Graphical abstract
Abbreviations: AMR, antimicrobial resistance; HMW, high molecular weight; LCA, lowest common ancestor; MAGs, Metagenome assembled genomes; NGS, next-generation sequencing; ONT, Oxford Nanopore Technologies; PAIs, pathogenicity islands; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; UMAP, Uniform Manifold Approximation and Projection
Keywords: Nanopore sequencing, Targeted sequencing, Metagenomics, Metatranscriptomics, Bioinformatics
Abstract
Since its introduction, nanopore sequencing has enhanced our ability to study complex microbial samples through the possibility to sequence long reads in real time using inexpensive and portable technologies. The use of long reads has allowed to address several previously unsolved issues in the field, such as the resolution of complex genomic structures, and facilitated the access to metagenome assembled genomes (MAGs). Furthermore, the low cost and portability of platforms together with the development of rapid protocols and analysis pipelines have featured nanopore technology as an attractive and ever-growing tool for real-time in-field sequencing for environmental microbial analysis. This review provides an up-to-date summary of the experimental protocols and bioinformatic tools for the study of microbial communities using nanopore sequencing, highlighting the most important and recent research in the field with a major focus on infectious diseases. An overview of the main approaches including targeted and shotgun approaches, metatranscriptomics, epigenomics, and epitranscriptomics is provided, together with an outlook to the major challenges and perspectives over the use of this technology for microbial studies.
1. Introduction
The study of microbial communities, including bacteria, viruses, archaea and fungi, is crucial to understand important aspects of the environment and/or human health. Over the last decades, there has been an important shift in the way microbial communities have been explored due to the introduction of sequencing technologies. In the late ’80s, scientists realized that the world of uncultured microorganisms outsized the cultured world, and the analysis of DNA sequences replaced culturing for the study of complex microbial communities [1], [2]. Since then, the advent of next-generation sequencing (NGS) technologies has unquestionably led to a real revolution in the area of microbiology, with major breakthroughs the complete characterization of the human gut microbiome [3] or the identification of novel phyla with undiscovered biology [4], to name a few. The introduction of third-generation sequencing represented another major turning point in the field, because it opened the possibility for real-time sequencing of long reads. The marketed technologies that currently dominate this field are single-molecule real-time sequencing (SMRT, commercialized by Pacific Biosciences) and nanopore sequencing (NS, commercialized by Oxford Nanopore Technologies (ONT)).
ONT nanopore sequencing allows single molecule sequencing based on bioengineered nanopores, which are embedded into an electrically resistant membrane where a voltage is applied. When a single stranded DNA/RNA fragment passes through the nanopore, it causes a change in electrical current through the membrane that is translated into a specific sequence of nucleotides using recurrent neural network (RNN)-based algorithms. The first marketed device to sequence by nanopores, the MinION sequencer, was introduced in 2014 and is a small and inexpensive device (starting pack available for 1000$) [5]. A single flow cell of MinION contains up to 2,048 nanopores which are controlled in groups of 512 via an application specific integrated circuit (ASIC). It can be directly plugged into a portable computer allowing real-time acquisition and analysis of data, making it the first sequencer to enable in-field sample genomic characterization [6]. Later on, other three devices were introduced by ONT, the GridION and the PromethION, which allow parallel running of up to five and 48 flow cells, respectively, and Flongle, a smaller adaptor device for running smaller experiments both on MinION and GridION.
Since nanopore sequencing does not need to occur in amplified DNA, very long reads (more than 2 Mb [7]) can be generated, with no theoretical limit in read length [8]. The possibility of obtaining these very long reads has allowed significant improvements in diverse applications such as de novo genome assembly [9], and in the deeper characterization of repetitive DNA elements [10], [11], which could not be resolved relying only on existing short-read NGS technologies. Furthermore, because sequencing is mediated by the translation of an electrical signal into a sequence of nucleotides, nanopore sequencing allows the identification of native base modifications [12] and the direct sequencing of RNA molecules [13]. The main drawback of this technology has been the high error rate [14]. However, this has been continuously improved over the years, with a current modal raw read accuracy of 97% with the latest released flow cell [15]. Furthermore, the continuous development of novel basecallers and bioinformatic tools for read error correction (polishing) and consensus generation has dramatically helped to further improve this aspect. As a consequence, the impact of nanopore read errors on taxonomic classification and other microbial analyses is limited. Sequencing throughput is also very variable. At the moment, around 30 Gb of data can theoretically be generated using a MinION flow cell, corresponding to the sequencing of 25 E. coli genomes with a coverage of 100×, and up to 200 Gb of data per flow cell using a PromethION system, allowing to sequence a minimum of one human genome with a coverage of around 30×. Thus, through parallel run of 48 flow cells, the acquisition of around 9600 Gb of DNA/RNA data is theoretically possible at the moment, corresponding to the sequencing of a minimum of 48 human genomes in a single run.
Box 1. Basic concepts.
Taxonomic profiling: type of analysis aiming at the identification of taxa present in a sample together with their relative abundances. It is typically performed using marker genes which enable the discrimination between different taxa. It answers the question: “who is there?”
Functional analysis: the study of the metabolic and other biological pathways related to the taxa present in a sample. It is usually performed by comparing gene sequences to functional databases. It answers the question: “what are they doing?”
Operational Taxonomic Unit (OTU): a group of closely related individuals which are arranged together based on the similarity of specific sequences (usually the 16S rRNA gene).
Basecalling: computational process to assign nucleotides to sequence from the raw electric signal data (squiggle) generated by a nanopore sequencing device.
Quality control: set of read filtering steps prior to analysis which usually consist of read-length and read quality filtering.
Contig: a series of overlapping sequences used to reconstruct the original DNA sequence of a genomic region.
Polishing: it refers to the analytical process aiming to improve the base accuracy of a contig.
k-mer: subsequences of length k that are contained within a sequence. k-mer frequencies encompass features that are characteristic of particular organisms and are suitable for taxonomic binning or microbial composition inference.
Genome or metagenomic assembly: computational process for reconstructing individual contigs from a genomic or metagenomic dataset that spans complete or nearly-complete microbial genes and genomes.
Assembly graph: graph representation of the final assembly of a genome or metagenome based on read overlap or k-mer information and including all possible paths for contig reconstruction.
2. Targeted 16S rRNA gene sequencing
Metataxonomics is defined as the process of characterization of the microbiota through the amplification and sequencing of conserved marker genes followed by the assignment of the generated reads to specific taxonomic levels, and/or the construction of a taxonomic tree [16]. Metataxonomics is commonly differentiated from metagenomics, which is defined as the study of the totality of genomes of the microbiota through shotgun sequencing techniques. An explanation of the basic concepts related to microbial study and bioinformatic analysis is given in Box 1. A general workflow of experimental and bioinformatics steps for targeted- and metagenomics studies is presented in Fig. 1. The most commonly used marker genes in metataxonomics are the 16S rRNA for bacteria and archaea, and the 18S rRNA for fungi, while there are no marker genes for viruses, even though they are an integral part of the microbiota. The 16S rRNA gene has been historically used in the taxonomic classification of known and new microbial taxa and in phylogenetic analysis [17], [18]. The 16S rRNA gene spans ~1500 bp including nine hypervariable segments (V1-V9) flanked by highly conserved regions which can be targeted by universal primers for amplification. The variable regions allow to distinguish between different taxa. A similarity threshold of 98.65% in the 16S rRNA gene sequences has been identified to discriminate between two species [19]. These characteristics make the 16S rRNA gene the gold standard for microbial classification [20]. Compared to the NGS technologies, where only short segments of the gene encompassing one or several hypervariable regions can be targeted, long-read sequencing allows amplification and analysis of the full-length 16S rRNA gene, which provides a more realistic representation of the taxa in a sample [21]. In fact, despite the higher error rate characterizing nanopore sequencing, the increased read length achieved through the full-length 16S rRNA gene sequencing allows species-level classification, improving taxa resolution over previous technologies [22], [23], [24], [25]. Furthermore, the use of clustering-based algorithms (e.g. NanoCLUST [25]) and other polishing tools allows overcoming the read error rate, with the generation of highly accurate consensus sequences of the 16S rRNA genes that are now able to discriminate between species.
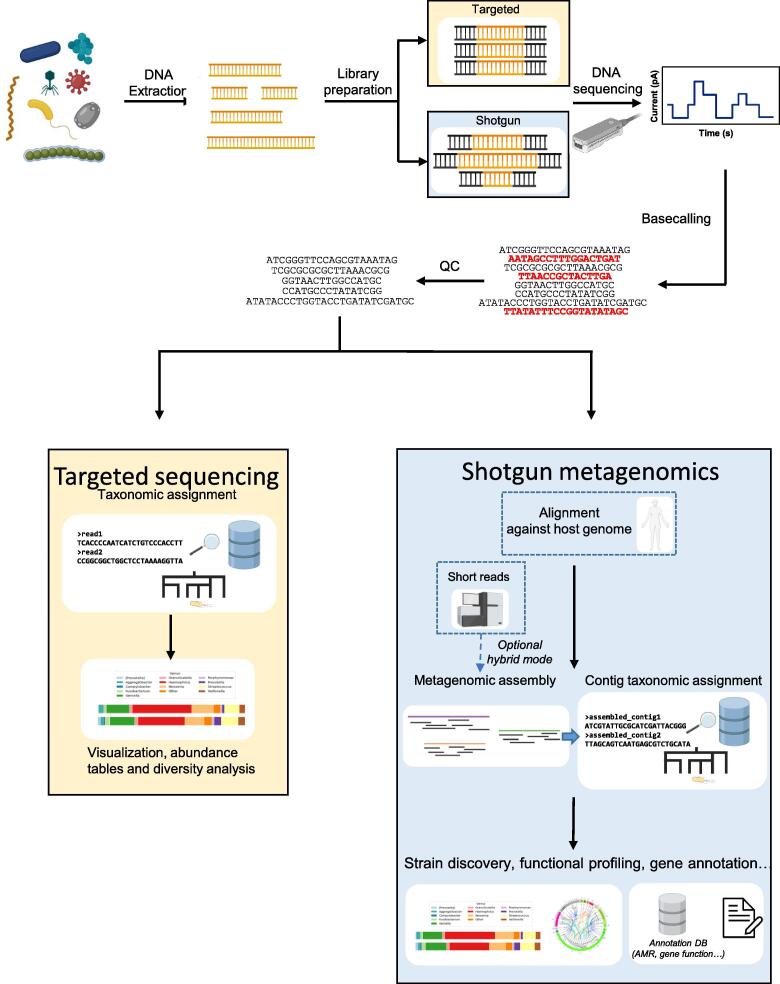
Fig. 1.
