Abstract
Glioblastoma is the most malignant tumor occurring in the human central nervous system with overall median survival time <14.6 months. Current treatments such as chemotherapy and radiotherapy cannot reach an optimal remission since tumor resistance to therapy remains a challenge. Glioblastoma stem cells are considered to be responsible for tumor resistance in treating glioblastoma. Previous studies reported two subtypes, proneural and mesenchymal, of glioblastoma stem cells manifesting different sensitivity to radiotherapy or chemotherapy. Mesenchymal glioblastoma stem cells, as well as tumor cells generate from which, showed resistance to radiochemotherapies. Besides, two metabolic patterns, glutamine or glucose dependent, of mesenchymal glioblastoma stem cells also manifested different sensitivity to radiochemotherapies. Glutamine dependent mesenchymal glioblastoma stem cells are more sensitive to radiotherapy than glucose-dependent ones. Therefore, the transition between proneural and mesenchymal subtypes, or between glutamine-dependent and glucose-dependent, might lead to tumor resistance to radiochemotherapies. Moreover, neural stem cells were also hypothesized to participate in glioblastoma stem cells mediated tumor resistance to radiochemotherapies. In this review, we summarized the basic characteristics, adaptive transition and implications of glioblastoma stem cells in glioblastoma therapy.
Subject terms: CNS cancer, Cancer stem cells
Background
Glioblastoma (GBM), classified as grade IV glioma, is a highly aggressive and heterogeneous tumor in the central nervous system. Standard treatments of GBM include maximal surgical resection and following radiochemotherapies, which is also known as the STUPP protocol.1 Nevertheless, the average overall survival time is still <14.6 months for newly diagnosed GBM patients and 6.9 months for recurrence GBM patients.2 Novel treatments such as anti-angiogenic therapy,3 immunotherapy4,5 and tumor-treating electric fields6 were proposed recently but their efficacies were still unsatisfied. GBM is categorized into four subtypes based on their molecular characteristics: proneural, neural, mesenchymal and classical gliomas,7–9 and patients’ prognoses varied among those subtypes. In primary GBM, the mesenchymal GBM is the most aggressive type while the proneural GBM is associated with a relatively better overall survival compared to other subtypes. Moreover, mostly primary GBM experiences the subtype switch at relapse, in which mesenchymal GBM is the most stable subtype.10 Therefore, this GBM subtype switch is reckoned as an adaptive transition considered as an underlying mechanism of tumor resistance to radiochemotherapies.11–13
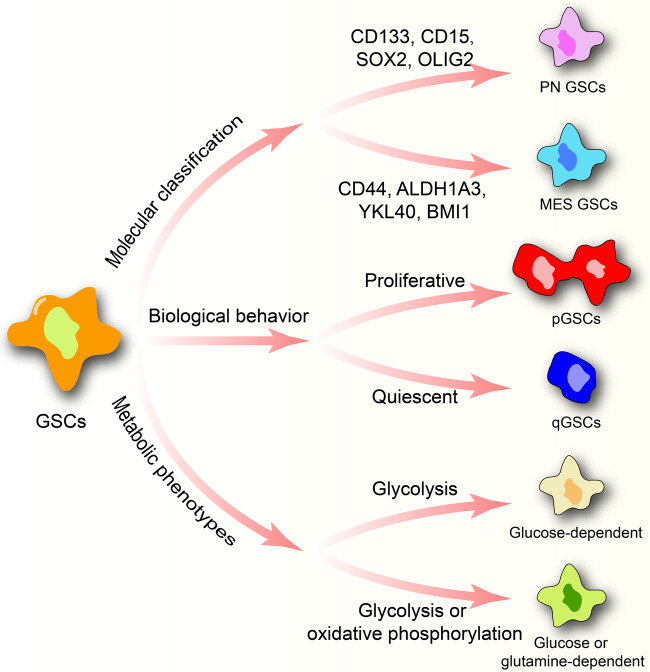
Cancer stem cells were first isolated from acute myeloid leukemia by Bonnet and Dick in 1997,14 which is defined as a cluster of undifferentiated cells with the ability of self-renewal and tumor initiation. Glioblastoma stem cells (GSCs) were isolated from GBM with the ability to develop GBM in the transplanted mouse.15,16 Several biomarkers were identified to distinguish GSCs from non-tumorigenic stromal cells including CD56+, SOX2+, SOX9+, CD133+, CD15+, CD248−, CD105−, αSMA− .17 Besides, GSCs were classified into different groups based on gene signatures,18–24 metabolic patterns25,26 and biological behaviors25,27 (Fig. 1). Those classifications were not isolated but were also closely connected. For example, GSCs are classified as proneural GSCs (PN GSCs) or mesenchymal GSCs (MES GSCs) based on gene signatures; in metabolic patterns, MES GSCs switches between glycolysis and oxidative phosphorylation (OXPHOS) whereas PN GSCs mainly dependent on glycolysis; PN GSCs and MES GSCs were characterized with potent proliferative and invasive abilities, respectively. The biological behavior classification, defining proliferative GSCs (pGSCs) and quiescent GSCs (qGSCs), is associate with neural stem cells (NSCs) but have no relationship with the molecular or metabolic classification.
Fig. 1.
GSCs classification based on molecular signatures, metabolic phenotypes and biological behavior. The molecular classification includes PN GSCs and MES GSCs. Two metabolic phenotypes of GSCs are identified. The former mostly dependent on glycolysis. The latter metabolizes glutamine to supply OXPHOS but it can switch to glycolysis when glutamine is insufficient. According to biological behavior, GSCs can be grouped into proliferative GSCs and quiescent GSCs
Moreover, those classifications are also linked with tumor resistance to radiotherapy or chemotherapy. For instance, MES GSCs and qGSCs manifest relatively higher resistance to radiotherapy compared to PN GSCs and pGSCs, respectively.28,29 Glutamine-dependent MES GSCs show resistance to radiochemotherapies.26,30,31 Besides, PN GSCs can be induced to transform into MES GSCs by treating with temozolomide.32 Similar phenomenon was also noticed between pGSCs and qGSCs.33 Given that, we summarize the characteristics of each classification of GSCs, explore their internal relationships and investigate their association with tumor resistance to radiochemotherapies. Consequently, the phenomenon that therapy sensitive GSCs switch to therapy-resistant GSCs is summarized with the term, ‘the adaptive transition of GSCs’. The role of NSCs and niche acts during GSCs adaptive transition are also discussed. Finally, the implication of GSCs adaptive transition to clinical treatment is investigated to provide novel potential strategies for future GBM treatment.
GSCs isolation
Currently, there are various methods to isolate GSCs including sphere-formation assay, side population assay, label-retention assay and flow cytometry.34,35 To verify the ability of self-renewal and tumor initiation, sphere-formation assay and GSCs allogeneic transplantation are required after isolation.36 Therefore, a regular protocol consists of isolation and verification. In order to obtain sufficient GSCs for verification, sphere-formation assay and flow cytometry assay are considered as qualified methods.37 In addition to those classical culture systems, 3D organoids system for GSCs was also proposed in recent years.38–40 Stem-like cells were also noticed during culturing GBM cells as 3D organoids in vitro. Moreover, high proliferative SOX2 positive GSCs enriched at the periphery of the organoids; while rare SOX2 positive GSCs were noticed at its hypoxic core. GSCs at the core of organoids exhibited worse proliferative ability by comparing with GSCs at the surface.39 Compared with the traditional sphere-forming assay, 3D organoids system highlighted the interaction between tumor cells and extracellular matrix (ECM) components.
In addition, proper culture medium selection is also critical to stem cells enrichment and the expression of surface markers on GSCs. For instance, serum-free medium can assist in isolating GSCs from tumor tissue and maintaining GSCs stemness.41,42 Cytokines like FGF has been proved with the ability of affecting surface marker of stem cells. Adding FGF into cell culturing medium affects the expression of Nestin and CD133, which are star biomarker of stem cells, and maintain the characteristics of GSCs.43,44 The Wnt signaling pathway can cross talk with FGF to influence cell surface marker expression, including CD133, CD44.45 Considering cell surface marker is critical to GSCs isolation and identification, we listed common hallmarks of GSCs along with their association with PN or MES GSCs. (Table 1).
Table 1.
Common hallmarks in GSCs isolation
| Hallmarks | Functions | Subtypes | Reference |
|---|---|---|---|
| CD133 | Cell cycle and tumor cell proliferation. | PN GSCs | 46 |
| CD15 | Cell proliferation, self-renewal, and multilineage differentiation. | PN GSCs | 51 |
| ITGA6 | Cell proliferation and adhesion. | PN GSCs | 56,57 |
| A2B5 | Cell proliferation, migration, clonogenicity, and tumorigenesis. | PN GSCs | 53 |
| CD44 | Cell invasion | MES GSCs | 19,55 |
| ALDH1 | Tumorigenesis, PMT transition, resistance to temozolomide, cell invasion, cell proliferation, glycolysis | MES GSCs | 58 |
| Nestin | A class VI intermediate filament protein | Not suitable | 105 |
| CD36 | A scavenger receptor, GSCs self-renewal and proliferation. | Not suitable | 60 |
| CD9 | Cell proliferation. | Not suitable | 61–63 |
| IL6R | Tumor growth | Not suitable | 64,65 |
| CXCR4 | Cell proliferation, self-renewal. | Not suitable | 66,67 |
CD133, also called prominin-1, is the most common hallmark be applied to GSCs isolation.46 CD133 is considered as a hallmark of PN GSCs while CD133 negative GSCs are considered as MES GSCs.47 A previous study reported that CD133 is associated with tumor angiogenesis, cell proliferation while CD133 negative GSCs lack the ability of self-renewal and forming sphere in vitro.48 Nevertheless, CD133 negative GSCs can form tumor in vivo and CD133 positive GSCs can be isolated from it.47 Therefore, isolation by targeting CD133 can obtain GSCs but may not be able to pure MES GSCs.
