Abstract
Multiplex families with a high prevalence of a psychiatric disorder are often examined to identify rare genetic variants with large effect sizes. In the present study, we analysed whether the risk for bipolar disorder (BD) in BD multiplex families is influenced by common genetic variants. Furthermore, we investigated whether this risk is conferred mainly by BD-specific risk variants or by variants also associated with the susceptibility to schizophrenia or major depression. In total, 395 individuals from 33 Andalusian BD multiplex families (166 BD, 78 major depressive disorder, 151 unaffected) as well as 438 subjects from an independent, BD case/control cohort (161 unrelated BD, 277 unrelated controls) were analysed. Polygenic risk scores (PRS) for BD, schizophrenia (SCZ), and major depression were calculated and compared between the cohorts. Both the familial BD cases and unaffected family members had higher PRS for all three psychiatric disorders than the independent controls, with BD and SCZ being significant after correction for multiple testing, suggesting a high baseline risk for several psychiatric disorders in the families. Moreover, familial BD cases showed significantly higher BD PRS than unaffected family members and unrelated BD cases. A plausible hypothesis is that, in multiplex families with a general increase in risk for psychiatric disease, BD development is attributable to a high burden of common variants that confer a specific risk for BD. The present analyses demonstrated that common genetic risk variants for psychiatric disorders are likely to contribute to the high incidence of affective psychiatric disorders in the multiplex families. However, the PRS explained only part of the observed phenotypic variance, and rare variants might have also contributed to disease development.
Subject terms: Bipolar disorder, Depression, Schizophrenia, Bipolar disorder, Depression
Introduction
Bipolar disorder (BD), characterised by alternating episodes of mania and depression, has a lifetime prevalence of ~1% and is a substantial contributor to disability throughout the world [1]. Nevertheless, reliable data concerning the aetiology of BD remain scarce. The heritability of BD is estimated to be above 70% [2–4], thus demonstrating an important genetic component in the development of the disorder. Genome-wide association studies (GWAS) in case/control samples have reported that single-nucleotide polymorphisms (SNP) with minor allele frequencies (MAF) of ≥ 1% explain a substantial proportion of the genetic risk for BD [5–12]: the heritability explained by such common variants (i.e., the SNP heritability) is estimated to be 0.17–0.23 on a liability scale [12]. Common variants also make a substantial contribution to the development of schizophrenia (SCZ) and major depressive disorder (MDD) [13, 14]. These three psychiatric disorders have a shared genetic component, whereby relatives of patients with BD have, in addition to BD, an increased risk for MDD and SCZ [15]. In fact, GWAS have shown that many genetic risk variants are associated with all three disorders [16–21].
Besides common variants with small individual effects, rare variants with larger effects may also contribute to BD development [22, 23]. In theory, such rare variants should be enriched in families with a high prevalence of illness, termed multiplex families, in comparison to unrelated BD cases. However, it remains unclear whether and to what extent disease incidence in multiplex families is caused by rare variants, a high load of common variants, or a combination of both.
To elucidate the molecular genetic causes of BD, we established the Andalusian Bipolar Family (ABiF) study in 1997, which recruited BD multiplex families [24–26]. In the present analyses, we first investigated whether common genetic variants make a significant contribution to the occurrence of BD in ABiF families. Next, we examined whether BD development was attributable to (a) BD-specific risk variants, (b) variants conferring risk to all three disorders BD, MDD, and SCZ, or, (c) a combination of both. To this end, polygenic risk scores (PRS) based on GWAS of BD, MDD, and SCZ were calculated for and compared between ABiF family members and unrelated BD cases and unrelated controls from the same population. Because of the strong genetic correlation between BD, SCZ, and MDD, standard PRS for BD cannot distinguish between BD-specific risks and factors shared between these disorders. To differentiate between genetic risk shared across and specific to any of the three disorders, we calculated PRS of disorder-specific risk variants using genome-wide inferred statistics (GWIS) and PRS of shared risk variants. To evaluate the possibility that population or technical differences between cohorts biased the results on psychiatric PRS, PRS for late-onset Alzheimer’s disease (LOAD) and simulated PRS were analysed as negative controls. Assuming a polygenic model with a contribution of common risk variants, we expected increased psychiatric PRS in the ABiF family members compared to unrelated samples and increased psychiatric PRS in patients compared to controls.
Materials and methods
Sample description
The ABiF study recruited BD multiplex families in Andalusia, Spain [24–26]. The present analyses included 395 members of 33 ABiF families. Diagnoses were assigned by two trained clinicians according to the Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV criteria using the best estimate approach [24]. Diagnoses comprised (Table 1 and Supplementary Table S1): BD, n = 166 (families (FAM)BD; BD type I (BD-I): n = 115; BD type II (BD-II): n = 41; not otherwise specified (NOS) BD: n = 10); MDD, n = 78 MDD (FAMMDD); no history of an affective disorder, n = 151 (FAMunaffected). Six unaffected individuals with a history of substance abuse were excluded from the analyses. Forty-four subjects married into the families and had no parent in the ABiF cohort (36 unaffected; 8 MDD). Furthermore, an independent, previously reported Spanish BD case/control (CC) sample was analysed. Here, BD cases (CCBD) were recruited from consecutive clinical admissions and BD was diagnosed, as in the ABiF families, using DSM-IV [9]; unrelated control individuals (CCcontrols) were recruited in the framework of the longitudinal European Community Respiratory Health Survey (ECRHS) study. Blood for genotyping was acquired at the ECRHS2 assessment in 2000–2001. After quality control (QC), the combined data set of both cohorts comprises data from 384 FAM (163 FAMBD, 73 FAMMDD, 142 FAMunaffected, and 6 FAMunaffected with a history of substance abuse) and 438 CC subjects (161 unrelated BD cases; BD-I: n = 156; BD-II: n = 5) and 277 unrelated controls. Of the 161 CCBD cases, 59 (36.6%) reported a family history of BD. However, in contrast to the data collection in the ABiF families, this information relied only on the self-report by the respective CCBD patient, and was not validated via an interview of further family members. BD diagnoses were not available for the unrelated controls, but the self-reported prevalence of current depression in this cohort was 3.3% at the time of genotyping and the self-reported prevalence of lifetime depression was 14.4% at the follow-up 10 years after genotyping, indicating that the cohort is fairly representative of a typical population in regard to the prevalence of depression [27].
Table 1.