General workflow for generation and analysis of data for targeted and shotgun metagenomics studies. Created with biorender.com. QC: quality control.
2.1. Protocols and libraries
ONT allows sequencing with portable devices, opening the possibility for the use of 16S rRNA gene sequencing for rapid in-field pathogen detection. In particular, ONT has developed two 16S barcoding kits for rapid sequencing of full-length 16S rRNA genes, which allow simultaneous sequencing of up to 12 or 24 samples (Table 1). The library preparation protocol for both kits has the advantage to be fast (< 2 h) and easy to perform, and consists of an amplification step where barcodes are added to the amplicons, followed by the attachment of adapters necessary to mediate molecule entrance into the nanopores on the flow cell. Alternatively, when multiplexing of more than 24 samples is needed, it is possible to use the standard PCR barcoding amplicon kit, which currently enables simultaneous sequencing of up to 96 samples in a single experiment.
Table 1.
Summary of available ONT library preparation kits for sequencing of microbial communities.
| ONT library preparation strategy | Input ng recommendation | Preparation time | Multiplexing | Application |
|---|---|---|---|---|
| 16S Rapid Barcoding Kit | < 10 ng gDNA | 10 min + PCR | Up to 12 or 24 samples | Targeted 16S rRNA gene sequencing |
| Rapid Sequencing Kit | ≥ 400 ng HMW DNA | 10 min | Up to 12 samples | Metagenomics and epigenomics, amplification-free |
| Rapid PCR Sequencing Kit | ≤ 10 ng gDNA | 15 min + PCR | Up to 12 samples | Metagenomics, requires amplification |
| Ligation Sequencing Kit | ≥ 1000 ng dsDNA | 60 min | Up to 96 samples | Metagenomics and epigenomics, amplification-free, high-throughput |
| PCR Sequencing Kit | ≤ 100 ng gDNA | 60 min + PCR | Up to 12 samples | Metagenomics, requires amplification, high-throughput |
| Direct cDNA Sequencing Kit | 100 ng poly-A+ RNA | 270 min | Up to 24 samples | Metatrascriptomics, requires retrotranscription |
| PCR cDNA Sequencing Kit | 1 ng poly-A+ or 50 ng total RNA | 165 min | Up to 12 samples | Metatranscriptomics, requires retrotranscription and amplification |
| Direct RNA Sequencing Kit | 500 ng poly-A+ RNA | 105 min | None | Metatrascriptomics and epitranscriptomics, retrotranscription- and amplification-free |
HMW: high-molecular weight.
A few considerations need to be made before embarking on a 16S rRNA gene sequencing experiment. First of all, only bacterial and archaea communities are identified using this approach, while viruses and fungi are missed. Alternatively, the mycobiome (the fungal microbiota) can be studied using the 18S rRNA gene or the internal transcribed spacer (ITS) as markers. However, ONT kits specifically targeting these regions have not yet been developed, and this may have limited the study of the fungal communities using this technology. Secondly, although the PCR step during library preparation increases the chances to detect low abundant taxa, it is well-known that the PCR introduces biases in taxonomic classification and estimation of relative abundances [26]. In fact, Kai et al. reported that Bifidobacterium is not detected by the ONT 16S rRNA library kit, due to the lack of annealing of the universal primers to the flanking regions of the 16S rRNA gene of this taxon [27]. They further addressed this bias by changing the reverse primer sequence to target all taxa present in their sample [28]. An additional source of bias to consider is the differential number of rrn operons in the genomes of different taxa, which often leads to inaccuracies in the estimation of the abundance profiles. Although algorithms for 16S rRNA gene copy number normalization have often been used to overcome this bias, it has recently been proved that they fail to provide a more reliable picture of the community composition in metataxonomics studies [29].
2.2. Bioinformatic analysis
There are a broad range of bioinformatics pipelines for metagenomics which are also valid for metataxonomic analysis (Table 2), including the widely used multi-purpose pipelines based on Operational Taxonomic Unit (OTU) picking and/or Amplicon Sequence Variants (ASV) analysis [30], [31]. Most of these were initially developed to work with short-read data (particularly for those from Illumina) and are not suitable for nanopore read lengths and error profiles, commonly leading to issues such as an overestimation of taxa diversity. Hence, the potential benefits of performing taxonomic classification with full-length 16S rRNA reads have not been extensively explored. Therefore, the rapid changes in the sequencing technology have outpaced the availability of specific tools and benchmark studies of nanopore 16S rRNA reads.
Table 2.
Main long-read bioinformatics tools for targeted and shotgun approaches.
| Type | Reference | Application | Brief description |
|---|---|---|---|
| Aligners/Alignment-based classifiers | |||
| BLAST, MEGABLAST | [58], [59] | Targeted; Shotgun | Gold-standard alignment tools for classification of nucleotide and protein sequences. Feature web-based version and multiple implementations for specific purposes. |
| minimap2 | [33] | Targeted; Shotgun | Versatile tool for fast read alignments against large reference databases. |
| Alignment-free classifiers | |||
| Kraken, Kraken2 | [35], [64] | Targeted; Shotgun | Taxonomic classification tool implementing an accurate and fast k-mer matching. |
| KrakenUniq | [65] | Shotgun | Classifier that combines Kraken classification tool with the assessment of the coverage of unique k-mers for better recall and precision. |
| Bracken | [66] | Targeted; Shotgun | Relative abundance estimation tool for single-level abundance using Kraken read classification output. |
| Metamaps | [69] | Shotgun | Read assignment and sample composition estimation for nanopore metagenomic datasets. |
| Centrifuge | [34] | Targeted; Shotgun | Read classification based on the Burrows-Wheeler transform (BWT) and the Ferragina-Manzini (FM) index that performs fast classification relying on small pre-computed index databases. |
| Mash | [72] | Targeted; Shotgun | Fast genome and metagenome distance estimation tool that computes distances between sequences using the MinHash algorithm. |
| Long-read assemblers | |||
| Canu | [90] | Shotgun | Assembly pipeline for long-reads that compute and process read overlaps for the generation of contigs and draft assemblies. |
| miniasm | [73] | Shotgun | Fast OLC-based assembler for long reads that builds assembly graphs from all-vs-all read mappings. |
| wtdbg2 | [91] | Shotgun | De-novo sequence assembler for uncorrected long-reads based on Fuzzy Bruijn graphs to compute contigs. |
| OPERA-MS | [95] | Shotgun | Hybrid metagenomic assembler that first performs a short-read assembly and then maps short and long reads to resolve contiguity of contigs. |
| MetaFlye | [96] | Shotgun | Metagenomic assembler from the Flye package featuring repeat graphs to compute high-quality metagenome assemblies. |
| MetaSPAdes | [74] | Shotgun | Metagenomic assembly module from SPAdes assembler that features a hybrid assembly option. |
| Sequence correction and polishing tools | |||
| Nanopolish | https://github.com/jts/nanopolish | Targeted; Shotgun | Signal-level analysis tool with modules that performs sequence polishing, base modification detection and variant calling. |
| Medaka | https://github.com/nanoporetech/medaka | Targeted; Shotgun | Neural network-based sequence correction and variant calling tool. |
| Metagenomic analysis pipelines | |||
| MEGAN-LR | [60] | Shotgun | Long-read implementation of the MEGAN workflow. Features taxonomic and functional analysis. |
| NanoCLUST | [25] | Targeted | Analysis pipeline for UMAP-based classification of amplicon-based full-length 16S rRNA nanopore reads. |
| Reticulatus | https://github.com/SamStudio8/reticulatus | Shotgun | Snakemake-based pipeline for assembly and polishing of long genomes from long nanopore reads. |
| MUFFIN | [70] | Shotgun | Metagenomics workflow for hybrid assembly, differential coverage binning, transcriptomics and pathway analysis. |
| NanoSPC | [71] | Shotgun | Metagenomic analysis pipeline that includes viral and bacterial pathogen identification, genome assembly and variant calling. |
| BusyBee | https://ccb-microbe.cs.uni-saarland.de/busybee/ | Shotgun | Web-based metagenomic analysis pipeline for long-reads and contigs that features taxonomic and functional annotation of AMR elements along with a comprehensive visualization of results. |
Computational methods for nanopore 16S rRNA analysis have been reviewed recently [32]. Depending on the chosen approach to classify the sequences, read classification techniques can be categorized into alignment-based and alignment-free methods. EPI2ME (ONT) is the most extensively used analysis pipeline for nanopore 16S rRNA. It covers end-to-end analysis of nanopore 16S rRNA data in a cloud-based environment and includes demultiplexing, quality filtering and taxonomic assignment using the BLAST tool against the NCBI database. The main drawback of using EPI2ME is its limited possibility to customize workflow parameters, such as reference databases and alignment options. Furthermore, this tool can only be accessed by ONT customers through a web application and the output data format is incompatible with other software for downstream analysis, highlighting the need for the development of alternatives based on available open-source tools. Simpler workflows proceed with the alignment of the input sequences using tools designed to work efficiently with long noisy reads, like minimap2 [33], against specifically designed 16S rRNA databases which contain only curated 16S rRNA sequences and their taxonomy (Table 3). Additionally, alignment-free methods like Centrifuge [34] or Kraken [35], [64] (described in more detail in following sections) have emerged as feasible options for taxonomic classification of nanopore 16S rRNA reads. A recently proposed approach, NanoCLUST, relies on the Uniform Manifold Approximation and Projection (UMAP) algorithm to cluster full-length 16S rRNA reads and then classifies a representative polished sequence from each cluster to deliver abundance profiles at different taxonomic levels [25].
Table 3.
