Abstract
Sporulation is a specialized developmental program employed by a diverse set of bacteria which culminates in the formation of dormant cells displaying increased resilience to stressors. This represents a major survival strategy for bacteria facing harsh environmental conditions, including nutrient limitation, heat, desiccation, and exposure to antimicrobial compounds. Through dispersal to new environments via biotic or abiotic factors, sporulation provides a means for disseminating genetic material and promotes encounters with preferable environments thus promoting environmental selection. Several types of bacterial sporulation have been characterized, each involving numerous morphological changes regulated and performed by non-homologous pathways. Despite their likely independent evolutionary origins, all known modes of sporulation are typically triggered by limited nutrients and require extensive membrane and peptidoglycan remodeling. While distinct modes of sporulation have been observed in diverse species, two major types are at the forefront of understanding the role of sporulation in human health, and microbial population dynamics and survival. Here, we outline endospore and exospore formation by members of the phyla Firmicutes and Actinobacteria, respectively. Using recent advances in molecular and structural biology, we point to the regulatory, genetic, and morphological differences unique to endo- and exospore formation, discuss shared characteristics that contribute to the enhanced environmental survival of spores and, finally, cover the evolutionary aspects of sporulation that contribute to bacterial species diversification.
Keywords: Firmicutes, Actinobacteria, endospore, exospore, regulation, structural biology, bacterial evolution
Introduction
Bacteria utilize diverse adaptations for perseverance and propagation under adverse environmental conditions: one such adaptive mechanism is sporulation. The process is characterized by enhanced resilience to stressors, which allows for preservation of genetic material for prolonged periods of time. Sporulation occurs through various modes, many of which remain to be thoroughly investigated and characterized, such as production of akinetes by Cyanobacteria, budding spores within the phylum Chloroflexi, and fruiting body formation in myxobacteria (Proteobacteria) (Kaplan-Levy et al., 2010; Yabe et al., 2010; Muñoz-Dorado et al., 2016). However, the two most extensively studied modes of sporulation are performed by members of the phyla Firmicutes (endospore formation) and Actinobacteria (exospore formation). Here, we discuss their main mechanisms of gene regulation, metabolic and structural transformations, evolutionary significance in the diversity of current bacterial species, as well as outline their shared and contrasting characteristics.
Bacteria of the phylum Firmicutes have been traditionally distinguished by possession of a low-G + C genome, Gram-positive (monoderm) cell envelope comprising a cytoplasmic membrane (CM) surrounded by a thick layer of peptidoglycan (PG). With recent developments in genome-based taxonomy, this notion has been challenged by the discovery of Firmicutes with relatively high-G + C genomes (e.g., Geobacillus), as well as Gram-negative, or diderm, cell envelopes comprising an inner membrane (IM), a thin layer of PG, and a typical outer membrane (OM, e.g., Acetonema longum, class Negativicutes) (Yutin and Galperin, 2013). Unsurprisingly, as a result of such variation, Firmicutes have been successful in populating diverse environments, such as soil, ocean and sediments, and colonize various hosts, including insects and mammals. Many Firmicutes are also major human pathogens, e.g., Clostridium difficile, Bacillus anthracis, and Streptococcus pneumoniae. Some Firmicutes, notably within the classes Bacilli and Clostridia, are capable of endospore formation, a process that is restricted to the phylum (Figure 1A) (Beskrovnaya et al., 2020). Endospore formation is characterized by asymmetric cell division of the mother cell, which results in the formation of a smaller compartment (prespore or forespore) that is then engulfed in a phagocytosis-like manner (Tan and Ramamurthi, 2014). The prespore undergoes further maturation through modifications in the cell envelope and the gradual halting of metabolism, ultimately followed by release into the environment with lysis of the mother cell. Endospores are highly resilient dormant life forms that can withstand exposure to extreme heat, desiccation, ultraviolet radiation and other factors, highlighting their significance in pathogenesis (Table 1) (Setlow, 2014).
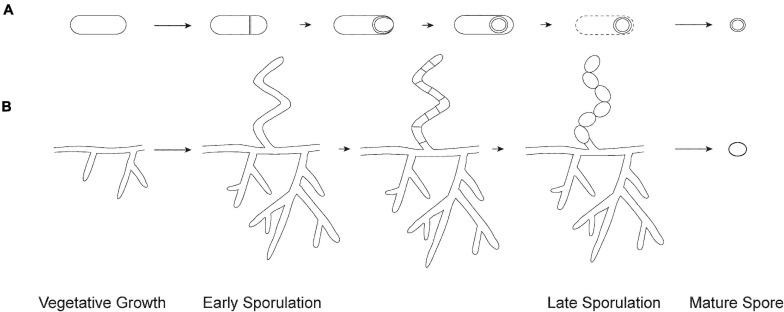
FIGURE 1.
Overview of sporulation. (A) Endospore formation in Firmicutes. The process begins with the formation of an asymmetric septum. Next, the larger compartment engulfs the smaller immature spore. The spore matures by the formation of protective layers and is released through lysis of the mother cell. (B) Exospore formation in Streptomyces. The process begins by aerial hyphae formation, which subsequently divide into numerous compartments. Each compartment matures into an exospore that gets released from the spore chain.
TABLE 1.
Effects of stressors on survival of endospores and exospores.
| Stressor | Endospore | Exospore |
| Wet heat (60°C) | Not determined | 60% survival after 10 min (Haiser et al., 2009) |
| Wet heat (80°C) | 50% survival after 30 h (Fox and Eder, 1969) | 0% survival after 10 min (Kawamoto et al., 1981) |
| UV 222 nm | 10% survival with 250 J/m2 (Clauss, 2006) | 10% survival with 127 J/m2 (Clauss, 2006) |
| UV 254 nm | 10% survival with 520 J/m2 (Clauss, 2006) | 10% survival with 85 J/m2 (Clauss, 2006) |
| Desiccation at 0% humidity | No effect on viability (Setlow, 2014) | 23% survival after 28 days at 25°C (McBride and Ensign, 1987) |
| Triple autoclaving (heat and pressure) | Survival at 121°C for 20 min, (O’Sullivan et al., 2015) | Not determined |
The Actinobacteria (high-G + C genomes) is another diverse phylum, including organisms with a variety of cell morphologies – from cocci and rods to more complex morphologies, such as differentiated mycelia. These bacteria live in a variety of environments, ranging from soil and marine environments to human skin, occupied by commensal organisms. A number of Actinobacteria are human, animal, or plant pathogens as well. Unlike in the Firmicutes, sporulation in Actinobacteria occurs by formation of exospores: new cells are produced through cell division, which then undergo morphological differentiation into mature spores (Hoskisson and Van Wezel, 2019). Speciation within the Actinobacteria has led to a wide variety of exospore formation modes, from single spores produced on undifferentiated mycelia, to the formation of sporangia. The best characterized form of sporulation within the Actinobacteria, however, is the long chains of exospores produced by the soil and ocean dwelling Streptomyces (McCormick and Flardh, 2012). Streptomyces grows vegetatively by tip extension and branching (Figure 1B). Specialized aerial hyphae extend upwards off the colony surface and undergo synchronous cell division to convert the multinucleate aerial hyphae into dozens of unigenomic spores (Szafran et al., 2020). These spores undergo maturation, which involves thickening of the spore wall and then entry into dormancy. Finally, exospores are presumably released from the spore chain by mechanical means. As is the case with endospores, exospores remain dormant in the environment until favorable growth conditions are sensed, at which point they germinate and resume vegetative growth (Sexton and Tocheva, 2020). Despite being a model system, less is known about the regulation and mechanics of sporulation in Streptomyces compared to Firmicutes.
Gene Regulation
The required set of environmental stimuli needed for initiating sporulation have eluded researchers for years. Generally, the main trigger for initiation of endo- and exospore formation appears to be depletion of nutrients – particularly of readily available carbon, nitrogen, and phosphorus sources (Higgins and Dworkin, 2012; McCormick and Flardh, 2012). However, individual species develop additional mechanisms for detection of niche-specific stimuli, such as exposure to oxygen in obligately anaerobic Firmicutes (Mearls et al., 2012). Although the environmental stimuli for sporulation remain somewhat comparable between Firmicutes and Actinobacteria, gene regulation is vastly different between the two phyla. Because sporulation involves extensive morphological transformations through the coordinated action of hundreds of proteins, it is a highly energetically costly process. Therefore, bacteria have evolved mechanisms to carefully monitor their environment and ensure the appropriate timing of each stage of the process.
Spo0A Is the Master Regulator in Activation of Endospore Formation
In Firmicutes, the ubiquitously distributed regulator protein Spo0A is recognized as the hallmark component of stress-activated gene expression machinery and, by extension, endospore formation (Ramos-Silva et al., 2019). Currently, over 80 genes, including spo0A, are known to be required for endospore formation (Galperin et al., 2021). Consistently, loss of spo0A from the genome correlates with the inability to sporulate across the phylum (Galperin et al., 2012). The Spo0A protein comprises two domains: a DNA-binding regulatory domain harboring a characteristic helix-turn-helix motif, and a sensory domain which accepts signal input through phosphorylation of a conserved aspartate residue by a cognate sensor kinase (Zhao et al., 2002). Several cell surface-associated and intracellular histidine kinases (Kin) implicated in direct and indirect phosphorylation of Spo0A have been identified in Bacilli and Clostridia (Jiang et al., 2000; Underwood et al., 2009; Steiner et al., 2011).
Unsurprisingly, despite possessing the structural and sequence features characteristic of histidine kinases, variability within the ligand-binding sites of their sensor domains makes it difficult to predict the exact signals recognized by each enzyme (Fabret et al., 1999). Therefore, the specific triggers and receptors that contribute to phosphorylation of Spo0A are not universally conserved between endospore formers. In Bacillus subtilis, for example, upon sensing extra- or intracellular signals, Kin proteins transfer a phosphoryl group onto the response regulator Spo0F, which in turn transfers the phosphate to the phosphotransferase Spo0B, which, lastly, phosphorylates Spo0A. Additionally, phosphatases present at each step in the phosphorelay system aid in finer regulation of phosphorylated levels of each protein and, ultimately, Spo0A (Fabret et al., 1999). In contrast, Clostridia, while relying on the same mechanism, make use of a simpler system lacking the intermediate components between Kin proteins and Spo0A (Stephenson and Hoch, 2002).
Elevated levels of Spo0A∼P are required for sporulation to occur (Fujita et al., 2005). Stochasticity is presumed to play a major role in driving the intracellular concentration of Spo0A∼P toward the level required for sporulation to commence (Maamar et al., 2007; Chastanet et al., 2010; de Jong et al., 2010). However, other endogenous stress response pathways that are similarly stimulated by the presence of Spo0A∼P simultaneously compete for control over cellular processes. This includes competence, biofilm formation and cannibalism (Maamar et al., 2007; Chastanet et al., 2010; de Jong et al., 2010). Here, cell fate is determined by the outcomes of co-occurring processes that indirectly affect phosphorylation of Spo0A at transcriptional, translational and post-translational levels.
Second Messenger c-di-GMP Regulates Major Checkpoints in Streptomyces Development
Rather than using a master regulator such as Spo0A, sporulation in Streptomyces is extensively regulated by levels of the nucleotide second messenger c-di-GMP. c-di-GMP is a cyclic dimer of two GMP molecules produced by diguanylate cyclases and degraded by phosphodiesterases containing EAL domains. While the signals regulating the activity of diguanylate cyclases and phosphodiesterases are unknown, the phenotypes associated with deletion of individual enzymes are different, suggesting that they respond to distinct environmental cues (Haist et al., 2020). Intracellular levels of c-di-GMP direct passage through two checkpoints in Streptomyces sporulation. The first is the initial production of sporulative hyphae, under the control of the master regulator of sporulation, BldD. Dimers of BldD are held together by a tetramer of c-di-GMP, allowing BldD to bind DNA to repress transcription of various genes involved in sporulation (Tschowri et al., 2014). It is currently unknown what causes the dissociation of BldD-c-di-GMP complexes to initiate sporulation; however, this likely involves a reduction in the concentration of intracellular c-di-GMP. Following the relief of transcriptional repression, the sigma factors σBldN, σH, and σWhiG are produced. These sigma factors regulate, either directly or indirectly through transcription factors, the expression of genes critical for the formation of aerial hyphae, as well as genes involved in cell division, chromosome segregation and condensation, and spore maturation. The second major checkpoint in sporulation is regulation of σWhiG, responsible for directing transcription of genes involved in chromosome segregation, cell division, spore maturation, and chromosome compaction (McCormick and Flardh, 2012). σWhiG is regulated by its cognate anti-sigma factor RsiG in a c-di-GMP dependent manner (Gallagher et al., 2020). High intracellular concentrations of c-di-GMP promote the formation of a heterodimer of σWhiG and RsiG, preventing σWhiG from interacting with RNA polymerase (Gallagher et al., 2020). When the concentration of c-di-GMP is reduced, the σWhiG and RsiG heterodimer dissociates, allowing σWhiG to bind to RNA polymerase (Gallagher et al., 2020). Similar to regulation by BldD, it is unknown how the concentration of c-di-GMP in the cell is reduced. Intriguingly, c-di-GMP is also used to regulate morphological differentiation in B. subtilis; however, it controls the transition from motility to biofilm formation (Chen et al., 2012).