Similar to CD133, CD15 (known as SSEA-1 or Lex) can be applied as a target in GSCs isolation. The expression of CD133 in CD15 positive GSCs is deceased during passage in vitro while the expression of CD15 remains stable. Notably, CD15 positive CD133 negative cells still maintain the characteristic of GSCs in vitro.49 However, no significant difference of phenotypic and genomic characteristics is observed between CD15 positive GSCs and CD15 negative GSCs, which both can develop a CD15 positive/negative mixed tumor in vivo.50 Therefore, some researches adopted CD15 and CD133 as isolation hallmarks simultaneously to reduce omission.51,52
A2B5 is recognized as an isolation hallmark of GSCs.53 The ability of tumor initiation of A2B5 positive CD133 negative GSCs is stronger than A2B5 negative CD133 negative GSCs.54 A study reported that A2B5 negative GSCs failed to form sphere in vitro or tumor in vivo.53 Therefore, A2B5 might be another compensate marker of CD133 to avoid GSCs isolation omission.
Several biomarkers are enriched in GSCs but rare studies adopted them for GSCs isolation. For instance, CD44 is a biomarker of MES GSCs.19,21 CD44 positive GSCs manifest stronger invasive ability but worse proliferative ability compared to CD133 positive GSCs.55 Integrin α6 co-expresses with CD133, and is associated with the ability of GSCs self-renewal and tumor initiation development.56,57 ALDH1 positive GSCs maintain the characteristics of GSCs including asymmetric division and sphere-formation in vitro.58 Nestin is expressed in both NSCs and CD133 positive GSCs.59 Other biomarkers like CD36,60 CD9,61–63 IL6R64,65 and CXCR466,67 are essential to the ability of sphere-formation of CD133 positive GSCs. But there is no enough evidences to support them as a qualified hallmark in GSCs isolation.
Proneural and mesenchymal GSCs
Basic characteristic of PN and MES GSCs
Based on gene signatures, GSCs can be categorized as MES or PN GSCs. MES GSCs are labeled with CD44, ALDH1A3, EGFR, YKL40, IDH1-wildtype, BMI1, and GFAP whereas PN GSCs are marked with CD133, CD15, DLL3, MAP2, SOX2, OLIG2, IDH1-mutant, and EZH2.11,19–24 The difference in splicing profiles between PN GSCs and MES GSCs affected cell cycle, DNA repair, splicing and cilium formation.68 The expression profile of long non-coding RNA between PN GSCs and MES GSCs was also analyzed and prognostic related long non-coding RNAs were identified.68 PN GSCs prefer peri-vascular niche while MES GSCs are mainly located at the necrotic niche.69 PN GSCs manifest high growth rates and are able to promote tumor angiogenesis.19 MES GSCs show strong invasive abilities, and tumor derived from which exhibit an aggressive growth pattern.28,70 However, it is more difficult for MES GSCs to generate tumor than PN GSCs.71,72 MES GSCs have a higher resistance to radiotherapy relative to PN GSCs, and PN GSCs can be induced to transform into MES GSCs.28,32 The comparison between PN and MES GSCs was summarized in Table 2 and introduced with Fig. 2.
Table 2.
The basic characteristics of PN GSCs and MES GSCs
| Difference | PN GSCs | MES GSCs |
|---|---|---|
| Hallmarks | CD133, CD15, MAP2, SOX2, OLIG2, IDH1-mutant and EZH2 | CD44, ALDH1A3, EGFR, YKL40, IDH1-wildtype, BMI1 and GFAP |
| Signaling pathways | The PDGF receptor-β mediated pathway; The Notch pathway; The Wnt pathway | The NF-κB pathway; FOXD-ALDH1A3 axis; glycolysis-mediated metabolism pathway |
| Niche | Peri-vascular niche or tumor edge tissue | Necrotic tissue |
| Lipid metabolism | Low | High |
| Glutamine utilization | Low | High |
| Metabolism preference | Glycolysis | Glycolysis or OXPHOS |
| Immunocytes infiltration | Natural killer cells | M2 macrophage; CD8+ cells and microglial |
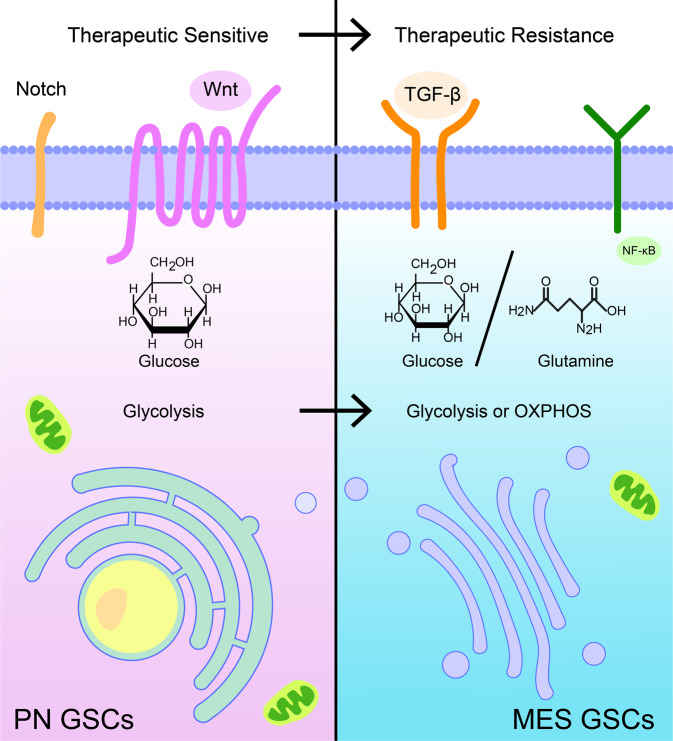
Fig. 2.
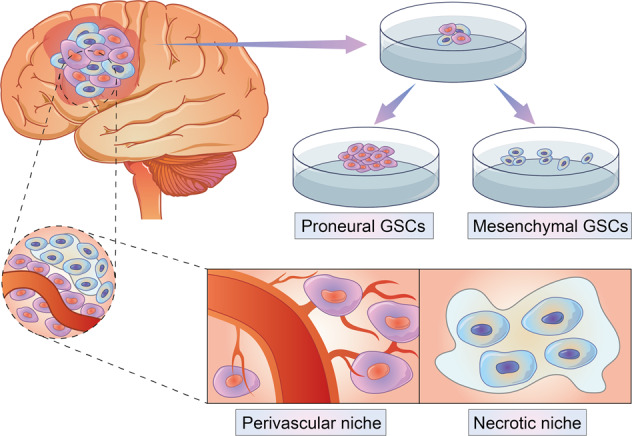
Difference between PN GSCs and MES GSCs. They manifest different growth pattern of sphere culture in vitro. PN GSCs tend to form bigger sphere and show higher growth rate than MES GSCs in vitro. In vivo, PN GSCs locate at perivascular niche while MES GSCs prefer necrotic tissue
The proneural-mesenchymal transition
Primary GBM tends to switch its subtype from proneural to mesenchymal at relapse and show resistance to radiochemotherapies.11,40 Similarly, PN GSCs can be transformed into MES GSCs during radiochemotherapies.12,28,29,73 The GSEA analysis was performed on differential expressed genes between MES GSCs and PN GSCs, and the result also supported that MES GSCs manifest higher resistance to tumor therapy relative to PN GSCs. Therefore, MES GSCs are able to survival from radiochemotherapies compared to PN SGCs and form therapy-resistance tumor eventually.13 Another study also reported, by treating GSCs with radio- or chemo-therapy, proneural related signatures (like CD133, OLIG2) in GSCs were decreased and mesenchymal-like gene signatures (like CD44, YKL40) were upregulated.73 Other than proneural or mesenchymal related signatures, several molecules also supported that the proneural-mesenchymal transition (PMT) in GSCs is associated with tumor resistance to tumor therapy. For instance, ZDHHC18 and ZDHHC23 are preferentially expressed in MES GSCs and PN GSCs, respectively.74 ZDHHC18 promotes the degradation of BMI1, an enzyme helps cells to survive under stressful environments, while ZDHHC23 assists to stabilize BMI1 expression.74
Differentially activated signaling pathways are also detected between PN GSCs and MES GSCs. The PDGF receptor-β mediated signaling pathway,75 the Notch pathway76 and the Wnt pathway76,77 are activated in PN GSCs; on the other hand, the TGF-β signaling pathway,20 the NF-κB signaling pathway,19,78 FOXD-ALDH1A3 axis28,79 and glycolysis-mediated metabolism pathway28 are activated in MES GSCs. Several pathways also were involved in the PMT and regulated cells sensitivity to therapy. The Notch signaling pathway is related to cells growth, differentiation and development. Inhibiting the activity of the Notch pathway can restore GSCs sensitivity to radiotherapy.80 The Wnt signaling pathway is associated with GSCs proliferation, self-renewal and tumor initiation. The dual inhibition of the Notch and Wnt pathway increased proneural related signatures in GSCs.77 Therefore, the Notch and Wnt pathway may be involved in the maintenance of the proneural phenotype. The activation of the NF-κB signaling pathway partly mediates the PMT in GBM.81,82 In GSCs, MLK4 is enriched in MES GSCs and can interact with the NF-κB signaling pathway to maintain this phenotype.78 On the other hand, radiotherapy increases the expression of STAT3 and C/EBP-β, the downstream of the NF-κB pathway, indicating the activation of the NF-κB pathway during the PMT.29,83,84 The hedgehog pathway is involved in the PMT in GBM but no research verifies its role in GSCs.85
In summary, molecular classification of GSCs clearly distinguishes GSCs sensitivity to radiochemotherapies. The PMT has been associated with GBM resistance to therapy, and the discovery of the PMT between GSCs subtypes might further support that theory. Therefore, the PMT can be viewed as the adaptive response of GSCs to unfavorable environment, and the inhibition of the PMT may improve patients’ overall survival outcome. Since multiple molecules or pathways related to the PMT have been identified, drugs are designed to target those mediators may improve tumor resistance to therapy.
Metabolic phenotype of GSCs
Tumor cells metabolic reprogramming, also known as the Warburg effect, refers to tumor cells preferring glycolysis rather than the tricarboxylic acid cycle even with adequate oxygen.86 In contrast to GBM cells, the metabolic profile of GSCs, including lower glycolytic, lower extracellular acidification rate, less oxygen consumption and maximal respiratory capacities,87 is more quiescent. It should be noted that recent studies reported different metabolic phenotypes of GSCs.
