Abstract
The highly-conserved Grainyhead-like (Grhl) transcription factors are critical regulators of embryogenesis that regulate cellular survival, proliferation, migration and epithelial integrity, especially during the formation of the craniofacial skeleton. Family member Grhl2 is expressed throughout epithelial tissues during development, and loss of Grhl2 function leads to significant defects in neurulation, abdominal wall closure, formation of the face and fusion of the maxilla/palate. Whereas numerous downstream target genes of Grhl2 have been identified, very little is known about how this crucial developmental transcription factor itself is regulated. Here, using in silico and in utero expression analyses and functional deletion in mice, we have identified a novel 2.4kb enhancer element (mm1286) that drives reporter gene expression in a pattern that strongly recapitulates endogenous Grhl2 in the craniofacial primordia, modulates Grhl2 expression in these tissues, and augments Grhl2-mediated closure of the secondary palate. Deletion of this genomic element, in the context of inactivation of one allele of Grhl2 (through generation of double heterozygous Grhl2+/−;mm1286+/− mice), results in a significant predisposition to palatal clefting at birth. Moreover, we found that a highly conserved 325 bp region of mm1286 is both necessary and sufficient for mediating the craniofacial-specific enhancer activity of this region, and that an extremely well-conserved 12-bp sequence within this element (CTGTCAAACAGGT) substantially determines full enhancer function. Together, these data provide valuable new insights into the upstream genomic regulatory landscape responsible for transcriptional control of Grhl2 during palatal closure.
Author Summary
The correct formation and integration of the bones, nerves, blood vessels, muscles and skin of the head and face is an exceptionally complex process. The synergy of multiple developmental genes is essential for correct craniofacial development. When these tightly-regulated processes go wrong – as they often do – the resultant craniofacial defects lead to substantial psychological, social and functional difficulties for affected individuals. By understanding how these developmental processes are regulated at the genetic level, we can form a more holistic understanding of how to tackle these debilitating disorders. Here we describe how a gene known to be critical for development of the head, face and jaws (Grainyhead-like 2; Grhl2) is regulated at the transcriptional level by a new craniofacial-specific enhancer region. This enhancer co-operates with Grhl2 to drive expression correctly within the developing face and jaws. Importantly, functional loss of this enhancer is a significant pre-disposing factor in the aetiology of palatal clefts in mice, and given the substantial sequence conservation across vertebrates, may also ultimately prove to be a factor in human craniofacial disorders.
Introduction
Correct formation of the craniofacial skeleton in vertebrates requires the precise synergy of cellular proliferation, migration, homing and fusion in order to establish the bones, muscles, nerves and vasculature of the skull, face and jaw. This incredibly tightly co-ordinated and complex process of development involves multiple transcription factors, migration cues, and patterning signals. Disruption of these genetic processes underpins craniofacial defects (CFD), disorders of development that affect 0.1–0.3% of all live births (Cordero et al., 2011).
Amongst the genes responsible for patterning the craniofacial complex, the highly conserved family of transcription factors, the Grainyhead-like (Grhl) genes, are emerging as critical master regulators of craniofacial fate in numerous species (Carpinelli et al., 2017; Dworkin et al., 2014; Peyrard-Janvid et al., 2013; Pyrgaki et al., 2011). Orthologues of the Drosophila gene grainyhead (Bray and Kafatos, 1991), the vertebrate Grhl factors are sub-functionalised into Grhl1–3, with mutations in both Grhl2 and Grhl3 implicated in the aetiology of human craniofacial malformations. Altered expression of Grhl3 leads to extreme developmental consequences. Human Grhl3 mutations lead to both syndromic (Van der Woude Syndrome; VWS (Peyrard-Janvid et al., 2014) and non-syndromic (Mangold et al., 2016) cleft palate, and Grhl3-null mice display neural tube defects (Ting et al., 2003), delayed palatal shelf elevation due to peridermal adhesions in the oral cavity (Peyrard-Janvid et al., 2014), and premature apposition of the frontal and parietal bones, analogous to the human condition craniosynostosis (Goldie et al., 2016). Grhl2 null mice show developmental retardation, facial clefting, exencephaly, and mid-gestational embryonic lethality (by embryonic day 11.5). Although implicated in deafness (Vona et al., 2013), age-related hearing loss (Van Laer et al., 2008) and ectodermal dysplasia (Petrof et al., 2014) in humans, thought to be due to various mutations in GRHL2 leading to reduced activity (Huang et al., 2010), GRHL2 has also been implicated in the aetiology of human CFD; five patients with CFD, harbouring microdeletions at chromosomal region 8q22.2q22.3, have been identified (Kuechler et al., 2011). These patients are hemizygous for a 1.9 Mb region containing nine genes, of which only GRHL2 has a known role in craniofacial development. Together, these data show that the Grhl family are highly-conserved genetic regulators of craniofacial formation.
Numerous genes that regulate craniofacial development are involved in the Grhl gene regulatory network. These include both upstream factors, such as IRF6 (Botti et al., 2011; de la Garza et al., 2013; Kousa et al., 2019) or FGF8 (Trumpp et al., 1999) and downstream targets such as Edn1 (Dworkin et al., 2014), TFAP2A (Kousa et al., 2019) or GSK3β (Liu et al., 2007). However, comparatively little is known about the genomic landscape that allows for regulation and correct spatiotemporal expression of Grhl factors themselves in development. In particular, little is known of putative enhancer elements that may be required for Grhl2 expression in the craniofacial primordia. Classically, enhancers are thought of as non-coding genomic regions located at some distance from the transcriptional start site (TSS) that allow binding of transcription factors and RNA polymerase II, and then through conformational DNA changes, serve to form a loop to facilitate interaction of these factors to the target gene promoter, allowing specific cellular and spatiotemporal regulation of transcriptional activity (Pennacchio et al., 2013). Enhancing elements located either upstream or downstream of the TSS (including within gene introns) can contribute to regulation of gene expression.
Mutations in enhancer regions are implicated in craniofacial disorders. Isolated Pierre Robin sequence, a syndrome involving cleft palate and micrognathia (Evans et al., 2011), is most commonly caused by a mutation near the SOX9 gene (Gordon et al., 2009). This mutation falls in an enhancer region and results in the downregulation of SOX9 activity (Gordon et al., 2009). In mice, the deletion of distant enhancing elements regulating Myc expression in craniofacial development was shown to cause facial dysmorphisms and low-penetrance CLP (Uslu et al., 2014). Currently, the nature of Grhl-gene enhancers is largely unknown, although a mutation (that underpins neural tube closure defects in the Grhl3-deficient curly-tail mouse) has been identified ~21kb upstream of the Grhl3 TSS (Gustavsson et al., 2008), and an intronic single nucleotide polymorphism (SNP) has been identified in GRHL2 that enhances susceptibility to age-related hearing loss (Van Laer et al., 2008).
Here through in silico prediction, in vivo expression analysis and functional deletion, we have interrogated the Grhl2 genomic landscape to identify an enhancer element that drives expression in the craniofacial primordia during embryogenesis, and moreover, regulates Grhl2 in the context of palatal development.
Results
Prediction of genomic Grhl2 craniofacial enhancers in mouse embryos
To identify in vivo enhancers involved in transcriptional regulation of Grhl2 in embryonic craniofacial tissues we screened the murine Grhl2 genomic locus (for interrogation purposes defined as the entire region of chromosome 15 between genes ZNF706 and NCALD, co-ordinates chr15:36,925,608–37,725,722, genome build mm9) for ChIP-Seq enrichment of the p300 transcriptional co-activator and displaying H3K27 acetylation (H3K27ac), the most frequently used epigenetic and epigenomic signatures to predict active enhancers (Osterwalder et al., 2018; Rada-Iglesias et al., 2011; Visel et al., 2009) (Fig. 1). Specifically, we used an unbiased list of 4,399 craniofacial enhancers predicted by ChIP-Seq analysis of non-coding genomic regions bound by the enhancer-associated transcriptional co-activator protein p300 (p300 ChIP-Seq) in the craniofacial primordia of E11.5 mouse embryos (Attanasio et al., 2013). To extend these enhancer predictions and to investigate the spatio-temporal activities of predicted enhancer regions we also used H3K27ac ChIP-Seq data from facial prominences at different developmental timepoints (available from www.encodeproject.org) (Fig. 1). Intersection of genome-wide p300 and H3K27ac enrichments revealed three high-confidence enhancer elements upstream of Grhl2 (−180kb, −97kb, −34kb), all of which are well-conserved in mammals and show predicted chromatin marks throughout craniofacial development (almost exclusively within the craniofacial primordia) from E10.5 to E15.5 (Fig. 1). In addition, relaxing the stringency of enhancer predictions in the Grhl2 upstream regulatory region, we defined additional craniofacial enhancer candidates 40kb and 23kb upstream of Grhl2, based on continuous temporal H3K27ac enrichment (Fig. 1) and specific activity in mandibular compartments at E11.5 (FaceBase H3K27ac), respectively.
