Abstract
Background
Nav1.5, which is encoded by the SCN5A gene, is the predominant voltage-gated Na+ channel in the heart. Several mutations of this gene have been identified and reported to be involved in several cardiac rhythm disorders, including type 3 long QT interval syndrome, that can cause sudden cardiac death. We analyzed the biophysical properties of 2 novel variants of the Nav1.5 channel (Q1491H and G1481V) detected in 5- and 12-week-old infants diagnosed with a prolonged QT interval.
Methods
The Nav1.5 wild-type and the Q1491H and G1481V mutant channels were reproduced in vitro. Wild-type or mutant channels were cotransfected in human embryonic kidney (HEK) 293 cells with the beta 1 regulatory subunit. Na+ currents were recorded using the whole-cell configuration of the patch-clamp technique.
Results
The Q1491H mutant channel exhibited a lower current density, a persistent Na+ current, an enhanced window current due to a +20-mV shift of steady-state inactivation, a +10-mV shift of steady-state activation, a faster onset of slow inactivation, and a recovery from fast inactivation with fast and slow time constants of recovery. The G1481V mutant channel exhibited an increase in current density and a +7-mV shift of steady-state inactivation. The observed defects are characteristic of gain-of-function mutations typical of type 3 long QT interval syndrome.
Conclusions
The 5- and 12-week-old infants displayed prolonged QT intervals. Our analyses of the Q1491H and G1481V mutations correlated with the clinical diagnosis. The observed biophysical dysfunctions associated with both mutations were most likely responsible for the sudden deaths of the 2 infants.
Résumé
Introduction
Le canal Nav1.5, codé par le gène SCN5A, est le canal Na+ dépendant du voltage prédominant dans le cœur. Plusieurs mutations de ce gène sont impliquées dans plusieurs anomalies du rythme cardiaque, dont le syndrome du QT long de type 3, qui peut provoquer la mort subite d’origine cardiaque. Nous avons analysé les propriétés biophysiques de deux nouveaux variants du canal Nav1.5 (Q1491H et G1481V) détectés chez deux bébés âgés respectivement de 5 et 12 semaines qui avaient une prolongation de l’intervalle QT.
Méthodes
Le canal Nav1.5 de type sauvage et les canaux mutants Q1491H et G1481V ont été reproduits in vitro. Les canaux de type sauvage ou mutants ont été co-transfectés dans les cellules des reins embryonnaires humains (REH) 293 avec la sous-unité régulatrice bêta 1. Les courants Na+ ont été enregistrés à partir de la configuration en cellule entière via la technique de patch-clamp.
Résultats
Le canal mutant Q1491H montre une densité de courant plus faible, un courant Na+ persistant, un courant fenêtre augmenté en raison d’un changement dép de +20 mV de l’inactivation à l’état stable, un changement de +10 mV de l’activation à l’état stable, une entrée plus rapide de l’inactivation lente et une récupération de l’inactivation rapide avec des constantes de temps rapides et lentes. Le canal mutant G1481V montre une augmentation de la densité de courant et un changement de +7 mV de l’inactivation à l’état stable. Les anomalies observées sont caractéristiques des mutations avec gain de fonction typiques du syndrome du QT long de type 3.
Conclusions
Les deux bébés âgés respectivement de cinq 5 et 12 semaines montraient une prolongation des intervalles QT. Nos analyses des mutations Q1491H et G1481V montrent une corrélation avec le diagnostic clinique. Les dysfonctions biophysiques observées qui sont associées aux deux mutations étaient fort probablement responsables des morts subites des deux bébés.
Voltage-gated Na+ channels are crucial for the amplitude and upstroke of cardiac action potentials (APs), which are important determinants for driving AP propagation and conduction velocity throughout the working myocardium.1 Mutations in SCN5A, the gene that encodes Nav1.5, the predominant cardiac Na+ channel, have been implicated in rare familial forms of cardiac arrhythmias, such as type 3 long QT interval syndrome (LQT3), Brugada syndrome, progressive cardiac conduction disorder (PCCD), atrial fibrillation, and sudden infant death syndrome (SIDS). Another SCN5A mutation has been reported recently to be involved in dilated cardiomyopathy, a structural heart disease.2,3 In addition to their role in changing gating characteristics, there is growing recognition that such mutations may also be associated with alterations in channel protein trafficking and expression levels. The SCN5A gene is also the only gene for which there is definitive evidence for clinical validity of Brugada syndrome.4
Long QT (interval) syndrome (LQTS) is an inherited cardiac channelopathy that may lead to syncope and even sudden cardiac death as a result of polymorphic ventricular tachycardia known as torsade de pointes. LQTS manifests as a prolonged corrected QT (QTc) interval exceeding 450 ms on 12-lead electrocardiograms. To date, 17 genes have been linked to inherited LQTS, but definitive evidence that they are a genetic cause for typical LQTS has been found for only 3 of these. Those genes are KCNQ1, KCNH2, and SCN5A.5
LQT3 is caused by mutations in SCN5A genes mapped to chromosome 3q21-24, which encodes the α-subunits of the cardiac Na+ channel.6,7 SCN5A mutations have been identified in about 10% of genotyped LQTS patients.8 When a patient presents with LQTS, genetic testing is used to identify the gene that is causing the disease. If the gene is identified, this tells the clinician what type of LQTS is involved (LQT1, LQT2, or LQT3).9 The development of arrhythmias is often associated with bradycardia during sleep or relaxation, when the QTc interval is prolonged. The most common mechanism for QT interval prolongation in LQT3 is due to a persistent or late Na+ current leading to an increase in AP plateau duration.6 This can be a substrate for early afterdepolarizations, which can potentially trigger ventricular tachyarrhythmias. Most mutations linked to LQT3 are located in exons 23, 26, and 28, which encode the III-IV linker or inactivation gate, the voltage sensor domain, and the C-terminus of the Na+ channel, all of which are involved in fast inactivation. These mutations impede the proper closure of the channel during this critical process, resulting in an increase in the number of channels that are unable to reach a stable inactivated state. This leads to a rise in persistent Na+ currents unlike the normal inactivation of wild-type (WT) Na+ channels.
Although SIDS is the leading cause of death in the first year of life, its cause is still unknown. The only recommended preventive measure is to avoid placing infants on their stomachs or sides for sleep.10 Several studies have linked SIDS to LQTS11 and have shown that 50% of infants who die of SIDS have a prolonged QTc interval and that a prolonged QTc interval over 440 ms in the first week of life increases the risk of SIDS by a factor of 41.12,13
We report here on 2 infants who presented with a prolonged QTc interval and who died suddenly, most likely after experiencing ventricular fibrillation. Although the 5- and 12-week-old infants were within the age range during which the incidence of SIDS peaks, their deaths were not attributed to SIDS. A sequencing analysis revealed a heterozygous G-to-T base substitution at position 4473 in exon 23 that resulted in a glutamate (E)-to-histidine (H) substitution at residue 1491 and a heterozygous G-to-T base substitution at position 4442 in exon 23 that resulted in a glycine (G)-to-valine (V) substitution at residue 1481. We recorded macroscopic Na+ currents in transfected mammalian cells using the whole-cell configuration of the patch-clamp technique. The data revealed a marked shift of inactivation to more positive potentials, the presence of a persistent Na+ current, and a large increase in the window current. All these characteristics point toward a gain-of-function due to the mutations.
