Abstract
Maintaining visual function is key to establishing improved longevity. However, the numbers of patients with diseases of the retina, the most important tissue for vision and the key to age-related blindness, are not declining due to the increase in the number of aging subjects worldwide and the technological advances in the delivery of premature infants. The primary treatment option for retinal diseases is still surgical intervention and includes laser or photocoagulation, which are associated with various complications and side effects. Many aspects of the pathogenesis of these retinal diseases are still unknown, thereby impeding drug discovery. This has led to an increase in the number of studies focused on the underlying pathogenic mechanisms of retinal diseases. Growing evidence suggests that non-coding RNAs play critical roles in the pathogenesis of retinal diseases. Herein, we have summarized the known functional roles of non-coding RNAs, emphasizing their contribution to the underlying pathogenesis of retinal diseases. In addition, we discuss the modulation of non-coding RNAs as potential therapeutics and the methods to control the non-coding RNAs for the treatment. We expect that targeting non-coding RNAs could be crucial for developing novel therapeutics for progressive diseases including diseases of the retina.
Keywords: non-coding RNA, retinopathy of prematurity, age-related macular degeneration, diabetic retinopathy, RNA therapy
Graphical abstract
Growing evidence suggests that non-coding RNAs play critical roles in the pathogenesis of retinal diseases. In this review, the functional role of non-coding RNAs underlying the pathogenesis of diverse retinal diseases is summarized, and the potential of modulating non-coding RNAs as therapeutics of retinal diseases is discussed.
Introduction: Diseases in the retina
The importance of maintaining our visual health is growing as the world continues to age, with vision being one of the most critical factors for improving the quality of life in older people. Retinal disease is a particular concern for maintaining vision, with the development of therapeutic interventions to slow retinal degeneration emerging as a research priority in the last few decades. However, the current treatment of retinal diseases relies on surgical interventions, which may have several unwanted side effects and pose unwanted risks to the patient.
Retinal diseases predominantly affect older people, but some diseases also occur in babies. Retinopathy of prematurity is a disease caused by abnormal blood vessel development in the retinas of premature babies.1,2 The major risk factors for retinopathy of prematurity include oxygen supplementation, low gestation periods, and lower birth weights. When a premature infant is born, before the retinal blood vessels are fully developed, the vascularization can be arrested as a result of their exposure to hyperoxic air. This means that when the retina is fully developed and its metabolic demands increase, hypoxic conditions are induced in the retina, resulting in pathological blood vessel growth. In severe cases, this fibrotic proliferation induces retinal detachment. Currently, retinopathy of prematurity is treated via various surgical interventions, including laser therapy, but it would be advantageous to develop less destructive approaches in the future.
Age-related macular degeneration is a leading cause of vision loss in the elderly; its incidence and prevalence rates are expected to increase as the numbers of elderly subjects increase worldwide.3,4 In the early stages of age-related macular degeneration, the cells in the retinal tissue lose their function and aggregate, creating deposits called drusen, which accumulate under the retina and produce an increasing burden on vision as they grow in size. In the later stages of age-related macular degeneration, geographic atrophy extends to the macula (dry or non-exudative type), or an abnormal choroidal vasculature is created (wet or exudative type), which seriously impairs retinal function and vision. Although several therapeutic options including anti-vascular endothelial growth factor (VEGF) and photodynamic therapy are available, complete treatment is not yet possible.
Diabetic retinopathy, a complication caused by circulatory disorders in the peripheral blood vessels of patients with diabetes, is another leading cause of blindness.5,6 In the non-proliferative stages of diabetic retinopathy, structural changes in the retinal blood vessels and resultant hemorrhages are observed. When non-proliferative diabetic retinopathy advances to proliferative diabetic retinopathy, new, fragile vasculature is formed in the retina, and this vasculature is easily damaged, resulting in serious bleeding and, eventually, blindness. Diabetic retinopathy is treated using a combination of diet control, to reduce blood sugar, and surgical intervention, including laser photocoagulation. Because this treatment is destructive and often has adverse effects, it is critical that new less destructive therapeutic options be developed.
Glaucoma is one of the most important causes of irreversible blindness. This disease is characterized by progressive degeneration of the optic nerve and retinal ganglion cells, which play important roles in the transmission of visual information between the eyes and the brain.7,8 The mechanism underlying the damage of the optic nerve in glaucoma is still unclear, and several mechanisms appear to work in combination to create this complex phenotype. One of the most important mechanisms of glaucoma is increased intraocular pressure, which leads to optic nerve damage. However, there are cases of glaucoma where the intraocular pressure is normal in which the patients present with optic nerve damage, highlighting the need for more advanced diagnostic interventions for this disease. Treatment options include the use of drugs to lower intraocular pressure or various surgical treatments, including laser-based therapies. Early detection is essential for a good prognosis, but this can be difficult because glaucoma is largely asymptomatic in its early stages.
Finally, retinoblastoma is rare but remains the most common form of malignant tumor of the retina in children.9 Hereditary retinoblastoma is caused by a mutation in the retinoblastoma 1 (RB1) tumor suppressor gene. In this case, there is also a high risk of developing cancer in other organs as well. Surgical removal of the eye is generally used to treat serious cases of retinoblastoma, but chemotherapy and radiation therapy are also used to treat less severely affected eyes.
Thus, the number of patients with various retinal diseases is constantly increasing, due to the increase in the number of aging subjects and the advance in the delivery technologies for prematurity. However, therapeutic intervention for these diseases remains limited, with most treatment strategies relying on laser treatment or surgical intervention, which may have significant adverse effects. In addition, drug development is hampered by our limited understanding of the pathogenesis of many of these retinal diseases, meaning that we need more studies evaluating the underlying pathological mechanisms of these diseases. In this review, we summarized the roles of non-coding RNAs (ncRNAs) as important regulators of the pathogenesis of retinal diseases. After explaining the types of ncRNAs and their biogenesis pathway and action mechanism, we introduce representative ncRNAs that have been confirmed to have important roles in each retinal disease. Finally, the treatment strategies targeting these ncRNAs are described.