Entry Into Sporulation
Cell Division and Cell Envelope Transformation During Sporulation
During vegetative growth, bacteria divide through a process called binary fission, where the cell divides symmetrically. Typically, this results in the formation of two daughter cells that are identical to the mother cell. Thus, vegetative septa in Firmicutes and Actinobacteria are formed by the pinching of the cytoplasmic membrane (CM in monoderm cells, Figures 2A,C), or the inner and outer membranes (IM and OM in diderm cells, Figure 2B). Alternatively, sporulation provides a different mode of cell division. In Firmicutes, the mother cell undergoes asymmetric division, generating two compartments of different sizes. The bigger one is the mother cell, whereas the smaller one becomes the mature endospore (Figures 1, 2D,E). Notably, sporulative septa in diderm bacteria are formed by invagination of the IM only (Figure 2E) (Tocheva et al., 2011). High-resolution imaging of thin sections in B. subtilis also reveals the presence of thinner septa in sporulating cells (∼23 nm vs ∼80 nm in vegetative cells) (Khanna et al., 2019). Despite the diversity in cell envelope architecture among the sporulating Firmicutes, the subsequent phagocytosis-like engulfment by the mother cell always generates an intracellular immature spore bound by two lipid bilayers, the inner (IsM) and outer (OsM) spore membranes (Figures 2G,H) (Tocheva et al., 2011, 2013) (whereas mature exospores of Actinobacteria are surrounded by one membrane only, Figure 2I). During the early stages of endospore formation, control over gene expression in both compartments is transferred from the master regulator Spo0A to the mother and daughter cell specific sigma factors, σF and σE, respectively. This transition toward σE-driven gene expression, as well as rapid autoproliferation, signifies the point of irreversible commitment to endospore formation, which coincides with engulfment of the prespore.
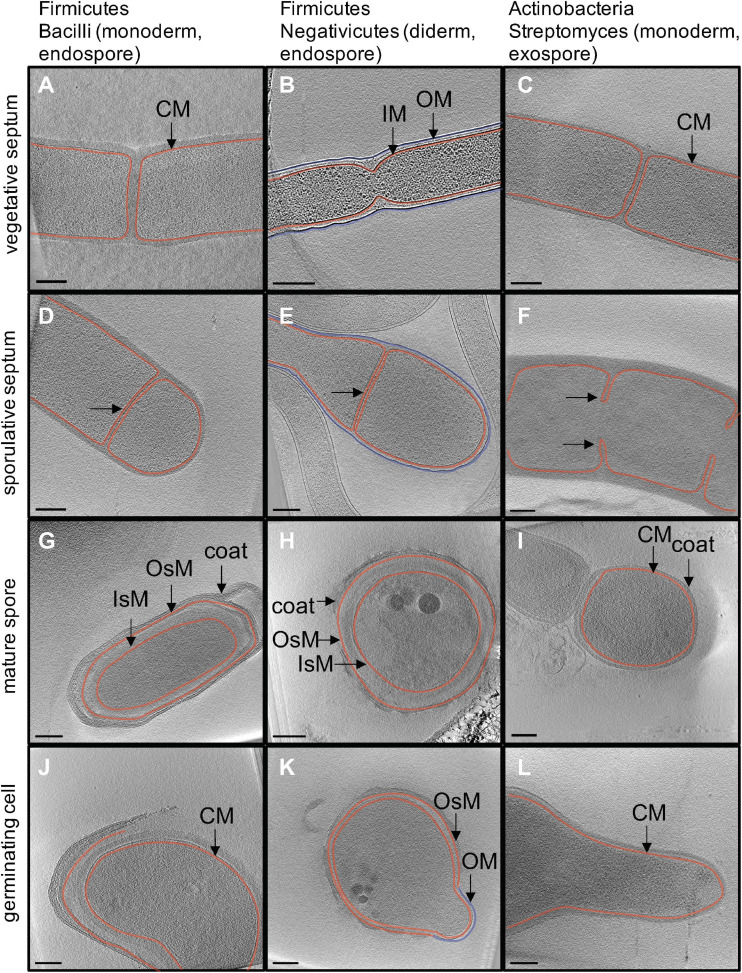
FIGURE 2.
In situ structural detail of sporulation revealed by cryo electron tomography. Each image is a 20-nm slice through a tomogram. Column one (A,D,G,J) outlines major stages of endospore formation in the model organism Bacillus subtilis. Column two (B,E,H,K) outlines endospore formation in Acetonema longum, a diderm Firmicute and member of the Negativicutes. Column three (C,F,I,L) outlines exospore formation in Streptomyces albus. Cytoplasmic (CM) and inner membranes (IM) are shown in red, outer membrane (OM) is shown in blue. The inner and outer spore membranes in Firmicutes (IsM and OsM) are both colored in red to show that they are derived from the CM/IM of the mother cell.
At the start of endospore formation, membrane-associated phosphatase SpoIIE, exclusive to endospore formers and previously implicated in septum positioning and thinning (Illing and Errington, 1991; Barák and Youngman, 1996; Khanna et al., 2019), recruits two filamentous cytoskeletal proteins FtsZ and FtsA. Here, FtsZ monomers polymerize into short filaments in a GTP dependent manner. These filaments are bundled together into rings which drive cell division through GTP hydrolysis and treadmilling to constrict the Z ring. Interestingly, unlike in vegetative cell division, FtsAZ filaments are restricted only to the mother cell face of the septum due to association with SpoIIE (Khanna et al., 2019). This pattern of co-localization is presumed to accommodate attachment of additional proteins for cell wall synthesis in early and late endospore formation, including PG remodeling by the SpoIIDMP machinery and formation of the SpoIIQ-SpoIIAH channel complex described further in this review (Bisson-Filho et al., 2017; Squyres et al., 2020).
The spore cortex is a thick layer of modified PG deposited between the IsM and OsM, which differs from vegetative PG by increased thickness and a reduced number of peptide crosslinks (Tocheva et al., 2013; Ojkic et al., 2016). During engulfment, PG remodeling is carried out by the SpoIIDMP complex consisting of two hydrolases, the amidase SpoIID and transglycosylase SpoIIP, as well as the transmembrane scaffolding protein SpoIIM. Both SpoIID and SpoIIP are crucial for this process in Bacillus and Clostridium, whereas SpoIIM is only required in Bacillus (Ribis et al., 2018). Surprisingly, bioinformatics analyses show that while SpoIIM is well-conserved across all endospore formers, SpoIID and SpoIIP demonstrate high primary sequence variability outside the key catalytic residues directly involved in cortex formation (Kelly and Salgado, 2019). Based on the compartmentalization of their respective regulons, DMP is presumed to at least partially anchor on the mother cell side in Bacilli and on the forespore side in Clostridia (Kelly and Salgado, 2019). The fully assembled complex localizes at the septal midpoint and moves to the leading edge of the engulfing membrane. This localization is dependent on SpoIIM in Bacillus, but not Clostridium (Dembek et al., 2018). Imaging studies in B. subtilis additionally demonstrate that during engulfment, septum PG exhibits homogenous thinning, likely due to increased turgor (Khanna et al., 2019). Moreover, cortex formation proceeds through PG synthesis at the leading edges of engulfing membranes, followed by sequential hydrolysis by SpoIIDP (Tocheva et al., 2013; Dembek et al., 2018). As SpoIID removes peptide crosslinks, SpoIIP follows with cleavage of “denuded” glycans leaving NAG-NAM disaccharides. Altogether, this process is suggested to drive membrane engulfment (Tocheva et al., 2013; Khanna et al., 2019). Cortex formation is finalized via activity of SpoV proteins: the putative membrane-associated lipid II flippase SpoVB, and the penicillin-binding proteins SpoVDE, produced by the mother cell (Popham and Stragier, 1991; Vasudevan et al., 2009; Cho et al., 2016; Bukowska-Faniband and Hederstedt, 2017). These proteins are essential for cortex formation and incorporate PG precursors provided by the mother cell. Experimental work in Bacillus reveals that absence of SpoVBDE blocks cortex formation and leads to accumulation of PG precursors (Vasudevan et al., 2007). In Clostridia, these proteins are conserved, but it is not known whether they are dispensable.
During engulfment, SpoIIQ, a protein under the control of σF in the daughter cell, is found throughout the daughter cell lipid membrane. Conversely, SpoIIIAH, a protein under the control of σE (spoIIIA operon) in the mother cell, is found on the mother cell lipid membrane. The two proteins form a complex via direct interactions between their extracellular domains in the intermembrane space, as the septal PG is degraded by the DMP complex. As multiple AH-Q multimers are formed at the interface between the mother and daughter cells, each of them further recruits additional proteins, such as SpoIIIAA-AG (also part of the spoIIIA operon), and GerM in Bacillus to assemble a multimeric complex (AH-Q channel) connecting the two compartments (Trouve et al., 2018). Although the resulting structure has not been observed directly in vivo with high resolution structural approaches, previous studies suggest that it comprises a channel ring, composed of IIIAG, IIIAH and IIQ oligomers, tethered to an ATPase and a basal body, with individual components displaying homology to proteins in type II, III, and IV secretion systems (Fimlaid et al., 2015b). Further, despite identification of the complex in both Bacilli and Clostridia, its structure and functional roles vary across the phylum. As such, cells lacking the channel show varying levels of compromised activity of σG, suggesting that it at least indirectly functions to transport as yet unknown molecules needed for σG activation prior to and after engulfment (Doan et al., 2009; Fimlaid et al., 2015b; Serrano et al., 2016). It has also been suggested that the complex could transport metabolites and small protein components required for spore maturation from inside the mother cell (Camp and Losick, 2009). The AH-Q channel also appears to play a major structural role, such as in the prevention of membrane deformation and spore collapse during engulfment by tethering of the mother and daughter cells, and in the facilitation of spore coat attachment in Clostridia (Doan et al., 2009; Fimlaid et al., 2015b; Ribis et al., 2018). Upon completion of engulfment, the AH-Q complex is disassembled with the help of the σG-controlled protease SpoIVB (Chiba et al., 2007).
In Firmicutes, septum formation is essential for both vegetative cell division and sporulation. In contrast, while cell division is not essential for Streptomyces vegetative growth, it is required for sporulation (McCormick et al., 1994). Streptomyces is hypothesized to use a positive selection mechanism to determine septum placement. The first protein that is known to localize to sites of future cell division is SsgA (sporulation of Streptomyces griseus). SsgA associates closely with the membrane but is not a membrane protein itself, so how SsgA localization is initially determined remains unknown (Noens et al., 2005; Traag and van Wezel, 2008). Because SsgA is limited to the streptomycetes (Girard et al., 2014), the mechanism for selection of cell division sites in other exospore forming actinomycetes also requires investigation. Following localization to sites of future cell division, SsgA is thought to recruit SsgB, an SsgA-like protein (SALP), which in turn recruits FtsZ (Willemse et al., 2011), which forms Z-rings in an evenly spaced ladder-like pattern along the length of aerial hyphae. Interestingly, Streptomyces lacks FtsA, ZipA and Zap proteins which traditionally anchor Z rings to the membrane and enhance Z ring stability. Instead, Streptomyces utilizes SepF homologs to anchor Z rings to the membrane during cell division in sporulation (Schlimpert et al., 2017). SepH, an Actinobacterial specific protein, is also integral for establishing the formation of Z rings (Ramos-Léon et al., 2020). However, it is unclear how SepH is initially localized and whether it interacts with other proteins for positioning of the Z rings. Z ring formation is also stabilized by DynA and DynB, two dynamin-like proteins which interact directly with SsgB and SepF2 during division (Schlimpert et al., 2017). PG synthesis and membrane invagination are required to drive cell division. This process is coordinated by FtsL, DivIC, FtsQ, FtsW, FtsI, and FtsEX. While many of these proteins are essential for viability of unicellular bacteria, deletion of these genes has no impact on the viability of Streptomyces species. Instead, deletion of these genes result in defects in cell division during sporulation ranging from no apparent effects for FtsE and FtsX (McCormick, 2009), to DivIC, FtsL, FtsW, and FtsI, which only have severe effects on completion of cell division under high osmolarity (Bennett et al., 2007, 2009). While many features of cell division are the same in Streptomyces and other unicellular bacteria, mechanisms for determining septum positioning, and the importance of proteins in the divisome to overall viability, are different.