Methods
Institutional Committee on Human Research
The study was conducted in accordance with the principles of the Declaration of Helsinki, and the protocol was approved by the local ethics committee. Parents provided written informed consent.
Cell cultures
Human embryonic kidney 293 (HEK 293) cells were used. The cells were grown in high-glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% streptomycin at 37 °C in a 5% CO2 atmosphere. The human Na+ channel β1-subunit and enhanced Green Fluorescence Protein (eGFP) were inserted in the pIRES bicistronic vector in the form of β1-pIRES-eGFP. The cells were transfected with the plasmid cloning DNA3.1 vector containing either WT Nav1.5 complementary DNA (1 μg) or Nav1.5/Q1491H and G1481V mutants with the pIRES2/EGFP vector containing β1 subunit complementary DNA (1 μg) in 10-cm cell culture dishes using the calcium phosphate method as previously reported.14
Whole-cell patch-clamp recordings
Na+ currents were recorded using low-resistance, fire-polished electrodes (≈1 MΩ) made from 8161 Corning borosilicate glass coated with HIPEC (Dow-Corning, Midland, MI) to minimize electrode capacitance. An Axopatch 200 amplifier and pClamp software (Molecular Devices, Sunnyvale, CA) were used to record Na+ currents. The series resistance was compensated to 80% to minimize voltage-clamp errors. The cells were allowed to stabilize for 5 minutes after the whole-cell configuration was established. The membrane potential was held at –140 mV before the currents were recorded. Sodium currents were filtered at 5 kHz and digitized at 83.33 kHz. The liquid junction potential was not corrected. All the experiments were performed at room temperature (22 °C).
Solutions
The intracellular solution was composed of 35 mM NaCl, 105 mM cesium floride, 10 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES). The pH was adjusted to 7.3 with 2 M CsOH. The first external solution (low Na+) was composed of 35 mM NaCl, 115 N-methyl-d-glucamine, 2 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES. The pH was adjusted to 7.4 with 2 M HCl. The second external solution (full Na+) used to record persistent Na+ currents was composed of 150 mM NaCl, 2 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES. The pH was adjusted to 7.4 with 2 M HCl. Tetrodotoxin (TTX; LATOXAN, Portes-lès-Valence, France) was diluted to 75 μM in 2% acetic acid.
Data analysis
The slope factor (K) and the midpoint (V1/2) for activation and inactivation were calculated using standard Boltzmann functions: 1/{1 + exp [(V1/2activation – V)/kactivation]} for activation and (1 – C)/{1 + exp [(V – V1/2inactivation)/kinactivation] + C} for inactivation. V is the voltage. The window current was obtained using equation 1: {1/[1 + exp ((V1/2activation – V)/kactivation)] x [(1 - C)/(1 + exp (V – V1/2inactivation)/kinactivation)] + C} x 100, which is the probability of having the channel in the open state.
Statistical analysis
Results are expressed as mean ± standard error of the mean. Statistical comparisons were performed using a 1-way analysis of variance in GraphPad Prism (La Jolla, CA) for statistical comparisons. Differences were considered significant at P < 0.05.
Results
Available clinical data and identification of the SCN5A mutations
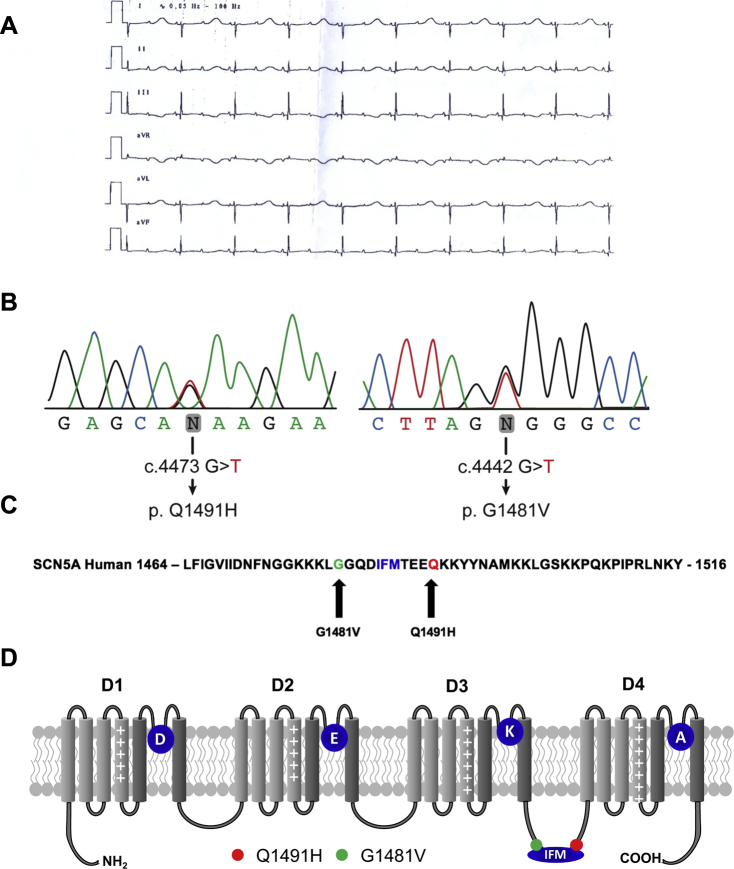
Patient #1 was an infant with a QTc of 700 ms and a 2:1 atrioventricular block (AVB) at birth (Fig. 1A). The 2:1 AVB was detected in utero at a gestational age of 31 weeks of amenorrhea. This infant received 2 mg/kg of propranolol, and a pacemaker was implanted a few days after birth. He experienced salvos of torsades de pointes. Flecainide and mexiletine were unsuccessful in preventing torsades. The patient died at 3 months of age after experiencing ventricular fibrillation.
Figure 1.
(A) A standard electrocardiogram was recorded soon after birth. Paper speed was 25 mm/s; 10 mm/1 mV. The QT interval was 720 ms; the QRS was 60 ms; and 2:1 atrioventricular block developed due to an extremely prolonged QT interval. (B) Sequence analysis of the G1481V and Q1491H mutations. (C) The Q1491H mutation resulted from a G-to-T substitution at position 4473, leading to a glutamine (Q)-to-histidine (H) substitution at residue 1491. The G1481V mutation resulted from a G-to -T substitution at position 4442, leading to a glycine (G)-to-valine (V) substitution at residue 1481. (D) Schematic representation of the 4 domains of the α-subunit of the Nav1.5 channel showing the locations of the Q1491 and G1481V mutations. DEKA represents the selectivity filter of the channel. The Q1491H mutation caused a glutamine-to-histidine substitution 4 amino acids downstream from the isoleucine, phenylalanine, and methionine motif. The G1481V mutation caused a glycine-to-valine substitution 4 amino acids upstream from the isoleucine, phenylalanine, and methionine motif.
Patient #2 had a QTc of 600 ms and a 2:1 AVB block at birth. The 2:1 AVB was detected in utero at a gestational age of 25 WA. A pacemaker was implanted a few days after birth, and 2 mg/kg of propranolol was started. The infant experienced several episodes of torsades de pointes and died at 1 month of age after experiencing ventricular fibrillation.