Introduction: ncRNAs
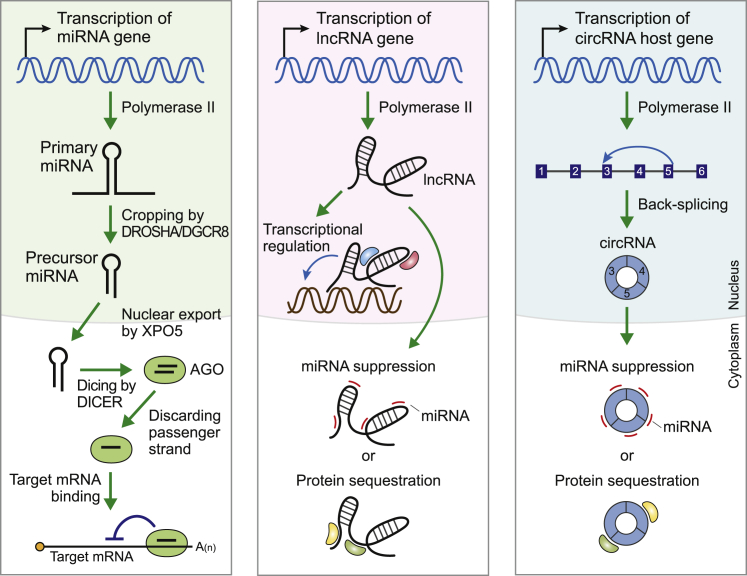
ncRNAs are a large class of non-coding genes that do not include any open reading frames allowing the production of a protein (Figure 1). MicroRNAs (miRNAs) are a well-established class of small ncRNAs that are ∼22 nt long; they are broadly conserved across mammalian species. They are produced by polymerase II transcription in the nucleus.10 This primary transcript is called primary miRNA (pri-miRNA) and is processed via the DROSHA-DGCR8 protein complex, which produces the precursor miRNA (pre-miRNA). This hairpin-shaped pre-miRNA is exported to the cytoplasm by EXPORTIN-5 and further processed by DICER. This produces a small duplex RNA that is incorporated into the ARGONAUTE (AGO) protein. Finally, one strand of the duplex is discarded from this protein, and the remaining strand, the mature miRNA, suppresses the expression of its target messenger RNAs (mRNAs) through sequence-specific binding.10 Diverse target genes have been described for each miRNA, and these miRNA-target gene relationships have been shown to be critical to various physiological processes. Therefore, it is not surprising that perturbations in these relationships can cause various diseases.11
Figure 1.
The biogenesis and working mechanisms of non-coding RNAs (ncRNAs)
The biogenesis and the main functional mechanism underlying the action of microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs) are shown. See the text for details.
Long ncRNAs (lncRNAs) are a large class of ncRNAs comprising transcripts exceeding 200 nt in length.12 Similar to mRNAs, lncRNAs have a 5′ cap structure and undergo splicing. Many lncRNAs are not polyadenylated, and the sequences of lncRNAs are less conserved, making them distinct from mRNA transcripts. lncRNAs exhibit different mechanisms of action based on their cellular localization. In the nucleus, lncRNAs modulate the transcription of other genes by binding to transcription factors,13 while in the cytoplasm, they regulate protein function by modulating protein availability. However, the most well-described mechanism of action for cytoplasmic lncRNAs is their role as miRNA “sponges”, i.e., these transcripts bind specific miRNAs, preventing their function. Similar to miRNAs, lncRNAs have been linked to a wide variety of human diseases, making them ideal therapeutic targets and potential effectors.
Circular RNAs (circRNAs) are single-stranded RNA transcripts that adopt a closed circular structure. In general, they are produced by the back-splicing reaction of host genes,14,15 and a recent study reported nearly 100,000 unique circRNA transcripts across several human cancer cell lines.16 circRNAs also demonstrate a diverse range of functional mechanisms, and, similar to lncRNAs, circRNAs can modulate the function of proteins by binding to them directly. However, circRNAs are best described as miRNA regulators, with many of these transcripts acting to sequester their target miRNAs and suppressing their function. Supporting this observation, most circRNAs exist in the cytoplasm, where they are likely to come into contact with their miRNA targets.17,18 Given the fact that miRNAs are broadly implicated in various diseases, it is safe to assume that the circRNAs that regulate them are also being widely evaluated.
In this review, we summarize the papers describing the functional mechanism of ncRNAs associated with diseases of the retina. We searched published papers in PubMed for the combination of disease name (age-related macular degeneration, diabetic retinopathy, and so on) with the name of the ncRNA group (miRNA, lncRNA, and circRNA). Among the searched papers, only the papers where ncRNA is the main subject of the study were included. In the case of the miRNA study, only those with confirmation experiments in each animal model were included due to a large number of published papers. Therefore, we note that not all papers related to the pathogenesis of retinal disease were included in this review. For more information on the published papers for each ncRNA group in retinal diseases, please refer to recent reviews (PMID: 31963809, 30927500, and 33015046). We have marked any ncRNA gene names that have been altered to conform with more general nomenclature rules where possible, specifically modifying names to include the strand information for any mature miRNAs (thus, −5p or −3p),19 and the prefix “circ” was added to the names of circRNAs.
ncRNAs and retinal diseases
Retinopathy of prematurity
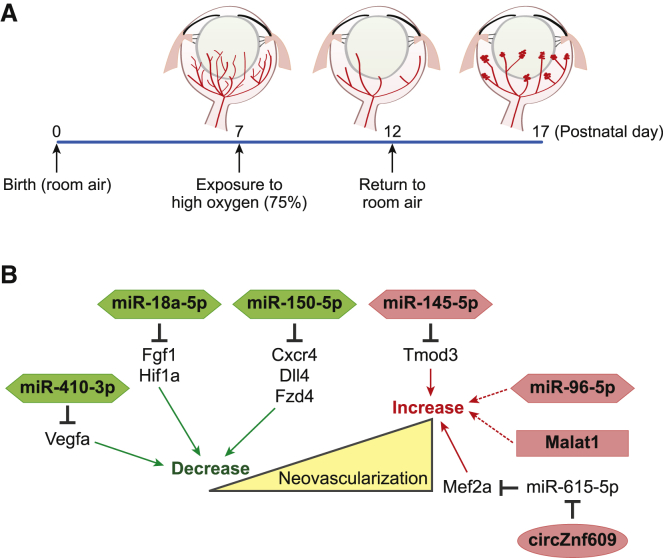
The mouse model of oxygen-induced retinopathy is widely used to facilitate the discovery of ncRNAs involved in the pathogenesis of retinopathy of prematurity.20 In this model, 1-week-old mouse pups (postnatal day [P]7), in which the retinal vasculature is undergoing active development, are exposed to hyperoxic conditions. This leads to the obliteration of the capillaries near the arteries at the center of the retina. Then, at P12, they are returned to room air, which causes hypoxia within the retina. As a result, neovascular tufts are formed, which are also a hallmark of retinopathy of prematurity (Figure 2). Transcriptome analysis of retinal samples from each stage of this mouse model allowed for the identification of several ncRNAs involved in vascular remodeling in the retina. To date, most studies using this system have focused on miRNA profiling, with most of these studies only describing the differentially expressed miRNAs. However, more recently a handful of studies have started to evaluate the therapeutic potential of targeting these miRNAs in order to treat retinopathy of prematurity (Figure 2).
Figure 2.
ncRNAs in retinopathy of prematurity
(A) Oxygen-induced retinopathy is primarily used as a model of retinopathy of prematurity. See the text for details. (B) The ncRNAs identified to be involved in retinopathy of prematurity and their functional mechanisms are summarized here. ncRNAs increasing or decreasing the neovascularization are indicated in red or green, respectively.
In the earliest studies, miR-410-3p was reported to prevent retinal neovascularization in the oxygen-induced retinopathy model.21 This miRNA was shown to suppress Vegfa expression by targeting the 3′ UTR of Vegfa mRNA. It is well established that the upregulation of VEGF under hypoxic conditions is important in the pathogenesis of retinopathy.22 When animals were treated with eye drops containing miR-410-3p, the authors were able to demonstrate that miR-410-3p suppresses Vegfa and prevents angiogenesis in the retina.
