with a sampling frequency of 62.5 MHz— here, we used a Verasonics L22-14 vX linear array transducer (20.2 MHz, 128 elements). The FOV was 12.8 × 19.12 mm (d × w), with a frame rate of 11 Hz.
Fig. 1.
Photoacoustic-ultrasound systems and experimental setup. (a) A laser-integrated photoacoustic-ultrasound system (Vevo LAZR, VisualSonics Inc.), an (b) LED-integrated photoacoustic ultrasound system (AcousticX, Cyberdyne Inc.), or (c) an ultrasound-only system (Vantage, Verasonics, Inc.) was used to collect 3D images by scanning each system’s linear array transducer via stepper motor and by hand. (d) Samples were first laterally scanned by a stepper motor as commonly performed. To induce motion artifacts in the motor-based imaging, the scan was repeated but the motor was perturbed by hand with lateral force at various intensities mimicking an imaging setup with imperfect subject-scanner immobilization. Samples were then manually scanned by hand to mimic the most convenient clinical imaging scenario.
The goal of this study was to validate the correction of motion artifacts for PA-US imaging conditions in which a common stepper motor was used or 3D sweeping was performed completely by hand. Freehand scanning is common and requires shaking corrections as does motor-based scanning when subject immobilization is imperfect [12]. Thus, we experimentally induced shaking motion artifacts by manually perturbing the stepper motor with periodic lateral force as the motor advanced [Fig. 1(d)]; this also caused shaking in the elevation and axial directions. This was done with varying intensities while maintaining similar periodicity to evaluate the tolerance of the algorithm to shaking amplitude. We also performed fully handheld scanning in which the operator’s elbow was stabilized but was otherwise subject to imperfect motion (i.e. not strictly 1-dimensional). The operator mimicked the scanning speed of the motor during imaging.
2.1.2. Phantom and ex vivo sample preparation
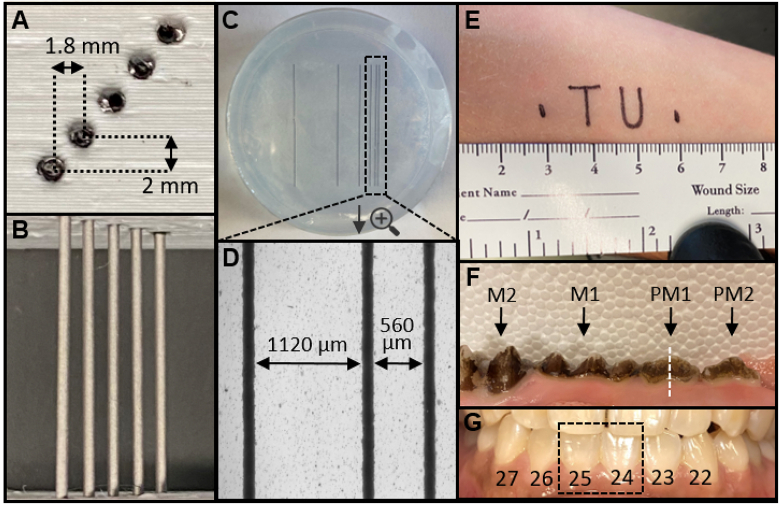
A photoacoustic depth phantom was prepared by 3D-printing (i3 MK3S, Prusa Research) a sample holder from polylactic acid (PLA) with holes for cylindrical samples at increasing depths [Fig. 2(a), 2(b), depth spacing: 2 mm, lateral spacing: 1.8 mm]. The holes were filled with 0.8 mm graphite cylinders. A photoacoustic lateral resolution phantom was prepared by first inkjet printing six parallel lines (130 µm thickness) at increasing spacing on optically transparent film [Fig. 2(c)]. The spacing distance [see Fig. 2(d)] was doubled between each sequential line (i.e., d1 = 560 µm, d2 = 1120 µm, d3 = 2240 µm, d4 = 4480 µm, and d5 = 8960 µm). This film was then embedded in 1% (w/v) agarose gel in a 5.5-cm petri dish [Fig. 2(c), 2(d)]. The phantoms were immersed in a water bath atop a height-adjustable stage.
Fig. 2.
Imaging phantoms and targets. (a) Photographic cross-section of depth phantom 3D-printed from PLA. The holes are 1-mm diameter and filled with 0.8 mm graphite pencil lead. The lateral spacing is 1.8 mm and the axial spacing is 2 mm. (b) Top-down photograph of depth phantom. (c) Top-down photograph of lateral resolution phantom, consisting of inkjet-printed lines on transparent film embedded in 1% agar within a 5.5-cm petri dish. The spacing between each line (width = 130 µm) doubles sequentially and was measured with brightfield microscopy. From right to left, d1 = 560 µm, d2 = 1120 µm, d3 = 2240 µm, d4 = 4480 µm, d5 = 8960 µm. (d) Spacing between the right-most (closest) lines is shown with brightfield microscopy. (e) Text printed with permanent marker on the supinated forearm to evaluate algorithm performance for a non-linear target. The total feature length was 3 cm. (f) Photograph of the ex vivo swine maxilla with teeth labeled: M2 (2nd molar), M1 (1st molar), PM1 (1st premolar), PM2 (2nd premolar). The white dashed line indicates the sagittal imaging plane generated from transverse B-mode images. (g) Photograph of the teeth and gingiva imaged in a healthy human subject labeled with the universal numbering system—the dashed box shows teeth imaged for deshaking.
Fresh swine jaws were acquired from an abattoir and prepared as previously described [11]. Briefly, the porcine head was sliced sagittally and the mandible was separated from the maxilla with a saw. The teeth and periodontal tissues were used as provided and immersed in a water bath for imaging [Fig. 2(f)].
