Abstract
Bone marrow-derived mesenchymal stem/stromal cells (BMSCs) are fundamental to bone regenerative therapies, tissue engineering, and postmenopausal osteoporosis. Donor variation among patients, cell heterogeneity, and unpredictable capacity for differentiation reduce effectiveness of BMSCs for regenerative cell therapies. The cell surface glycoprotein CD24 exhibits the most prominent differential expression during osteogenic versus adipogenic differentiation of human BMSCs. Therefore, CD24 may represent a selective biomarker for subpopulations of BMSCs with increased osteoblastic potential. In undifferentiated human BMSCs, CD24 cell surface expression is variable among donors (range: 2%–10%) and increased by two to fourfold upon osteogenic differentiation. Strikingly, FACS sorted CD24pos cells exhibit delayed mineralization and reduced capacity for adipocyte differentiation. RNAseq analysis of CD24pos and CD24neg BMSCs identified a limited number of genes with increased expression in CD24pos cells that are associated with cell adhesion, motility, and extracellular matrix. Downregulated genes are associated with cell cycle regulation, and biological assays revealed that CD24pos cells have reduced proliferation. Hence, expression of the cell surface glycoprotein CD24 identifies a subpopulation of human BMSCs with reduced capacity for proliferation and extracellular matrix mineralization. Functional specialization among BMSCs populations may support their regenerative potential and therapeutic success by accommodating cell activities that promote skeletal tissue formation, homeostasis, and repair.
Keywords: bone marrow mesenchymal stem cells, bone, osteoblast, differentiation
Introduction
Populations of bone marrow-derived mesenchymal stromal cells (BMSCs) contain tissue-specific progenitor cells and represent an attractive biological source for regenerative therapies and tissue engineering applications [1–4]. BMSCs have multilineage potential that can differentiate into cells that express cartilage, fat, and bone markers. Moreover, BMSC populations are very heterogeneous and variable between donors, while there is a lack of specific cell surface markers that can identify defined BMSC subpopulations with distinct biological properties [5–7]. The heterogeneity of these cells and variable differentiation capacity together limit their application in regenerative medicine and tissue engineering [8].
Due to the large differences between isolates of BMSCs and isolates from different anatomical locations (ie, bone marrow, adipose tissue, placenta, Wharton's jelly, and many others), several criteria have been defined that identify isolated BMSCs from various tissues [9,10]. First, BMSCs are plastic adherent non-hematopoietic progenitor cells that express CD71, CD90, and CD150, and lack the expression of CD11b, CD14, CD19, CD34, CD45, and HLA-DR surface molecules. Furthermore, these cells have tri-lineage potential and are able to differentiate into cells that express markers characteristic of osteoblasts, adipocytes, and chondrocytes in cell culture. Although each of these markers can enrich for CFU-F from BMSC, the percentage of cells with properties of progenitor cells is highly variable between different donors [8]. Since the majority of established surface markers are defined for BMSCs, they may be present in different amounts on stromal cells derived from different anatomical locations [10–13] and dependent on the proliferative status [12,14].
Cell surface molecules may support enrichment of BMSC subpopulations with increased stemness and/or osteochondral differentiation potential [15], including CD146/MCAM [16], Stro-1 [17], CD271/LNGFR [18], and SSEA-4 [19,20]. Additionally, their expression is very heterogeneous among different MSC sources [20]. In-depth characterization of cell surface proteins may define select subpopulations of BMSCs with distinct biological properties independent of donor variation.
We have previously identified early regulators of osteogenic and adipogenic lineage commitment based on transcriptome analysis at high temporal density that clarify transcriptional regulatory events during the early stages of osteogenic and adipogenic differentiation of human BMSCs [21]. These gene expression profiles of differentiating BMSCs also revealed that the cell surface protein CD24 [22] is specifically upregulated during osteogenic differentiation of BMSCs. Here, we further investigated the expression of CD24 in human BMSCs and show that CD24 is present on a small subset of ex vivo expanded BMSCs. We characterize this population using gene expression analyses, flow cytometry, and RNA-seq. The main finding of our study is that CD24 positive cells have reduced differentiation potential and proliferative capacity relative to CD24 negative cells. The functional specialization of CD24 positive and negative cells may support different biological activities in regenerative therapies.
Materials and Methods
Cell culture
Bone marrow-derived human mesenchymal stromal cells from healthy individuals were obtained from Lonza (Basel, Switzerland) and differentiated as previously described [23]. Briefly, cells were expanded in Mesenchymal Stem Cell Growth Media (Lonza, Belgium) and 5 × 10 [3] vital cells/cm2 were seeded in 12-well cell culture plates or 175 cm2 cell culture flasks in basic growth media consisting of αMEM (Fisher Scientific, the Netherlands) supplemented with 10% heat inactivated FCS (Fisher Scientific, the Netherlands), 20 mM HEPES (Sigma-Aldrich, the Netherlands), 1.8 mM CaCl2 (Sigma-Aldrich, the Netherlands) and adjusted to pH 7.5. Two days after seeding, cells were differentiated into adipocytes using basic growth media supplemented with 100 nM dexamethasone (Sigma-Aldrich, the Netherlands), 60 μM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich, the Netherlands) and 500 μM indomethacin (Sigma-Aldrich, the Netherlands), or differentiated into osteoblast using basic growth media supplemented with 100 nM dexamethasone (Sigma-Aldrich, the Netherlands) and 10 mM b-glycerophosphate (Sigma-Aldrich, the Netherlands) and cultured at 37°C and 5% CO2 in a humidified atmosphere. Differentiation media was replaced every 3–4 days. Histology of extracellular matrix mineralization (Alizarin Red) and lipid droplets [Oil Red O (ORO)] were performed as previously described [23]. Subsequently to ORO staining, DAPI [4′,6-diamidino-2-phenylindole, final concentration 0.1 μg/mL in phosphate buffered saline (PBS)] was added to stain nuclei. Five independent images were taken from 2 different wells (10 in total) and the total number of cells (ie, stained nuclei DAPI) and ORO positive cells were calculated by using ImageJ and the percentage of adipocytes were calculated.
