Abstract
Background
Viral genomic surveillance is vital for understanding the transmission of COVID-19. In Hong Kong, breakthrough outbreaks have occurred in July (third wave) and November (fourth wave) 2020. We used whole viral genome analysis to study the characteristics of these waves.
Methods
We analyzed 509 SARS-CoV-2 genomes collected from Hong Kong patients between 22nd January and 29th November, 2020. Phylogenetic and phylodynamic analyses were performed, and were interpreted with epidemiological information.
Findings
During the third and fourth waves, diverse SARS-CoV-2 genomes were identified among imported infections. Conversely, local infections were dominated by a single lineage during each wave, with 96.6% (259/268) in the third wave and 100% (73/73) in the fourth wave belonging to B.1.1.63 and B.1.36.27 lineages, respectively. While B.1.1.63 lineage was imported 2 weeks before the beginning of the third wave, B.1.36.27 lineage has circulated in Hong Kong for 2 months prior to the fourth wave. During the fourth wave, 50.7% (37/73) of local infections in November was identical to the viral genome from an imported case in September. Within B.1.1.63 or B.1.36.27 lineage in our cohort, the most common non-synonymous mutations occurred at the helicase (nsp13) gene.
Interpretation
Although stringent measures have prevented most imported cases from spreading in Hong Kong, a single lineage with low-level local transmission in October and early November was responsible for the fourth wave. A superspreading event or lower temperature in November may have facilitated the spread of the B.1.36.27 lineage.
Funding
Richard and Carol Yu, Michael Tong, and the Government Consultancy Service (see acknowledgments for full list).
Keywords: COVID19, SARS-CoV-2, Phylogenetic, Phylodynamic, Viral genome, Next generation sequencing, Outbreak
Research in Context.
Evidence before this study
Whole viral genome sequence analysis has played an important role in the investigation of SARS-CoV-2 outbreaks. We searched PubMed without language restrictions on 13th December 2020 for articles using the terms “COVID-19″ or “SARS-CoV-2″ and the terms “phylogenetic”, or “phylogenomic”. Most of the studies using whole viral genome on outbreak investigations were performed in areas with high incidence. Very few were performed in low incidence areas.
Added value of this study
We analyzed 509 whole viral genomes from specimens collected between January and November 2020. We focused the analysis on viral genomes that were collected during the third and the fourth waves in Hong Kong, which occurred after the relaxation of restriction measures. Although multiple genetic lineages were found in imported cases, most of the locally-acquired cases belong to single lineages within the third and fourth waves which suggested that the stringent border control has prevented the transmission of SARS-CoV-2 from imported cases into the local community. The fourth wave was caused by the B.1.36.27 lineage that has been circulating in Hong Kong for 2 months with little genetic changes. The sudden increase of cases in the fourth wave was related to a dancing cluster which suggested the possibility of a superspreading event, and the lower temperature in November may have contributed to the rapid spread of the infection.
Implications of all available evidence
Due to the efficient person-to-person transmission of SARS-CoV-2, sudden outbreak of COVID-19 can easily occur in low incidence area even when there are few sources of infection. Whole viral genome analysis plays a pivotal role in understanding the characteristics of each outbreak, which can guide public health measures. The transmissibility of SARS-CoV-2 with the change in weather should be further investigated.
Alt-text: Unlabelled box
1. Introduction
SARS-CoV-2 is characterized by efficient person-to-person transmission [1]. Within only 12 months since the first report of SARS-CoV-2 human infections, over 70 million cases have been reported globally. Seroprevalence studies suggested that the true burden of infections could be much higher [2]. The successful control of COVID-19 requires a coordinated public health effort that should be guided by scientific evidence, and viral genomic analysis plays an essential role in understanding the transmission dynamics.
Genomic epidemiology studies have demonstrated multilineage introduction of SARS-CoV-2 into Europe and America during the early stage of the COVID-19 pandemic [3], [4], [5]. Hong Kong has a relatively low incidence of COVID-19 with about 0.1% of the population having laboratory-confirmed infections at the time of writing. During the early stage of the pandemic (first and second waves between January and May 2020), most of the COVID-19 patients were travelers or their close contacts, and their virus genomes were genetically diverse [6]. After stepping up control measures such as universal mandatory mask wearing, social distancing policies, and border controls, the incidence of COVID-19 cases was reduced [7].
However, unlike nearby regions that have largely eliminated local transmission of SARS-CoV-2, Hong Kong continues to have locally-acquired cases. The third wave, which began in early July 2020, was different from the first two waves in that most cases were acquired locally, and did not have direct contact with imported cases. We previously reported that the early cases of the third wave belong to a single lineage B.1.1.63 (previously designated as genetic cluster HK1), which is within the Global Initiative on Sharing All Influenza Data (GISAID) clade GR and Nextstrain clade 20B [6]. The lineage B.1.1.63 was newly introduced into Hong Kong, and was most closely related to viral genomes of travelers from the Philippines in late June [6]. Since this third wave was likely related to travelers, the Hong Kong government has stepped up measures to prevent transmission of SARS-CoV-2 from imported cases, including the requirement of all inbound travelers from designated high risk areas to provide negative SARS-CoV-2 nucleic acid test report before departure, and reducing the number of individuals that can be exempted from mandatory quarantine. With these public restrictions in place, the third wave peaked in late July, and the number of locally-acquired cases reduced substantially in September.
