Graphical abstract
Abbreviations: PCBs, polychlorinated biphenyls; NAFLD, nonalcoholic fatty liver disease; TASH, toxicant-associated steatohepatitis; AhR, arylhydrocarbon receptor; CAR, constitutive androstane receptor; ACHS, anniston community healthy survey; HFD, high fat diet; PXR, pregnane-xenobiotic receptor; WT, wildtype; OTU, operational taxonomic unit; LEfSe, linear discriminant analysis effect size; LDA, linear discriminant analysis; Tjp1, tight junction protein 1; Ocln, occludin; Cdh5, adhesion molecule VE-cadherin; Cldn, claudin; Muc, mucin; Reg3g, regenerating islet-derived protein 3-gamma; Camp, cathelicidin anti-microbial peptide; Tff3, trefoil factor 3; Fgf15, fibroblast growth factor 15; Tnfa, tumor necrosis factor; Fasn, fatty acid synthase; Pck1, phosphoenolpyruvate carboxykinase 1; Ppara, peroxisome-proliferator activated receptor alpha; HOMA, homeostasis model assessment; RER, respiratory exchange rate; NASH, nonalcoholic steatohepatitis; IBD, inflammatory bowel diseases
Keywords: PCBs, Aroclor1260, Gut-liver, NAFLD, TASH, Microbiome
Highlights
-
•
Aroclor1260 worsened diet-induced steatohepatitis in CAR and PXR knockout mice.
-
•
CAR and PXR ablation increased gut microbiome diversity and modified composition.
-
•
Improved bacterial diversity did not protect mice from PCB-mediated toxicity.
-
•
Toxicant-nuclear receptor interactions modulate the gut-liver axis.
Abstract
Polychlorinated biphenyls (PCBs) are persistent organic pollutants associated with non-alcoholic fatty liver disease (NAFLD). Previously, we demonstrated that the PCB mixture, Aroclor1260, exacerbated NAFLD, reflective of toxicant-associated steatohepatitis, in diet-induced obese mice, in part through pregnane-xenobiotic receptor (PXR) and constitutive androstane receptor (CAR) activation. Recent studies have also reported PCB-induced changes in the gut microbiome that consequently impact NAFLD. Therefore, the objective of this study is to examine PCB effects on the gut-liver axis and characterize the role of CAR and PXR in microbiome alterations. C57Bl/6 (wildtype, WT), CAR and PXR knockout mice were fed a high fat diet and exposed to Aroclor1260 (20 mg/kg, oral gavage, 12 weeks). Metagenomics analysis of cecal samples revealed that CAR and/or PXR ablation increased bacterial alpha diversity regardless of exposure status. CAR and PXR ablation also increased bacterial composition (beta diversity) versus WT; Aroclor1260 altered beta diversity only in WT and CAR knockouts. Distinct changes in bacterial abundance at different taxonomic levels were observed between WT and knockout groups; however Aroclor1260 had modest effects on bacterial abundance within each genotype. Notably, both knockout groups displayed increased Actinobacteria and Verrucomicrobia abundance. In spite of improved bacterial diversity, the knockout groups however failed to show protection from PCB-induced hepato- and intestinal- toxicity including decreased mRNA levels of ileal permeability markers (occludin, claudin3). In summary, CAR and PXR ablation significantly altered gut microbiome in diet-induced obesity while Aroclor1260 compromised intestinal integrity in knockout mice, implicating interactions between PCBs and CAR, PXR on the gut-liver axis.