2.1.3. Human periodontal and epidermal imaging
The study enrolled one male and one female healthy adult. These subjects provided written informed consent and all work was conducted with approval from the UCSD Institutional Review Board and was in accordance with the ethical guidelines for human subject research set forth by the Helsinki Declaration of 1975. To evaluate a non-linear imaging target, a fine-tip permanent marker was used to inscribe the text “TU” on the supinated forearm of the male subject [Fig. 2(e)]; this is called the “TU experiment” throughout the paper. The forearm was immersed in a water bath and imaging was performed as described above.
PA-US images of the periodontium were collected from a healthy female subject as previously described [12]. Briefly, the subject was seated in front of the laser-based PA-US system and the subject’s head was immobilized upon a chin-level platform using a medical grade head immobilizer. The 40-MHz transducer was positioned at the gingiva perpendicular to the long axes of the central mandibular incisors. Sterile US gel was used for coupling. The stepper motor was then initiated to collect frames spanning the gingiva to the apical edge of teeth 24 and 25 (universal numbering system [see Fig. 2(g)]).
2.2. Image registration
In this paper, the reconstructed images are deshaken by the method introduced in [19] where computed tomography (CT) and magnetic resonance images MRI images of lungs were registered. It is based on the self-similarity concept used for noise removal [20]. The noise removal equation is as follows:
| (1) |
where is the weight of each pixel (voxel in 3D) and the criterion for self-similarity, v is the is the noisy image, and is the denoised image [19]. First, a descriptor insensitive to the imaging modality (e.g., CT and MRI) and noise are defined, as follows:
| (2) |
Here, we need to consider a search area of R in which the distance between the patch (i.e., sub-image) I. centered at x and another patch centered at is denoted by . Term n is a constant coefficient to normalize the equation, V. is the variance of the patch, and is the modality independent neighborhood descriptor used for registration.
After finding the descriptor for each pixel of the images, the similarity term for each pixel in both the patches ( and ) are calculated as follows:
| (3) |
The metric S can be adapted in any registration algorithm. To obtain a better convergence, the Gauss–Newton optimization technique was used in this study. Readers are referred to [19,20] for more information. We refer to the deshaking technique by MIND through the rest of the paper.
3. Results
3.1. Phantom experiments
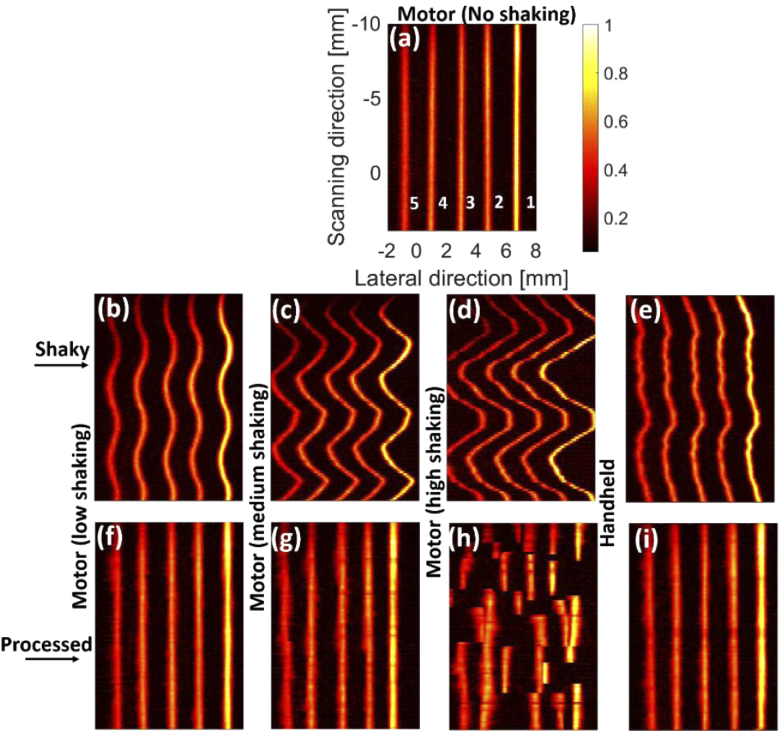
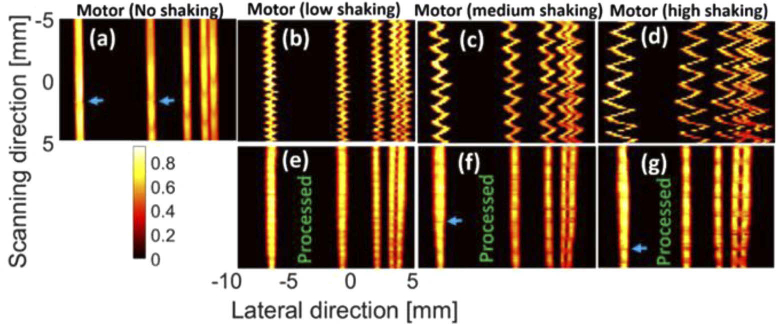
The maximum intensity projections (MIPs) of the reconstructed images using the depth phantom [see Figs. 2(a), 2(b)] are shown in Fig. 3. The intensity decreases from the target 1 to 5 [see Fig. 3(a)] due to a lower laser fluence in depth. The effects of the shaking caused by the tapping (in three levels) and movement of the hand can be seen in Figs. 3(b)–3(e); the peak-to-peak distance (PPD) of the most intense target in Figs. 3(b)–3(d) is 0.7 mm, 1.9 mm, and 3.9 mm, respectively. MIND fails to correct the shaking in Fig. 3(h) because the level of shaking is too high [see Fig. 3(d)]. For the other two levels and also the handheld sweeping, MIND compensates for the shaking and leads to an image visually close to the ground truth [Fig. 3(a)]. We expect no shaking artifacts in the ground truth because the phantom is stable and the minor shaking of the motor in such an ideal condition can be ignored.
Fig. 3.
The photoacoustic maximum intensity projection (MIP) of the depth phantom generated by (a-d) motor with no, low, medium, and high shaking, respectively, as well as (e) hand sweeping. (e-i) The processed (i.e., deshaken) MIPs of (b-e), respectively. The processing was performed on the B-mode images and the MIPs were then generated. The LED-based imaging system was used for data acquisition.
