Abstract
Augmenting shoot multiplication through genetic engineering is an emerging biotechnological application desirable in optimizing regeneration of genetically modified plants on selection medium and rapid clonal propagation of elite cultivars. Here, we report the improved shoot multiplication in transgenic banana lines with overexpression of MusaSNAC1, a drought-associated NAC transcription factor in banana. Overexpression of MusaSNAC1 induces hypersensitivity of transgenic banana lines toward 6-benzylaminopurine ensuing higher shoot number on different concentrations of 6-benzylaminopurine. Altered transcript levels of multiple genes involved in auxin signaling (Aux/IAA and ARFs) and cytokinin signaling pathways (ARRs) in banana plants overexpressing MusaSNAC1 corroborate the hypersensitivity of transgenic banana plants toward 6-benzylaminopurine. Modulation in expression of ARRs reported to be involved in ABA-hypersensitivity and closure of stomatal aperture correlates with the function of MusaSNAC1 as a drought-responsive NAC transcription factor. Present study suggests a prospective cross talk between shoot multiplication and drought responses coordinated by MusaSNAC1 in banana plants.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02744-5.
Keywords: ARRs, Aux/IAA, Banana, NAC transcription factor, Shoot multiplication, SNAC1
Introduction
In vitro shoot proliferation is an important factor for augmenting the banana productivity globally. This is of paramount importance in case of species propagated through vegetative means as establishment of propagules is generally prolonged and uncertain in nature. Auxin and cytokinin have pivotal roles in the plant development including shoot regeneration (Skoog and Miller 1957). In particular, high cytokinin-to-auxin ratios are aptly established to induce shoot formation as substantiated by roles of cytokinin in formation of shoot apical meristem (Skoog and Miller 1957; Kurakawa et al. 2007). Auxin are known to suppress shoot branching, while cytokinin stimulates shoot multiplication (Leyser 2009). Cytokinin mediated signals are conveyed as multifarious phosphorelay cascades consisting of a complex two component system having two contrasting types of response regulators, commonly known as ARRs or Arabidopsis response regulators (Heyl and Schmülling 2003). Experimental evidences point toward negative roles of type-A ARRs (ARR3-ARR9, ARR15-ARR17, ARR22, and ARR24) in shoot regeneration, while type-B ARRs (ARR1-ARR2, ARR10–ARR14, ARR18–ARR21, and ARR23) are demonstrated to augment shoot proliferation in plants (Hwang and Sheen 2001; Hutchison and Kieber 2002). Auxin initiates its signaling through degradation of Auxin/Indole-3-acetic acid (Aux/IAAs) proteins which relieves their repressive dimerization with auxin response factors (ARFs) proteins (Liscum and Reed 2002; Powers and Strader 2020). Auxin acts as “molecular glue” between Aux/IAAs and F-box E3 ubiquitin ligase allowing polyubiqutination followed by degradation of Aux/IAA proteins (Tan et al. 2007). In Arabidopsis, 29 Aux/IAAs are predicted and many of them appears to be redundant in their functions (Liscum and Reed 2002). Despite available information on hormonal signaling in shoot proliferation, the knowledge in particular about independent regulators of shoot branching appears to be patchy. Transcription factors such as LAS (LATERAL SUPPRESSOR) from Arabidopsis, MOC1 (MONOCULM1), and TAD1 (TILLERING AND DWARF1) from rice belongs to GRAS family transcription factors with important roles in lateral branching (Li et al. 2003; Greb et al. 2003; Xu et al. 2012). Besides shoot branching, MOC1 and TAD1 also control plant height which, hence, can be utilized as potential tool for improving plant productivity through optimization of height and branching pattern in plants (Li et al. 2003; Xu et al. 2012; Mathan et al. 2016). Other transcription factors involved in direct or indirect regulation of lateral shoot multiplication such as BA1 (BARREN STALK1) in maize, BL (BLIND) in tomato, and TB1 (TEOSINTE BRANCHED1) in rice and maize have also been reported (Takeda et al. 2003; Schmitz et al. 2002; Gallavotti et al. 2004).