While there were sporadic locally-acquired cases from September to early November, the number of local cases remained relatively low with <10 cases per day. During this period, a novel viral genome in GISAID GH clade was found among local cases, which was most closely related to imported cases from Nepal [8]. However, since mid-November, a large outbreak occurred in Hong Kong (fourth wave), which was epidemiologically linked to dancing venues where people take dancing lessons [9]. Here, we sought to perform detailed phylogenetic analysis and compare the third (July) and fourth (November) waves in Hong Kong.
2. Methods
2.1. COVID-19 cases data
The epidemic curve data was constructed based on the information from the Centre for Health Protection, Department of Health, the Government of the Hong Kong Special Administrative Region [10].
2.2. Study design and participants
In this study, we performed whole viral genome sequencing on clinical specimens from COVID-19 patients, and analyzed the viral genomes together with viral genomes we reported previously [6,[11], [12], [13], [14]]. The archived clinical specimens sequenced in this study were from patients with laboratory-confirmed COVID-19 who were admitted to Queen Mary Hospital, Queen Elizabeth Hospital or Princess Margaret Hospital in Hong Kong. The study was approved by the Institutional Review Board of the University of Hong Kong/ Hospital Authority Hong Kong West Cluster (UW 13-372, UW 20-292), the Kowloon West Cluster REC (KW/EX-20-038[144-26]), and the Kowloon Central/Kowloon East Cluster REC (KC/KE-20-0321/ER-2). Written informed consent was waived.
2.3. Nanopore sequencing
Library preparation, nanopore sequencing and bioinformatic analysis were performed as we described previously [6]. Briefly, nanopore sequencing was performed following the Nanopore protocol - PCR tiling of COVID-19 (Version: PTC_9096_v109_revH_06Feb2020) according to the manufacturer's instructions with modifications (Oxford Nanopore Technologies). Briefly, extracted RNA was first reverse transcribed to cDNA either using SuperScript™ IV reverse transcriptase (ThermoFisher Scientific, Waltham, MA, USA) or LunaScript® RT SuperMix Kit (New England Biolabs, Ipswich, Massachusetts, USA). PCR amplification was then performed using the hCoV-2019/nCoV-2019 Version 3 Amplicon Set [Integrated DNA Technologies (IDT), Coralville, IA, USA]. End preparation and native barcode ligation were performed according to the PCR tiling of COVID-19 virus protocol (EXP-NBD196, Oxford Nanopore Technologies). Barcoded and pooled libraries were then ligated to sequencing adapter and sequenced with the Oxford Nanopore MinION device using R9.4.1 or R10.3 flow cells for 24–48 h.
Bioinformatic analysis was performed according to the ARTIC-nCoV network workflow [15] with minor modifications for converting raw data into the consensus sequences using the Medaka pipeline. The only modifications were (1) reducing the minimum length at the guppyplex step to 350 to allow potential deletions to be detected, and (2) increasing the –normalize value to 999,999 to incorporate all the sequenced reads.
2.4. Illumina sequencing
For Illumina sequencing, extracted RNA was first reverse transcribed to cDNA using SuperScript™ IV reverse transcriptase, followed by PCR amplification using the ARTIC network nCoV-2019 version 3 primer set (Integrated DNA Technologies). DNA libraries were then prepared using the Illumina DNA Prep kit (Illumina, San Diego, CA, USA) and IDT for Illumina DNA/RNA UD Indexes sets (Illumina). Briefly, DNA fragments with indexed adaptors were generated by tagmentation, and then amplified and pooled according to manufacturer's instructions. The quality of the libraries was validated using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed on the iSeq 100 (Illumina) to generate paired-end 151-bp reads.
The Illumina reads data was then processed according to the Utah DoH ARTIC/Illumina Bioinformatic Workflow. Detailed steps of the workflow are available at https://github.com/CDCgov/SARS-CoV-2_Sequencing/tree/master/protocols/BFX-UT_ARTIC_Illumina.
2.5. Phylogenetic and phylodynamic analysis
Multiple sequence alignment was performed using MAFFT [16]. The maximum-likelihood whole genome phylogenetic tree construction and phylodynamic analysis were performed using IQ-TREE [17] and TreeTime [18] (Please refer to supplementary methods for details). For the construction of the phylogenetic tree, 1000 replicates were used, and the option -czb was used to mask the unrelated substructure of the tree with branch length representing mutation count of less than 1. Furthermore, we applied a masking scheme to avoid biases caused by homoplastic and highly ambiguous sites as suggested [19]. Phylogenetic network was constructed using SplitsTree4 [20]. We described the genetic information using GISAID [21], Nextstrain [22], and PANGO lineage [23] nomenclatures. Nucleotide position was numbered according to the reference genome Wuhan-Hu-1 (GenBank accession number MN908947.3). For the maximum likelihood phylodynamic analysis, in addition to the described homoplastic position masking, we also removed the highly diverged sequences suggested by the TreeTime program. The evolutionary rate of B.1.1.63 lineage was estimated using root-to-tip (RtT) regression analysis. The consensus sequences have been deposited into GISAID (Supplementary Table S1).