RNA isolation, cDNA synthesis and quantative PCR
RNA isolation and cDNA synthesis was performed as previously described [23]. Briefly, BMSCs were expanded in Mesenchymal Stem Cell Growth Media (Lonza) and CD24neg and CD24pos were sorted with flow cytometry (BD FACSJazz). After FACSort, cells were immediately spun down (5 min, 1,500 rpm) and cell pellets were resuspended in 500 μL TRIzol (Fisher Scientific, the Netherlands) and further processed using Chloroform and Isopropanol precipitation according to manufacturer's protocol (Fisher Scientific, the Netherlands). Next, 100 ng total RNA was used for the generation of cDNA. All quantitative PCR experiments were generated on an Applied Biosystems' 7500 Real-Time PCR System and relative gene expression levels were calculated using GAPDH as a housekeeping gene. Primers used for the gene expression analyses were as follows: hGAPDH-for: CCG CAT CTT TTG CGT CG; hGAPDH-rev: CCC AAT ACG ACC AAA TCC GTT G; hCD24-for: ACC GAC GGA GGG GAC ATG GG; hCD24-rev: GCG TGG GTA GGA GCA GTG CC; hHTR2A-for: GTG GAC CCT GAA GAC AAA TGA CA; hHTR2A-rev: TTC TCA CCA AAC CGA GGA CA; hFBLN1-for: GGA GAC CGG AGA TTT GGA TGT; hFBLN1-rev: TCA GAT ATG GGT CCT CTT GTT CCT; hALPL-for: TAA AGCA GGT CTT GGG GTG C; hALPL-rev: GGG TCT TTC TCT TTC TCT GGC A; hCCNA2-for: GCG GTA CTG AAG TCC GGG AA; hCCNA2-rev: GTG CAA CCC GTC TCG TCT TC; hCDC20-for: TGG CTG AAC TCA AAG GTC ACA; hCDC20-rev: CAA AAC AGC GCC ATA GCC TC.
RNAseq and gene expression data analyses
The microarray gene expression from Fig. 1 is publicly available and can be retrieved from the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) under the accession number GSE80614 [21]. RNA sequencing was performed as previously described [14]. All differentially expressed cell surface proteins were selected and a hierarchical cluster tree was generated using R. RNAseq data analysis was carried out using the DESeq2 package and limma [24,25].
FIG. 1.
CD24 is induced immediately upon osteoblast differentiation. (A) Heatmap representing the expression changes of 192 differential expressed cell surface proteins (represented by 251 different probes) during osteoblast differentiation compared to adipocyte differentiating BMSC. Red is higher expressed in osteoblasts differentiating BMSC, Green is higher expressed in adipocyte differentiation BMSC. (1 donor, n = 3 per time point). (B) Next generation RNA sequencing data of CD24 mRNA expression in osteoblast differentiating human mesenchymal stromal cells from two different donors (n = 1 per donor). (C) qPCR analyses of CD24 mRNA expression in osteoblast differentiating BMSCs in two different donors illustrate that CD24 is quickly induced upon osteogenic differentiation and reduce to basal levels just prior extracellular matrix mineralization (n = 2 per donor). BMSC, bone marrow-derived mesenchymal stem/stromal cell; qPCR, quantitative PCR. Color images are available online.
Flow cytometry analysis and sorting
After trypsinization, cells were washed with 1% PBA (PBS with 1% Bovine Serum Albumin Fraction V, Roche), resuspended in 1% PBA, and incubated with mouse αCD24 (human) conjugated with PE or FITC (1:50, clone ML5; BD Biosciences) for 30 min on ice. Cells were washed twice and samples were analyzed on a flow cytometer (BD Accuri c6; BD Bioscience, USA). Flow Cytometer assisted cell sorting experiments were carried out on a FACSAria II or FACSjazz (BD Bioscience, USA). Cells were gated in the forward- and side-scatter, Doublets were excluded and subsequently gated for CD24 positive events (Fig. 2A). For the Ki67 proliferation experiments, BMSCs cultured in basic growth media were trypsinized and fixed with 70% EtOH. After 30 min incubation at −20°C, cells were washed in 1% PBA (PBS with 1% Bovine Serum Albumin Fraction V, Roche) and stained for 30 min with αKi67-Alexa488 (#561165; BD Bioscience, USA) and αCD24-PE (#555428; BD Bioscience, USA). Stained cells were washed once with 1% PBA and analyzed on a Accuri C6 FACS (BD Bioscience). For the analyses of cell proliferation using EdU incorporation (Click-iT™ EdU Alexa Fluor™ 647 Flow Cytometry Assay, #C10424; Invitrogen, the Netherlands) cells were incubated for 24 h with Click-it EdU (final concentration 5 μM), trypsinized, and fixed with Click-iT Saponin–based permeabilization. After Click-iT reaction with Alexa647, cells were stained with CD24-PE for 30 min and analyzed on a Accuri C6 FACS.
