Dear Editor,
In this Journal, Tang and colleagues recently commented on the introduction of the South African SARS-CoV-2 variant 501Y.V2 into the United Kingdom1. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are not always related to increased threat to human health because the virus acquires genomic diversity during the course of its life cycle2. However, some of these mutations have been shown to be associated with attenuation of the neutralizing activity of antibodies3. During the ongoing evolution of SARS-CoV-2, newly emerging lineages are likely to be circulating in the human population and genomic surveillance will be important for evaluating the emergence, spread, vaccine efficacy, and transmissibility of these lineages.
Currently, an emergent D614G mutation in the spike glycoprotein of SARS-CoV-2 is prevalent globally4. More recently, new emerging lineages with spike protein mutations have been discovered in the United Kingdom (B.1.1.7 lineage, 20I/501Y.V1, also named VOC 202,012/01)5, South Africa (B.1.351 lineage, 20H/501Y.V2)6, and Brazil (P.1 lineage, 20 J/501Y.V3)7 , 8. All of these lineages have a N501Y mutation in the receptor binding domain (RBD), which directly binds to the angiotensin converting enzyme 2 (ACE2) receptor of the host cell, contributing to increased transmissibility. Both B.1.351 and P.1 lineages also have additional K417N/T and E484K mutations. K417N/T, E484K, and N501Y confer reduced neutralizing activity of convalescent and mRNA vaccine-elicited serum3.
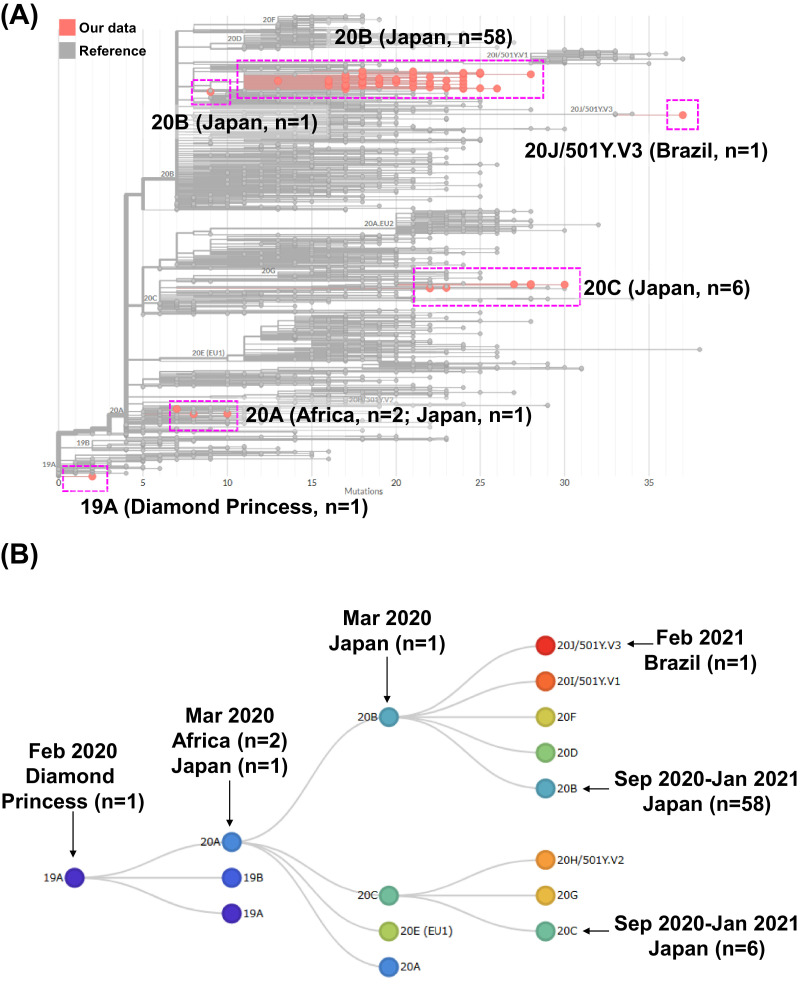
To determine the genomic characteristics of the SARS-CoV-2 variant identified in the Kofu, Japan, we started whole genome sequencing analysis using the Ion Torrent Genexus System (Thermo Fisher Scientific) on January 8, 2021. By February 15, 2021, a total of 136 samples from COVID-19-positive patients were obtained in our hospital from which 70 were subjected to analysis9 , 10. The sequence data were subjected to phylogenetic analysis using Nextclade, identifying five types of clades: 19A (n = 1), 20A (n = 3), 20B (n = 59), 20C (n = 6), and 20 J/501Y.V3 (n = 1) (Fig. 1 ). The SARS-CoV-2 strain from the Diamond Princess cruise ship patient was classified into clade 19A (Fig. 1), and four patients admitted at the end of March 2020 were classified into 20A (n = 3) and 20B (n = 1). For the patients admitted from September 2020–January 2021, 64 patients were classified into 20B clade (n = 58) and 20C (n = 6) (Fig. 1). The newly confirmed patient was classified as 20 J/501Y.V3 (P.1 lineage) on February 10, 2021 (Accession No. EPI_ISL_978917).