1. Introduction
Polychlorinated biphenyls (PCBs) are a class of persistent organic pollutants associated with numerous diseases in exposed populations, including non-alcoholic fatty liver disease (NAFLD) and toxicant-associated steatohepatitis (TASH) [1,2]. Prior to being banned, these man-made chemicals had a variety of applications due to their thermo-dynamic stability and were widely used as dielectric fluids [3]. Although PCB production and use has been prohibited in the United States by Congress (1970s) and globally at the Stockholm Convention (2001), PCBs continue to persist in our environment and ecosystem due to their resistance to bio-degradation. In humans, the general route of exposure is through ingestion of PCB-laden food; being lipophilic in nature, PCBs bioaccumulate in living organisms primarily in the adipose tissue, and levels gradually increase along trophic levels of the food chain [4,5]. Based on the chlorine atom substitutions in the biphenyl ring, PCBs are broadly classified as either “coplanar” or “non-coplanar” congeners [6]. Coplanar congeners are deemed “dioxin-like” in nature due to their ability to activate the arylhydrocarbon receptor (AhR), similar to dioxin and constitute mostly lower molecular weight congeners; while non-coplanar congeners are deemed “phenobarbital-like” or “non-dioxin-like” due to their ability to activate the constitutive androstane receptor (CAR), similar to phenobarbital, and constitute mostly higher molecular weight congeners with higher number of chlorine substituents [6,7]. In North America, PCBs were primarily produced at the Monsanto plant located in Anniston, AL and sold under the trade name “Aroclor” [3]. Aroclor1260 was one of Monsanto’s first-generation commercial PCB mixtures, containing 60 % chlorine by weight and, due to its toxicity, was later replaced by second-generation or “Late” Aroclors with lower chlorine content [3]. Composition-wise, Aroclor1260 consisted of highly chlorinated, non-coplanar congeners with higher molecular weights that are resistant to metabolism, and therefore bioaccumulate, and are relevant to human exposure patterns [7].
Previously, our group reported the prevalence of liver disease in the Anniston Community Healthy Survey (ACHS) population which has a well-known history of exposures to PCBs [2]. Furthermore, studies from our group have also demonstrated that exposures to Aroclor1260 at doses resembling human PCB levels exacerbated NAFLD endpoints and TASH in male mice fed a high fat diet (HFD) [8]. NAFLD encompasses a broad spectrum of pathological disorders in the liver, and is initially characterized by lipid accumulation in the liver (steatosis), often accompanied by inflammation (steatohepatitis) and can further progress to fibrosis/scarring and cirrhosis. The term TASH reflects a form of NAFLD caused by exposure to toxic pollutants such as PCBs [[9], [10], [11]]. Indeed, Arolcor1260 exposure worsened hepatic inflammation, increased hepatic pro-fibrotic markers’ expression, induced phosphoprotein signaling disruption and perturbed overall energy homeostasis in these diet-induced obese mice [8,12]. Mechanistically, because Aroclor1260 constituted predominantly of non-coplanar PCBs, it was postulated that it elicited its actions though activation of hepatic nuclear receptors, namely CAR and the pregnane-xenobiotic receptor (PXR) [8]. Using CAR and PXR knockout mice, we demonstrated that some but not all the observed PCB effects were CAR and PXR-driven [13].
CAR and PXR are xenobiotic receptors whose main function is detoxification of drugs and chemicals in the liver through induction of cytochrome P450 enzymes. However, these receptors also play important roles in regulating energy metabolism and modulating steatosis, obesity, insulin resistance and inflammatory responses [14]. Importantly, CAR and PXR play a significant role in regulating the gut microbiome, specifically bile acid-metabolizing bacteria and modulating bile acid homeostasis [15]. Regulation of the microbiome in liver health and disease by various factors, including environmental chemicals, has become increasingly researched and disruption of the gut-liver axis is now recognized as a major determinant in liver disease development and progression [10,11,16]. Recent studies have also demonstrated PCBs’ capability in inducing gut microbiome alterations such as decreased alpha diversity and increased Firmicutes/Bacteroidetes ratio in mice [10,[17], [18], [19]]. Certainly, alterations in microbiome abundance and disruption of the gut-liver axis are emerging modes of action in PCB-mediated liver and metabolic diseases [10]. However, the role of CAR and PXR on the gut-liver axis in PCB-mediated TASH is still largely unknown. Therefore, the objective of the current study was to evaluate i) effects of the PCB mixture, Aroclor1260, on gut microbiome in HFD-fed mice; ii) role of CAR and PXR in regulating gut microbiome in presence of HFD; and iii) if microbiota alterations are related to the observed phenotypic characteristics.