FIG. 2.
CD24 is expressed in a subset of BMSC and induced upon osteogenic differentiation. (A) Flow cytometry gating of CD24 positive cells. After gating cells, the CD24 positive cells (αCD24-PE, Fl-2) are gated against autofluorescent signals (Fl-1). Example of (1) Isotype (Upper right), (2) undifferentiated BMSC (lower left), and (3) 7 days osteogenic differentiated BMSCs (lower right). (B) Percentage of CD24pos cells determined by flow cytometry. Analyses of cell surface expression of CD24 before and after 7 days of osteoblast differentiation in three different BMSC donors and different passages. (C) Fold change increase in the number of CD24pos cells after 7 days of osteogenic differentiating BMSCs compared to undifferentiated BMSCs. Fold changes are derived from the FACS experiments in Fig. 2B. (D) CD24 cell surface expression during osteogenic and adipogenic differentiating BMSC (two different BMSC donors, n = 2 per time point). Color images are available online.
Results
CD24 expression is induced early upon osteogenic differentiation of BMSC
Building on transcriptome data that revealed transcriptional changes during the early stages of osteoblast and adipocyte differentiation of human BMSC [21], we have identified genes encoding cell surface proteins that are induced in early differentiating osteoblasts. We hypothesized that genes coding for cell surface proteins that are differentially expressed during BMSC differentiation may identify human BMSC subpopulations with increased bone regenerative capacity in undifferentiated heterogeneous BMSCs. Therefore, we compared the expression levels of all cell surface expressed proteins after induction of either osteoblastic or adipocytic differentiation. The cell surface protein CD24, a glycoprotein that was initially identified on the surface of B lymphocytes [26], was among the most strongly induced genes during the first 4 days of osteogenic differentiation (Fig. 1A). RNA-seq data from two independent BMSC donors (#3520 and #4266) and similar time course and treatment corroborated the finding that CD24 expression is upregulated and reaches maximal levels during osteogenic differentiation between days 4 and 14 (Fig. 1B). Quantitative PCR analysis of CD24 expression during osteoblast differentiation validated that CD24 was reproducibly induced upon osteogenic differentiation and back at basal levels just prior extracellular matrix mineralization at day 17 (Fig. 1C).
CD24 cell surface expression is heterogeneously present in differentiating BMSC
To assess whether changes in mRNA levels of CD24 (Fig. 1) translate into cell surface expression, we performed fluorescence-activated flow cytometry (FACS) analysis using a CD24 antibody with human BMSCs that are either proliferating or subjected to induction of osteogenic differentiation (Fig. 2A–C). In proliferating cells (day 0), expression of CD24 was observed in 3%–15% of the cells among three different donors, and this fraction increased to 12%–35% after 7 days of osteogenic differentiation (Fig. 2B). While the initial fraction of CD24pos cells varied before the induction of differentiation by as much as fivefold (Fig. 2B), the fold increase in the percentage of 7 days osteogenic differentiated CD24pos cells was comparable among donors and varies by two to four fold (Fig. 2C). Because CD24 expression was induced in differentiating osteoblasts by day 7 (Fig. 1), we compared its expression in a longer time course during nondifferentiating, osteogenic and adipogenic differentiation using fluorescence-activated flow cytometry (Fig. 2D). The CD24 cell surface expression is dynamic and induced within 3 days of osteogenic differentiation of BMSCs, but not upon stimulation of adipogenic differentiation or in undifferentiated cells at this time point. During osteoblastic differentiation of BMSCs, total expression of CD24 within cell cultures increases further at days 7 and 10, while a modest elevation was observed during adipogenic differentiation and CD24 levels remained low in undifferentiated cell cultures (Fig. 2D). These results indicate that CD24 is only present on the surface of a relatively small fraction of human BMSCs, is heterogeneously expressed, exhibits significant donor to donor to variation, and is consistently stimulated upon osteogenic differentiation while reaching maximal cell surface expression after 7–10 days of osteogenic differentiation.
CD24 positive sorted cells have reduced capacity for adipogenic differentiation in vitro
Because CD24 is specifically induced during osteogenesis of BMSCs, we hypothesized that these cells may have an important function during osteoblast differentiation and the formation of a mineralized extracellular matrix. Therefore, we investigated the biological properties of FACS-sorted proliferating BMSCs for CD24pos cells (Fig. 3A, B) and monitored the ability of the sorted cells to differentiate in vitro along the osteoblastic (Fig. 3C) or adipocytic (Fig. 3D, E) lineage. Analysis of CD24 mRNA levels in unsorted and sorted BMSC populations 2 days after the sort and just prior differentiation shows that flow sorting based on CD24 cell surface expression is effective in separating cells that either express CD24 mRNA or not (Fig. 3B, fivefold increase in CD24 expression after sorting). RT-qPCR shows that expression of CD24 also increases in CD24neg BMSCs after 7 days of osteoblast differentiation (Fig. 3B). Thus, CD24neg BMSCs remain capable of inducing CD24 expression during osteogenic differentiation. Interestingly, osteogenic differentiation of CD24pos was reduced whereas CD24neg and non-sorted (NS) BMSCs each produced a mineralized extracellular matrix after 21 days (Fig. 3C). However, in contrast to CD24neg and NS cells, adipogenic differentiation of CD24pos BMSCs resulted in a significantly lower number of ORO positive adipocytes and decreased cell number (Fig. 3D, E). These findings suggest that CD24pos cells have decreased differentiation potential, cell proliferation, and/or cell survival relative to other BMSCs in the population.