Chromosome Segregation
During chromosome segregation in Firmicutes, intracellular levels of Spo0A∼P control the activity of DnaA, a bacterial DNA replication protein involved in cell division, via the regulator protein SirA, which also plays a role in polar localization of chromosomes in endospore formation (Jameson and Wilkinson, 2017). Prior to septum formation, Soj and Spo0J proteins tether chromosomes by their oriC region at each pole of the cell (Duan et al., 2016). Soj and Spo0J are homologues of the ParAB system: Soj/ParA is a Walker type ATPase protein which undergoes ATP hydrolysis to translocate foci of Spo0J/ParB bound to the chromosome (Soh et al., 2019; Jalal et al., 2020). Thus, asymmetric division leads to entrapment of one third of the chromosome in the prespore, necessitating utilization of DNA transport machinery for complete segregation. For this purpose, the protein SpoIIIE oligomerizes into a hexameric channel and uses the energy from ATP hydrolysis for DNA transport (Burton et al., 2007; Fiche et al., 2013; Cattoni et al., 2014). Chromosomal DNA then traverses the two lipid membranes into the prespore in a directional manner by binding of non-coding recognition sequences at the SpoIIIE γ domain, resulting in activation of ATPase (Besprozvannaya et al., 2013; Besprozvannaya and Burton, 2014; Bose et al., 2016). Consistently, SpoIIIE shows homology to another type of DNA pump involved in vegetative cell division, FtsK (Besprozvannaya and Burton, 2014). Super-resolution microscopy reveals that SpoIIIE first localizes at the leading edge of the septum, but eventually moves towards the septum midpoint (Fiche et al., 2013). The in vivo stoichiometry and mechanism of DNA translocation during endospore formation remain unknown.
In Streptomyces, chromosome segregation during sporulation occurs simultaneously with selection of numerous cell division sites. Chromosomes are segregated predominantly using the parABS system. ParA is thought to form mitotic spindle like filaments in S. coelicolor aerial hyphae during the early stages of segregation (Jakimowicz et al., 2007). ATP hydrolysis by ParA drives translocation of ParB foci into position between forming Z rings (Jakimowicz et al., 2005; Jakimowicz et al., 2007), suggesting that there is an as yet undiscovered interaction between chromosome partitioning and the selection of sites for future cell division. Furthermore, deletion of parA affects both chromosome segregation and septum placement (Jakimowicz et al., 2007). ParJ, a protein specific to the Actinobacteria, interacts directly with ParA to regulate its polymerization (Ditkowski et al., 2010). In par mutants, almost 90% of spores still contain at least one copy of the chromosome (Kim et al., 2000), suggesting that there are additional mechanisms for chromosome segregation. FtsK, a SpoIIIE homolog, is positioned on the leading edge of the invaginating membrane and may assist with resolving the ends of the linear chromosome to prevent truncation of the arms (Dedrick et al., 2009). Streptomyces chromosomes contain additional FtsK-like proteins. One of these, SffA, is localized to closing septa during sporulation by SmeA (Ausmees et al., 2007). Deletion of sffA and smeA results in a fivefold increase in the number of anucleate spores compared to the wildtype, although it remains unclear if SffA functions as a DNA pump, similar to SpoIIIE, or if it promotes chromosome segregation in a different manner (Ausmees et al., 2007). Links between chromosome segregation and cell division during sporulation are still undefined and offer many exciting avenues for future research.
Spore Maturation
Endospore maturation begins during prespore engulfment and culminates in lysis-mediated spore release. Here, control over gene expression is gradually transferred to the late-stage sigma factors of the prespore, σG, and the mother cell, σK. Maturation involves finalization of cortex formation, assembly of the protein coat and in some cases, an exosporium, dehydration of the endospore core, and DNA compaction. Altogether, these morphological changes account for immense resistance to biotic and abiotic stressors allowing for preservation of endospores in harsh environments for extended periods of time.
Unlike in endospore formers, engulfment of the spore does not occur in Actinobacteria and the mature spore is bound by one membrane, rather than two (Figure 2I). Following septation (Figure 2F), cells undergo division via rapid mechanical separation, or “V snapping” (Zhou et al., 2016). Immature spores remain associated with each other in the spore chain as they undergo maturation, likely held in place by the rodlet ultrastructure that forms a sheath on the surface of aerial hyphae. Maturation is accompanied by an increase in spore wall thickness (Figure 2I), presumably directed by MreB and other components of the spore wall synthesizing complex, consisting of proteins from the mre gene cluster along with RodZ, FtsI, and SCO3901 (Heichlinger et al., 2011; Kleinschnitz et al., 2011; Girard et al., 2014; Sexton and Tocheva, 2020). In addition to increased thickness, the PG in the spore wall features an increase in 3–4 crosslinks and a decrease in 3–3 crosslinks (van der Aart et al., 2018), and multiple layers are visible in the spore wall after spore maturation (Sexton and Tocheva, 2020). It is unclear if the spore wall layers feature the same PG structure, and what the biological significance of modified PG crosslinking is in the spore wall. Several cell wall lytic enzymes, including carboxypeptidase, endopeptidases, and lytic transglycosylases, have been implicated in PG remodeling during exospore maturation (Haiser et al., 2009; Sexton et al., 2015; Rioseras et al., 2016). While these enzymes are critical for spore PG architecture, the modifications they perform and their impact on spore integrity require further investigation.
Metabolic Adaptations for Environmental Resilience of Spores
Mature endospores are characterized by a unique phase-bright appearance when imaged with phase-contrast light microscopy, owing to their dehydrated state. A higher solids content and lower water content in the spore generate a higher refractive index thus resulting in phase reversion from dark to bright. Dehydration is achieved by several mechanisms. Initially, ATP hydrolysis and loss of K+ ions between the mother cell and the spore generate osmotic dehydration. Subsequently, simultaneous replacement of water with dipicolinic acid (DPA) and minerals is thought to decrease hydration even further (Marquis, 1998). DPA is found in all endospore formers, constitutes up to 15% dry weight of mature endospores and has been linked to increased heat resistance (Powell, 1953; Church and Halvorson, 1959; Setlow et al., 2006). Synthesis of DPA occurs in the mother cell under the control of σK in late stages of sporulation, and is mediated by the enzymes DpaA and DpaB of the spoVF operon, through transformation of L-2,3-dihydrodipicolinate, an intermediate of the lysine biosynthesis pathway (Daniel and Errington, 1993; Takahashi et al., 2015). Interestingly, an alternative pathway for DPA synthesis, possibly of ancient origin, has been identified in Clostridia and is performed by the protein EtfA using the product of DpaA activity (Orsburn et al., 2010; Donnelly et al., 2016). DPA relies on a two-step transport pathway: it is taken up by the predicted SpoVV transporter in the OsM for transport into the intermembrane space, then by the multicomponent SpoVA transport system through the IsM (Li et al., 2012; Ramírez-Guadiana et al., 2017). Once inside the spore, DPA associates with calcium ions. Eventually, spore water content decreases to a level that is no longer supportive of unrestricted movement of macromolecules, causing metabolic dormancy.
Similar to endospores, exospores produced by Streptomyces species undergo dramatic changes to protect cytoplasmic contents during dormancy. Several proteins and mRNAs are deposited in the spore cytoplasm prior to dormancy for use during germination (Mikulik et al., 2002; Strakova et al., 2013). In order to preserve these proteins and mRNAs, trehalose, a dimer of two glucose molecules joined through an α,α-1,1-glycosidic linkage, is deposited in large aggregates in the spore cytoplasm. Depending on growth conditions, trehalose can comprise up to 25% of the dry spore biomass (McBride and Ensign, 1987). Trehalose interacts with the cytoplasmic contents of the spore through hydrogen bonding, allowing it to protect a wide variety of macromolecules from denaturation and aggregation during periods of excessive dehydration, heat, freezing, radiation and oxidative damage. Additionally, trehalose likely serves as an energy source to be used during the initial stages of germination (McBride and Ensign, 1987). During spore maturation, the spore pigment produced by the whiE gene cluster is also deposited inside cells. The exact function of the spore pigment has not yet been determined; however, it seems reasonable to assume that it functions to protect against UV damage. Deletion of the genes responsible for spore pigment in S. griseus result in a slight increase in susceptibility to UV irradiation (Funa et al., 2005). Mature Streptomyces spores display decreased resistance to a variety of abiotic stressors, including heat and desiccation, when compared to endospores (Table 1). Intriguingly, Streptomyces exospores exhibit low level metabolic activity (Constant et al., 2008; Liot and Constant, 2016), which may contribute to reduced tolerance to abiotic stresses in the environment when compared to Firmicutes.
DNA Compaction
At the end of spore maturation, the chromosome is condensed, likely offering an additional layer of protection for genetic material during dormancy. This is accomplished through the action of nucleoid associated proteins, DNA supercoiling, molecular crowding, and dehydration of the spore. In Firmicutes, dormant endospores exhibit resilience to UV damage due to protection of DNA by DNA-binding proteins called small acid-soluble proteins, or SASPs. SASPs display a range of sizes from 5 to 7 kDa and are produced in the forespore, from ssp genes primarily under the control of the late sporulation sigma factor σG (Setlow, 1988; Wetzel and Fischer, 2015). Evidence suggests that all endospore formers harbor SASPs of the α/β type, whereas small amounts of the non-DNA binding γ SASP have only been identified in Bacillus (Setlow, 1988; Vyas et al., 2011). Despite the variety of SASPs produced across all endospore-forming species, each organism displays a major preference toward one or two α/β type SASPs for DNA packaging. SASPs are well-conserved at the DNA-binding helix-turn-helix domains, but can otherwise show sequence variability, particularly between aerobes and anaerobes (Setlow, 2007; Vyas et al., 2011; Wetzel and Fischer, 2015). SASPs restrict access of mutagens to DNA strands by physical protection. However, harmful UV rays, heat and mutagens can still damage DNA while the cell is metabolically dormant. For this, endospores rely on pre-packaged DNA repair enzymes to revert the damage in germination, before cell division resumes (Setlow, 2007).
In Streptomyces, chromosome compaction is much more elaborate and involves the action of several classes of nucleoid associated proteins rather than SASPs. SMC complexes promote chromosome compaction and segregation, potentially through an interaction with ParB (Dedrick et al., 2009; Kois et al., 2009). In S. coelicolor, this complex has two additional interacting partners: ScpA and ScpB, which both promote chromosome compaction (Dedrick et al., 2009; Kois et al., 2009). sIHF and the HU-like protein HupS function as analogs for histones during sporulation to enhance chromosome compaction (Salerno et al., 2009; Swiercz et al., 2013). DbdA has a histone-like domain which is involved in compacting the DNA, which enhances resistance to oxidative stress (Aldridge et al., 2013). A Dps homolog, DpsA, interacts with the chromosome in a sequence-independent manner to promote compaction. Deletion of dpsA results in irregularly long spores which often contain multiple copies of the chromosome, suggesting an interplay between chromosome segregation and septation (Facey et al., 2009). However, dpsA is not widely conserved in Streptomyces species (Szafran et al., 2020). Dps-like proteins also oxidize iron and deposit ferric oxide, which may protect DNA from oxidative damage, but it is unclear whether other Dps homologs would fulfill this function in other streptomycetes and in Actinobacteria in general. Collectively, these nucleoid-associated proteins condense the chromosome and shield it from damage that may occur during dormancy.