One study identifies two clusters of GSCs manifesting different metabolic phenotypes, Clone A and Clone B, in the murine GBM model. Cells in Clone A are glycolysis dependent while the metabolic phenotype of Clone B can switch between mitochondrial respiration and glycolysis.88 One research divide GSCs into GLN-low and GLN-high GSCs based on glutamine consumption.30 GLN-high GSCs metabolize more glutamine to sustain its mitochondrial respiration, and the reduction of glutamine can weaken its ability in proliferation or self-renewal. Another study clusters GSCs into GSf-like GSCs and GSr-like GSCs based on metabolism profile. Cells in the former group show metabolic feature with low mobile lipids and high glutamine while cells in the latter group show the opposite.18 GSf-like GSCs and GSr-like GSCs also express proneural and mesenchymal related signatures, respectively. Notably, metabolic phenotype of GSCs corresponds with molecular classification. GSf-like GSCs express proneural related signatures while mesenchymal related signatures are enriched in GSr-like GSCs. Another study reported that GSCs with activated lipid metabolism and reduced glucose consumption are resistance to radiochemotherapies.89 Activated glutamine metabolism is also associated with GSCs resistance to radiotherapy.26 Therefore, therapy-resistant GSCs consume less glucose, with activated glutamine and lipid metabolism by contrasting with therapy sensitive GSCs.
In summary, there are two metabolic phenotypes of GSCs. The first phenotype is GSCs dependent on aerobic glycolysis. This type of GSCs metabolizes glucose to supply cells proliferation. The other phenotype is more complicated. GSCs can switch between glycolysis and OXPHOS according to extracellular stimulation. Instead of consuming glucose, GSCs in this type prefer to metabolize glutamine to initiate OXPHOS. Glycolysis is only activated when the supply of glutamine is insufficient.30 Besides, this type of GSCs contains more mobile lipids in cytoplasm, which indicates the activated lipid metabolism-related pathways. As aforementioned, metabolic phenotype of GSCs affects its sensitivity to cancer therapy. More effort on exploration about the mechanisms of how abnormal metabolic pattern affects cells resistance to therapy remains to be urgently needed.
Biological behavior classification
Classification based on GSCs biological behavior classifies GSCs into qGSCs or pGSCs.25,27 ‘Quiescent cells’ refers to cells with slow cell cycle relative to normal cells, and cells are able to quit this state when necessary.33,90,91 Label-retaining assay is able to distinguish quiescent GSCs from tumor.27,92 Apart from that, recent studies reported that isolation of qGSCs by marking the promoter of nuclear receptor tailless of GSCs with GFP93 or based on GSCs’ sensitivity to a different chemical compound.94 In 3D organoids culturing system, stem-like cells at the periphery showed strong proliferative ability while cells in the hypoxic core more quiescence.39 However, the accuracy of qGSCs isolation between those protocols is lack of comparison.
The proliferation ratio of pGSCs is significantly quicker than qGSCs but there are no specific molecular hallmarks to distinguish them.25,33 Differentially expressed genes profile identifies SAT1 and ID1 upregulated in qGSCs while EGFR enriched in pGSCs.95 pGSCs are mostly located in the perivascular niche, which is similar to PN GSCs, while qGSCs are located in necrotic niche.33,96 BMP and TGF-β signaling pathways are selectively activated in qGSCs and pGSCs respectively.95 Besides, biofunction prediction suggests that dysregulated genes in qGSCs are related to tumor immune landscape and tumor resistance to therapy, while genes in pGSCs are associated with cell proliferation.95
Potential regulators of biological behavior of GSCs
The expression profile of cell cycle-related genes reveals the mechanism of different biological behavior of GSCs. For instance, cyclin B1, CDKN1A and G0S2 expression are dysregulated in qGSCs.33 Accumulation of p27 at G0 phase in qGSCs is associated with the maintenance of cells quiescence.97 Factors like Ca2+ influx related genes expression (like CACNB1, CAPS, CACNA2D1, PKD2 and ORAI2),98 the activity of Notch signaling pathway,99 mitochondrial shape96 and hypoxia and acidic niche39 are also raised for their role in quiescence state.
Other potential regulators involved in the biological behavior of GSCs are also summarized. DOCK4 and β-catenin affect GSCs proliferative ability through influencing GSK3-β activity.100 NGF and its receptors control GSCs proliferation.101 The proliferation rate of GSCs can be inhibited by silencing the expression of STAT3 and integrin α6.102,103 Those regulators can affect the proliferation ability of GSCs but their role in pGSCs or qGSCs remain elusive.
Biological behavior transition
The proliferation-quiescence transition is termed as pGSCs entering the ‘quiescence’ status. This transition can be induced by hypoxia or an acidic environment through altering mitochondrial shape and cytometric calcium concentration of GSCs.33 Notably, the ratio of qGSCs in tumor is positively correlated with tumor recurrence times.33 The population of qGSCs is increased after treating GBM with RTK inhibitors, and the activity of the Notch pathway and KDM expression is also increased.104 In general, pGSCs can be transformed to qGSCs under the stimulation of unfavorable environment or radiochemotherapies, and this transition could be a novel mechanism of tumor resistance to therapy.25,92,93,105
Since the ‘quiescence’ state is a reversible state, pGSCs can also generate from qGSCs.93 The GINS complex (comprise of SLD5, PSF1, PSF2, and PSF3) re-initiates cell cycle in qGSCs by altering cell-cycle-related genes expression.106 Evidence supporting the quiescence-proliferation transition is insufficient, and this transition might be related to tumor recurrence.
Biological behavior of GSCs also sheds light on the mechanism of GSCs sensitivity to therapy. For instance, qGSCs can survive from an unfavorable environment and develop tumor by quitting the quiescence state. pGSCs can enter the quiescence state when the environment is not favorable for survival. This dual-transition highlights the mechanism of tumor recurrence and tumor resistance to therapy.
Internal connection between different classifications of GSCs
Previous studies subdivided GSCs into PN GSCs or MES GSCs based on molecular classification. Notably, two metabolic phenotypes of GSCs are also associated with PN GSCs or MES GSCs. According to the metabolic profile and molecule signatures, cells in Clone A, GSf-like GSCs and GLN-low GSCs are PN GSCs while cells in another group (Clone B, GSr-like GSCs and GLN-high GSCs) are MES GSCs. Therefore, PN GSCs depend on aerobic glycolysis while the metabolic phenotype of MES GSCs is more flexible.26
MES GSCs consume glutamine and glucose to supply OXPHOS and glycolysis, respectively. Since glutamine can replenish lipid biosynthesis precursors and supply mitochondrial respiration,26 glutamine and lipid metabolism-related pathways are also activated in MES GSCs.18 Multiple studies supported that activated glutamine and lipid metabolism are involved in tumor resistance to therapy.31,107,108 Thus, the mechanism of MES GSCs shows resistance to therapy might relate to this metabolic phenotype.
On the other hand, PN GSCs and MES GSCs manifest stronger ability in proliferation and migration, respectively. The connection between molecular classification and GSCs biological behavior is unclear. Given the slow cell cycle of qGSCs, pGSCs might be a group of cells containing PN GSCs and MES GSCs simultaneously. However, pGSCs and PN GSCs both are located at perivascular niche while qGSCs and MES GSCs prefer necrotic niche.
Three classifications, molecular, biological behavior, and metabolic phenotype all elaborate only one feature of GSCs. Within each group, GSCs can also be grouped as therapy sensitive or resistant cells. Transition restricted to each classification clearly map the response of GSCs to therapy or unfavorable environment. Several studies have confirmed the connection between molecular classification and metabolic phenotype. However, their association with GSCs biological behavior is unclear. Figuring out the internal connection between different classifications can reveal the feature of therapy-resistant GSCs and promote clinical management (Fig. 3).
Fig. 3.
The adaptive transition within GSCs classifications. Therapeutic sensitive GSCs like PN GSCs can transform into MES GSCs which show resistance to cancer treatment. PN GSCs mostly dependent on glycolysis, and MES GSCs switch its metabolism between glycolysis and OXPHOS. PN GSCs metabolize glucose while MES GSCs can switch between glucose and glutamine. The Notch and Wnt pathway are preferentially activated in PN GSCs; the activation of TGF-β pathway and NF-κB pathway are mostly observed in MES GSCs
The relationship between NSCs and GSCs
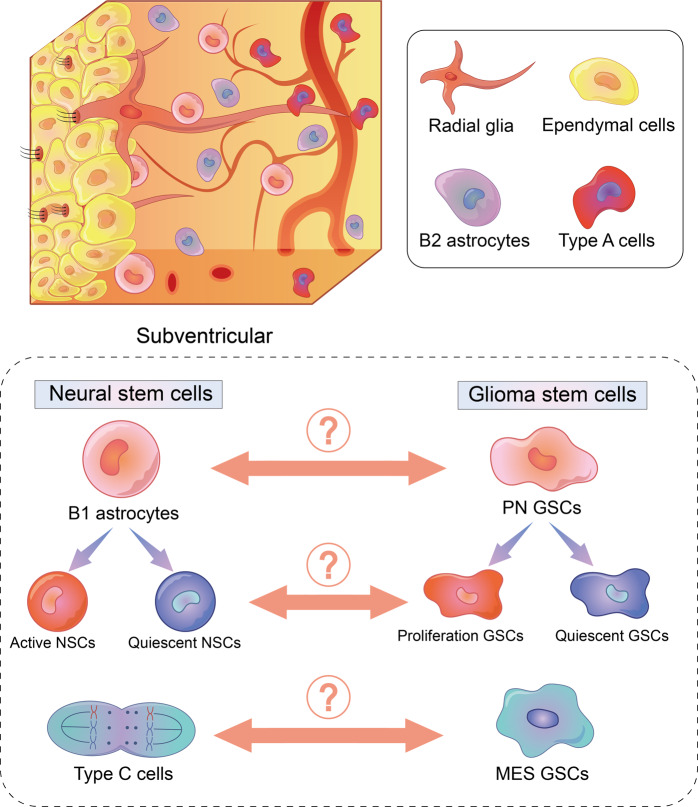
Subventricular NSCs (also called as astrocyte-like NSCs or type B cells) contain two groups of cells, B1 astrocytes and B2 astrocytes. B2 astrocytes are non-neurogenic astrocytes. B1 astrocytes asymmetrically split into type C cells (also known as transit-amplifying progenitor cells) which will differentiate into type A cells (also known as neuroblasts) or oligodendrocyte precursor cells in the end. In the meantime, B1 astrocytes can be subdivided into quiescent NSCs (qNSCs) and active NSCs (aNSCs) based on their biological behavior. Type A cells can form neurons, and oligodendrocyte precursor cells differentiate into oligodendrocytes or astrocytes.109,110
Several studies reported that GSCs are derived from subventricular NSCs,111–113 and the fact that by engineering p53,114 EGFR115 or H-RasV12 88 in NSCs can induce the formation of GSCs. Gnomically, NSCs and GSCs share common gene signatures including SOX2, NESTIN, OLIG2, CD133, YKL40, et al.116,117 CD133 and Nestin are both expressed in B1 astrocytes and PN GSCs; EGFR is mainly enriched in type C cells and MES GSCs118,119 (Table 3). This hallmark similarity implies the association between NSCs and GSCs.