Characteristics of 389 individuals from the 33 ABiF families, the 161 unrelated bipolar cases (CCBD), and the 277 unrelated controls (CCcontrols)
| FAMBD (n = 166) |
FAMMDD (n = 78) |
FAMunaffected (n = 145) |
CCBD (n = 161) |
CCcontrols (n = 277) |
|
|---|---|---|---|---|---|
| Median age at interview (MAD) | 40.5 (12.5) | 44.5 (11.5) | 44 (15)d | 44 (11)d |
43 (6)b,c Missing = 3 |
| Median age at onset (MAD) |
20 (5)c Missing = 3 |
26 (8)a Missing = 1 |
23 (6)a | ||
| Female sex n (%) | 103 (62.1)b,d | 54 (69.2)b,d | 51 (35.2)a,c,d | 91 (56.5)b | 132 (47.7)a,b |
| Married-in n (%) | 0 (100.0) | 8 (10.0) | 36 (24.8) | ||
| Educational level | |||||
| Primary school | 118 (71.5)c | 53 (68.0) | 102 (70.3)c | 88 (54.7)a,b | |
| Secondary school | 39 (23.6)c | 18 (23.1)c | 36 (24.8)c | 58 (36.0)a,b | |
| University degree | 8 (4.9) | 7 (9.0) | 7 (4.8) | 15 (9.3) | |
| Missing = 1 | |||||
| Severe impairment during disorder |
105 (65.6)c Missing = 6 |
4 (5.6)a,c Missing = 6 |
160 (99.4)a | ||
| History of psychosis | 110 (66.3)c | 4 (5.1)a,c | 159 (98.8)a | ||
| History of suicide attempts | 41 (24.7)b | 2 (2.6)a,c |
1 (0.7)a,c Missing = 1 |
40 (24.8)b | |
Six unaffected individuals with a history of substance abuse were excluded from the analyses and are not shown in this table. Age and age at onset were analysed using Mann–Whitney U-tests; median and median absolute deviation (MAD) are shown. Categorical values were analysed using chi-squared (χ²) tests with two degrees (education) or one degree (other) of freedom; number (n) and percentage (%) of subjects are shown. Missing: number of individuals with missing data. All subjects passed QC in the FAM sample (numbers as shown in the table), but 11 family members were excluded during QC of the joint sample, therefore reported numbers differ slightly between comparisons. Note that the unaffected, married-in family members were excluded from analyses of the combined data set (FAM + CC sample) unless specified otherwise. Differences between the following groups were at least nominally significant (for details and p-values adjusted for multiple testing see Supplementary Table 1)
aDifferent from FAMBD
bDifferent from FAMunaffected
cDifferent from CCBD
dDifferent from CCcontrols
Note that, while all subjects passed QC in the family-only sample, 11 family members were excluded during QC of the joint sample because they showed significant differences in autosomal heterozygosity from the mean. Reported numbers of subjects thus differ slightly for different comparisons. The joint data set contained 35 unaffected, married-in family members who were excluded from analyses using the combined sample (unless specified otherwise). A detailed description of QC procedures is provided in the Supplementary Methods.
The study was approved by the respective local ethics committees (Comités de ética de la investigación provincial de Cádiz, Córdoba, Granada, Jaén and Málaga), and all participants provided written informed consent. For five adolescents (age 15–17 years), written informed consent was also obtained from the parents.
Genotyping and imputation
Genome-wide genotyping of the FAM sample was carried out using the Illumina Infinium PsychArray BeadChip (PsychChip). QC and population substructure analyses were performed in PLINK v1.9 [28], as described in the Supplementary Methods. Genotyping and basic QC of the CC sample were conducted previously and are described elsewhere [9]. The study used two genotype data sets: Analyses of family members by themselves used variants genotyped on the PsychChip. For analyses on the combined FAM + CC sample, the genotype data of the CC data set were, for the variants genotyped in both samples, merged with the genotype data of the FAM sample. Both genotype data sets (family-only and combined) were imputed independently to the 1000 Genomes phase 3 reference panel using SHAPEIT and IMPUTE2 [29–31]. After imputation and post-imputation QC, the combined data set of both cohorts contained 6,862,461 variants with an INFO metric of ≥ 0.8 and a MAF of ≥ 1%. The imputed FAM data set without the CC subjects contained 8,628,089 variants.
Calculation of polygenic risk scores
PRS were calculated in R v3.3 [32] using imputed genetic data. For each PRS, the effect sizes of variants below a selected p-value threshold, both obtained from large GWAS (training data), were multiplied by the imputed SNP dosage in the test data and then summed to produce a single PRS per threshold. Test statistics and alleles in the GWAS training data were flipped so that effect sizes were always positive. Thus, the PRS represent cumulative, additive risk. PRS were scaled to represent the relative risk load (minimum possible cumulative risk load = 0, maximum = 1). For each disorder, ten PRS based on different GWAS p-value thresholds (<5 × 10−8, <1 × 10−7, <1 × 10−6, <1 × 10−5, <1 × 10−4, <0.001, <0.01, <0.05, <0.1, <0.2) were calculated. The number of SNPs used for each PRS is shown in Supplementary Table S2. For additional details, see the Supplementary Methods.
For BD, MDD, and SCZ diagnoses, summary statistics of GWAS by the Psychiatric Genomics Consortium (PGC) were used as training data. For BD, the data freeze contained 20,352 cases and 31,358 controls [12]. As selected index patients from the ABiF families and the unrelated Spanish BD case/control data set were part of this BD GWAS, we recalculated summary statistics for this PGC GWAS without these Spanish samples, to prevent false-positive results caused by sample overlap between training and test samples. For MDD and SCZ, published data sets were used. These contained 130,664 cases and 330,470 controls for MDD [14] and 33,640 cases and 43,456 controls for SCZ [13]. There was no overlap between the subjects included in those GWAS and the ABiF and Spanish case/control samples. Variants with an INFO metric of < 0.6 in the GWAS summary statistics were removed.
Shared psychiatric PRS were generated using all variants showing an association at p < 0.05 in the GWAS of BD, SCZ, and MDD and for which effect sizes pointed in the same direction across studies. For this shared set of variants, p-values and effect sizes, used as weights in the PRS, were obtained using random-effects meta-analysis. PRS were then calculated using the meta-analysis summary statistics. We generated disorder-specific summary statistics to assess genetic risk unique to each disorder. To this end, genome-wide inferred statistics (GWIS) were calculated as explained in detail elsewhere [33]. For example, we calculated BD GWAS summary statistics corrected for the MDD GWAS results (BD-MDD). These BD-MDD GWIS results are similar to results obtained from a conditional analysis for BD corrected for MDD. They represent a genetic unique BD liability, which is estimated based on the heritability of BD and the coheritability of BD and MDD, both estimated using LD score regression [34]. As recommended for this method, variants with an INFO metric of <0.9 or >1.1 were removed. Disorder-specific PRS, e.g., BD-MDD PRS, were then calculated based on the corresponding GWIS summary statistics.
To confirm whether family members and BD cases had an increased PRS specifically for the tested psychiatric disorders but not because of population or technical differences between cohorts, PRS for late-onset Alzheimer’s disease (LOAD) were calculated as a negative control, based on a GWAS by the International Genomics of Alzheimer’s Project (IGAP) with 17,008 cases and 37,154 controls [35]. For additional details, see the Supplementary Methods. Furthermore, 10,000 simulated PRS for each of the ten p-value thresholds were calculated as negative controls. To this end, random variants from across the genome were drawn, using the same number of variants as for the BD PRS at each threshold and random effect sizes from the pool of all available BD, SCZ, and MDD effects. The code for simulating PRS is available at: https://gitlab.com/tillandlauer/abif-prs-analyses/.
Statistical analysis
PRS analyses on binary variables (e.g., diagnoses and comparisons between cohorts) were conducted in R with the function glmm.wald of the package GMMAT, using a logistic mixed model, fitted by maximum likelihood using Nelder–Mead optimisation [36] to account for family structure. For logistic models, PRS underwent Z-score standardisation to generate comparable odds ratios (OR). Family structure was modelled as a random effect, with a genetic relationship matrix calculated on pruned genotype data in GEMMA [37].
Linear mixed models (LMMs) taking family structure into account were calculated using the function polygenic of the package GenABEL [38] for analyses of quantitative variables (anticipation and age at onset). In these analyses, test statistics, including 95% confidence intervals (CI), were calculated using bootstrapping (package boot [39, 40]) and p-values were validated using permutation analysis (10,000 permutations). In these permutation analyses, the null distribution of test statistics was empirically determined by repeating regression analyses 10,000 times with random sampling of phenotype data. To calculate a p-value, the number of tests were counted where a model with a random genotype-phenotype association showed the same or a more extreme p-value than the correct, non-randomised model and this number was divided by the total number of tests (10,000).