Summary of the most widely used reference databases for metataxonomics and metagenomics analysis.
| Database name | Reference | Description |
|---|---|---|
| Metataxonomics databases | ||
| GreenGenes | [77] | 16S rRNA database from Genbank sequences, manually curated and modified by the user community. |
| SILVA | [78] | Small and large rRNA subunits database including 16S rRNA sequences from the European Nucleotide Archive. |
| The Ribosomal Database Project (RDP) | [79] | 16S rRNA taxonomically annotated sequence collection from the INSDC database. |
| RefSeqTargeted Loci Project | (https://www.ncbi.nlm.nih.gov/refseq/targetedloci/) | BLAST specific marker gene databases for Bacteria (16S/23S) and Fungi (28S/18S) extracted and curated from GenBank sequences. |
| Metagenomics databases | ||
| nt/nr | [58] | Default database for BLAST sequence searches including RefSeq RNA and GenBank sequences. |
| RefSeq | [75] | Non-redundant and NCBI curated and annotated database based on Genbank sequences. |
| GenBank | [76] | Main NCBI nucleotide database with the largest complete and draft microbial genomes sequence collection. |
| Annotation and functional databases | ||
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | [81] | Manually curated set of 18 databases for annotating cellular and organism-level functions from nucleotide sequences. |
| Integrated reference catalog of the human gut microbiome (IGC) | [86] | Gut-specific annotated microbial genes from KEGG functional databases. |
| Comprehensive Antibiotic Resistance Database (CARD) | [82] | Bioinformatic resources and database for the annotation of antimicrobial resistance genes (AMR) and mutations from genomic sequences. |
| DeepARG-DB | [87] | Antibiotic resistance genes database generated by a deep-learning prediction algorithm trained with ARG from other sequence collections. |
| MEGARes | [83] | Hand-curated database containing AMR genes optimized for use with high-throughput sequencing data. |
2.3. Potential utility in clinical applications
ONT nanopore sequencing has opened the possibility for the first time to sequence and analyse data in real-time at competitive costs. A major application in hospital settings is the rapid diagnostics of infectious diseases to allow prompt patient management and appropriate treatment decisions. A summary of the successful clinical applications of ONT nanopore sequencing in infectious diseases is given in Table 4. Proof-of-principle studies have been conducted leveraging ONT 16S rRNA targeted sequencing for the diagnosis of bacterial infections. In an early study, Mitsuhashi et al. developed a protocol for rapid characterization of bacterial composition using a mock community, which was then evaluated on a pleural effusion sample from a patient with empyema [36]. The protocol was based on 16S rRNA gene sequencing followed by analysis with BLAST-based searching or Centrifuge classification. They efficiently characterized the mock community at the species level using BLAST against the GenomeSync database, while Centrifuge missed the identification of one of the species. By comparing results at different times during the sequencing run, they concluded that 5 min of sequencing time was enough to obtain a sufficient amount of data to identify all bacteria taxa in the sample, and to reach a sensitivity >90% using BLAST. However, when the rapid protocol was tested on the clinical sample, a longer sequencing time was necessary to identify low abundant taxa. Later studies adopted a similar analysis protocol (replacing BLAST by minimap2) to conduct point-of-care diagnostics, both in developed [37] and in resource-poor countries [38]. In particular, this last study was conducted on cerebrospinal fluid samples from eleven patients with bacterial meningitis in Zambia, where a portable sequencing-based system was set up. Although they could successfully confirm the results from culture-based methods on four samples, two positive samples showed different bacterial compositions using the MinION sequencer, and these results were not confirmed using any additional method. Furthermore, culture-negative samples were positive using MinION, suggesting higher sensitivity of the sequencing method compared to the traditional culturing. However, no further validation with complementary methods was carried out.
Table 4.
Summary of main successful clinical applications of NS.
| Clinical application of NS | Approach used | Reference |
|---|---|---|
| Rapid pathogen identification in clinical samples | 16S rRNA targeted | [36], [37], [38] |
| RNA sequencing | [118] | |
| Rapid identification of pathogens and AMR genes | Shotgun metagenomics | [97], [98] |
| Surveillance of pathogens and AMR in hospital settings | Shotgun metagenomics | [99] |
| Genomic surveillance for viral outbreaks | RNA sequencing | [119], [120], [124], [125] |
For the adoption of the MinION sequencer for rapid diagnostics of infectious diseases in clinical settings, it is necessary for future studies to be conducted with standardized protocols on large sample size, followed by appropriate cross-validation of the results. This issue was partially addressed by Neuenschwander et al. who developed LORCAN, a standardized laboratory protocol and automated pipeline for taxonomic identification of bacterial mixtures based on the ONT 16S rRNA sequencing [39]. The library consists of PCR amplicons from regions of the 16S rRNA gene (length between 500 and 1000 bp) and the pipeline generates consensus sequences that improve the accuracy of taxonomic classification. The workflow was tested on culture isolates and artificial mock communities from read or amplicon mixing. Analysis of samples using LORCAN generated consensus sequences with a sequence identity of 99.6% to their corresponding Sanger-obtained sequences and a turnaround time from raw amplicons to reports of about 8 h. While this workflow has the potential to be used in the clinics, it has not been tested yet on clinical samples, leaving open the question of whether it could truly be adopted in real settings.
3. Metagenomics
Metagenomics allows deep investigations of microbial communities, usually encompassing taxonomic analysis, functional profiling and whole-genome assembly. In contrast to targeted approaches, the entire genomic content of a microbial sample is sequenced, providing greater genomic information [40]. In fact, strain-level information can be accurately recovered with the metagenomic approach, and helps to find the associations between phylogeny and function [41]. Furthermore, metagenomics enables the identification of antimicrobial resistance (AMR) genes or other virulence elements that can be assigned to specific pathogens through genome assembly. It also helps to identify viral communities that are missed using targeted sequencing approaches since their genomes lack consensus sequences that are necessary for universal primer attachments during the library preparation stage [42]. Nanopore sequencing has led to important improvements in the metagenomic analysis because reads can span extended areas of the genome. In fact, long reads can resolve complex genomic structures such as repetitive elements [43], and allow to identify the position and organization of bacterial pathogenicity islands (PAIs) encoding virulence factors [44]. This long-range genomic information has been particularly relevant for de novo and metagenomics assemblies where it enabled to resolve areas of the genomes where short reads failed. For example, ONT sequencing had allowed to localize AMR gene positions in Klebsiella pneumoniae [45], to reconstruct plasmid genomes in Enterobacteriaceae isolates [46], [47], and to circularize the genome of Bordetella pertussis isolates [48], as well as to assemble the human gut microbiome [43], [49]. The accurate and complete reconstruction of genomes is important for the identification of gene-genome associations in functional and evolutionary studies that, for example, aim to shed light on the genomic organization of metabolic pathways or on the horizontal gene transfer.
3.1. Protocols and libraries
Currently, there are four ONT library preparation kits available for metagenomic studies that differ in the input DNA quantity, preparation time, and throughput (Table 1). The Rapid Sequencing Kit allows fast library preparation (~10 min) and requires an input of > 400 ng of high molecular weight (HMW) genomic DNA (> 30 kb). Multiplexing up to 12 samples can be achieved using the Rapid Barcoding Kit. When a lower starting amount of DNA is available (< 10 ng), the alternative Rapid PCR Barcoding Kit can be used, which includes a PCR step for the amplification of the target DNA and for the attachment of barcodes for multiplexing (up to 12 samples). By using this kit, library preparation requires ~15 min added to the time for the PCR step, and read length distribution is centred at around 2 kb. These library preparation kits use a transposase for fragmentation of the HMW genomic DNA and the attachment of transposase adapters which can then be used as anchors for sequencing adapters or as binding sites for PCR primers in the Rapid PCR Barcoding Kit. When higher throughput is needed, ONT suggests the use of other two library preparation kits. The Ligation Sequencing Kit, which requires ~1 μg of starting HMW DNA and consists of a protocol lasting ~60 min, allowing the maximum throughput achievable while retaining the possibility to call base modifications. DNA molecules are nick-repaired and dA tailed, and then sequencing adapters are ligated onto the prepared ends. This kit can be combined with upstream processes such as target enrichment by capture, size selection, or whole genome amplification (when < 1 ng of original DNA is available), and multiplexing of up to 96 samples can be achieved by using the Native Barcoding Expansion kits. When lower starting material is accessible or sample purity is compromised, the PCR Sequencing Kit can be adopted. In this protocol, the original DNA is fragmented, sheared ends are repaired and dA tailed, adapters containing primer binding sites are ligated and an amplification step follows using primers containing tags for ligase-free attachment of rapid sequencing adapters. The mean read length distribution of the protocol is larger than that obtained by the Rapid PCR Sequencing Kit and is limited only by the processivity of the DNA polymerase. This kit requires around 60 min of hands-on time, added to that for the PCR step, which is variable depending on the number of cycles, template length, and polymerase speed. Multiplexing of up to 12 samples is possible by integrating the PCR Barcoding Kit in the protocol, which uses barcoded primers during the PCR step.
The sequencing data yield will depend on the experimental aim and on the sample analysed. Since the recommended coverage varies depending on the aim of the study – the recommended minimal depth of coverage is 10× for detection of taxa, 20× for taxonomic assignation and AMR gene analysis, and 30× for genome assembly [50]- the sequencing data necessary to achieve that specific goal is also variable. As an example, to assemble a genome of 3 Mb, 90 Mb of data from that genome is needed. If 1 Gb of data is sequenced, it will be possible to assemble genomes representing up to 9% of the total data. In addition, the presence of host DNA in the sample is also likely to reduce the amount of data related to the metagenome itself, requiring a larger amount of sequencing data. Host DNA depletion protocols can overcome this issue, which is especially relevant in clinical samples (e.g. respiratory specimens and swabs) where up to 95% of reads could be host-derived [51]. Examples of host depletion protocols include saponin, molYsis kits (Molzym, Germany), or kits for rRNA depletion. In particular, Charalampous et al. have recently optimized a metagenomic protocol for the detection of lower respiratory infections which includes a saponin-based depletion step removing up to the 99.99% of host nucleic acids and enables profiling of pathogen and AMR genes within 6 h [52]. They achieved a sensitivity of 96.6% and a limit of detection similar to that of culture-based methods.