Table 3.
Common gene signatures of NSCs
| Cells | GFAP | Nestin | CD133 | EGFR | CD15 |
|---|---|---|---|---|---|
| qNSCs | + | − | + | − | + |
| aNSCs | + | + | + | + | + |
| B2 astrocytes | + | + | − | + | − |
| Type C cells | − | + | − | + | + |
| Type A cells | − | − | − | − | − |
| PN GSCs | − | + | + | − | + |
| MES GSCs | − | − | − | + | + |
Metabolically, NSCs depend on glycolysis to maintain its stemness, but its differentiation is involved in the activation of several metabolic pathways including elevated fatty acid consumption, increased lipogenesis, decreased glycolysis and activated OXPHOS.120–122 This metabolic phenotype transition during NSCs differentiation is similar to the PMT. Glycolysis dependent type C cells show tolerance to the hypoxia environment,123 but its proliferation still relies on absorbing extracellular fatty acid and activating de novo lipogenesis.124 The metabolic patterns transition during the differentiation of NSCs is similar to GSCs adaptive transition.
The biological behavior of qNSCs and aNSCs is similar to that of qGSCs and pGSCs, respectively.125,126 Besides, aNSCs and pGSCs both show sensitivity to temozolomide but qNSCs and qGSCs can survive from it.127 Therefore, the nature of NSCs may also affect GSCs sensitivity to therapy.
Another study pioneered exploring the association between GSCs and NSCs by performing single-cell RNA sequencing analysis.128 They proved that the apex of GBM hierarchy is progenitor cancer cells, and most of them carry with proneural signature while few of them are classified as mesenchymal or classical. They also identify an un classified type of GSCs which show similarity with progenitor cancer cells.
Taken together, similarity in transcriptomic signature, metabolic profile, biological behavior and single-cell RNA sequencing analysis highlighted the internal correlation of GSCs and NSCs. Furthermore, it may be hypothesized that GSCs are derived from B1 astrocytes, and the PMT is the glioma version of the B1 astrocytes differentiation. Nevertheless, evidences from some studies make different voice. For instance, CD44, hallmarks of MES GSCs, is expressed on astrocyte-restricted precursors that do not express on NSCs. Metabolic profile of PN GSCs is similar to type C cells instead of B1 astrocyte.88 The origin of IDH wildtype glioma and IDH mutant glioma might different.129 Since differentiation of NSCs is a complicated, precise, dynamic process, their internal relationship with GSCs still needs more investigation (Fig. 4).
Fig. 4.
Association between subventricular NSCs and GSCs. Similarities, including genomic characteristics and metabolism pattern, are found between PN GSCs and B1 astrocytes, MES GSCs and type C cells. The biological behavior of B1 astrocytes is similar to GSCs. However, the explicit relationship between NSCs and GSCs remains unknown
Niche and GSCs
Niche is a special microenvironment where stem cells are preserved. The niche of GSCs comprises of multiple components including endothelial cells (ECs), arterioles, immunocytes, fibroblasts, NSCs, pericytes, et al.130 Several studies reported five types niches in GBM, including peri-vascular niches,131 peri-arteriolar niches,132 peri-hypoxic niches,133 peri-immune niches134, and ECM niches,135 based on their unique traits. Those niches share similar features and interrelate with each other. Therefore, a comprehensive model integrating those niches called hypoxic peri-arteriolar niche was proposed.136 More importantly, this dynamic model simplifies the catalog of GSCs niches and improves the understanding of the interconnection between niches and GSCs. However, discussion about the relationship between MES GSCs and this model is not mentioned.
Blood vessels and vasculogenic mimicry
Blood vessels distribution in GBM varies from normal brain tissue. ECs, pericytes and smooth muscle cells are constituents of blood vessels, and they both affect GSCs. For instance, ECs are associated with the maintenance of GSCs stemness by activating the Notch pathway.137,138 On the other hand, GSCs can transdifferentiate into ECs, pericytes and smooth muscle cells, and involve in the formation of vasculogenic mimicry.139–141 GSCs-derived pericytes contribute to tumor resistance to therapy by altering the permeability of the blood-brain barrier.142–144 Besides, several molecules are involved in this process, including Flk-1,145 CDH5,146 YKL40,147 KDR148, and VEGF.149 In summary, GSCs are closely associated with tumorigenesis and vasculogenic mimicry.
Hypoxia and acidic tumor microenvironment
Hypoxia and acidic are critical characteristics of tumor microenvironment.150,151 The survival probability of PN in hypoxia and acid environment is lower than MES GSCs.74 HIF-2α is involve in maintaining the stemness of GSCs and contributes to the PMT.55 Hypoxia can activate glutamine metabolism-related pathway in tumor cells.152,153 Besides, hypoxia and acidic tumor microenvironment affect the proliferation-quiescence transition of GSCs.33 Together, those results indicate that niche is involved in GSCs resistance to therapy.
Immunocytes infiltration
Immunocytes infiltration of PN and MES GSCs is different. For instance, qGSCs upregulate the expression of T cell targeted antigen and are infiltrated with more T cells than pGSCs.154 MES GSCs have higher infiltration of CD8 positive T cells and microglial than PN GSCs.155 Tumor-associated macrophages are derived from bone marrow-derived monocytes, microglial cells and GSCs.156 PN GSCs induce the formation of tumor-associated macrophages and recruit M2 tumor-associated macrophages.36,157
Compared with MES GSCs, PN GSCs increase the expression of MHC I, CD40 and CD86 and downregulate the expression of MHC II and CD80. B7-H1, an inhibitory molecule of T cells, is also increased in PN GSCs.158 Hypoxia microenvironment promotes PN GSCs to release immunosuppressive cytokines.159 PN GSCs show resistance to TGF-β stimulation and its low TGF-β expression indicates that TGF-β acts an immunosuppressive role in PN GSCs.155 Together, those results reveal lower immunocytes infiltration in PN GSCs than MES GSCs, and this difference might relate to the efficacy of GSCs sensitivity to immunotherapy.
The comprehensive model of niche allows a clearer view of the relationship between niche and GSCs. The components of the niche are complicated and dynamic. GSCs affect the formation of niche, in turn, niche components like pericytes or characteristics like hypoxia influence the subtype of GSCs as well as GSCs sensitivity to therapy. Currently, whether niche contributes to GSCs resistance to therapy is still unclear.
Implications on treatments
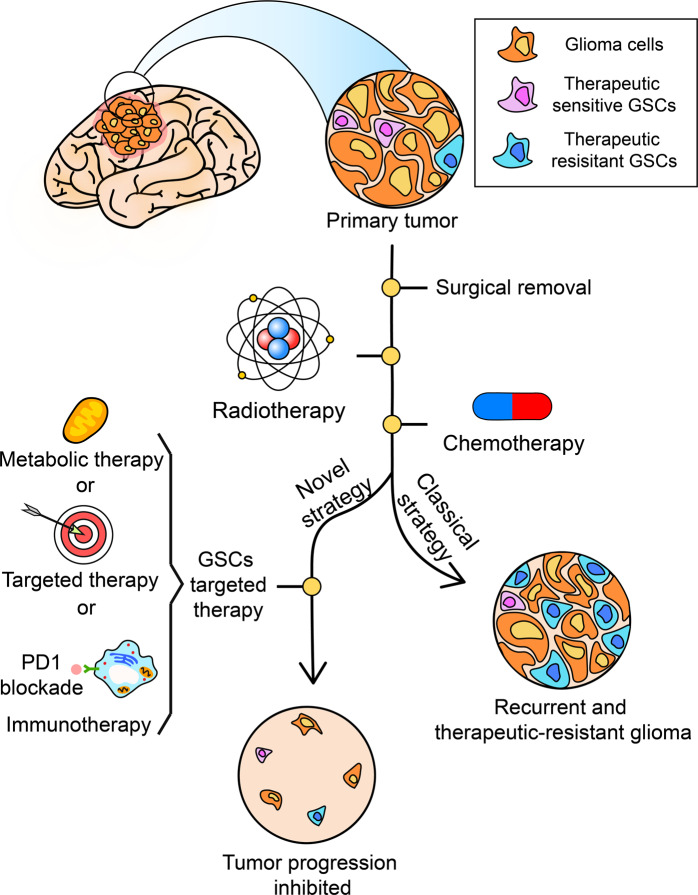
As aforementioned, a different subtype of GSCs carry with different characteristics and show different sensitivity to tumor therapy. Therefore, targeting to GSCs selectively may be an option. In recent years, progresses in selectively targeting to GSCs subtype have been reported, and the section summarized those progresses (Fig. 5).
Fig. 5.
Therapy resistant GSCs like MES GSCs can survive from a classical strategy like chemo- or radio-therapy. In the meantime, the combination of GSCs targeted therapy and classical strategy may improve treatment efficiency
Radiotherapy
PN GSCs can transform into MES GSCs by treating with radiotherapy and show resistance to radiotherapy.28 Multiple studies revealed that interfering the PMT related pathway can restore GBM sensitivity to radiotherapy.160–164 However, the efficacy of inhibiting the PMT in GSCs is not verified. The inhibition of relevant metabolic pathways in GSCs might also be a novel treatment to restore GSCs sensitivity to radiotherapy.
Chemotherapy
The relationship between GSCs and chemotherapy is more complicated. Temozolomide is the most common and efficient chemotherapeutic agent in clinical application to treat GBM. In primary GBM, PN GSCs are resistant to multiple chemotherapeutic agents including temozolomide.165,166 Another study reported temozolomide can inhibit GSCs proliferation with still a small group of GSCs survived.167 Besides, the expression of MGMT, a biomarker to predict GBM sensitivity to temozolomide, can also predict GSCs sensitivity to temozolomide regardless of molecular signatures.167 Notably, the combination of temozolomide and perillyl alcohol has a lethal effect on PN GSCs derived and MES GSC derived GBM.168 On the contrary, MES GSCs derived GBM show resistance to temozolomide and gradually lost its mesenchymal related signatures during treating with temozolomide.32 In the meantime, tumor sensitivity to radiotherapy is restored.32 In general, MES GSCs have a higher expression of several therapy resistance-related genes compared to PN GSCs. But their sensitivity to chemotherapy do not show no significant difference, and MGMT is still a qualified biomarker to predict GSCs sensitivity to temozolomide.