Assembly and Synthesis of the Spore Coat and Exosporium
The endospore coat is a multilayer protein structure that encapsulates the OsM and provides protection from lytic enzymes, heat, physical disruption, UV radiation, predation and other factors (Figures 2G,H) (Riesenman and Nicholson, 2000; Klobutcher et al., 2006; Setlow, 2006; Alves Feliciano et al., 2019). The coat is a highly complex structure made up of dozens of proteins, many of which are unique to individual species. For example, the B. subtilis spore coat is made up of over 70 proteins, but only a small fraction of them have orthologs in other Firmicutes (Abhyankar et al., 2013). The majority of these conserved proteins are morphogenic, meaning that they play important roles in the initial assembly of the coat. One such example of a morphogenic protein is SpoVM, a 3 kDa-sized amphipathic helix produced shortly upon completion of septum formation and during engulfment, which is under the control of σE in the mother cell. SpoVM is driven to the OsM due to its increased preference for lipid membranes with increased curvature (Levin et al., 1993; Kim et al., 2017). Here, it presumably utilizes a “dash-and-recruit” mechanism of rapid accumulation of SpoVM monomers and their subsequent polymerization (Kim et al., 2017). Then, SpoIVA attaches to SpoVM and mediates assembly of the coat base layer, or “platform,” through ATP hydrolysis. Experimental evidence shows that while both of these proteins are essential for coat assembly in Bacillus, in Clostridia the vital role of SpoVM is diminished and instead carried out by the functionally homologous SipL (Stevens et al., 1992; Levin et al., 1993; Ribis et al., 2017). Intriguingly, deficiencies in these coat platform proteins also inhibit formation of the cortex (Ebmeier et al., 2012; Benito de la Puebla et al., 2020). Additional spore coat morphogenesis proteins of Bacilli include SafA and CotE, which are essential for formation of the inner and outer coat layers, respectively (Zheng et al., 1988; Takamatsu et al., 1999; Abhyankar et al., 2013; Stelder et al., 2018). In Clostridia, a recently identified protein, CotL, has been deemed important for assembly of the coat, although its function is poorly understood (Alves Feliciano et al., 2019). Altogether, the endospore coat usually comprises multiple layers, each characterized by distinct morphogenic and structural proteins (McKenney et al., 2010; Stelder et al., 2018). While morphogenic proteins are relatively conserved, structural proteins can vary by organism and are tailored to its respective environmental niche (Abhyankar et al., 2013). Despite its thickness and a significant role in protection from various stressors, the coat is not dense enough to fully restrict movement of molecules, such as germinants, into the intramembrane space and core (Knudsen et al., 2016). Germination (Ger) receptors, are loaded from the mother cell during maturation and reside in the IsM. Lytic proteins involved in germination and degradation of the endospore coat and cortex are also embedded in the IsM. Finally, the proteinaceous coat also provides a mechanism for core expansion during rehydration through expansion of its layers (Sahin et al., 2012).
Some endospore forming species, such as B. anthracis and A. longum, have an additional outer layer composed of proteins, lipids and carbohydrates called the exosporium. The appearance of the exosporium differs by organism, can exhibit various thickness and includes appendage-like structures (Abhyankar et al., 2013; Pizarro-Guajardo et al., 2016). Although the exosporium provides a barrier for entry of large molecules, its exact functions are not known (Knudsen et al., 2016). Some proteins previously identified in the exosporia of Bacillus and Clostridium species could be relevant in pathogenesis, attachment or endospore dissemination (Abhyankar et al., 2013; Stewart, 2015; Pizarro-Guajardo et al., 2016).
On the other hand, the Actinobacterial exospore coat is characterized by a simpler architecture. For example, a thin spore coat has been observed on the surface of the spore wall of Streptomyces spores (Sexton and Tocheva, 2020). However, the composition and function of this coat in spore survival remain to be characterized. Some Streptomyces species produce elaborate coatings on the surface reminiscent of the exosporium produced by Firmicutes (Dietz and Mathews, 1970), likely in order to promote attachment and dispersal. It is unknown if these elaborate coats offer additional protection to the spore.
Spore Release
Endospores are released through lysis of the mother cell. However, little is known about the regulation and dynamics of programmed death of the mother cell. Time-lapse fluorescence microscopy of B. subtilis reveals that the endospore is released from the cell pole by rupturing of the mother cell membrane at both poles, characterized by an intense signal from the membrane dye FM4-64 at the mother cell poles and then inside the cytosol (Hosoya et al., 2007). Simultaneously, decreased fluorescence from DNA dyes is observed in the mother cell. This suggests that lipid membranes become ruptured, and DNA gets degraded inside the mother cell prior to cell wall collapse and complete release of the endospore (Hosoya et al., 2007). Autolysins such as CwlB, CwlC, and CwlH, under the control of σK, have been shown to play major roles in cell wall collapse and the subsequent lysis of the mother cell through PG degradation in B. subtilis (Smith and Foster, 1995; Nugroho et al., 1999). In diderm endospore formers, such as Negativicutes, this would likely be insufficient and would require additional players for degradation of the OM. Cryo-electron tomography imaging of the diderm endospore former A. longum reveals loosening of the mother cell OM in endospore maturation, suggesting that the OM likely “blebs off” during spore maturation (Tocheva et al., 2011). However, mother cell lysis is not well described outside the model organism B. subtilis.
In contrast, spore release occurs in a more direct manner for exospore formers. Exospores are released from the spore chain when the rodlet ultrastructure on the cell surface is mechanically disrupted (Di Berardo et al., 2008). This could be accomplished by biotic factors, such as passing insects, or abiotic factors, such as wind or ocean currents.
Endospores ultimately have enhanced resistance to a variety of environmental stressors compared to exospores (Table 1). This includes heat tolerance, where 60% of Streptomyces spores are viable after heating to 60°C for 10 min, but no spores are viable after heating to 80°C for 10 min (Kawamoto et al., 1981; Haiser et al., 2009). In contrast, endospores produced by Bacillus are well adapted to tolerate extended heat treatment. At 80°C in a humid environment, 50% of Bacillus spores remain viable after 30 h of exposure (Setlow, 2014). DPA, core dehydration, and heat shock proteins are proposed to protect the cell against damage from wet heat (Setlow, 2014). Endospores are also significantly more resistant to desiccation and UV damage compared to exospores (Table 1). Both desiccation and UV impact spore viability through DNA damage (Setlow, 2014), which suggests that DPA and SASPs found in endospores are a more robust mechanism for DNA protection than trehalose and the combination of nucleoid associated proteins found in exospores (Setlow, 2014).
Germination
Germination stimuli are diverse, vary between species, and include free amino acids, PG molecules, organic acids, as well as chemicals specifically tailored to the ecological niche (e.g., bile salts for the intestinal pathogen C. difficile) (Bhattacharjee et al., 2016). In Firmicutes, germination receptors (Ger) residing in the IsM sense signals from the environment via interaction with small molecules (germinants) penetrating the spore cell wall. Ger type receptors are conserved across endospore formers, but show variability in the types of signals they receive, meaning that structure alone cannot be used to predict cognate germinants (Ross and Abel-Santos, 2010). Ger receptors form membrane-spanning complexes in the IsM and are often implicated in recognition of free amino acids (Ross and Abel-Santos, 2010). Preliminary investigations into the structure of Ger receptors reveal that they are similar to inner membrane-associated small molecule transporters and signal transducers, possibly forming clusters (Li et al., 2014, 2019). Small PG fragments produced during spore germination stimulate germination of B. subtilis spores by binding to PrkC, a Ser/Thr kinase containing a peptidoglycan binding PASTA domain (Shah et al., 2008). PrkC is conserved in Clostridia (Shah et al., 2008), so it may play a broad role in stimulating germination of spores produced by Firmicutes. In B. subtilis, PrkC is proposed to phosphorylate the elongation factors EF-G and EF-Tu to promote ribosome assembly and translation elongation (Shah et al., 2008; Absalon et al., 2009; Pompeo et al., 2012). It is likely that PrkC phosphorylates additional targets to promote germination, although these are yet to be defined. Additionally, Clostridia possess Csp-type receptors indirectly connected to the OsM via lipoproteins and coupled to the cortex lytic enzyme SleC, which is activated by cleavage upon recognition of germinant (Fimlaid et al., 2015a; Bhattacharjee et al., 2016; Kochan et al., 2018; Donnelly et al., 2020). Organisms such as C. difficile lack Ger-type receptors altogether and instead rely on Csp proteins, such as CspC (Bhattacharjee et al., 2016). Structural studies of Csp-type receptors reveal that they function as subtilisin-like proteases in activation of SleC, but the complete mechanism for signal recognition and integration remains unelucidated (Adams et al., 2013; Rohlfing et al., 2019).
Stimulants activate a signal cascade triggering germination. In B. subtilis, a cluster of proteins called the germinosome is hypothesized to mediate this process. The germinosome comprises germination receptors, scaffolding proteins and other components, such as the SpoVA protein involved in DPA transport (Griffiths et al., 2011). Fluorescence visualization studies reveal that the germinosome localizes at one pole of the dormant endospore, then disperses as germination progresses (Troiano et al., 2015; Breedijk et al., 2020; Wang et al., 2020). The germination cascade is then followed by degradation of the spore cell wall by lytic enzymes, increasing permeability and allowing Ca2+-DPA complexes and ions to escape the endospore in order to be replaced with water from the environment (Griffiths et al., 2011; Wang et al., 2015). Cortex hydrolysis in the Bacilli is mediated by the conserved enzymes SleB and CwlJ (Setlow, 2003). While many Clostridia similarly encode and utilize these enzymes, they also rely on an alternative mechanism of cortex lysis by the enzyme SleC activated by the Csp complex (Kochan et al., 2018; Shen et al., 2019). Mechanism for protein coat degradation remains to be investigated, but possibly involves OsM or coat-associated proteases which initiate thinning from within (Santo and Doi, 1974; Jagtap et al., 2006; Plomp et al., 2007). As the spore swells rapidly, rehydration and changes in core pH allow for resumption of metabolic activity (Setlow, 2003; Liang et al., 2014). Subsequently, DNA-binding SASPs become degraded and provide building blocks for biosynthesis, as well as allow for gene expression upon chromosome release (Jagtap et al., 2006; Swarge et al., 2020). Core mRNA, prepackaged in maturation, is also degraded in this manner and likely not used for protein expression (Setlow and Kornberg, 1970; Swarge et al., 2020). Transcriptomic and proteomic studies in Bacillus and Clostridium endospores reveal that the first genes to become expressed upon resumption of metabolic activity include those for transcription regulation, biosynthesis, DNA repair, and nutrient uptake pathways. Cell growth, elongation and cell wall remodeling become activated during the next stage (outgrowth). Cell division, motility and biofilm formation are activated last upon resumption of vegetative growth (Keijser et al., 2007; Bassi et al., 2013; Dembek et al., 2013; Swarge et al., 2020; Xing and Harper, 2020).
Upon endospore coat shedding, cortex degradation, and dissolution of the OsM, monoderm Firmicutes, such as Bacillus, emerge as cells surrounded by a cytoplasmic membrane and a layer of vegetative-like PG, termed the germ cell wall, pre-deposited under the cortex and serving as a scaffold for growth of additional PG (Figure 2J) (Tocheva et al., 2013; Sella et al., 2014). In contrast, germination and outgrowth in diderm bacteria, such as the Negativicute A. longum, require extensive cortex hydrolysis and the remodeling of their inner-membrane like OsM into a typical OM (containing lipopolysaccharide, LPS and β-barrel OMPs) (Figure 2K). The mechanism of this novel membrane remodeling is not known (Tocheva et al., 2011).
Similar to endospores, Streptomyces spores remain dormant in the environment until they are stimulated to germinate. Streptomyces species do not contain any known homologs of Ger receptors for signaling germination, and PASTA domain containing kinases which sense PG fragments do not function to stimulate germination (Sexton et al., 2020). Instead, divalent cations such as calcium are known to function as germinants (Eaton and Ensign, 1980), although the mechanism underlying this stimulation is unclear. Following the signal to initiate germination, the spore swells and switches from phase bright to phase dark, and outgrowth occurs. This stage relies on several of the macromolecules deposited in the spore during preparation for dormancy, including proteins and mRNAs used for protein synthesis (Strakova et al., 2013). Outgrowth relies on the activity of cell wall lytic enzymes to make space for new polar growth to occur (Sexton et al., 2020). The innermost layer of the spore wall is continuous with the new vegetative cell wall, and the coat and outer layer of the spore wall peel away from the germinating exospore (Figure 2L) (Sexton and Tocheva, 2020). Tip extension and branching is then used to produce the vegetative mycelium which colonizes the environment.