Fig. 1.
Phylogenetic tree analysis of SARS-CoV-2. (A) Sequencing data were uploaded to Nextclade (https://clades.nextstrain.org/) for phylogenetic analysis. The boxed regions show the sequencing data obtained in this analysis. (B) Evolution of SARS-CoV-2 clades over time. Schematic tree from Nextstrain showing clade evolution in Japan since February 2020. The arrows indicate the clades into which the 70 patients identified in our analysis were classified.
This patient was a 46-year-old man who entered our hospital in early February 2021 with a fever at 38.9 °C and with a history of staying in Brazil. RT-qPCR indicated a high viral load (7.1 log10/µL) and low cycle threshold (Ct) value of 15. The patient had displayed mild symptoms upon returning to Japan 4 days earlier and was admitted to another hospital. However, he was declared negative for SARS-CoV-2 during quarantine.
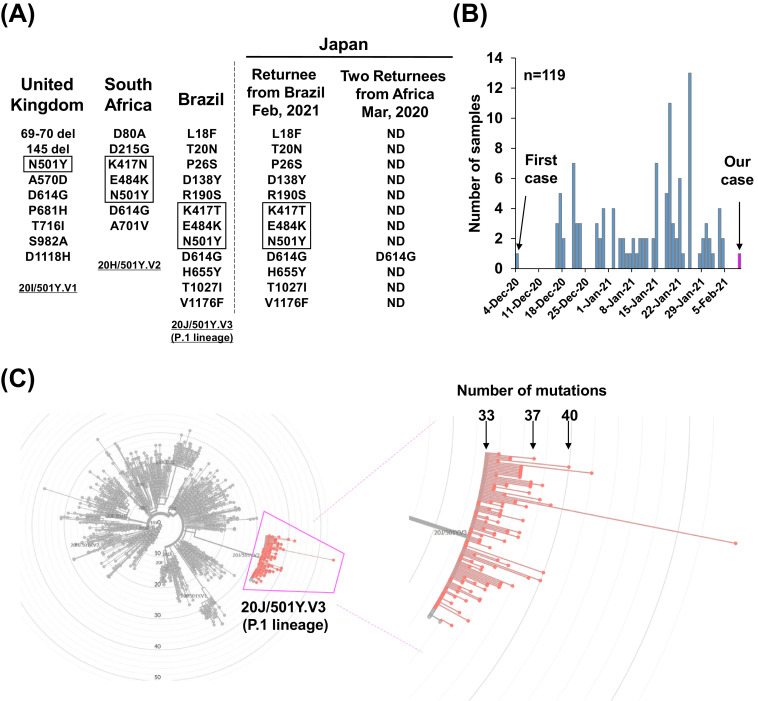
Sequencing analysis revealed that the SARS-CoV-2 variant 20 J/501Y.V3 (P.1 lineage) had 37 mutations, including 22 missense, 10 synonymous, three intergenic, one frameshift, and one in-frameshift mutation. In the spike protein, we observed 12 missense mutations (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I and V1176F). These mutations matched perfectly with the mutations in the P.1 variant previously discovered in Brazil7 (Fig. 2 A). In the RBD of the spike protein, three mutations (K417T, E484K and N501Y) were identified. These results indicated that we had identified a variant related to 20 J/501Y.V3 (P.1 lineage) in Japan.