The genomic DNAs of the 2 infants were screened using specific primers for variant changes in several genes associated with arrhythmia syndrome (see Supplemental Table S1), in particular in all 28 SCN5A exons. Molecular screening by automatic sequencing analysis of the genomic DNAs revealed 2 novel mutations in the SCN5A gene (G1481V and Q1491H), namely, c. 4473 G>T, which resulted in a Glu1491His substitution, and c. 4442 G>T, which resulted in a Gly1481Val substitution (Fig. 1B). No mutations were detected in other genes. Applying the American College of Medical Genetics and Genomics guidelines15 revealed unclear initial results. This is illustrated and confirmed by Varsome (https://varsome.com) and CardioClassifier (https://www.cardioclassifier.org), which respectively returned both variations as being likely pathogenic (class 4) or of uncertain significance (class 3). This initial variant classification consequently precluded their use in future presymptomatic diagnosis. This warranted further detailed biophysical characterization (see below). The 2 substitutions are highly conserved in the Na+ channels of many species and are located in the III-IV linker, which plays a role in channel inactivation in all voltage-gated Na+ channels. It was hypothesized that these 2 mutations were responsible for the sudden death events. Family screening documented normal phenotypes in both parents of the 2 infants.
Biophysical characteristics of Nav1.5/Q1491H and Nav1.5/G1481V
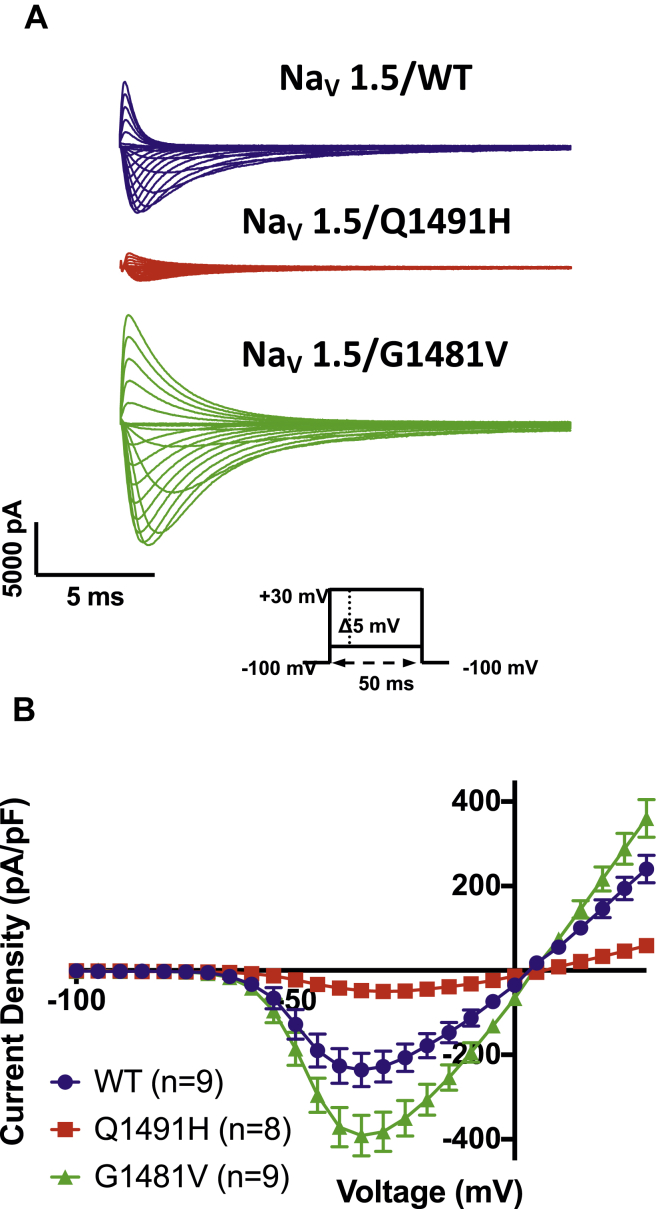
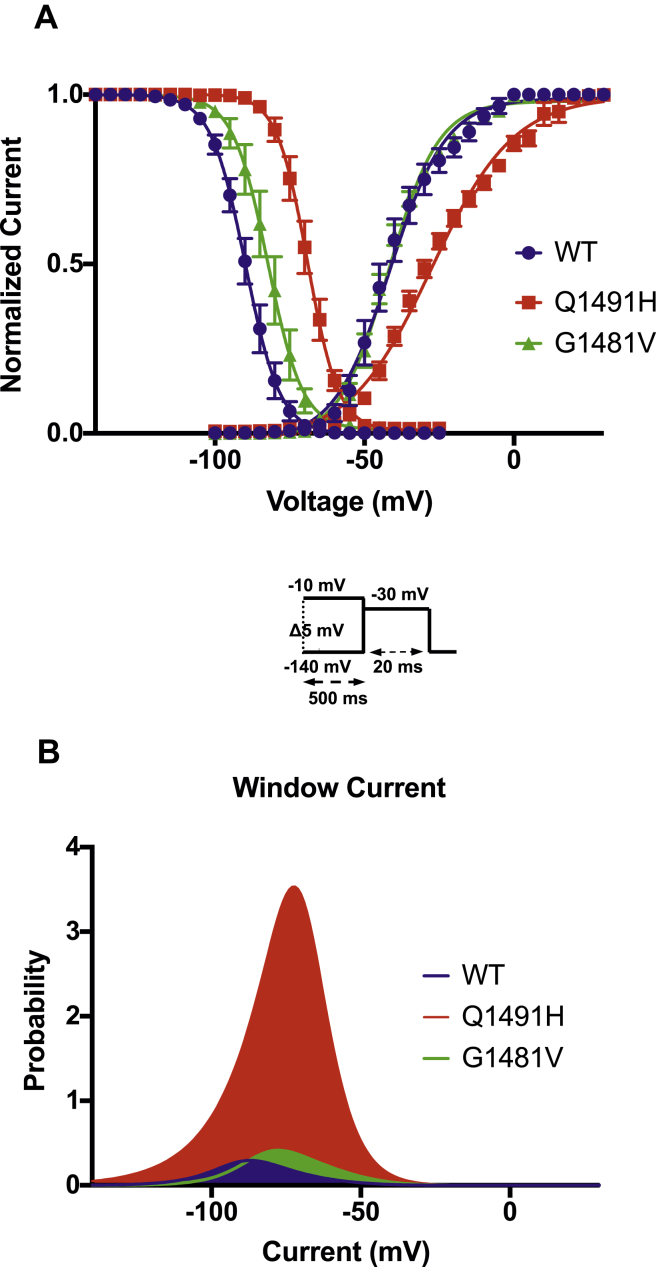
Currents were elicited by sequential depolarizing steps of the cell membrane from –100 mV to +30 mV in 5-mV increments (Fig. 2A), with the low Na+ external solution (see Methods). The I–V curves were constructed by measuring the peak of each Na+ current and were normalized to the membrane capacitance to obtain current densities (pA/pF). The current density of the Q1491H mutant channel was lower than that of the WT channel, whereas the current density of the G1481V mutant channel was higher than that of the WT channel. These results suggest that the expression levels of the Q1491H and G1481V mutant channels were different than that of the WT channel. The potential of the maximum peak current amplitude of the Q1491H channel was shifted toward more positive potentials compared to the WT channel (Fig. 2B). To study the activation and inactivation parameters, we first calculated the G–V curves (steady-state activation), which were fitted with a standard Boltzmann function. The V1/2 of the Q1491H mutant channel was positively shifted by 9 mV, with significant differences in k values. There was no difference between the V1/2 values of the WT channel and the G1481V mutant channel (Fig. 3A; Table 1).
Figure 2.