Fig. 1: In silico prediction strategy to identify putative enhancing elements of Grhl2 in craniofacial development:
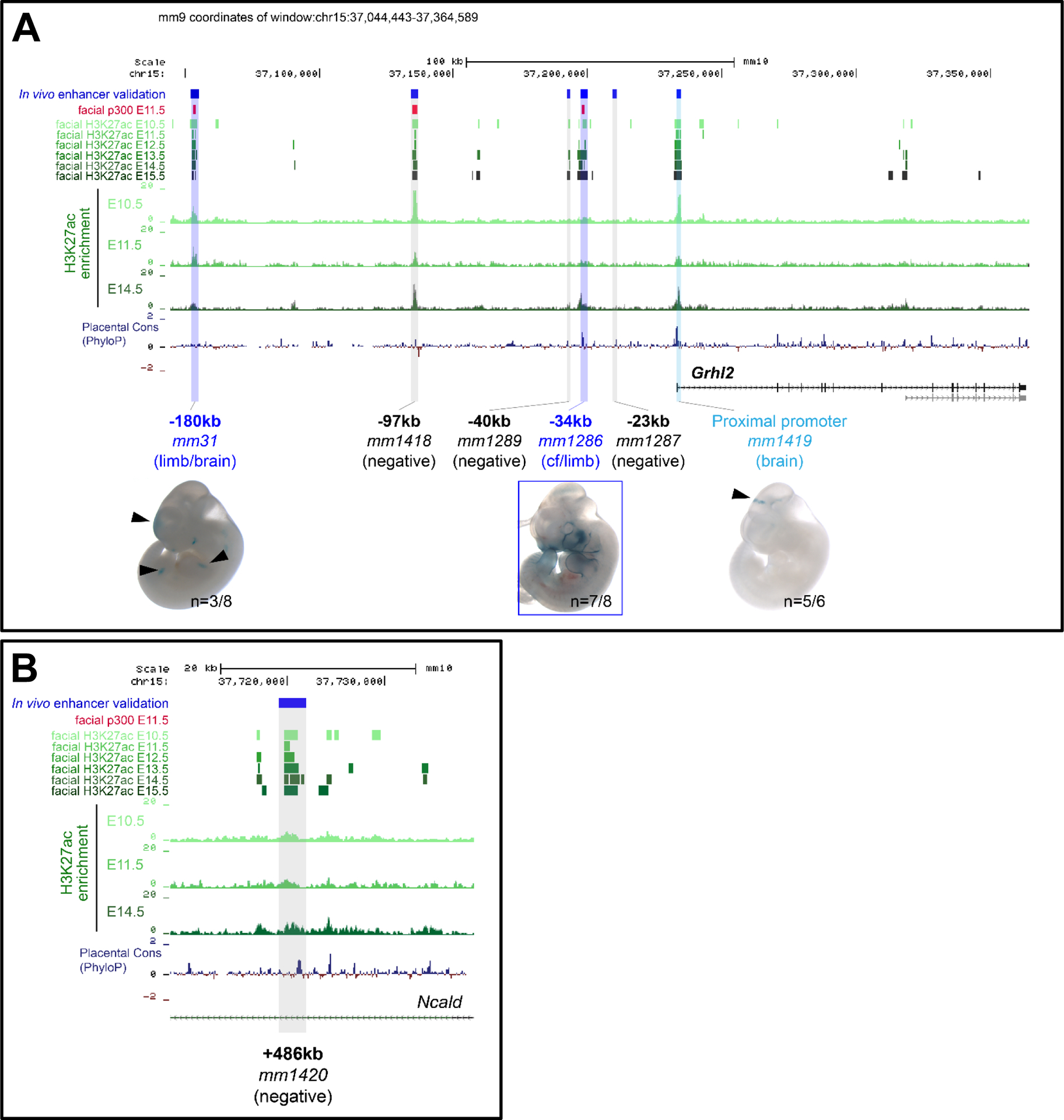
UCSC genome browser tracks showing ENCODE datasets as predictive analyses showing location of predicted enhancing elements within the murine Grhl2 genomic locus, defined as being situated between the Zfp706 and Ncald genes, at co-ordinates chr15:36,925,608–37,725,722 (A-B). Putative functional elements (relative to transcriptional start site; TSS) were identified at −180kb (mm31), −97kb (mm1418), −40kb (mm1289), −34kb (mm1286), −23kb (mm1287; A) and +486kb (mm1420; B). Of these, only the mm31 (limb/brain) and mm1286 (craniofacial primordia and limb) showed reproducible enhancer activities. The Grhl2 proximal promoter alone (mm1419) drove weak expression within the brain, at the level of the midbrain/hindbrain boundary.
Screening the non-coding regulatory domain downstream of the Grhl2 gene body (Ncald intronic regions) revealed another putative enhancer element (not shown) located 486kb downstream of the Grhl2 TSS hallmarked by continuous H3K27ac enrichment in craniofacial tissues from E10.5 to E15.5. Collectively, these analyses identified six predicted enhancer regions (−180kb, −97kb, −40kb, −34kb, −23kb and +486kb relative to TSS) potentially relevant for regulation of Grhl2 in craniofacial tissues. Of these, the element 180kb upstream of Grhl2 had been previously screened at E11.5, and did not drive any reproducible expression pattern following a LacZ reporter assay (Vista enhancer browser, ID: mm31; Visel et al., 2007) and displays reproducible activity in the developing brain and limbs (at E11.5), but not in craniofacial domains.
The Grhl2 proximal promoter region is also enriched for H3K27ac in facial prominences throughout major stages of development. Therefore, we used a well-established transgenic LacZ assay (Attanasio et al., 2013) to assess whether this promoter element itself is sufficient to drive craniofacial expression in mouse embryos at E11.5. However, while this element was able to drive reproducible activity in the diencephalon, at the level of the midbrain-hindbrain boundary (Fig. 1), no craniofacial activities were detected. Therefore, our analyses suggested that one or more of the predicted enhancer elements was necessary for ensuring spatiotemporal expression of Grhl2 within the developing craniofacial region.
Identification of an epithelial-specific Grhl2 enhancer active in craniofacial domains
In order to determine whether any of the remaining five elements could drive gene expression in the context of craniofacial morphogenesis, we used our transgenic LacZ assay to determine in vivo transcriptional enhancer activities in mouse embryos at E11.5. Somewhat surprisingly, the majority of the predicted enhancer elements (−97kb, −40kb, −23kb and +486kb; Table S1) did not drive reproducible LacZ reporter expression at E11.5. In contrast, the element located −34kb upstream of the Grhl2 TSS (Vista enhancer ID mm1286) was able to drive highly reproducible and specific reporter expression in craniofacial, skin and limb bud epithelia at E11.5, as seen in both whole-mount expression and confirmed by histological analysis of haematoxylin and eosin (H&E)-stained coronal sections of transgenic embryos (Fig. 2A–B). This expression, particularly strong in the dorsal epithelium surrounding the fronto-nasal process (FNP), and the maxillary and mandibular prominences (MXP and MDP respectively) closely recapitulated endogenous expression of Grhl2 (shown by autoradiographic ISH; Fig. 2C and (Brouns et al., 2011). Furthermore, sagittal sections at E14.5 revealed enhancer activity in the oral and maxillary epithelium and in the surface ectoderm (developing skin; Fig. 2D–F). Together, these data indicated that the mm1286 element could drive gene expression specifically within the craniofacial region.