However, miR-150-5p, another miRNA, was reported to suppress pathological neovascularization.23 miR-150-5p was shown to be highly expressed in quiescent retinal blood vessels and suppressed in the retina of animals undergoing oxygen-induced proliferative retinopathy. Several angiogenic genes, including C-X-C chemokine receptor type 4 (Cxcr4), delta-like ligand 4 (Dll4), and frizzled-4 (Fzd4), were all determined to be direct targets of miR-150-5p, and intravitreal injection of synthetic miR-150-5p decreased blood vessel production in the retinas of mice, suggesting the therapeutic potential of these miRNAs in the treatment of retinopathy.
miR-145-5p was reported to be upregulated during the neovascularization phases of the oxygen-induced retinopathy model, and treatment with a synthetic miR-145-5p inhibitor decreased neovascularization in the retina.24 Tropomodulin 3 (Tmod3), the actin-capping protein, was confirmed to be a direct target of miR-145-5p and is known to be involved in the creation of the cytoskeletal structures in endothelial cells.
In a recent study, miR-18a-5p was reported to be highly expressed in the retina of mouse pups, and its expression was shown to decrease during retinal development.25 However, the expression of this miRNA was not decreased during pathogenic neovascularization. In addition, intravitreal injection of synthetic miR-18a-5p suppressed retinal neovascularization in the oxygen-induced retinopathy model. Several vascular growth factors, including fibroblast growth factor 1 (Fgf1) and hypoxia-inducible factor 1-alpha (Hif1a), were verified as targets of miR-18a-5p.
miR-96-5p maintains a high expression level during vascular development but is downregulated during vascular degeneration in the rat model of oxygen-induced retinopathy.26 Intravitreal supplementation with miR-96-5p prevented the obliteration of the retinal vessels and facilitated their revascularization in this model. Because the direct target of miR-96-5p was not identified, future studies are required to identify and describe the regulatory network of miR-96-5p in the retina and expand its potential applications in therapeutic intervention.
Taken together, this shows that several miRNAs have been linked to the pathogenesis of retinopathy of prematurity using the oxygen-induced retinopathy model, but in the case of lncRNAs and circRNAs, this network is less clear, as there are only a few studies evaluating their effects in this model. A recent study demonstrated that metastasis-associated lung adenocarcinoma transcript 1 (Malat1) is dysregulated in the retina of mice with oxygen-induced retinopathy.27 Importantly, the intravitreal injection of short interfering RNAs (siRNAs) against Malat1 efficiently reduces the degree of retinopathy in this model. Because the mechanism underlying the action of Malat1 was not reported in this study, further analysis is required to identify the regulatory network underpinning these observations.
The circRNA generated from ZNF609 (circZNF609) is the only circRNA reported to be involved in the pathogenesis of retinopathy of prematurity.28 The expression of circZNF609 was decreased in hyperoxia but increased during hypoxia. Silencing the expression of this circRNA protected the endothelial cells of the retina from oxidative and hypoxic stress in the oxygen-induced retinopathy model. Researchers went on to verify that circZNF609 sequesters miR-615-5p, and this miRNA represses the expression of myocyte-specific enhancer factor 2A (Mef2a).
In summary, most studies designed to identify ncRNAs involved in the pathogenesis of retinopathy of prematurity used the mouse/rat oxygen-induced retinopathy model. In addition, to date, these studies have identified a larger number of miRNAs than lncRNAs and circRNAs, and because of this, future research should be focused on identifying these novel ncRNAs. Moreover, there are no translational studies confirming the regulatory networks described above in humans, possibly due to the difficulty in obtaining these types of samples. However, because a similar change in vascularization is observed in other models of retinopathy, including wet-type age-related macular degeneration and diabetic retinopathy, and the fact that human samples are more readily obtained for these diseases, it may be possible to confirm the regulatory networks of these ncRNAs in other pathologies, as described below.
Age-related macular degeneration
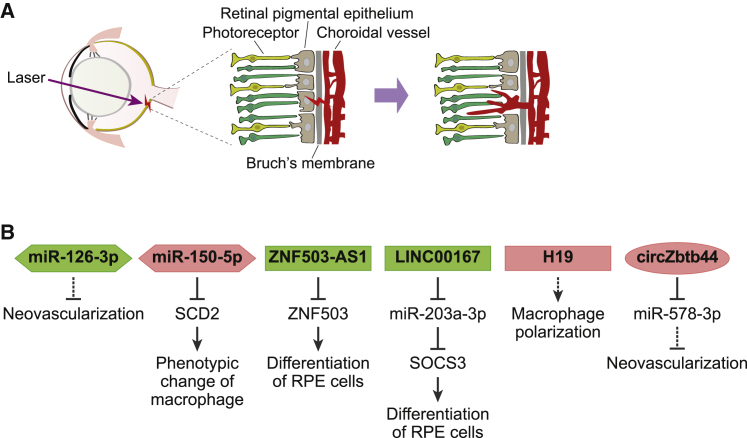
In general, studies describing the ncRNAs involved in age-related macular degeneration use either human retinal tissues, which are more readily available than those for retinopathy of prematurity, or an animal model of laser-induced choroidal neovascularization. This animal model is primarily used to evaluate the exudative form of age-related macular degeneration (Figure 3).29 In this model, the Bruch’s membrane (also called the vitreous lamina) of the mouse is perforated with a laser, resulting in the recruitment of new blood vessels to the subretinal region, mimicking the pathogenesis of the exudative form of human age-related macular degeneration.
Figure 3.
ncRNAs in age-related macular degeneration
(A) Laser-induced choroidal neovascularization as a commonly used model of age-related macular degeneration. See the text for details. (B) Several ncRNAs identified to be involved in age-related macular degeneration and their functional mechanisms are summarized here. ncRNAs aggravating or alleviating disease progression are indicated in red or green, respectively.
This model was used to identify several ncRNAs, including miR-126-3p,30 involved in age-related macular degeneration (Figure 3). Further evaluation demonstrated that miR-126-3p decreased in the retinal pigment epithelium (RPE) and choroid following the development of laser-induced choroidal neovascularization and that intravitreal injection of synthetic miR-126-3p significantly reduced the size of the neovascular lesion in this model.
A recent study demonstrated that miR-150-5p is highly upregulated in human peripheral blood mononuclear cells from patients with age-related macular degeneration.31 This miRNA regulates the transition of macrophages from the normal to angiogenesis-promoting phenotypes by targeting stearoyl-coenzyme A (CoA) desaturase-2 (SCD2). Given the fact that an increase in the number of choroidal macrophages has been reported to coincide with the development of age-related macular degeneration,32 miR-150-5p could be an important regulator for macrophage-mediated changes in age-related macular degeneration.
Zinc finger protein 505 (ZNF503) antisense RNA 1 (ZNF503-AS1) is one of a handful of lncRNAs that have been identified in patients with age-related macular degeneration.33 Research has demonstrated that ZNF503 inhibits RPE differentiation, while its antisense lncRNA, ZNF503-AS1, exerts the opposite effect via its targeted regulation of ZNF503. ZNF503-AS1 was downregulated in patients with age-related macular degeneration, suggesting that ZNF503-AS1 may be a potential therapeutic target for the treatment of this disease.