NAC-domain containing proteins belong to one of the largest family of transcription factors in plants with pivotal roles such as developmental regulation, stress responses, and regulation of multiple signaling pathways (Olsen et al. 2005; Kim et al. 2007; Negi et al. 2018b; Singh et al. 2021). Overexpressing SNAC1 gene of rice can significantly improves drought and salinity tolerance by stimulating photosynthesis rate and boosting relative water content of transgenic ramie plants under stress (An et al. 2015). Some NAC transcription factors influence the developmental parameter such as improved growth of different plant organs (Xie et al. 2000; Negi et al. 2016). Increased transcript level of NAC1 transcription factor from Arabidopsis modulates auxin signaling pathway culminating in bigger plants with improved leaf area, stem girth, frequency of lateral roots, and weight of transgenic lines (Xie et al. 2000). Overexpressing NAC68 transcription factor from banana enhances height and root biomass of transgenic lines besides reducing secondary wall thickness of xylem vessels and changing the expression of ARFs (Auxin response factors) and Aux/IAA genes (Negi et al. 2016, 2019). NAC transcription factor also regulates shoot apical meristem (SAM) and boundary formation for organ separation at embryonic, floral, and vegetative phases (Souer et al.1996; Aida et al. 1997; Sablowski and Meyerowitz 1998; Vroemen et al. 2003). Despite having enormous roles in plant development and architecture, only few of the NAC transcription factors have been assigned functions in lateral shoot multiplication. OsNAC2, a NAC transcription factor of rice augments shoot branching in transgenic rice lines and thus can be prospectively utilized for improving photosynthetic efficiency leading to increased crop yield (Mao et al. 2007). Another report delineates positive effect of overexpressing CUC1 and CUC2 transcription factors on frequency of adventitious shoot formation on calli derived from hypocotyl explants (Daimon et al. 2003). Recently, RNA-Seq analysis of rice plants overexpressing SNAC1 gene indicates significant alteration in expression of a plethora of genes ranging from those involved in stress responses, transcriptional regulation, and auxin responsiveness suggesting the roles of SNAC1 in regulating complex mechanisms in rice plants (Li et al. 2019).
In a previous study, we reported the improved drought tolerance linked with pronounced stomatal closure by H2O2 generation in guard cells of transgenic banana lines overexpressing MusaSNAC1 cells (Negi et al. 2018a). MusaSNAC1 regulates multiple stress responsive genes and influences their transcription after direct binding to their promote region (Negi et al. 2018a). In the present work, we report the improved shoot multiplication in transgenic banana lines observed after overexpressing SNAC1 transcription factor of banana. Our work suggest that overexpressing MusaSNAC1 induced hypersensitivity of banana plants toward 6-benzylaminopurine (6-BAP). Transgenic banana plants with overexpression of MusaSNAC1 displayed altered expression of multiple genes involved in auxin signaling pathway (Aux/IAA and ARFs) and cytokinin signaling pathway (ARRs). The present study points toward functions of MusaSNAC1 transcription factor in auxin and cytokinin signaling pathways for a tighter control over shoot multiplication in banana plants.
Material and methods
Tissue culture conditions and plant material
Transgenic lines of Musa cv. Rasthali (AAB genome) were regenerated and characterized in a previous report (Negi et al. 2018a). The shoot multiplication medium composed of MS medium (Murashige and Skoog) supplemented with 30 gm L−1 sucrose, 2 mg L−1 6-BAP, and 30 mg L−1 adenine sulphate. Individual shoots were elongated and rooted on MS medium supplemented with 30 gm L−1 sucrose and NAA (1 mg L−1). Tissue culture plants were maintained in control conditions of a culture facility maintaining 25 ± 2 °C with a 16 h light and 8 h dark regime. Following rooting, the elongated shoots of transgenic lines were hardened in sterile soil in pots in ambient conditions of green house facility.
Analysis of hypersensitivity of transgenic lines toward 6-benzylaminopurine (6-BAP)
Single shoot of similar age and size from control plant and transgenic lines was cultured on 6-BAP-free MS medium or MS medium supplemented with either 0.2 mg L−1 or 0.5 mg L−1 of 6-benzylaminopurine. The number of shoots obtained after 1 month of culture from each plant on different treatment was individually isolated and counted. The experiment was conducted with three replications.
Isolation of total RNA and synthesis of first-strand cDNA
Leaves collected from 2 month old plants in green house were used for total RNA isolation. Leaves were finely grounded to powder using liquid nitrogen in a mortar pestle and the powder was used with Concert plant RNA reagent (Invitrogen, USA) and RNeasy plant mini kit (Qiagen, Hilden, Germany) as described earlier (Tak et al. 2017). The traces of genomic DNA contamination in RNA preparation were eliminated with the help of on-column DNAase digestion (Qiagen, Hilden, Germany). Integrity of RNA was analyzed on 1% agarose gel and then first-strand cDNA was synthesized using 2 μg total RNA and thermoscript AMV-RT (Invitrogen, USA) following the kit manufacturer’s instructions. The cDNA was diluted 1:50 with milliQ water and used for expression analysis of target genes.