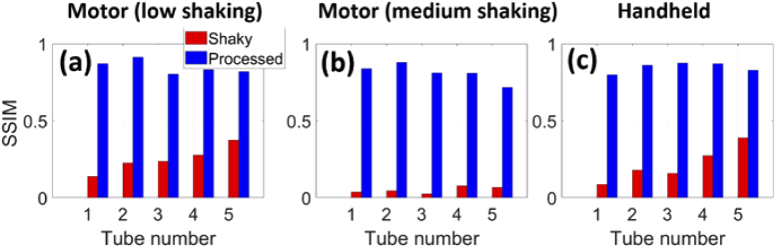
The structural similarity index (SSIM) was used to quantitatively evaluate the performance of MIND [21]. The Structural Similarity Index (SSIM) is a metric for evaluating the similarity between two images. It works structurally and does not perform any point-to-point comparison. The maximum value of this index is 1 and occurs when the two images are exactly the same. This metric is available by the ssim command in Matlab. The calculated SSIMs for all the tubes in Fig. 4(a) are higher than those in Fig. 4(b) due to the fact that the level of shaking in Fig. 3(b) is lower than that of Fig. 3(c). The MIND corrects the movements and results in a SSIM of about 0.8 in average for both the shaking levels and handheld scenario [see Fig. 4(a)-(c)]. The high shaking image presented in Fig. 3(d) was not evaluated because MIND failed to correct the shakiness [Fig. 3(h)]. We also evaluated the contrast in Fig. 3. The ratio of brightness between a region on the most intense line and the background is 15.3 for both the shaky and processed MIP images.
Fig. 4.
The structural similarity index (SSIM) for measuring the image quality for (a,b) motor with low and medium shaking, respectively, and (c) handheld sweeping. The results obtained with the depth phantom [presented in Fig. 1] were used.
We conducted the same experiment with the Verasonics system, and the results are presented in Fig. 5. Even though a higher frequency probe was used for imaging, MIND could correct the shaky images [see Fig. 5(b), 5(c)] with an average SSIM of 0.83.
Fig. 5.
The ultrasound maximum intensity projection (MIP) of the depth phantom generated by (a-c) motor with and without the shaking and hand sweeping, respectively. (d-e) The processed MIPs of (b,c), respectively. The processing was performed on the B-mode images and the MIPs were then generated. The Verasonics imaging system was used for data acquisition.
The MIPs of the lateral resolution phantom [see Fig. 2(c), 2(d)] are presented in Fig. 6. Even though we used low, medium and high level of shaking in both Fig. 3 and Fig. 6, it does not mean that the applied forces to the motor were the same in these figures. The PPD caused by the shaking is about 1 mm, 1.7 mm and 2.4 mm for Fig. 6(b)–6(d), respectively. The first two lines from right side [see Fig. 2(d)] were not detected in the MIP images due to the resolution of the imaging system. The intensity of all the targets is the same due to the same laser fluence incident on the targets positioned at the same depth. Shaking causes the tubes to spatially mix together [see Fig. 6(b)–6(d); more specifically, the first two lines in the right side]. However, MIND still compensates for the motion artifacts and leads to MIPs [see Fig. 6(e)–6(g)] that are structurally close to the ground truth [Fig. 6(a)]. It should be noted that the even though a high shaking was applied in Fig. 6(d), its PPD is still 1.5 mm lower than Fig. 3(d), which is why a reasonable image is obtained in Fig. 6(g). Even though the structure of the lines are preserved with MIND, there are some discontinuities [see the blue arrows] in the deshaken images [also visible in Fig. 6(a) at the scanning distance of 2 mm]. These issues could be due to the combined effects of the step motor shaking, inhomogeneities in the fluence, and lower performance of the MIND on those regions.
Fig. 6.
The photoacoustic maximum intensity projection (MIP) of the lateral resolution phantom generated by (a-d) motor with no, low, medium, and high shaking, respectively. (e-g) The processed MIPs of (b-d), respectively. The processing was performed on the B-mode images and then the MIPs were generated. The LED-based imaging system was used for data acquisition.
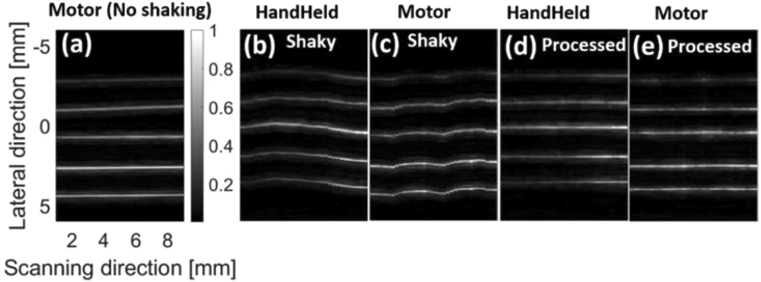
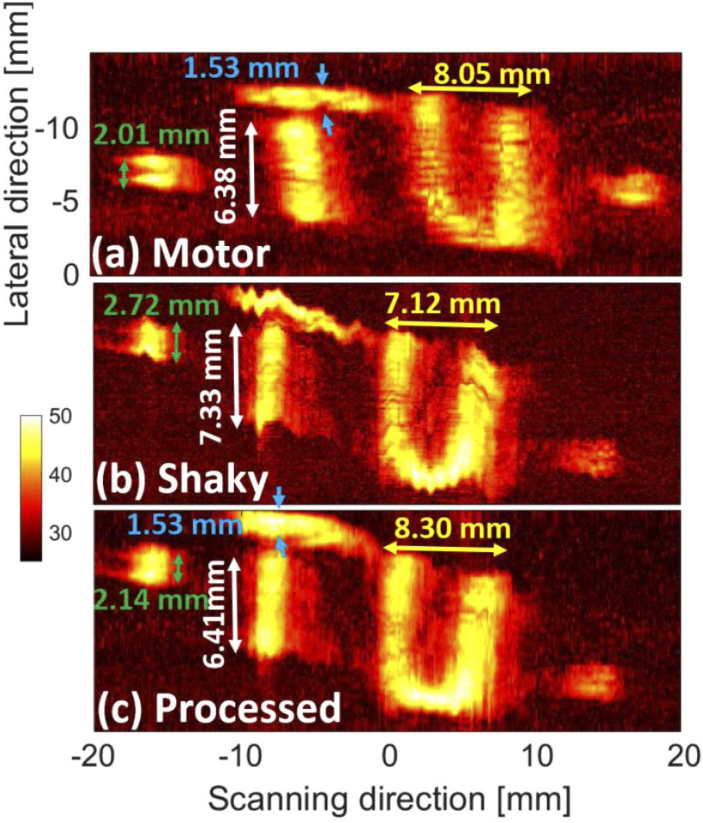
The MIPs of the TU experiment are provided in Fig. 7 where the shaking reduces the image quality in Fig. 7(b), but the deshaken image [Fig. 7(c)] has a quality comparable with Fig. 7(a) where no shaking was applied. Figure 7(b) shows that the shaking causes up to 1 mm error in the lateral and scanning directions. The lateral size of some structures [such as what is shown with the blue arrows in Fig. 7(a)] are not even measurable in Fig. 7(b). MIND reduces the error to about 0.1 mm and 0.25 mm in the lateral and scanning directions, respectively [compare Fig. 7(a) and (c)]. Figure 7(a) is slightly tilted due to the movement and relative direction of handheld scanning in our experiment. This effect becomes more noticeable after deshaking in Figs. 7(b),7(c).