Metabolic therapy
Activated glutamine metabolism in MES GSCs is associated with GSCs resistance to radiochemotherapies and GSCs proliferation.89 EGCG, an inhibitor of transglutaminase, can restore GSCs sensitivity to temozolomide and inhibit GSCs proliferation.30,169 Dichloroacetate, the inhibitor of pyruvate dehydrogenase kinase inhibitor, can increase GSCs sensitivity to radiotherapy.170 In MES GSCs, glutamine serves as a metabolic substrate of OXPHOS, and pyruvate dehydrogenase kinase is also critical to supply OXPHOS. Therefore, the activation of mitochondrial in MES GSCs might be connected with its resistance to therapy. Metformin, an inhibitor of mitochondrial complex I, can affect tumor cells resistance to therapy but its role in GSCs is not confirmed.171
Immunotherapy
Immunotherapy targeting GSCs or adopting GSCs as therapeutic methods to treat GBM has made some progress in recent years.172,173 Besides, immune check point genes and antigen presentation genes are differentially expressed on PN GSCs and MES GSCs as aforementioned.158 Immunocytes infiltration difference is identified in molecular classification and biological behavior classification.154,155 Metabolic phenotypes also affect GSCs sensitivity to immunotherapy. For instance, ‘metabolic check point’like glucose depletion and hypoxia affect the function of tumor infiltrated immunocytes.174 Together, those results suggest that the response of GSCs to immunotherapy might also differ from each other.154,175 However, the association between GSCs and immunotherapy is not clear. Notably, a recent study reported tumor immune escape can be inhibited by blocking glutamine metabolism-related pathways, indicating that a similar strategy could be applied to GSCs.176
Other treatments
Anti-VEGF therapy is a novel strategy to treat cancer but its efficacy in GBM is not optimistic.3,177 Bevacizumab, VEGF inhibitor, promotes vasculogenic mimicry formation by PN GSCs148 which might be associated with its failure of treating primary GBM.178
Tumor-treating fields inhibit tumor cells proliferation to prevent GBM progression. It can also be applied to treat tumor derived from GSCs.179–182 However, the association between GSCs resistance and tumor-treating fields is not clear.
The previous study reported GSCs to respond differently to targeted therapy.183 Since various strategies such as nanoparticles184,185 and Zika virus186–188 can be applied to target GSCs, it is critical to treat GSCs with combined therapeutic options to improve treatment efficacy. A recent study summarized the efficacy of strategy that by targeting SOX2 on stem-like cells can inhibit tumor progression.189
Clinical trials targeting GSCs
Apart from traditional treatments of GBM, several clinical trials proposed a novel strategy in recent years. All information about clinical trials is obtained from public clinical trial databases (https://clinicaltrials.gov/). Seventy-eight results in total are obtained by setting ‘glioma stem cells’ as the keyword, and nineteen of them are about targeting GSCs or adopting NSCs as therapeutic means (Table 4). Two out of six completed clinical trials have published their results. The efficacy of therapy adopting NSCs as therapeutic means can be evaluated by MRI.190 Dendritic cells loaded with GSCs-derived mRNA can inhibit GBM growth.191 Only one trial which used GSCs as the antigen source of the vaccine was terminated due to limit efficacy and extreme toxicity (ClinicalTrial.gov Identifier: NCT01400672). However, few clinical trials take the difference in GSCs into account.
Table 4.
Clinical trials target on GSCs or adopt NSCs as therapeutic means
| NCT number | Title | Status | Target or medium | Therapy |
|---|---|---|---|---|
| NCT02039778 | Stem cell radiotherapy and temozolomide for newly diagnosed high-grade glioma | Terminated | GSCs | Radio- and chemo-therapy |
| NCT03072134 | Neural stem cell-based virotherapy of newly diagnosed malignant glioma | Completed | NSCs | Virotherapy |
| NCT01872221 | Study of the capacity of the MRI spectroscopy to define the tumor area enriched in glioblastoma stem cells. Proof of concept study | Completed | GSCs | Radio- and chemo-therapy |
| NCT02192359 | Carboxylesterase-expressing allogeneic neural stem cells and irinotecan hydrochloride in treating patients with recurrent high-grade gliomas | Recruiting | NSCs | Genetically modified therapy |
| NCT01172964 | A pilot feasibility study of oral 5-fluorocytosine and genetically modified neural stem cells expressing E. Coli cytosine deaminase for treatment of recurrent high-grade gliomas | Completed | NSCs | Genetically modified therapy |
| NCT02010606 | Phase I study of a dendritic cell vaccine for patients with either newly diagnosed or recurrent glioblastoma | Active, not recruiting | GSCs | Immunotherapy |
| NCT02055196 | Genetically modified stem cells and irinotecan hydrochloride in treating patients with recurrent high-grade gliomas | Withdrawn | NSCs | Genetically modified therapy |
| NCT02015819 | Genetically modified neural stem cells, flucytosine, and leucovorin for treating patients with recurrent high-grade gliomas | Active, not recruiting | NSCs | Genetically modified therapy |
| NCT01171469 | Vaccination with dendritic cells loaded with brain tumor stem cells for progressive malignant brain tumor | Completed | GSCs | Immunotherapy |
| NCT03956706 | Study of stereotactic radiosurgery to the subventricular zone in malignant gliomas | Recruiting | NSCs | Radiotherapy |
| NCT01567202 | Study of DC vaccination against glioblastoma | Recruiting | GSCs | Immunotherapy |
| NCT00473408 | The effect of radiotherapy and temozolomide on the tumor vasculature and stem cells in human high-grade astrocytomas | Terminated | GSCs | Radio- and chemo-therapy |
| NCT03632135 | Standard chemotherapy vs. chemotherapy guided by cancer stem cell test in recurrent glioblastoma | Recruiting | GSCs | Chemotherapy |
| NCT02654964 | Cancer stem cell high-throughput drug screening study | Unknown status | GSCs | Chemotherapy |
| NCT03548571 | Dendritic cell immunotherapy against cancer stem cells in glioblastoma patients receiving standard therapy | Recruiting | GSCs | Immunotherapy |
| NCT00846456 | Safe study of dendritic cell (DC) based therapy targeting tumor stem cells in glioblastoma | Completed | GSCs | Immunotherapy |
| NCT00890032 | Vaccine therapy in treating patients undergoing surgery for recurrent glioblastoma multiforme | Completed | GSCs | Immunotherapy |
| NCT01400672 | Imiquimod/brain tumor initiating cell (BTIC) vaccine in brain stem glioma | Terminated | GSCs | Radio- and immune-therapy |
| NCT02177578 | Subventricular zone (SVZ) and temozolomide in glioblastoma multiforme | Recruiting | NSCs | Radio- and chemo-therapy |
Conclusion and prospection
In this review, different classifications of GSCs are summarized and integrated. However, there are several questions about GSCs classification. First, several genes signatures of the proneural or mesenchymal subtype are identified nowadays, but few of them can be applied to GSCs isolation. Considering the PMT in GSCs, a precise method to isolate GSCs can bring about a more accurate result. Second, the inner relationship between biological behavior classification and the other two classifications is not clear. pGSCs seem to be connected with PN GSCs but no similarity is found between qGSCs and other subtypes of GSCs. Third, the metabolic phenotype of GSCs requires more attention. One study which subdivided GSCs into three groups (Cluster1a, Cluster1b and Cluster2) reported that the molecular signatures of Cluster1a are similar to those of PN GSCs whereas cells in the other two groups are similar to those in MES GSCs.192 Notably, Cluster1 (including Cluster1a and Cluster1b) manifests a flexible metabolic phenotype while Cluster2 mainly depends on glycolysis. Obviously, this conclusion is non consistent with previous results.18,30,72,88 Each classification mentioned above merely reveals one feature of GSCs. An integrative analysis of those classifications will provide a better understanding of GSCs.
Multiple studies proved tumor cell adaptive survival from anti-tumor therapy, and this process was viewed as tumor therapeutic response193 Therapy sensitive or resistant GSCs are also identified in each classification. For instance, MES GSCs, glutamine dependent GSCs and qGSCs show nature resistance to cancer therapy. Transition restricted to each classification like the PMT and the proliferative-quiescent transition is highly associated with GSCs resistance to cancer therapy. Besides, MES GSCs can transform their metabolic pattern according to the context, indicating that MES GSCs are hard to be affected by constraining its nutrition supply. All kinds of GSCs adaptive transition not only reveal the mechanisms of tumor recurrence and tumor resistance to cancer therapy but also highlight multiple potential targets for future research. Therefore, molecular signatures, pathways or metabolic pattern involved in GSCs adaptive transition can be served as potential targets to improve therapeutic efficacy.
The inner relationship between NSCs and GSCs is not clear. Multiple studies suggested that NSCs are the derivation of GSCs, in the meantime, other studies further confirmed the similarity between different GSCs subtypes and NSCs differentiation lineage. Therefore, GSCs adaptive transition might share common features with NSCs differentiation lineage. Niche is another factor that affecting GSCs adaptive transition, and GSCs influence the formation of the niche in turn. Besides, immunocytes infiltration and vasculogenic mimicry can affect tumor response to cancer therapy. Therefore, NSCs, niche and GSCs interact with each other. But more in-depth mechanisms remain to be revealed. In general, GSCs is a novel breakpoint for understanding tumor recurrence and tumor resistance to cancer therapy.
Acknowledgements
This work was supported by the National Nature Science Foundation of China (NO.81873635, NO.82073893, NO.81703622); the China Postdoctoral Science Foundation (NO.2018M63302), Hunan Provincial Natural Science Foundation of China (NO.2018JJ3838, 2018SK2101), Hunan Provincial Health Committee Foundation of China (C2019186). Xiangya Hospital Central South University postdoctoral foundation. The author would like to thank Prof. Liyang Zhang from Xiangya Hopital Central South University for the help with this article.