Expression of auxin and cytokinin responsive genes
Multiple genes involved in auxin signaling (Aux/IAA and ARFs) and cytokinin signaling pathways (histidine kinases, ARRs, and histidine-containing phosphotransfer proteins) were identified from NCBI (National Center for Biotechnology Information) database and genome sequence database (https://banana-genome-hub.southgreen.fr/) of DH-Pahang, a doubled haploid Musa acuminata genotype (AA) and their expression were quantified by quantitative RT-PCR assay. Expression levels of total 23 auxin signaling pathway genes and 30 cytokinin signaling pathway genes were analyzed in qRT-PCR experiments. Quantitative RT-PCR was performed on a rotor gene-Q RT-PCR instrument (Qiagen, Germany) using KAPA SYBR fast universal qPCR Master Mix (2X) (Sigma, USA; Catalogue number: KK4601). The cycling conditions were: 94 °C (5 min) followed by 30 cycles of 94 °C (25 s), 56 °C (25 s), and 72 °C (25 s). The specificity of primer pair annealing was monitored by introducing a melting curve analysis in qRT-PCR run. During each qPCR run, expression of MusaEF1α gene was also monitored using primers: FP:5′-CCGATTGTGCTGTCCTCATT-3′ and RP:5′-TTGGCACGAAAGGAATCTTCT-3′. The Ct values of target genes obtained after the qPCR run were normalized by Ct values of MusaEF1α gene, and then, fold value change above control values was estimated with the help of 2−ΔΔCt as described in the comparative Ct method previously (Schmittgen and Livak 2008). The locus identifier and primer pairs of the genes analyzed are provided as in the supplementary information.
Results
Overexpression of MusaSNAC1 increases shoot proliferation
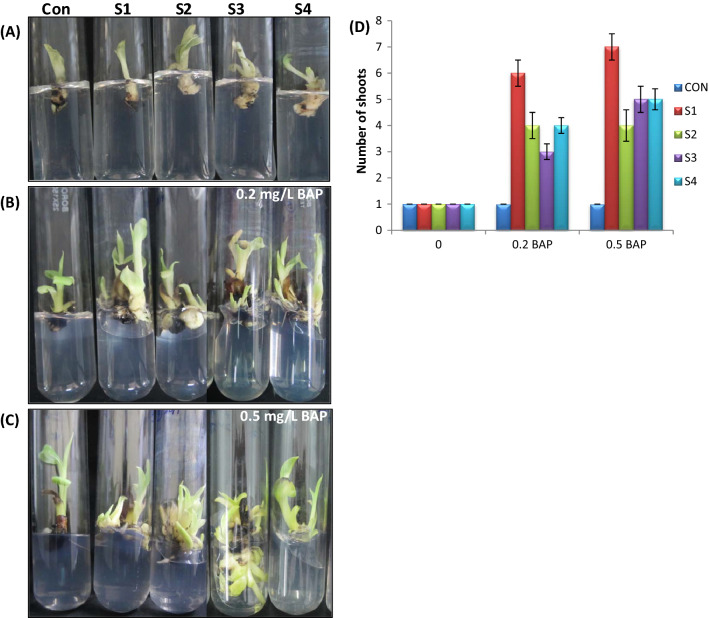
Our observation suggested that transgenic lines overexpressing MusaSNAC1 generate more number of shoots than control plants from a single shoot explant on shoot multiplication medium-containing BAP (2 mg L−1). Subsequent culturing of transgenic plants also gave similar kind of observation, suggesting that MusaSNAC1 may have a probable function in shoot proliferation of banana plants (Fig. 1).
Fig. 1.
Overexpression of MusaSNAC1 augments shoot proliferation. Transgenic lines generate more number of shoots compared to control plant on shoot multiplication medium. Figure shows shoot abundance obtained from culture of single shoot of control plant and transgenic lines on medium with 2 mg L−1 BAP. S1–S4; Transgenic lines
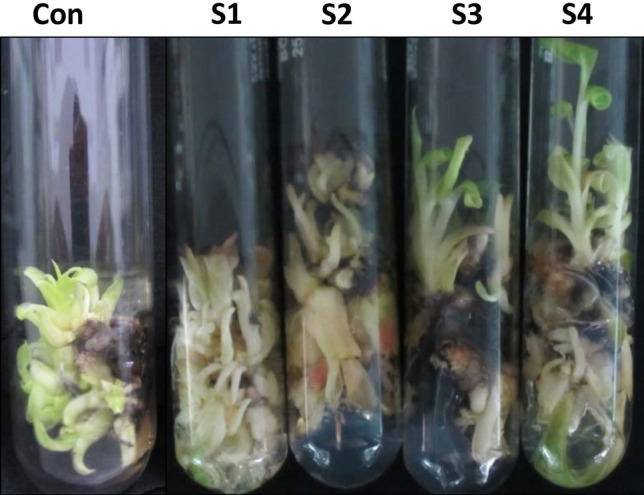
Overexpression of MusaSNAC1-induced hypersensitivity toward 6-BAP
As transgenic lines were generating more number of shoots on 6-BAP supplemented medium, we analyzed this response with different concentrations of 6-BAP. 6-BAP is the most widely utilized plant cytokinin and regulates cell division and shoot multiplication in plants. Single shoot explant of transgenic lines and control plants were cultured on medium supplemented with either 0.2 mg L−1 or 0.5 mg L−1 concentration of 6-BAP for 1 month for shoot regeneration (Fig. 2a). Under both the concentrations of 6-BAP, the control explants failed to regenerate additional shoots, while all the transgenic lines regenerates remarkably higher number of shoots (Fig. 2b–d). Therefore overexpression of MusaSNAC1 induce hypersensitivity of banana plants toward 6-BAP which suggest potential roles of MusaSNAC1 in regulating cytokinin signaling pathway and shoot multiplication in banana plants. In medium devoid of 6-BAP, both control and transgenic lines failed to regenerate additional shoots (Fig. 2d). The transgenic lines have 8–12-fold expression of MusaSNAC1 with respect to control and line S2 has lowest level of MusaSNAC1 overexpression (Negi et al. 2018a). The varied level of shoot multiplication over control among transgenic lines might be due to factors such as fold change in expression of MusaSNAC1, insertion effects of T-DNA in genome, and other unexplored contributing factors.