Analysis of whole-cell Na+ currents recorded from human embryonic kidney (HEK) 293 cells expressing Nav1.5 wild type, Q1491H, and G1481V. (A) Example of raw traces from Nav1.5/WT, Q1491H, and G1481V were obtained using depolarizing pulses from –100 mV to +30 mV in 5-mV increments. (B) Current–voltage relationship of wild-type • (n = 9); Q1491H ▪ (n = 9); and G1481V ▴ (n = 9). The current amplitude was normalized to the membrane capacitance to obtain the current density (pA/pF).
Figure 3.
Gating properties of steady-state activation and inactivation and window currents. (A) Voltage-dependence of steady-state activation and inactivation in wild type (• activation, n = 9; inactivation, n = 9); Q1491H (▪ activation, n = 9; inactivation, n = 7); and G1481V (▲; activation, n = 9; and inactivation, n = 7). The inactivated currents were generated using the protocol described in the insets, and the activated currents were obtained from the IV recording. The resulting data were fitted to a standard Boltzmann function. (B) The predicted window current was obtained using equation 1 (see Methods).
Table 1.
Biophysical properties of the Nav1.5 WT and mutant channels
| Nav1.5/WT | Nav1.5/Q1491H | Nav1.5/G1481V | |
|---|---|---|---|
| Peak current density (pA/pF) | –235.8 ± 39.8 (n = 9) | –49.2 ± 7.3 (n = 9) | –391.2 ± 48.3 (n = 9) |
| Steady-state inactivation | |||
| V1/2 (mV) | –89.6 ± 1.6 (n = 9) | –69.4 ± 1.5§ (n = 7) | –82.6 ± 2.3∗ (n = 7) |
| K (mV) | –5.1 ± 0.1 (n = 9) | –4.5 ± 0.3 (n = 7) | –4.6 ± 0.2 (n = 7) |
| Steady-state activation | |||
| V1/2 (mV) | –41.4 ± 2.5 (n = 9) | –32.0 ± 1.5† (n = 9) | –42.2 ± 1.2 (n = 9) |
| K (mV) | 7.6 ± 1.0 (n = 9) | 14.2 ± 1.3‡ (n = 9) | 8.0 ± 0.6 (n = 9) |
| Recovery from slow inactivation | |||
| tfast (ms) | 4.2 ± 0.6 (n = 7) | 0.8 ± 0.1§ (n = 8) | 1.9 ± 0.2‡ (n = 7) |
| tslow (ms) | - | 55.9 ± 5.0 (n = 8) | 88.2 ± 12.2 (n = 8) |
| Afast | 91.0 ± 1.1% (n = 5) | 56.0 ± 8.7%† (n = 8) | 83.2 ± 4.5% (n = 7) |
| Aslow | 9.0 ± 1.1% (n = 5) | 44.0 ± 8.7%† (n = 8) | 16.8 ± 4.5% (n = 7) |
| Onset of slow inactivation | |||
| Current plateau | 0.9 ± 0.1 (n = 6) | 0.7 ± 0.1‡ (n = 6) | 0.8 ± 0.1 (n = 5) |
| Closed state of activation | |||
| Current plateau | 0.8 ± 0.1 (n = 8) | 1.1 ± 0.1§ (n = 10) | 1.0 ± 0.1‡ (n = 7) |
A, fraction of recovery component; K, slope factor for activation or inactivation; t, time constant; V1/2, midpoint for activation or inactivation.
P < 0.05.
P < 0.01.
P < 0.001.
P < 0.0001
Voltage-dependent steady-state inactivation was assessed by applying 500-ms pre-pulses ranging from –140 mV to –30 mV to allow the channels to enter the inactivated state, followed by a test pulse at –30 mV to assess the number of functional channels. The current amplitude of the test pulse was normalized to the maximum current recorded during the pre-pulse and was plotted vs the voltage of the pre-pulse to obtain the voltage-dependent inactivation curve, which was then fitted to a standard Boltzmann function. V1/2 and k were generated by fitting each data set with a standard Boltzmann function (see values in Table 1). A +20-mV shift for Q1491H and a +7-mV shift for G1481V were observed, but k was not significantly affected (Fig. 3A).
Window currents were determined and were used to assess the open probability. The total areas were 1.4- and 11-fold larger, respectively, for G1481V and Q1491H than for the WT channel and were shifted to more-positive voltages (Fig. 3B).
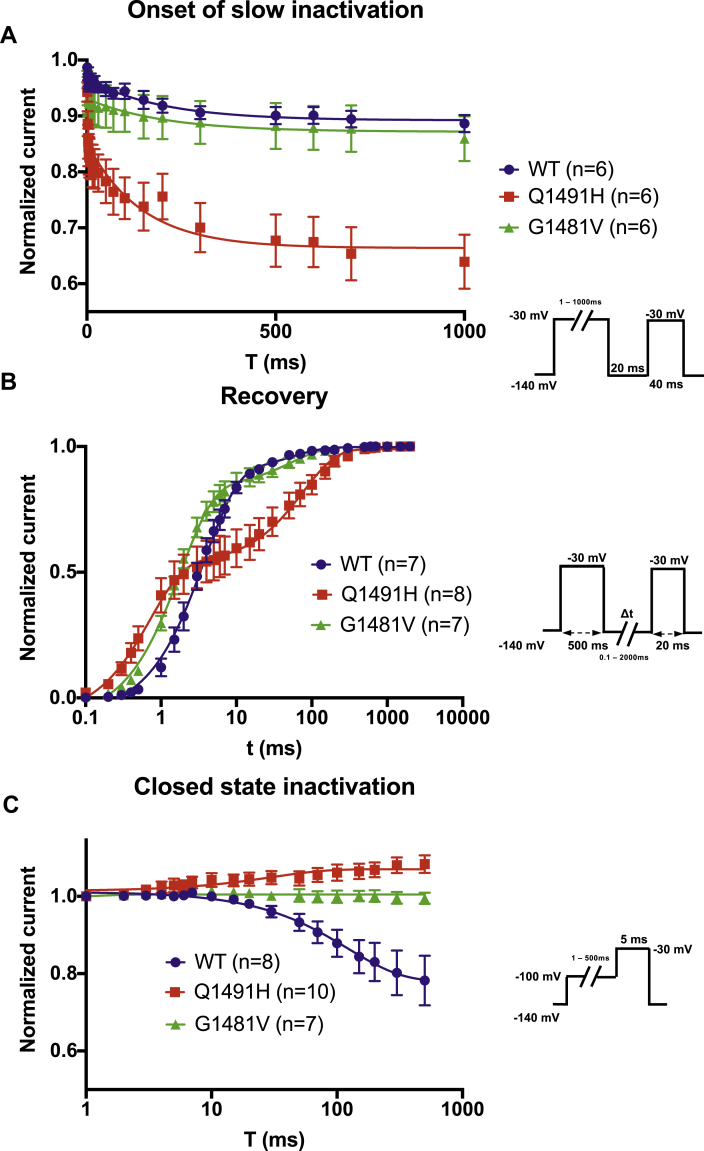
Slow inactivation was measured using a 2-pulse protocol, with an initial conditioning pulse (pre-pulse) and a final test pulse. The current was normalized to the current amplitude during the pre-pulse. There was no difference between the G1481V and WT channels in terms of the kinetics of entry into slow inactivation. However, the Q1491H mutant channel entered into slow activation faster than the WT channel (Fig. 4A). Recovery from slow inactivation was measured using a 2-pulse protocol. The channels were first inactivated by a 500-ms pre-pulse and were then allowed to recover from inactivation. The peak currents were normalized to the maximum peak current. The Q1491H mutant exhibited a complex recovery with 2 (fast and slow) time constants (Table 1). The G1481V mutant also exhibited a 2–time constant recovery compared to the WT channel (Fig. 4B). The currents of the mutant channels (Q1491H and G1481V) did not decrease during closed-state inactivation. As the WT channel exhibited only a fast time constant, slow is not given in Table 1. Closed-state inactivation was assessed using a 2-pulse protocol. The currents were normalized to the first pulse. Neither mutated channel was affected, unlike the WT channel, which showed closed-state inactivation (Fig. 4C).