2. Materials and methods
2.1. Animal model
The animal protocol was approved by the University of Louisville Institutional Animal Care and Use Committee. Wild type male C57Bl/6 mice (WT), Car-/- and Pxr-/- mice (8-weeks old, Taconic, Hudson, NY) were separated into 6 experimental groups (n = 10) based on Aroclor1260 exposure utilizing a 2 × 3 design. The knockout mice were developed by Taconic in collaboration with CXR Biosciences. Generating the Car-/- and Pxr-/- mice involved crossing a CAR and a PXR humanized mouse line, respectively, with a PhiC31 deleter mouse [13]. All mice were fed a high fat diet (HFD, 42 % kCal from fat; TD.88137 Harlan Teklad) during this one-time 12-week study, based on previous findings that Aroclor1260 exacerbated steatohepatitis in HFD-fed mice [8]. Mice were housed in a temperature- and light-controlled room (12 h light; 12 h dark) with food and water ad libitum. Aroclor1260 (AccuStandard, New Haven, CT) dissolved in corn oil was administered by oral gavage (vs. corn oil alone) at 20 mg/kg on week 1. This previously-reported dose was designed to mimic the highest human PCB levels seen in the PCB-exposed ACHS cohort [8]. After the 12-week period, mice were euthanized using ketamine/ xylazine (100/20 mg/kg body weight, i.p.); blood and tissue samples were harvested for further analysis. Thus, six different groups were evaluated in the study; WT, WT + Aroclor1260 (WTAr), Car-/- (CARko), Car-/-+Aroclor1260 (CARkoAr), Pxr-/- (PXRko), Pxr-/-+Aroclor1260 (PXRkoAr). It is important to note that the wildtype mice (WT) which were considered the experimental control group in the study were not littermate controls of the knockout groups; and this is a limitation of the study.
2.2. 16S Metagenomic sequencing library preparation and sequencing run
Microbial genomic DNA was extracted from frozen cecal samples using the PowerSoil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) according to the manufacturer’s instruction. Cecal microbiota composition was analyzed using Illumina MiSeq technology targeting the variable V3 and V4 regions of 16S ribosomal RNA. Libraries were prepared using Illumina’s 16S library preparation guide and Illumina’s Nextera Index Kit (FC-121-1012). Briefly, quantitation of microbial genomic DNA was performed using Qubit Broad Range (BR) assay (Thermo Fisher Scientific, Waltham, MA) in a Qubit 2.0 Fluorometer. Amplicon PCR was carried out using primers that were complementary upstream and downstream of the region of interest with overhang adapters. The 16S variable region was amplified using 12.5 ng of microbial genomic DNA. Amplicon PCR Clean‐Up was carried out using AMPure XP beads. Index PCR was then performed to attach dual indices and Illumina sequencing adapters using the Nextera XT Kit (FC-121-1012). After Clean-Up, sequencing libraries were aliquoted and mixed to make pooled libraries and concentration determined on the Bioanalyzer. After normalization, the pooled library was denatured and mixed with PhiX control library. Sequencing was then performed on a Nano-300 cycle test chip (MS-103-1001) to confirm sample concentration followed by Illumina MiSeq Reagents kit v3 (600 cycles) (MS-102-3003) at 9 pM and 30 % PhIX.
2.3. Sequencing data analysis
Quality control of the raw sequence data was performed using FastQC (version 0.10.1); sequence data were trimmed and further analyzed using QIIME 2 (version 2019.4) [20]. The sequences reads were then demultiplexed and denoised into amplicon sequence variants or clustered into operational taxonomic units (OTUs) by using Divisive Amplicon Denoising Algorithm (DADA2) [21]. A tree was generated for phylogenetic diversity analyses using the representative sequences and core diversity analyses of feature table including alpha and beta diversity were performed. Taxonomic analysis was based on the pre-trained classifiers with sequences that were assigned to OTUs at 99 % similarity to the Greengenes database (v13.8) from the V3V4 regions of sequences [22]. The Emperor tool was used to explore the principal covariant analysis plot in the context of sample metadata which is the distance between samples (beta diversity) using weighted and unweighted UniFrac. Linear discriminant analysis (LDA) effect size (LEfSe) method was used to find the most differentially abundant enriched microbial taxa between the groups. The analysis was performed on the Galaxy platform [23] using taxa results from QIIME 2 and the outputs generated were illustrated by histogram and cladogram with LDA score 2 and significance α < 0.05 as determined by Kruskal-Wallis test.