Fig. 7.
The photoacoustic maximum intensity projection (MIP) of the TU experiment generated by (a) motor (no shaking), (b) handheld without processing, and (c) handheld with processing. The processing was performed on the B-mode images and then the MIPs were generated. The LED-based imaging system was used for data acquisition.
3.2. Ex vivo swine experiments
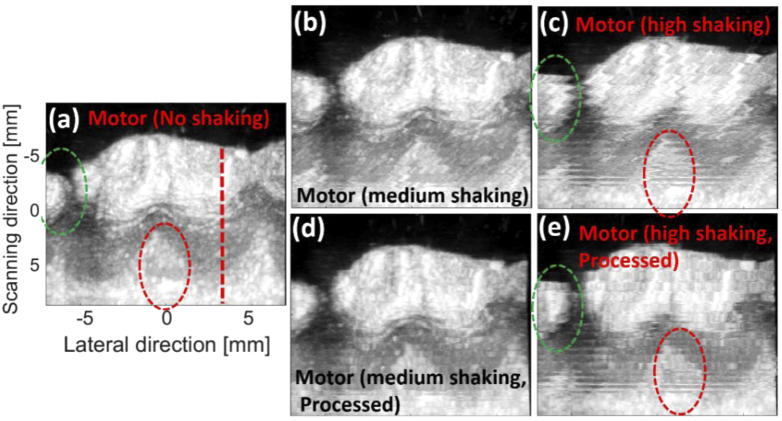
The MIPs of the ultrasound images generated by the ex vivo swine experiments are provided in Fig. 8. The effects of motor shaking are visible in Fig. 8(b), 8(c). MIND provides better and more accurate structural information (compare the green and red dashed-ovals). Table 1 indicates that SSIM of the processed MIP images are higher than the shaky ones (the mean SSIM for the regions indicated by the green and red dashed circles are presented).
Fig. 8.
The ultrasound maximum intensity projection (MIP) of the ex vivo experiments generated by (a-c) motor with no, medium, and high shaking, respectively. (d,e) The processed MIPs of (b,c), respectively. The processing was performed on the B-mode images, and the MIPs were then generated. A Vevo imaging system was used for data acquisition.
Table 1. SSIM measurement for the shaky and processed images with three levels of shaking [presented in Fig. 8].
| SSIM (%) | ||
|---|---|---|
| Motion artifact level | Shaky | Processed |
| Medium | 19 | 50 |
| High | 15 | 47 |
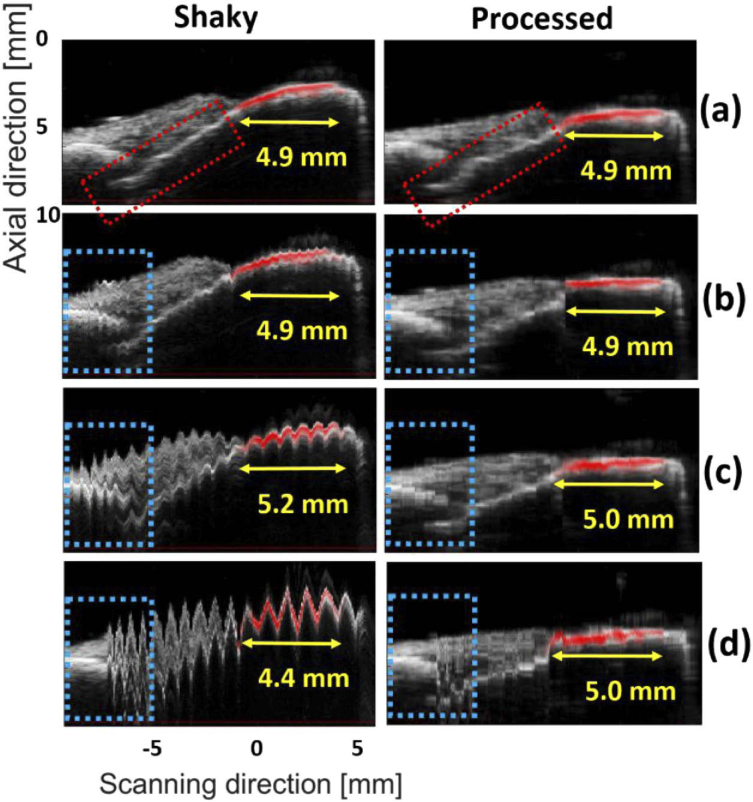
To better understand the improvements, the sagittal planes of the MIP images [the red dashed-line in Fig. 8(a)] are presented in Fig. 9 where the extent of enamel staining is measured by the photoacoustic imaging modality. The gray and red colormaps indicate the ultrasound and photoacoustic images, respectively. The image generated with the motor without shaking [Fig. 9(a)] was given to the MIND to evaluate the bias of the deshaking method. In the deshaken version [Fig. 9(a), second column], the image is smooth compared to the Fig. 9(a), first column. However, the calculus depth is estimated at 4.9 mm in both images, which proves the accuracy of our deshaking method.