2.6. Role of the funding source
The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
3. Results
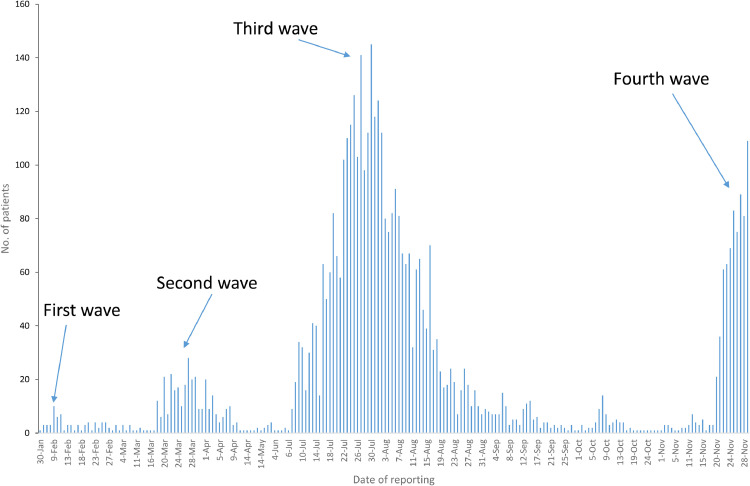
In Hong Kong, the third wave started in early July, peaked in late July, and almost returned to baseline level in September (Fig. 1). Between 16th September and 19th November 2020 (except 8th October), the number of locally-acquired cases per day fell below 10. However, the number of locally-acquired cases suddenly increased to 21 on 20th November, signifying the beginning of the fourth wave.
Fig. 1.
Number of locally-acquired cases in Hong Kong between 22nd January and 29th November 2020. Data were adapted from the center for Health Protection [10].
In total, we have analyzed 509 SARS-CoV-2 genome sequences from 508 patients, which represented 8.1% (509/6239 episodes) of COVID-19 cases in Hong Kong (Supplementary Table S2). We have added 380 new whole genome sequences from specimens that were collected between 24th June and 29th November 2020 (Supplementary Table S1). The remaining 121 viral genomes from the first, second and the beginning of the third waves have been reported previously [6,[11], [12], [13], [14]]. For one patient with reinfection, the viral genomes in both episodes (March and August) were included [11]. Eight viral genomes were previously published by Siu et al. [8].
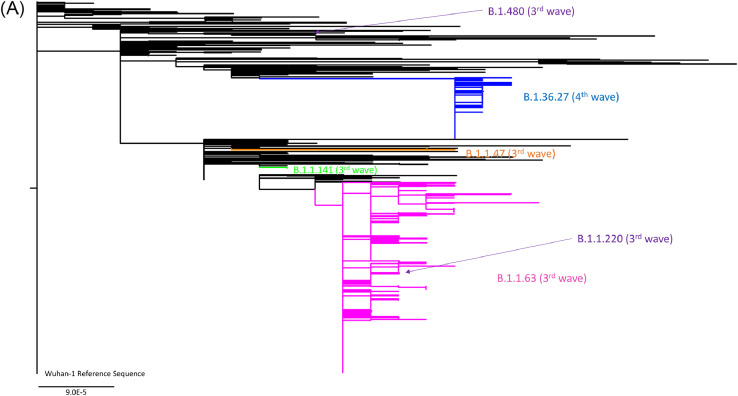
Phylogenetic analysis showed that during the third wave, 96.6% (259/268) of the locally-acquired cases belong to the PANGO lineage B.1.1.63 (Fig. 2a, Supplementary Fig. S1 and Supplementary Fig. S2). These were collected between 7th July and 27th September 2020. During this period, 3 locally-acquired cases were found in the PANGO lineage B.1.1.141 (previously designated as genetic clusters HK2 [6]), 3 in the PANGO lineage B.1.1.47, 2 in the PANGO lineage B.1.1.220 and 1 in the PANGO lineage B.1.480 (Fig. 2a). During the third wave, imported cases were genetically diverse (Fig. 2a, Fig. 2c, and Supplementary Figure S1).
Fig. 2.
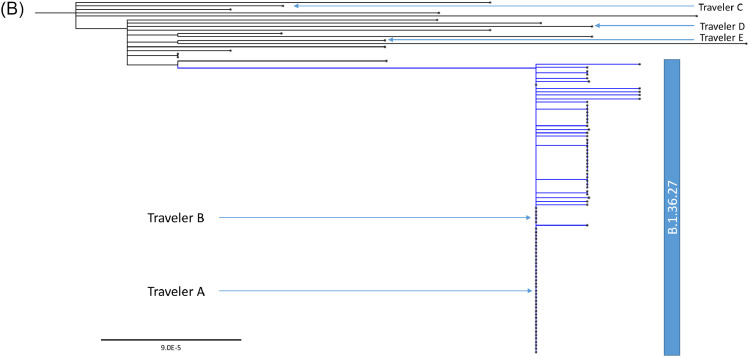
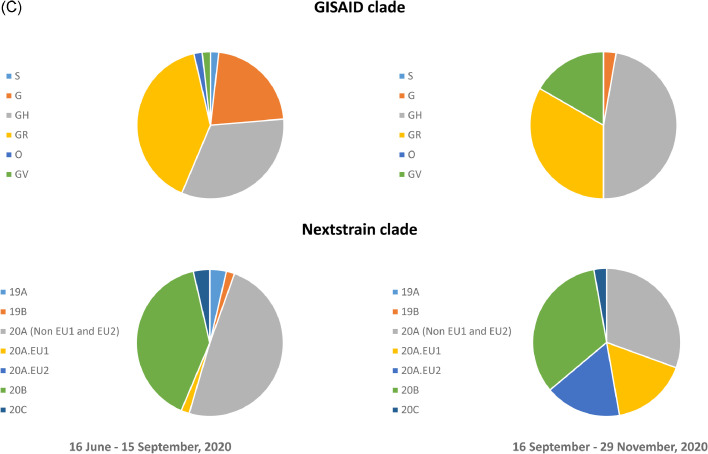
Whole genome phylogenetic analysis of 509 viral genomes showing the relationship between the genomes from locally-acquired and imported COVID-19 cases in Hong Kong from January to November 2020. The trees were constructed by maximum likelihood method with IQTree and Treetime. The reference genome Wuhan-Hu-1 (GenBank accession number MN908947.3) was used as the root of the tree. The substitution model GTR+F+I was used. (a) The entire phylogenetic tree. The blue branch indicates B.1.36.27 lineage from fourth wave. Pink, green and orange branches indicate B.1.1.63, B.1.1.141 and B.1.1.47 lineages from third wave, respectively. (b) A magnified figure focusing on the B.1.36.27 lineage. Blue triangles indicate Travelers C, D and E from Nepal, for whom the viral genomes are phylogenetically distinct to B.1.36.27 lineage. (c) GISAID and Nextstrain clade distribution of imported cases in Hong Kong. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Among the 73 locally-acquired cases in November (collected up to 29th November 2020), all were closely related to the viral genomes from 2 imported cases who had returned to Hong Kong from Nepal (Travelers A and B; Specimens collected on 19th September 2020 and on 25th September 2020, respectively) (PANGO lineage B.1.36.27, GISAID clade GH and Nextstrain 20A) (Fig. 2a and 2b). Out of these 73 cases, 37 (50.7%) had identical genomes, 32 (43.8%) had only one nucleotide difference, and 4 (5.5%) had 2 nucleotide difference from travelers A and B (Fig. 2b). Ten locally-acquired cases in October, including 8 cases that were released in GISAID previously [8], also belong to the B.1.36.27 lineage. These 10 locally-acquired cases in October were highly similar to the viral genomes from Travelers A and B, with 4 genomes being identical, 5 genomes having 1 nucleotide difference and 2 genomes had 2 nucleotide differences. Since 16th September 2020, genetically diverse genomes in different GISAID or Nextstrain clades were found among imported cases (Fig. 2c). Apart from Travelers A and B, we have also obtained the SARS-CoV-2 genomes of 3 other imported cases from Nepal (Travelers C, D and E), which were collected on 21st and 26th of October and 16th November 2020. Genomes from Travelers C-E were phylogenetically distinct from the genomes from Travelers A and B (Fig. 2b). Furthermore, the viral genomes from Travelers C-E differs from each other by 10–14 single nucleotide polymorphisms.
Sensitivity analysis was also performed to verify our conclusions by constructing phylogenetic trees using different substitution models. The topological similarities of the phylogenetic trees based on the 3 best substitution models ranged from 0.75117 to 0.784555 (Supplementary Table S3), suggesting that different models would still result in the same conclusion. Furthermore, there was no difference in the conclusions inferred from the maximum-likelihood and Bayesian phylogenetic trees (Fig. 2a and Supplementary Figure S2).
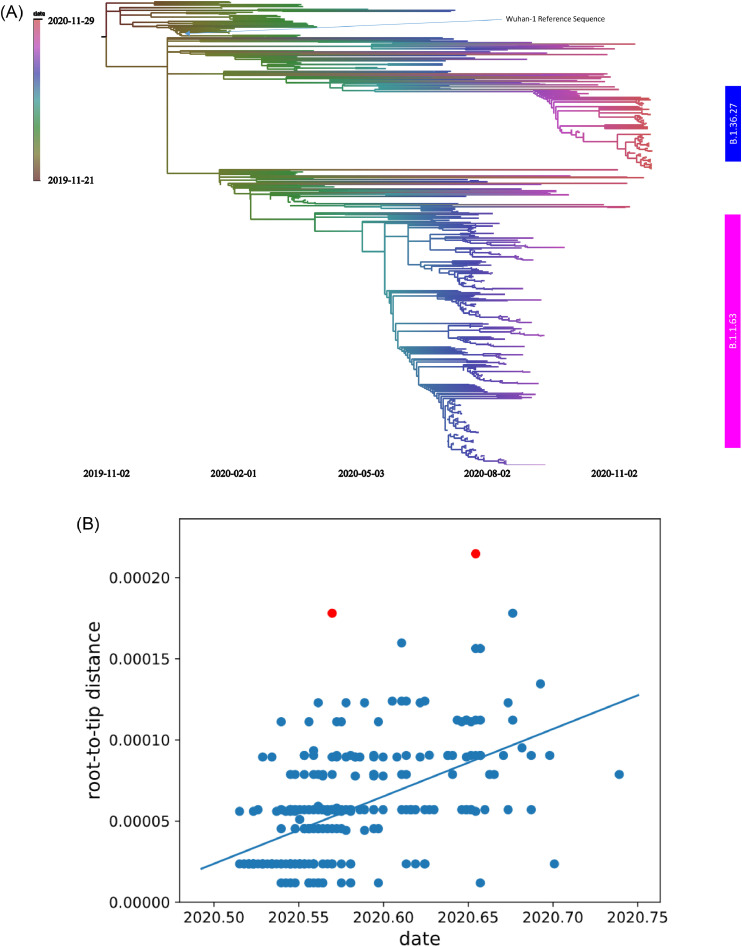
Time-resolved phylogenetic tree was constructed to show the evolution of the virus (Fig. 3a). Ten highly diverse genomes were removed as suggested by TreeTime. Therefore, in total, we analyzed 499 sequences, including 259 from the B.1.1.63 lineage and 85 from the B.1.36.27 lineage. The estimated divergent date of third wave B.1.1.63 lineage is 21st May 2020. The evolutionary rate of B.1.1.63 lineage is estimated to be 4.15 × 10−4 substitutions per site per year (r2: 0.3) (Fig. 3b). Since there are only 1–2 nucleotide changes within the fourth wave B.1.36.27 lineage, we considered the difference not reaching the phylodynamic threshold to estimate the evolutionary rate [24].