3.2. Bioinformatic analysis
Determining the taxonomic entities present in a sample from a metagenomic sequencing dataset is a key step in metagenomics studies. The assignment of taxonomic labels to the sequencing reads and the subsequent inference of the composition of a microbial community are increasingly popular research areas due to the growing use of high-throughput technologies demanding more accurate and efficient tools for metagenomic analyses (Table 2). Generally, long reads enable better taxonomic and functional analysis than short reads due to the higher information content enclosed in the sequence. Yet, most of the widely adopted metagenomic classification tools or pipelines often rely on algorithms built on short reads which, by default, do not scale well with long-read datasets – ranging from 13 kb up to 2 Mb – and/or do not account for the higher error profile of nanopore reads. The inclusion of long-read datasets in benchmarking studies and bioinformatic software updates have provided some guidance for metagenomic tool suitability and performance with nanopore reads [53]. Furthermore, long-read specific tools are being continuously developed [54], including error correction methods [55] and hybrid approaches to overcome read error-related issues [56], [57].
Traditional read classification methods are based on the detection of similarities between sequencing reads and genomes from known organisms through an initial alignment against databases containing taxonomic information. Of these, the most popular are classification tools based on the classic BLAST algorithm [58], which remains the gold standard for the taxonomic assignment task. Apart from the classic nucleotide BLAST, more recently developed methods like MEGABLAST [59] provide faster alignments. Alternative methods built upon BLAST or other alignment tools have also been proposed to improve classification results combining sequence alignment of input reads with machine learning techniques for taxonomic resolution at different levels. For example, MEGAN-LR [60] expands the functionality of the interactive metagenomics pipeline featuring long-read approaches for taxonomic and functional analysis. It adopts alignment-based comparisons using LAST aligner [61] to compute frameshift-aware DNA-to-protein alignments and applies a custom lowest common ancestor (LCA) algorithm to resolve taxonomy and deliver classification results.
Alignment-based techniques also output useful information for results interpretation such as genomic locations and qualities of alignments. This feature usually comes at the cost of an increase in required computing resources and time when analyzing long-read datasets. Because of this, in the last few years, alignment-free classification methods have become popular for the analysis of short- and long-read datasets. These methods mainly rely on a k-mer based classification against precomputed indexes and guarantee the efficient search and storage of sequence databases. As a result, most of these tools are capable of classifying millions of reads per minute with a relatively small memory footprint and enable the analysis of extensive long-read datasets. A major limitation of k-mer based methods is that they are sensitive to low error rates and may lead to misclassifications when used with error-prone long reads, especially when classifying similar organisms at the species level or organisms that have high sequence identity. However, the continuous chemistry updates and the release of novel basecalling algorithms, such as Bonito (https://github.com/nanoporetech/bonito) [62], [63], have improved the raw read accuracy, leading to an overall increase in the performance of downstream analysis tools. Despite that, further inspection and post hoc analysis of k-mer based classification outputs have been suggested in order to limit possible misclassifications [58], [59], [60]. An example of an alignment-free classification tool is Kraken [64], which uses exact k-mer matches for each read against a k-mer-to-LCA records database. These records are generated from sequence databases and indexed in time-efficient data structures that enable faster look-up searches. However, this process is memory-intensive, an issue that was addressed by the development of Kraken2 [35], characterized by an enhanced database efficiency and improved k-mer-based read analysis. Using the same Kraken k-mer based classification technique, KrakenUniq [65] improves precision and recall by assessing the coverage of unique k-mers of each taxon that is present in the dataset. An additional development leveraging the Kraken classification output is Bracken [66], a statistical method to compute the abundance estimation of a sample at any given taxonomic level from Kraken/Kraken2 classifications for each read. Another software, Centrifuge (Kim et al., 2016), builds a data structure based on the Ferragina-Manzini index, a technique based on widely used read aligners, i.e. the Burrows-Wheeler Aligner (BWA) algorithm [67] and Bowtie [68]. This data structure provides efficient storage of database sequences and the classification is also performed by k-mer matching against the pre-built index. Metamaps [69] is a long-read specific approach featuring taxonomic assignment and sample composition estimations at strain-level along with an output that includes per read positional and quality information. In addition, a number of pipelines for analysis of metagenomic data have been developed, which integrate previously mentioned tools into easy-to-deploy workflows and generate comprehensive outputs for result interpretation (Table 2) [25], [60], [70], [71].
3.3. Reference databases
Taxonomic classification methods use pre-computed or indexed reference databases (Table 3). While some tools are designed to work with specific databases, most tools allow a variety of sequence collections to be indexed. Thus, the database choice is important for metagenomic workflows. Popular reference databases include the NCBI RefSeq collection of complete genomes [75], encompassing both prokaryotic and eukaryotic genomes, and the nt BLAST database built from more than 50 million high-quality nucleotide sequences. The GenBank database [76] includes a wider collection of complete genomes albeit with lower quality standards than RefSeq collections. Other databases that are better suited for metataxonomics include GreenGenes [77], SILVA [78], RDP [79] for 16S rRNA gene sequencing, and the NCBI RefSeqTargeted Loci Project database containing 16S/23S and 18S/28S rRNA genes from the GenBank database (https://www.ncbi.nlm.nih.gov/refseq/targetedloci/). These databases contain partial and full-length 16S rRNA gene sequences providing a more lightweight and comprehensive sequence collection for metataxonomic analysis. Other purpose databases are Prokka [80], for the annotation of assembled genomes, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [81], which includes annotated genes and genomes for functional annotation profiling. Regarding AMR-specific databases, CARD [82] and MEGARes [83] contain well-curated gene sequences and annotations for integration in the AMR analysis and detection workflows.
The size of microbial genome databases grows exponentially every year and can reach 100s of GBs for some of the large collections, e.g., RefSeq. Despite that, their size increase does not guarantee the successful classification of every generated read of the experiment [84]. False negatives are common due to undiscovered and yet to be sequenced microorganisms. Modifications or updates of databases include changes in the structure of the taxonomy tree and the inclusion of new sequences or re-sequenced strains. The increase in the number of total sequences added to a database (through updates) is not representative of a gain on species richness. Some of these additions tend to create redundancy of certain genera and species in the database. Hence, the use of databases created at different times and, therefore, containing different sequence content and taxonomy can affect the results and profoundly confound the software benchmarkings [85]. These issues emphasize the importance of performing continuous comparisons and benchmarkings of widely adopted tools using varied testing datasets and databases, necessary for result interpretation and evaluation of computational requirements and performance.
3.4. Metagenomic assembly
In addition to the improvements in the taxonomic analysis when relying on long reads, it has been shown that nanopore sequencing data can produce contiguous and highly accurate assemblies, including full-length plasmids and viruses, without any preprocessing step, such as initial read binning [88]. These assemblies are in the range of 95% of completeness when applied to whole-genome sequencing combined with posterior assembly polishing using error correction tools [55]. Long-read metagenomic assembly has the potential not only to improve contiguity over short-read assemblies but also to enable strain resolution, to sequence novel plasmids and viruses, and to enhance the power of identifying horizontal gene transfer. Despite the advantages over short-read sequencing data, complete contiguous assemblies are still constrained by the relatively high error rate of nanopore reads, and the quality of the metagenome assembly is related to the coverage of the different species present in the sample, which in turn depends on the experiment throughput. In practice, the read-length advantage of nanopore enables nearly complete assemblies even for low abundance strains, provided that they are covered at a minimal level [89].
Metagenomic assembly requires the development of specific-purpose algorithms to overcome the limitations of classic assemblers which assume that the depth of coverage is approximately uniform across the genome. Some long-read assemblers have been used for metagenomic assembly even if not specifically designed for the task (Table 2). For example, Canu [90] is one of the first and most popular assemblers for long-read data and generates contigs using an adaptive k-mer weighting strategy to produce an assembly with high coverage long-read data. wtdbg2 [91] adopts a fast all-versus-all read alignment and a layout algorithm based on fuzzy-Bruijn graphs for sequence assembly. A recent development is Raven [92], a specific long-read assembler that features a faster read overlap step and assembly graph building. While these approaches have been successfully used and benchmarked for metagenomics assembly [20], [93], [94], novel assemblers that were built specifically for metagenomics datasets have recently been developed. An example is OPERA-MS [95], a hybrid assembler that leverages the strengths of both short-read and long-read sequencing approaches using first a short-read assembler to create contigs and then the long-read information to create an assembly graph for all genomes that are distinguished in a coverage-based clustering. Another software is MetaFlye [96], which selects high-frequency k-mers from the dataset to detect read overlaps and creates error-prone contigs used for developing the final assembly graph.
3.5. Expanding the view of metagenomics by nanopore sequencing
Nanopore sequencing has been used in a number of clinically relevant metagenomics applications due to the possibility of real-time sequencing (Table 4). One example is the rapid species and AMR profiling to guide proper antibiotic treatment, which is of particular importance for the current worldwide AMR threat. A recent study used nanopore sequencing for the characterization of pathogenic bacteria and AMR genes in gut-associated microbial communities in preterm infants [97]. To do this, they used the NanoOK RT software, where reads are aligned to bacteria and AMR databases on the fly while they are generated in the run, resulting in a sequencing turnaround time of 1 h. Furthermore, they successfully linked AMR genes to the harboring pathogens, suggesting that nanopore sequencing coupled with bioinformatic analysis can tailor antibiotic therapies. Similarly, Břinda et al. have recently developed an innovative method, called genomic neighbor typing, which accelerates pathogen detection and AMR typing [98]. The method is based on the assumption that resistance elements are genetically linked to the rest of the genome and that it is possible to define the antimicrobial susceptibility by only inferring the bacterial strains that are present in the sample. It relies on a two-step algorithm where the sequence is first compared to a reference database, and then the most probable phenotype (drug resistance or susceptibility) of the sample is determined from the phenotype of its nearest genomic neighbor. They were able to identify the correct lineage, strain, and antibiotic susceptibility of pneumococcal and gonococcal isolates in < 10 min. They also compared it to the AMR gene-based approach, which required 25 min for single copies to be detected. They further validated the method by successfully confirming resistance in pneumococcus in six sputum samples from patients suffering lower respiratory infections. The main limitation of this method is that strain and AMR detection are based on the information provided by the database. Therefore, it is crucial that the constructed database includes genomic sequences and resistance metadata of the strains encountered in the clinical samples analysed. As a consequence, further applications of this method may include pathogen diagnostics and surveillance, provided that the target microbe and AMR are known.