LINC00167 is another lncRNA downregulated in RPE-choroid tissues from patients with age-related macular degeneration.34 It was reported that reduced expression of LINC00167 resulted in the dedifferentiation of RPE and that this lncRNA exerts its function via the suppression of miR-203a-3p expression, thereby increasing the expression of its target suppressor of cytokine signaling 3 (SOCS3).
A recent lncRNA profiling study identified lncRNA H19 as a significantly upregulated transcript in the aqueous humor of patients with the exudative form of human age-related macular degeneration and the RPE-choroid-sclera complexes isolated from the laser-induced choroidal neovascularization model.35 H19 was shown to regulate the polarization of macrophages in the animal model, although the regulatory target of H19 was not identified in this study.
To date, only one circRNA has been identified as a regulator in the pathogenesis of age-related macular degeneration. This circRNA is generated from locus ZBTB44 (circZBTB44) and was significantly upregulated in the mouse model of laser-induced choroidal neovascularization.36 The suppression of circZBTB44 in this model retarded the development of neovascularization via the sequestering of miR-578-3p. Interestingly, increased circZBTB44 expression was also noted in the aqueous humor of eyes from patients with the exudative form of human age-related macular degeneration, raising the possibility of targeting this circRNA in novel therapeutic strategies in the future.
Diabetic retinopathy
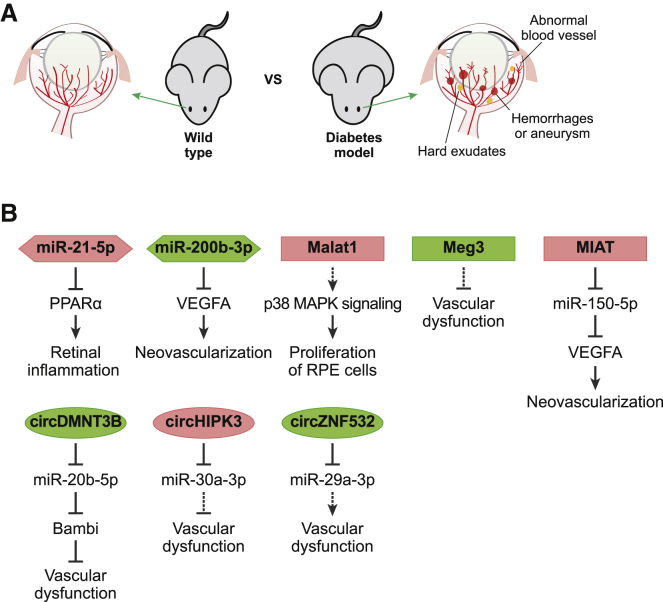
Because retina samples from patients with diabetic retinopathy are more readily available, and the fact that there are several animal models that mimic diabetes, including rats with streptozotocin-induced type 1 diabetes and db/db type 2 diabetic mice, which are commonly used, there are several studies describing the dysregulation of various ncRNAs in diabetic retinopathy (Figure 4).37
Figure 4.
ncRNAs in diabetic retinopathy
(A) Diabetic animal models are commonly used for studying diabetic retinopathy. In rodents with diabetes, diverse pathological problems including abnormal blood vessels, hemorrhages, aneurysms, and hard exudates are observed. (B) Several ncRNAs identified to be involved in diabetic retinopathy have been described together with their functional mechanism and are summarized here. ncRNAs aggravating or alleviating diabetic retinopathy are indicated in red or green, respectively.
Initial miRNA profiling showed that miR-200b-3p was downregulated in the retina of rats with streptozotocin-induced diabetes and patients with diabetes,38 and the intravitreal injection of synthetic miR-200b-3p prevented diabetes-induced increases in the expression of Vegfa, the direct target of miR-200b-3p. Alternatively, the overexpression of miR-21-5p, observed in the retina of db/db mice,39 induced the repression of peroxisome proliferator-activated receptor alpha (Ppar-α), the target gene of miR-21-5p, in the same samples. Researchers then went on to confirm that the intravitreal injection of miR-21-5p inhibitors attenuated retinal inflammation by targeting Ppar-α.
Several studies have identified specific lncRNAs in diabetic retinopathy, with an initial study demonstrating that the expression of the lncRNA Malat1 is increased in the retinal tissues of rats with streptozotocin-induced diabetes and in db/db mice.40 In addition, the intraocular injection of short hairpin RNA (shRNA) against Malat1 ameliorated diabetic retinopathy by decreasing the proliferation of retinal endothelial cells via a p38 mitogen-activated protein kinase signaling-dependent mechanism. A later study showed that the expression of inflammation-related transcripts was significantly decreased in the retina of Malat1 knockout mice, and MALAT1 expression was also shown to be increased in the vitreous humor of patients with diabetes, suggesting that MALAT1 may be a target in diabetic retinopathy.41
Myocardial infarction-associated transcript (MIAT, also known as retinal ncRNA 2 [RNCR2]) is another lncRNA overexpressed in the retina of diabetic rats and patients with diabetes.42 MIAT expression is also known to be significantly increased in the plasma of patients with diabetic retinopathy; MIAT exerts its function by sequestering miR-150-5p.43 Accordingly, MIAT knockdown ameliorates microvascular dysfunction in diabetic retinopathy by releasing miR-150-5p, which in turn suppresses the expression of VEGF, the direct target of this miRNA, attenuating the pathogenesis of retinopathy.42
It has also been reported that the expression of lncRNA maternally expressed gene 3 (Meg3) is decreased in streptozotocin-induced diabetic mice and that the intra-vitreous addition of a shRNA against Meg3 aggravated vascular dysfunction in the retina.44 This suggests that restoring Meg3 expression may present a possible therapeutic approach for the treatment of diabetic retinopathy.
circHIPK3 (the circRNA produced from HIPK3) was the first circRNA reported to be highly upregulated in the retina of mice with streptozotocin-induced diabetes, and circHIPK3 inhibition by a targeted shRNA efficiently alleviated the vascular dysfunction in the retinas of these animals.45 Subsequent studies identified miR-30a-3p as an endogenous target of circHIPK3, and the addition of synthetic miR-30a-3p exhibited similar effects as circHIPK3 suppression on the pathogenesis of diabetic retinopathy. Interestingly, the increased expression of circHIPK3 was only observed in the aqueous humor of patients with diabetes, suggesting that this may be a unique regulator in human diabetic retinopathy.
A recent study reported an increased expression of miR-20b-5p in the retina of diabetic rats and identified that circDMNT3B (circRNA produced from DMNT3B locus), the expression of which was decreased in the retina of diabetic patients, could sequester miR-20b-5p and block its function.46 BMP and activin membrane-bound inhibitor (Babmi) were identified as the targets of miR-20b-5p, and overexpression of circDNMT3B alleviated the vascular dysfunction of diabetic rats via its ability to regulate the miR-20b-5p-Bambi pathway.
circZNF532 (circRNA produced from ZNF532) was recently identified in diabetic retinopathy and shown to be upregulated in the vitreous humor of patients with diabetes, the retinal vessels of mice with streptozotocin-induced diabetes, and pericytes under diabetic conditions.47 The suppression of circZNF532 expression in retinal pericytes by shRNA or the introduction of synthetic miR-29a-3p, the identified target of circZNF532, aggravated the vascular dysfunction in mice with streptozotocin-induced diabetes. In contrast, the overexpression of circZNF532 or the suppression of miR-29a-3p expression ameliorated retinal vascular dysfunction, suggesting the therapeutic potential of these ncRNAs.