Fig. 3.
(a) Time-resolved phylogenetic tree of 499 viral genomes from December 2019 to November 2020. The scale bars indicate the substitution rates per site per year. (b) Evolutionary rate estimate using root-to-tip (RtT) regression analysis for B.1.1.63 lineage in third wave.
Fourth wave B.1.36.27 differs from the reference Wuhan-Hu-1 strain by 18 nucleotides, including 8 non-synonymous mutations in nsp2, nsp3, nsp12, spike, ORF3a and N genes (Table 1). The B.1.36.27 differs from the most closely related cluster in 6 nucleotide positions (G3431T, T5653C, G5950A, C6255T, C7504T, T24175C), including 2 non-synonymous mutations (nsp3 V238L and nsp3 A1179V).
Table 1.
Mutations identified in the third wave B.1.1.63 and the fourth wave B.1.36.27.
| Gene | Nucleotide | Amino acid | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Positiona | Wuhan-Hu-1 MN908947 | Third wave B.1.1.63 | Fourth wave B.1.36.27 | Position | Wuhan-Hu-1 MN908947 | Third wave B.1.1.63 | Fourth wave B.1.36.27 | ||
| Clade defining mutation | Untranslated region | 241 | C | T | T | – | – | – | – |
| Nsp3 | 3037 | C | T | T | 106 | F | F | F | |
| Nsp12 (RdRP) | 14,408 | C | T | T | 323 | P | L | L | |
| Spike | 23,403 | A | G | G | 614 | D | G | G | |
| ORF3a | 25,563 | G | G | T | 57 | Q | Q | H | |
| N | 28,881 | G | A | G | 203 | R | K | R | |
| 28,882 | G | A | G | 203 | R | K | R | ||
| 28,883 | G | C | G | 204 | G | R | G | ||
| Fourth wave B.1.36.27 defining mutation | Nsp2 | 922 | G | G | A | 39 | L | L | L |
| 1947 | T | T | C | 381 | V | V | A | ||
| Nsp3 | 3431 | G | G | T | 238 | V | V | L | |
| 5653 | T | T | C | 978 | Y | Y | Y | ||
| 5950 | G | G | A | 1077 | K | K | K | ||
| 6255 | C | C | T | 1179 | A | A | V | ||
| 7504 | C | C | T | 1595 | Y | Y | Y | ||
| Nsp14 | 18,877 | C | C | T | 280 | L | L | L | |
| Spike | 22,444 | C | C | T | 294 | D | D | D | |
| 24,175 | T | T | C | 871 | A | A | A | ||
| ORF3a | 26,060 | C | C | T | 223 | T | T | I | |
| M | 26,735 | C | C | T | 71 | Y | Y | Y | |
| N | 28,854 | C | C | T | 194 | S | S | L | |
| Third wave B.1.1.63 defining mutation | Nsp3 | 2973 | C | T | C | 85 | A | V | A |
| Nsp15 | 20,312 | C | T | C | 231 | A | V | A | |
| Spike | 21,597 | C | T | C | 12 | S | F | S | |
| N | 28,308 | C | G | C | 12 | A | G | A | |
| 29,144 | C | T | C | 291 | L | L | L | ||
Abbreviations: M, membrane; N, nucleoprotein; RdRp, RNA-dependent RNA polymerase.
The nucleotide position is numbered according the reference genome Wuhan-Hu-1 (Genbank accession number MN908947.3).
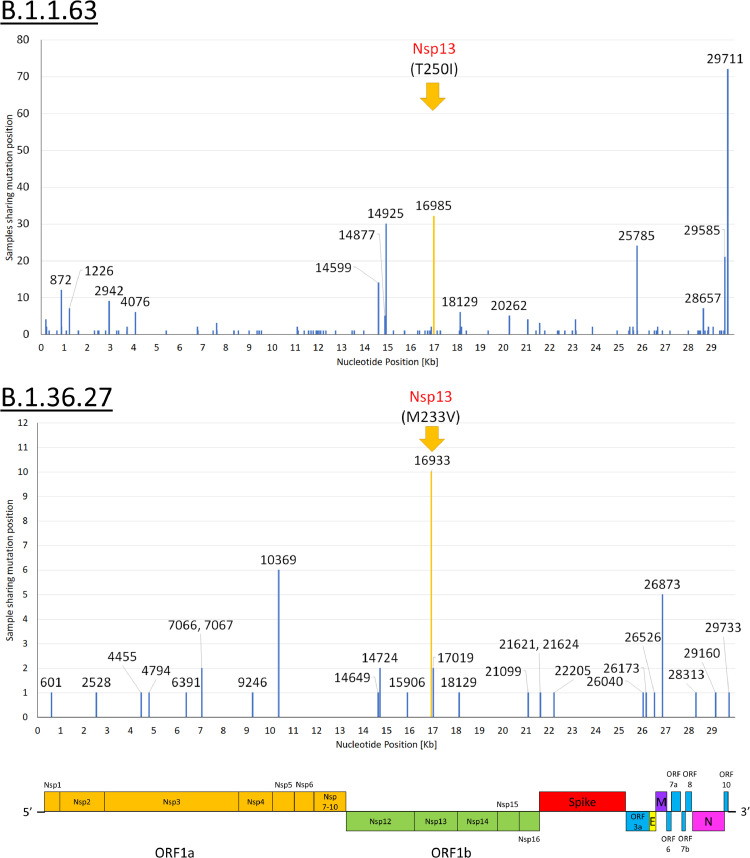
Within lineages B.1.1.63 and B.1.36.27 in third and fourth wave, single nucleotide polymorphisms can be found throughout the entire genome (Fig. 4). The most common non-synonymous mutations were found in the nsp13 gene (helicase). For the third wave B.1.1.63, 12.4% (32/259) contain the C16985T mutation (nsp13 T250I). For the fourth wave B.1.36.27, 11.8% (10/85) contain A16933G mutation (nsp13 M233V).