Fig. 2: In vivo functional validation of craniofacial and limb-specific element mm1286:
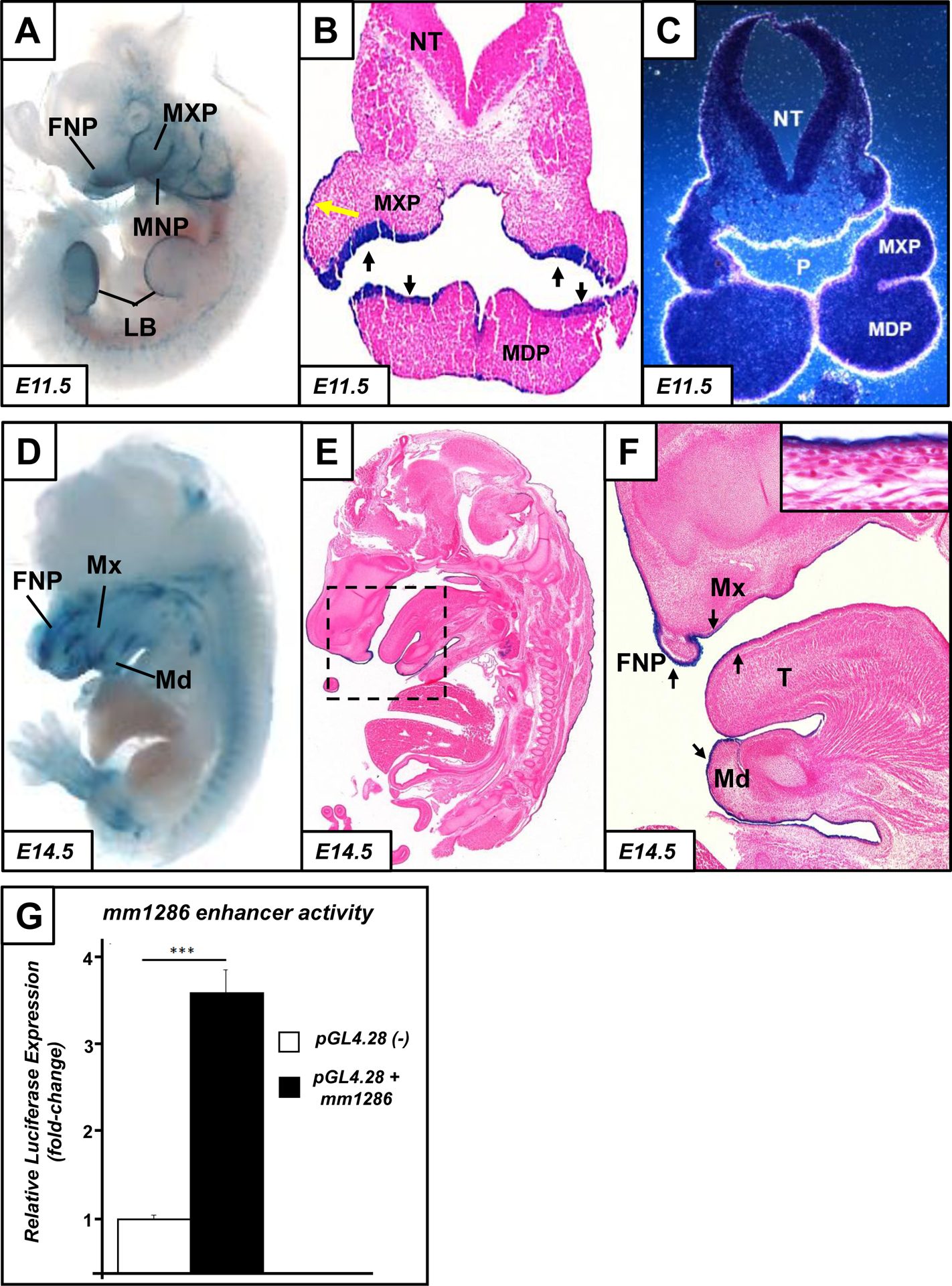
At embryonic day (E) 11.5, the mm1286 enhancer drives LacZ expression within the craniofacial primordia, specifically the frontonasal prominence (FNP), maxillary prominence (MXP) and mandibular prominence (MNP), as well as the limb buds (LB; A). Coronal sections at E11.5 (B) confirm that enhancer activity (as shown by positive LacZ staining, blue) is restricted to the epithelium of the maxillary (MXP) and mandibular (MDP) prominences. Expression is also visible in the external skin at E11.5 and E14.5 (yellow arrow, B), although expression is absent from the neural tube. Autoradiographic in-situ hybridisation (ISH) of endogenous Grhl2 expression at E11.5 (C) shows a highly-similar expression pattern to that of mm1286, particularly in the pharyngeal epithelium (P) of the MXP and MDP and the external surface ectoderm/developing skin, as well as absence of expression in the neural tube (NT). mm1286 continues to drive expression at E14.5 (D); sagittal H&E-stained histological sections (E-F) confirm expression in the now-fused FNP, Maxilla (Mx), dorsal and ventral epithelium of the mandible (Md) and tongue (T; arrows) and back-skin (inset, F). In vitro luciferase activity assay (G) showing that the mm1286 element enhances expression driven by a minimal promoter (in the pGL4.28 vector) approximately 3.5-fold over expression driven by the minimal promoter alone (n=3; ***p<0.001 by student’s t-test).
The mm1286 element is 2,741bp in length and located at mm9:chr15:37,127,220–37,129,961 (Table S1). As a secondary measure of transactivation, we cloned this region and performed luciferase assays in HEK-293 cells. This enhancer was able to transactivate a minimal promoter in the pGL4.28 vector approximately 3.5-fold over control vector with minimal promoter only (n=3; p<0.05 by Student’s t-test), indicative of transcription-enhancing function (Fig. 2G). Taken together, our results indicate that element mm1286 is likely to be a key driver of Grhl2 expression in the context of craniofacial development.
Enhancer element mm1286 regulates expression and function of Grhl2
In order to determine the function of the mm1286 enhancer in normal craniofacial development, we deleted this region in mice using CRISPR-Cas9 genome engineering. We generated two independent mouse lines (founder line 1 and founder line 2; Fig. S1), comprising deletions of 3,745bp and 3,740bp respectively, encompassing the entire enhancer element (Fig. S1 and Table S2). Timed intercrosses of mm1286+/− mice of either line resulted in mm1286−/− E18.5 embryos at approximately Mendelian frequency (Table S5), and without any observable gross size/weight or morphological defects.
As craniofacial enhancers are frequently known to subtly influence craniofacial morphology (Attanasio et al., 2013), and the Grhl-family member Grhl3 had previously been shown to influence the size and suture width of the skull (Goldie et al., 2016), we performed X-ray Micro-computed Tomography (μCT) analysis in order to definitively interrogate skull formation (Fig. 3A–F). We performed analyses as described previously (Goldie et al., 2016; Motch Perrine et al., 2014), paying particular attention to the size and overall length/width ratios of the entire skull, the length and articulation angle of the mandibles, and the overall size and closure of the upper palate. Despite careful analysis of numerous landmark measurements, we found no significant differences in skull size (Fig. S3–S4 and Table S3–S4), indicating that the mm1286 enhancer alone is not required to influence the length, width, height or depth of facial and skull bone development, and nor does it play a role in premature suture fusion between the major bones of the skull.
Fig. 3. Characterisation of craniofacial development in the mm1286−/− deletion mouse:
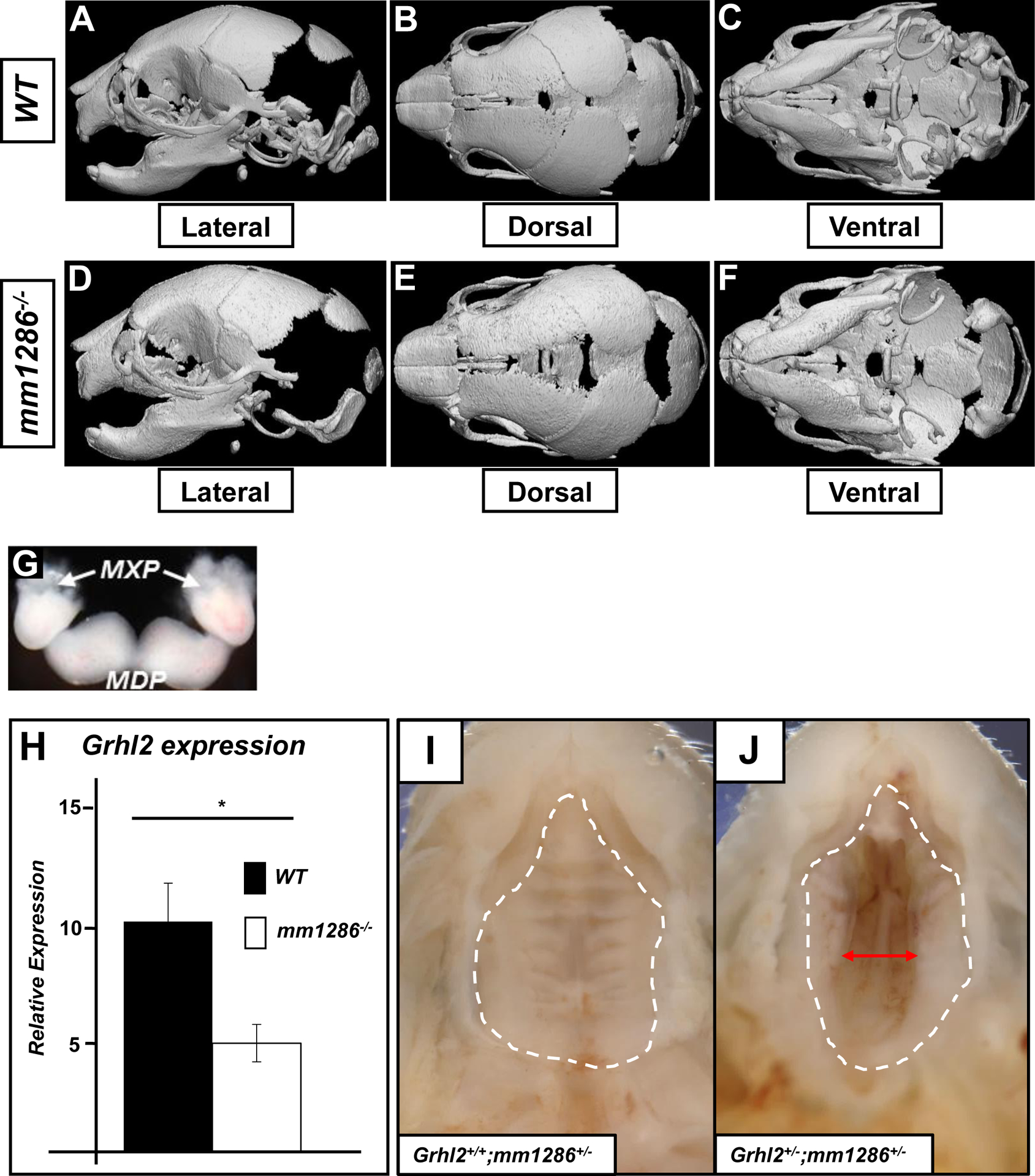
X-Ray computed microtopography (Micro-CT) images of representative P0 skulls from wild-type (A-C) and mm1286−/− (D-F) mice (n=5 for each) shown in lateral (A,D), dorsal (B, E) and ventral (C,F) views. No gross developmental defects were observed in skull width, length, depth, height, suture width or patency, or overall morphology. Dissection of craniofacial primordia (maxillary prominence; MXP and mandibular prominence; MDP) at E14.5 (G) followed by Q-RT-PCR (H) shows that expression of Grhl2 (relative to housekeeping gene Actb) in the craniofacial primordia is reduced by approximately 50% in mice lacking the mm1286 enhancer element (n=5; *p<0.05). Photographs of the secondary palate of newborn mice taken from the oral (ventral) side (I-J). White dashed lines outline primary and secondary palate. Grhl2+/+;mm1286+/− mice did not display any palatal defects (0/39 embryos; white dashed lines, I), whereas 13% of Grhl2+/−;mm1286+/− mice presented with a secondary palatal cleft with a closed lip and primary palate (3/18 embryos; red arrow, J, p=0.011 by Fisher’s exact test).