The roles of several ncRNAs in diabetic retinopathy have been evaluated in various diabetic models, and transcriptional profiling has shown that their expression is reasonably well conserved between the retina and the vitreous humor of patients with diabetes and in the animal models. Thus, it is possible to predict a regulatory pathway in these patients based on the experimental results from the diabetic animal models and test their therapeutic potential in the same models. Because diverse diabetic animal models are available,48 important discoveries around ncRNAs in diabetic retinopathy are expected soon.
One thing to note is that the same ncRNA can be commonly involved in several types of retinal diseases. For example, as described above, miR-150-5p is associated with the progression of not only diabetic retinopathy, but also retinopathy of prematurity and age-related macular degeneration.23,31,42 Since these retinal diseases have a common feature of vascular abnormalities, if a specific ncRNA was discovered in a type of retinal disease, the same ncRNA would likely be involved in other types of retinal diseases. Thus, it would be necessary to test the ncRNA discovered in a type of retinal disease to other retinal diseases.
Glaucoma
Due to the complex pathophysiology of glaucoma, there are no well-established animal models that can be easily used for the evaluation of this disease.49 In general, glaucoma models are produced by inducing high degrees of intraocular pressure, with various methods being used in different studies.50 These include episcleral vein cauterization, which is used to establish chronic high intraocular pressure. Using this technique, researchers were able to describe the role of lncRNA Malat1 in glaucoma.51 It was shown that the overexpression of Malat1 could inhibit the apoptosis of retinal ganglion cells in a high intraocular pressure model of glaucoma.
Microbead injection, another method used to generate glaucoma models, results in the blockage of the aqueous humor trabecular meshwork, preventing its drainage.52 In a recent microbead-based model, circZRANB1 (the circRNA produced from ZRANB1 locus) was shown to experience increased transcription with increasing intraocular pressure, while knockdown of circZRANB1 increased the survival rate of retinal ganglion cells.53 Further investigation identified miR-217-5p as the target of circZRANB1 and, interestingly, this circRNA was also shown to increase in the aqueous humor of patients with glaucoma.
Given the difficulties in producing reliable experimental models for glaucoma, it is not surprising that the regulatory roles of ncRNAs in glaucoma are less understood. Given the diversity of the methods used to produce these models, the identification of common ncRNAs among these models could be a good starting point in identifying important ncRNAs needed for the pathogenesis of glaucoma.
Retinoblastoma
Retinoblastoma is a relatively rare cancer, but there are several studies describing its pathogenesis because there are readily available retinoblastoma cell lines, making this an attractive area of study. Several studies have described the expression profiling of retinoblastoma tissues, with these studies identifying a number of differentially expressed ncRNAs. In addition, other studies have described changes in the level of circulating miRNAs in serum and plasma samples from patients with retinoblastoma, suggesting that miRNAs might be applied as useful biomarkers in this cancer.54 For example, miR-361-3p was found to be downregulated in retinoblastoma tissues and sera from patients with retinoblastoma, and the overexpression of miR-361-3p suppressed the proliferation of the retinoblastoma cell line via its regulation of glioma-associated oncogenes 1 and 3 (GLI1 and GLI3), both hedgehog signaling effectors.55 miR-758-3p was also shown to be downregulated in retinoblastoma tissues, and its overexpression decreased the proliferation of retinoblastoma cells.56 This miRNA exerted its effect by targeting paired box protein 6 (PAX6), an important regulator of ocular development.57
In addition, lncRNA BANCR (Braf-activated non-protein coding RNA),58 AFAP1-AS1 (actin filament-associated protein 1 AS1),59 and PVT1 (plasmacytoma variant translocation 1)60 were all shown to be upregulated in retinoblastoma tissues. Increased expression levels of BANCR, AFAP1-AS1, or PVT1 were all identified as poor prognostic factors for this cancer, and patients with high levels of these transcripts were shown to exhibit poor survival rates. Cell line experiments showed that the suppression of these lncRNAs inhibited the proliferation of retinoblastoma and that miR-488-3p was identified as the target of PVT1,60 although the functional mechanisms for BANCR and AFAP1-AS1 were not identified.
circSHPRH (the circRNA produced from SHPRH; named circ_0001649 in the original study) was reported to be downregulated in retinoblastoma tissues, and patients with low circSHPRH levels were shown to present with poor survival rates.61 Modulation of circSHPRH expression affected cell growth in the retinoblastoma cell line and components of the AKT (protein kinase B)/mTOR (mammalian target of rapamycin) pathway were affected by changes in the expression levels of this circRNA, although the detailed mechanism of these interactions remains unknown.
In summary, while the differential expression levels of ncRNAs during the pathogenesis of diverse diseases of the retina have been widely studied, relatively little is known about their mechanism of action, especially in comparison to their protein-coding counterparts. Given that there are many more ncRNAs than coding RNAs,62 it is probable that there are many more ncRNAs that may play key roles in the progression of diseases of the retina and more studies are required.
Targeting ncRNAs as therapy
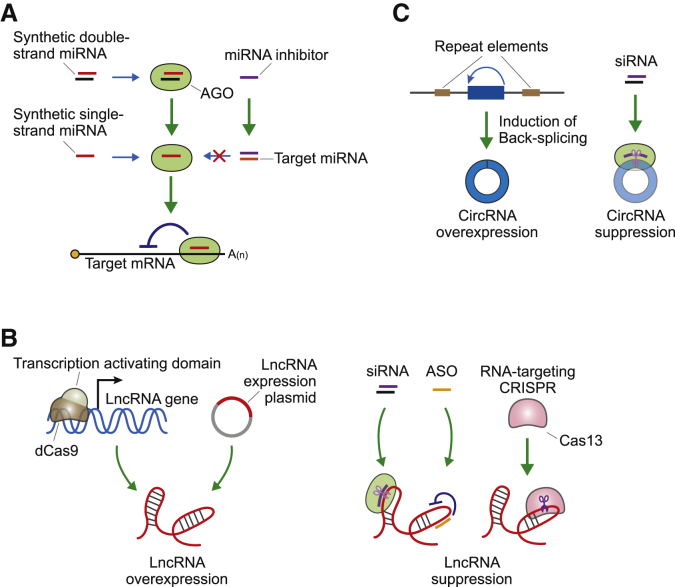
Diverse ncRNAs have been identified in the pathogenesis of retinal diseases, and given their critical functions in disease progression, the modulation of their expression could likely facilitate the development of novel therapeutics. Given this, it is not surprising that the modulation of ncRNAs is a growing field in the development of novel therapeutics for various diseases, including diseases of the retina. In general, RNA therapy uses RNA molecules to target RNA transcripts or proteins in the cells with varying degrees of success.63 Herein, we summarize the techniques that have been used to target ncRNAs as potential therapeutic approaches for the treatment of retinal disease (Figure 5).
Figure 5.