FIG. 3.
CD24 marks a subset of hMSC with reduced osteogenic and adipogenic differentiation capacity. (A) To investigate the functional differences of the CD24 cell populations in BMSCs, we sorted the cells using flow cytometry. Three populations were sorted: CD24pos, CD24neg, and unsorted cells (NS), and differentiated into adipocytes and osteoblasts. (B) CD24 expression determined 2 days after sorting for CD24 cell surface expression. Gene expression levels are relative to GAPDH. (n = 2). (C) Differentiation of the sorted population resulted in a reduced mineralization. In the CD24 positive sorted cells. (D) CD24pos sorted cells have a reduced adipogenic differentiation potential. Cells were fixed after 18 days of differentiation and stained with DAPI (nuclei, total cell number) and ORO (Adipocytes). For each condition, five independent images were analyzed (from two different wells). Quantification shows that only 12% of the CD24pos cells were differentiated into ORO positive adipocytes whereas the 25%–30% of the unsorted and CD24neg cells were differentiated into ORO positive adipocytes. (E) Quantification (n = 10) of the data represented in Fig. 3D. Left panel total nr cells (counted by the DAPI positive nuclei). Right panel: percentage of ORO positive adipocytes. DAPI, 4′,6-diamidino-2-phenylindole; NS, non-sorted; ORO, Oil Red O. Color images are available online.
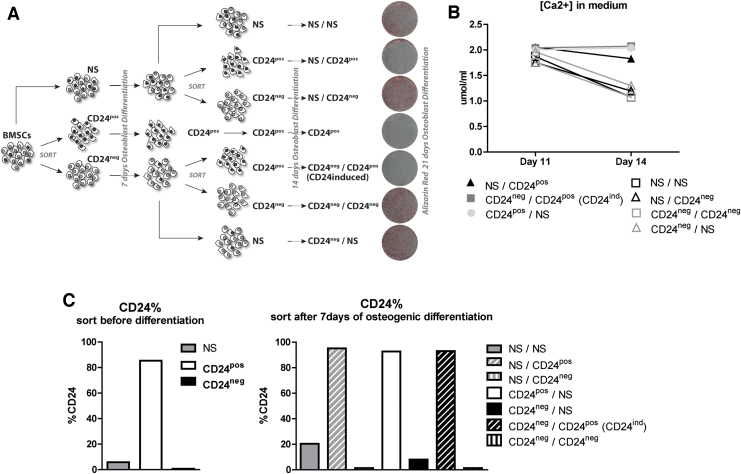
Both CD24pos and CD24ind cells have reduced osteogenic capacity
The induction of CD24pos cells from CD24neg cells upon osteoblast differentiation indicates the presence of an induced population of CD24pos cells (ie, CD24ind) that develops within the first week of differentiation. To investigate the differences between CD24pos and the CD24ind cells, we sorted the CD24pos and CD24neg cells from the proliferating BMSCs and performed a second sort in both populations after 7 days of osteogenic differentiation (Fig. 4A). FACS analyses illustrate that CD24 cell surface expression in CD24pos cells isolated from proliferating cells is stable and does not change appreciably after 7 days of osteogenic differentiation (Fig. 4C, compare CD24pos in left panel and CD24pos/NS in right panel). In contrast, populations of CD24neg cells from the proliferating BMSC population developed a small but noticeable subset of CD24pos cells (up to ∼10%) after osteogenic differentiation (Fig. 4C, compare CD24neg in left panel with CD24neg/NS in right panel). This subset is lower than the subset of CD24pos cells (20%) that is derived from the cells that were not sorted (Fig. 4C, compare NS in left panel with NS/NS in right panel). The FACS results collectively indicate that there are at least three distinct populations present in undifferentiated human BMSCs: (1) CD24pos cells, (2) CD24neg cells, and (3) CD24neg cells that become CD24pos after osteogenic differentiation (ie, CD24ind). Remarkably, both CD24pos and CD24ind cells exhibit reduced and/or delayed mineralization of the extracellular matrix after 21 days of differentiation (Fig. 4A), whereas the CD24neg cells and control sorted cells remain competent to produce a mineralized extracellular matrix. Quantitative analyses of Ca2+ depletion from de media corroborated the results of Alizarin Red staining (Fig. 4A, B). Taken together, although CD24 is strongly induced upon osteogenic differentiation, CD24pos cells have reduced matrix mineralization potential.
FIG. 4.
CD24pos population derived from proliferating and osteogenic differentiated cells have lower differentiation capacity. (A) Double sorting strategy to obtain and investigate CD24pos cells that are present in undifferentiated human BMSCs and that are formed from CD24neg cells upon osteoblast differentiation. Proliferating human BMSCs were sorted in three populations: CD24pos, CD24neg, and unsorted cells (NS). All populations were osteogenic differentiated for 7 days, trypsinized, and a second round of cell sorting was applied from the CD24neg and unsorted population (NS) to obtain another CD24neg, CD24pos and unsorted population NS. The cells were seeded and cultured in osteogenic differentiation media for another 14 days. Cells were fixed and the extracellular matrix mineralization was stained with Alizarin Red (B) Calcium concentration in the media after differentiation of the different populations of Fig. 4A. (C) Percentage of CD24pos cells after sorting of proliferating cells (left panel) or after the second sort of osteogenic differentiated populations (right panel). Color images are available online.