Given that long reads facilitate accessing repetitive elements and structural variants, nanopore sequencing has improved our capacity for de novo assembly of genomes, metagenomes, and plasmids, which in turn allow gathering information on the localization of resistance and virulence factors, such as AMR genes and PAIs. An example of this is a recent study that collected the most comprehensive characterization of opportunistic pathogens and their resistomes colonizing tertiary hospital environments [99]. The workflow consisted of culturing, antibiotic selection, metagenomic sequencing, and OPERA-MS-based genome assembly of environmental samples collected from 179 sites associated with 45 beds. They obtained genomes for 69 species, 16% of them from novel species. Furthermore, they reconstructed plasmids and phages, of which more than 90% were uncharacterized. By doing this, they were able to identify novel associations between AMR genes, characterize chromosomal cassettes and AMR gene combinations in plasmids, and detect the persistence of multidrug-resistant organisms in hospital environments over years. This study highlighted the importance of monitoring environmental pathogens in clinical settings, which can be responsible for occasional outbreaks in hospitals. More recently, another research group has introduced a novel long-read assembly-based approach for metagenome-assembled genomes (MAGs), called Lathe [43]. By coupling Lathe to a newly developed experimental protocol for HMW DNA extraction, they assembled seven genomes (out of 12) from a mock sample into single contigs, obtaining circular genomes, while three more genomes were assembled into four or fewer contigs. They validated their protocol on 13 human stool samples and demonstrated that Lathe generated better assembly contiguity than those from short reads and the read cloud assembler (hybrid). Notably, they were able to resolve the circular genome of Prevotella copri, known to be characterized by a high degree of repetitive sequences. By resolving complete circularized genomes, it is possible to optimally study microbial phenomena, such as horizontal gene transfers, and to investigate how inter-strain structural variants may be linked to a specific phenotype of a microbial community [100].
Because of their reduced length (typically within 3–300 kb), viral genomes can be sequenced as a single molecule using nanopore sequencing. Beaulaurier et al. discovered more than 1,800 phage genomes in seawater samples [101]. The analytical method consisted at first of filtering those sequences containing the direct terminal regions, which are characteristic of the virus genome termini in dsDNA-tailed phages. Then, the analysis consisted of a step of dimensionality reduction and clustering, followed by polishing to create high-quality draft phage genomes. By adopting this method, they were able to identify viral microheterogeneity, otherwise very difficult to detect using short-read sequencing. Furthermore, this approach allowed inferring the phage packaging strategy and identifying concatemers of sequences similar to the phage-inducible chromosomal islands, revealing the utility of this approach to identify repeat sequences derived from phage-induced mobile elements.
4. Metatranscriptomics and viral RNA sequencing
The study of the microbiome can be approached through metatranscriptomics, i.e. the study of the totality of transcripts in a sample. Nanopore sequencing enables obtaining full-length transcripts in a single read, facilitating transcriptome analysis by avoiding the challenging steps necessary for short-read transcriptomics. Furthermore, ONT technologies can directly sequence the RNA molecules eliminating the biases introduced by the reverse transcription or the amplification step, given that all transcripts do not amplify with the same efficiency [13], [102]. In addition, the processes of retrotranscription and amplification erase the epitranscriptomic information, which is known to have a role in modulating transcript activity and stability. Viral RNA genomes can also be sequenced as native RNA molecules or as cDNA after retrotranscription using nanopore sequencing. This is of particular importance for many emerging human viral diseases, such as Ebola, severe acute respiratory syndrome (SARS), and the coronavirus disease 2019 (COVID-19), all of them caused by RNA viruses.
4.1. Protocols and libraries
ONT offers three main sequencing library preparation kits for the analysis of transcriptome and viral RNA genomes (Table 1). Two of them (the Direct cDNA Sequencing Kit and the cDNA PCR Sequencing Kit) are based on a retrotranscription step, followed by either digestion of the RNA strand and ligation of sequencing primers, or by a PCR step with rapid attachment primers when the initial target RNA does not reach the minimum required amount of 100 ng. A third library preparation kit, the Direct RNA Sequencing Kit, is based on a DNA primer annealing and ligation to the RNA strand, followed by an optional retrotranscription step (necessary only to stabilize the RNA molecule) and by the attachment of sequencing adapter at the RNA 3′ end. This library preparation protocol is faster (< 2 h) because it does not include cDNA synthesis. However, it requires a higher amount of initial RNA (~500 ng) and has lower throughput compared to the cDNA kits. However, ONT is continuously modifying the chemistry of these kits in order to improve current accuracy and throughput.
Although ONT successfully enabled gaining insights into eukaryotic messenger RNA [103] and viral RNA with a poly-A tail [104], [105], the study of the prokaryotic transcriptome has been hindered by the lack of a poly-A tail required for the attachment of primers during library construction. A way to overcome this issue would be by adding a step of polyadenylation of prokaryotic transcriptomes in the experimental protocol to make them recognizable and modifiable by the ONT Direct RNA Sequencing Kit [106]. Another possibility is to design custom adapters to ligate the 3′ end of the transcripts, the tRNAs, or the rRNAs of interest. For example, Smith et al. designed adapters containing a 20-nucleotide overhanging sequence targeting the conserved anti-Shine Dalgarno region, present in prokaryotic 16S rRNA, to study how canonical and non-canonical base modifications affect antimicrobial susceptibility in Escherichia coli strains [107]. A similar approach was employed by Keller et al. to sequence, for the first time, the complete RNA genome of the influenza A virus, by designing adapters targeting the highly conserved genome termini of the virus [108].
4.2. Bioinformatic analysis
Whereas some well-known pipelines for short-read metagenomics, such as MEGAN [30] and MG-RAST [109], can be used with RNA reads for taxonomic assignment, the availability of specific tools to analyze long-read RNA profiles is limited. Metataxonomic workflows, as described elsewhere [110], can be performed alternatively by extracting rRNA sequences, such as the small subunits (16S/18S) and large subunits (23S/28S), using specialized software, e.g., METAXA2 [111]. Functional analysis is performed with BLASTx or Magic-BLAST [112] to align the RNA sequences to a protein database in order to assign either Gene Ontology terms (GO) with Blast2GO [113] or metabolic pathway annotations according to KEGG [114]. Recent examples of specific tools for long-read transcriptomics are Poreplex (https://github.com/hyeshik/poreplex), a signal-level processor for ONT direct RNA sequencing data that features real-time basecalling, quality filtering, 3′ adapter trimming, and alignment to reference transcriptomes. Bambu [115] is an R software package for multi-sample transcript discovery and quantification from long-read RNA data. Regarding alignment-free methods, a recent development is isONclust [116], a tool for de novo transcript reconstruction with a cluster-based approach that accounts for large dataset scaling.
4.3. Metatranscriptomics and emerging infectious diseases through nanopore sequencing
The study of the transcriptome for metagenomics can be adopted to address several issues which cannot be tackled by DNA sequencing. In fact, RNA sequencing provides additional information, such as the functionality of AMR genes, allowing to identify a situation where the resistance gene is present but not transcribed, and therefore does not generate a resistant phenotype [106]. Metatranscriptomics is also very useful for the identification of viable pathogens since DNA-based approaches are unable to differentiate between viable and unviable bacterial cells [117]. This approach is particularly important in the detection of food pathogens, where food processing and storage often kill bacteria cells without removing their genomic DNA. Direct RNA sequencing has recently been compared to multiplex real-time PCR amplicon sequencing for this purpose. Results suggest it to be especially applicable to complex microbiomes because it does not require assay customization for specific biohazards when a complete database is used during the bioinformatic analysis step [117]. Other applications of metatranscriptomics using nanopore sequencing also include pathogen detection from clinical samples, which is particularly useful for diseases caused by RNA viruses (Table 4). For example, nanopore RNA sequencing has been used for differential diagnosis of dengue and chikungunya viruses, two single-stranded positive RNA viruses circulating in the same geographical areas and causing diseases with similar symptomatology [118]. Despite that, transcriptomics is still a very immature application of nanopore sequencing for microbial studies.
In recent years, emerging RNA viruses have become a threat to global health, and viral genome sequencing has turned into an essential tool for outbreak identification and monitoring of transmission patterns. Nanopore technology has been demonstrated to be an exceptionally valuable tool for this purpose because it can produce data in real-time, directly in-field and under extreme conditions thanks to inexpensive portable devices. For this reason, nanopore technology has been adopted for genomic surveillance during the Ebola outbreak in West Africa [119], the Zika outbreak in Brazil and the Americas [120], and the ongoing COVID-19 pandemic. Since the first identification of a novel coronavirus in December 2019 [121], thousands of SARS-CoV-2 genomes have been sequenced using the ARTIC [122] or alternative [123] protocols developed for fast sequencing of the virus using nanopore technology, allowing to gather information on virus biology, transmission, and viral dynamics. For example, an important study performed by Fauver et al. coupled genomic data with domestic and international travel patterns in the USA, tracking down the SARS-CoV-2 transmission dynamics in early March 2020 [124]. In this study, nine viral genomes from early cases in Connecticut were sequenced within 24 h and used to build a phylogenetic tree. When compared to other 168 publicly available genomes at that time, seven out of nine genomes clustered into one clade containing sequences from other USA samples, suggesting domestic transmission of SARS-CoV-2 in the USA early in the first wave of the pandemic. This information was further confirmed by estimating the SARS-CoV-2 travel importation risk into Connecticut using airline travel data and epidemiological dynamics in regions where travel routes came from. Nanopore sequencing of SARS-CoV-2 has also been adopted in a prospective genomic surveillance study aiming to identify healthcare-associated infections in a hospital in the UK [125]. Around 1,000 genomes were sequenced within five months and results were compared on the basis of the ward location data of patients or healthcare workers in order to unravel transmission patterns within the hospital. This information was transmitted almost in real-time to the hospital management team and allowed to identify risk factors for transmission in clinical settings. Importantly, this study supports the adoption of combined epidemiological and genomic data for the implementation of infection control measures and highlights the importance of genomic epidemiology to guide decision-making on a local, national and international level.