Diverse strategies designed to target ncRNAs
(A) Methods to modulate miRNAs. Both single- and double-stranded synthetic miRNAs are introduced into the cells and incorporated into ARGONAUTE (AGO) proteins, suppressing the expression of their target mRNAs. Conversely, miRNA inhibitors can be designed to recognize and bind to their complementary target miRNAs and suppress their function. (B) Methods to modulate the functions of lncRNAs. lncRNAs can be overexpressed by fusing a catalytically dead Cas9 to a transcription-activating domain, which when introduced into cells will activate the transcription of a specific lncRNA. Alternatively, the researcher could transfect cells with an expression plasmid encoding the full sequences or functionally important domains of the target lncRNAs. lncRNA expression can also be suppressed by the addition of short interfering RNAs (siRNAs) or antisense oligonucleotides (ASOs). The Cas13 system, which cleaves specific RNA transcripts, can also be used for the same purpose. (C) Methods to modulate circRNA expression. An expression plasmid containing circRNA sequences at the exonic region and repeat elements at the neighboring intronic regions can be used to overexpress circRNAs. Alternatively, siRNAs targeting the back-splicing junction can be used to suppress the expression of circRNAs without affecting their host genes.
The upregulation of miRNA activity can be simply achieved by introducing a synthetic form of the target miRNA. To increase the stability of these synthetic miRNAs, the backbone or sugars from the nucleotides are modified.64 Moreover, a specific moiety can be conjugated to these miRNAs allowing for tissue-specific targeting of specific organs. This is exemplified by the addition of N-acetyl galactosamine (GalNAc) to siRNA-based drugs.65 Downregulation of miRNA activity can be achieved by adding nucleotide-based inhibitors that bind stably and specifically to the miRNA. Modifications of the backbone or sugars of the miRNA inhibitors increase the thermodynamic stability of the interactions between these inhibitors and their targets, improving inhibition and preventing miRNA-mediated translational regulation.
lncRNA expression can be modulated using similar techniques to those used to change mRNA expression. siRNAs can be designed to degrade lncRNAs, and the same siRNA structures used as RNA-based drug-targeting mRNAs can be used in these settings.63 However, the key siRNA machinery, RNA-induced silencing complex (RISC), mainly works in the cytoplasm. This means that a large proportion of nuclear lncRNAs can escape siRNA-mediated silencing. To mitigate this, antisense nucleotides that induce ribonuclease (RNase) H-mediated cleavage can be used to target and degrade nuclear lncRNAs.63 Alternatively, the clustered regularly interspaced short palindromic repeats (CRISPR)-based RNA targeting method could be used to degrade and suppress target lncRNAs.66
The overexpression of lncRNAs is relatively difficult. Although plasmid or viral vectors can be used for experimental purposes, this delivery method is not reliable in therapeutic settings due to the large size of the lncRNAs. For the same reason, the production of synthetic lncRNAs is also not that feasible. However, only some parts of the lncRNA sequences, such as the protein-targeting domain or those sequences that form the secondary structure, are essential for specific functions.67, 68, 69 Given this, truncated versions of lncRNAs could be introduced into certain systems to generate the desired response. Another option is the application of CRISPR activators.70 In this case, the CRISPR activator complex binds to the promoter of the lncRNA gene and induces its transcription.
The silencing of circRNAs can be achieved using siRNA-based RNAi because most circRNAs exist in the cytoplasm.17,18 If one targets the back-splicing junction of the circRNA, siRNAs can target circRNA transcripts without affecting their linear host gene transcripts, preventing the disruption of the host gene function. Upregulation of circRNAs can be accomplished by creating expression plasmids that induce the circularization of the exon sequence and thus produce exogenous copies of the specific circRNA transcripts. These constructs rely on the presence of the repeat elements in the neighboring introns to induce the circularization of the exon located between those introns.71
Similar to the case of proteins, one ncRNA can regulate many different target genes. In particular, it is well known that one miRNA regulates hundreds of mRNAs.72 In these cases, it may be possible to consider a way to specifically block the regulatory relationship between a specific ncRNA and its target molecule. For example, for a sequence within an mRNA to which a specific miRNA binds, a decoy oligonucleotide may be designed to interfere with the binding of miRNA to mRNA. Also, to block the interaction between lncRNA (or circRNA) and its targeting miRNA, an oligonucleotide may be prepared for a sequence within a lncRNA (or circRNA) to which the miRNA binds. These methods may be applied to the treatment more specifically than the methods of directly modulating ncRNAs as described above.
In order to apply the various substances described above to the treatment of retinal diseases, proper drug delivery techniques are required. It is possible that the same ncRNA can function not only in the retina but also in other tissues or systems. However, in the treatment of retinal diseases, there is an advantage of being able to deliver the drugs locally through direct injection, and the possibility of affecting other organs is relatively small due to the confined compartment of the eye. Intravitreal injection, which is currently the most frequently used drug delivery method in the eye, is relatively less invasive than surgical treatment such as laser coagulation.73 Various substances described above can be directly injected, or they can be injected into the vitreous body by packaging them in liposomes or lipid nanoparticles. Several clinical trials have been conducted to treat retinal diseases based on siRNAs that can inhibit the expression of protein-coding genes.74 However, the intravitreal injection may also cause side effects with low frequency, including endophthalmitis, retinal detachment, and increased intraocular pressure. To overcome these side effects, various kinds of drug delivery technologies, such as topical installation in the form of eye drops, are currently being developed.73,75
In summary, there are a number of diverse methods that can be used to induce changes in the expression of each class of ncRNAs, and although some of these methods are already used in the production of commercially available drugs,63 many techniques are still only used in the laboratory setting. Given that many studies are currently underway, there is no doubt that there will be even more methods for the development of new ncRNA-targeting drugs developed in the foreseeable future.
Future perspectives
Diverse ncRNAs have been described and linked to various essential functions in the pathogenesis of retinal diseases. Although the initial studies only profiled the changes in their expression in the retina, recent studies have identified several ncRNAs as key regulators in retinal diseases and detailed their unique mechanisms of action. In addition, diverse studies are ongoing to develop novel methods for the efficient targeting and modulation of these ncRNAs. These studies should facilitate a better understanding of retinal diseases and may provide insights into their pathogenesis, allowing for improved therapeutic interventions, ultimately creating a better quality of life for elderly patients.
Acknowledgments
This work was supported by grants from the Basic Science Research Program of the National Research Foundation of Korea (NRF; NRF-2019R1F1A1054111 to J.S. and NRF-2021R1A2B5B02001501 and NRF-2019R1A4A1028534 to Y.-K.K.).