Fig. 9.
The sagittal view of the images generated for the ex vivo experiment: (a-d) motor with no, low, medium and high shaking, respectively. The right-hand side images show the processed (i.e., deshaken) MIPs. The yellow arrows show the extent of the staining. The blue and red dotted-boxes can be used for comparison of the structural information. The US and PA images are shown in gray and hot colormaps, respectively.
For the low-shaking dataset, there is no difference in the calculus depth measured by the two images [see Fig. 9(b)]. In the medium and high shaking levels, the calculus depth is over and under-estimated by about 0.3 mm and 0.5 mm, respectively, but the MIND lowers the measurement error to 0.1 mm. For all levels of shaking, better structural information is obtained with MIND [compare the dotted-boxes in the left and right side of Fig. 9].
3.3. In vivo human experiments
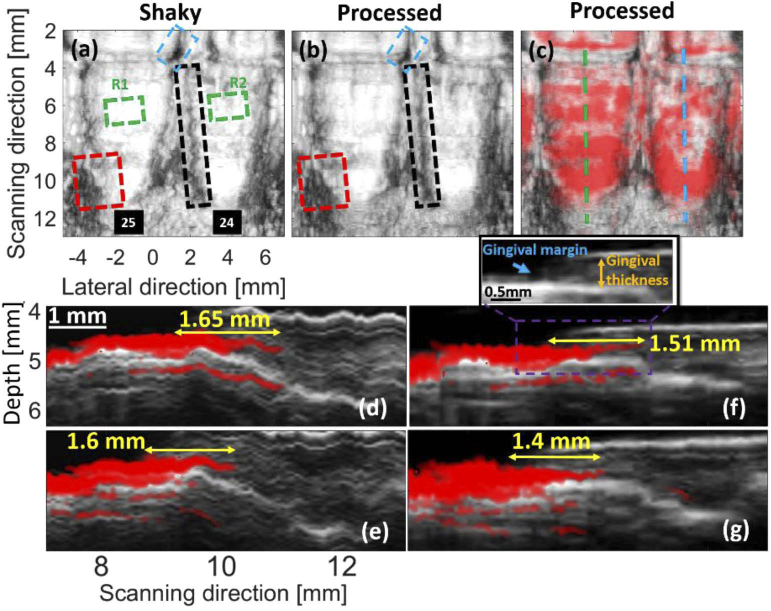
The results obtained for the in vivo experiment are presented in Fig. 10. In Fig. 10(a), no intentional motion artifact was applied. However, we expect to see motion artifacts [the dashed boxes in Fig. 10(a)] due to the movement of the subject (e.g., breathing and minor head movements). These artifacts are addressed in Fig. 10(b) with MIND [compare the boxes in Fig. 10(a) and Fig. 10(b)].
Fig. 10.
The results of the in vivo experiment. Target sweeping was conducted with a motor. (a) The MIP generated with multiple shaky US B-mode images. (b) Processed (i.e., deshaken) version of (a). (c) The overlapped processed US-PA MIPs. The sagittal cross section shown with the blue and green dashed-lines in (c) are presented in (d,e) and (f,g), respectively, where (e,g) are the processed version of (d,f), respectively. The blue, black and red dashed-boxes can be used for comparison of the structural information. The green box is used for statistical brightness analysis (R stands for region). The yellow arrows indicate the pocket depth. The US and PA images are shown in gray and hot colormaps, respectively. The Vevo imaging system was used here for data acquisition.
US and PA imaging modalities were used to detect the gingival margin and pocket depth, respectively [see Fig. 10(f) and its zoomed version]. Figure 10(d), 10(e) shows the sagittal cross-section indicated with the blue and green dashed-lines in Fig. 10(c), respectively, where the shaking reduces the image quality. Figure 10(f), 10(g) shows sagittal cross-section of the processed MIPs where the images are deshaken, and the pocket depth is well estimated in agreement with our prior study [12].
Figure 11 shows the statistical brightness analysis conducted on R1 and R2 [see Fig. 10(a)]; R, P and S stand for region, processed and shaky, respectively. The mean brightness only reduces for about 3%, which could be due to displacement of pixels within the regions. The proposed deshaking method does not modify the brightness of pixels and only uses brightness as one of the features to look for similarities.
Fig. 11.
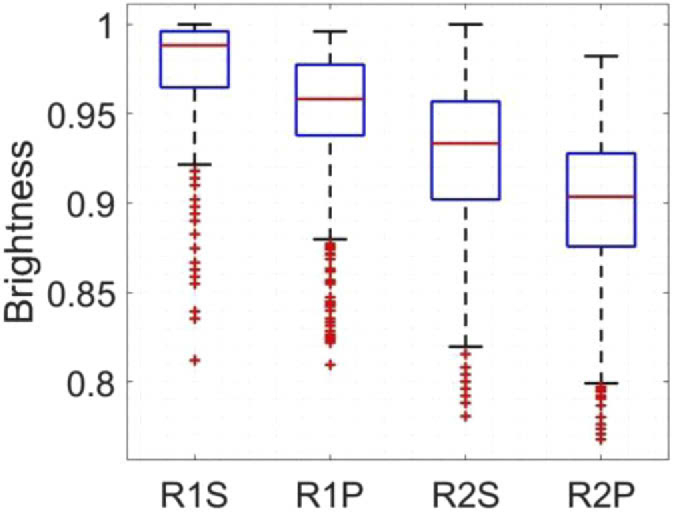
The statistical brightness analysis for R1 and R2 shown in Fig. 10. R, P and S stand for region, processed and shaky, respectively. The US images were used for this comparison.