Figure 4.
The gating properties of slow inactivation, recovery from slow inactivation, and closed-state inactivation. (A) Slow inactivation in wild type (•, n = 6), Q1491H (▪, n = 6), and G1481V (▲, n = 6). The 2-pulse protocol described in the inset was used to generate the currents. (B) Time courses of recovery from slow inactivation in WT (•, n = 7), Q1491H (▪, n = 8), and G1481V (▲, n = 7). (C) Closed-state inactivation in WT (•, n = 8), Q1491H (▪, n = 10), and G1481V (▲, n = 7).
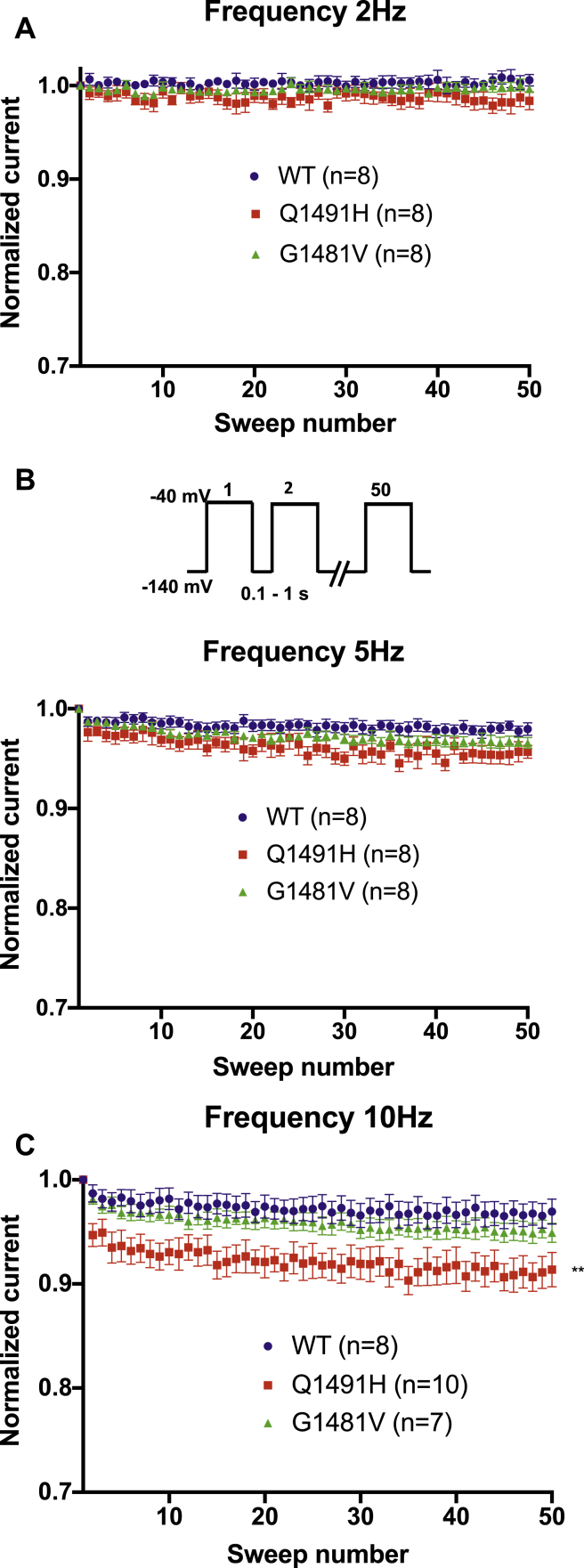
We recorded the frequency dependence of the Na+ currents in order to determine the channels that enter the inactivated state by applying a series of 50 depolarizing pulses at –40 mV. To evaluate current inhibition during rapid pulsing, the channels were pulsed 50 times at 2 Hz, 5 Hz, and 10 Hz. There was a significant difference between the WT channel and the Q1491H mutant channel at 10 Hz, with the mutant channel displaying a reduced current during rapid pulsing. There was no difference between the G1481V mutant channel and the WT channel (Fig. 5).
Figure 5.
Frequency dependence of wild type (•, n = 8), Q1491H (▪, n = 8), and G1481V (▲, n = 8). Currents were evaluated at (A) 2 Hz, (B) 5 Hz, and (C) 10 Hz. Fifty pulses were applied at –40 mV from a holding potential of –140 mV. Peak currents were normalized to the first peak current and were plotted vs the pulse number.
Persistent Na+ currents in cells transfected with the Q1491H mutant channel
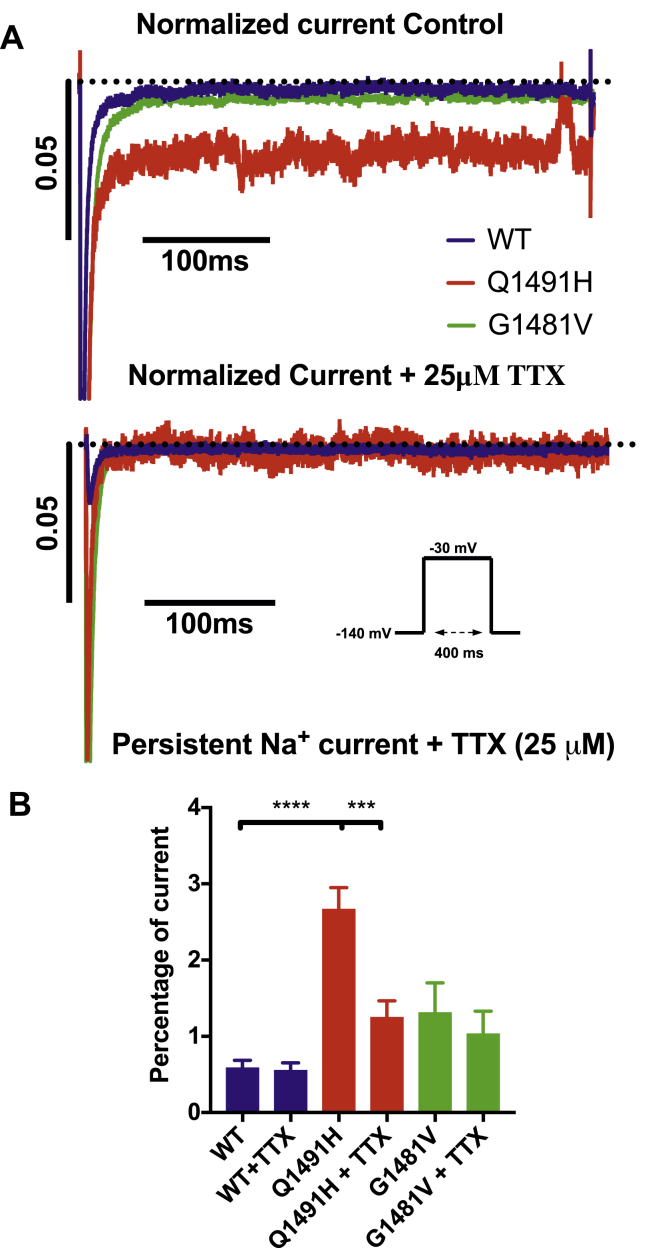
Persistent non-inactivating Na+ currents were recorded using a 400-ms depolarization pulse from –140 mV to –30 mV (see protocol inset) and were normalized to the peak. The current was inhibited using 25 μM tetrodotoxin (TTX). Only the Q1491H mutant channel displayed a significant decrease in current (2.66 ± 0.29 to 1.25 ± 0.22), indicating the presence of a persistent current (Fig. 6). TTX had no effect on the G1481V mutant and WT currents, which is indicative of a nonspecific leak.