Author contributions
J.S. and Y.-K.K. analyzed the available literature and prepared the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Chen J., Smith L.E. Retinopathy of prematurity. Angiogenesis. 2007;10:133–140. doi: 10.1007/s10456-007-9066-0. [DOI] [PubMed] [Google Scholar]
- 2.Hellström A., Smith L.E., Dammann O. Retinopathy of prematurity. Lancet. 2013;382:1445–1457. doi: 10.1016/S0140-6736(13)60178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jager R.D., Mieler W.F., Miller J.W. Age-related macular degeneration. N. Engl. J. Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 5.Cheung N., Mitchell P., Wong T.Y. Diabetic retinopathy. Lancet. 2010;376:124–136. doi: 10.1016/S0140-6736(09)62124-3. [DOI] [PubMed] [Google Scholar]
- 6.Lechner J., O’Leary O.E., Stitt A.W. The pathology associated with diabetic retinopathy. Vision Res. 2017;139:7–14. doi: 10.1016/j.visres.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Quigley H.A. Glaucoma. Lancet. 2011;377:1367–1377. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- 8.Weinreb R.N., Aung T., Medeiros F.A. The pathophysiology and treatment of glaucoma: A review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimaras H., Corson T.W. Retinoblastoma, the visible CNS tumor: A review. J. Neurosci. Res. 2019;97:29–44. doi: 10.1002/jnr.24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Mendell J.T., Olson E.N. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santer L., Bär C., Thum T. Circular RNAs: A novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019;27:1350–1363. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 16.Ruan H., Xiang Y., Ko J., Li S., Jing Y., Zhu X., Ye Y., Zhang Z., Mills T., Feng J. Comprehensive characterization of circular RNAs in ∼ 1000 human cancer cell lines. Genome Med. 2019;11:55. doi: 10.1186/s13073-019-0663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr. Protoc. Bioinformatics. 2010;Chapter 12 doi: 10.1002/0471250953.bi1209s29. Unit 12.9.1–10. [DOI] [PubMed] [Google Scholar]
- 20.Scott A., Fruttiger M. Oxygen-induced retinopathy: a model for vascular pathology in the retina. Eye (Lond.) 2010;24:416–421. doi: 10.1038/eye.2009.306. [DOI] [PubMed] [Google Scholar]
- 21.Chen N., Wang J., Hu Y., Cui B., Li W., Xu G., Liu L., Liu S. MicroRNA-410 reduces the expression of vascular endothelial growth factor and inhibits oxygen-induced retinal neovascularization. PLoS ONE. 2014;9:e95665. doi: 10.1371/journal.pone.0095665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurihara T., Westenskow P.D., Friedlander M. Hypoxia-inducible factor (HIF)/vascular endothelial growth factor (VEGF) signaling in the retina. Adv. Exp. Med. Biol. 2014;801:275–281. doi: 10.1007/978-1-4614-3209-8_35. [DOI] [PubMed] [Google Scholar]
- 23.Liu C.H., Sun Y., Li J., Gong Y., Tian K.T., Evans L.P., Morss P.C., Fredrick T.W., Saba N.J., Chen J. Endothelial microRNA-150 is an intrinsic suppressor of pathologic ocular neovascularization. Proc. Natl. Acad. Sci. USA. 2015;112:12163–12168. doi: 10.1073/pnas.1508426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C.H., Wang Z., Huang S., Sun Y., Chen J. MicroRNA-145 regulates pathological retinal angiogenesis by suppression of TMOD3. Mol. Ther. Nucleic Acids. 2019;16:335–347. doi: 10.1016/j.omtn.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan J.T., Li X.X., Peng D.W., Zhang W.M., Qu J., Lu F., D’Amato R.J., Chi Z.L. MicroRNA-18a-5p administration suppresses retinal neovascularization by targeting FGF1 and HIF1A. Front. Pharmacol. 2020;11:276. doi: 10.3389/fphar.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjarlais M., Wirth M., Rivera J.C., Lahaie I., Dabouz R., Omri S., Ruknudin P., Borras C., Chemtob S. MicroRNA-96 promotes vascular repair in oxygen-induced retinopathy—A novel uncovered vasoprotective function. Front. Pharmacol. 2020;11:13. doi: 10.3389/fphar.2020.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Wang X., Wang Y.X., Ma Y., Di Y. Effect and mechanism of the long noncoding RNA MALAT1 on retinal neovascularization in retinopathy of prematurity. Life Sci. 2020;260:118299. doi: 10.1016/j.lfs.2020.118299. [DOI] [PubMed] [Google Scholar]
- 28.Liu C., Yao M.D., Li C.P., Shan K., Yang H., Wang J.J., Liu B., Li X.M., Yao J., Jiang Q., Yan B. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert V., Lecomte J., Hansen S., Blacher S., Gonzalez M.L., Struman I., Sounni N.E., Rozet E., de Tullio P., Foidart J.M. Laser-induced choroidal neovascularization model to study age-related macular degeneration in mice. Nat. Protoc. 2013;8:2197–2211. doi: 10.1038/nprot.2013.135. [DOI] [PubMed] [Google Scholar]
- 30.Wang L., Lee A.Y., Wigg J.P., Peshavariya H., Liu P., Zhang H. miR-126 regulation of angiogenesis in age-related macular degeneration in CNV mouse model. Int. J. Mol. Sci. 2016;17:895. doi: 10.3390/ijms17060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin J.B., Moolani H.V., Sene A., Sidhu R., Kell P., Lin J.B., Dong Z., Ban N., Ory D.S., Apte R.S. Macrophage microRNA-150 promotes pathological angiogenesis as seen in age-related macular degeneration. JCI Insight. 2018;3:e120157. doi: 10.1172/jci.insight.120157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLeod D.S., Bhutto I., Edwards M.M., Silver R.E., Seddon J.M., Lutty G.A. Distribution and quantification of choroidal macrophages in human eyes with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2016;57:5843–5855. doi: 10.1167/iovs.16-20049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Jiang C., Qin B., Liu G., Ji J., Sun X., Xu M., Ding S., Zhu M., Huang G. lncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017;8:e3046. doi: 10.1038/cddis.2017.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., Sun R., Yang D., Jiang C., Liu Q. LINC00167 regulates RPE differentiation by targeting the miR-203a-3p/SOCS3 axis. Mol. Ther. Nucleic Acids. 2020;19:1015–1026. doi: 10.1016/j.omtn.2019.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P., Lu B., Xu F., Wang C., Zhang R., Liu Y., Wei C., Mei L. Analysis of long noncoding RNAs in choroid neovascularization. Curr. Eye Res. 2020;45:1403–1414. doi: 10.1080/02713683.2020.1748659. [DOI] [PubMed] [Google Scholar]