Fig. 2.
SARS-CoV-2 spike protein mutation in emerging lineages and global distribution. (A) Mutations in the SARS-CoV-2 spike protein. (Left) Amino acid changes identified in the emerging strains reported in the United Kingdom, South Africa, and Brazil. (Right) Results of the current analysis: the patient who returned from Brazil on February 2021 had the same mutation as 20 J/501Y.V3 (P.1 lineage); the two patients who returned from Africa on March 2020 had only a D614G mutation in the spike protein. The mutations highlighted by boxes indicate those in the receptor binding domain. ND, not detected. (B) The number of 20 J/501Y.V3 (P.1 lineage) strains deposited in GISAID by February 14, 2021. The first case was identified on December 4, 2020 and our case on February 10, 2021. (C) A total of 119 sequencing data were analyzed on Nextclade. (Left) Radial phylogenetic tree showing the location of 20 J/501Y.V3 (P.1 lineage). (Right) Magnified view of boxed area showing the P.1 lineage. The total numbers of mutations denoted are with respect to the SARS-CoV-2 strain from Wuhan, China.
To examine the global prevalence of 20 J/501Y.V3 (P.1 lineage), we referred to sequence data deposited in GISAID (https://www.gisaid.org/). By February 14, 2021, 121 sequence data were available, 119 of which were derived from patients. Variant 20 J/501Y.V3 (P.1 lineage) was first discovered in a sample collected from Manaus, Amazonas, Brazil on December 4, 2020, and has subsequently been identified from numerous other samples (Fig 2B). Almost all 20 J/501Y.V3 (P.1 lineage) samples sequenced have 33–40 mutations compared with original strain reported from Wuhan, China; our identified strain has 37 mutations (Fig. 2C). Of the 119 patients infected with this variant, 82 (68.9%) were identified in Brazil, five (4.2%) in Japan, 20 (16.8%) in Europe, and three (2.5%) in the USA (Supplemental Table 1), suggesting global prevalence.
In summary, we have confirmed the emergence of five clades over time. Consecutive analysis identified SARS-CoV-2 variant 20 J/501Y.V3 (P.1 lineage) in a patient and detected mutations that are identical to those of the original P.1 variant discovered in Brazil7. This is the first report of 20 J/501Y.V3 (P.1 lineage) in Kofu, Japan.
Declaration of Competing Interest
None.
Acknowledgments
Funding
This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science (JSPS) KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), a Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.).
Acknowledgments
We thank the researchers who deposited the SARS-CoV-2 sequencing data in the GISAID. We also thank Masato Kondo, Ryota Tanaka, and Kazuo Sakai (Thermo Fisher Scientific) for technical help, all of the medical and ancillary hospital staff.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.03.013.
Appendix. Supplementary materials
Reference
- 1.Tang J.W., Toovey O.T.R., Harvey K.N., Hui D.D.S.: Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. [DOI] [PMC free article] [PubMed]
- 2.van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11(1):5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021 doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Centre for Disease Prevention and Control: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. 2020:1–13.
- 6.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020;2020 2012.2021.20248640. [Google Scholar]
- 7.Faria NR., Claro I.M., Candido D., Moyses Franco LA., Andrade PS., Coletti TM., Silva CA.M., Sales FC., Manuli ER., Aguiar RS., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2020;(20–01–2021) https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 [Google Scholar]
- 8.Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, Costa Á, Duarte D, Pessoa K, Gonçalves L, et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. https://virologicalorg/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585 2021.
- 9.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci. Rep. 2020;10(1):18899. doi: 10.1038/s41598-020-76043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Prospective study of 1,308 nasopharyngeal swabs from 1,033 patients using the LUMIPULSE SARS-CoV-2 antigen test: comparison with RT-qPCR. Int. J. Infect. Dis. 2021 doi: 10.1016/j.ijid.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.