Figure 6.
Persistent Na+ currents in wild type, Q1491H, and G1481V inhibited by tetrodotoxin (TTX). Recordings were performed using a –30-mV depolarizing pulse (see protocol in inset). (A) Currents recorded without TTX (left) and with 25 μM TTX (right). (B) Currents normalized to the maximum current, without TTX (left) and with 25 μM TTX (right). (C) Histograms of persistent Na+ currents. The persistent currents accounted for 0.58 ± 0.10 of the peak current amplitude for wild type without TTX (n = 6), 2.66 ± 0.29 for Q1491H without TTX (n = 13), 1.25 ± 0.22 for Q1491H with 25 μM TTX (n = 13), and 1.33 ± 0.35 for G1481V without TTX (n = 11) at –30 mV (∗∗P < 0.01, ∗∗∗P < 0.001).
Discussion
Q1491H and G1481V are SCN5A heterozygous mutations identified in 2 infants who died from long QT (Q1491H, 700 ms AVB 2:1; G1481V, 600 ms AVB 2:1) at 5 (G1481V) and 12 (Q1491H) weeks of age, respectively. The mutations were located in the Nav1.5 intracellular loop between domains III and IV near the isoleucine, phenylalanine, and methionine (IFM) motif (Fig. 1), which is involved in fast inactivation.16 Such mutations in the inactivation gate are often associated with changes in inactivation of voltage-gated Na+ channels. Both mutations were identified in clinical practice. The SCN5A gene is known as a clinically actionable gene (https://search.clinicalgenome.org). The variants’ pathogenic status is thus critical to inform the family appropriately and for their use in presymptomatic diagnosis.
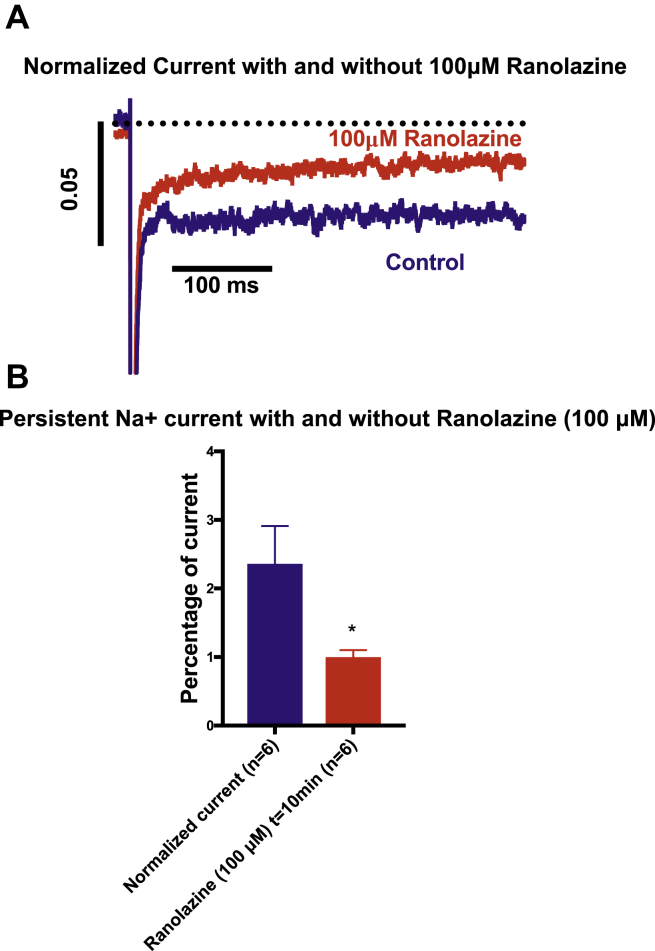
The Q1491H mutation was located 4 amino acids downstream from the IFM motif. Glutamine has a polar uncharged side chain, whereas histidine has electrically charged side chains. A biophysical characterization revealed that this mutation has a marked effect on the normal function of the NaV1.5 channel. We observed a loss-of-function for peak current density that could be due to a consequence of a modulatory effect. We also observed a loss-of-function for recovery time (a slow time constant: 56.01 ± 8.73%; Table 1), which led to a decreased inward Na+ current and a longer recovery from inactivation time. An increase in the window current may lead to an increase in Na+ influx, which can increase the risk of fatal ventricular arrhythmias. This increase was correlated with the depolarized shift of steady-state inactivation (+20.27 mV) and activation (+9.36 mV) that drove channel activity to more-positive potentials. We also showed that the Q1491H mutant channel displays a persistent current that is inhibited by 25 μM TTX (Fig. 6) and ranolazine (Fig. 7), which is characteristic of LQT3. This mutation also displayed pronounced frequency-dependent inactivation at 10 Hz, suggesting that the Nav1.5/Q1491H channel has an unstable inactivated state. These results provide a rationale for linking this mutation and the clinical phenotype, especially for the long QTc interval (700 ms), measured in the infant prior to his sudden death. Clearly, Q1491H resulted in several biophysical defects, which may contribute to the deleterious effect of the mutation. Obviously, some of the parameters are loss-of-function effects, but what is uniquely determinant is the persistent current.
Figure 7.
Effects of ranolazine on persistent sodium current. (A) Persistent Na+ current traces using the same protocol as shown in Figure 6 showing the effect of 100 μM ranolazine. (B) Bar graph of persistent Na+ currents inhibition by ranolazine (∗P < 0.05).
The G1481V mutation was located 4 amino acids upstream from the IFM motif. Given that valine and glycine are both uncharged, this may explain the more modest effect on the function of the Nav1.5/G1481V channel. The G1481V mutation resulted in an increase in the current density (Fig. 2) as well as a +7.03-mV depolarized shift in inactivation, which increased by 1.4-fold the window current (Fig. 3). Furthermore, the recovery from the inactivation was also faster, indicating that the sodium channel will recover faster from inactivation. All these changes represent a gain of function of G1481V mutant sodium channels.
The biophysical characterization of both Q1491H and G1481V SCN5A mutations thus revealed a functional defect. The manual patch-clamp is not validated as a well-established method in a variant classification perspective, considering the clear biophysical defects, a “Pathogenic Strong 3” (PS3) criterion in the American College of Medical Genetics and Genomics classification process.17,18 Interestingly, both variants were reclassified as either likely pathogenic (class 4) or pathogenic (class 5), indicating that with the support of the biophysical characterization, both SNC5A variants can now be used for presymptomatic diagnosis.