- 36.Zhou R.M., Shi L.J., Shan K., Sun Y.N., Wang S.S., Zhang S.J., Li X.M., Jiang Q., Yan B., Zhao C. Circular RNA-ZBTB44 regulates the development of choroidal neovascularization. Theranostics. 2020;10:3293–3307. doi: 10.7150/thno.39488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson R., Barathi V.A., Chaurasia S.S., Wong T.Y., Kern T.S. Update on animal models of diabetic retinopathy: From molecular approaches to mice and higher mammals. Dis. Model. Mech. 2012;5:444–456. doi: 10.1242/dmm.009597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McArthur K., Feng B., Wu Y., Chen S., Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011;60:1314–1323. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q., Qiu F., Zhou K., Matlock H.G., Takahashi Y., Rajala R.V.S., Yang Y., Moran E., Ma J.X. Pathogenic role of microRNA-21 in diabetic retinopathy through downregulation of PPARα. Diabetes. 2017;66:1671–1682. doi: 10.2337/db16-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J.Y., Yao J., Li X.M., Song Y.C., Wang X.Q., Li Y.J., Yan B., Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biswas S., Thomas A.A., Chen S., Aref-Eshghi E., Feng B., Gonder J., Sadikovic B., Chakrabarti S. MALAT1: An epigenetic regulator of inflammation in diabetic retinopathy. Sci. Rep. 2018;8:6526. doi: 10.1038/s41598-018-24907-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan B., Yao J., Liu J.Y., Li X.M., Wang X.Q., Li Y.J., Tao Z.F., Song Y.C., Chen Q., Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015;116:1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 43.Li Q., Pang L., Yang W., Liu X., Su G., Dong Y. Long non-coding RNA of myocardial infarction associated transcript (lncRNA-MIAT) promotes diabetic retinopathy by upregulating transforming growth factor-β1 (TGF-β1) signaling. Med. Sci. Monit. 2018;24:9497–9503. doi: 10.12659/MSM.911787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu G.Z., Tian W., Fu H.T., Li C.P., Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016;471:135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 45.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 46.Zhu K., Hu X., Chen H., Li F., Yin N., Liu A.L., Shan K., Qin Y.W., Huang X., Chang Q. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. 2019;49:341–353. doi: 10.1016/j.ebiom.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Q., Liu C., Li C.P., Xu S.S., Yao M.D., Ge H.M., Sun Y.N., Li X.M., Zhang S.J., Shan K. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Invest. 2020;130:3833–3847. doi: 10.1172/JCI123353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olivares A.M., Althoff K., Chen G.F., Wu S., Morrisson M.A., DeAngelis M.M., Haider N. Animal models of diabetic retinopathy. Curr. Diab. Rep. 2017;17:93. doi: 10.1007/s11892-017-0913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouhenni R.A., Dunmire J., Sewell A., Edward D.P. Animal models of glaucoma. J. Biomed. Biotechnol. 2012;2012:692609. doi: 10.1155/2012/692609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswas S., Wan K.H. Review of rodent hypertensive glaucoma models. Acta Ophthalmol. 2019;97:e331–e340. doi: 10.1111/aos.13983. [DOI] [PubMed] [Google Scholar]
- 51.Li H.B., You Q.S., Xu L.X., Sun L.X., Abdul Majid A.S., Xia X.B., Ji D. Long non-coding RNA-MALAT1 mediates retinal ganglion cell apoptosis through the PI3K/Akt signaling pathway in rats with glaucoma. Cell. Physiol. Biochem. 2017;43:2117–2132. doi: 10.1159/000484231. [DOI] [PubMed] [Google Scholar]
- 52.Morgan J.E., Tribble J.R. Microbead models in glaucoma. Exp. Eye Res. 2015;141:9–14. doi: 10.1016/j.exer.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 53.Wang J.J., Shan K., Liu B.H., Liu C., Zhou R.M., Li X.M., Dong R., Zhang S.J., Zhang S.H., Wu J.H., Yan B. Targeting circular RNA-ZRANB1 for therapeutic intervention in retinal neurodegeneration. Cell Death Dis. 2018;9:540. doi: 10.1038/s41419-018-0597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Golabchi K., Soleimani-Jelodar R., Aghadoost N., Momeni F., Moridikia A., Nahand J.S., Masoudifar A., Razmjoo H., Mirzaei H. MicroRNAs in retinoblastoma: Potential diagnostic and therapeutic biomarkers. J. Cell. Physiol. 2018;233:3016–3023. doi: 10.1002/jcp.26070. [DOI] [PubMed] [Google Scholar]
- 55.Zhao D., Cui Z. MicroRNA-361-3p regulates retinoblastoma cell proliferation and stemness by targeting hedgehog signaling. Exp. Ther. Med. 2019;17:1154–1162. doi: 10.3892/etm.2018.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J., You X. MicroRNA-758 inhibits malignant progression of retinoblastoma by directly targeting PAX6. Oncol. Rep. 2018;40:1777–1786. doi: 10.3892/or.2018.6563. [DOI] [PubMed] [Google Scholar]
- 57.Shaham O., Menuchin Y., Farhy C., Ashery-Padan R. Pax6: A multi-level regulator of ocular development. Prog. Retin. Eye Res. 2012;31:351–376. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Su S., Gao J., Wang T., Wang J., Li H., Wang Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol. 2015;36:7205–7211. doi: 10.1007/s13277-015-3413-3. [DOI] [PubMed] [Google Scholar]
- 59.Hao F., Mou Y., Zhang L., Wang S., Yang Y. lncRNA AFAP1-AS1 is a prognostic biomarker and serves as oncogenic role in retinoblastoma. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180384. BSR20180384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X.Z., Cui H.P., Lv H.J., Feng L. Knockdown of lncRNA PVT1 inhibits retinoblastoma progression by sponging miR-488-3p. Biomed. Pharmacother. 2019;112:108627. doi: 10.1016/j.biopha.2019.108627. [DOI] [PubMed] [Google Scholar]
- 61.Xing L., Zhang L., Feng Y., Cui Z., Ding L. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed. Pharmacother. 2018;105:326–333. doi: 10.1016/j.biopha.2018.05.141. [DOI] [PubMed] [Google Scholar]
- 62.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim Y.K. RNA therapy: Current status and future potential. Chonnam Med. J. 2020;56:87–93. doi: 10.4068/cmj.2020.56.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baumann V., Winkler J. miRNA-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014;6:1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang X., Leroux J.C., Castagner B. Well-defined multivalent ligands for hepatocytes targeting via asialoglycoprotein receptor. Bioconjug. Chem. 2017;28:283–295. doi: 10.1021/acs.bioconjchem.6b00651. [DOI] [PubMed] [Google Scholar]
- 66.Kim V.N. RNA-targeting CRISPR comes of age. Nat. Biotechnol. 2018;36:44–45. doi: 10.1038/nbt.4054. [DOI] [PubMed] [Google Scholar]
- 67.Flintoft L. Non-coding RNA: Structure and function for lncRNAs. Nat. Rev. Genet. 2013;14:598. doi: 10.1038/nrg3561. [DOI] [PubMed] [Google Scholar]
- 68.Yamazaki T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H., Bond C.S., Nakagawa S., Pierron G., Hirose T. Functional domains of NEAT1 architectural lncRNA Induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70:1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 69.Zampetaki A., Albrecht A., Steinhofel K. Long non-coding RNA structure and function: Is there a link? Front. Physiol. 2018;9:1201. doi: 10.3389/fphys.2018.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 73.Gote V., Sikder S., Sicotte J., Pal D. Ocular drug delivery: Present innovations and future challenges. J. Pharmacol. Exp. Ther. 2019;370:602–624. doi: 10.1124/jpet.119.256933. [DOI] [PubMed] [Google Scholar]
- 74.Guzman-Aranguez A., Loma P., Pintor J. Small-interfering RNAs (siRNAs) as a promising tool for ocular therapy. Br. J. Pharmacol. 2013;170:730–747. doi: 10.1111/bph.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Joseph R.R., Venkatraman S.S. Drug delivery to the eye: what benefits do nanocarriers offer? Nanomedicine (Lond.) 2017;12:683–702. doi: 10.2217/nnm-2016-0379. [DOI] [PubMed] [Google Scholar]