Author contributions
Z.Y.W.: manuscript preparation. H.Z.: reference collection and manuscript revision. S.C.X.: manuscript revision. Q.C. and Z.X.L.: final approval for publication and offer revision advice.
Competing interests
The authors declare no competing interests.
Contributor Information
Zhixiong Liu, Email: zhixiongliu@csu.edu.cn.
Quan Cheng, Email: chengquan@csu.edu.cn.
References
- 1.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Cheng Q, et al. Role of miR-223/paired box 6 signaling in temozolomide chemoresistance in glioblastoma multiforme cells. Mol. Med. Rep. 2017;15:597–604. doi: 10.3892/mmr.2016.6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert MR, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iorgulescu JB, Reardon DA, Chiocca EA, Wu CJ. Immunotherapy for glioblastoma: going viral. Nat. Med. 2018;24:1094–1096. doi: 10.1038/s41591-018-0142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson CM, Choi J, Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat. Immunol. 2019;20:1100–1109. doi: 10.1038/s41590-019-0433-y. [DOI] [PubMed] [Google Scholar]
- 6.Fonkem E, Wong ET. NovoTTF-100A: a new treatment modality for recurrent glioblastoma. Expert Rev. Neurother. 2012;12:895–899. doi: 10.1586/ern.12.80. [DOI] [PubMed] [Google Scholar]
- 7.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neftel C, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849.e821. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Halliday J, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc. Natl Acad. Sci. USA. 2014;111:5248–5253. doi: 10.1073/pnas.1321014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedele M, et al. Proneural-Mesenchymal Transition: Phenotypic Plasticity to Acquire Multitherapy Resistance in Glioblastoma. Int. J. Mol. Sci. 2019;20:2746. doi: 10.3390/ijms20112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 15.Ignatova TN, et al. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 16.Kappadakunnel M, et al. Stem cell associated gene expression in glioblastoma multiforme: relationship to survival and the subventricular zone. J. Neurooncol. 2010;96:359–367. doi: 10.1007/s11060-009-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behnan J, et al. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene. 2017;36:570–584. doi: 10.1038/onc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marziali G, et al. Metabolic/proteomic signature defines two glioblastoma subtypes with different clinical outcome. Sci. Rep. 2016;6:21557. doi: 10.1038/srep21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat KPL, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lottaz C, et al. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 21.Jin X, et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 2017;23:1352–1361. doi: 10.1038/nm.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, et al. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 2019;9:1708–1719. doi: 10.1158/2159-8290.CD-19-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayanan A, et al. The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Cell Death Differ. 2019;26:1813–1831. doi: 10.1038/s41418-018-0248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park NI, et al. ASCL1 reorganizes chromatin to direct neuronal fate and suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell. 2017;21:209–224 e207. doi: 10.1016/j.stem.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Lan X, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549:227–232. doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garnier D, et al. Glioblastoma stem-like cells, metabolic strategy to kill a challenging target. Front. Oncol. 2019;9:118. doi: 10.3389/fonc.2019.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deleyrolle LP, et al. Evidence for label-retaining tumour-initiating cells in human glioblastoma. Brain. 2011;134:1331–1343. doi: 10.1093/brain/awr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao P, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl Acad. Sci. USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minata M, et al. Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 2019;26:1893–1905 e1897. doi: 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oizel K, et al. Efficient mitochondrial glutamine targeting prevails over glioblastoma metabolic plasticity. Clin. Cancer Res. 2017;23:6292–6304. doi: 10.1158/1078-0432.CCR-16-3102. [DOI] [PubMed] [Google Scholar]
- 31.Fu S, et al. Glutamine synthetase promotes radiation resistance via facilitating nucleotide metabolism and subsequent DNA damage repair. Cell Rep. 2019;28:1136–1143.e1134. doi: 10.1016/j.celrep.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Garnier D, et al. Divergent evolution of temozolomide resistance in glioblastoma stem cells is reflected in extracellular vesicles and coupled with radiosensitization. Neuro Oncol. 2018;20:236–248. doi: 10.1093/neuonc/nox142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aulestia FJ, et al. Quiescence status of glioblastoma stem-like cells involves remodelling of Ca(2+) signalling and mitochondrial shape. Sci. Rep. 2018;8:9731. doi: 10.1038/s41598-018-28157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirino V, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27:13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig K, Kornblum HI. Molecular markers in glioma. J. Neurooncol. 2017;134:505–512. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lathia JD, et al. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broadley KW, et al. Side population is not necessary or sufficient for a cancer stem cell phenotype in glioblastoma multiforme. Stem Cells. 2011;29:452–461. doi: 10.1002/stem.582. [DOI] [PubMed] [Google Scholar]
- 38.Azzarelli ROrganoid. Models of glioblastoma to study brain tumor stem cells. Front Cell Dev. Biol. 2020;8:220. doi: 10.3389/fcell.2020.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubert CG, et al. A Three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76:2465–2477. doi: 10.1158/0008-5472.CAN-15-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yachi K, et al. miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition. Signal Transduct. Target Ther. 2018;3:33. doi: 10.1038/s41392-018-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim EJ, et al. Glioma stem cells and their non-stem differentiated glioma cells exhibit differences in mitochondrial structure and function. Oncol. Rep. 2018;39:411–416. doi: 10.3892/or.2017.6075. [DOI] [PubMed] [Google Scholar]
- 42.Chesnelong C, Restall I, Weiss S. Isolation and Culture of Glioblastoma Brain Tumor Stem Cells. Methods Mol. Biol. 2019;1869:11–21. doi: 10.1007/978-1-4939-8805-1_2. [DOI] [PubMed] [Google Scholar]
- 43.Podergajs N, et al. Expansive growth of two glioblastoma stem-like cell lines is mediated by bFGF and not by EGF. Radio. Oncol. 2013;47:330–337. doi: 10.2478/raon-2013-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Katoh M. Canonical and non-canonical WNT signaling in cancer stem cells and their niches: Cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review) Int J. Oncol. 2017;51:1357–1369. doi: 10.3892/ijo.2017.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemke D, et al. Primary glioblastoma cultures: can profiling of stem cell markers predict radiotherapy sensitivity? J. Neurochem. 2014;131:251–264. doi: 10.1111/jnc.12802. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int. J. Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 48.Lathia JD, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao XG, et al. Brain tumor stem-like cells identified by neural stem cell marker CD15. Transl. Oncol. 2009;2:247–CD257. doi: 10.1593/tlo.09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kenney-Herbert E, et al. CD15 expression does not identify a phenotypically or genetically distinct glioblastoma population. Stem Cells Transl. Med. 2015;4:822–831. doi: 10.5966/sctm.2014-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son MJ, et al. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan Y, et al. Autofluorescence of NADH is a new biomarker for sorting and characterizing cancer stem cells in human glioma. Stem Cell Res Ther. 2019;10:330. doi: 10.1186/s13287-019-1467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tchoghandjian A, et al. A2B5 cells from human glioblastoma have cancer stem cell properties. Brain Pathol. 2010;20:211–221. doi: 10.1111/j.1750-3639.2009.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogden AT, et al. Identification of A2B5+CD133- tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62:505–514. doi: 10.1227/01.neu.0000316019.28421.95. [DOI] [PubMed] [Google Scholar]
- 55.Brown DV, et al. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS ONE. 2017;12:e0172791. doi: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lathia JD, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corsini NS, Martin-Villalba A. Integrin alpha 6: anchors away for glioma stem cells. Cell Stem Cell. 2010;6:403–404. doi: 10.1016/j.stem.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Soehngen E, et al. Hypoxia upregulates aldehyde dehydrogenase isoform 1 (ALDH1) expression and induces functional stem cell characteristics in human glioblastoma cells. Brain Tumor Pathol. 2014;31:247–256. doi: 10.1007/s10014-013-0170-0. [DOI] [PubMed] [Google Scholar]
- 59.Jin X, et al. Cell surface Nestin is a biomarker for glioma stem cells. Biochem. Biophys. Res. Commun. 2013;433:496–501. doi: 10.1016/j.bbrc.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 60.Hale JS, et al. Cancer stem cell-specific scavenger receptor CD36 drives glioblastoma progression. Stem Cells. 2014;32:1746–1758. doi: 10.1002/stem.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Podergajs N, et al. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget. 2016;7:593–609. doi: 10.18632/oncotarget.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y, et al. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ. 2017;24:167–180. doi: 10.1038/cdd.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li, D. et al. Glioma-associated human endothelial cell-derived extracellular vesicles specifically promote the tumourigenicity of glioma stem cells via CD9. Oncogene38, 6958 (2019). [DOI] [PubMed]
- 64.Erhart F, et al. Gliomasphere marker combinatorics: multidimensional flow cytometry detects CD44+/CD133+/ITGA6+/CD36+ signature. J. Cell Mol. Med. 2019;23:281–292. doi: 10.1111/jcmm.13927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells. 2009;27:2393–2404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ehtesham M, Mapara KY, Stevenson CB, Thompson RC. CXCR4 mediates the proliferation of glioblastoma progenitor cells. Cancer Lett. 2009;274:305–312. doi: 10.1016/j.canlet.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gatti M, et al. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology. 2013;314:209–220. doi: 10.1016/j.tox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Guardia GDA, et al. Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles. NPJ Genom. Med. 2020;5:2. doi: 10.1038/s41525-019-0108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Behnan J, Finocchiaro G, Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142:847–866. doi: 10.1093/brain/awz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conroy S, et al. IL-8 associates with a pro-angiogenic and mesenchymal subtype in glioblastoma. Oncotarget. 2018;9:15721–15731. doi: 10.18632/oncotarget.24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie Y, et al. The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine. 2015;2:1351–1363. doi: 10.1016/j.ebiom.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakano I. Stem cell signature in glioblastoma: therapeutic development for a moving target. J. Neurosurg. 2015;122:324–330. doi: 10.3171/2014.9.JNS132253. [DOI] [PubMed] [Google Scholar]