Fig. 2.
Overexpression of MusaSNAC1-induced hypersensitivity of banana plants toward 6-benzylaminopurine (BAP). a Single shoot of control plant and transgenic lines was cultured on medium-containing either 0.2 mg L−1 or 0.5 mg L−1 concentration of 6-benzylaminopurine. b–c Transgenic lines produce more shoots than control plant on both 0.2 mg L−1 or 0.5 mg L−1 concentration of 6-benzylaminopurine. The visual results were photographed and a representative picture is shown. d Number of shoots of control plant and transgenic lines after culturing in medium with different concentration of BAP. Substantially higher number of shoots were obtained with transgenic lines. Results are presented as mean ± SD
QPCR analysis of auxin-responsive genes
Increased regeneration of shoots due to overexpression of MusaSNAC1 prompted us to analyze the expression of auxin signaling pathway genes as shoot multiplication is outcome of interplay between concentrations of auxin and cytokinin. We performed transcript level analysis of Aux/IAA (Indole acetic acid induced) and ARFs (auxin response factors) type auxin-responsive genes in transgenic banana lines and control plants. Among 23 auxin signaling pathways genes analyzed, expression of at least four Aux/IAA encoding genes (GSMUA_Achr4T21020_001, GSMUA_Achr4T22520_001, GSMUA_Achr9T24330_001, and GSMUA_Achr9T25550_001) and one ARF encoding gene (GSMUA_Achr8T13620_001) was remarkably elevated over control expression (Fig. 3). The identities of these differentially expressed auxin signaling pathway genes as annotated in banana genome sequence database and their closest homologue in Arabidopsis are provided in Table 1.
Fig. 3.
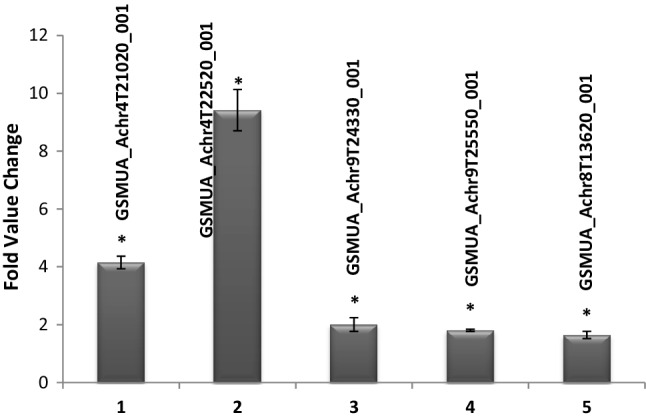
Expression analysis of Aux/IAA (indole acetic acid induced) and ARFs (auxin response factors) type auxin-responsive genes. The transcript abundance of different genes was estimated after quantitative RT-PCR in transgenic banana plants with overexpression of MusaSNAC1. The data values obtained after normalization with the expression of EF1α gene of banana spp. were represented as fold change over control values. Identity of different genes is represented as banana genome locus identifier on the top of each bar. Each data point represented mean ± SD of at least three independent replications. Statistically significant difference for at least 5% (P ≤ 0.05) is denoted with an asterisk (*)
Table 1.