2.4. Real-time PCR of ileal samples
Mouse ileal samples were homogenized and total RNA was extracted using the RNA-STAT 60 protocol (Tel-Test, Austin, TX). The Nanodrop (ND-1000, Thermo Scientific, Wilmington, DE) was employed to assess RNA purity and quantity using the ND-1000 V3.8.1 software. cDNA was synthesized from total RNA using qScript cDNA Synthesis Kit (Quantabio, Beverly, MA). RT-PCR was performed on the CFX384 TM Real-Time System (BioRad, Hercules, CA) using iTaq Universal Probes Supermix and Taqman probes purchased from Thermo Fisher Scientific. Gene expression levels were calculated according to the 2−ΔΔCt method. Levels of mRNA were normalized relative to housekeeping genes and mean expression levels in unexposed, wild type mice were set at 1.
2.5. Statistical analyses
Statistical comparisons were performed using GraphPad Prism version 8 (GraphPad Software Inc., La Jolla, CA). Due to the exploratory nature of the study, initially, One-Way analysis of variance (ANOVA) was conducted between groups based on genotype or Aroclor1260, as previously described [13]. In addition, effects of exposure on different groups were analyzed using 2 sample t-test for comparisons. Results were declared statistically significant at significance level of 5%. Kruskal-Wallis test was used for pairwise comparisons between two groups for core metric analysis, and an FDR adjusted p-value (q-value) was used to infer significance (5%). For principal covariant analysis, the mean distance between two groups was compared using multivariate analysis of variance (PERMANOVA), and nonparametric p-values were calculated using 999 Monte Carlo permutations.
3. Results
3.1. Aroclor1260 and CAR, PXR ablation resulted in modified bacterial diversity
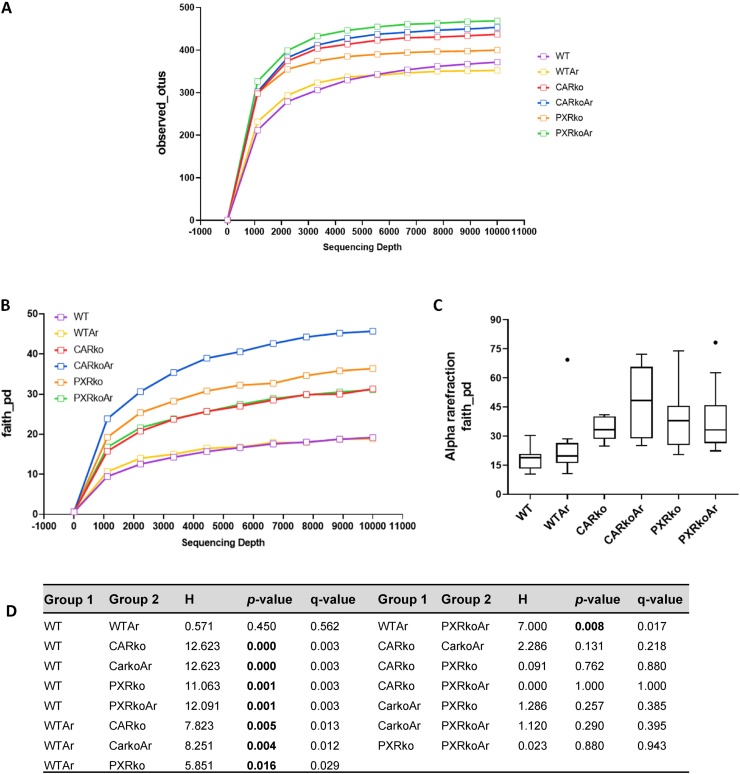
A total of 2,101,788 reads were obtained from a total of 60 cecal samples with a mean of 35,029.8 reads per sample. Alpha diversity, which reflects diversity of species within the given samples, was measured using two QIIME metrics, namely, observed_species and faith_pd tree measure. A snapshot of the alpha rarefaction curves for the number of OTUs showed that Aroclor1260-exposed, PXR knockout mice had the highest rarefaction curve, followed closely by exposed, CAR knockout mice, while exposed, WT mice had the lowest (Fig. 1A). Rarefaction curves generated using faith_pd showed that exposed, CAR knockout mice had the highest rarefaction curve while both WT groups showed the lowest rarefaction curves (Fig. 1B). Boxplots displaying distribution of the number of species were plotted (Fig. 1C). The distribution of diversity of species in the CAR and PXR knockout mice was markedly different from the WT groups. Additionally, q-values obtained from the Kruskal-Wallis test between WT versus knockout mice indicated that the diversity of species between these groups were significantly different, while the diversity of species between CAR and PXR groups with or without Aroclor1260 exposure were similar (Fig. 1D).