Next, if the mm1286 element was functional in vivo, and if it impacted on Grhl2 expression specifically, then deletion of this element should result in decreased abundance of Grhl2 mRNA in the craniofacial primordia at E11.5. In order to determine whether Grhl2-expression was reduced in the craniofacial primordia of mm1286−/− embryos, we extracted the E11.5 maxillary and mandibular prominence (Fig. 3G) and performed Q-RT-PCR on these tissues from WT and mm1286−/− embryos. We found a significant, ~50% reduction in Grhl2 mRNA expression within the craniofacial primordia in mm1286−/− embryos (Fig. 3H), indicating that this element is a positive regulator of Grhl2 in this context. Such expression levels of Grhl2 are commensurate with those in previously described Grhl2+/− mice that similarly do not present with any craniofacial phenotype (Rifat et al., 2010), explaining the lack of gross phenotypes in mm1286−/− embryos.
Developmental enhancers with subtle contributions to target gene expression have been shown to provide a mechanism of genetic robustness and their deletion in the presence of reduced target gene dosage often leads to overt phenotypic alterations (Dickel et al., 2013; Osterwalder et al., 2018). In order to further reduce Grhl2 expression specifically within the craniofacial primordia, we intercrossed the mm1286+/− and Grhl2+/− mice to generate Grhl2+/−;mm1286+/− mice. These mice lack one functional copy of Grhl2 on one allele and are devoid of the mm1286 enhancer on the other allele, resulting in a predicted reduction of Grhl2 mRNA in the craniofacial region to approximately one-quarter of wild-type. Grhl2+/−;mm1286+/− mice were present at expected Mendelian frequencies at P0 (Table S5B). However, although occurring at low penetrance, 3 out of 18 Grhl2+/−;mm1286+/− (but 0/59 WT, Grhl2+/+;mm1286+/− or Grhl2+/−;mm1286+/+) littermates (Fig. 3I–J and Table S5) displayed a cleft secondary palate. These results demonstrate that deletion of the mm1286 enhancer in the presence of reduced Grhl2 gene dosage predisposes mice to palatal clefting and indicates its’ requirement for epithelial Grhl2 expression to promote closure of the secondary palate in the absence of genetic robustness.
Identification of a functional 325-bp conserved region within the mm1286 enhancer element
To explore the level of sequence homology and conservation of the mm1286 element amongst animals, we aligned the 2,741bp mouse sequence to orthologous sequences in mammals, birds, reptiles and amphibians, lobe-finned fishes (those that have defined articulated appendages, e.g. coelacanth), cartilaginous and (bony) ray-finned fishes (those that lack defined articulated appendages, e.g. zebrafish, mandarin fish, elephant shark, tilapia, sea bass) and invertebrates (Drosophila, honeybee) using ClustalW. Using BLAST, we determined that a very-highly conserved 325bp sequence (mm9:chr15: 37,127,755–37,128,079; Fig. 4A) was present at >80% homology in all tetrapods and lobe-finned fish but was missing in cartilaginous and ray-finned fishes and invertebrates, suggesting that it arose in evolution at approximately the same ancestral time-point as bony vertebrate limbs. Furthermore, we found that a 12bp sequence (CTGTCAAACAGGT; Fig. 4A and Fig. S5) within this region was identical amongst all tetrapods examined (as well as coelacanth).
Fig. 4: The highly conserved 325bp region within the −34 kb mm1286 enhancer largely recapitulates full-length mm1286 craniofacial activity:
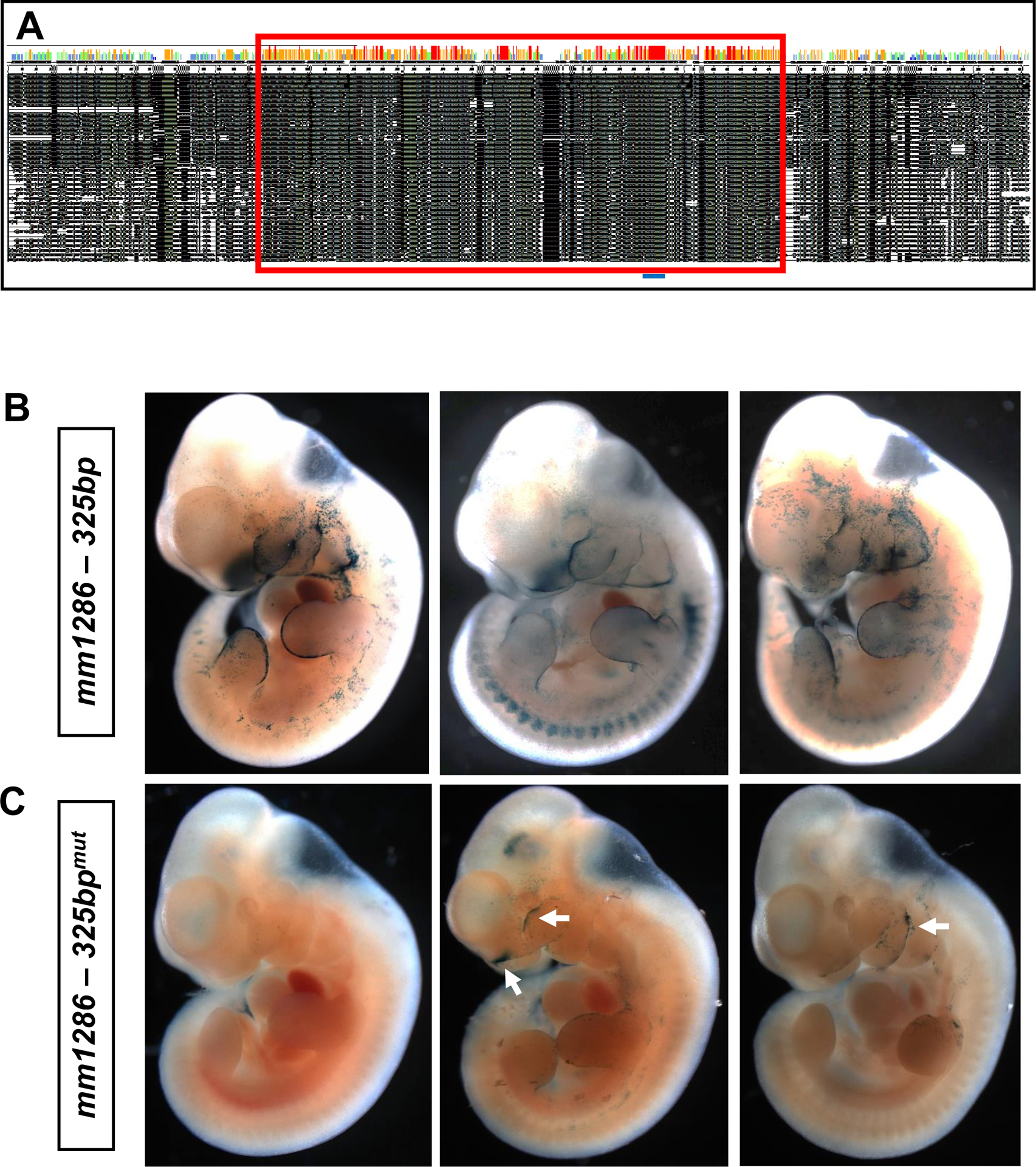
Nucleotide alignment (black regions; A) of the first 620bp of the mm1286 enhancer (mm9:chr15:37,127,220–37,127,840) in 39 different animal species, comprising mammals, reptiles, amphibians, fish and birds. Species were broadly ordered according to phylogenetic distance from the human sequence, with those at the top of the table largely comprising a lesser evolutionary distance. The red box highlights a 325 bp region that is exceptionally well-conserved, additionally comprising a 12 bp invariant sequence (blue line; A) that was identical amongst all species examined. Injection of this conserved 325bp region linked to a LacZ transgene into fertilised oocytes that were subsequently implanted into pseudopregnant mothers (B) largely resulted in a recapitulation of mm1286 activity (especially within the craniofacial primordia and limb buds, n=3/5) at E11.5. Mutation of the invariant 12 bp sequence (CTGTCAAACAGGT) within the 325 bp region (to CCGGGAAACAGGT), eliminating the homeobox recognition site, resulted in either complete abrogation of enhancer-driven LacZ activity, or greatly reduced restricted expression in isolated areas of the frontonasal prominence, maxillary and mandibular epithelia (arrows; n=6/7, three representative embryos shown, C).