Apart from the above-described protocols, which require a retrotranscription step, direct RNA sequencing has also been used for SARS-CoV-2 sequencing [126], [127], [128]. This method allowed to sequence regions spanning almost the entire viral genome (~30 kb), although the coverage was found to be extremely variable along the genome, ranging from 34× to >160,000×, and biased towards the poly-A 3′ end [128]. The reason for this is the abundance of subgenomic mRNAs carrying these regions and the directional nanopore sequencing from the poly-A 3′ end. Nonetheless, this study allowed gaining insights into the transcriptome and epitranscriptome of the virus, with the identification of eight major transcripts and 42 positions with 5′ methyl-cytosine (5mC) modifications. Furthermore, by direct RNA sequencing, Taiaroa et al. were able to estimate the evolutionary rate of the virus, which is important for epidemiological studies [128].
5. Epigenomics and epitranscriptomics
Epigenetic modifications of DNA in bacteria and DNA/RNA in viruses are responsible for several biological functions, such as the regulation of DNA/RNA replication and repair, control of gene expression, and protection from external pathogens [129]. So far, methylation is the only nucleotide modification known in bacterial DNA, with three forms of methylation identified: 5mC, N4-methylcytosine (4mC), and N6-methyladenine (6 mA), the latter being the most prevalent form [130]. Each of these types of epimodifications occurs in a highly motif-driven manner, where every occurrence of the motif is methylated. Nanopore sequencing allows direct detection of the native modified bases on the nucleic acid during its passage through the nanopore. In fact, the characteristic ionic current observed when a certain sequence passes through the pore is altered by the presence of a methylated base, generating a distinctive current pattern that can be distinguished from the non-methylated DNA/RNA. While ONT amplification-free libraries can be easily generated via standard ONT kits, the bottleneck for the study of nucleotide modifications in nanopore sequencing is still the basecalling process, where the presence of multiple new current signals generated by one or multiple methylated bases in the k-mer passing the pore causes a considerable computational challenge [12], [131]. Multiple research groups have tried to face this by developing tools to detect methylated bases. For example, Stoiber et al. presented a method based on the statistical comparison between ionic current signals from native and methylated sequences [132], which has then evolved into the current ONT Tombo platform (https://github.com/nanoporetech/tombo). Other methods use pre-trained classification models to capture epigenomic modifications such as Nanopolish [131] and SignalAlign [12], which use Hidden-Markov models, or the recently developed mCaller [133], Deepsignal [134], and Deepmod [135] which adopt neural network classifiers. However, these tools are characterised by detection accuracies that vary based on the methylation type and the target motif, and the capability to detect de novo methylated motifs is limited by the training data [136]. A recent study [137] tried to address this issue by generating a large training dataset for de novo methylation typing and mapping of all three forms of DNA methylation and applied it to individual bacteria and mouse gut microbiome samples. In this work, Tourancheau et al. also developed a novel approach for methylation binning of metagenome contigs and demonstrated how methylation patterns may assist in the process of metagenome assembly. Although this method enabled de novo methylation typing and fine mapping, accuracy is still highly dependent on the type and position of the methylated base which remains an issue to be addressed in the future.
6. Summary and outlook
Nanopore technology has improved many aspects of microbial analysis and has the potential to be adopted routinely in clinical settings in the near future. In fact, many proof-of-concept studies have demonstrated that nanopore sequencing can be adopted for infectious disease diagnostics and for monitoring the human microbiome, which can be a useful tool in clinical medicine. For example, dysbiosis of the lung microbiome has been shown to have a prognostic value for mortality in patients with non-pulmonary sepsis in intensive care units [138], and nanopore sequencing was proposed to be used as a prognostic tool for real-time monitoring of this dysbiosis. Similarly, nanopore sequencing could be used to monitor changes in the gut microbiome over time, before or after antibiotic treatments [97], or to assess species engraftment after faecal microbiota transplantation [139]. Although there are many possible applications for nanopore sequencing in the field of metagenomics, there are still numerous challenges that need to be addressed. For example, standardized protocols for microbial characterization are needed in order to use this technology in clinical settings. Furthermore, novel and efficient HMW DNA extraction protocols from microbial samples are required to produce high-quality long reads, and library preparation protocols need to be simplified, especially for in-field and educational settings. For this purpose, VolTRAX has been released by ONT as a system for automated library preparation to provide high reproducibility and portability to the library preparation step. However, the cartridge used by the system can hold up a maximum of ten barcoded samples, which is far too low for experiments requiring multiplexing of 96 samples. Another aspect that needs to be considered is the read error rate. This issue is continuously addressed by ONT through the constant improvement of the flow cell and the release of more rapid and accurate basecalling methods. However, both read and consensus accuracy are limited by the organisms chosen for model training, and they are drastically reduced when the basecaller is used for less frequently sequenced microbial species [140]. This issue can be addressed by developing more custom-trained basecallers, built-on taxon-specific training data so that the users can choose which basecaller most closely matches the organisms present in their samples [140], [141]. Sequencing data throughput by single flow cell is also continuously improving, enabling the detection and sequencing of DNA/RNA from microbes even when present in very low abundance. Furthermore, real-time selective sequencing [142], or read until, is also becoming popular among nanopore users and consists of extruding specific molecules from the pores, such as host DNA or other non-interesting molecules. It has the potential to increase the efficiency of the run by reducing the time taken to complete an experiment, to enrich the data with the less represented genomes in the sample and, at the same time, to simplify library preparation protocols by eliminating host DNA depletion or target enrichment steps [143], [144].
Regarding the analysis of metagenomic data, improvements are expected in taxonomic analysis and sequence comparison software in order to achieve better resolution of closely related strains and higher classification accuracy, along with the development of efficient indexing techniques for metagenomics databases. Metagenomic assembly software and hybrid techniques using both short and long reads have the potential to enhance the analysis of complex samples improving the detection of unknown organisms and enabling the assembly of mobile elements and resistance genes, which are crucial for the characterization of complex microbial environments such as the human microbiome [43], [145]. Advances in nanopore technologies, such as direct RNA sequencing [13] and the detection of epimodifications [12], have highlighted the need for novel bioinformatic tools enabling accurate characterization or discovery of transcriptomes and the determination of the type and position of modified bases on a sequence and their functional impact. Furthermore, benchmarking studies of metagenomics tools for the analysis of long reads are lacking, together with long-read metagenomic datasets representing complex microbial communities to be used during software development and tool assessments. Future advancements in metagenomic analysis tools and workflows for long reads will need to follow the quick pace in modifications and updates of the sequencing technology and also account for software efficiency and scalability in order to enable the analysis of sequence data from high-throughput devices such as the GridION and PromethION.
Funding
This work was supported by the Ministerio de Ciencia e Innovación (RTC-2017-6471-1; AEI/FEDER, UE) and the Instituto de Salud Carlos III (PI14/00844, PI17/00610, FI18/00230), which were co-financed by the European Regional Development Funds ‘A way of making Europe’ from the European Union; Fundación Canaria Instituto de Investigación Sanitaria de Canarias (PIFUN48/18); Cabildo Insular de Tenerife (CGIEU0000219140 and “Apuestas científicas del ITER para colaborar en la lucha contra la COVID-19”); and by agreement OA17/008 with the Instituto Tecnológico y de Energías Renovables (ITER) to strengthen scientific and technological education, training, research, development, and innovation in genomics, personalized medicine, and biotechnology. The sponsors had no involvement in the review conceptualization, the manuscript writing and the decision to submit the article for publication.
CRediT authorship contribution statement
Laura Ciuffreda: Conceptualization, Visualization, Writing - original draft, Writing - review & editing. Héctor Rodríguez-Pérez: Conceptualization, Visualization, Writing - original draft, Writing - review & editing. Carlos Flores: Conceptualization, Supervision, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Laura Ciuffreda, Email: lciuffreda.bio@gmail.com.
Héctor Rodríguez-Pérez, Email: alu0100774429@ull.edu.es.
Carlos Flores, Email: cflores@ull.edu.es.
References
- 1.Lane D.J., Pace B., Olsen G.J., Stahl D.A., Sogin M.L., Pace N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pace NR, Stahl DA, Lane DJ, Olsen GJ. The Analysis of Natural Microbial Populations by Ribosomal RNA Sequences, Springer, Boston, MA; 1986, p. 1–55. 10.1007/978-1-4757-0611-6_1. [DOI]
- 3.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C.T., Hug L.A., Thomas B.C., Sharon I., Castelle C.J., Singh A. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 5.Jain M., Olsen H.E., Paten B., Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu H., Giordano F., Ning Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics, Proteomics Bioinforma. 2016;14:265–279. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne A, Holmes N, Rakyan V, Loose M. BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 2019;35:2193–8. 10.1093/bioinformatics/bty841. [DOI] [PMC free article] [PubMed]
- 8.Leggett R.M., Clark M.D. A world of opportunities with nanopore sequencing. J Exp Bot. 2017;68:5419–5429. doi: 10.1093/jxb/erx289. [DOI] [PubMed] [Google Scholar]
- 9.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 10.De Roeck A., De Coster W., Bossaerts L., Cacace R., De Pooter T., Van Dongen J. NanoSatellite: Accurate characterization of expanded tandem repeat length and sequence through whole genome long-read sequencing on PromethION. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giesselmann P., Brändl B., Raimondeau E., Bowen R., Rohrandt C., Tandon R. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat Biotechnol. 2019;37:1478–1481. doi: 10.1038/s41587-019-0293-x. [DOI] [PubMed] [Google Scholar]
- 12.Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E., Akeson M. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods. 2017;14:411–413. doi: 10.1038/nmeth.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M. Highly parallel direct RN A sequencing on an array of nanopores. Nat Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- 14.Rang F.J., Kloosterman W.P., de Ridder J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oxford Nanopore Technologies. Nanopore sequencing accuracy 2019. https://nanoporetech.com/accuracy (accessed February 3, 2021).