4. Discussion
The availability of a noninvasive imaging method to comprehensively profile oral anatomy is of great benefit for periodontal health monitoring. The feasibility of a dual modality PA-US imaging system to this end was previously presented by our group [11,12]. In this follow up study, we aimed to improve our imaging system. Different central frequencies will be used for different imaging scales, and the experiments described here were conducted with four probes with different central frequencies (i.e., 10, 20.2 and 40 MHz). Motion artifacts were induced by hand in different levels. An image registration algorithm based on MIND was used to deshake the images and provide more accurate structural information. The images of the depth phantom were deshaken with 80% similarity to the ground truth. The structural errors of about 1 mm were reduced to 0.1 mm and 0.25 mm in the lateral and scanning directions, respectively. Finally, the extent of enamel staining was measured with 0.1 mm error, and the in vivo results agreed with our prior study [22].
To deshake the images, the first image is defined as the reference, and image +1 is registered on the image i. If registering and aligning error occurs between image i. and i. +1, then it affects all the subsequent steps. For applications with many images to be registered (e.g., our application), this makes the image deshaking very critical. Here, the MIND algorithm was used mainly due to the its simultaneous sensitivity to corners, textures, and edge features as well as its capability to perform deformable registration. Another advantage of this algorithm is that it operates regionally, which increases the processing speed and reduces the incidence of fundamental errors. The direction of the motion artifact does not influence MIND as discussed in [19].
The SSIM calculated for the experiments conducted with Vantage (F0 = 20.2 MHz) and AcousticX (F0 = 10 MHz) systems were almost the same (about 80%). This analogy can be used to generalize the error reduction obtained with MIND in AcousticX (0.1 mm and 0.25 mm in the lateral and scanning directions, respectively) to the Vantage system. The measurement error in the Vevo system (F0 = 40 MHz) was also reduced to 0.1 mm. Therefore, a measurement error of about 0.1 mm can be expected in all the three investigated central frequencies. The performance of the proposed deshaking method might be independent of the central frequency of the probe, but further study is needed to confirm this.
Even though there is no shaking applied in Fig. 9(a), MIND smoothens some of the structural variations especially the region showing the calculus depth (PA signals). It is not clear to us whether this is due to the minor internal shaking caused by the step motor or the fact that our deshaking technique is biased. The structures indicated by the red dotted-box are not smoothened in the second column of Fig. 9(a), and we believe that it should be the minor internal shaking of the step motor. This of course needs further investigation, but if biased, one solution can be to use machine learning and different datasets to teach MIND to prevent the smoothening of realistic structural variation. The extent of enamel staining measured in Fig. 9(a) does not change after applying the MIND, which demonstrates the preservation of the structural information after deshaking.
The beamformer used for image formation and the contrast of the images along with the signal-to-noise (SNR) affect the performance of deshaking. In our study, a delay-and-sum (DAS) beamformer was used for image formation. More advanced image formation techniques ([22–26]) can be used to improve the contrast of the image and improve the performance of MIND because it locally processes features and sidelobes that might add unrealistic features. Machine learning and deep learning can further improve the performance of the deshaking by providing a better SNR [27,28]. The number of pixels used in the reconstruction of each image could also affect the performance of the MIND. Our investigation (not presented in this paper) showed that a higher number reduced deshaking error.
In the ex vivo and in vivo experiments, the ultrasound probe was horizontal, and vertical scanning was conducted. To measure the staining, we could perform the imaging with a vertically-held transducer, which would provide an artifact-free sagittal plane of the tooth. However, if a complete profile of the tooth is needed, then image deshaking is necessary to correctly align the sagittal planes and provide an artifact-free image. In our study, the reconstructed B-mode images were used as the input of our deshaking method. This makes our method suitable to be coupled with commercial US/PA imaging systems without any need for RF data. This is an advantage as RF data is only available in research-based imaging systems, which are expensive to be used in clinics. The proposed motion compensation technique could be used in other applications such as imaging spinal curvatures [29], wound staging [30], carotid imaging [31–33] and generally free-hand imaging systems [34,35] where the 1D ultrasound probe is used handheld (without any sensor to track the trajectory of the hand) for sweeping the imaging medium and making 3D images.
Care must be taken to set the parameters of the deshaking method properly for different imaging scenarios. The PPD was a key parameter in our application and could be controlled by to the level of applied force to the motor in our study. A higher value of the PPD led to a higher possibility of failure. Our method failed in Fig. 3(h) mainly due to a large PPD (about 3.9 mm at the central frequency of 10 MHz). Following the fact that the same measurement error of 0.1 mm was obtained in different frequencies (10, 20 and 40 MHz), a PPD of 3.9 mm most probably led to failure in other frequencies as well. Our evaluation showed that the maximum PPD that our deshaking method can handle is 3.6 mm.
One solution to this failure is to increase the search region [parameter R in (2i t)]. However, if a large R is selected, then more complex images such as those presented in the original work (see [19]) might fail due to non-rigid deformation leading to poor image quality. Here, R was equal to 4, but using an R of 48 could still deshake Fig. 3(h) with a high similarity to the ground truth. Of course, all other results would be different as well.
A larger R also imposes a higher computational complexity. The boundary at which the MIND algorithm fails is not entirely clear and was also not discussed in prior work [19]. However, the clinicians using this approach have fine motor skills and will move the probe in a relatively straight trajectory. Thus, the expected tolerance of the hand will be well within the capacity of the proposed deshaking algorithm to improve the images; we have shown this in Figs. 3(e), 5(i) and Figs. 5(b), 5(d).