Interestingly, although the patient with the G1481V mutation presented a long QTc interval of 600 ms, the increase in the persistent current was not statistically significant. In addition, there was no significant difference in the time constants of inactivation for all conditions (data not shown).
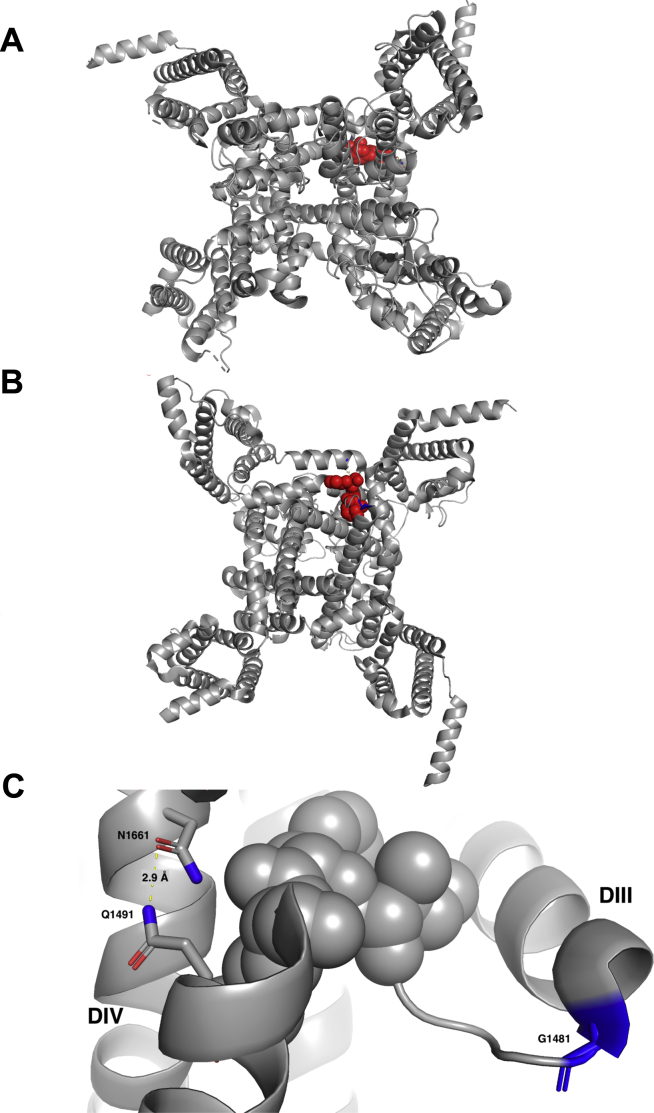
The III–IV linker acts as a lid in which the inactivation particle IFM amino acid trio occludes the inner pore by binding to the docking site like a latch. We used the recently crystalized rat Nav1.5 to visualize the position of the 2 residues and their interacting sites.19 The IFM motif is highly conserved in all Na+ channels. The sites of the Q and G mutations investigated in the present study were located up- and downstream, respectively, from the IFM. They are highly conserved in several Na+ channels, underlining their importance during the process of inactivation. The structure revealed that Q1493 residue interacts with N1661 (N1659 in hNav1.5) and is one of the key residues for optimum IFM docking. It has been shown previously that mutating N1661 to alanine abolishes fast inactivation.20 The recently published rat Nav1.5 structure showed that Q1493 at the C-terminal of the DIII–DIV linker (Q1491 in hNav1.5) forms a strong hydrogen bond with N1661 (Fig. 8). Q1491H mutation may lead to a weaker interaction with N1659 because of a histidine shorter imidazole side chain. This weakened interaction will impair fast inactivation and lead to a gain-of-function effect. This may explain the appearance of persistent current and the rightward shift of the steady-state inactivation curve of the Q1491H mutant. This could impede this interaction and contribute to change in pore closure and the slowing of slow inactivation. On the other hand, G1481, which is located at the tight turn of DIII S6 and the beginning DIII–DIV linker, is important for keeping the flexibility of movement of the IFM motif. A valine substitution, with its bulky side chain, at this position will lead to less flexibility of the IFM motif movement. This structural insight explains the mild slow inactivation kinetics of the G1481V mutant compared to the wild-type channel. However, the effects of this mutation are expected to be less drastic compared to those of the Q1491H mutation, just as the effects of G1481V mutation on channel function are limited.
Figure 8.
Open state structure of the human Nav1.5 sodium channel. (A) Top view of the cryogenic electron microscopy (Cryo-EM) structure of rat Nav1.5 channel showing the IFM motif in red spheres. (B) Bottom view of the Cryo-EM structure of the rat Nav1.5 channel showing the isoleucine, phenylalanine, and methionine (IFM) motif in red spheres. (C) Close-up view of the IFM motif flanked by G1481 on the N-terminal and Q1491 on the C-terminal. A strong hydrogen bond between N1661 and Q1491 (2.9A) ensures the proper docking of the IFM motif. DII, sodium channel domain 2; DIV, sodium channel domain 4.
Many SCN5A mutations, especially in the III–IV linker and the voltage sensor domain, have been linked to BrS and LQT3. The difference between BrS and LQT3 is that BrS appears with age.21 The first mutation to be associated with LQT3 was a 3-amino-acid deletion (Lys1505-Pro1506-Gln1507, ΔKPQ). This mutation shares the same deletion (Q1507) as the delQKP deletion at position 1507-1509. Both are located in the III–IV linker region and are associated with a gain-of-function and a late persistent Na+ current.22,23 This region is a hotspot of mutations that induce inactivation disturbances, including a depolarized shift in inactivation that enhances the window current, as is the case for Q1491H and G1481V. This same effect was observed with F1486del, which involves a deletion of a phenylalanine on the IFM motif.24 This mutation alters lidocaine sensitivity, which is a local anesthetic that blocks late currents associated with LQT3 mutations. Although IFM/QQQ (Q for Glutamine) mutants also display an alteration in lidocaine sensitivity,25 this effect is not seen if the phenylalanine (F) is replaced by a glutamine (Q).26 Other mutations have been shown to induce a long QTc interval, including N1774H, which is located in the C-terminal.27 Like the Q1491H mutations, N1774H results in a loss of function in peak current density and an increase in the late current. The N1774H mutation was detected in a 19-year-old woman. The S1333Y mutation on the S4-5 linker results in an enhanced window current and a persistent current. The S1333Y mutation was detected in a 25-day-old infant.28 Although all these mutations lead to LQT3, they are located in different parts of the Nav1.5 channel that all play a role in inactivation. The ineffectiveness of mexiletine and flecainide in case 1, and propranolol in case 2, is not very clear, especially given that these drugs all have some sodium channel–blocking properties. The possibility of interindividual pharmacokinetic and pharmacodynamic differences could explain the nonuniform clinical response to these drugs.
Conclusion
The Q1491H and G1481V mutations are both located near the IFM motif, which is involved in fast inactivation. These mutations changed the inactivation state, shifting it to a more positive voltage. They also increased the window current, which can result in heart malfunctions such as LQT3. Full biophysical characterization constitutes a strong benefit, providing a useful supplemental criterion in the variant classification procedure. These 2 infants might have benefited from class 1 antiarrhythmic drug therapy, but the lack of effect of these drugs is not well understood and warrants further study.
Funding Sources
This research was supported by the Canadian Institutes of Health Research (grants MOP-111072 and MOP-130373 to MC) and the Association Française contre les Myopathies—Téléthon (grant AFM19962 to MC).
Disclosures
All authors have no conflicts of interest to disclose.