- 73.Segerman A, et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17:2994–3009. doi: 10.1016/j.celrep.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 74.Chen X, et al. DHHC protein family targets different subsets of glioma stem cells in specific niches. J. Exp. Clin. Cancer Res. 2019;38:25. doi: 10.1186/s13046-019-1033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim Y, et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26:1247–1262. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saito N, et al. A high Notch pathway activation predicts response to gamma secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells. 2014;32:301–312. doi: 10.1002/stem.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajakulendran N, et al. Wnt and Notch signaling govern self-renewal and differentiation in a subset of human glioblastoma stem cells. Genes Dev. 2019;33:498–510. doi: 10.1101/gad.321968.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim SH, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-kappaB-dependent manner. Cancer Cell. 2016;29:201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng P, et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76:7219–7230. doi: 10.1158/0008-5472.CAN-15-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moreno M, et al. GPR56/ADGRG1 inhibits mesenchymal differentiation and radioresistance in glioblastoma. Cell Rep. 2017;21:2183–2197. doi: 10.1016/j.celrep.2017.10.083. [DOI] [PubMed] [Google Scholar]
- 82.Cai J, et al. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-kappaB pathway. J. Neurooncol. 2018;140:225–236. doi: 10.1007/s11060-018-2951-0. [DOI] [PubMed] [Google Scholar]
- 83.Stanzani E, et al. Radioresistance of mesenchymal glioblastoma initiating cells correlates with patient outcome and is associated with activation of inflammatory program. Oncotarget. 2017;8:73640–73653. doi: 10.18632/oncotarget.18363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hua, T. N. M. et al. Peroxisome proliferator-activated receptor gamma as a theragnostic target for mesenchymal-type glioblastoma patients. Exp. Mol. Med. 52, 629–642 (2020). [DOI] [PMC free article] [PubMed]
- 85.Nanta R, et al. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol. Cell Biochem. 2019;454:11–23. doi: 10.1007/s11010-018-3448-z. [DOI] [PubMed] [Google Scholar]
- 86.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spehalski EI, et al. The quiescent metabolic phenotype of glioma stem cells. J. Proteom. Bioinform. 2019;12:96–103. doi: 10.35248/0974-276X.19.12.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saga I, et al. Integrated analysis identifies different metabolic signatures for tumor-initiating cells in a murine glioblastoma model. Neuro Oncol. 2014;16:1048–1056. doi: 10.1093/neuonc/nou096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ye F, et al. Protective properties of radio-chemoresistant glioblastoma stem cell clones are associated with metabolic adaptation to reduced glucose dependence. PLoS ONE. 2013;8:e80397. doi: 10.1371/journal.pone.0080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vallette FM, et al. Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer. Biochem. Pharm. 2019;162:169–176. doi: 10.1016/j.bcp.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 91.van Velthoven CTJ, Rando TA. Stem cell quiescence: dynamism, restraint, and cellular idling. Cell Stem Cell. 2019;24:213–225. doi: 10.1016/j.stem.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Azari H, Deleyrolle LP, Reynolds BA. Using carboxy fluorescein succinimidyl ester (CFSE) to identify quiescent glioblastoma stem-like cells. Methods Mol. Biol. 2018;1686:59–67. doi: 10.1007/978-1-4939-7371-2_4. [DOI] [PubMed] [Google Scholar]
- 93.Zhu Z, et al. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell. 2014;15:185–198. doi: 10.1016/j.stem.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Dong J, et al. Bisacodyl and its cytotoxic activity on human glioblastoma stem-like cells. Implication of inositol 1,4,5-triphosphate receptor dependent calcium signaling. Biochim Biophys. Acta Mol. Cell Res. 2017;1864:1018–1027. doi: 10.1016/j.bbamcr.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 95.Sachdeva R, et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci. Rep. 2019;9:14569. doi: 10.1038/s41598-019-51270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ishii A, et al. Histological characterization of the tumorigenic “peri-necrotic niche” harboring quiescent stem-like tumor cells in glioblastoma. PLoS ONE. 2016;11:e0147366. doi: 10.1371/journal.pone.0147366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oki T, et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci. Rep. 2014;4:4012. doi: 10.1038/srep04012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Neant I, et al. Ca(2+)-dependent transcriptional repressors KCNIP and regulation of prognosis genes in glioblastoma. Front. Mol. Neurosci. 2018;11:472. doi: 10.3389/fnmol.2018.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamada K, et al. Essential gene pathways for glioblastoma stem cells: clinical implications for prevention of tumor recurrence. Cancers. 2011;3:1975–1995. doi: 10.3390/cancers3021975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Debruyne DN, et al. DOCK4 promotes loss of proliferation in glioblastoma progenitor cells through nuclear beta-catenin accumulation and subsequent miR-302-367 cluster expression. Oncogene. 2018;37:241–254. doi: 10.1038/onc.2017.323. [DOI] [PubMed] [Google Scholar]
- 101.Forsyth PA, et al. p75 neurotrophin receptor cleavage by alpha- and gamma-secretases is required for neurotrophin-mediated proliferation of brain tumor-initiating cells. J. Biol. Chem. 2014;289:8067–8085. doi: 10.1074/jbc.M113.513762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krebsbach PH, Villa-Diaz LG. The role of integrin alpha6 (CD49f) in stem cells: more than a conserved biomarker. Stem Cells Dev. 2017;26:1090–1099. doi: 10.1089/scd.2016.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liau BB, et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell. 2017;20:233–246 e237. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kimura T, et al. Induced expression of GINS complex is an essential step for reactivation of quiescent stem-like tumor cells within the peri-necrotic niche in human glioblastoma. J. Cancer Res. Clin. Oncol. 2019;145:363–371. doi: 10.1007/s00432-018-2797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cao Y. Adipocyte and lipid metabolism in cancer drug resistance. J. Clin. Invest. 2019;129:3006–3017. doi: 10.1172/JCI127201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jin H, et al. Targeting lipid metabolism to overcome EMT-associated drug resistance via integrin beta3/FAK pathway and tumor-associated macrophage repolarization using legumain-activatable delivery. Theranostics. 2019;9:265–278. doi: 10.7150/thno.27246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martinez-Cerdeno V, Noctor SC. Neural progenitor cell terminology. Front. Neuroanat. 2018;12:104. doi: 10.3389/fnana.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fuentealba LC, Obernier K, Alvarez-Buylla A. Adult neural stem cells bridge their niche. Cell Stem Cell. 2012;10:698–708. doi: 10.1016/j.stem.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Altmann, C., Keller, S. & Schmidt, M. H. H. The role of SVZ stem cells in glioblastoma. Cancers11, 448 (2019). [DOI] [PMC free article] [PubMed]
- 112.Lee JH, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560:243–247. doi: 10.1038/s41586-018-0389-3. [DOI] [PubMed] [Google Scholar]
- 113.Okawa S, et al. Proteome and secretome characterization of glioblastoma-derived neural. Stem Cells Stem Cells. 2017;35:967–980. doi: 10.1002/stem.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/S1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 116.Brochner CB, Mollgard K. SSEA-4 and YKL-40 positive progenitor subtypes in the subventricular zone of developing human neocortex. Glia. 2016;64:90–104. doi: 10.1002/glia.22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Batista KMP, et al. Mesenchymal/proangiogenic factor YKL-40 related to glioblastomas and its relationship with the subventricular zone. Folia Neuropathol. 2017;55:14–22. doi: 10.5114/fn.2017.66709. [DOI] [PubMed] [Google Scholar]
- 118.Daynac M, et al. Quiescent neural stem cells exit dormancy upon alteration of GABAAR signaling following radiation damage. Stem Cell Res. 2013;11:516–528. doi: 10.1016/j.scr.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 119.Llorens-Bobadilla E, et al. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 120.Ferreira AC, et al. Metabolism and adult neurogenesis: towards an understanding of the role of lipocalin-2 and iron-related oxidative stress. Neurosci. Biobehav Rev. 2018;95:73–84. doi: 10.1016/j.neubiorev.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 121.Maffezzini, C., Calvo-Garrido, J., Wredenberg, A. & Freyer, C. Metabolic regulation of neurodifferentiation in the adult brain. Cell. Mol. Life Sci. 77, 2483–2496 (2020). [DOI] [PMC free article] [PubMed]
- 122.Cavallucci V, Fidaleo M, Pani G. Neural stem cells and nutrients: poised between quiescence and exhaustion. Trends Endocrinol. Metab. 2016;27:756–769. doi: 10.1016/j.tem.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 123.Candelario KM, Shuttleworth CW, Cunningham LA. Neural stem/progenitor cells display a low requirement for oxidative metabolism independent of hypoxia inducible factor-1alpha expression. J. Neurochem. 2013;125:420–429. doi: 10.1111/jnc.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Knobloch M. The role of lipid metabolism for neural stem cell regulation. Brain Plast. 2017;3:61–71. doi: 10.3233/BPL-160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ponti G, et al. Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc. Natl Acad. Sci. USA. 2013;110:E1045–E1054. doi: 10.1073/pnas.1219563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Daynac, M. et al. Cell sorting of neural stem and progenitor cells from the adult mouse subventricular zone and live-imaging of their cell cycle dynamics. J. Vis. Exp. 2015, 53247 (2015). [DOI] [PMC free article] [PubMed]
- 127.Mich JK, et al. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife. 2014;3:e02669. doi: 10.7554/eLife.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Couturier CP, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 2020;11:3406. doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tirosh I, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schiffer, D. et al. Glioblastoma: MIcroenvironment and Niche Concept. Cancers11, 5 (2018). [DOI] [PMC free article] [PubMed]
- 131.Calabrese C, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 132.Hira VVV, Aderetti DA, van Noorden CJF. Glioma stem cell niches in human glioblastoma are periarteriolar. J. Histochem. Cytochem. 2018;66:349–358. doi: 10.1369/0022155417752676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kokubu Y, et al. Induction of protumoral CD11c(high) macrophages by glioma cancer stem cells through GM-CSF. Genes Cells. 2016;21:241–251. doi: 10.1111/gtc.12333. [DOI] [PubMed] [Google Scholar]