Fig. 1.
Effects of Aroclor1260 and CAR/PXR ablation on alpha diversity. Alpha diversity was measured using two QIIME metrics, namely observed_species and faith_pd tree measure. Rarefaction curves were generated for (A) observed_species and (B) faith_pd. (C) Boxplots displaying distribution of the number of species were plotted for faith_pd. (D) Kruskal-Wallis test was performed for pairwise comparison between different groups for faith_pd. q-value <0.05 denotes statistical significance in bacterial diversity between the two groups.
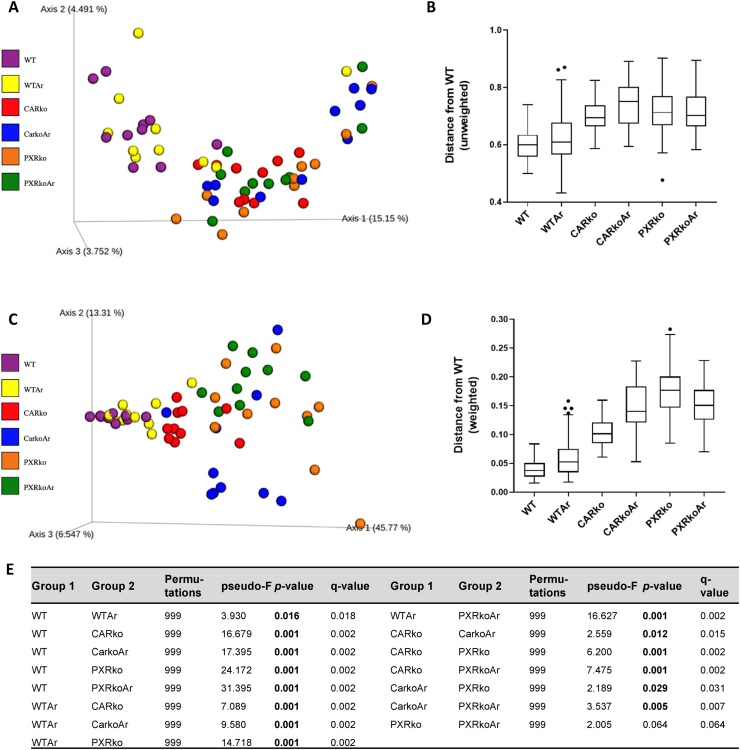
Beta diversity was computed to compare how each group of samples differed from the other. Both weighted and unweighted variants of UniFrac were used to measure distance between pairs of samples. The matrix for the weighted and unweighted UniFrac were visualized with principal covariant analysis resulting in plots containing axes for three principal coordinates (Fig. 2A and C). Boxplots showing the distribution of the unweighted and weighted UniFrac distance in the six groups were also plotted (Fig. 2B and D). Results from the PERMANOVA pairwise test for the weighted UniFrac distance matrix were obtained (Fig. 2E). Unweighted UniFrac analysis showed that the WT groups, irrespective of Aroclor1260 exposure, were clustered together, and different from the knockout groups (Fig. 2A & B), while the knockout groups, irrespective of exposure, more closely resembled each other. Weighted UniFrac analysis also showed a distinction between the WT and knockout groups (Fig. 2C & D). Additionally, the analysis also showed that Aroclor1260 exposure led to different diversities in the WT and CAR knockout groups but not in the PXR knockout group.
Fig. 2.