Genetic Differences and Evolution
With regards to classification of endospore formers, phylogeny remains a point of controversy. Historically, endospore formation genes have been shown to differ between two groups within Firmicutes: bacteria of the class Bacilli, and a large taxonomic group (hereby referred to as the Clostridia-like spore-formers) which initially comprised bacteria typically assigned to the class Clostridia but was later expanded to include other Firmicutes carrying homologous sporulation genes, such as the Negativicutes, Peptococcaceae, and Halanaerobiales. Although our analysis based on the current data from GTDB indicates otherwise (Figure 3), some studies propose placement of these taxonomic groups within the class Clostridia (Yutin and Galperin, 2013; Davidson et al., 2018). Despite the questionable taxonomic placement, spore-forming Negativicutes display higher similarity in sporulation genes and proteins of typical Clostridia than Bacilli, pointing to their common evolutionary origins (Ramos-Silva et al., 2019). Altogether, such evidence suggests that diversification of endospore formation machinery between the classes Bacilli and Clostridia occurred early in the evolutionary timeline, preceding the emergence of Negativicutes. However, because the spore-forming Negativicutes are not yet genetically tractable, functional studies of Clostridia-like sporulation machinery are limited to the canonical representatives of the class Clostridia.
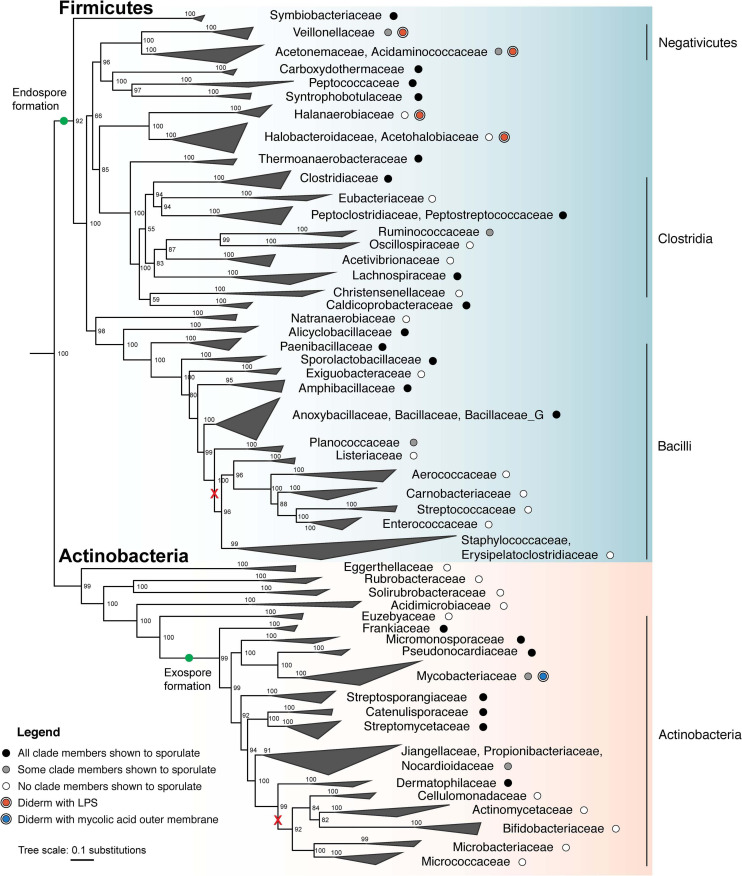
FIGURE 3.
Maximum likelihood phylogeny of sporulating and non-sporulating Firmicutes and Actinobacteria. Tree was constructed using an alignment of 120 single-copy marker gene sequences from several hundred representative genomes from each phylum using GTDB-Tk (Parks et al., 2018; Chaumeil et al., 2019). Whole genome phylogeny was determined using the concatenated marker gene alignment and IQ-TREE (v. 2.0.3), with the substitution model LG + G4 and 1000 ultrafast bootstraps (Minh et al., 2020). The tree was subsequently down-sampled and collapsed to show major families. Several representative Cyanobacteria genomes served as an outgroup. Tree was visualized using ggtree (Yu, 2020). Green dots indicate that endospore formation was likely present in the last universal ancestor of all Firmicutes, whereas exospore formation appears to have evolved after phylum differentiation. Black and gray dots represent the demonstrated ability to sporulate in all members and some members, respectively, whereas white dots represent the lack of sporulation ability. Major losses involving multiple families are shown (red x). Clades which include diderm bacteria are indicated for possession of either an LPS-containing OM (red dots) or a mycolic acid-containing OM (blue dot). Classes, as defined by the GTDB, are labeled on the right.
Comparative genomics and functional studies reveal several trends in sporulation mechanisms of Bacilli and the Clostridia-like spore formers. As such, despite the overall conservation of Spo0A and the associated phosphorylation mechanisms for initiation of sporulation in both groups, members of the class Clostridia lack the intermediate proteins Spo0F, Spo0B, as well as the associated phosphatases bridging the histidine kinases to the master regulator (Galperin et al., 2012; Abecasis et al., 2013; Al-Hinai et al., 2015). Moreover, the regulation of gene expression in endospore formation and maturation by the sporulation-specific sigma factors, despite the same chronological progression, occurs in a semi-independent manner in Clostridia (Al-Hinai et al., 2015). In general, all sporulation-associated machinery, including those involved in cell division, DNA transport and compaction, cortex formation, the AA-AH complex, DPA synthesis, spore coat formation and germination, is conserved between all endospore formers (Galperin et al., 2012; Abecasis et al., 2013; Ramos-Silva et al., 2019). However, minor differences are observed with occurrence of supplementary pathways (such as DPA synthesis by EtfA, and Csp-mediated triggering of germination in the Clostridia-like spore formers; Orsburn et al., 2010; Rohlfing et al., 2019), replacement of individual components by their functional homologues (assembly of the spore coat base via SpoVM in Bacillus and SipL in Clostridium; Ribis et al., 2017), losses of intermediate components (modification of Spo0A via a phosphorelay system in Bacilli and a two-component system in Clostridia), and slight discrepancies in co-regulation and co-dependence of cellular processes (e.g., influence of deficiencies in spore coat assembly on cortex formation; Ebmeier et al., 2012; Benito de la Puebla et al., 2020).
Analysis of sporulation-associated genes also aids in elucidating the evolutionary history of Firmicutes as a phylum. Due to the extensive, multi-step nature of sporulation, horizontal transfer of sporulation genes would likely not be successful. The observation that endospore formation is widely spread throughout the phylum in both monoderm and diderm members indicates that the ability to sporulate was an ancient feature (Figure 3). Intriguingly, diderm Firmicutes possess an OM that is identical to that of classical Gram-negative bacteria of the phylum Proteobacteria. The recent discovery that two independent diderm lineages (Negativicutes and Halanaerobiales) exist in Firmicutes that share OM features with Proteobacteria, argues against horizontal gene transfer of OM genes and further points to a diderm ancestor of Firmicutes (Figure 3) (Megrian et al., 2020; Taib et al., 2020). Altogether, data show that the ability to sporulate and the presence of a true OM in Firmicutes were present in the last common ancestor of the phylum (Figure 3) (Tocheva et al., 2016). Subsequent loss of either sporulation or the OM could explain the observed diversity in modern species. For example, among the Negativicutes class, both spore-forming (e.g., A. longum) and non-spore forming (e.g., Veillonella parvula) diderm organisms have been described (Tocheva et al., 2011), indicating of loss of sporulation. Analogously, monoderm and diderm sporulators (B. subtilis and A. longum, respectively) have also been described, indicating OM loss in monoderm species. Gene losses in endospore and/or OM formation pathways along the evolutionary timeline would thus result in the diversity observed in Firmicutes today, yielding monoderm non-spore formers (e.g., Lactobacillus sp.), monoderm spore-formers (e.g., B. subtilis), diderm non-spore formers (e.g., V. parvula) and diderm spore-formers (e.g., A. longum).
Within the phylum Actinobacteria, sporulation is restricted to a single branch (class Actinobacteria, Figure 3) which evolved roughly 1.7 billion years ago. Because all sporulating Actinobacteria have evolved from a single lineage, it is thought that sporulation evolved once within the phylum, and speciation has resulted in the diversity of modes for producing exospores (green dot, Figure 3) (Chandra and Chater, 2014). While many of the regulatory networks controlling sporulation in the Actinobacteria are conserved, the genes within the regulons are not. This is likely due to the fact that sporulation within the Actinobacteria ranges from production of single exospores on vegetative mycelia to elaborate sporangia containing spores. As a result, the core set of genes responsible for exospore formation within the Actinobacteria is still poorly defined. However, bldD is conserved in all sporulating Actinobacteria, indicating that intracellular c-di-GMP levels govern the initiation of sporulation across the diverse modes of sporulation. whiG is not conserved outside of Actinobacteria which produce long chains of exospores (Chandra and Chater, 2014), suggesting that this level of regulation is unique to this specific type of exospore production. SsgB is conserved in most sporulating Actinobacteria, although its interacting partner SsgA is only found in the streptomycetes (Chandra and Chater, 2014). MreB is not conserved in all sporulating Actinobacteria, along with other members of the mre gene cluster including MreC, MreD, PBP2, and Sfr (Chandra and Chater, 2014), suggesting that there may be alternative mechanisms for synthesizing the spore wall. Finally, genes for trehalose biosynthesis are broadly conserved in all sporulating Actinobacteria, suggesting that all exospores produced by Actinobacteria use trehalose to conserve their cytoplasmic contents during dormancy. Notably, sporulation and filamentous growth are inextricably linked within the Actinobacteria, as all filamentous organisms produce exospores. Filamentous growth and sporulation have been lost independently several times throughout the Actinobacteria, such as in Mycobacterium, Corynebacterium, Propionibacterium, and Rhodococcus. The inability of these organisms to sporulate is correlated with a loss of several genes associated with exospore formation (Chandra and Chater, 2014). Importantly, the evolution of the mycobacterial OM occurred independently of the LPS-containing OM within Firmicutes (Vincent et al., 2018).
Concluding Remarks
Despite numerous parallels between endospore and exospore formation, such as similar environmental stimuli and spore envelope architecture, it is clear that these processes have distinct regulatory, structural, and evolutionary origins, and thus represent an example of convergent evolution.
Nutrient limitation drives sporulation in both Firmicutes and Actinobacteria, and both endo- and exospore formation are controlled by a central transcription factor, albeit in distinct ways (activation by Spo0A for endospore formers, and repression by BldD for exospore formers). The response to yet-to-be-determined environmental cues impacts the DNA-binding ability of these regulators, triggering a downstream cascade of transcription and sigma factors, which sequentially direct the expression of genes required for sporulation. Both endo- and exospore formation involve cell division and chromosome segregation by homologous machinery – however, the results of these processes differ vastly between the two modes of sporulation (production and engulfment of a daughter cell in the Firmicutes, and simultaneous generation of multiple non-engulfing spores in Actinobacteria). Later stages of endo- and exospore formation, including cell envelope transformations and metabolic adaptations for initiation of dormancy, occur through superficially similar, but evolutionarily independent pathways. However, both Firmicutes and Actinobacteria utilize the mechanisms of cell wall thickening, DNA compaction and spore core dehydration for enhanced resilience under stressful environmental conditions. Finally, reintroduction of dormant spores into a more favorable environment results in germination in both endo- and exospore formers, but their distinct ecological niches, as well as the difference in overall cell cycles, necessitate utilization of vastly independent processes for resumption of vegetative growth.
Author Contributions
PB, DS, MG, AH, and ET wrote the text and prepared the figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Work in the ET laboratory was supported by the Natural Sciences and Engineering Research Council of Canada Discovery Grant (RGPIN 04345).