For each analysis of PRS, all ten PRS p-value thresholds were analysed. In analyses of the combined FAM and CC data set, sex was used as a covariate. In the analysis of FAM data alone, sex and age at the time of the interview were used as fixed effects covariates; whether an individual had married into the family was incorporated as a second random effect. Following the hypothesis that family members or subjects with a psychiatric diagnosis have increased PRS for psychiatric disorders, one-sided p-values were calculated for all PRS-based analyses. In all analyses, p-values below the significance threshold α = 0.05 were considered as nominally significant. Unless otherwise stated, this threshold was corrected for 10 × 6 = 60 tests using the Bonferroni method (α = 0.05/60 = 8.33 × 10−4). For further details, see the Supplementary Methods.
To determine whether population or technical differences might have influenced the observed effects independently of diagnosis groups, simulated PRS, generated as described above, were analysed. For each model, association statistics of the 10,000 simulated PRS were calculated for the ten p-value thresholds; the disorder PRS at the threshold showing the lowest mean association p-value was analysed further: The number of simulated PRS at this threshold that showed the same or a stronger association was counted and compared to the association of the disorder PRS. This count was used as the number of successes in a binomial test to estimate the probability of success. For computational efficiency, models were fitted using restricted maximum likelihood estimation and the average information optimisation algorithm for this analysis.
Results
Figures 1 and 2 show the test statistics for the PRS with the training GWAS p-value threshold pPRS that showed the strongest association per PRS type. Full results for all ten pPRS per PRS type calculated using logistic mixed models are provided in Supplementary Figs. S1–S12 and in Supplementary Tables S3–S11.
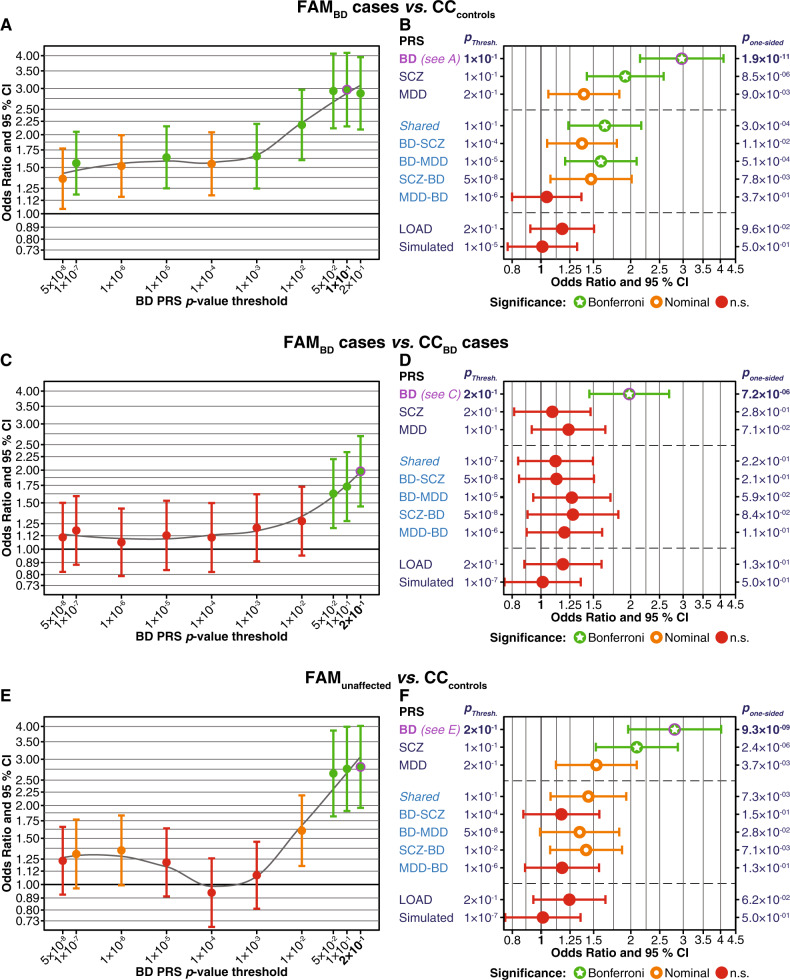
Fig. 1.
Comparison of PRS between FAM and CC samples. Married-in family members were excluded from these analyses. The plots show one-sided p-values, following the hypothesis that family members have higher PRS than individuals from the CC samples. All PRS have been normalised using Z-score standardisation. a, b Comparison of FAMBD cases to CCcontrols. a FAMBD cases had higher BD PRS across all ten pPRS thresholds. The plot shows odds ratios (OR, y-axis, filled circles) and 95% confidence intervals (CI); pPRS thresholds are shown on the x-axis. Results for each threshold are coloured by their degree of significance (one-sided p-values): red = not significant, orange = nominally significant, green = significant after Bonferroni correction for multiple testing (α = 0.05/60 = 0.00083). The top-associated PRS (pPRS = 0.1) is indicated in bold font and was marked by a magenta circle (also in b). b For ten different PRS, this plot shows association statistics for the top-associated pPRS thresholds. The x-axis shows ORs. BD, SCZ, MDD: Standard PRS using the respective PGC GWAS summary statistics. Shared: Shared psychiatric PRS (SNPs with BD, MDD, SCZ p < 0.05, random-effects meta-analysis). BD-SCZ, BD-MDD: BD-specific GWIS PRS corrected for SCZ and MDD, respectively. SCZ-BD and MDD-BD: GWIS PRS for SCZ and MDD, each corrected for BD. LOAD: PRS for late-onset Alzheimer’s disease. Simulated: Mean and CI of the 10,000 simulated PRS at the pPRS with the lowest mean association p-value of all simulated PRS. The column to the left of the plot: pPRS with the strongest association. Supplementary Fig. S2 shows plots for all pPRS. Column to the right: pone-sided = one-sided p-value. For full association test statistics, see Supplementary Table S3. Bonferroni = significant after Bonferroni correction for multiple testing; nominal = nominally significant (p < 0.05); n.s. = not significant. c, d Comparison of FAMBD cases and unrelated CCBD cases. See Supplementary Fig. S3 and Table S4 for more detailed plots and full association test statistics. e, f Comparison of FAMunaffected and CCcontrols. See Supplementary Fig. S4 and Table S5 for more detailed plots and full association test statistics
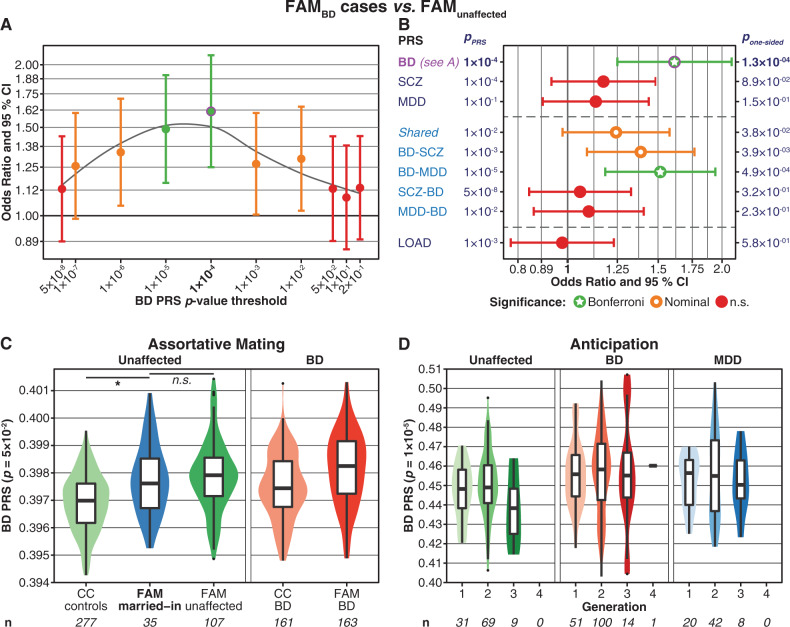
Fig. 2.