Transcriptome analyses of CD24 sorted populations indicates reduced cell proliferation in CD24 positive cells
Although CD24 is specifically induced upon osteoblast differentiation, sorted CD24pos cells are not able to produce a normal mineralized extracellular matrix in contrast to the CD24neg population. To determine the molecular and transcriptional differences between the CD24pos and CD24neg cells in proliferating human BMSCs, we sorted the two different populations of three different BMSCs donors and performed next generation RNA sequencing (Fig. 5A). CD24 was the most differentially expressed gene (∼40-fold, Fig. 5B, C) between the CD24pos and CD24neg cells reflecting the purity of the sorted population. Moreover, this finding suggests that surface expression of CD24 correlates with the mRNA expression in the cells. Surprisingly, only a limited number of genes were differentially expressed between the CD24pos and CD24neg cells (Fig. 5A). We found 69 genes that have significantly higher and 111 genes significantly lower gene expression levels of which only 27 and 16 genes are modulated by more than twofold, respectively (Fig. 5B, Supplementary Data S1).
FIG. 5.
Differential expressed genes in CD24pos cells have reduced expression of genes involved in mitotic cell cycle. (A) FACSorting strategy of proliferating BMSCs that express CD24 on surface for the RNAseq. (B) Gene expression analyzes comparing the CD24pos and CD24neg BMSCs. In red the genes that are significantly (FDR 1%) differentially expressed between the two populations. In brackets the number of genes significantly different (1%FDR) with a fold change = 2 (n = 3 donors). (C) Validation of genes higher (red) or lower (green) expressed in CD24pos cells compared to CD24neg cells. Left panel gene expression data from the RNAseq analyses and right panel analyses of the same genes by qPCR in independently sorted cells (n = 3 donors). (D) Venn diagram of the genes associated with extracellular matrix, cell adhesion and cell motility and enriched in the CD24pos population. qPCR, quantitative PCR. Color images are available online.
Validation of some up- and down-regulated genes using RT-qPCR of independent FACS sorted cells confirmed the RNAseq data (Fig. 5C). Gene ontology analyses indicated that beyond very general functional groups, the CD24pos cells are enriched for genes associated with extracellular matrix organization (GO:0044421, GO:0030198, GO0043062), programmed cell death (GO:0008219, GO:0012501), and cell differentiation (GO:0030154) (Supplementary Table S1). Genes that were enriched in CD24neg cells are mainly associated with processes of cell cycle and mitosis (GO:0051301, GO:0000278, GO:0007049) (Supplementary Table S2). Interestingly, we found 20 out 69 genes with increased expression that belong to 3 gene ontology terms: cell adhesion (GO:0007155; P = 7.5 × 10−4), motility (GO:0048870; P = 5.9 × 10−3) and extracellular matrix (GO:0031012; P = 1.3 × 10−4) (Fig. 5D). Furthermore, these results indicate that CD24pos cells have a reduced proliferative capacity that agrees with the decreased number of cells when cells were differentiated into adipocytes (Fig. 3D). Although not significant, cell proliferation analyses illustrated reduced proliferative capacity of the CD24pos population just prior osteogenic differentiation (Fig. 6A, B upper panel) or after 7 days of osteogenic differentiation (Fig. 6B lower panel).
FIG. 6.
CD24 positive cells have lower proliferative capacity. (A) Ki67 analyses in proliferating BMSCs. Proliferating BMSC double stained with Ki67-Alexa488 and CD24-PE and analyzed on a BD Accuri. The CD24 positive and negative cells were gated (panel 2) and analyzed for the percentage Ki67pos cells (n = 4, 2 donors). (B) EDU analyses of proliferating BMSCs and after 7 days osteogenic differentiation. Proliferating and osteogenic differentiating (7 days) BMSCs were pulse labeled with EDU and stained with CD24-PE and analyzed on a BD Accuri. The CD24 positive and negative cells were gated (panel 2) and analyzed for the percentage EDUpos cells (n = 6, 1 donor). Color images are available online.
Discussion
Mesenchymal stromal cells are the ideal source in regenerative therapies for skeletal defects. However, a priori prediction of mesenchymal stromal cell behavior, expansion capacity, and differentiation potential is important for efficient use in the clinic. Although it is well known that the mesenchymal stromal cell population is very heterogeneous, the selection of a homogeneous cell population with reproducible expansion and differentiation capacity is still difficult due to the lack of well characterized cell surface molecules.
Here, we have investigated a cell surface expressed protein, CD24, which is induced upon osteogenic differentiation and present only on a subset of the proliferating and differentiated BMSCs. We observed that CD24 expression is strongly induced upon osteogenic differentiation of MSCs. Cell sorting experiments revealed that the CD24pos cells, which were derived from cultures undergoing proliferative expansion, were not able to generate a mineralized extracellular matrix after osteoblastic differentiation in vitro. These cells also exhibited a reduced ability for adipogenic differentiation. Furthermore, RNA-seq data showed that genes involved in mitosis and cell cycle progression are expressed at lower levels in these sorted CD24pos cells. Hence, CD24pos cells appear to have less proliferative potential and reduced capacity for either osteogenic or adipogenic differentiation.