Fig. 4.
Single nucleotide mutations within B.1.1.63 in third wave and B.1.36.27 in fourth wave. The most common non-synonymous mutation are highlighted in yellow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Despite stringent control measures, Hong Kong experienced a large COVID-19 outbreak in summer (third wave) and is now facing another outbreak which started in November (fourth wave). Both waves were mainly comprised of locally-acquired cases. This study analyzed these two waves by phylogenetic, phylodynamic and single nucleotide variant analyses using whole viral genome sequences. Several similarities and differences between these two waves were identified. First, the third and fourth waves were caused by distinct viruses from different lineages, suggesting that the local transmission of the third wave has subsided. Second, both the third and fourth waves were each dominated by a single genetic lineage related to import cases. While cases belonging to the B.1.1.63 lineage is phylogenetically most related to imported cases from the Philippines, cases belonging to the lineage B.1.36.27 is identical to two viral genomes obtained from imported cases from Nepal. Third, unlike the third wave in which the imported B.1.1.63 virus was only found 2 weeks before the local outbreak, the B.1.36.27 lineage has been imported and circulated in the community for about 2 months before the start of the fourth wave. Our results have important implications for the control of COVID-19 outbreaks in areas with low incidence, such as China, Singapore, Australia and New Zealand.
Unlike the third wave which was caused by the B.1.1.63 lineage that was newly introduced into Hong Kong shortly before the wave, the fourth wave was caused by the B.1.36.27 lineage that was introduced into Hong Kong two months before the outbreak. Furthermore, there was little genetic difference between the viruses collected in November and the first virus within the B.1.36.27 lineage that was collected in September. Hence, the fourth wave is not related to viral mutations that emerge during the circulation in Hong Kong. There are several possibilities for the sudden onset of the fourth wave in November. First, the early cases in the fourth wave were traced to dancing groups, and the efficient transmission in these early cases was likely related to the clustering of people in an indoor environment without wearing masks. The lack of genomic diversity suggests the possibility of a superspreading event. Another possible explanation for the fourth wave is the change in weather. Morris et al. showed that the survival of SARS-CoV-2 was longer when the temperature decreases from 27 °C to 22 °C [25]. In Hong Kong, the mean daily temperature decreased from 28.4 °C in September to 23.5 °C in November [26]. Therefore, the lower temperature in November rendered the virus more stable in the environment, facilitating the spread of the virus. However, the contribution of weather change remains to be determined as many outbreaks in the world, such as those in South Africa, Brazil and India, occurred during the summer.
Multiple studies in other countries have shown that imported cases are an important source of local outbreaks. Without restrictions on imported cases, there would be simultaneous introductions of different genetic lineages into the local community [27]. Although B.1.1.63 dominated the third wave, four other lineages were also identified, suggesting multiple sources of imported cases. However, all locally-acquired cases during the fourth wave belonged to B.1.36.27 lineage, despite the continuous detection of other lineages among travelers from September to November. Our results suggest that the stepping up of control measures for travelers during and after the third wave, including the tightening of testing and quarantine arrangements for sea crew and air crew members, has indeed prevented the spread of infection from most imported cases.
There are several possibilities for the introduction of the new viruses into Hong Kong from incoming travelers despite stringent control measures. First, some patients may have the virus detected only after the quarantine period. Studies have shown that the incubation period can be longer than 14 days [28]. Second, there may have been transmission from the index case to the community via either direct contact with the hotel staff, or from indirect contact via inanimate objects. To prevent imported cases to cause local transmission, the Hong Kong government has implemented mandatory quarantine for all incoming travelers since March 2020, which has tightened gradually. At the time of writing, all incoming travelers, except those from mainland China, Macau and Taiwan, must be quarantined at designated hotels for 21 days. SARS-CoV-2 testing is performed on arrival, and then on day 12 and 19 after arrival. Between 22nd June 2020 and 29th November 2020, there were a total of 922 imported cases in Hong Kong [10], but we only identified 4 lineages related to imported cases during this period.
Fourth wave B.1.36.27 has 8 amino acid differences from the reference strain Wuhan-Hu-1. Spike protein D614G, which is present in all the GISAID clade G, GR or GH viruses, has been shown to confer better viral replication and transmissibility [29]. ORF3a Q57H, which is present in GISAID clade GH, has been proposed to affect the protein structure and binding affinity of ORF3a to S or ORF8 proteins [30]. Two mutations were located in nsp3, including V238L (or ORF1ab V1056L) and A1179V (or ORF1ab A1997V). Nsp3 is a papain-like protease which is involved in polyprotein processing, de-ADP ribosylation, deubiquitination, double membrane vesicle formation, and interferon antagonism [31]. The N protein S194L may enhance the interaction between N and E protein but may decrease the interaction between N and M protein [30]. Further studies are needed to clarify the impact of these viral mutations on the virulence and transmissibility of SARS-CoV-2.