Footnotes
Ethics Statement: The study was conducted in accordance with the principles of the Declaration of Helsinki, and the protocol was approved by the local ethics committee. Parents provided written informed consent.
See page 266 for disclosure information.
To access the supplementary material accompanying this article, visit CJC Open at https://www.cjcopen.ca/ and at https://doi.org/10.1016/j.cjco.2020.09.023.
Supplementary Material
References
- 1.Coraboeuf E., Deroubaix E., Coulombe A. Effect of tetrodotoxin on action potentials of the conducting system in the dog heart. Am J Physiol—Heart Circ Physiol. 1979;236:H561–H567. doi: 10.1152/ajpheart.1979.236.4.H561. [DOI] [PubMed] [Google Scholar]
- 2.Moreau A., Gosselin-Badaroudine P., Delemotte L., Klein M.L., Chahine M. Gating pore currents are defects in common with two Nav1.5 mutations in patients with mixed arrhythmias and dilated cardiomyopathy. J Gen Physiol. 2015;145:93–106. doi: 10.1085/jgp.201411304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gosselin-Badaroudine P., Keller D.I., Huang H. A proton leak current through the cardiac sodium channel is linked to mixed arrhythmia and the dilated cardiomyopathy phenotype. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosseini S.M., Kim R., Udupa S. Reappraisal of reported genes for sudden arrhythmic death: evidence-based evaluation of gene validity for Brugada Syndrome. Circulation. 2018;138:1195–1205. doi: 10.1161/CIRCULATIONAHA.118.035070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adler A., Novelli V., Amin A.S. An international, multicentered, evidence-based reappraisal of genes reported to cause congenital long QT syndrome. Circulation. 2020;141:418–428. doi: 10.1161/CIRCULATIONAHA.119.043132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George A.L. Assignment of the human heart tetrodotoxin-resistant voltage-gated Na+ channel α-subunit gene (SCN5A) to band 3p21. Cytogenet Cell Genet. 1995;68:67–70. doi: 10.1159/000133892. [DOI] [PubMed] [Google Scholar]
- 7.Bennett P.B., Yazawa K., Makita N., George A.L. Molecular mechanisms for an inherited cardiac arrhythmia. Nature. 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 8.Tan B.-H., Pundi K.N., Van Norstrand D.W. Sudden infant death syndrome–associated mutations in the sodium channel beta subunits. Heart Rhythm. 2010;7:771–778. doi: 10.1016/j.hrthm.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gollob M.H., Blier L., Brugada R. Recommendations for the use of genetic testing in the clinical evaluation of inherited cardiac arrhythmias associated with sudden cardiac death: Canadian Cardiovascular Society/Canadian Heart Rhythm Society joint position paper. Can J Cardiol. 2011;27:232–245. doi: 10.1016/j.cjca.2010.12.078. [DOI] [PubMed] [Google Scholar]
- 10.Dwyer T., Ponsonby A.L., Blizzard L., Newman N.M., Cochrane J.A. The contribution of changes in the prevalence of prone sleeping position to the decline in sudden infant death syndrome in tasmania. Obstet Gynecol Surv. 1995;50:704–705. [PubMed] [Google Scholar]
- 11.Schwartz P.J., Priori S.G., Dumaine R. A molecular link between the sudden infant death syndrome and the long-QT syndrome. New Eng J Med. 2000;343:262–267. doi: 10.1056/NEJM200007273430405. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz P.J., Stramba-Badiale M., Segantini A. Prolongation of the QT interval and the sudden infant death syndrome. N Engl J Med. 1998;338:1709–1714. doi: 10.1056/NEJM199806113382401. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz P.J., Stramba-Badiale M. Repolarization abnormalities in the newborn. J Cardiovasc Pharmacol. 2010;55:539–543. doi: 10.1097/FJC.0b013e3181d86525. [DOI] [PubMed] [Google Scholar]
- 14.Huang H., Priori S.G., Napolitano C., O’Leary M.E., Chahine M. Y1767C, a novel SCN5A mutation, induces a persistent Na+ current and potentiates ranolazine inhibition of Nav1.5 channels. Am J Physiol Heart Circ Physiol. 2011;300:288–299. doi: 10.1152/ajpheart.00539.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chahine M., Deschene I., Chen L.Q., Kallen R.G. Electrophysiological characteristics of cloned skeletal and cardiac muscle sodium channels. Am J Physiol Heart Circ Physiol. 1996;271:498–506. doi: 10.1152/ajpheart.1996.271.2.H498. [DOI] [PubMed] [Google Scholar]
- 17.Kanavy D.M. Comparative analysis of functional assay evidence use by ClinGen Variant Curation Expert Panels. Genome Med. 2019;11:77. doi: 10.1186/s13073-019-0683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brnich S.E., Tayoun A.N.A., Couch F.J. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12:3. doi: 10.1186/s13073-019-0690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D., Shi H., Tonggu L. Structure of the cardiac sodium channel. Cell. 2020;180:122–134.e10. doi: 10.1016/j.cell.2019.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPhee J.C., Ragsdale D.S., Scheuer T., Catterall W.A. A critical role for the S4-S5 intracellular loop in domain IV of the sodium channel α-subunit in fast inactivation. J Biol Chem. 1998;273:1121–1129. doi: 10.1074/jbc.273.2.1121. [DOI] [PubMed] [Google Scholar]
- 21.Beaufort-Krol G.C.M., van den Berg M.P., Wilde A.A.M. Developmental aspects of long QT syndrome type 3 and Brugada syndrome on the basis of a single SCN5A mutation in childhood. J Am Coll Cardiol. 2005;46:331–337. doi: 10.1016/j.jacc.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 22.Keller D.I., Acharfi S., Delacretaz E. A novel mutation in SCN5A, delQKP 1507-1509, causing long QT syndrome: role of Q1507 residue in sodium channel inactivation. J Mol Cell Cardiol. 2003;35:1513–1521. doi: 10.1016/j.yjmcc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q., Shen J., Splawski I. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 24.Song W., Xiao Y., Chen H. The human Nav1.5 F1486 deletion associated with long QT syndrome leads to impaired sodium channel inactivation and reduced lidocaine sensitivity. J Physiol. 2012;590:5123–5139. doi: 10.1113/jphysiol.2012.235374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett P.B., Valenzuela C., Chen L.Q., Kallen R.G. On the molecular nature of the lidocaine receptor of cardiac Na+ channels modification of block by alterations in the α-subunit III-IV interdomain. Circ Res. 1995;77:584–592. doi: 10.1161/01.res.77.3.584. [DOI] [PubMed] [Google Scholar]
- 26.Balser J.R., Nuss H.B., Romashko D.N., Marban E., Tomaselli G.F. Functional consequences of lidocaine binding to slow-inactivated sodium channels. J Gen Physiol. 1996;107:643–658. doi: 10.1085/jgp.107.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neubauer J., Wang Z., Rougier J.S. Functional characterization of a novel SCN5A variant associated with long QT syndrome and sudden cardiac death. Int J Legal Med. 2019;133:1733–1742. doi: 10.1007/s00414-019-02141-x. [DOI] [PubMed] [Google Scholar]
- 28.Huang H., Millat G., Rodriquez-Lafrasse C. Biophysical characterization of a new SCN5A mutation S1333Y in a SIDS infant linked to long QT syndrome. FEBS Lett. 2009;583:890–896. doi: 10.1016/j.febslet.2009.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.