In the mid 1990s, Liu et al. investigated individual osteoblast populations by PCR and immunohistochemistry and already concluded that individual osteoblast colonies were different in osteoblast marker expression and were able to divide in less mature or more mature osteoblast colonies [27]. Nevertheless, osteoblast biomarker expression within single colonies was very variable. These studies advanced the notion that osteoblasts exhibit a considerable degree of heterogeneity and there may not be a single unique osteoblast phenotype, but rather a flexible pattern of osteoblast gene expression [28]. More recently, quantitative measurements of gene expression levels in chondrocytes and chondrogenically induced MSCs showed that these cells exhibit substantial mRNA expression heterogeneity [29]. RNA FISH experiments in single cells indicated that differentiation markers in sister cell pairs have high levels of mRNA variability and that marker gene expression in chondrocytes is not heritable. Hence, sorting of subpopulations in chondrocytes based on cartilage markers may only marginally enrich for progenitor populations that are suitable for therapeutic applications [29].
Our data show that BMSCs sorted for the CD24 cell surface marker exhibit a transcriptome that is quite similar to the original starting population. Thus, rather than representing a unique subpopulation of BMSCs, these CD24pos cells may represent a dynamic subpopulation that is part of a continuum of molecular phenotypes in MSCs that apparently display a level of plasticity that is comparable to that initially described for osteoblastic cells by the Aubin laboratory [28]. While others found a difference of 60-fold in CD24 mRNA expression in in vitro BMSCs cultures [30], we show that CD24 was differentially present on the cell surface (3%–15%) of different BMSCs donors and illustrating the large heterogeneity in CD24 expression among different donors.
CD24 is a sialoglycoprotein and anchored via a glycosyl phosphatidylinositol (GPI) link to the cell surface [31] and first identified on B lymphocytes. Moreover, CD24 has been described to be involved in many different downstream signaling networks and pathways during neural development [32]. Interestingly, lineages tracing studies in mice have shown that CD24pos cells can generate a CD24neg population in vivo that express late markers of adipogenesis [33]. Others showed that CD24 is significantly increased in cultures rich in mesenchymal stem cells and suggests that CD24 marks cells with stem cell properties within human bone marrow and breast adipose tissue [34]. Our results illustrate that the CD24pos population is stable after sorting, does not change CD24 cell surface expression but has a reduced adipogenic and osteogenic differentiation capacity. Suggesting that CD24pos population here is representing a different pool than was described earlier [33,34], or its expression varies among different compartments [35] and may be the result of differences between human and mice cell surface expression [36,37]. Moreover, our results suggest that the sorted CD24pos by its own have an impaired osteogenic and adipogenic differentiation capacity that is dependent on other subpopulations of BMSCs.
Our flow cytometry cell sorting on CD24 expression was very efficient and the phenotypic outcomes (eg, reduced osteogenic differentiation) were very reproducible within and among BMSCs from different donors. RNAseq data indicate that only very few genes are co-expressed within the different populations (eg, CD24neg cells and CD24pos cells). Beyond the expected robust differences in CD24 mRNA expression, we observed only very modest quantitative changes in gene expression for the few genes that were modulated in the two sorted populations. The strong glycosylation of CD24 on the cell surface have been involved in signaling of cell–cell interactions suggest additional role in cell–cell communication in osteogenic differentiating BMSCs and beyond the intracellular signaling and is explained by the overrepresentation of genes involved in cell adhesion and motility [38,39].
Although CD24pos cells and the parental BMSC populations have very similar transcriptomes, cell surface expression of CD24 identifies a subpopulation with reduced proliferative potential and differentiation capacity. The key question remains whether this subset of BMSCs has a unique biological function or may appear as part of a dynamic heterogeneous population [40,41]. Because MSCs in general are known to have trophic functions, it is possible that these CD24pos cells have an auxiliary role as “helper cells” in the overall BMSC population as was previously suggested for CD24pos Paneth cells in the intestinal crypt [42]. Furthermore, CD24 is a well defined negative surface marker for breast cancer stem cells [43], however, a recent report suggests that CD24 can be used as a positive marker for osteosarcoma tumor-initiating cells [44]. Zhou et al. illustrate that the invasive and migration ability of osteosarcoma cells were significantly enhanced after upregulating CD24. Others demonstrated that downregulation of CD24 suppresses bone metastasis of lung cancer cells [45]. Interestingly, 20 out 69 genes upregulated in CD24pos cells are associated with cell adhesion, extracellular matrix, and cell motility. Hence, CD24pos cells may support the overall activity of adjacent MSCs through cell/cell contact, production of an extracellular matrix, or secretion of paracrine factors.
Conclusions
Heterogeneity of cultured BMSCs is highly relevant for their use in regenerative medicine, because these cells have individual properties with unique biological functions that upon sorting may permit use as specialized BMSC subtypes and as a population they have collective properties as mutually supportive cells that may communicate by juxtacrine or paracrine signaling. Although we find CD24 is upregulated in osteogenic differentiation of BMSCs, the CD24 population by itself was not able to increase ECM mineralization in vitro. In addition, it is unclear from our study how expression of CD24 alters the biological properties of BMSCs, and hence what the physiological relevance of these cells could be in vivo. Our data indicate that this population exists in cultured BMSC, and our RNA-seq data revealed changes in the transcriptomes of CD24 positive cells compared to CD24 negative cells related to cell migration and adhesion similar to findings on CD24 positive cells in other contexts [46].