Within both B.1.1.63 and B.1.36.27 in third and fourth waves, the most common non-synonymous single nucleotide polymorphism was found in the nsp13 gene, also known as the helicase. Nsp13 is important for viral replication, and participates in the cap synthesis during mRNA translation [32]. Inhibition of nsp13 with bismuth has been shown to reduce viral replication and disease severity in our SARS-CoV-2 hamster model [33]. Nsp13 is also an interferon antagonist [34]. It remains to be determined whether these nsp13 mutations confer higher virulence or transmissibility of the virus.
We have taken several measures to ensure the accuracy of the phylodynamic analysis. First, we excluded viral genomes that have been considered not suitable for analysis by the TreeTime, the phylodynamic program used by Nextstrain. Second, we have masked the locations which were suspected to be problematic, before phylogenetic tree construction. Turakhia et al. have demonstrated that some nucleotide mutations are potential systemic errors generated by specific protocols, and may mislead phylogenetic analysis [19]. None of our genome sequences contain these changes. The estimated evolutionary rate of third wave B.1.1.63, 4.15 × 10−4 substitutions/site/year, is slightly slower than the previously reported rates inferred from genomes during the early pandemic. Duchene et al. reported an evolutionary rate of 1.1 × 10−3 substitutions/site/year using genomes before 2nd February 2020 [24], while Leung et al. reported an evolutionary rate of 3.04 × 10−3 substitutions/site/year for genomes on or before 28th February 2020 [35]. The slightly slower evolution rate of B.1.1.63 is consistent with the fact that SARS-CoV-2 has already circulated in humans for over at least 6 months when B.1.1.63 lineage appears in our population [36].
There are several limitations to this study. First, sequencing failed on some clinical specimens due to low viral load. Second, at the time of submission, the fourth wave has not ended yet. Therefore, the further evolution of B.1.36.27 lineage and whether other lineages will emerge during the fourth wave remains to be determined. Third, we cannot exclude the possibility that multiple sources carrying identical genomes have led to the community outbreak.
In conclusion, our genomic analysis uncovered the similarities and differences between the third and fourth waves of COVID-19 in Hong Kong, highlighting the successes and limitations of the current control measures. While restriction measures on inbound travelers have prevented local transmission of SARS-CoV-2 from most imported cases, even a single imported source can result in a large outbreak. There was a long period of low-level transmission of the B.1.36.27 lineage in Hong Kong prior to the fourth wave. A coordinated strategy to curb transmission through improved diagnostic testing and public health measures are required. Continued genomic surveillance of locally-acquired cases is pivotal in detecting novel lineages that enters Hong Kong.
Declaration of Competing Interest
All authors declare no conflict of interest.
Acknowledgments
Acknowledgement
We gratefully acknowledge the originating and submitting laboratories who contributed sequences to GISAID (Supplementary Table S4).
Author's contribution
DCL, IFNH, KYY and KKWT had roles in study design, data collection, data analysis, data interpretation, literature search and writing of the manuscript. ART, MYWK, YSY, WSL, TSHC, WKT, ACKN, CCYY, RWSP, KHC, VCCC and IFNH had roles in recruitment, data collection, and/or clinical management. WMC, JDI, AWHC, HT, SCYW and GKYC had roles in performing the experiments, data collection, data analysis, and/or data interpretation. XL and KHC had roles in data interpretation and writing of the manuscript. All authors interpreted the data, revised the manuscript critically for important intellectual content and approved the final report.
Funding
This work was supported by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for the Department of Health of the HKSAR Government, HMRF Commissioned Research on Control of Infectious Disease (Phase IV) (CID-HKU1-2), and donations of Richard Yu and Carol Yu, May Tam Mak Mei Yin, the Shaw Foundation Hong Kong, Michael Seak-Kan Tong, Lee Wan Keung Charity Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, Kai Chong Tong, Tse Kam Ming Laurence, Foo Oi Foundation Limited, Betty Hing-Chu Lee, and Ping Cham So. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Data sharing
Data are available upon reasonable request.
Editor note: The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Footnotes
Funding: Richard and Carol Yu, Michael Tong, and the Government Consultancy Service (see acknowledgments for full list).
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.lanwpc.2021.100130.
Appendix. Supplementary materials
Phylogenetic networks showing the relationship of 509 viral genomes from locally-acquired and imported COVID-19 cases in Hong Kong from January to November 2020. The phylogenetic network was constructed by SplitsTree4. The reference genome Wuhan-Hu-1 (GenBank accession number MN908947.3) was used as the root of the tree. The blue box indicates B.1.36.27 lineage from fourth wave. Pink box indicates B.1.1.63 lineage from third wave.
Bayesian phylogenetic tree of 510 SARS-CoV-2 genomes, including Wuhan-Hu-1. The tree was constructed by MrBayes with model propinv. The blue branch indicates B.1.36.27 lineage from fourth wave. Pink, green and orange branches indicate B.1.1.63, B.1.1.141 and B.1.1.47 lineages from third wave, respectively. The scale bar indicates the expected substitution rates per site.
GISAID Accession number of sequences used in this study.
Number of COVID-19 patients in Hong Kong during the study period.
Topological similarities among the three phylogenetic tree constructed using different substitution models.
Acknowledgement table for GISAID sequences used in this study.