Musa genome locus identifier and primer sequences of genes displaying altered expression in MusaSNAC1 overexpressing lines
| S. No | Musa genome locus identifier | Forward primer and reverse primer | Annotation in banana genome sequence database | Closest homologues in Arabidopsis (accession number) |
|---|---|---|---|---|
| 1 | GSMUA_Achr3T08680_001 |
5′-GTGATCATGTCATCCGAAAA-3’ 5′-GTCTCGGTCTTCTCCTCTCA-3’ |
Two-component response regulator ARR9 |
ARR8 (XP_020884524.1); ARR9 (NP_001325622.1) |
| 2 | GSMUA_Achr5T08230_001 |
5′-ACAGGGTATGACCTCCTCAA-3’ 5′-CTGCTGATGATGATGGTGTT-3’ |
Two-component response regulator ARR9 |
ARR8 (XP_020884524.1); ARR9 (NP_001325622.1) |
| 3 | GSMUA_Achr10T06040_001 |
5′-AAACCGAGTTCGTCCACTTA-3’ 5′-TATTGCATCAACATCCAACC-3’ |
Putative two-component response regulator |
Arabidopsis response regulator 12 (NP_180090.6); ARR10 (XP_002867269.1) |
| 4 | GSMUA_Achr1T17800_001 |
5′-GCAGCTCCAAGTACAGAGTG-3’ 5′-GGCTTGAGCAGGAAATCTAC-3’ |
Two-component response regulator ARR9 |
ARR4 (NP_001321924.1); ARR9 (NP_001325622.1) |
| 5 | GSMUA_Achr11T08310_001 |
5′-CTCTCAGCAGAAATGGGATT-3’ 5′-GAGCCGTATTCACCCTTAAA-3’ |
Putative two-component response regulator |
Arabidopsis response regulator 12 |
| 6 | GSMUA_Achr2T05130_001 |
5′-CACCTCTGAGTGATCCAACA-3’ 5′-CATTCTCTGATGCCTGACAC-3’ |
Two-component response regulator ARR12 |
Arabidopsis response regulator 12 |
| 7 | GSMUA_Achr4T21020_001 |
5′-ACTGTGGCTCTCAAGGAATGA-3’ 5′-TTGCACTTCTCCATTGCTCTT-3’ |
Auxin-responsive protein IAA30 |
Indoleacetic acid-induced protein 14 (NP_193191.2); IAA9 (OAO96506.1) |
| 8 | GSMUA_Achr4T22520_001 |
5′-CTGGCTGTCCAATCTGAGAAG-3’ 5′-ACCCACAGCTTGATGAAACAC-3’ |
Indoleacetic acid-induced protein 30 |
Indoleacetic acid-induced protein 16 |
| 9 | GSMUA_Achr9T24330_001 |
5′-TGTAGCCAAGAAACAGGTCGT-3’ 5′-TCCTTCTGCACCACCTAAAGA-3’ |
Auxin-induced protein 22D |
Indoleacetic acid-induced protein 4 |
| 10 | GSMUA_Achr9T25550_001 |
5′-AGGGGGTTCGCTGAGACTAT-3’ 5′-CCTTGCTTCCCTTCTCAGAGT-3’ |
Indoleacetic acid-induced protein 16 |
Indoleacetic acid-induced protein 16 |
| 11 | GSMUA_Achr8T13620_001 |
5′-CCCTGAAGGGAACACTGACC-3’ 5′-GCAGATGCATCACCGAGAGT-3’ |
Auxin response factor 12 |
Auxin response factor 8 |
QPCR analysis of cytokinin responsive genes
Cytokinin are key players for lateral branching in plants, hence, to further obtain an insight in the hypersensitivity of transgenic lines toward 6-BAP, we examined transcript levels of multiple cytokinin signaling pathway genes. Among 3 histidine kinases, 22 ARRs and 5 histidine-containing phosphotransfer protein coding genes, expression of at least 6 ARRs coding genes (GSMUA_Achr3T08680_001, GSMUA_Achr5T08230_001, GSMUA_Achr10T06040_001, GSMUA_Achr1T17800_001, GSMUA_Achr11T08310_001, and GSMUA_Achr2T05130_001) were found to be significantly reduced suggesting an imperative role of MusaSNAC1 in cytokinin signaling pathways (Fig. 4). The identities of these differentially expressed cytokinin signaling pathway genes as annotated in DH-Pahang genome sequence database and their closest homologue in Arabidopsis is provided in Table 1.
Fig. 4.
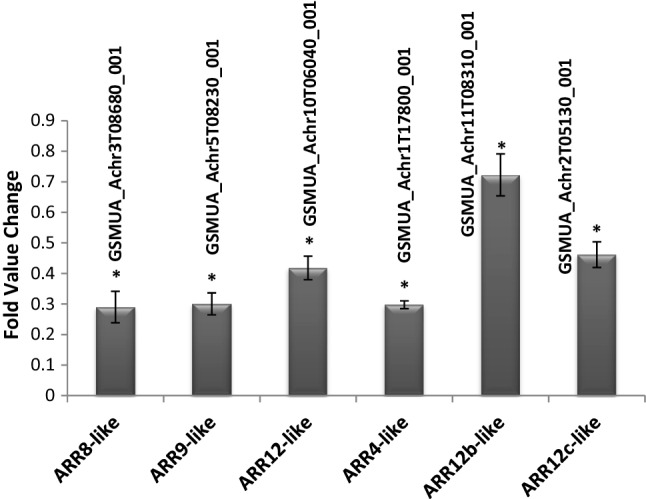
Quantitative RT-PCR analysis for transcript abundance of cytokinin signaling pathway genes. Transcript abundance of different genes was estimated after quantitative real-time RT-PCR of transgenic banana plants overexpressing MusaSNAC1. The data values obtained after normalization with the expression of EF1α gene of banana spp. were represented as fold change over control values. Identity of different genes is represented as banana genome locus identifier on the top of each bar. Each data point represented mean ± SD of at least three independent replications. Statistically significant difference for at least 5% (P ≤ 0.05) is denoted with an asterisk (*)
Discussion
Auxin and cytokinin plays key roles in multifarious aspects of plant growth and development such as formation of lateral organs and meristem development (Waldie and Leyser 2018). Auxins and cytokinins have antagonistic activities in control of shoot and root formation (Kurepa et al. 2019; Müller and Leyser 2011). Auxin negatively impacts shoot multiplication by inhibiting development of axillary buds, while cytokinin promotes shoot multiplication (Leyser 2009; Müller and Leyser 2011). This striking antagonistic effect of auxins and cytokinins on plant development was established nearly half a decade back by pioneering work of Skoog and Miller (1957) which demonstrates shoot induction in the presence of high cytokinin-to-auxin ratio. Previous reports have analyzed the effects of silencing Aux/IAA and ARF genes in Arabidopsis and results have demonstrated the redundant roles of different member of these genes (Okushima et al. 2005; Overvoorde et al. 2005). Single mutants of ARFs in Arabidopsis failed to trigger a phenotypic aberration, albeit it was observed in case of few ARFs; however, increased transcript levels of ARFs can potentially lead to auxin-mediated phenotypic abnormalities (Okushima et al. 2005; Tian et al. 2004). Transcript level of one ARF (GSMUA_Achr8T13620_001) with close homology with Arabidopsis ARF8 was elevated in banana plants overexpressing MusaSNAC1. ARF8 is an important auxin-responsive gene for vegetative and