- 135.Herrera-Perez M, Voytik-Harbin SL, Rickus JL. Extracellular matrix properties regulate the migratory response of glioblastoma stem cells in three-dimensional culture. Tissue Eng. Part A. 2015;21:2572–2582. doi: 10.1089/ten.tea.2014.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aderetti DA, Hira VVV, Molenaar RJ, van Noorden CJF. The hypoxic peri-arteriolar glioma stem cell niche, an integrated concept of five types of niches in human glioblastoma. Biochim Biophys. Acta Rev. Cancer. 2018;1869:346–354. doi: 10.1016/j.bbcan.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 137.Zhu TS, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–6072. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Charles N, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chiao MT, et al. CD133+ glioblastoma stem-like cells induce vascular mimicry in vivo. Curr. Neurovasc Res. 2011;8:210–219. doi: 10.2174/156720211796558023. [DOI] [PubMed] [Google Scholar]
- 140.Guerra-Rebollo M, et al. Targeting of replicating CD133 and OCT4/SOX2 expressing glioma stem cells selects a cell population that reinitiates tumors upon release of therapeutic pressure. Sci. Rep. 2019;9:9549. doi: 10.1038/s41598-019-46014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guelfi S, et al. Vascular transdifferentiation in the CNS: a focus on neural and glioblastoma stem-like cells. Stem Cells Int. 2016;2016:2759403. doi: 10.1155/2016/2759403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fidoamore A, et al. Glioblastoma stem cells microenvironment: the paracrine roles of the niche in drug and radioresistance. Stem Cells Int. 2016;2016:6809105. doi: 10.1155/2016/6809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guerra DAP, et al. Targeting glioblastoma-derived pericytes improves chemotherapeutic outcome. Angiogenesis. 2018;21:667–675. doi: 10.1007/s10456-018-9621-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cheng L, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Scully S, et al. Transdifferentiation of glioblastoma stem-like cells into mural cells drives vasculogenic mimicry in glioblastomas. J. Neurosci. 2012;32:12950–12960. doi: 10.1523/JNEUROSCI.2017-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mao XG, et al. CDH5 is specifically activated in glioblastoma stemlike cells and contributes to vasculogenic mimicry induced by hypoxia. Neuro Oncol. 2013;15:865–879. doi: 10.1093/neuonc/not029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shao R, Taylor SL, Oh DS, Schwartz LM. Vascular heterogeneity and targeting: the role of YKL-40 in glioblastoma vascularization. Oncotarget. 2015;6:40507–40518. doi: 10.18632/oncotarget.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu HB, et al. Autophagy-induced KDR/VEGFR-2 activation promotes the formation of vasculogenic mimicry by glioma stem cells. Autophagy. 2017;13:1528–1542. doi: 10.1080/15548627.2017.1336277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yao X, et al. Vascular endothelial growth factor receptor 2 (VEGFR-2) plays a key role in vasculogenic mimicry formation, neovascularization and tumor initiation by Glioma stem-like cells. PLoS ONE. 2013;8:e57188. doi: 10.1371/journal.pone.0057188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Teng J, et al. Dissecting inherent intratumor heterogeneity in patient-derived glioblastoma culture models. Neuro Oncol. 2017;19:820–832. doi: 10.1093/neuonc/now253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dirkse A, et al. Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 2019;10:1787. doi: 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014;19:285–292. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Y, et al. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 2019;10:201. doi: 10.1038/s41467-018-08033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rapp C, et al. Identification of T cell target antigens in glioblastoma stem-like cells using an integrated proteomics-based approach in patient specimens. Acta Neuropathol. 2017;134:297–316. doi: 10.1007/s00401-017-1702-1. [DOI] [PubMed] [Google Scholar]
- 155.Beier CP, et al. The cancer stem cell subtype determines immune infiltration of glioblastoma. Stem Cells Dev. 2012;21:2753–2761. doi: 10.1089/scd.2011.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liebelt BD, et al. Glioma stem cells: signaling, microenvironment, and therapy. Stem Cells Int. 2016;2016:7849890. doi: 10.1155/2016/7849890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou W, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wei J, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol. Cancer Ther. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wei J, et al. Hypoxia potentiates glioma-mediated immunosuppression. PLoS ONE. 2011;6:e16195. doi: 10.1371/journal.pone.0016195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lau J, et al. STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res. 2015;75:4302–4311. doi: 10.1158/0008-5472.CAN-14-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gouaze-Andersson V, et al. FGFR1/FOXM1 pathway: a key regulator of glioblastoma stem cells radioresistance and a prognosis biomarker. Oncotarget. 2018;9:31637–31649. doi: 10.18632/oncotarget.25827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Goffart N, et al. CXCL12 mediates glioblastoma resistance to radiotherapy in the subventricular zone. Neuro Oncol. 2017;19:66–77. doi: 10.1093/neuonc/now136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hu B, et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell. 2016;167:1281–1295.e1218. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bhuvanalakshmi G, et al. Secreted frizzled-related protein 4 inhibits glioma stem-like cells by reversing epithelial to mesenchymal transition, inducing apoptosis and decreasing cancer stem cell properties. PLoS ONE. 2015;10:e0127517. doi: 10.1371/journal.pone.0127517. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 165.Liu G, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kang MK, Kang SK. Tumorigenesis of chemotherapeutic drug-resistant cancer stem-like cells in brain glioma. Stem Cells Dev. 2007;16:837–847. doi: 10.1089/scd.2007.0006. [DOI] [PubMed] [Google Scholar]
- 167.Beier D, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68:5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 168.Jhaveri N, et al. A novel drug conjugate, NEO212, targeting proneural and mesenchymal subtypes of patient-derived glioma cancer stem cells. Cancer Lett. 2016;371:240–250. doi: 10.1016/j.canlet.2015.11.040. [DOI] [PubMed] [Google Scholar]
- 169.Zhang Y, et al. EGCG inhibits properties of glioma stem-like cells and synergizes with temozolomide through downregulation of P-glycoprotein inhibition. J. Neurooncol. 2015;121:41–52. doi: 10.1007/s11060-014-1604-1. [DOI] [PubMed] [Google Scholar]
- 170.Morfouace M, et al. Comparison of spheroids formed by rat glioma stem cells and neural stem cells reveals differences in glucose metabolism and promising therapeutic applications. J. Biol. Chem. 2012;287:33664–33674. doi: 10.1074/jbc.M111.320028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wheaton WW, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Esparza R, et al. Glioblastoma stem cells and stem cell-targeting immunotherapies. J. Neurooncol. 2015;123:449–457. doi: 10.1007/s11060-015-1729-x. [DOI] [PubMed] [Google Scholar]
- 173.Finocchiaro G, Pellegatta S. Immunotherapy with dendritic cells loaded with glioblastoma stem cells: from preclinical to clinical studies. Cancer Immunol. Immunother. 2016;65:101–109. doi: 10.1007/s00262-015-1754-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kouidhi S, Ben Ayed F, Benammar Elgaaied A. Targeting tumor metabolism: a new challenge to improve immunotherapy. Front Immunol. 2018;9:353. doi: 10.3389/fimmu.2018.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Neidert MC, et al. The natural HLA ligandome of glioblastoma stem-like cells: antigen discovery for T cell-based immunotherapy. Acta Neuropathol. 2018;135:923–938. doi: 10.1007/s00401-018-1836-9. [DOI] [PubMed] [Google Scholar]
- 176.Leone RD, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Castro BA, Aghi MK. Bevacizumab for glioblastoma: current indications, surgical implications, and future directions. Neurosurg. Focus. 2014;37:E9. doi: 10.3171/2014.9.FOCUS14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mei X, et al. Glioblastoma stem cell differentiation into endothelial cells evidenced through live-cell imaging. Neuro Oncol. 2017;19:1109–1118. doi: 10.1093/neuonc/nox016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rick J, Chandra A, Aghi MK. Tumor treating fields: a new approach to glioblastoma therapy. J. Neurooncol. 2018;137:447–453. doi: 10.1007/s11060-018-2768-x. [DOI] [PubMed] [Google Scholar]
- 180.Ballo MT, et al. Correlation of tumor treating fields dosimetry to survival outcomes in newly diagnosed glioblastoma: a large-scale numerical simulation-based analysis of data from the phase 3 EF-14 randomized trial. Int. J. Radiat. Oncol. Biol. Phys. 2019;104:1106–1113. doi: 10.1016/j.ijrobp.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 181.Stupp R, et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314:2535–2543. doi: 10.1001/jama.2015.16669. [DOI] [PubMed] [Google Scholar]
- 182.Clark PA, et al. The effects of tumor treating fields and temozolomide in MGMT expressing and non-expressing patient-derived glioblastoma cells. J. Clin. Neurosci. 2017;36:120–124. doi: 10.1016/j.jocn.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kanabur P, et al. Patient-derived glioblastoma stem cells respond differentially to targeted therapies. Oncotarget. 2016;7:86406–86419. doi: 10.18632/oncotarget.13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li M, Deng H, Peng H, Wang Q. Functional nanoparticles in targeting glioma diagnosis and therapies. J. Nanosci. Nanotechnol. 2014;14:415–432. doi: 10.1166/jnn.2014.8757. [DOI] [PubMed] [Google Scholar]
- 185.Pohlmann ES, et al. Real-time visualization of nanoparticles interacting with glioblastoma stem cells. Nano Lett. 2015;15:2329–2335. doi: 10.1021/nl504481k. [DOI] [PubMed] [Google Scholar]
- 186.Iannolo G, et al. Zika virus infection induces MiR34c expression in glioblastoma stem cells: new perspectives for brain tumor treatments. Cell Death Dis. 2019;10:263. doi: 10.1038/s41419-019-1499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhu Z, et al. Correction: Zika virus has oncolytic activity against glioblastoma stem cells. J. Exp. Med. 2017;214:3145. doi: 10.1084/jem.2017109309122017c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhu Z, et al. Zika virus targets glioblastoma stem cells through a SOX2-Integrin alphavbeta5 Axis. Cell Stem Cell. 2020;26:187–204.e110. doi: 10.1016/j.stem.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang S, Xiong X, Sun Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct. Target Ther. 2020;5:135. doi: 10.1038/s41392-020-00242-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sahoo P, et al. Early changes in tumor perfusion from T1-weighted dynamic contrast-enhanced MRI following neural stem cell-mediated therapy of recurrent high-grade glioma correlate with overall survival. Stem Cells Int. 2018;2018:5312426. doi: 10.1155/2018/5312426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vik-Mo EO, et al. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol. Immunother. 2013;62:1499–1509. doi: 10.1007/s00262-013-1453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grande S, et al. Metabolic heterogeneity evidenced by MRS among patient-derived glioblastoma multiforme stem-like cells accounts for cell clustering and different responses to drugs. Stem Cells Int. 2018;2018:3292704. doi: 10.1155/2018/3292704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Qin S, et al. Emerging role of tumor cell plasticity in modifying therapeutic response. Signal Transduct. Target Ther. 2020;5:228. doi: 10.1038/s41392-020-00313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]