Effects of Aroclor1260 and CAR/PXR ablation on beta diversity. Beta diversity was computed by measuring distance between pairs of samples using both weighted and unweighted variants of UniFrac. Using the Emperor tool, the matrix for the (A) unweighted UniFrac was visualized with principal covariant analysis and (B) boxplots showing the distribution of the unweighted UniFrac distance was plotted. Likewise, (C) the matrix for weighted UniFrac was visualized with principal covariant analysis and (D) boxplots showing the distribution of weighted UniFrac distance in the six groups were plotted. (E) Table depicting results obtained from the PERMANOVA pairwise test for the weighted UniFrac distance matrix. q-value <0.05 denotes statistical significance in bacterial diversity between the two groups.
3.2. Effects of Aroclor1260, CAR and PXR on bacterial abundance at higher taxonomic levels
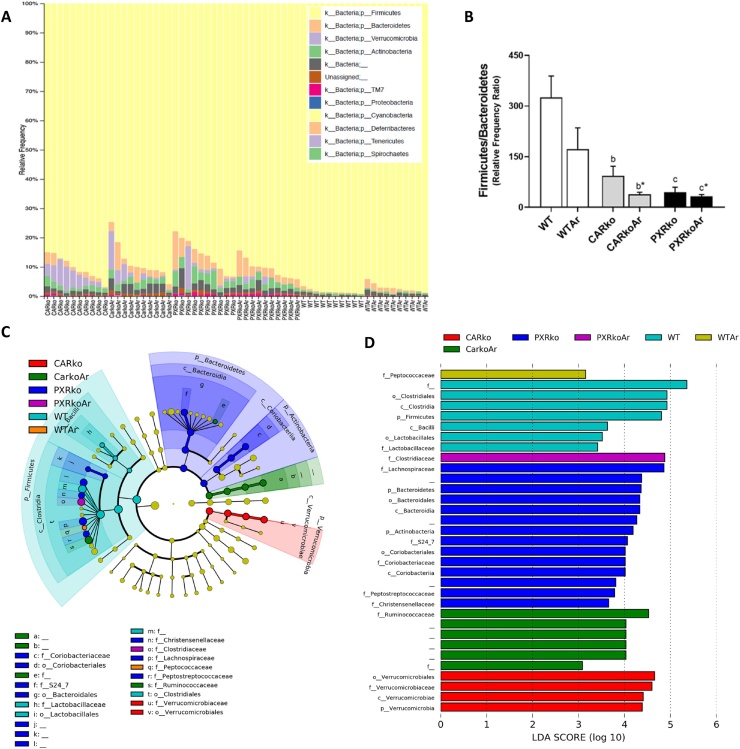
At the phylum level, stark differences were observed between the WT and knockout groups; Aroclor1260 exposure, however, had modest effects on this taxon as seen by the relative abundance of bacteria for each sample (Fig. 3A). Relative abundance of Firmicutes was highest in WT mice, while Bacteroidetes was highest in PXR knockout mice (Fig. 3A & Supplemental Fig. 1). Compared to WT mice, both CAR and PXR knockout mice had decreased Firmicutes/Bacteroidetes ratio, irrespective of Aroclor1260 exposure (Fig. 3B). Additionally, abundance of Actinobacteria and TM7 were robustly increased in PXR knockout mice, while Aroclor1260-exposed, CAR knockouts had increased Actinobacteria levels compared to their unexposed, WT counterparts (Fig. 3A & Supplemental Fig. 1). In contrast, Verrucomicrobia abundance was highest in CAR knockouts, followed by PXR knockouts, while WT mice had no detectable levels (Fig. 3A & Supplemental Fig. 1). In terms of exposure, Aroclor1260 increased Bacteroidetes and decreased Firmicutes in WT mice, while increasing Firmicutes in PXR knockouts (Fig. 3A & Supplemental Fig. 1). In order to obtain further information on the effects of the different groups on each taxa, LEfSe was performed and a plot cladogram and LDA scores were generated (Fig. 3C & D). At the class level, WT mice exhibited increased abundance for Clostridia and Bacilli, PXR knockout mice increased Bacteroidia and Coriobacteriia and CAR knockout mice increased Verrucomicrobiae, irrespective of exposure. Aroclor1260 effects were observed at the family level, where exposure increased abundance for Peptococcaceae in WT mice, Ruminococcaceae in CAR knockout mice and Clostridiaceae in PXR knockout mice. Other prominent observations at the family level include increased Lactobacillaceae in WT mice; increased Lachnospiraceae and Coriobacteriaceae in PXR knockouts; and increased Verrucomicrobiaceae in CAR knockouts.