a, b Comparison of PRS between FAMBD cases and FAMunaffected. The plots show one-sided p-values, following the hypothesis that BD cases have higher PRS than unaffected individuals. Further details of the plots are as described in the legend for Fig. 1. See Supplementary Fig. S5 and Table S6 for more detailed plots and full association test statistics. c, d Analyses of assortative mating (c) and anticipation (d). These plots were not adjusted for covariates; n = sample size. The y-axis shows the PRS values. c: Assortative mating. The plot shows violin- and boxplots of the BD PRS (pPRS = 0.05), comparing unaffected, married-in individuals with no parent among the ABiF families to other FAM and CC subjects. At pPRS = 0.05, married-in family members showed the highest BD PRS compared to CCcontrols (p = 6.5 × 10−5, Supplementary Fig. S6A and Table S7). The BD PRS of married-in individuals was not significantly higher than the PRS of FAMunaffected at any pPRS (p ≥ 0.167, Supplementary Fig. S6B and Table S7). Covariate used: sex. One-sided p-values were calculated, following the hypothesis that married-in individuals have higher PRS than other unaffected subjects. Note that, in the context of assortative mating, the boxplots of affected BD cases are displayed for reference only and have not been included in the analysis. d Anticipation: the BD PRS did not increase across generations. The plot shows violin- and boxplots of the BD PRS (pPRS = 1 × 10−5) across different generations of the FAM sample for the three diagnosis groups. At pPRS = 1 × 10−5, the association of the BD PRS with generation was strongest but not significant (p = 0.45; Supplementary Fig. S7A and Table S8). Married-in family members were excluded from this analysis. Covariates used: sex, age at the interview, diagnostic group. One-sided p-values were calculated, following the hypothesis that the PRS increase across generations
FAMBD cases had higher psychiatric PRS than controls from the general population
On average, familial FAMBD cases had higher BD PRS than unrelated CCcontrols across the pPRS thresholds (Fig. 1a, b; Supplementary Figs. S1 and S2 and Supplementary Table S3). The most substantial support for an increased BD PRS was found with the threshold pPRS = 0.1 (OR = 2.97, one-sided p = 1.9 × 10−11). FAMBD cases also had significantly higher SCZ PRS than CCcontrols; the increase of the MDD PRS was nominally significant (Fig. 1b).
Shared PRS generated from the variants associated jointly with BD, SCZ, and MDD were significantly increased at pPRS ≥ 0.01 in FAMBD cases compared to CCcontrols. The GWIS BD-MDD PRS—the BD PRS corrected for associations shared with MDD—were significantly increased in FAMBD cases compared to CCcontrols. All other disorder-specific GWIS PRS were not significantly higher in FAMBD cases after correction for multiple testing.
No significant increase was found for the negative-control PRS for late-onset Alzheimer’s disease (LOAD) and associations of the PRS for BD and SCZ and of the Shared PRS were significantly stronger than simulated PRS in FAMBD compared to CCcontrols (Table 2).
Table 2.
The psychiatric disorder PRS can distinguish better between groups than simulated PRS
| Group | Disorder PRS | Simulated PRS min. pPRS | N simulated PRS with p ≤ p of disorder PRS | Prob. of success | 95% CI |
|---|---|---|---|---|---|
| FAMBD vs. CCcontrols | BD | 1 × 10−5 | 0 | <1 × 10−4 | 0–0 |
| SCZ | 1 × 10−5 | 0 | <1 × 10−4 | 0–0 | |
| MDD | 1 × 10−5 | 91 | 0.0091 | 0.007–0.011 | |
| Shared | 1 × 10−5 | 2 | 0.0002 | 0–0.001 | |
| FAMBD vs. CCBD | BD | 1 × 10−7 | 0 | <1 × 10−4 | 0–0 |
| SCZ | 1 × 10−7 | 2858 | 0.2858 | 0.277–0.295 | |
| MDD | 1 × 10−7 | 744 | 0.0744 | 0.069–0.080 | |
| Shared | 1 × 10−7 | 2229 | 0.2229 | 0.215–0.231 | |
| FAMunaffected vs. CCcontrols | BD | 1 × 10−7 | 0 | <1 × 10−4 | 0–0 |
| SCZ | 1 × 10−7 | 0 | <1 × 10−4 | 0–0 | |
| MDD | 1 × 10−7 | 37 | 0.0037 | 0.003–0.005 | |
| Shared | 1 × 10−7 | 74 | 0.0074 | 0.006–0.009 | |
| FAMMDD vs. CCcontrols | BD | 2 × 10−1 | 0 | <1 × 10−4 | 0–0 |
| SCZ | 2 × 10−1 | 409 | 0.0409 | 0.037–0.045 | |
| MDD | 2 × 10−1 | 2 | 0.0002 | 0–0.001 | |
| Shared | 2 × 10−1 | 125 | 0.0125 | 0.010–0.015 |
In binomial tests with 10,000 trials, the number of successes was the number of simulated PRS that showed the same or a stronger association than the disorder PRS (one-sided p-values). The 10,000 simulated PRS with ten p-value thresholds each were calculated by drawing random variants from across the genome, using the same number of variants as for the BD PRS at each threshold and random effect sizes from the pool of all available BD, SCZ, and MDD effects. For the present test, the pPRS of the simulated PRS showing the lowest mean association p-value was chosen. Prob. = binomial test probability estimate of success; CI = confidence interval of the probability estimate, both calculated using the R package binom (method: exact). Significance threshold: 0.05/16 = 0.003125, comparisons surpassing this threshold are shown in bold font
FAMBD cases had higher BD PRS than unrelated CCBD cases
The BD PRS was significantly higher in FAMBD than in CCBD cases at pPRS ≥ 0.05, but no other type of PRS was increased in FAMBD compared to CCBD cases (Fig. 1c, d). The association of the BD PRS was significantly stronger than simulated PRS (Table 2).
Unaffected family members showed higher psychiatric PRS than CC controls
In the comparison of FAMunaffected to CCcontrols, PRS for BD and SCZ were significantly higher in unaffected family members (Fig. 1e, f). The increases of the MDD, Shared, BD-MDD, and SCZ-BD GWIS PRS were nominally significant. The associations of BD and SCZ PRS were significantly stronger than the associations of simulated PRS (Table 2).
FAMBD cases had an increased PRS specifically for BD
In comparison to FAMunaffected, the BD PRS and the BD-MDD disorder-specific PRS were significantly higher in FAMBD (Fig. 2a, b). The Shared and the BD-SCZ PRS were increased at nominal significance.
Effects of assortative mating on BD PRS in family members
Eight of the 44 individuals who had married into the families had a diagnosis of MDD and none of BD (Table 1). While the unaffected married-in individuals had higher BD PRS than CCcontrols (p = 6.5 × 10−5), their BD PRS was not higher than the PRS of other FAMunaffected (Fig. 2c; Supplementary Fig. S6 and Supplementary Table S7). We also examined possible anticipation of BD in the families: neither did the BD PRS increase significantly over generations nor did the age at onset decrease over time (Fig. 2d; Supplementary Fig. S7 and Supplementary Table S8).
FAMMDD cases had higher psychiatric PRS than CCcontrols
In comparison to CCcontrols, FAMMDD cases had significantly higher BD and MDD PRS, increases of the Shared and SCZ PRS were nominally significant (Supplementary Figs. S8 and S9 and Supplementary Table S9). Both the BD and MDD PRS were increased at nominal significance when comparing FAMMDD to FAMunaffected (Supplementary Fig. S10 and Supplementary Table S10). Notably, in both comparisons, FAMMDD showed a nominal increase in SCZ-MDD PRS, but not in SCZ-BD PRS.