References
- Abecasis A. B., Serrano M., Alves R., Quintais L., Pereira-Leal J. B., Henriques A. O. (2013). A genomic signature and the identification of new sporulation genes. J. Bacteriol. 195 2101–2115. 10.1128/jb.02110-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhyankar W., Hossain A. H., Djajasaputra A., Permpoonpattana P., Ter Beek A., Dekker H. L., et al. (2013). In pursuit of protein targets: proteomic characterization of bacterial spore outer layers. J. Proteome Res. 12 4507–4521. 10.1021/pr4005629 [DOI] [PubMed] [Google Scholar]
- Absalon C., Obuchowski M., Madec E., Delattre D., Holland I. B., Seror S. J. (2009). CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155 932–943. 10.1099/mic.0.022475-0 [DOI] [PubMed] [Google Scholar]
- Adams C. M., Eckenroth B. E., Putnam E. E., Doublié S., Shen A. (2013). Structural and functional analysis of the CspB protease required for clostridium spore germination. PLoS Pathog. 9:e1003165. 10.1371/journal.ppat.1003165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge M., Facey P., Francis L., Bayliss S., Del Sol R., Dyson P. (2013). A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res. 41 4813–4824. 10.1093/nar/gkt180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hinai M. A., Jones S. W., Papoutsakis E. T. (2015). The Clostridium sporulation programs: diversity and preservation of endospore differentiation. Microbiol. Mol. Biol. Rev. 79 19–37. 10.1128/mmbr.00025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves Feliciano C., Douché T., Giai Gianetto Q., Matondo M., Martin-Verstraete I., Dupuy B. (2019). CotL, a new morphogenetic spore coat protein of Clostridium difficile. Environ. Microbiol. 21 984–1003. 10.1111/1462-2920.14505 [DOI] [PubMed] [Google Scholar]
- Ausmees N., Wahlstedt H., Bagchi S., Elliot M. A., Buttner M. J., Flardh K. (2007). SmeA, a small membrane protein with multiple functions in streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol. Microbiol. 65 1458–1473. 10.1111/j.1365-2958.2007.05877.x [DOI] [PubMed] [Google Scholar]
- Barák I., Youngman P. (1996). SpoIIE mutants of Bacillus subtilis comprise two distinct phenotypic classes consistent with a dual functional role for the SpoIIE protein. J. Bacteriol. 178 4984–4989. 10.1128/jb.178.16.4984-4989.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi D., Cappa F., Cocconcelli P. S. (2013). Array-based transcriptional analysis of Clostridium sporogenes UC9000 during germination, cell outgrowth and vegetative life. Food Microbiol. 33 11–23. 10.1016/j.fm.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Benito de la Puebla H., Giacalone D., Cooper A., Shen A. (2020). Role of SpoIVA ATPase motifs during Clostridioides difficile sporulation. J. Bacteriol. 202 e320–e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. A., Aimino R. M., Mccormick J. R. (2007). Streptomyces coelicolor genes ftsL and divIC play a role in cell division but are dispensable for colony formation. J. Bacteriol. 189 8982–8992. 10.1128/jb.01303-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. A., Yarnall J., Cadwallader A. B., Kuennen R., Bidey P., Stadelmaier B., et al. (2009). Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J. Bacteriol. 191 661–664. 10.1128/jb.01048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beskrovnaya P., Fakih D., Morneau I., Hashimi A., Bello D. G., Xing S., et al. (2020). No endospore formation confirmed in members of the phylum Proteobacteria. Appl. Environ. Microbiol. 2020:219022. 10.1128/AEM.02312-02320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besprozvannaya M., Burton B. M. (2014). Do the same traffic rules apply? directional chromosome segregation by SpoIIIE and FtsK: directional chromosome segregation by SpoIIIE/FtsK. Mol. Microbiol. 93 599–608. 10.1111/mmi.12708 [DOI] [PubMed] [Google Scholar]
- Besprozvannaya M., Pivorunas V. L., Feldman Z., Burton B. M. (2013). SpoIIIE protein achieves directional DNA translocation through allosteric regulation of ATPase activity by an accessory domain. J. Biol. Chem. 288 28962–28974. 10.1074/jbc.m113.484055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee D., Mcallister K. N., Sorg J. A. (2016). Germinants and their receptors in clostridia. J. Bacteriol. 198 2767–2775. 10.1128/jb.00405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho A. W., Hsu Y.-P., Squyres G. R., Kuru E., Wu F., Jukes C., et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355 739–743. 10.1126/science.aak9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose B., Reed S. E., Besprozvannaya M., Burton B. M. (2016). Missense mutations allow a sequence-blind mutant of SpoIIIE to successfully translocate chromosomes during sporulation. PLoS One 11:e0148365. 10.1371/journal.pone.0148365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedijk R. M. P., Wen J., Krishnaswami V., Bernas T., Manders E. M. M., Setlow P., et al. (2020). A live-cell super-resolution technique demonstrated by imaging germinosomes in wild-type bacterial spores. Sci. Rep. 10:5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowska-Faniband E., Hederstedt L. (2017). Transpeptidase activity of penicillin-binding protein S po VD in peptidoglycan synthesis conditionally depends on the disulfide reductase S to A. Mol. Microbiol. 105 98–114. 10.1111/mmi.13689 [DOI] [PubMed] [Google Scholar]
- Burton B. M., Marquis K. A., Sullivan N. L., Rapoport T. A., Rudner D. Z. (2007). The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell 131 1301–1312. 10.1016/j.cell.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp A. H., Losick R. (2009). A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23 1014–1024. 10.1101/gad.1781709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoni D. I., Thakur S., Godefroy C., Le Gall A., Lai-Kee-Him J., Milhiet P.-E., et al. (2014). Structure and DNA-binding properties of the Bacillus subtilis SpoIIIE DNA translocase revealed by single-molecule and electron microscopies. Nucleic Acids Res. 42 2624–2636. 10.1093/nar/gkt1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra G., Chater K. F. (2014). Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol. Rev. 38 345–379. 10.1111/1574-6976.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet A., Vitkup D., Yuan G.-C., Norman T. M., Liu J. S., Losick R. M. (2010). Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. 107 8486–8491. 10.1073/pnas.1002499107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P. A., Mussig A. J., Hugenholtz P., Parks D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36 1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chai Y., Guo J. H., Losick R. (2012). Evidence for cyclic Di-GMP-mediated signaling in Bacillus subtilis. J. Bacteriol. 194 5080–5090. 10.1128/jb.01092-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Coleman K., Pogliano K. (2007). Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J. Biol. Chem. 282 2576–2586. 10.1074/jbc.m606056200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Wivagg C. N., Kapoor M., Barry Z., Rohs P. D. A., Suh H., et al. (2016). Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1:16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church B. D., Halvorson H. (1959). Dependence of the heat resistance of bacterial endospores on their dipicolinic acid content. Nature 183 124–125. 10.1038/183124a0 [DOI] [PubMed] [Google Scholar]
- Clauss M. (2006). Higher effectiveness of photoinactivation of bacterial spores, UV-resistant vegetative bacteria and mold spores with 222 nm compared to 254 nm wavelength. Acta Hydroch. Hydrob. 34 525–532. 10.1002/aheh.200600650 [DOI] [Google Scholar]
- Constant P., Poissant L., Villemur R. (2008). Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J. 2 1066–1076. 10.1038/ismej.2008.59 [DOI] [PubMed] [Google Scholar]
- Daniel R. A., Errington J. (1993). Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J. Mol. Biol. 232 468–483. 10.1006/jmbi.1993.1403 [DOI] [PubMed] [Google Scholar]
- Davidson P., Eutsey R., Redler B., Hiller N. L., Laub M. T., Durand D. (2018). Flexibility and constraint: evolutionary remodeling of the sporulation initiation pathway in Firmicutes. PLoS Genet. 14:e1007470. 10.1371/journal.pgen.1007470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong I. G., Veening J.-W., Kuipers O. P. (2010). Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J. Bacteriol. 192 2053–2067. 10.1128/jb.01484-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick R. M., Wildschutte H., Mccormick J. R. (2009). Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J. Bacteriol. 191 320–332. 10.1128/jb.00858-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek M., Kelly A., Barwinska-Sendra A., Tarrant E., Stanley W. A., Vollmer D., et al. (2018). Peptidoglycan degradation machinery in Clostridium difficile forespore engulfment: engulfment machinery in C. difficile. Mol. Microbiol. 110 390–410. 10.1111/mmi.14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek M., Stabler R. A., Witney A. A., Wren B. W., Fairweather N. F. (2013). Transcriptional Analysis of Temporal Gene Expression in Germinating Clostridium difficile 630 Endospores. PLoS One 8:e64011. 10.1371/journal.pone.0064011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Berardo C., Capstick D. S., Bibb M. J., Findlay K. C., Buttner M. J., Elliot M. A. (2008). Function and redundancy of the chaplin cell surface proteins in aerial hypha formation, rodlet assembly, and viability in Streptomyces coelicolor. J. Bacteriol. 190 5879–5889. 10.1128/jb.00685-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz A., Mathews J. (1970). Classification of streptomyces spore surfaces into five groups. Appl. Microbiol. 21 527–533. 10.1128/aem.21.3.527-533.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditkowski B., Troc P., Ginda K., Donczew M., Chater K. F., Zakrzewska-Czerwinska J., et al. (2010). The actinobacterial signature protein ParJ (SCO1662) regulates ParA polymerization and affects chromosome segregation and cell division during Streptomyces sporulation. Mol. Microbiol. 78 1403–1415. 10.1111/j.1365-2958.2010.07409.x [DOI] [PubMed] [Google Scholar]
- Doan T., Morlot C., Meisner J., Serrano M., Henriques A. O., Moran C. P., et al. (2009). Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5:e1000566. 10.1371/journal.pgen.1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. L., Fimlaid K. A., Shen A. (2016). Characterization of Clostridium difficile spores lacking either spoVAC or dipicolinic acid synthetase. J. Bacteriol. 198 1694–1707. 10.1128/jb.00986-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. L., Forster E. R., Rohlfing A. E., Shen A. (2020). Differential effects of ‘resurrecting’ Csp pseudoproteases during Clostridioides difficile spore germination. Biochem. J. 477 1459–1478. 10.1042/bcj20190875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Huey J. D., Herman J. K. (2016). The DnaA inhibitor SirA acts in the same pathway as Soj (ParA) to facilitate oriC segregation during Bacillus subtilis sporulation. Mol. Microbiol. 102 530–544. 10.1111/mmi.13477 [DOI] [PubMed] [Google Scholar]
- Eaton D., Ensign J. C. (1980). Streptomyces viridochromogenes spore germination initiated by calcium ions. J. Bacteriol. 143 377–382. 10.1128/jb.143.1.377-382.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier S. E., Tan I. S., Clapham K. R., Ramamurthi K. S. (2012). Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol. Microbiol. 84 682–696. 10.1111/j.1365-2958.2012.08052.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C., Feher V. A., Hoch J. A. (1999). Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181 1975–1983. 10.1128/jb.181.7.1975-1983.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facey P. D., Hitchings M. D., Saavedra-Garcia P., Fernandez-Martinez L., Dyson P. J., Del Sol R. (2009). Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 73 1186–1202. 10.1111/j.1365-2958.2009.06848.x [DOI] [PubMed] [Google Scholar]
- Fiche J.-B., Cattoni D. I., Diekmann N., Langerak J. M., Clerte C., Royer C. A., et al. (2013). Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol. 11:e1001557. 10.1371/journal.pbio.1001557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimlaid K. A., Jensen O., Donnelly M. L., Francis M. B., Sorg J. A., Shen A. (2015a). Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog. 11:e1005239. 10.1371/journal.ppat.1005239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimlaid K. A., Jensen O., Donnelly M. L., Siegrist M. S., Shen A. (2015b). Regulation of Clostridium difficile spore formation by the SpoIIQ and SpoIIIA proteins. PLoS Genet. 11:e1005562. 10.1371/journal.pgen.1005562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K., Eder B. D. (1969). Comparison of survivor curves of Bacillus subtilis spores subjected to wet and dry heat. Food Sci. 34 518–521. 10.1111/j.1365-2621.1969.tb12075.x [DOI] [Google Scholar]
- Fujita M., González-Pastor J. E., Losick R. (2005). High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187 1357–1368. 10.1128/jb.187.4.1357-1368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funa N., Funabashi M., Ohnishi Y., Horinouchi S. (2005). Biosynthesis of hexahydroxyperylenequinone melanin via oxidative aryl coupling by cytochrome P-450 in Streptomyces griseus. J. Bacteriol. 187 8149–8155. 10.1128/jb.187.23.8149-8155.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K. A., Schumacher M. A., Bush M. J., Bibb M. J., Chandra G., Holmes N. A., et al. (2020). c-di-GMP Arms an anti-sigma to control progression of multicellular differentiation in streptomyces. Mol. Cell. 77:e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Mekhedov S. L., Puigbo P., Smirnov S., Wolf Y. I., Rigden D. J. (2012). Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ. Microbiol. 14 2870–2890. 10.1111/j.1462-2920.2012.02841.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Wolf Y. I., Makarova K. S., Vera Alvarez R., Landsman D., Koonin E. V. (2021). COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49 D274–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G., Willemse J., Zhu H., Claessen D., Bukarasam K., Goodfellow M., et al. (2014). Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces. Antonie Van. Leeuwenhoek 106 365–380. 10.1007/s10482-014-0209-1 [DOI] [PubMed] [Google Scholar]