Database interrogation of transcription factor binding sites using MotifMap suggests that this 12bp region may correspond to a conserved “homeodomain” site, predicted to serve as a recognition sequence for homeobox transcription factors (TFs) including Prep1, Meis1, Meis2, MRG2 and TGIF2 (Fig. S5). In order to determine whether the 325 bp conserved region (as well as the 12 bp invariant sequence) was necessary and sufficient for enhancer-driven LacZ expression in vivo, we performed transgenic LacZ reporter analyses using the isolated 325bp region with an intact or inactivated 12bp predicted homeodomain binding site (Fig. 4B and 4C). We found that spatiotemporal expression of the full-length enhancer within the epithelia of the pharyngeal arches and limb buds was precisely recapitulated in 3 out of 5 embryos carrying the wild-type 325 bp transgene (Fig. 4B), whereas this expression pattern was either completely absent (4/7) or substantially reduced (2/7) in 6/7 embryos expressing the mutant 325 bp transgene (Fig. 4C). These data suggest than not only are the sequences responsible for driving craniofacial enhancer activity contained within the 325 bp conserved element, but moreover, the 12 bp invariant homeobox recognition sequence performs essential roles in directing, driving and/or fine-tuning Grhl2 expression within the craniofacial primordia.
Taken together, our study indicates that the mm1286 genomic region is a very-highly conserved, functional enhancer in the context of palatal closure during embryogenesis. This study marks characterisation of the first-known enhancer that drives expression of Grhl2.
Discussion
Our study used a combination of sequence conservation, H3K27Ac marks and regions bound by p300 to predict six putative enhancer elements within the Grhl2 genomic locus in mouse. Following in vivo confirmation that one of these enhancer elements, mm1286, drove gene expression in the craniofacial primordia of developing embryos, in a pattern strikingly similar to that of endogenous Grhl2, genetic deletion confirmed an interaction with Grhl2 in the context of palatal closure. Moreover, further interrogation of this enhancer region showed that it comprises a highly conserved (325bp) region, and an invariant 12bp sequence (CTGTCAAACAGGT) amongst tetrapods that may serve as a binding site for several homeobox transcription factors. Functional mutation and subsequent in vivo reporter experiments indicated that this 12bp region was essential for driving correct reporter expression in the craniofacial region. Taken together, our work has identified the first known functional enhancer of mammalian Grhl2.
The novel enhancer we have identified and characterised here is required for craniofacial-specific expression of Grhl2, at E11.5 and also drives expression at E14.5. The other predicted elements do not drive Grhl2 in the craniofacial primordia at this timepoint, however our future studies will comprise analysis of these elements in other tissues, such as heart and lungs (Kersbergen et al., 2018; Pyrgaki et al., 2011) where Grhl2 function is required for development. Moreover, our future work is also focussed on interrogating expression and function of these putative elements at later developmental timepoints (beyond E14.5) or in the adult, where these elements may additionally contribute to regulation of Grhl2. As Grhl2 is implicated in numerous human cancers (Cieply et al., 2012; Nishino et al., 2017; Tanaka et al., 2008; Werner et al., 2013), further work will be required to determine whether these other elements may influence the transcription of Grhl2 in other disease contexts, outside of embryogenesis.
One of the other elements we tested, mm1419, weakly drove expression in the dorsal skin at the level of the midbrain-hindbrain boundary (MHB) at E11.5. This is significant for two reasons. Firstly, a number of Grhl2-loss of function models present with exencephaly and/or impaired neural tube closure at the level of the cranial neural tube (Menke et al., 2015; Pyrgaki et al., 2011), suggesting that this enhancer may help to drive neurulation at this closure point. Secondly, we have previously shown that the zebrafish Grhl2-orthologue (grhl2b) regulates both maintenance and morphogenesis of the MHB (Dworkin et al., 2012), and both grhl2b in fish and Grhl2 in mouse (Menke et al., 2015) also regulate cellular survival within the brain, specifically at the level of the MHB. Together, these data suggest that enhancer mm1419 may be involved in regulating Grhl2 expression at the MHB, although further experimentation, such as functional enhancer deletion and compound Grhl2/mm1419 heterozygosity (as we have performed here for Grhl2/mm1286) will be necessary to test this hypothesis.
We observed no difference in the survival, viability, size, weight or gross morphology of mm1286−/− embryos, and these embryos were present at expected Mendelian frequencies at both E14.5 and P0 (Table S5). Our detailed MicroCT analyses did not indicate any significant variability in the length, width, depth, height, suture patency or bone integrity of either the skull or facial bones in the mm1286−/− P0 embryos. In particular, our thorough interrogation of the developing palate did not indicate any morphological defects or failed palatal fusion in mice lacking a functional mm1286 enhancer. This finding was not particularly surprising, as numerous studies have demonstrated that deletion of putative functional enhancers in isolation (even those that are exceptionally well-conserved) leads to few differences in survival or viability, and, at most, subtle morphological phenotypes during embryogenesis (Ahituv et al., 2007; Dickel et al., 2018; Osterwalder et al., 2018). Although we did not perform μCT analysis of the Grhl2+/−;mm1286+/− doubly heterozygous mice, save for the three pups that presented with a cleft secondary palate, we did not observe any gross morphological deformities in either the skull or facial bones.
We also did not observe any defects in limb or digit formation in either mm1286−/−; or Grhl2+/−;mm1286+/−doubly heterozygous mice, even though both Grhl2 and mm1286 are strongly expressed in the limb buds, and deletion of the highly-conserved 325bp region of mm1286 led to abrogated LacZ expression in the apical ectodermal ridge (AER) of the limb buds. This is consistent with findings from other Grhl2-loss of function models, that likewise have not identified a role for Grhl2 in limb and digit development (Menke et al., 2015; Pyrgaki et al., 2011; Rifat et al., 2010), despite robust Grhl2 expression being detected. Similarly, despite robust mm1286-driven LacZ expression being visible in the epidermis, again consistent with Grhl2 expression (Auden et al., 2006; Brouns et al., 2011), no qualitative epidermal defects were observed in either mm1286−/−; or Grhl2+/−;mm1286+/−doubly heterozygous mice. It is likely that functional redundancy between Grhl2 and other Grhl-family members, or interacting partner proteins, is sufficient to compensate for loss of Grhl2 expression in the AER and epidermis. Although beyond the scope of this study, it is likely that differential histone modification states (and by extension, differential recruitment of disparate transcription-factor complexes) will either repress or activate Grhl2 expression from this enhancer during development (Nakagawa et al., 2018). Taken together, our data show that loss of mm1286 alone does not overtly affect development or morphogenesis in mouse.
Although complete deletion of Grhl2 leads to substantial neural tube and craniofacial defects, palatal clefting, even at low penetrance, had never been reported previously in numerous disparate Grhl2+/− (heterozygous) mice, generated by several different deletion strategies (Brouns et al., 2011; Menke et al., 2015; Pyrgaki et al., 2011; Rifat et al., 2010). Given the fact that Grhl2+/;mm1286+/− embryos did display palatal clefting, albeit at low (yet statistically significant) penetrance, together with the reduction in Grhl2 mRNA expression in the mm1286−/− P0 embryos seen by Q-RT-PCR, we believe that we have identified a novel genetic mechanism by which Grhl2 drives and shapes palatal development and fusion.
We are confident that palatal clefting in our model is exclusively due to the functional interaction between loss of mm1286 and Grhl2 heterozygosity. However, as a single enhancer can on occasion regulate multiple genes (Pennacchio et al., 2013), we cannot empirically rule out an interaction between mm1286 and other genes on murine chromosome 15 (human chromosome 8). However, this is extremely unlikely. Firstly, human chromosomal analyses have identified five patients with chr8:q22 chromosomal micro-deletions that present with craniofacial morphogenesis defects (Kuechler et al., 2011). The common micro-deletion in all 5 patients encompasses loss of 9 genes – GRHL2, RRM2B, NCALD, ZNF706, UBR5, ODF1, KLF10, YWHAZ and AZIN1. Of these, only Grhl2 is implicated in palatal clefting (or indeed any other craniofacial defect), suggesting that Grhl2 heterozygosity may underpin the subtle craniofacial morphological aberrations in these patients. Secondly, outside this micro-deleted region, the only gene within 5Mb of the Grhl2 locus on murine chromosome 15 (human chromosome 8) that has known roles in craniofacial development, is Osr2, a gene that is located ~2Mb upstream of GRHL2, at genomic position chr15:35,296,112–35,303,305. However, this gene is expressed only in neural-crest derived mesenchyme of the palatal shelves, and is not expressed in any craniofacial epithelia, suggesting it is not normally regulated by mm1286. Conversely, as multiple enhancers are also frequently required for correct spatiotemporal expression of a single gene (Dickel et al., 2013; Dickel et al., 2018; Osterwalder et al., 2018), it is quite possible that deletion of other predicted candidate enhancer elements in the mm1286 mouse model may uncover further synergies in enhancer function regulating Grhl2.