Discussion
Genome-wide association studies in large samples of unrelated patients and controls have unravelled the polygenic nature of BD, i.e., many common variants, each with a small effect size, contribute to BD. It has also been consistently shown that BD, MDD, and SCZ share many risk-conferring variants. The aim of the present study was to investigate whether common variants also contribute to BD in families with a high density of the disorder and if so, whether these variants are specific to BD.
We found that, compared to CCcontrols, unrelated subjects from the general population unscreened for BD, affected and unaffected ABiF family members had an elevated genetic risk for the tested psychiatric disorders, mainly for BD but also for SCZ. FAMBD cases were characterised by a particularly high load of BD-specific risk variants: The strongest association observed across all comparisons was the increase of the BD PRS in FAMBD compared to CCcontrols. In addition, the BD but not the SCZ and MDD PRS of FAMBD were significantly higher than the PRS of unrelated CCBD cases and unaffected family members. Together with the disorder-specific GWIS PRS, these results support the major contribution of BD-associated variants to the high density of the disorder in the investigated families.
An increased polygenic psychiatric risk has also been described in other studies of BD multiplex families [41–43]. However, the scope and results of these studies differed from the present study to some extent: Fullerton et al. [41] described an increased BD PRS in affected family members compared to unrelated controls and, when selecting families with a high polygenic BD risk load, also to unaffected family members. They constructed PRS only based on a small set of 32 SNPs from an older GWAS [10], and no other PRS were investigated. De Jong et al. [43] focused their analyses in a large Brazilian family with BD and MDD on assortative mating and anticipation and found BD and SCZ PRS to be increased at nominal significance in affected compared to unaffected members. In a large Swedish pedigree with mainly BD but also some SCZ cases, Szatkiewicz et al. [42] reported increased SCZ PRS in affected family members compared to family-level and population controls, as well as BD PRS increased at nominal significance in affected family members compared to family controls. However, no differences were observed between unaffected family members and population controls. Of note, none of these studies investigated differences in PRS between families and unrelated BD cases.
Compared to the CCBD in our study, FAMBD displayed, apart from an earlier age at onset, signs of a less severe clinical picture, i.e., less frequent impairment and less psychosis. This could be explained by the fact that CCBD cases were almost all BD-I patients recruited from consecutive admissions to a hospital, while most of the FAMBD cases were reached through other family members in the context of the study. Apart from this, the FAMBD did not display any striking differences in clinical features compared to the CCBD. Thus, we consider it likely that the increased PRS in the FAMBD is linked to the familial aggregation and not to clinical characteristics.
It appears striking that none of the ABiF family members have been diagnosed with SCZ. However, this can most likely be attributed to ascertainment bias as the recruitment strategy focused on BD multiplex families. With respect to this lack of SCZ diagnoses in the ABiF families, it is of interest that the family members showed not only an increased BD PRS but also increased SCZ and Shared PRS compared to unrelated controls. This increase could be an indirect consequence of the genetic correlation between BD and SCZ [14, 16, 18–21]. Furthermore, affected family members also had higher Shared PRS than CCcontrols. Of the psychiatric disorder GWAS data sets (i.e., SCZ, BD, and MDD) used in the present analysis, the SCZ GWAS both identified the largest amount of risk loci (108, 30, and 44, respectively) and the corresponding PRS explained the highest amount of case/control variance (7%, 4%, and 2% on a liability scale, respectively) [12–14]. Taking this and the genetic correlations between the disorders into account, the SCZ PRS might have included more cross-disorder signals with smaller effects than the PRS of BD and MDD. If family members had an increased Shared risk burden, this cross-disorder risk might have rendered them vulnerable to psychiatric disorders in general, with the high BD PRS then shaping the final BD diagnosis outcome. Of note, the analyses of FAMMDD cases are discussed in the Supplementary Data.
Our study furthermore indicates that assortative mating may have contributed to the increased BD PRS in the ABiF families: in their study, de Jong et al. [43] found no increased PRS in married-in subjects, but an increase of polygenic risk and a decrease in age at onset over generations. We observed that individuals who married into the ABiF families had higher BD PRS than CCcontrols, and their BD risk load was similar to other FAMunaffected. At the time of the interview, none of the married-in family members had a diagnosis of BD. Nevertheless, their increased BD PRS suggests that assortative mating may have occurred. Unaffected individuals with an above average BD PRS may display sub-threshold characteristics of BD, such as a broader range of emotions [44–46]. Consistent with the observation that married-in subjects did not have higher BD PRS than the other FAMunaffected, no increase in BD PRS was found across generations. However, assortative mating may have contributed to the establishment and maintenance of a high genetic risk load for BD in these families. Furthermore, assortative mating may have already occurred in previous generations, for which no DNA was available. Of note, DNA was not available for all ABiF family members of the current generations, limiting the scope of the analysis of assortative mating.
Although both the FAM and CC samples were recruited in Spain [9], minor population differences may have influenced the present results. Even if such minor differences existed, it is unlikely that they caused the highly significant associations observed for the psychiatric PRS, given that the pairwise genetic relationship matrix was used as a random effect in the association analyses. Additionally, results from three further analyses support our assumption that systematic differences between the genotype data of FAM, CCcontrols, and CCBD samples did not distort our findings: First, we did not find significant differences between the cohorts in a population substructure analysis (see Supplementary Fig. S11 and Supplementary Methods). Second, PRS for LOAD were not significantly increased in family members in any analysis. Since LOAD shows no genetic correlation with BD, MDD, or SCZ [14, 47, 48], this result further supports the specificity of our analyses. Third, when a psychiatric disorder PRS was significantly increased in family members, this association was stronger than for simulated PRS. While these findings cannot entirely exclude the influence of unknown confounders on our results, we consider them as strong evidence that the high psychiatric PRS observed in family members compared to controls cannot be attributed to population or technical differences between the cohorts.
The lower a pPRS threshold in the GWAS training data, the fewer SNPs were included in the calculation of the corresponding PRS. In most cases, significant differences between groups were not observed for these low pPRS but the higher thresholds based on thousands of variants. This is commonly observed and in line with the polygenic nature of psychiatric disorders as complex disorders, with genome-wide significant SNPs only accounting for a small share of the polygenic signal. The training GWAS used for BD, SCZ, and MDD, the largest available for these phenotypes, differ in the number of included subjects, their statistical power, and the number of identified signals. Therefore, the derived PRS also differ in the number of SNPs used in the calculation of each threshold (see Supplementary Table S2). However, even though the BD GWAS was based on the smallest number of subjects and contained the lowest number of genome-wide-associated loci among the three GWAS, the BD PRS showed the strongest associations with BD case status or family membership, underlining the substantial contribution of BD risk variants to the development of BD in the ABiF families.
One limitation of the study is that the subjects of the unrelated control cohort were not systematically screened for psychiatric disorders. The lifetime prevalence of unipolar depression in this cohort (up to 14.4% until the time of the interview) was in line with typically observed numbers [27], the prevalence of BD was not assessed. However, as BD has a lifetime prevalence of ~1%, we expect up to three BD cases among the 277 controls, a number we consider unlikely to have markedly influenced our results. Moreover, using controls unscreened for BD instead of “super-healthy” controls as a comparison to family members and unrelated BD cases represents a conservative approach and thereby strengthens the observed group differences in psychiatric PRS.
Similarly, around one third of the CCBD reported a family history of BD. The CCBD thus do not represent a sample of truly sporadic BD cases. However, the aim of our study was to investigate how members of multiplex BD families differ from typical BD cases regarding the polygenic contribution to their disorder. The observation that ABiF multiplex cases showed a higher polygenic psychiatric risk than CCBD, despite part of the CC cases also reporting a family history for BD, thus rather strengthens the validity of our findings.