- 16.Marchesi J.R., Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarridge J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo P.C.Y., Lau S.K.P., Teng J.L.L., Tse H., Yuen K.-Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14:908–934. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 20.Bharti R., Grimm D.G. Current challenges and best-practice protocols for microbiome analysis. Brief Bioinform. 2019;22:178–193. doi: 10.1093/bib/bbz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson J.S., Spakowicz D.J., Hong B.-Y., Petersen L.M., Demkowicz P., Chen L. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10 doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benítez-Páez A., Portune K.J., Sanz Y. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinIONTM portable nanopore sequencer. GigaScience. 2016;5:4. doi: 10.1186/s13742-016-0111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin J., Lee S., Go M.-J., Lee S.Y., Kim S.C., Lee C.-H. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep. 2016;6 doi: 10.1038/srep29681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nygaard A.B., Tunsjø H.S., Meisal R., Charnock C. A preliminary study on the potential of Nanopore MinION and Illumina MiSeq 16S rRNA gene sequencing to characterize building-dust microbiomes. Sci Rep. 2020;10:3209. doi: 10.1038/s41598-020-59771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Pérez H., Ciuffreda L., Flores C. NanoCLUST: a species-level analysis of 16S rRNA nanopore sequencing data. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa900. btaa900. [DOI] [PubMed] [Google Scholar]
- 26.Bonk F, Popp D, Harms H, Centler F. PCR-based quantification of taxa-specific abundances in microbial communities: Quantifying and avoiding common pitfalls. J Microbiol Methods 2018;153:139–47. 10.1016/j.mimet.2018.09.015. [DOI] [PubMed]
- 27.Kai S., Matsuo Y., Nakagawa S., Kryukov K., Matsukawa S., Tanaka H. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinIONTM nanopore sequencer. FEBS Open Bio. 2019;9:548–557. doi: 10.1002/2211-5463.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuo Y, Komiya S, Yasumizu Y, Yasuoka Y, Mizushima K, Takagi T, et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinIONTM nanopore sequencing confers species-level resolution. BioRxiv 2020:2020.05.06.078147. 10.1101/2020.05.06.078147. [DOI] [PMC free article] [PubMed]
- 29.Starke R., Pylro V.S., Morais D.K. 16S rRNA gene copy number normalization does not provide more reliable conclusions in metataxonomic surveys. Microb Ecol. 2021;81:535–539. doi: 10.1007/s00248-020-01586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos A, van Aerle R, Barrientos L, Martinez-Urtaza J. Computational methods for 16S etabarcoding studies using Nanopore sequencing data. Comput Struct Biotechnol J 2020;18:296–305. 10.1016/j.csbj.2020.01.005. [DOI] [PMC free article] [PubMed]
- 33.Li H. Sequence analysis Minimap2 : pairwise alignment for nucleotide sequences 2018;34:3094–100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed]
- 34.Kim D., Song L.i., Breitwieser F.P., Salzberg S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitsuhashi S., Kryukov K., Nakagawa S.o., Takeuchi J.S., Shiraishi Y., Asano K. A portable system for rapid bacterial composition analysis using a nanopore-based sequencer and laptop computer. Sci Rep. 2017;7 doi: 10.1038/s41598-017-05772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka H., Matsuo Y., Nakagawa S., Nishi K., Okamoto A., Kai S. Real-time diagnostic analysis of MinIONTM-based metagenomic sequencing in clinical microbiology evaluation: a case report. JA Clin Reports. 2019;5:24. doi: 10.1186/s40981-019-0244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa S.o., Inoue S., Kryukov K., Yamagishi J., Ohno A., Hayashida K. Rapid sequencing-based diagnosis of infectious bacterial species from meningitis patients in Zambia. Clin Transl Immunol. 2019;8 doi: 10.1002/cti2.v8.1110.1002/cti2.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuenschwander S.M., Terrazos Miani M.A., Amlang H., Perroulaz C., Bittel P., Casanova C. A sample-to-report solution for taxonomic identification of cultured bacteria in the clinical setting based on nanopore sequencing. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00060-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 41.Riesenfeld C.S., Schloss P.D., Handelsman J.o. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 42.Kristensen D.M., Mushegian A.R., Dolja V.V., Koonin E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss E.L., Maghini D.G., Bhatt A.S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat Biotechnol. 2020;38:701–707. doi: 10.1038/s41587-020-0422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashton P.M., Nair S., Dallman T., Rubino S., Rabsch W., Mwaigwisya S. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 45.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genomics. 2017;3 doi: 10.1099/mgen.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Judge K., Hunt M., Reuter S., Tracey A., Quail M.A., Parkhill J. Comparison of bacterial genome assembly software for MinION data and their applicability to medical microbiology. Microb Genomics. 2016;2 doi: 10.1099/mgen.0.000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George S, Pankhurst L, Hubbard A, Votintseva A, Stoesser N, Sheppard AE, et al. Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb Genomics 2017;3:e000118. 10.1099/mgen.0.000118. [DOI] [PMC free article] [PubMed]
- 48.Bouchez V., Baines S.L., Guillot S., Brisse S., Bruno V. Complete genome sequences of bordetella pertussis clinical isolate FR5810 and reference strain tohama from combined oxford nanopore and illumina sequencing. Microbiol Resour Announc. 2018;7 doi: 10.1128/MRA.01207-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maghini D.G., Moss E.L., Vance S.E., Bhatt A.S. Improved high-molecular-weight DNA extraction, nanopore sequencing and metagenomic assembly from the human gut microbiome. Nat Protoc. 2021;16:458–471. doi: 10.1038/s41596-020-00424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oxford Nanopore Technologies. Metagenomic sequencing with Oxford Nanopore 2020. https://nanoporetech.com/sites/default/files/s3/literature/metagenomic-sequencing-guide.pdf (accessed February 3, 2021).
- 51.Cookson W.O.C.M., Cox M.J., Moffatt M.F. New opportunities for managing acute and chronic lung infections. Nat Rev Microbiol. 2018;16:111–120. doi: 10.1038/nrmicro.2017.122. [DOI] [PubMed] [Google Scholar]
- 52.Charalampous T., Kay G.L., Richardson H., Aydin A., Baldan R., Jeanes C. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 53.Leidenfrost R.M., Pöther D.C., Jäckel U., Wünschiers R. Benchmarking the MinION: evaluating long reads for microbial profiling. Sci Rep. 2020;10:5125. doi: 10.1038/s41598-020-61989-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makałowski W., Shabardina V. Bioinformatics of nanopore sequencing. J Hum Genet. 2020;65:61–67. doi: 10.1038/s10038-019-0659-4. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H., Jain C., Aluru S. A comprehensive evaluation of long read error correction methods. BMC Genomics. 2020;21:1–15. doi: 10.1186/s12864-020-07227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu S., Wang A., Au K.F. A comparative evaluation of hybrid error correction methods for error-prone long reads. Genome Biol. 2019;20:1–17. doi: 10.1186/s13059-018-1605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z., Erickson D.L., Meng J. Benchmarking long-read assemblers for genomic analyses of bacterial pathogens using oxford nanopore sequencing. Int J Mol Sci. 2020;21:1–27. doi: 10.3390/ijms21239161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y., Ye W., Zhang Y., Xu Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015;43:7762–7768. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huson D.H., Albrecht B., Bağcı C., Bessarab I., Górska A., Jolic D. MEGAN-LR: new algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biol Direct. 2018;13 doi: 10.1186/s13062-018-0208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kielbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konishi H., Yamaguchi R., Yamaguchi K., Furukawa Y., Imoto S. Halcyon: an accurate basecaller exploiting an encoder–decoder model with monotonic attention. Bioinformatics. 2020:1–7. doi: 10.1093/bioinformatics/btaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Silvestre-Ryan J., Holmes I. Pair consensus decoding improves accuracy of neural network basecallers for nanopore sequencing. Genome Biol. 2021;22:38. doi: 10.1186/s13059-020-02255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood D.E., Salzberg S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Breitwieser F.P., Baker D.N., Salzberg S.L. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018;19:198. doi: 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 2017;3:e104. 10.7717/peerj-cs.104. [DOI]
- 67.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B., Salzberg S. Bowtie2. Nat Methods. 2013;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dilthey A.T., Jain C., Koren S., Phillippy A.M. Strain-level metagenomic assignment and compositional estimation for long reads with MetaMaps. Nat Commun. 2019;10:3066. doi: 10.1038/s41467-019-10934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Damme R, Hölzer M, Viehweger A, Müller B, Bongcam-Rudloff E, Brandt C. Metagenomics workflow for hybrid assembly, differential coverage binning, transcriptomics and pathway analysis (MUFFIN). BioRxiv 2020:2020.02.08.939843. 10.1101/2020.02.08.939843. [DOI] [PMC free article] [PubMed]
- 71.Xu Y., Yang-Turner F., Volk D., Crook D. NanoSPC: a scalable, portable, cloud compatible viral nanopore metagenomic data processing pipeline. Nucleic Acids Res. 2020;48:W366–W371. doi: 10.1093/nar/gkaa413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haft DH, Dicuccio M, Badretdin A, Brover V, Chetvernin V, Neill KO, et al. RefSeq : an update on prokaryotic genome annotation and curation 2018;46:851–60. 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed]
- 76.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J. GenBank. Nucleic Acids Res. 2013;41:36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 2013;41:590–6. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed]
- 79.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 81.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 2020;48:D517–25. 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed]