floral development and it negatively control the turnover of free IAA in plants (Nagpal et al. 2005; Tian et al. 2004). Overexpression of ARF8 reduces free IAA content in hypocotyl and roots causing auxin deprivation symptoms leading to stunted hypocotyl and diminished abundance of lateral roots (Tian et al. 2004). This suggests that increased transcript level of GSMUA_Achr8T13620_001 could potentially leads to auxin depletion phenotype which in part explains the augmented shoot multiplication observed in transgenic lines. We did not notice any phenotypic variations in the transgenic banana lines overexpressing MusaSNAC1 as reported by Tian et al. (2004) about varied hypocotyl length in Arabidopsis plants with altered expression of ARF8. This might be due to the fact that the banana cultivar Rasthali is a vegetatively propagated crop which makes the observation of hypocotyl length impossible. Moreover, the stem is underground in banana and aerial pseudostem in transgenic lines appeared similar to control. Unlike ARFs, even triple mutants of Aux/IAA genes in Arabidopsis failed to develop auxin-mediated phenotypic anomalies despite that dominant mutations leading to gain of function are reported to induce severe phenotypic peculiarities (Overvoorde et al. 2005; Leyser et al. 1996). In the present study transcript level of four Aux/IAA encoding genes were escalated, three (GSMUA_Achr4T21020_001, GSMUA_Achr4T22520_001, and GSMUA_Achr9T25550_001) of them having maximum elevation exhibits high-sequence similarities with functionally characterized IAA14 and IAA16 of Arabidopsis (Table 1). Dominant gain-of-function mutation in IAA16 in Arabidopsis drastically diminished auxin responses such as fewer lateral roots and reduced plant height (Rinaldi et al. 2012). Gain of function studies of IAA14 in Arabidopsis also results in loss of auxin-mediated effects such as reduced lateral root abundance and lower auxin-induced gene expression (Fukaki et al. 2002). These reports indicate that increased expressions of GSMUA_Achr4T21020_001, GSMUA_Achr4T22520_001, and GSMUA_Achr9T25550_001 in transgenic banana have potential to overcome auxin-mediated effects such as suppression of shoot multiplication which is consistent with our observations in the present study. Cytokinin-induced signals are transmitted by response regulators (ARRs) and type-A ARRs negatively regulate shoot multiplication (Heyl and Schmülling 2003; Hwang and Sheen 2001). In the present study, expressions of 6 ARR coding genes were significantly reduced in transgenic banana lines. These aforementioned 6 ARR (GSMUA_Achr3T08680_001, GSMUA_Achr5T08230_001, GSMUA_Achr10T06040_001, GSMUA_Achr1T17800_001, GSMUA_Achr11T08310_001, and GSMUA_Achr2T05130_001) exhibits high-sequence similarities with ARR8, ARR9, ARR12, and ARR4 of Arabidopsis. ARR4, ARR8, and ARR9 of Arabidopsis are type-A ARRs and hence negatively regulates shoot multiplication, while ARR12 is type-B ARR and promotes shoot regeneration in Arabidopsis (Hwang and Sheen 2001; Hutchison and Kieber 2002). Overexpression of ARR4, ARR5, ARR6, ARR7, and ARR9 induce cytokinin resistance which was demonstrated by increased root growth in the presence of benzyladenine (BA) in a root elongation assay (To et al. 2007). The aforementioned study and other reports indicate that reduced transcript levels of GSMUA_Achr3T08680_001, GSMUA_Achr5T08230_001, and GSMUA_Achr1T17800_001 can augment cytokinin-mediated phenotypes including shoot multiplication as observed in the present study (To et al. 2007; Hwang and Sheen 2001). This become more evident with repression of cytokinin induced genes and diminished shoot multiplication observed after overexpression of ARR8 of Arabidopsis (Osakabe et al. 2002). Despite that, our study also found repression in expression of three ARRs (GSMUA_Achr10T06040_001, GSMUA_Achr11T08310_001, and GSMUA_Achr2T05130_001) with high-sequence similarity with Arabidopsis ARR12 and ARR10 which are type-B ARRs and acts as positive mediators of cytokinin-induced effects. Overexpressing ARR12 of Arabidopsis improves shoot regeneration, while arr12 mutants have impaired cytokinin responsiveness and shoot regeneration (Dai et al. 2017). Despite that, arr1, arr10, arr12 mutants were reported to have superior drought tolerance which was attributed in part to ABA-hypersensitivity and reduced stomatal aperture establishing the negative function of ARR1, ARR10, and ARR12 in drought tolerance (Nguyen et al. 2016). In line with report by Nguyen et al. (2016), repression in ARR10 and ARR12 like ARRs in banana is consistent with improved drought tolerance due to higher relative water content and increased drought induced stomatal closure of transgenic lines overexpressing MusaSNAC1 (Negi et al. 2018a). Despite that, the improved shoot multiplication due to overexpression of MusaSNAC1 is an outcome of finely tuned balance between auxin and cytokinin signaling pathways meticulously orchestrated by a stress inducible NAC transcription factor MusaSNAC1 in banana plants. Moreover, a recent report indicates important roles of SMALL AUXIN UP RNA (SAUR) proteins in stomatal movements strengthening the potential roles of auxin signaling under drought conditions (Wong et al. 2021).