- Griffiths K. K., Zhang J., Cowan A. E., Yu J., Setlow P. (2011). Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81 1061–1077. 10.1111/j.1365-2958.2011.07753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser H. J., Yousef M. R., Elliot M. A. (2009). Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J. Bacteriol. 191 6501–6512. 10.1128/jb.00767-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haist J., Neumann S. A., Al-Bassam M. M., Lindenberg S., Elliot M. A., Tschowri N. (2020). Specialized and shared functions of diguanylate cyclases and phosphodiesterases in Streptomyces development. Mol. Microbiol. 114 808–822. 10.1111/mmi.14581 [DOI] [PubMed] [Google Scholar]
- Heichlinger A., Ammelburg M., Kleinschnitz E. M., Latus A., Maldener I., Flardh K., et al. (2011). The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J. Bacteriol. 193 1533–1542. 10.1128/jb.01100-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D., Dworkin J. (2012). Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskisson P. A., Van Wezel G. P. (2019). Streptomyces coelicolor. Trends Microbiol. 27 468–469. [DOI] [PubMed] [Google Scholar]
- Hosoya S., Lu Z., Ozaki Y., Takeuchi M., Sato T. (2007). Cytological analysis of the mother cell death process during sporulation in Bacillus subtilis. J. Bacteriol. 189 2561–2565. 10.1128/jb.01738-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N., Errington J. (1991). Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J. Bacteriol. 173 3159–3169. 10.1128/jb.173.10.3159-3169.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap P., Michailidis G., Zielke R., Walker A. K., Patel N., Strahler J. R., et al. (2006). Early events of Bacillus anthracis germination identified by time-course quantitative proteomics. PROTEOMICS 6 5199–5211. 10.1002/pmic.200600314 [DOI] [PubMed] [Google Scholar]
- Jakimowicz D., Gust B., Zakrzewska-Czerwinska J., Chater K. F. (2005). Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 187 3572–3580. 10.1128/jb.187.10.3572-3580.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D., Zydek P., Kois A., Zakrzewska-Czerwinska J., Chater K. F. (2007). Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65 625–641. 10.1111/j.1365-2958.2007.05815.x [DOI] [PubMed] [Google Scholar]
- Jalal A. S., Tran N. T., Le T. B. (2020). ). ParB spreading on DNA requires cytidine triphosphate in vitro. Elife 9:e53515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson K. H., Wilkinson A. J. (2017). Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes (Basel) 8 1–32. 10.1007/978-3-319-23534-9_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Shao W., Perego M., Hoch J. A. (2000). Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38 535–542. 10.1046/j.1365-2958.2000.02148.x [DOI] [PubMed] [Google Scholar]
- Kaplan-Levy R. N., Hadas O., Summers M. L., Rücker J., Sukenik A. (2010). “Akinetes: Dormant Cells of Cyanobacteria,” in Dormancy and Resistance in Harsh Environments, eds Lubzens E., Cerda J., Clark M. (Berlin, Heidelberg: Springer; ), 5–27. 10.1007/978-3-642-12422-8_2 [DOI] [Google Scholar]
- Kawamoto I., Tetsuo O., Nara T. (1981). Spore resistance of Micromonospora olivoasterospora, Micromonospora sagamiensis and related organisms. Agric. Biol. Chem. 46 221–231. 10.1271/bbb1961.46.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser B. J. F., Ter Beek A., Rauwerda H., Schuren F., Montijn R., Van Der Spek H., et al. (2007). Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J. Bacteriol. 189 3624–3634. 10.1128/jb.01736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A., Salgado P. S. (2019). The engulfasome in C. difficile: variations on protein machineries. Anaerobe 60:102091. 10.1016/j.anaerobe.2019.102091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K., Lopez-Garrido J., Zhao Z., Watanabe R., Yuan Y., Sugie J., et al. (2019). The molecular architecture of engulfment during Bacillus subtilis sporulation. Elife 8:e45257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Tyndall E. R., Huang K. C., Tian F., Ramamurthi K. S. (2017). Dash-and-recruit mechanism drives membrane curvature recognition by the small bacterial protein SpoVM. Cell. Syst. 5 518–526. 10.1016/j.cels.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Calcutt M. J., Schmidt F. J., Chater K. F. (2000). Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182 1313–1320. 10.1128/jb.182.5.1313-1320.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschnitz E. M., Heichlinger A., Schirner K., Winkler J., Latus A., Maldener I., et al. (2011). Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3(2) cooperate in spore wall synthesis. Mol. Microbiol. 79 1367–1379. 10.1111/j.1365-2958.2010.07529.x [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Ragkousi K., Setlow P. (2006). The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. U S A. 103 165–170. 10.1073/pnas.0507121102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S. M., Cermak N., Feijó Delgado F., Setlow B., Setlow P., Manalis S. R. (2016). Water and small-molecule permeation of dormant Bacillus subtilis spores. J. Bacteriol. 198 168–177. 10.1128/jb.00435-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan T. J., Foley M. H., Shoshiev M. S., Somers M. J., Carlson P. E., Hanna P. C. (2018). Updates to Clostridium difficile Spore germination. J. Bacteriol. 200:e00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kois A., Swiatek M., Jakimowicz D., Zakrzewska-Czerwinska J. (2009). SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J. Bacteriol. 191 310–319. 10.1128/jb.00513-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P. A., Fan N., Ricca E., Driks A., Losick R., Cutting S. (1993). An unusually small gene required for sporulation by Bacillus subtilis. Mol. Microbiol. 9 761–771. 10.1111/j.1365-2958.1993.tb01736.x [DOI] [PubMed] [Google Scholar]
- Li Y., Davis A., Korza G., Zhang P., Li Y.-Q., Setlow B., et al. (2012). Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194 1875–1884. 10.1128/jb.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Jin K., Ghosh S., Devarakonda P., Carlson K., Davis A., et al. (2014). Structural and functional analysis of the GerD spore germination protein of Bacillus species. J. Mol. Biol. 426 1995–2008. 10.1016/j.jmb.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Jin K., Perez-Valdespino A., Federkiewicz K., Davis A., Maciejewski M. W., et al. (2019). Structural and functional analyses of the N-terminal domain of the A subunit of a Bacillus megaterium spore germinant receptor. Proc. Natl. Acad. Sci. U S A. 116 11470–11479. 10.1073/pnas.1903675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Zhang P., Setlow P., Li Y.-Q. (2014). High-precision fitting measurements of the kinetics of size changes during germination of individual bacillus spores. Appl. Environ. Microbiol. 80 4606–4615. 10.1128/aem.01204-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liot Q., Constant P. (2016). Breathing air to save energy–new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis. Microbiologyopen 5 47–59. 10.1002/mbo3.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H., Raj A., Dubnau D. (2007). Noise in gene expression determines cell fate in Bacillus subtilis. Science 317 526–529. 10.1126/science.1140818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis R. E. (1998). “Bacterial spores - resistance, dormancy and water status,” in The Properties of Water in Foods ISOPOW 6, ed. Reid D. S. (Boston, MA: Springer; ). [Google Scholar]
- McBride M. J., Ensign J. C. (1987). Effects of intracellular trehalose content on Streptomyces griseus spores. J. Bacteriol. 169 4995–5001. 10.1128/jb.169.11.4995-5001.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J. R. (2009). Cell division is dispensable but not irrelevant in Streptomyces. Curr. Opin. Microbiol. 12 689–698. 10.1016/j.mib.2009.10.004 [DOI] [PubMed] [Google Scholar]
- McCormick J. R., Flardh K. (2012). Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36 206–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J. R., Su E. P., Driks A., Losick R. (1994). Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14 243–254. 10.1111/j.1365-2958.1994.tb01285.x [DOI] [PubMed] [Google Scholar]
- McKenney P. T., Driks A., Eskandarian H. A., Grabowski P., Guberman J., Wang K. H., et al. (2010). A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr. Biol. 20 934–938. 10.1016/j.cub.2010.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mearls E. B., Izquierdo J. A., Lynd L. R. (2012). Formation and characterization of non-growth states in Clostridium thermocellum: spores and L-forms. BMC Microbiol. 12:180. 10.1186/1471-2180-12-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megrian D., Taib N., Witwinowski J., Beloin C., Gribaldo S. (2020). One or two membranes? diderm firmicutes challenge the gram-positive/gram-negative divide. Mol. Microbiol. 113 659–671. 10.1111/mmi.14469 [DOI] [PubMed] [Google Scholar]
- Mikulik K., Bobek J., Bezouskova S., Benada O., Kofronova O. (2002). Expression of proteins and protein kinase activity during germination of aerial spores of Streptomyces granaticolor. Biochem. Biophys. Res. Commun. 299 335–342. 10.1016/s0006-291x(02)02606-2 [DOI] [PubMed] [Google Scholar]
- Minh B. Q., Hahn M. W., Lanfear R. (2020). New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 37 2727–2733. 10.1093/molbev/msaa106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Dorado J., Marcos-Torres F. J., García-Bravo E., Moraleda-Muñoz A., Pérez J. (2016). Myxobacteria: moving, killing, feeding, and surviving together. Front. Microbiol. 7:781. 10.3389/fmicb.2016.00781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noens E. E., Mersinias V., Traag B. A., Smith C. P., Koerten H. K., Van Wezel G. P. (2005). SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 58 929–944. 10.1111/j.1365-2958.2005.04883.x [DOI] [PubMed] [Google Scholar]
- Nugroho F. A., Yamamoto H., Kobayashi Y., Sekiguchi J. (1999). Characterization of a new sigma-K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J. Bacteriol. 181 6230–6237. 10.1128/jb.181.20.6230-6237.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N., López-Garrido J., Pogliano K., Endres R. G. (2016). Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. eLife 5:e18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsburn B. C., Melville S. B., Popham D. L. (2010). EtfA catalyses the formation of dipicolinic acid in Clostridium perfringens. Mol. Microbiol. 75 178–186. 10.1111/j.1365-2958.2009.06975.x [DOI] [PubMed] [Google Scholar]
- O’Sullivan L. A., Roussel E. G., Weightman A. J., Webster G., Hubert C. R., Bell E., et al. (2015). Survival of Desulfotomaculum spores from estuarine sediments after serial autoclaving and high-temperature exposure. ISME J. 9 922–933. 10.1038/ismej.2014.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D. H., Chuvochina M., Waite D. W., Rinke C., Skarshewski A., Chaumeil P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36 996–1004. 10.1038/nbt.4229 [DOI] [PubMed] [Google Scholar]
- Pizarro-Guajardo M., Calderón-Romero P., Castro-Córdova P., Mora-Uribe P., Paredes-Sabja D. (2016). Ultrastructural variability of the exosporium layer of Clostridium difficile spores. Appl. Environ. Microbiol. 82 2202–2209. 10.1128/aem.03410-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomp M., Leighton T. J., Wheeler K. E., Hill H. D., Malkin A. J. (2007). In vitro high-resolution structural dynamics of single germinating bacterial spores. Proc. Natl. Acad. Sci. 104 9644–9649. 10.1073/pnas.0610626104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompeo F., Freton C., Wicker-Planquart C., Grangeasse C., Jault J. M., Galinier A. (2012). Phosphorylation of CpgA protein enhances both its GTPase activity and its affinity for ribosome and is crucial for Bacillus subtilis growth and morphology. J. Biol. Chem. 287 20830–20838. 10.1074/jbc.m112.340331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D. L., Stragier P. (1991). Cloning, characterization, and expression of the spoVB gene of Bacillus subtilis. J. Bacteriol. 173 7942–7949. 10.1128/jb.173.24.7942-7949.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. F. (1953). Isolation of dipicolinic acid (pyridine-2:6-dicarboxylic acid) from spores of Bacillus megatherium. Biochem. J. 54 210–211. 10.1042/bj0540210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Guadiana F. H., Meeske A. J., Rodrigues C. D. A., Barajas-Ornelas R. D. C., Kruse A. C., Rudner D. Z. (2017). A two-step transport pathway allows the mother cell to nurture the developing spore in Bacillus subtilis. PLoS Genet. 13:e1007015. 10.1371/journal.pgen.1007015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Léon F., Bush M. J., Sallmen J. W., Chandra G., Richardson J., Findlay K. C., et al. (2020). A conserved cell division protein directly regulates FtsZ dynamics in filamentous and unicellular actinobacteria. bioRxiv 2020:322578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Silva P., Serrano M., Henriques A. O. (2019). From root to tips: sporulation evolution and specialization in Bacillus subtilis and the intestinal pathogen Clostridioides difficile. Mol. Biol. Evol. 36 2714–2736. 10.1093/molbev/msz175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribis J. W., Fimlaid K. A., Shen A. (2018). Differential requirements for conserved peptidoglycan remodeling enzymes during Clostridioides difficile spore formation: regulation of Clostridium difficile engulfment. Mol. Microbiol. 110 370–389. 10.1111/mmi.14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribis J. W., Ravichandran P., Putnam E. E., Pishdadian K., Shen A. (2017). The conserved spore coat protein SpoVM is largely dispensable in Clostridium difficile spore formation. mSphere 2 e00315–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenman P. J., Nicholson W. L. (2000). Role of the spore coat layers in Bacillus subtilis spore resistance to hydrogen peroxide, artificial UV-C, UV-B, and solar UV radiation. Appl. Environ. Microbiol. 66 620–626. 10.1128/aem.66.2.620-626.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioseras B., Yague P., Lopez-Garcia M. T., Gonzalez-Quinonez N., Binda E., Marinelli F., et al. (2016). Characterization of SCO4439, a D-alanyl-D-alanine carboxypeptidase involved in spore cell wall maturation, resistance, and germination in Streptomyces coelicolor. Sci. Rep. 6:21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing A. E., Eckenroth B. E., Forster E. R., Kevorkian Y., Donnelly M. L., Benito De La Puebla H., et al. (2019). The CspC pseudoprotease regulates germination of Clostridioides difficile spores in response to multiple environmental signals. PLoS Genet. 15:e1008224. 10.1371/journal.pgen.1008224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C., Abel-Santos E. (2010). The Ger receptor family from sporulating bacteria. Curr. Issues Mol. Biol. 12 147–158. [PMC free article] [PubMed] [Google Scholar]