Supplementary Material
Acknowledgments
We thank the members of our laboratories, including Marijke Schreuders-Koedam for technical assistance, as well as Chris Paradise and Roman Thaler for stimulating discussions.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the Dutch Institute for Regenerative medicine (NIRM: grant no. FES0908), Erasmus MC Stem Cell and Regenerative Medicine Institute and Erasmus Medical Center (EMC-MM-01-39-02), and European Commission FP7 Program INTERBONE Grant PIRSES-GA-2011–295181. This work was also supported in part by NIH grant R01 AR049069 (to AJvW).
Supplementary Material
References
- 1. Steinert AF, Rackwitz L, Gilbert F, Nöth U and Tuan RS. (2012). Concise review: the clinical application of mesenchymal stem cells for musculoskeletal regeneration: current status and perspectives. Stem Cells Transl Med 1:237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernández Vallone VB, Romaniuk MA, Choi H, Labovsky V, Otaegui J and Chasseing NA. (2013). Mesenchymal stem cells and their use in therapy: what has been achieved? Differentiation 85:1–10 [DOI] [PubMed] [Google Scholar]
- 3. Murphy MB, Moncivais K and Caplan AI. (2013). Mesenchymal stem cells: environmentally responsive therapeutics for regenerative medicine. Exp Mol Med 45:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Samsonraj RM, Raghunath M, Nurcombe V, Hui JH, van Wijnen AJ and Cool SM. (2017). Concise review: multifaceted characterization of human mesenchymal stem cells for use in regenerative medicine. Stem Cells Transl Med 6:2173–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and Marshak DR. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147 [DOI] [PubMed] [Google Scholar]
- 6. Strioga M, Viswanathan S, Darinskas A, Slaby O and Michalek J. (2012). Same or not the same? Comparison of adipose tissue-derived versus bone marrow-derived mesenchymal stem and stromal cells. Stem Cells Dev 21:2724–2752 [DOI] [PubMed] [Google Scholar]
- 7. Billing AM, Ben Hamidane H, Dib SS, Cotton RJ, Bhagwat AM, Kumar P, Hayat S, Yousri NA, Goswami N, et al. (2016). Comprehensive transcriptomic and proteomic characterization of human mesenchymal stem cells reveals source specific cellular markers. Sci Rep 6:21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sivasubramaniyan K, Lehnen D, Ghazanfari R, Sobiesiak M, Harichandan A, Mortha E, Petkova N, Grimm S, Cerabona F, et al. (2012). Phenotypic and functional heterogeneity of human bone marrow- and amnion-derived MSC subsets. Ann N Y Acad Sci 1266:94–106 [DOI] [PubMed] [Google Scholar]
- 9. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini FC, Krause DS, Deans RJ, Keating A, Prockop DJ and Horwitz EM. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8:315–317 [DOI] [PubMed] [Google Scholar]
- 10. Mushahary D, Spittler A, Kasper C, Weber V and Charwat V. (2018). Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytometry A 93:19–31 [DOI] [PubMed] [Google Scholar]
- 11. Rojewski MT, Weber BM and Schrezenmeier H. (2008). Phenotypic characterization of mesenchymal stem cells from various tissues. Transfus Med Hemother 35:168–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Camilleri ET, Gustafson MP, Dudakovic A, Riester SM, Garces CG, Paradise CR, Takai H, Karperien M, Cool S, et al. (2016). Identification and validation of multiple cell surface markers of clinical-grade adipose-derived mesenchymal stromal cells as novel release criteria for good manufacturing practice-compliant production. Stem Cell Res Ther 7:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Samsonraj RM, Paradise CR, Dudakovic A, Sen B, Nair AA, Dietz AB, Deyle DR, Cool SM, Rubin J and van Wijnen AJ. (2018). Validation of osteogenic properties of cytochalasin D by high-resolution RNA-sequencing in mesenchymal stem cells derived from bone marrow and adipose tissues. Stem Cells Dev 27:1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, et al. (2014). High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem 115:1816–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Busser H, Najar M, Raicevic G, Pieters K, Velez Pombo R, Philippart P, Meuleman N, Bron D and Lagneaux L. (2015). Isolation and characterization of human mesenchymal stromal cell subpopulations: comparison of bone marrow and adipose tissue. Stem Cells Dev 24:2142–2157 [DOI] [PubMed] [Google Scholar]
- 16. Baksh D, Yao R and Tuan RS. (2007). Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cells 25:1384–1392 [DOI] [PubMed] [Google Scholar]
- 17. Ning H, Lin G, Lue TF and Lin C-S. (2011). Mesenchymal stem cell marker Stro-1 is a 75kd endothelial antigen. Biochem Biophys Res Commun 413:353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuci S, Kuci Z, Kreyenberg H, Deak E, Putsch K, Huenecke S, Amara C, Koller S, Rettinger E, et al. (2010). CD271 antigen defines a subset of multipotent stromal cells with immunosuppressive and lymphohematopoietic engraftment-promoting properties. Haematologica 95:651–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gang EJ, Bosnakovski D, Figueiredo CA, Visser JW and Perlingeiro RCR. (2007). SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood 109:1743–1751 [DOI] [PubMed] [Google Scholar]
- 20. Lv F-J, Tuan RS, Cheung KMC and Leung VYL. (2014). Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells 32:1408–1419 [DOI] [PubMed] [Google Scholar]