- 83.Lakin S.M., Dean C., Noyes N.R., Dettenwanger A., Ross A.S., Doster E. MEGARes: An antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017;45:D574–D580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nasko D.J., Koren S., Phillippy A.M., Treangen T.J. RefSeq database growth influences the accuracy of k-mer-based lowest common ancestor species identification. Genome Biol. 2018;19:1–21. doi: 10.1186/s13059-018-1554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Q., Zobel J., Verspoor K. Duplicates, redundancies and inconsistencies in the primary nucleotide databases: A descriptive study. Database. 2017;2017:1–16. doi: 10.1093/database/baw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 87.Arango-Argoty G., Garner E., Pruden A., Heath L.S., Vikesland P., Zhang L. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018;6:23. doi: 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nicholls SM, Quick JC, Tang S, Loman NJ. Ultra-deep, long-read nanopore sequencing of mock microbial community standards. Gigascience 2019;8. 10.1093/gigascience/giz043. [DOI] [PMC free article] [PubMed]
- 89.Hu Y.u., Fang L.i., Nicholson C., Wang K. Implications of error-prone long-read whole-genome shotgun sequencing on characterizing reference microbiomes. IScience. 2020;23:101223. doi: 10.1016/j.isci.2020.101223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruan J., Li H. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 2020;17:155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vaser R, Šikić M. Raven: a de novo genome assembler for long reads. BioRxiv 2020:2020.08.07.242461. 10.1101/2020.08.07.242461. [DOI]
- 93.Stewart R.D., Auffret M.D., Warr A., Walker A.W., Roehe R., Watson M. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat Biotechnol. 2019;37:953–961. doi: 10.1038/s41587-019-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Latorre-Pérez A., Villalba-Bermell P., Pascual J., Vilanova C. Assembly methods for nanopore-based metagenomic sequencing: a comparative study. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-70491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertrand D., Shaw J., Kalathiyappan M., Ng A.H.Q., Kumar M.S., Li C. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat Biotechnol. 2019;37:937–944. doi: 10.1038/s41587-019-0191-2. [DOI] [PubMed] [Google Scholar]
- 96.Kolmogorov M., Bickhart D.M., Behsaz B., Gurevich A., Rayko M., Shin S.B. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods. 2020;17:1103–1110. doi: 10.1038/s41592-020-00971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leggett R.M., Alcon-Giner C., Heavens D., Caim S., Brook T.C., Kujawska M. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat Microbiol. 2020;5:430–442. doi: 10.1038/s41564-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Břinda K., Callendrello A., Ma K.C., MacFadden D.R., Charalampous T., Lee R.S. Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat Microbiol. 2020;5:455–464. doi: 10.1038/s41564-019-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chng K.R., Li C., Bertrand D., Ng A.H.Q., Kwah J.S., Low H.M. Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat Med. 2020;26:941–951. doi: 10.1038/s41591-020-0894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zeevi D., Korem T., Godneva A., Bar N., Kurilshikov A., Lotan-Pompan M. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–48. doi: 10.1038/s41586-019-1065-y. [DOI] [PubMed] [Google Scholar]
- 101.Beaulaurier J., Luo E., Eppley J.M., Uyl P.D., Dai X., Burger A. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 2020;30:437–446. doi: 10.1101/gr.251686.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu H., Begik O., Lucas M.C., Ramirez J.M., Mason C.E., Wiener D. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun. 2019;10 doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8 doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tombácz D., Moldován N., Balázs Z., Gulyás G., Csabai Z., Boldogkői M. Multiple long-read sequencing survey of herpes simplex virus dynamic transcriptome. Front Genet. 2019;10 doi: 10.3389/fgene.2019.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Price AM, Hayer KE, Depledge DP, Wilson AC, Weitzman MD. Novel splicing and open reading frames revealed by long-read direct RNA sequencing of adenovirus transcripts. BioRxiv 2019:2019.12.13.876037. 10.1101/2019.12.13.876037. [DOI]
- 106.Pitt ME, Nguyen SH, Duarte TPS, Teng H, Blaskovich MAT, Cooper MA, et al. Evaluating the genome and resistome of extensively drug-resistant Klebsiella pneumoniae using native DNA and RNA Nanopore sequencing. Gigascience 2020;9:giaa002. 10.1093/gigascience/giaa002. [DOI] [PMC free article] [PubMed]
- 107.Smith A.M., Jain M., Mulroney L., Garalde D.R., Akeson M., Wieden H.-J. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS ONE. 2019;14:e0216709. doi: 10.1371/journal.pone.0216709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Keller M.W., Rambo-Martin B.L., Wilson M.M., Ridenour C.A., Shepard S.S., Stark T.J. Direct RNA sequencing of the coding complete influenza a virus genome. Sci Rep. 2018;8 doi: 10.1038/s41598-018-32615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keegan KP, Glass EM, Meyer F. MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol. Biol., vol. 1399, Humana Press Inc.; 2016, p. 207–33. 10.1007/978-1-4939-3369-3_13. [DOI] [PubMed]
- 110.Semmouri I., De Schamphelaere K.A.C., Mees J., Janssen C.R., Asselman J. Evaluating the potential of direct RNA nanopore sequencing: Metatranscriptomics highlights possible seasonal differences in a marine pelagic crustacean zooplankton community. Mar Environ Res. 2020;153:104836. doi: 10.1016/j.marenvres.2019.104836. [DOI] [PubMed] [Google Scholar]
- 111.Bengtsson-Palme J., Hartmann M., Eriksson K.M., Pal C., Thorell K., Larsson D.G.J. METAXA2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour. 2015;15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 112.Boratyn G.M., Thierry-Mieg J., Thierry-Mieg D., Busby B., Madden T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinf. 2019;20:1–19. doi: 10.1186/s12859-019-2996-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Conesa A., Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:1–12. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ying C., Goeke J., Wan Y.K. GoekeLab/bambu: bambu release version 0.3.0 (Version v0.3.0) Zenodo. 2020 doi: 10.5281/zenodo.3963706. [DOI] [Google Scholar]
- 116.Sahlin K., Medvedev P. De novo clustering of long-read transcriptome data using a greedy, quality value-based algorithm. J Comput Biol. 2020;27:472–484. doi: 10.1089/cmb.2019.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang M., Cousineau A., Liu X., Luo Y., Sun D., Li S. Direct metatranscriptome RNA-seq and multiplex RT-PCR amplicon sequencing on nanopore MinION – promising strategies for multiplex identification of viable pathogens in food. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kafetzopoulou LE, Efthymiadis K, Lewandowski K, Crook A, Carter D, Osborne J, et al. Assessment of metagenomic Nanopore and Illumina sequencing for recovering whole genome sequences of chikungunya and dengue viruses directly from clinical samples. Eurosurveillance 2018;23. 10.2807/1560-7917.ES.2018.23.50.1800228. [DOI] [PMC free article] [PubMed]
- 119.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Quick J. nCoV-2019 sequencing protocol v3 (LoCost). ProtocolsIo 2020. 10.17504/protocols.io.bdp7i5rn. [DOI]
- 123.Freed N.E., Vlková M., Faisal M.B., Silander O.K. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and oxford nanopore rapid barcoding. Biol Methods Protoc. 2020;5 doi: 10.1093/biomethods/bpaa014. bpaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.021. 990-996.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Viehweger A., Krautwurst S., Lamkiewicz K., Madhugiri R., Ziebuhr J., Hölzer M. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29:1545–1554. doi: 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Taiaroa G, Rawlinson D, Featherstone L, Pitt M, Caly L, Druce J, et al. Direct RNA sequencing and early evolution of SARS-CoV-2. BioRxiv 2020:2020.03.05.976167. 10.1101/2020.03.05.976167. [DOI]
- 129.Casadesús J., Low D. Epigenetic Gene Regulation in the Bacterial World. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Beaulaurier J., Schadt E.E., Fang G. Deciphering bacterial epigenomes using modern sequencing technologies. Nat Rev Genet. 2019;20:157–172. doi: 10.1038/s41576-018-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 132.Stoiber M, Quick J, Egan R, Eun Lee J, Celniker S, Neely RK, et al. De novo identification of DNA modifications enabled by genome-guided Nanopore signal processing. BioRxiv 2017:94672. 10.1101/094672. [DOI]
- 133.McIntyre A.B.R., Alexander N., Grigorev K., Bezdan D., Sichtig H., Chiu C.Y. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ni P, Huang N, Zhang Z, Wang D-P, Liang F, Miao Y, et al. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 2019;35:4586–95. 10.1093/bioinformatics/btz276. [DOI] [PubMed]
- 135.Liu Q., Fang L., Yu G., Wang D., Xiao C.-L., Wang K. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat Commun. 2019;10:2449. doi: 10.1038/s41467-019-10168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tourancheau A, Mead EA, Zhang X-S, Fang G. Discovering and exploiting multiple types of DNA methylation from individual bacteria and microbiome using nanopore sequencing. BioRxiv 2020:2020.02.18.954636. 10.1101/2020.02.18.954636. [DOI] [PMC free article] [PubMed]
- 138.Guillen-Guio B., Hernandez-Beeftink T., Ciuffreda L., Rodríguez-Pérez H., Domínguez D., Baez-Ortega A. Could lung bacterial dysbiosis predict ICU mortality in patients with extra-pulmonary sepsis? A proof-of-concept study. Intensive Care Med. 2020;46:2118–2120. doi: 10.1007/s00134-020-06190-4. [DOI] [PubMed] [Google Scholar]
- 139.Benítez-Páez A, Hartstra A V, Nieuwdorp M, Sanz Y. Strand-wise and bait-assisted assembly of nearly-full rrn operons applied to assess species engraftment after faecal microbiota transplantation. BioRxiv 2020:2020.09.11.292896. 10.1101/2020.09.11.292896. [DOI]
- 140.Wick R.R., Judd L.M., Holt K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vereecke N., Bokma J., Haesebrouck F., Nauwynck H., Boyen F., Pardon B. High quality genome assemblies of Mycoplasma bovis using a taxon-specific Bonito basecaller for MinION and Flongle long-read nanopore sequencing. BMC Bioinf. 2020;21(1) doi: 10.1186/s12859-020-03856-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loose M., Malla S., Stout M. Real-time selective sequencing using nanopore technology. Nat Methods. 2016;13:751–754. doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kovaka S., Fan Y., Ni B., Timp W., Schatz M.C. Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bao Y, Wadden J, Erb-Downward JR, Ranjan P, Dickson RP, Blaauw D, et al. Real-Time, Direct Classification of Nanopore Signals with SquiggleNet. BioRxiv 2021:2021.01.15.426907. 10.1101/2021.01.15.426907. [DOI] [PMC free article] [PubMed]
- 145.van der Helm E., Imamovic L., Hashim Ellabaan M.M., van Schaik W., Koza A., Sommer M.O.A. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1328. [DOI] [PMC free article] [PubMed] [Google Scholar]