The present study generated substantial evidence that members of the ABiF families, including unaffected subjects, carried a higher risk burden of common genetic risk variants than an unrelated control sample mainly for the psychiatric disorders BD and SCZ and, at least the FAMMDD cases, for MDD. In line with previous theoretical assumptions [49] and preliminary results from a pilot study in a single ABiF family [26], our results suggest that a high polygenic load of common risk variants is a major contributor to the increased risk for BD and MDD in families with a high density of BD. However, given the modest effect sizes of the PRS, they explained only a fraction of the phenotypic variance, and rare mutations such as copy number variants [50] or rare single-nucleotide variants likely also play an important role in each of the families. Sequencing studies carried out in multiplex families have suggested rare variants are involved in the aetiology of BD [51–53]. To date, however, it has proven difficult to identify replicable causal associations between rare variants and BD susceptibility. In a pilot study that analysed a single ABiF pedigree, we did not identify any rare causal variants for BD [26]. The analysis of rare variants in the remaining ABiF families using next-generation sequencing technologies is envisioned for the future, including integrative analyses in international consortia such as the Bipolar Sequencing Consortium [54]. Of note, the present analyses did not assess single families separately, but integrated PRS associations across all examined 33 ABiF families. Thus, the degree to which common and rare variants shaped the emergence of psychiatric disorders may vary between families.
Furthermore, PRS are commonly based on and applied to sets of unrelated individuals, and polygenic risk might act differently in the case of familial genetic background. Moreover, a broad range of environmental factors have been shown to influence the risk of psychiatric disorders and might act on top of the increased genetic risk in these families. However, environmental factors have not been systematically assessed in the present study. To further enhance our understanding regarding the aetiology of BD, integrated analyses of common and rare variants, as well as of environmental risk in the ABiF families are warranted in the future.
Supplementary information
Acknowledgements
The study was supported by the German Federal Ministry of Education and Research (BMBF), through the Integrated Network IntegraMent, under the auspices of the e:Med programme (grants 01ZX1314A to MMN and SC; 01ZX1314G to MR; 01ZX1614J to BMM) through grants 01EE1406C to MR and 01EE1409C to MR and SHW, and through ERA-NET NEURON, “SynSchiz—Linking synaptic dysfunction to disease mechanisms in schizophrenia—a multilevel investigation” (01EW1810 to MR) and BMBF grants 01EE1409C and 01EE1406C to MR and SHW; by the German Research Foundation (DFG grants FOR2107; RI908/11-2 to MR; NO246/10-2 to MMN; MU1315/8-2 to BMM; WI 3439/3-2 to SHW), by the Andalusian regional Health and Innovation Government (grants PI-0060-2017, RC-0006-2015 the Nicolas Monarde Programme for YDO and CTS-546) and by the Swiss National Science Foundation (SNSF grant 156791 to SC). MMN is a member of the DFG-funded cluster of excellence ImmunoSensation. The PGC has received major funding from the US National Institute of Mental Health and the US National Institute of Drug Abuse (U01 MH109528 and U01 MH1095320). We thank the research participants and employees of 23andMe, Inc. for their contribution to the MDD meta-analysis published in [14]. We thank the International Genomics of Alzheimer's Project (IGAP) for providing summary results data for the present analyses. See the Supplementary Data for extended Acknowledgements.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Joint last authors: Andreas J. Forstner, Markus M. Nöthen, Marcella Rietschel
Joint first authors: Till F. M. Andlauer, Jose Guzman-Parra, Fabian Streit
Members of Bipolar and Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium are listed in the Supplementary Material
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marcella Rietschel, Email: Marcella.Rietschel@zi-mannheim.de.
Bipolar Disorder Working Group of the Psychiatric Genomics Consortium:
Eli A Stahl, Gerome Breen, Andreas J Forstner, Andrew McQuillin, Stephan Ripke, Vassily Trubetskoy, Manuel Mattheisen, Yunpeng Wang, Jonathan R I Coleman, Héléna A Gaspar, Christiaan A de Leeuw, Stacy Steinberg, Jennifer M Whitehead Pavlides, Maciej Trzaskowski, Tune H Pers, Peter A Holmans, Liam Abbott, Esben Agerbo, Huda Akil, Diego Albani, Ney Alliey-Rodriguez, Thomas D Als, Adebayo Anjorin, Verneri Antilla, Swapnil Awasthi, Judith A Badner, Marie Bækvad-Hansen, Jack D Barchas, Nicholas Bass, Michael Bauer, Richard Belliveau, Sarah E Bergen, Carsten Bøcker Pedersen, Erlend Bøen, Marco Boks, James Boocock, Monika Budde, William Bunney, Margit Burmeister, Jonas Bybjerg-Grauholm, William Byerley, Miquel Casas, Felecia Cerrato, Pablo Cervantes, Kimberly Chambert, Alexander W Charney, Danfeng Chen, Claire Churchhouse, Toni-Kim Clarke, William Coryell, David W Craig, Cristiana Cruceanu, Piotr M Czerski, Anders M Dale, Simone de Jong, Franziska Degenhardt, Jurgen Del-Favero, J Raymond DePaulo, Srdjan Djurovic, Amanda L Dobbyn, Ashley Dumont, Torbjørn Elvsåshagen, Valentina Escott-Price, Chun Chieh Fan, Sascha B Fischer, Matthew Flickinger, Tatiana M Foroud, Liz Forty, Josef Frank, Christine Fraser, Nelson B Freimer, Louise Frisén, Katrin Gade, Diane Gage, Julie Garnham, Claudia Giambartolomei, Marianne Giørtz Pedersen, Jaqueline Goldstein, Scott D Gordon, Katherine Gordon-Smith, Elaine K Green, Melissa J Green, Tiffany A Greenwood, Jakob Grove, Weihua Guan, José Guzman Parra, Marian L Hamshere, Martin Hautzinger, Urs Heilbronner, Stefan Herms, Maria Hipolito, Per Hoffmann, Dominic Holland, Laura Huckins, Stéphane Jamain, Jessica S Johnson, Anders Juréus, Radhika Kandaswamy, Robert Karlsson, James L Kennedy, Sarah Kittel-Schneider, James A Knowles, Manolis Kogevinas, Anna C Koller, Ralph Kupka, Catharina Lavebratt, Jacob Lawrence, William B Lawson, Markus Leber, Phil H Lee, Shawn E Levy, Jun Z Li, Chunyu Liu, Susanne Lucae, Anna Maaser, Donald J