The present study provides vital information on genetic control of shoot regeneration and multiplication using banana as a model system. This is of great importance in rapid clonal propagation and genetic transformation of plant species such as those belonging to Malvaceae and Chenopodiaceae, wherein the regeneration is quite clumsy and arduous (Mustafa 2012). These genetic differences toward in vitro shoot regeneration despite augmenting cytokinin concentration in culture medium point toward potential in ability of such species to respond to cytokinin (Hill and Schaller 2013). This becomes evident when enhanced expression of ARR10 augmented plant regeneration by inducing hypersensitivity to cytokinin supplementation (Hill et al. 2013; Hill and Schaller 2013). The present study reports MusaSNAC1-induced hypersensitivity of banana plants toward 6-BAP leading to enhanced shoot multiplication. Improved shoot multiplication of transgenic lines is corroborated with differential expression of auxin and cytokinin signaling pathway genes. However, further work with emphasis on deletion analysis of Aux/IAA and ARFs in banana will lead to more insights in regulation of shoot branching in banana plants. The present study also points toward a potential cross talk between shoot multiplication and drought stress responses, wherein the SNAC1 functions at the cross roads of these imperative aspects of plant functions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors thank, Head, Nuclear Agriculture and Biotechnology Division, BARC for support and encouragement. SN thanks “Department of Science and Technology” (DST), New Delhi for DST INSPIRE Faculty award.
Author contributions
SN, HT, and TG conceived and designed research. SN and HT conducted experiments and analyzed data. SN, HT, and TG wrote the manuscript. All authors read and approved the manuscript.
Funding
The work was supported from funding of Department of Atomic Energy, Government of India.
Availability of data and materials
All the relevant data are contained within the manuscript.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Sanjana Negi and Himanshu Tak contributed equally for this study.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An X, Liao Y, Zhang J, Dai L, Zhang N, Wang B, Liu L, Peng D. Overexpression of rice NAC gene SNAC1 in ramie improves drought and salt tolerance. Plant Growth Regul. 2015;76:211–223. [Google Scholar]
- Dai X, Liu Z, Qiao M, Li J, Li S, Xiang F. ARR12 promotes de novo shoot regeneration in Arabidopsis thaliana via activation of WUSCHEL expression. J Integr Plant Biol. 2017;59:747–758. doi: 10.1111/jipb.12567. [DOI] [PubMed] [Google Scholar]
- Daimon Y, Takabe K, Tasaka M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 2003;44:113–121. doi: 10.1093/pcp/pcg038. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pe ME, Schmidt RJ. The role of barren stalk1 in the architecture of maize. Nature. 2004;432:630–635. doi: 10.1038/nature03148. [DOI] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17:1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Schmülling T. Cytokinin signal perception and transduction. Curr Opin Plant Biol. 2003;6:480–488. doi: 10.1016/s1369-5266(03)00087-6. [DOI] [PubMed] [Google Scholar]
- Hill K, Schaller GE. Enhancing plant regeneration in tissue culture: a molecular approach through manipulation of cytokinin sensitivity. Plant Signal Behav. 2013;8:10. doi: 10.4161/psb.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Mathews DE, Kim HJ, Street IH, Wildes SL, Chiang YH, Mason MG, Alonso JM, Ecker JR, Kieber JJ, Schaller GE. Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol. 2013;162:212–224. doi: 10.1104/pp.112.208736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ. Cytokinin signaling in Arabidopsis. Plant Cell. 2002;14:S47–S59. doi: 10.1105/tpc.010444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Kim HS, Park BO, Yoo JH, et al. Identification of a calmodulin binding NAC protein as a transcriptional repressor in Arabidopsis. J Biol Chem. 2007;282:36292–36302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Kurepa J, Shull TE, Smalle JA. Antagonistic activity of auxin and cytokinin in shoot and root organs. Plant Direct. 2019;3:e00121. doi: 10.1002/pld3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. The control of shoot branching: an example of plant information processing. Plant Cell Environ. 2009;32:694–703. doi: 10.1111/j.1365-3040.2009.01930.x. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F, et al. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- Li X, Chang Y, Ma S, Shen J, Hu H, Xiong L. Genome-wide identification of SNAC1-targeted genes involved in drought response in Rice. Front Plant Sci. 2019;10:982. doi: 10.3389/fpls.2019.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- Mao C, Ding W, Wu Y, Yu J, He X, Shou H, Wu P. Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol. 2007;176:288–298. doi: 10.1111/j.1469-8137.2007.02177.x. [DOI] [PubMed] [Google Scholar]