5. Conclusion
In this follow up study, we used an image registration technique to correctly align different B-mode PA-US images and create an artifact-free 2D profile of the tooth and its periodontium. The experimental results obtained with the depth phantom showed that 80% similarity to ground truth could be obtained. An error of about 1 mm in the TU experiment was reduced to 0.1 mm and 0.25 mm in the lateral and scanning directions, respectively. The results of the ex vivo experiment showed that the depth of calculus could be measured with 0.1 mm error. The deshaking technique shows potential for clinical collection of motion artifact-free oral PA-US images in both stepper motor and handheld configurations. This reduces the burden of technical skill on the operator and the need for stringent head immobilization.
Acknowledgments
Authors acknowledge funding from the National Institutes of Health under grants R21 DE029025, R21 DE029917, 1R21 AG065776, DP2 137187, and S10 OD021821, as well as funding from a joint grant from the Netherlands Organisation for Scientific Research (NWO)/the Netherlands Organisation for Health Research and Development (ZonMw) and the Department of Biotechnology (Government of India) under the program Medical Devices for Affordable Health (MDAH) as Project Imaging Needles (Grant Number 116310008). This publication was supported in part by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650112. C.M. also acknowledges the generous support of the ARCS (Achievement Reward for College Scientists) Foundation.
Funding
Netherlands Organisation for Health Research and Development/Nederlandse Organisatie voor Wetenschappelijk Onderzoek10.13039/501100003246 (116310008); National Institutes of Health10.13039/100000002 (1R21AG065776, DP2 137187, R21 DE029025, R21 DE029917, S10 OD021821); National Science Foundation10.13039/100000001 (DGE-1650112).
Disclosures
J.V.J. is a co-founder of StyloSonics, LLC.
References
- 1.Eke P. I., Dye B., Wei L., Thornton-Evans G., Genco R., “Prevalence of periodontitis in adults in the United States: 2009 and 2010,” J. Dent. Res. 91(10), 914–920 (2012). 10.1177/0022034512457373 [DOI] [PubMed] [Google Scholar]
- 2.Leng W.-D., Zeng X.-T., Kwong J. S., Hua X.-P., “Periodontal disease and risk of coronary heart disease: An updated meta-analysis of prospective cohort studies,” Int. J. Cardiol. 201, 469–472 (2015). 10.1016/j.ijcard.2015.07.087 [DOI] [PubMed] [Google Scholar]
- 3.Stewart R., West M., “Increasing evidence for an association between periodontitis and cardiovascular disease,” (Am Heart Assoc, 2016). [DOI] [PubMed] [Google Scholar]
- 4.Tonetti M. S., Van Dyke T. E., “Periodontitis and atherosclerotic cardiovascular disease: consensus report of the Joint EFP/AAPWorkshop on Periodontitis and Systemic Diseases,” J. Periodontol. 84(4-s), S24–S29 (2013). 10.1902/jop.2013.1340019 [DOI] [PubMed] [Google Scholar]
- 5.Michaud D., Kelsey K., Papathanasiou E., Genco C., Giovannucci E., “Periodontal disease and risk of all cancers among male never smokers: an updated analysis of the Health Professionals Follow-up Study,” Ann. Oncol. 27(5), 941–947 (2016). 10.1093/annonc/mdw028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y. T., Lee H. C., Hu C. J., Huang L. K., Chao S. P., Lin C. P., Su E. C. Y., Lee Y. C., Chen C. C., “Periodontitis as a modifiable risk factor for dementia: a nationwide population-based cohort study,” J. Am. Geriatr. Soc. 65(2), 301–305 (2017). 10.1111/jgs.14449 [DOI] [PubMed] [Google Scholar]
- 7.Kornman K. S., “Mapping the pathogenesis of periodontitis: a new look,” J. Periodontol. 79(8s), 1560–1568 (2008). 10.1902/jop.2008.080213 [DOI] [PubMed] [Google Scholar]
- 8.Chan H.-L., Sinjab K., Chung M.-P., Chiang Y.-C., Wang H.-L., Giannobile W. V., Kripfgans O. D., “Non-invasive evaluation of facial crestal bone with ultrasonography,” PLoS One 12(2), e0171237 (2017). 10.1371/journal.pone.0171237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen K.-C. T., Le L. H., Kaipatur N. R., Major P. W., “Imaging the cemento-enamel junction using a 20-MHz ultrasonic transducer,” Ultrasound Med. Biol. 42(1), 333–338 (2016). 10.1016/j.ultrasmedbio.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 10.Moore C., Jokerst J. V., “Photoacoustic Ultrasound for Enhanced Contrast in Dental and Periodontal Imaging,” in Dental Ultrasound in Periodontology and Implantology (Springer, 2020), pp. 215–230. [Google Scholar]
- 11.Lin C., Chen F., Hariri A., Chen C., Wilder-Smith P., Takesh T., Jokerst J., “Photoacoustic imaging for noninvasive periodontal probing depth measurements,” J. Dent. Res. 97(1), 23–30 (2018). 10.1177/0022034517729820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore C., Bai Y., Hariri A., Sanchez J. B., Lin C.-Y., Koka S., Sedghizadeh P., Chen C., Jokerst J. V., “Photoacoustic imaging for monitoring periodontal health: A first human study,” Photoacoustics 12, 67–74 (2018). 10.1016/j.pacs.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukashevich P., Zalesky B., Ablameyko S., “Medical image registration based on SURF detector,” Pattern Recognit. Image Anal. 21(3), 519–521 (2011). 10.1134/S1054661811020696 [DOI] [Google Scholar]