MacIntyre, Pamela B Mahon, Wolfgang Maier, Lina Martinsson, Steve McCarroll, Peter McGuffin, Melvin G McInnis, James D McKay, Helena Medeiros, Sarah E Medland, Fan Meng, Lili Milani, Grant W Montgomery, Derek W Morris, Thomas W Mühleisen, Niamh Mullins, Hoang Nguyen, Caroline M Nievergelt, Annelie Nordin Adolfsson, Evaristus A Nwulia, Claire O’Donovan, Loes M Olde Loohuis, Anil P S Ori, Lilijana Oruc, Urban Ösby, Roy H Perlis, Amy Perry, Andrea Pfennig, James B Potash, Shaun M Purcell, Eline J Regeer, Andreas Reif, Céline S Reinbold, John P Rice, Alexander L Richards, Fabio Rivas, Margarita Rivera, Panos Roussos, Douglas M Ruderfer, Euijung Ryu, Cristina Sánchez-Mora, Alan F Schatzberg, William A Scheftner, Nicholas J Schork, Cynthia Shannon Weickert, Tatyana Shehktman, Paul D Shilling, Engilbert Sigurdsson, Claire Slaney, Olav B Smeland, Janet L Sobell, Christine Søholm Hansen, Anne T Spijker, David St Clair, Michael Steffens, John S Strauss, Fabian Streit, Jana Strohmaier, Szabolcs Szelinger, Robert C Thompson, Thorgeir EThorgeirsson, Jens Treutlein, Helmut Vedde, Weiqing Wang, Stanley J Watson, Thomas W Weickert, Stephanie H Witt, Simon Xi, Wei Xu, Allan H Young, Peter Zandi, Peng Zhang, Sebastian Zollner, Rolf Adolfsson, Ingrid Agartz, Martin Alda, Lena Backlund, Bernhard T Baune, Frank Bellivier, Wade H Berrettini, Joanna M Biernacka, Douglas H R Blackwood, Michael Boehnke, Anders D Børglum, Aiden Corvin, Nicholas Craddock, Mark J Daly, Udo Dannlowski, Tõnu Esko, Bruno Etain, Mark Frye, Janice M Fullerton, Elliot S Gershon, Michael Gill, Fernando Goes, Maria Grigoroiu-Serbanescu, Joanna Hauser, David M Hougaard, Christina M Hultman, Ian Jones, Lisa A Jones, René S Kahn, George Kirov, Mikael Landén, Marion Leboyer, Cathryn M Lewis, Qingqin S Li, Jolanta Lissowska, Nicholas G Martin, Fermin Mayoral, Susan L McElroy, Andrew M McIntosh, Francis J McMahon, Ingrid Melle, Andres Metspalu, Philip B Mitchell, Gunnar Morken, Ole Mors, Preben Bo Mortensen, Bertram Müller-Myhsok, Richard M Myers, Benjamin M Neale, Vishwajit Nimgaonkar, Merete Nordentoft, Markus M Nöthen, Michael C O’Donovan, Ketil J Oedegaard, Michael J Owen, Sara A Paciga, Carlos Pato, Michele T Pato, Danielle Posthuma, Josep Antoni Ramos-Quiroga, Marta Ribasés, Marcella Rietschel, Guy A Rouleau, Martin Schalling, Peter R Schofield, Thomas G Schulze, Alessandro Serretti, Jordan W Smoller, Hreinn Stefansson, Kari Stefansson, Eystein Stordal, Patrick F Sullivan, Gustavo Turecki, Arne E Vaaler, Eduard Vieta, John B Vincent, Thomas Werge, John I Nurnberger, Naomi R Wray, Arianna Di Florio, Howard J Edenberg, Sven Cichon, Roel A Ophoff, Laura J Scott, Ole A Andreassen, John Kelsoe, and Pamela Sklar
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium:
Naomi R Wray, Stephan Ripke, Manuel Mattheisen, Maciej Trzaskowski, Enda M Byrne, Abdel Abdellaoui, Mark J Adams, Esben Agerbo, Tracy M Air, Till F M Andlauer, Silviu-Alin Bacanu, Marie Bækvad-Hansen, Aartjan T F Beekman, Tim B Bigdeli, Elisabeth B Binder, Julien Bryois, Henriette N Buttenschøn, Jonas Bybjerg-Grauholm, Na Cai, Enrique Castelao, Jane Hvarregaard Christensen, Toni-Kim Clarke, Jonathan R I Coleman, Lucía Colodro-Conde, Baptiste Couvy-Duchesne, Nick Craddock, Gregory E Crawford, Gail Davies, Ian J Deary, Franziska Degenhardt, Eske M Derks, Nese Direk, Conor V Dolan, Erin C Dunn, Thalia C Eley, Valentina Escott-Price, Farnush Farhadi Hassan Kiadeh, Hilary K Finucane, Jerome C Foo, Andreas J Forstner, Josef Frank, Héléna A Gaspar, Michael Gill, Fernando S Goes, Scott D Gordon, Jakob Grove, Lynsey S Hall, Christine Søholm Hansen, Thomas F Hansen, Stefan Herms, Ian B Hickie, Per Hoffmann, Georg Homuth, Carsten Horn, Jouke-Jan Hottenga, David M Hougaard, David M Howard, Marcus Ising, Rick Jansen, Ian Jones, Lisa A Jones, Eric Jorgenson, James A Knowles, Isaac S Kohane, Julia Kraft, Warren W. Kretzschmar, Zoltán Kutalik, Yihan Li, Penelope A Lind, Donald J MacIntyre, Dean F MacKinnon, Robert M Maier, Wolfgang Maier, Jonathan Marchini, Hamdi Mbarek, Patrick McGrath, Peter McGuffin, Sarah E Medland, Divya Mehta, Christel M Middeldorp, Evelin Mihailov, Yuri Milaneschi, Lili Milani, Francis M Mondimore, Grant W Montgomery, Sara Mostafavi, Niamh Mullins, Matthias Nauck, Bernard Ng, Michel G Nivard, Dale R Nyholt, Paul F O’Reilly, Hogni Oskarsson, Michael J Owen, Jodie N Painter, Carsten Bøcker Pedersen, Marianne Giørtz Pedersen, Roseann E Peterson, Erik Pettersson, Wouter J Peyrot, Giorgio Pistis, Danielle Posthuma, Jorge A Quiroz, Per Qvist, John P Rice, Brien P. Riley, Margarita Rivera, Saira Saeed Mirza, Robert Schoevers, Eva C Schulte, Ling Shen, Jianxin Shi, Stanley I Shyn, Engilbert Sigurdsson, Grant C B Sinnamon, Johannes H Smit, Daniel J Smith, Hreinn Stefansson, Stacy Steinberg, Fabian Streit, Jana Strohmaier, Katherine E Tansey, Henning Teismann, Alexander Teumer, Wesley Thompson, Pippa A Thomson, Thorgeir E Thorgeirsson, Matthew Traylor, Jens Treutlein, Vassily Trubetskoy, André G Uitterlinden, Daniel Umbricht, Sandra Van der Auwera, Albert M van Hemert, Alexander Viktorin, Peter M Visscher, Yunpeng Wang, Bradley T. Webb, Shantel Marie Weinsheimer, Jürgen Wellmann, Gonneke Willemsen, Stephanie H Witt, Yang Wu, Hualin S Xi, Jian Yang, Futao Zhang, Volker Arolt, Bernhard T Baune, Klaus Berger, Dorret I Boomsma, Sven Cichon, Udo Dannlowski, E J C de Geus, J Raymond DePaulo, Enrico Domenici, Katharina Domschke, Tõnu Esko, Hans J Grabe, Steven P Hamilton, Caroline Hayward, Andrew C Heath, Kenneth S Kendler, Stefan Kloiber, Glyn Lewis, Qingqin S Li, Susanne Lucae, Pamela A F Madden, Patrik K Magnusson, Nicholas G Martin, Andrew M McIntosh, Andres Metspalu, Ole Mors, Preben Bo Mortensen, Bertram Müller-Myhsok, Merete Nordentoft, Markus M Nöthen, Michael C O’Donovan, Sara A Paciga, Nancy L Pedersen, Brenda W J H Penninx, Roy H Perlis, David J Porteous, James B Potash, Martin Preisig, Marcella Rietschel, Catherine Schaefer, Thomas G Schulze, Jordan W Smoller, Kari Stefansson, Henning Tiemeier, Rudolf Uher, Henry Völzke, Myrna M Weissman, Thomas Werge, Cathryn M Lewis, Douglas F Levinson, Gerome Breen, Anders D Børglum, and Patrick F Sullivan
Supplementary information
The online version of this article (10.1038/s41380-019-0558-2) contains supplementary material, which is available to authorised users.