- Mathan J, Bhattacharya J, Ranjan A. Enhancing crop yield by optimizing plant developmental features. Development. 2016;143:3283–3294. doi: 10.1242/dev.134072. [DOI] [PubMed] [Google Scholar]
- Müller D, Leyser O. Auxin, cytokinin and the control of shoot branching. Ann Bot. 2011;107:1203–1212. doi: 10.1093/aob/mcr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa Y. The prerequisite of the success in plant tissue culture: high frequency shoot regeneration. In: Leva A, Rinaldi L, editors. Recent advances in plant in vitro culture. IntechOpen; 2012. pp. 63–90. [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Negi S, Tak H, Ganapathi TR. Expression analysis of MusaNAC68 transcription factor and its functional analysis by overexpression in transgenic banana plants. Plant Cell Tiss Org Cult. 2016;125:59–70. [Google Scholar]
- Negi S, Tak H, Ganapathi TR. A banana NAC transcription factor (MusaSNAC1) impart drought tolerance by modulating stomatal closure and H2O2 content. Plant Mol Biol. 2018;96:457–471. doi: 10.1007/s11103-018-0710-4. [DOI] [PubMed] [Google Scholar]
- Negi S, Tak H, Ganapathi TR. Xylem specific activation of 5' upstream regulatory region of two NAC transcription factors (MusaVND6 and MusaVND7) in banana is regulated by SNBE-like sites. PLoS ONE. 2018;13:e0192852. doi: 10.1371/journal.pone.0192852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Tak H, Ganapathi TR. Overexpression of MusaNAC68 reduces secondary wall thickness of xylem tissue in banana. Plant Biotechnol Rep. 2019;13:151–160. [Google Scholar]
- Nguyen KH, Ha CV, Nishiyama R, Watanabe Y, Leyva-González MA, Fujita Y, Tran UT, Li W, Tanaka M, Seki M, Schaller GE, Herrera-Estrella L, Tran LS. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc Natl Acad Sci USA. 2016;113:3090–3095. doi: 10.1073/pnas.1600399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana:unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Miyata S, Urao T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem Biophys Res Commun. 2002;293:806–815. doi: 10.1016/S0006-291X(02)00286-3. [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, Smith A, Yu G, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17:3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Strader LC. Regulation of auxin transcriptional responses. Dev Dyn. 2020;249:483–495. doi: 10.1002/dvdy.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi MA, Liu J, Enders TA, Bartel B, Strader LC. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol Biol. 2012;79:359–373. doi: 10.1007/s11103-012-9917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes Apetala3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA. 2002;99:1064–1069. doi: 10.1073/pnas.022516199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Koyama H, Bhati KK, Alok A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J Plant Res. 2021 doi: 10.1007/s10265-021-01270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–130. [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The NO APICAL MERISTEM gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Tak H, Negi S, Ganapathi TR. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma. 2017;254:803–816. doi: 10.1007/s00709-016-0991-x. [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003;33:513–520. doi: 10.1046/j.1365-313x.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tian CE, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004;40:333–343. doi: 10.1111/j.1365-313X.2004.02220.x. [DOI] [PubMed] [Google Scholar]
- To JP, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19:3901–3914. doi: 10.1105/tpc.107.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldie T, Leyser O. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 2018;177:803–818. doi: 10.1104/pp.17.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Klejchová M, Snipes SA, Nagpal P, Bak G, Wang B, Dunlap S, Park MY, Kunkel EN, Trinidad B, Reed JW, Blatt MR, Gray WM. SAUR proteins and PP2C.D phosphatases regulate H+-ATPases and K+ channels to control stomatal movements. Plant Physiol. 2021;185:256–273. doi: 10.1093/plphys/kiaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang Y, Yu Y, Duan J, Liao Z, Xiong G, Meng X, Liu G, Qian Q, Li J. Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering. Nat Commun. 2012;3:750. doi: 10.1038/ncomms1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the relevant data are contained within the manuscript.