As the palatal clefting defect observed in Grhl2+/−;mm1286+/− mice at P0 was not fully penetrant, it is likely that even substantially-reduced Grhl2-expression is usually sufficient for palate closure. Compensatory differential gene-dosage of other family members (Grhl1 and Grhl3) could be one potential strategy by which an embryo may rescue substantial Grhl2 loss. Additionally, other factors such as differential expression of other (target) genes or co-factors, subtle differences in cell-cell contact and interaction during development, minor variations in genetic background, or even environmental factors affecting certain embryos in utero will also impinge on palatal closure. This latter hypothesis is particularly intriguing, given that retinoic acid, hydroxyurea, mitomycin C and hyperthermia (Seller et al., 1979; Seller and Perkins-Cole, 1987; Seller and Perkins, 1983, 1986) have previously been shown to increase the penetrance of neural tube defects in the curly-tail mouse, a mouse harbouring a mutation in an enhancer of the closely related family member Grhl3 (Gustavsson et al., 2007). Given the known high interaction between environmental factors (such as maternal obesity, alcohol consumption and micro-organism infection) and genetic insufficiency on the prevalence and severity of craniofacial defects (Dixon et al., 2011), it is possible that mutations in the mm1286 enhancer in humans may ultimately prove to be a risk factor for the development of palatal clefts.
The mm1286 enhancer is active in maxillary process epithelium at both E11.5 and E14.5, when the secondary palatal shelves are growing, first downwards adjacent to the tongue and later towards the midline (Bush and Jiang, 2012). Given its role in suppressing epithelial-mesenchymal transition (Ray and Niswander, 2016), we hypothesise that Grhl2 maintains the epithelial phenotype of palatal shelves. Furthermore, this enhancer drives expression in the surface ectoderm (developing skin at E14.5). It is clear that Grhl2 is required for normal skin development because children homozygous for GRHL2 hypomorphic point mutations present with palmoplantar keratoderma (Petrof et al., 2014). As such, further research into the factors that drive Grhl2 expression during skin development may uncover further, non-neural ectoderm specific roles for mm1286 in embryogenesis, adult skin maintenance, skin barrier formation and function, and potentially, also in the development of epidermal cancers.
Our analysis of sequence conservation across forty tetrapod species identified a highly-conserved 325bp element within the mm1286 enhancer that was largely-sufficient for driving reporter (lacZ) expression within the craniofacial primordia at E11.5. Moreover, our discovery of a seemingly critical, invariant 12bp sequence (CTGTCAAACAGGT; Fig. 4A and Fig. S5) within this element paves the way for future studies on the specific genetic mechanisms that physically interact with the mm1286 element through direct binding and formation of transcriptional machinery to recruit RNA polymerase II and augment transcription. As the prevailing model of enhancer-mediated transcription holds that activation by the enhancer and recruitment of the transcriptional complex may drive conformational change to physically localise these proteins to the promoter (Nakagawa et al., 2018), identification of these binding factors will go some way to understanding the nature of upstream signals that drive Grhl2 expression. Our predictive interrogation of putative binding sites has identified a number of candidate homeobox transcription factors - Prep1, Meis1, Meis2, MRG2 and TGIF2 – that may bind to this 12bp region based on known binding site homology. Mutating this sequence substantially abrogated the craniofacial specific activity of the mm1286-325bp element at E11.5, suggesting that binding by one (or more) of these homeobox factors is critical for mediating Grhl2 expression in this region. Functional studies, such as genetic complementarity analyses in mice, in vitro luciferase and in vivo LacZ reporter analyses, and analysis of embryos in which the predicted homeobox transcription factors have been deleted (for synergies with Grhl2) will further advance our knowledge of the pathways by which Grhl2 drives palate closure.
Materials and Methods
Ethics declaration
All animal experimentation procedures, housing and maintenance were performed under Animal Ethics Committee approval from Monash University (E/1200/2012/M) and La Trobe University (#16–72). All animal work was reviewed and approved by the Lawrence Berkeley National Laboratory (LBNL) Animal Welfare Committee. All mice used in this study were housed at the Animal Care Facility (ACF) at LBNL. Mice were monitored daily for food and water intake, and animals were inspected weekly by the Chair of the Animal Welfare and Research Committee and the head of the animal facility in consultation with the veterinary staff. The LBNL ACF is accredited by the American Association for the Accreditation of Laboratory Animal Care (AAALAC). Transgenic mouse assays were performed in Mus musculus FVB background mice. Mm1286 enhancer knockouts were generated in Mus musculus C57Bl/6 strain mice.
Functional validation of putative enhancer elements
Putative enhancer elements (see text and Table S1 for details) were amplified from mouse genomic DNA (Clontech) and cloned into the hsp68-LacZ vector (Kothary et al., 1989; Visel et al., 2007). Pronuclear injections into fertilised eggs, implantation into pseudopregnant mothers, and embryo harvest at embryonic days 11.5 (E11.5) and 14.5 (E14.5) were performed as described previously (Attanasio et al., 2013). Enhancer activity was determined by LacZ staining followed by both whole-mount visualisation and histology and defined by reproducible activity patterns in ≥3 discrete embryos, as described previously (Attanasio et al., 2013). Analysis of Grhl2 expression by autoradiographic in-situ hybridisation (ISH) was conducted as described previously (Auden et al., 2006). Transgenic results for the six tested elements have been deposited in the VISTA enhancer browser (Visel et al., 2007).
Luciferase assays
The mm1286 and homologous human element (termed hs1286) were first cloned into pCRII-TOPO, and next sub-cloned into the pGL4.28 luciferase vector containing a minimal promoter (Promega) using EcoRV/KpnI digestion via standard methods. Next, empty pGL4.28, pGL4.28+mm1286 and pGL4.28+hs1286 (0.5–1μg) were transfected into HEK-293 cells (selected as these cells largely lack endogenous GRHL activity) using TransIT-LT1 according to manufacturer’s instructions, together with the pRL-TK Renilla vector (Promega) to normalise subsequent luciferase expression readings for transfection efficiency. Luciferase assays were conducted using the Dual Luciferase Reporter System (Promega) using a Fluostar Optima Luminometer.
Generation of the mm1286 murine deletion model.
The mm1286 enhancer deletion in mice was generated using in vivo CRISPR/Cas9 editing, as previously described, with only minor modifications (Yang et al., 2014; Yang et al., 2013). Pairs of single guide RNAs (sgRNAs) targeting genomic sequence 5’ and 3’ of the sequence to be deleted were designed using CHOPCHOP (Montague et al., 2014) (see Table S2 for sgRNA sequences and coordinates of deleted regions). Knockout mice were engineered as described previously (Kvon et al., 2016) using a mix containing Cas9 mRNA (final concentration of 100 ng/μl) and two sgRNAs (25 ng/μl each) in injection buffer (10 mM Tris, pH 7.5; 0.1 mM EDTA). This mix was injected into the cytoplasm of single-cell C57Bl/6 mouse embryos. Founder (F0) mice were genotyped using PCR with High Fidelity Platinum Taq Polymerase (Thermo Fisher) to identify those with the desired non-homologous end joining (NHEJ)-generated deletion breakpoints (Fig. S1; Table S2). Sanger sequencing was used to identify and confirm deletion breakpoints in F0 and F1 mice (Fig. S1). Two founder lines were established and F1 or F2 mice were intercrossed. All expected genotypes (mm1286+/+, mm1286+/−, mm1286−/−) were observed at expected Mendelian frequencies. Genotyping was conducted using standard genotyping PCR techniques, with forward and reverse primers located 5’ and 3’ to the mm1286 enhancer respectively, and an enhancer-specific reverse primer to determine presence/absence of enhancer sequence (Fig. S2; forward primer 5’-ggaccagttgtggagaactaactt-3’; reverse primer 5’-ctgcagagctatactgagcaagag-3’; enhancer-specific primer 5’-ttctagaatagctaaccagggct-3’). Grhl2+/−;mm1286+/− embryos were generated by crossing the mm1286+/− mice with the Grhl2+/− mouse line described previously (Rifat et al., 2010).
X-Ray Micro Computed Tomography (μCT)
MicroCT analysis was conducted as described previously (Goldie et al., 2016). Briefly, MicroCT scans were performed on nine mice at post-natal day 0 (P0) from four mm1286+/+ and five mm1286+/− mice, using the Bruker SkyScan 1276 at a pixel resolution of approximately 6.5μm. In these scans, the locations of key skeletal landmarks were identified (Fig. S3, Table S3–S4) based off the set of P0 landmarks (Motch Perrine et al., 2014), and the distances between landmarks were measured (Fig. S3). Landmarks were identified on 3D images from the μCT scan using 3-dimensional (3D) image processing software Mimics (Materialise). Each measurement was taken twice, independently and blindly as to mouse genotype and used to determine mean and standard error. A total of 27 measurements were taken (Fig. S4, Table S3–S4).
Generation of hsp68-lacZ-325bp wild-type and hsp68-lacZ-325bp-mut mutant plasmids for in vivo testing of Grhl2 enhancer activity
The hsp68-lacZ plasmid was enzymatically digested with AscI and XhoI and the 7,185-nucleotide plasmid backbone isolated by gel purification. The 325bp conserved region of the Grhl2 epithelial enhancer (see text for details) was amplified from C57BL/6J mouse genomic DNA using the forward primer 5’-TAAGCACTCGAGTATTTATTTTGCTTCTTTACCTAGC-3’ and reverse primer 5’-TAAGCAGGCGCGCCAAGGTCAAAGGTTTGCAGGTTTAA-3’. This amplicon was subsequently purified using the QIAGEN PCR purification kit according to manufacturer’s instructions. Following digestion with AscI and XhoI, this PCR product was ligated into hsp68-LacZ using T4 DNA ligase and standard protocols to create hsp68-lacZ-325bp. Sanger sequencing was performed to confirm the fidelity of the cloned region. The hsp68-lacZ-325bp-mut plasmid was generated using the Agilent Technologies Quick Change II site-directed mutagenesis kit using forward (5’-CTAATTGTTACCCGGGAAACAGGTCGA-3’) and reverse (5’-TCGACCTGTTTCCCGGGTAACAATTAG-3’) primers. These primers led to a mutation within the homeobox site from CTGTCA to CCGGGA, while simultaneously creating an XmaI restriction site. DNA was isolated from ampicillin-resistant clones and screened by XmaI/XhoI restriction digest, whereby specific 246bp and 126bp restriction digest products were visible; mutagenesis was also confirmed by Sanger sequencing.