References
- 1.Merikangas KR, Jin R, He J-P, Kessler RC, Lee S, Sampson NA, et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch Gen Psychiatry. 2011;68:241–51. doi: 10.1001/archgenpsychiatry.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381:1654–62. doi: 10.1016/S0140-6736(13)60855-7. [DOI] [PubMed] [Google Scholar]
- 3.Edvardsen J, Torgersen S, Røysamb E, Lygren S, Skre I, Onstad S, et al. Heritability of bipolar spectrum disorders. Unity or heterogeneity? J Affect Disord. 2008;106:229–40. doi: 10.1016/j.jad.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 4.McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 5.Charney AW, Ruderfer DM, Stahl EA, Moran JL, Chambert K, Belliveau RA, et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiatry. 2017;7:e993. doi: 10.1038/tp.2016.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landén M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–94. doi: 10.1093/hmg/ddw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2017;511:421. doi: 10.1038/mp.2016.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MTM, Chen C-H, Lee CS, Chen CC, Chong MY, Ouyang WC, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry. 2011;16:548–56. doi: 10.1038/mp.2010.43. [DOI] [PubMed] [Google Scholar]
- 9.Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:3339. doi: 10.1038/ncomms4339. [DOI] [PubMed] [Google Scholar]
- 10.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–81. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. [DOI] [PMC free article] [PubMed]
- 14.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J, Bergen SE, Kuja-Halkola R, Larsson H, Landén M, Lichtenstein P. Bipolar disorder and its relation to major psychiatric disorders: a family-based study in the Swedish population. Bipolar Disord. 2015;17:184–93. doi: 10.1111/bdi.12242. [DOI] [PubMed] [Google Scholar]
- 16.Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Schizophrenia Working Group of Psychiatric Genomics Consortium et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014;19:1017–24. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forstner AJ, Hecker J, Hofmann A, Maaser A, Reinbold CS, Mühleisen TW, et al. Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS ONE. 2017;12:e0171595. doi: 10.1371/journal.pone.0171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address: douglas.ruderfer@vanderbilt.edu, Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173:1705–.e16. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, Zhu Z, et al. Genome wide meta-analysis identifies genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. bioRxiv. 2019:528117. [DOI] [PMC free article] [PubMed]
- 22.McClellan J, King M-C. Genomic analysis of mental illness: a changing landscape. JAMA. 2010;303:2523–4. doi: 10.1001/jama.2010.869. [DOI] [PubMed] [Google Scholar]
- 23.Kerner B. Toward a deeper understanding of the genetics of bipolar disorder. Front Psychiatry. 2015;6:105. doi: 10.3389/fpsyt.2015.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzman-Parra J, Rivas F, Strohmaier J, Forstner A, Streit F, Auburger G, et al. The Andalusian Bipolar Family (ABiF) Study: Protocol and sample description. Rev Psiquiatr Salud Ment. 2017. 10.1016/j.rpsm.2017.03.004. [DOI] [PubMed]
- 25.Schumacher J, Kaneva R, Jamra RA, Díaz GO, Ohlraun S, Milanova V, et al. Genomewide scan and fine-mapping linkage studies in four European samples with bipolar affective disorder suggest a new susceptibility locus on chromosome 1p35-p36 and provides further evidence of loci on chromosome 4q31 and 6q24. Am J Hum Genet. 2005;77:1102–11. doi: 10.1086/498619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins AL, Kim Y, Szatkiewicz JP, Bloom RJ, Hilliard CE, Quackenbush CR, et al. Identifying bipolar disorder susceptibility loci in a densely affected pedigree. Mol Psychiatry. 2013;18:1245–6. doi: 10.1038/mp.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler RC, Bromet EJ. The epidemiology of depression across cultures. Annu Rev Public Health. 2013;34:119–38. doi: 10.1146/annurev-publhealth-031912-114409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 32.R Core Team. R: A Language and Environment for Statistical Computing. R Core Team: Vienna, Austria; 2017.
- 33.Nieuwboer HA, Pool R, Dolan CV, Boomsma DI, Nivard MG. GWIS: genome-wide inferred statistics for functions of multiple phenotypes. Am J Hum Genet. 2016;99:917–27. doi: 10.1016/j.ajhg.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–66. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11:407–9. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belonogova NM, Svishcheva GR, van Duijn CM, Aulchenko YS, Axenovich TI. Region-based association analysis of human quantitative traits in related individuals. PLoS ONE. 2013;8:e65395. doi: 10.1371/journal.pone.0065395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canty A, Ripley BD. Boot: Bootstrap R (S-Plus) functions. comprehensive R archive network (CRAN), 2017. https://cran.r-project.org/web/packages/boot/.
- 40.Davison AC, Hinkley DV. Bootstrap methods and their application. Cambridge University Press, Cambridge, 1997.
- 41.Fullerton JM, Koller DL, Edenberg HJ, Foroud T, Liu H, Glowinski AL, et al. Assessment of first and second degree relatives of individuals with bipolar disorder shows increased genetic risk scores in both affected relatives and young At-Risk Individuals. Am J Med Genet B Neuropsychiatr Genet. 2015;168:617–29. doi: 10.1002/ajmg.b.32344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szatkiewicz J, Crowley JJ, Adolfsson AN, Åberg KA, Alaerts M, Genovese G, et al. The genomics of major psychiatric disorders in a large pedigree from Northern Sweden. Transl Psychiatry. 2019;9:60. doi: 10.1038/s41398-019-0414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Jong S, Diniz MJA, Saloma A, Gadelha A, Santoro ML, Ota VK, et al. Applying polygenic risk scoring for psychiatric disorders to a large family with bipolar disorder and major depressive disorder. Commun Biol. 2018;1:163. doi: 10.1038/s42003-018-0155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peyrot WJ, Robinson MR, Penninx BW, Wray NR. Exploring boundaries for the genetic consequences of assortative mating for psychiatric traits. JAMA Psychiatry. 2016;73:1189–95. doi: 10.1001/jamapsychiatry.2016.2566. [DOI] [PubMed] [Google Scholar]
- 45.Nordsletten AE, Larsson H, Crowley JJ, Almqvist C, Lichtenstein P, Mataix-Cols D. Patterns of nonrandom mating within and across 11 major psychiatric disorders. JAMA Psychiatry. 2016;73:354–61. doi: 10.1001/jamapsychiatry.2015.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18:953–5. doi: 10.1038/nn.4040. [DOI] [PubMed] [Google Scholar]
- 47.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brainstorm Consortium. Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, Visscher PM, Wray NR. Sporadic cases are the norm for complex disease. Eur J Hum Genet. 2010;18:1039–43. doi: 10.1038/ejhg.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green EK, Rees E, Walters JTR, Smith K-G, Forty L, Grozeva D, et al. Copy number variation in bipolar disorder. Mol Psychiatry. 2016;21:89–93. doi: 10.1038/mp.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goes FS, Pirooznia M, Parla JS, Kramer M, Ghiban E, Mavruk S, et al. Exome Sequencing of familial bipolar disorder. JAMA Psychiatry. 2016;73:590–7. doi: 10.1001/jamapsychiatry.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruceanu C, Schmouth J-F, Torres-Platas SG, Lopez JP, Ambalavanan A, Darcq E, et al. Rare susceptibility variants for bipolar disorder suggest a role for G protein-coupled receptors. Mol Psychiatry. 2017;3:577. doi: 10.1038/mp.2017.223. [DOI] [PubMed] [Google Scholar]
- 53.Ament SA, Szelinger S, Glusman G, Ashworth J, Hou L, Akula N, et al. Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci USA. 2015;112:3576–81. doi: 10.1073/pnas.1424958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shinozaki G, Potash JB. New developments in the genetics of bipolar disorder. Curr Psychiatry Rep. 2014;16:493. doi: 10.1007/s11920-014-0493-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.