References
- 1.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.To K.K., Cheng V.C., Cai J.P., Chan K.H., Chen L.L., Wong L.H. Seroprevalence of SARS-CoV-2 in Hong Kong Special Administrative Region and our returnees evacuated from Hubei province of China: a multi-cohort study. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worobey M., Pekar J., Larsen B.B., Nelson M.I., Hill V., Joy J.B. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370:564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., O'Toole A., Haverkate M., Mollers M. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat. Med. 2020;26:1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 5.Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To K.K., Chan W.M., Ip J.D., Chu A.W., Tam A.R., Liu R. Unique SARS-CoV-2 clusters causing a large COVID-19 outbreak in Hong Kong. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1119. 10.1093/cid/ciaa1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam H.Y., Lam T.S., Wong C.H., Lam W.H., Leung C.M.E., Au K.W.A. The epidemiology of COVID-19 cases and the successful containment strategy in Hong Kong-January to May 2020. Int. J. Infect. Dis. 2020;98:51–58. doi: 10.1016/j.ijid.2020.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siu G.K., Lee L.K., Leung K.S., Leung J.S., Ng T.T., Chan C.T. Will a new clade of SARS-CoV-2 imported into the community spark a fourth wave of the COVID-19 outbreak in Hong Kong? Emerg. Microbes. Infect. 2020 doi: 10.1080/22221751.2020.1851146:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Government of the Hong Kong Special Administrative Region. CHP investigates 43 additional confirmed cases of COVID-19. Available at https://www.info.gov.hk/gia/general/202011/21/P2020112100738p.htm. Accessed on 25th November 2020.
- 10.Centre for Health Protection. Latest situation of cases of COVID-19. Available at https://www.chp.gov.hk/files/pdf/local_situation_covid19_en.pdf. Accessed on 2nd December 2020.
- 11.To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan W.M., Ip J.D., Chu A.W., Yip C.C., Lo L.S., Chan K.H. Identification of nsp1 gene as the target of SARS-CoV-2 real-time RT-PCR using nanopore whole-genome sequencing. J. Med. Virol. 2020;92(11):2725–2734. doi: 10.1002/jmv.26140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quick J. Artic Network-nCoV 2019 sequencing protocol. 2020. Available at https://artic.network/ncov-2019. Accessed on July 22, 2020.
- 16.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagulenko P., Puller V., Neher R.A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turakhia Y., De Maio N., Thornlow B., Gozashti L., Lanfear R., Walker C.R. Stability of SARS-CoV-2 phylogenies. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 21.GISAID. Clade and lineage nomenclature aids in genomic epidemiology studies of active hCoV-19 viruses. Available at https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/. Accessed on July 19, 2020. 2020
- 22.Hodcroft E.B., Hadfield J., Neher R.A., Bedford T. Year-letter Genetic Clade Naming for SARS-CoV-2 on Nextstain.org. Available at https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming. Accessed on July 25, 2020.
- 23.Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris D.H., Yinda K.C., Gamble A., Rossine F.W., Huang Q., Bushmaker T. The effect of temperature and humidity on the stability of SARS-CoV-2 and other enveloped viruses. bioRxiv. 2020 doi: 10.1101/2020.10.16.341883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Observatory H.K. Monthly weather summary. Available at https://www.hko.gov.hk/en/wxinfo/pastwx/mws/mws.htm. Accessed on 7th December 2020. 2020
- 27.Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M.J., Ciferri B., Alshammary H., Obla A. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 2020;369:297–301. doi: 10.1126/science.abc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie X., Fan L., Mu G., Tan Q., Wang M., Xie Y. Epidemiological characteristics and incubation period of 7015 Confirmed Cases With Coronavirus Disease 2019 Outside Hubei Province in China. J. Infect. Dis. 2020;222:26–33. doi: 10.1093/infdis/jiaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S., Tian C., Liu P., Guo D., Zheng W., Huang X. Effects of SARS-CoV-2 mutations on protein structures and intraviral protein-protein interactions. J. Med. Virol. 2020 doi: 10.1002/jmv.26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2020 doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan S., Wang R., Chan J.F., Zhang A.J., Cheng T., Chik K.K. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat. Microbiol. 2020;5:1439–1448. doi: 10.1038/s41564-020-00802-x. [DOI] [PubMed] [Google Scholar]
- 34.Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes. Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung K.S., Ng T.T., Wu A.K., Yau M.C., Lao H.Y., Choi M.P. Territorywide study of early coronavirus disease outbreak, Hong Kong, China. Emerg. Infect. Dis. 2021;27:196–204. doi: 10.3201/eid2701.201543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elena S.F., Miralles R., Cuevas J.M., Turner P.E., Moya A. The two faces of mutation: extinction and adaptation in RNA viruses. IUBMB Life. 2000;49:5–9. doi: 10.1080/713803585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic networks showing the relationship of 509 viral genomes from locally-acquired and imported COVID-19 cases in Hong Kong from January to November 2020. The phylogenetic network was constructed by SplitsTree4. The reference genome Wuhan-Hu-1 (GenBank accession number MN908947.3) was used as the root of the tree. The blue box indicates B.1.36.27 lineage from fourth wave. Pink box indicates B.1.1.63 lineage from third wave.
Bayesian phylogenetic tree of 510 SARS-CoV-2 genomes, including Wuhan-Hu-1. The tree was constructed by MrBayes with model propinv. The blue branch indicates B.1.36.27 lineage from fourth wave. Pink, green and orange branches indicate B.1.1.63, B.1.1.141 and B.1.1.47 lineages from third wave, respectively. The scale bar indicates the expected substitution rates per site.
GISAID Accession number of sequences used in this study.
Number of COVID-19 patients in Hong Kong during the study period.
Topological similarities among the three phylogenetic tree constructed using different substitution models.
Acknowledgement table for GISAID sequences used in this study.