Supplementary Material
Supplementary Fig. S1: CRISPR/Cas9-mediated deletion of the mm1286 enhancer element. Two separate mouse lines were generated via CRISPR/Cas9-mediated genomic deletion (A-B). Sanger sequencing chromatograms confirming the deletion breakpoints in each mouse line, together with the size of the genomic element deleted (3,745bp in founder line 1 and 3,740bp in founder line 2 respectively).
Supplementary Fig. S2: Genotyping of mm1286 mouse lines. Schematic diagram of the Grhl2 genomic locus comprising the mm1286 enhancer in WT or mm1286−/− mice (A), showing primerbinding locations relative to the Grhl2 transcription start site (TSS) and expected amplicon size following PCR. (B) representative gel electrophoresis image showing mm1286 nullizygous, heterozygous and non-deleted, wild-type control mice.
Supplementary Fig. S3: Landmarks utilised for quantitation of skull morphometry. Landmarks annotated on a representative μCT skull image, shown in lateral (A), dorsal (B) and ventral (C) views. Ventral view showing detailed landmark measurements of the developing hard palate (D). A listing of landmark points is presented in Supplementary Table 3.
Supplementary Fig. S4: Morphometric analysis of skull shape and size in WT and mm1286−/− (KO) pups at P0 by μCT analysis. (A-C) Quantitation of landmarks (mean ± SEM) in the developing face (A), palate (B) and overall skull (C) show no significant differences in craniofacial development in mice lacking the mm1286-enhancer element.
Supplementary Fig. S5: Genomic region and sequence surrounding the 12bp invariant region of mm1286 from the 40 species shown in Fig. 4. The 12bp sequence (red line) comprises a homeobox region and putative binding sites for the Prep1, Meis1, Meis2, MRG2 and TGIF2 transcription factors.
Supplementary Table S1: Cloning and analysis details for each of the putative Grhl2 enhancer elements.
Supplementary Table S2: CRISPR-Cas9 single guide RNA details for generation of the mm1286−/− mouse.
Supplementary Table S3: Nomenclature and location of skull landmarks for μCT analysis.
Supplementary Table S4: Skull measurements used for μCT analysis.
Supplementary Table S5: Genotypes of embryos derived from mm1286+/− and Grhl2+/;mm1286+/− mouse crosses. (A) Genotypes of embryos derived from mm1286+/− heterozygous parents at either embryonic day 14.5 post-conception (E14.5) or post-natal day 0 (P0), showing no differences from expected Mendelian ratios. (B) Genotypes of embryos derived from crossing Grhl2+/− heterozygous mice with mm1286+/− heterozygous mice at post-natal day 0 (P0), again showing no differences from expected Mendelian ratios. Although no apparent differences in craniofacial (or other) development were seen in wild-type, Grhl2+/− or mm1286+/− pups, 3/18 Grhl2+/−;mm1286+/− presented with a cleft secondary palate (^).
Acknowledgements:
The authors would like to thank Dr. Jose Gonzalez and Dr. Sarbin Ranjitkar for technical assistance. A.V. and M.O. were supported by National Institutes of Health grants U01DE024427 and R01DE028599. S.D. was partially supported by La Trobe UD RFA grant #2000003053. Part of this research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM, 2007. Deletion of ultraconserved elements yields viable mice. PLoS Biol 5, e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrimsson B, Pennacchio LA, Visel A, 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. Science (New York, N.Y 342, 1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM, 2006. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns 6, 964–970. [DOI] [PubMed] [Google Scholar]
- Botti E, Spallone G, Moretti F, Marinari B, Pinetti V, Galanti S, De Meo PD, De Nicola F, Ganci F, Castrignano T, Pesole G, Chimenti S, Guerrini L, Fanciulli M, Blandino G, Karin M, Costanzo A, 2011. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proceedings of the National Academy of Sciences of the United States of America 108, 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ, Kafatos FC, 1991. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes & development 5, 1672–1683. [DOI] [PubMed] [Google Scholar]
- Brouns MR, De Castro SC, Terwindt-Rouwenhorst EA, Massa V, Hekking JW, Hirst CS, Savery D, Munts C, Partridge D, Lamers W, Kohler E, van Straaten HW, Copp AJ, Greene ND, 2011. Over-expression of Grhl2 causes spina bifida in the Axial defects mutant mouse. Hum Mol Genet 20, 1536–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Jiang R, 2012. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development (Cambridge, England) 139, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpinelli MR, de Vries ME, Jane SM, Dworkin S, 2017. Grainyhead-like Transcription Factors in Craniofacial Development. J Dent Res 96, 1200–1209. [DOI] [PubMed] [Google Scholar]
- Cieply B, Riley P.t., Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM, 2012. Suppression of the Epithelial-Mesenchymal Transition by Grainyhead-Like-2. Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M, Helms JA, 2011. Cranial neural crest cells on the move: their roles in craniofacial development. American journal of medical genetics 155A, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, Butcher S, Mansour TA, Kousa YA, Fukazawa CF, Houston DW, Manak JR, Schutte BC, Wagner DS, Cornell RA, 2013. Interferon Regulatory Factor 6 Promotes Differentiation of the Periderm by Activating Expression of Grainyhead-Like 3. J Invest Dermatol 133, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Visel A, Pennacchio LA, 2013. Functional anatomy of distant-acting mammalian enhancers. Philos Trans R Soc Lond B Biol Sci 368, 20120359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, Khin YS, Fukuda-Yuzawa Y, Plajzer-Frick I, Pickle CS, Lee EA, Harrington AN, Pham QT, Garvin TH, Kato M, Osterwalder M, Akiyama JA, Afzal V, Rubenstein JLR, Pennacchio LA, Visel A, 2018. Ultraconserved Enhancers Are Required for Normal Development. Cell 172, 491–499 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC, 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet 12, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin S, Darido C, Georgy SR, Wilanowski T, Srivastava S, Ellett F, Pase L, Han Y, Meng A, Heath JK, Lieschke GJ, Jane SM, 2012. Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development (Cambridge, England) 139, 525–536. [DOI] [PubMed] [Google Scholar]
- Dworkin S, Simkin J, Darido C, Partridge DD, Georgy SR, Caddy J, Wilanowski T, Lieschke GJ, Doggett K, Heath JK, Jane SM, 2014. Grainyhead-like 3 regulation of endothelin-1 in the pharyngeal endoderm is critical for growth and development of the craniofacial skeleton. Mechanisms of development. [DOI] [PubMed] [Google Scholar]
- Evans KN, Sie KC, Hopper RA, Glass RP, Hing AV, Cunningham ML, 2011. Robin Sequence: From Diagnosis to Development of an Effective Management Plan. Pediatrics 127, 936948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldie SJ, Arhatari BD, Anderson P, Auden A, Partridge DD, Jane SM, Dworkin S, 2016. Mice lacking the conserved transcription factor Grainyhead-like 3 (Grhl3) display increased apposition of the frontal and parietal bones during embryonic development. BMC developmental biology 16, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Tan TY, Benko S, FitzPatrick D, Lyonnet S, Farlie PG, 2009. Long-range regulation at the SOX9 locus in development and disease. Journal of medical genetics 46, 649–656. [DOI] [PubMed] [Google Scholar]
- Gustavsson P, Copp AJ, Greene ND, 2008. Grainyhead genes and mammalian neural tube closure. Birth defects research 82, 728–735. [DOI] [PubMed] [Google Scholar]
- Gustavsson P, Greene ND, Lad D, Pauws E, de Castro SC, Stanier P, Copp AJ, 2007. Increased expression of Grainyhead-like-3 rescues spina bifida in a folate-resistant mouse model. Hum Mol Genet 16, 2640–2646. [DOI] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME, 2010. Characterising and Predicting Haploinsufficiency in the Human Genome. PLoS Genet 6, e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersbergen A, Best SA, Dworkin S, Ah-Cann C, de Vries ME, Asselin-Labat ML, Ritchie ME, Jane SM, Sutherland KD, 2018. Lung morphogenesis is orchestrated through Grainyhead-like 2 (Grhl2) transcriptional programs. Developmental biology 443, 1–9. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J, 1989. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development (Cambridge, England) 105, 707–714. [DOI] [PubMed] [Google Scholar]
- Kousa YA, Zhu H, Fakhouri WD, Lei Y, Kinoshita A, Roushangar RR, Patel NK, Agopian AJ, Yang W, Leslie EJ, Busch TD, Mansour TA, Li X, Smith AL, Li EB, Sharma DB, Williams TJ, Chai Y, Amendt BA, Liao EC, Mitchell LE, Bassuk AG, Gregory S, Ashley-Koch A, Shaw GM, Finnell RH, Schutte BC, 2019. The TFAP2A-IRF6GRHL3 genetic pathway is conserved in neurulation. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler A, Buysse K, Clayton-Smith J, Le Caignec C, David A, Engels H, Kohlhase J, Mari F, Mortier G, Renieri A, Wieczorek D, 2011. Five patients with novel overlapping interstitial deletions in 8q22.2q22.3. American journal of medical genetics 155A, 1857–1864. [DOI] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissieres V, Pickle CS, Plajzer-Frick I, Lee EA, Kato M, Garvin TH, Akiyama JA, Afzal V, Lopez-Rios J, Rubin EM, Dickel DE, Pennacchio LA, Visel A, 2016. Progressive Loss of Function in a Limb Enhancer during Snake Evolution. Cell 167, 633–642 e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT, 2007. Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature 446, 79–82. [DOI] [PubMed] [Google Scholar]