- 21. van de Peppel J, Strini T, Tilburg J, Westerhoff H, van Wijnen AJ and van Leeuwen JP. (2017). Identification of three early phases of cell-fate determination during osteogenic and adipogenic differentiation by transcription factor dynamics. Stem Cell Reports 8:947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aigner S, Sthoeger ZM, Fogel M, Weber E, Zarn J, Ruppert M, Zeller Y, Vestweber D, Stahel R, Sammar M and Altevogt P. (1997). CD24, a mucin-type glycoprotein, is a ligand for P-selectin on human tumor cells. Blood 89:3385–3395 [PubMed] [Google Scholar]
- 23. Bruedigam C, van Driel M, Koedam M, van de Peppel J, van der Eerden BCJ, Eijken and J Mvan Leeuwen PTM. (2011). Basic techniques in human mesenchymal stem cell cultures: differentiation into osteogenic and adipogenic lineages, genetic perturbations, and phenotypic analyses. Curr Protoc Stem Cell Biol Chapter 1:Unit1H..3. [DOI] [PubMed] [Google Scholar]
- 24. Love MI, Huber W and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W and Smyth GK. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duperray C, Boiron JM, Boucheix C, Cantaloube JF, Lavabre-Bertrand T, Attal M, Brochier J, Maraninchi D, Bataille R and Klein B. (1990). The CD24 antigen discriminates between pre-B and B cells in human bone marrow. J Immunol 145:3678–3683 [PubMed] [Google Scholar]
- 27. Liu F, Malaval L, Gupta AK and Aubin JE. (1994). Simultaneous detection of multiple bone-related mRNAs and protein expression during osteoblast differentiation: polymerase chain reaction and immunocytochemical studies at the single cell level. Dev Biol 166:220–234 [DOI] [PubMed] [Google Scholar]
- 28. Liu F, Malaval L and Aubin JE. (1997). The mature osteoblast phenotype is characterized by extensive plasticity. Exp Cell Res 232:97–105 [DOI] [PubMed] [Google Scholar]
- 29. Cote AJ, McLeod CM, Farrell MJ., McClanahan PD, Dunagin MC, Raj A and Mauck RL. (2016). Single-cell differences in matrix gene expression do not predict matrix deposition. Nat Commun 7:10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schäck LM, Buettner M, Wirth A, Neunaber C, Krettek C, Hoffmann A and Noack S. (2016). Expression of CD24 in human bone marrow-derived mesenchymal stromal cells is regulated by TGFβ3 and induces a myofibroblast-like genotype. Stem Cells Int 2016:1319578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kay R, Rosten PM and Humphries RK. (1991). CD24, a signal transducer modulating B cell activation responses, is a very short peptide with a glycosyl phosphatidylinositol membrane anchor. J Immunol 147:1412–1416 [PubMed] [Google Scholar]
- 32. Gilliam DT, Menon V, Bretz NP and Pruszak J. (2017). The CD24 surface antigen in neural development and disease. Neurobiol Dis 99:133–144 [DOI] [PubMed] [Google Scholar]
- 33. Berry R and Rodeheffer MS. (2013). Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol 15:302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wetzig A, Alaiya A, Al-Alwan M, Pradez CB, Pulicat MS, Al-Mazrou A, Shinwari Z, Sleiman GM, Ghebeh H, et al. (2013). Differential marker expression by cultures rich in mesenchymal stem cells. BMC Cell Biol 14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subramanian A, Fong C-Y, Biswas A and Bongso A. (2015). Comparative characterization of cells from the various compartments of the human umbilical cord shows that the Wharton's Jelly compartment provides the best source of clinically utilizable mesenchymal stem cells. PLoS One 10:e0127992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mestas J and Hughes CCW. (2004). Of mice and not men: differences between mouse and human immunology. J Immunol 172:2731. [DOI] [PubMed] [Google Scholar]
- 37. Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J and Rao MS. (2004). Differences between human and mouse embryonic stem cells. Dev Biol 269:360–380 [DOI] [PubMed] [Google Scholar]
- 38. Ohl C, Albach C, Altevogt P and Schmitz B. (2003). N-glycosylation patterns of HSA/CD24 from different cell lines and brain homogenates: a comparison. Biochimie 85:565–573 [DOI] [PubMed] [Google Scholar]
- 39. Barkeer S, Chugh S, Batra SK and Ponnusamy MP. (2018). Glycosylation of cancer stem cells: function in stemness, tumorigenesis, and metastasis. Neoplasia 20:813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schellenberg A, Stiehl T, Horn P, Joussen S, Pallua N, Ho AD and Wagner W. (2012). Population dynamics of mesenchymal stromal cells during culture expansion. Cytotherapy 14:401–411 [DOI] [PubMed] [Google Scholar]
- 41. Duscher D, Rennert RC, Januszyk M, Anghel E, Maan ZN, Whittam AJ, Perez MG, Kosaraju R, Hu MS, et al. (2014). Aging disrupts cell subpopulation dynamics and diminishes the function of mesenchymal stem cells. Sci Rep 4:7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M and Clevers H. (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469:415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ and Clarke MF. (2003). Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Z, Li Y, Kuang M, Wang X, Jia Q, Cao J, Hu J, Wu S, Wang Z and Xiao J. (2020). The CD24+ cell subset promotes invasion and metastasis in human osteosarcoma. EBioMedicine 51:102598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Okabe H, Aoki K, Yogosawa S, Saito M, Marumo K and Yoshida K. (2018). Downregulation of CD24 suppresses bone metastasis of lung cancer. Cancer Sci 109:112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sonoyama W, Liu Y, Fang D, Yamaza T, Seo B-M, Zhang C, Liu H, Gronthos S, Wang C-Y, Wang S and Shi S. (2006). Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS One 1:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.