- 14.Sotiras A., Davatzikos C., Paragios N., “Deformable medical image registration: A survey,” IEEE Trans. Med. Imaging 32(7), 1153–1190 (2013). 10.1109/TMI.2013.2265603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen G.-J., Lv J.-j., Yu W.-x., Sensing R., “A high-performance feature-matching method for image registration by combining spatial and similarity information,” IEEE Trans. Geosci. Remote Sens. 46(4), 1266–1277 (2008). 10.1109/TGRS.2007.912443 [DOI] [Google Scholar]
- 16.Bay H., Tuytelaars T., Van Gool L., “Surf: Speeded up robust features,” in European conference on computer vision, (Springer, 2006), 404–417. [Google Scholar]
- 17.Lowe D. G., “Distinctive image features from scale-invariant keypoints,” Int. J. Comput. Vis. 60, 91–110 (2004). 10.1023/B:VISI.0000029664.99615.94 [DOI] [Google Scholar]
- 18.Matas J., Chum O., Urban M., Pajdla T., “Robust wide-baseline stereo from maximally stable extremal regions,” Image Vis Comput. 22(10), 761–767 (2004). 10.1016/j.imavis.2004.02.006 [DOI] [Google Scholar]
- 19.Heinrich M. P., Jenkinson M., Bhushan M., Matin T., Gleeson F. V., Brady M., Schnabel J. A., “MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration,” Med. Image Anal. 16(7), 1423–1435 (2012). 10.1016/j.media.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 20.Buades A., Coll B., Morel J.-M., “A non-local algorithm for image denoising,” in 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05), (IEEE, 2005), 60–65. [Google Scholar]
- 21.Wang Z., Bovik A. C., Sheikh H. R., Simoncelli E. P., “Image quality assessment: from error visibility to structural similarity,” IEEE Trans. on Image Process. 13(4), 600–612 (2004). 10.1109/TIP.2003.819861 [DOI] [PubMed] [Google Scholar]
- 22.Mozaffarzadeh M., Mahloojifar A., Orooji M., Kratkiewicz K., Adabi S., Nasiriavanaki M., “Linear-array photoacoustic imaging using minimum variance-based delay multiply and sum adaptive beamforming algorithm,” J. Biomed. Opt. 23(2), 026002 (2018). 10.1117/1.JBO.23.2.026002 [DOI] [PubMed] [Google Scholar]
- 23.Matrone G., Savoia A. S., Caliano G., Magenes G., “The delay multiply and sum beamforming algorithm in ultrasound B-mode medical imaging,” IEEE Trans. Med. Imaging 34(4), 940–949 (2015). 10.1109/TMI.2014.2371235 [DOI] [PubMed] [Google Scholar]
- 24.Mozaffarzadeh M., Mahloojifar A., Orooji M., Adabi S., Nasiriavanaki M., “Double-stage delay multiply and sum beamforming algorithm: Application to linear-array photoacoustic imaging,” IEEE Trans. Biomed. Eng. 65(1), 31–42 (2018). 10.1109/TBME.2017.2690959 [DOI] [PubMed] [Google Scholar]
- 25.Mozaffarzadeh M., Hariri A., Moore C., Jokerst J. V., “The double-stage delay-multiply-and-sum image reconstruction method improves imaging quality in a led-based photoacoustic array scanner,” Photoacoustics 12, 22–29 (2018). 10.1016/j.pacs.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozaffarzadeh M., Varnosfaderani M. H., Sharma A., Pramanik M., de Jong N., Verweij M. D., “Enhanced contrast acoustic-resolution photoacoustic microscopy using double-stage delay-multiply-and-sum beamformer for vasculature imaging,” J. Biophotonics 12(11), e201900133 (2019). 10.1002/jbio.201900133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hariri A., Alipour K., Mantri Y., Schulze J. P., Jokerst J. V., “Deep learning improves contrast in low-fluence photoacoustic imaging,” Biomed. Opt. Express 11(6), 3360–3373 (2020). 10.1364/BOE.395683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farnia P., Najafzadeh E., Hariri A., Lavasani S. N., Makkiabadi B., Ahmadian A., Jokerst J. V., “Dictionary learning technique enhances signal in LED-based photoacoustic imaging,” Biomed. Opt. Express 11(5), 2533–2547 (2020). 10.1364/BOE.387364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H., Zheng R., Lou E., Ta D., “Imaging Spinal Curvatures of AIS Patients using 3D US Free-hand Fast Reconstruction Method,” in 2019 IEEE International Ultrasonics Symposium (IUS), (IEEE, 2019), 1440–1443. [Google Scholar]
- 30.Hariri A., Chen F., Moore C., Jokerst J. V., “Noninvasive staging of pressure ulcers using photoacoustic imaging,” Wound Rep. Reg. 27(5), 488–496 (2019). 10.1111/wrr.12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mozaffarzadeh M., Soozande M., Fool F., Pertijs M. A., Vos H. J., Verweij M. D., Bosch J. G., de Jong N., “Receive/transmit aperture selection for 3D ultrasound imaging with a 2D matrix transducer,” Appl. Sci. 10(15), 5300 (2020). 10.3390/app10155300 [DOI] [Google Scholar]
- 32.van Knippenberg L., van Sloun R. J., Shulepov S., Bouwman A., Mischi M., “Towards flow estimation in the common carotid artery using free-hand cross-sectional doppler,” in 2019 IEEE International Ultrasonics Symposium (IUS), (IEEE, 2019), 1322–1325. [DOI] [PubMed] [Google Scholar]
- 33.Chung S.-W., Shih C.-C., Huang C.-C., “Freehand three-dimensional ultrasound imaging of carotid artery using motion tracking technology,” Ultrasonics 74, 11–20 (2017). 10.1016/j.ultras.2016.09.020 [DOI] [PubMed] [Google Scholar]
- 34.Mozaffari M. H., Lee W.-S., “Freehand 3-D ultrasound imaging: a systematic review,” Ultrasound Med Biol. 43(10), 2099–2124 (2017). 10.1016/j.ultrasmedbio.2017.06.009 [DOI] [PubMed] [Google Scholar]
- 35.Solberg O. V., Lindseth F., Torp H., Blake R. E., Hernes T. A. N., “Freehand 3D ultrasound reconstruction algorithms—a review,” Ultrasound Med Biol. 33(7), 991–1009 (2007). 10.1016/j.ultrasmedbio.2007.02.015 [DOI] [PubMed] [Google Scholar]