- Mangold E, Bohmer AC, Ishorst N, Hoebel AK, Gultepe P, Schuenke H, Klamt J, Hofmann A, Golz L, Raff R, Tessmann P, Nowak S, Reutter H, Hemprich A, Kreusch T, Kramer FJ, Braumann B, Reich R, Schmidt G, Jager A, Reiter R, Brosch S, Stavusis J, Ishida M, Seselgyte R, Moore GE, Nothen MM, Borck G, Aldhorae KA, Lace B, Stanier P, Knapp M, Ludwig KU, 2016. Sequencing the GRHL3 Coding Region Reveals Rare Truncating Mutations and a Common Susceptibility Variant for Nonsyndromic Cleft Palate. Am J Hum Genet 98, 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke C, Cionni M, Siggers T, Bulyk ML, Beier DR, Stottmann RW, 2015. Grhl2 is required in nonneural tissues for neural progenitor survival and forebrain development. Genesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E, 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42, W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motch Perrine SM, Cole TM 3rd, Martinez-Abadias N, Aldridge K, Jabs EW, Richtsmeier JT, 2014. Craniofacial divergence by distinct prenatal growth patterns in Fgfr2 mutant mice. BMC developmental biology 14, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Yoneda M, Higashi M, Ohkuma Y, Ito T, 2018. Enhancer function regulated by combinations of transcription factors and cofactors. Genes Cells 23, 808–821. [DOI] [PubMed] [Google Scholar]
- Nishino H, Takano S, Yoshitomi H, Suzuki K, Kagawa S, Shimazaki R, Shimizu H, Furukawa K, Miyazaki M, Ohtsuka M, 2017. Grainyhead-like 2 (GRHL2) regulates epithelial plasticity in pancreatic cancer progression. Cancer Med-Us 6, 2686–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissieres V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE, Visel A, Pennacchio LA, 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G, 2013. Enhancers: five essential questions. Nat Rev Genet 14, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof G, Nanda A, Howden J, Takeichi T, McMillan JR, Aristodemou S, Ozoemena L, Liu L, South AP, Pourreyron C, Dafou D, Proudfoot LE, Al-Ajmi H, Akiyama M, McLean WH, Simpson MA, Parsons M, McGrath JA, 2014. Mutations in GRHL2 result in an autosomal-recessive ectodermal Dysplasia syndrome. Am J Hum Genet 95, 308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, Rautio J, Pegelow M, Karsten A, Basel-Vanagaite L, Gordon W, Andersen B, Svensson T, Murray JC, Cornell RA, Kere J, Schutte BC, 2013. Dominant Mutations in GRHL3 Cause Van der Woude Syndrome and Disrupt Oral Periderm Development. Am J Hum Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, Rautio J, Pegelow M, Karsten A, Basel-Vanagaite L, Gordon W, Andersen B, Svensson T, Murray JC, Cornell RA, Kere J, Schutte BC, 2014. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet 94, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrgaki C, Liu A, Niswander L, 2011. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Developmental biology 353, 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J, 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray HJ, Niswander LA, 2016. Grainyhead-like 2 downstream targets act to suppress epithelialto-mesenchymal transition during neural tube closure. Development (Cambridge, England) 143, 1192–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y, Parekh V, Wilanowski T, Hislop NR, Auden A, Ting SB, Cunningham JM, Jane SM, 2010. Regional neural tube closure defined by the Grainy head-like transcription factors. Developmental biology 345, 237–245. [DOI] [PubMed] [Google Scholar]
- Seller MJ, Embury S, Polani PE, Adinolfi M, 1979. Neural tube defects in curly-tail mice. II. Effect of maternal administration of vitamin A. Proceedings of the Royal Society of London. Series B, Biological sciences 206, 95–107. [DOI] [PubMed] [Google Scholar]
- Seller MJ, Perkins-Cole KJ, 1987. Hyperthermia and neural tube defects of the curly-tail mouse. J Craniofac Genet Dev Biol 7, 321–330. [PubMed] [Google Scholar]
- Seller MJ, Perkins KJ, 1983. Effect of hydroxyurea on neural tube defects in the curly-tail mouse. J Craniofac Genet Dev Biol 3, 11–17. [PubMed] [Google Scholar]
- Seller MJ, Perkins KJ, 1986. Effect of mitomycin C on the neural tube defects of the curly-tail mouse. Teratology 33, 305–309. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S, Yoshida H, Kawabe T, Yokosuka O, Ogawa S, Omata M, 2008. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol 49, 746–757. [DOI] [PubMed] [Google Scholar]
- Ting SB, Wilanowski T, Auden A, Hall M, Voss AK, Thomas T, Parekh V, Cunningham JM, Jane SM, 2003. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nature medicine 9, 1513–1519. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR, 1999. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes & development 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, Spitz F, 2014. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nature genetics 46, 753–758. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Van Eyken E, Fransen E, Huyghe JR, Topsakal V, Hendrickx JJ, Hannula S, Maki-Torkko E, Jensen M, Demeester K, Baur M, Bonaconsa A, Mazzoli M, Espeso A, Verbruggen K, Huyghe J, Huygen P, Kunst S, Manninen M, Konings A, Diaz-Lacava AN, Steffens M, Wienker TF, Pyykko I, Cremers CW, Kremer H, Dhooge I, Stephens D, Orzan E, Pfister M, Bille M, Parving A, Sorri M, Van de Heyning PH, Van Camp G, 2008. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum Mol Genet 17, 159–169. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA, 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA, 2007. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35, D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona B, Nanda I, Neuner C, Muller T, Haaf T, 2013. Confirmation of GRHL2 as the gene for the DFNA28 locus. American journal of medical genetics 161A, 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U, Pantel K, Assmann V, 2013. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. The Journal of biological chemistry 288, 2299323–008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Jaenisch R, 2014. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9, 1956–1968. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R, 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1: CRISPR/Cas9-mediated deletion of the mm1286 enhancer element. Two separate mouse lines were generated via CRISPR/Cas9-mediated genomic deletion (A-B). Sanger sequencing chromatograms confirming the deletion breakpoints in each mouse line, together with the size of the genomic element deleted (3,745bp in founder line 1 and 3,740bp in founder line 2 respectively).
Supplementary Fig. S2: Genotyping of mm1286 mouse lines. Schematic diagram of the Grhl2 genomic locus comprising the mm1286 enhancer in WT or mm1286−/− mice (A), showing primerbinding locations relative to the Grhl2 transcription start site (TSS) and expected amplicon size following PCR. (B) representative gel electrophoresis image showing mm1286 nullizygous, heterozygous and non-deleted, wild-type control mice.
Supplementary Fig. S3: Landmarks utilised for quantitation of skull morphometry. Landmarks annotated on a representative μCT skull image, shown in lateral (A), dorsal (B) and ventral (C) views. Ventral view showing detailed landmark measurements of the developing hard palate (D). A listing of landmark points is presented in Supplementary Table 3.
Supplementary Fig. S4: Morphometric analysis of skull shape and size in WT and mm1286−/− (KO) pups at P0 by μCT analysis. (A-C) Quantitation of landmarks (mean ± SEM) in the developing face (A), palate (B) and overall skull (C) show no significant differences in craniofacial development in mice lacking the mm1286-enhancer element.
Supplementary Fig. S5: Genomic region and sequence surrounding the 12bp invariant region of mm1286 from the 40 species shown in Fig. 4. The 12bp sequence (red line) comprises a homeobox region and putative binding sites for the Prep1, Meis1, Meis2, MRG2 and TGIF2 transcription factors.
Supplementary Table S1: Cloning and analysis details for each of the putative Grhl2 enhancer elements.
Supplementary Table S2: CRISPR-Cas9 single guide RNA details for generation of the mm1286−/− mouse.
Supplementary Table S3: Nomenclature and location of skull landmarks for μCT analysis.
Supplementary Table S4: Skull measurements used for μCT analysis.
Supplementary Table S5: Genotypes of embryos derived from mm1286+/− and Grhl2+/;mm1286+/− mouse crosses. (A) Genotypes of embryos derived from mm1286+/− heterozygous parents at either embryonic day 14.5 post-conception (E14.5) or post-natal day 0 (P0), showing no differences from expected Mendelian ratios. (B) Genotypes of embryos derived from crossing Grhl2+/− heterozygous mice with mm1286+/− heterozygous mice at post-natal day 0 (P0), again showing no differences from expected Mendelian ratios. Although no apparent differences in craniofacial (or other) development were seen in wild-type, Grhl2+/− or mm1286+/− pups, 3/18 Grhl2+/−;mm1286+/− presented with a cleft secondary palate (^).