- Sahin O., Yong E. H., Driks A., Mahadevan L. (2012). Physical basis for the adaptive flexibility of Bacillus spore coats. J. Royal Soc. Interface 9 3156–3160. 10.1098/rsif.2012.0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno P., Larsson J., Bucca G., Laing E., Smith C. P., Flardh K. (2009). One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J. Bacteriol. 191 6489–6500. 10.1128/jb.00709-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo L. Y., Doi R. H. (1974). Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J. Bacteriol. 120 475–481. 10.1128/jb.120.1.475-481.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlimpert S., Wasserstrom S., Chandra G., Bibb M. J., Findlay K. C., Flardh K., et al. (2017). Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc. Natl. Acad. Sci. U S A. 114 E6176–E6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella S. R. B. R., Vandenberghe L. P. S., Soccol C. R. (2014). Life cycle and spore resistance of spore-forming Bacillus atrophaeus. Microbiol. Res. 169 931–939. 10.1016/j.micres.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Serrano M., Crawshaw A. D., Dembek M., Monteiro J. M., Pereira F. C., Pinho M. G., et al. (2016). The SpoIIQ-SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis: the SpoIIQ-SpoIIIAH complex of C. difficile. Mol. Microbiol. 100 204–228. 10.1111/mmi.13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow B., Atluri S., Kitchel R., Koziol-Dube K., Setlow P. (2006). Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-Protective α/β-type small acid-soluble proteins. J. Bacteriol. 188 3740–3747. 10.1128/jb.00212-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. (1988). Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Ann. Rev. Microbiol. 42 319–338. 10.1146/annurev.mi.42.100188.001535 [DOI] [PubMed] [Google Scholar]
- Setlow P. (2003). Spore germination. Curr. Opin. Microbiol. 6 550–556. [DOI] [PubMed] [Google Scholar]
- Setlow P. (2006). Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101 514–525. 10.1111/j.1365-2672.2005.02736.x [DOI] [PubMed] [Google Scholar]
- Setlow P. (2007). I will survive: DNA protection in bacterial spores. Trends Microbiol. 15 172–180. 10.1016/j.tim.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Setlow P. (2014). Spore resistance properties. Microbiol. Spectr. 2:2012. [DOI] [PubMed] [Google Scholar]
- Setlow P., Kornberg A. (1970). Biochemical studies of bacterial sporulation and germination. 23. Nucleotide metabolism during spore germination. J. Biol. Chem. 245 3645–3652. [PubMed] [Google Scholar]
- Sexton D. L., Herlihey F. A., Brott A. S., Crisante D. A., Shepherdson E., Clarke A. J., et al. (2020). Roles of LysM and LytM domains in resuscitation-promoting factor (Rpf) activity and Rpf-mediated peptidoglycan cleavage and dormant spore reactivation. J. Biol. Chem. 295 9171–9182. 10.1074/jbc.ra120.013994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton D. L., St-Onge R. J., Haiser H. J., Yousef M. R., Brady L., Gao C., et al. (2015). Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J. Bacteriol. 197 848–860. 10.1128/jb.02464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton D. L., Tocheva E. I. (2020). Ultrastructure of exospore formation in streptomyces revealed by Cryo-Electron tomography. Front. Microbiol. 11:581135. 10.3389/fmicb.2020.581135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. (2008). A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135 486–496. 10.1016/j.cell.2008.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A., Edwards A. N., Sarker M. R., Paredes-Sabja D. (2019). Sporulation and germination in clostridial pathogens. Microbiol. Spectr. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. J., Foster S. J. (1995). Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J. Bacteriol. 177 3855–3862. 10.1128/jb.177.13.3855-3862.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh Y. M., Davidson I. F., Zamuner S., Basquin J., Bock F. P., Taschner M., et al. (2019). Self-organization of parS centromeres by the ParB CTP hydrolase. Science 366 1129–1133. 10.1126/science.aay3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squyres G. R., Holmes M. J., Barger S. R., Pennycook B. R., Ryan J., Yan V. T., et al. (2020). Dynamics of bacterial cell division: Z ring condensation is essential for cytokinesis. bioRxiv 2020:180737. [Google Scholar]
- Steiner E., Dago A. E., Young D. I., Heap J. T., Minton N. P., Hoch J. A., et al. (2011). Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol. Microbiol. 80 641–654. 10.1111/j.1365-2958.2011.07608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelder S. K., Benito De Moya C., Hoefsloot H. C. J., De Koning L. J., Brul S., De Koster C. G. (2018). Stoichiometry, absolute abundance, and localization of proteins in the Bacillus cereus spore coat insoluble fraction determined using a QconCAT approach. J. Proteome Res. 17 903–917. 10.1021/acs.jproteome.7b00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson K., Hoch J. A. (2002). Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 46 297–304. 10.1046/j.1365-2958.2002.03186.x [DOI] [PubMed] [Google Scholar]
- Stevens C. M., Daniel R., Illing N., Errington J. (1992). Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174 586–594. 10.1128/jb.174.2.586-594.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. C. (2015). The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol. Mol. Biol. Rev. 79 437–457. 10.1128/mmbr.00050-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakova E., Bobek J., Zikova A., Rehulka P., Benada O., Rehulkova H., et al. (2013). Systems insight into the spore germination of Streptomyces coelicolor. J. Proteome Res. 12 525–536. [DOI] [PubMed] [Google Scholar]
- Swarge B., Abhyankar W., Jonker M., Hoefsloot H., Kramer G., Setlow P., et al. (2020). Integrative analysis of proteome and transcriptome dynamics during Bacillus subtilis spore revival. mSphere 5 e00420–e00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz J. P., Nanji T., Gloyd M., Guarne A., Elliot M. A. (2013). A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 41 4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran M. J., Jakimowicz D., Elliot M. A. (2020). Compaction and control - the role of chromosome organizing proteins in Streptomyces. FEMS Microbiol. Rev. 44 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taib N., Megrian D., Witwinowski J., Adam P., Poppleton D., Borrel G., et al. (2020). Genome-wide analysis of the firmicutes illuminates the diderm/monoderm transition. Nat. Ecol. Evol. 4 1661–1672. [DOI] [PubMed] [Google Scholar]
- Takahashi F., Sumitomo N., Hagihara H., Ozaki K. (2015). Increased dipicolinic acid production with an enhanced spoVF operon in Bacillus subtilis and medium optimization. Biosci. Biotechnol. Biochem. 79 505–511. [DOI] [PubMed] [Google Scholar]
- Takamatsu H., Kodama T., Nakayama T., Watabe K. (1999). Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181 4986–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I. S., Ramamurthi K. S. (2014). Spore formation in Bacillus subtilis. Environ. Microbiol. Rep. 6 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E. I., Lopez-Garrido J., Hughes H. V., Fredlund J., Kuru E., Vannieuwenhze M. S., et al. (2013). Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol. 88 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E. I., Matson E. G., Morris D. M., Moussavi F., Leadbetter J. R., Jensen G. J. (2011). Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E. I., Ortega D. R., Jensen G. J. (2016). Sporulation, bacterial cell envelopes and the origin of life. Nat. Rev. Microbiol. 14 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag B. A., van Wezel G. P. (2008). The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van. Leeuwenhoek 94 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troiano A. J., Zhang J., Cowan A. E., Yu J., Setlow P. (2015). Analysis of the dynamics of a Bacillus subtilis spore germination protein complex during spore germination and outgrowth. J. Bacteriol. 197 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouve J., Mohamed A., Leisico F., Contreras-Martel C., Liu B., Mas C., et al. (2018). Structural characterization of the sporulation protein GerM from Bacillus subtilis. J. Struc. Biol. 204 481–490. [DOI] [PubMed] [Google Scholar]
- Tschowri N., Schumacher M. A., Schlimpert S., Chinnam N. B., Findlay K. C., Brennan R. G., et al. (2014). Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158 1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood S., Guan S., Vijayasubhash V., Baines S. D., Graham L., Lewis R. J., et al. (2009). Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 191 7296–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aart L. T., Spijksma G. K., Harms A., Vollmer W., Hankemeier T., Van Wezel G. P. (2018). High-resolution analysis of the peptidoglycan composition in Streptomyces coelicolor. J. Bacteriol. 200 e218–e290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P., Mcelligott J., Attkisson C., Betteken M., Popham D. L. (2009). Homologues of the Bacillus subtilis SpoVB protein are involved in cell wall metabolism. J. Bacteriol. 191 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P., Weaver A., Reichert E. D., Linnstaedt S. D., Popham D. L. (2007). Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol. Microbiol. 65 1582–1594. [DOI] [PubMed] [Google Scholar]
- Vincent A. T., Nyongesa S., Morneau I., Reed M. B., Tocheva E. I., Veyrier F. J. (2018). The Mycobacterial cell envelope: a relict from the past or the result of recent evolution? Front. Microbiol. 9:2341. 10.3389/fmicb.2018.02341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J., Cox J., Setlow B., Coleman W. H., Setlow P. (2011). Extremely variable conservation of -type small, acid-soluble proteins from spores of some species in the bacterial order Bacillales. J. Bacteriol. 193 1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Setlow P., Li Y.-Q. (2015). Slow leakage of Ca-dipicolinic acid from individual Bacillus spores during initiation of spore germination. J. Bacteriol. 197 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., De Boer R., Vischer N., Van Haastrecht P., Setlow P., Brul S. (2020). Visualization of germination proteins in putative bacillus cereus germinosomes. Int. J. Mol. Sci. 21:5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel D., Fischer R.-J. (2015). Small acid-soluble spore proteins of Clostridium acetobutylicum are able to protect DNA in vitro and are specifically cleaved by germination protease GPR and spore protease YyaC. Microbiology 161 2098–2109. [DOI] [PubMed] [Google Scholar]
- Willemse J., Borst J. W., De Waal E., Bisseling T., Van Wezel G. P. (2011). Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes. Dev. 25 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Harper W. F. (2020). Bacillus spore awakening: recent discoveries and technological developments. Curr. Opin. Biotechnol. 64 110–115. [DOI] [PubMed] [Google Scholar]
- Yabe S., Aiba Y., Sakai Y., Hazaka M., Yokota A. (2010). A life cycle of branched aerial mycelium- and multiple budding spore-forming bacterium Thermosporothrix hazakensis belonging to the phylum Chloroflexi. J. Gen. Appl. Microbiol. 56 137–141. [DOI] [PubMed] [Google Scholar]
- Yu G. (2020). Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinformatics 69:e96. [DOI] [PubMed] [Google Scholar]
- Yutin N., Galperin M. Y. (2013). A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol. 15 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Msadek T., Zapf J., Madhusudan, Hoch J. A., Varughese K. I. (2002). DNA complexed structure of the key transcription factor initiating development in sporulating bacteria. Structure 10 1041–1050. [DOI] [PubMed] [Google Scholar]
- Zheng L. B., Donovan W. P., Fitz-James P. C., Losick R. (1988). Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Amp. Dev. 2 1047–1054. [DOI] [PubMed] [Google Scholar]
- Zhou X., Halladin D. K., Theriot J. A. (2016). Fast mechanically driven daughter cell separation is widespread in actinobacteria. mBio 7 e916–e952. [DOI] [PMC free article] [PubMed] [Google Scholar]