Fig. 3.
Effects of Aroclor1260, CAR and PXR on bacterial abundance at higher taxonomic levels. (A) The relative abundance or relative frequency of bacterial samples at the phyla level for each sample was plotted. (B) The ratio of the relative abundance of Firmicutes to Bacteroidetes was calculated. A linear discriminant analysis effect size (LEfSe) analysis was performed at the phyla, class, order and family levels and a (C) plot cladogram and (D) LDA scores were generated.
3.3. Effects of Aroclor1260, CAR and PXR on bacterial abundance at the genus level
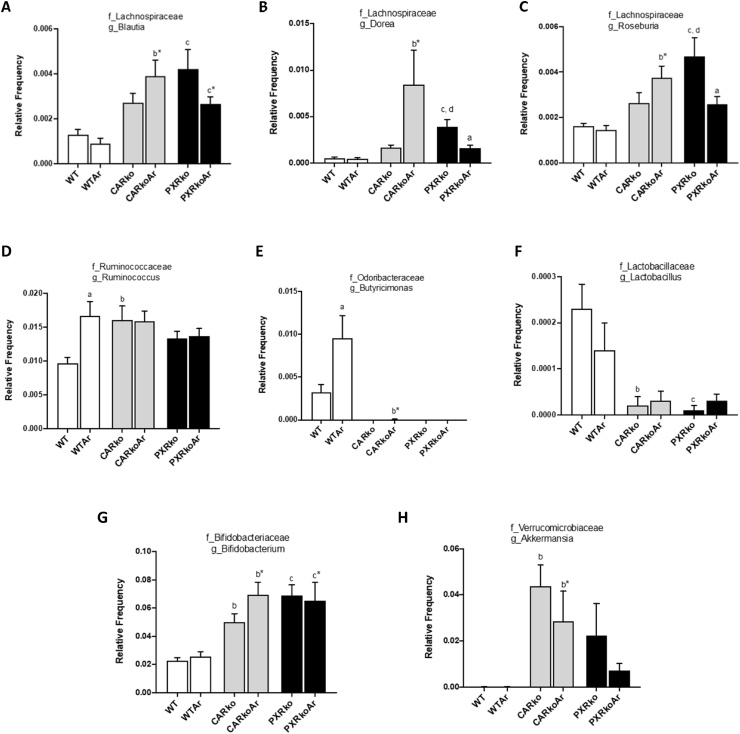
At the genus level, 51 bacteria were identified and 16 were significantly altered between different groups. Distinct differences in bacterial abundance were observed between the WT and knockout groups; Aroclor1260 exposure also modified abundance of certain bacteria (Fig. 4 & Supplemental Fig. 2). From the Lachnospiraceae family, Blautia, Dorea and Roseburia were all increased in exposed, CAR knockout mice (Fig. 4A–C). While PXR ablation also increased abundance of these bacteria versus WT, Aroclor1260 however decreased both Dorea and Roseburia in PXR knockouts. In contrast, Aroclor1260 increased abundance of Ruminococcus and Butyricimonas in WT mice, while the knockout groups have no detectable Butyricimonas (Fig. 4D & E). Both knockout groups also had decreased Lactobacillus while Bifidobacterium was increased (Fig. 4F & G). Additionally, irrespective of exposure, CAR knockouts had increased Akkermansia (Fig. 4H). Other notable observations include higher levels of Adlercruetzia, Collinsella, Odoribacter and Anaerotruncus in PXR knockouts; increased Ruminococcus (Lachnospiraceae family) in both CAR and PXR knockouts; and increased Allobaculum and Gemmiger only in exposed, CAR knockouts (Supplemental Fig. 2). Furthermore, exposed, CAR knockout mice also had increased Faecalibacterium abundance while it was decreased in exposed, PXR knockouts (Supplemental Fig. 2). To summarize, the changes in bacterial abundance for Ruminococcus (Ruminococcaceae family) and Butyricimonas were dependent on Arcolor1260 exposure; Dorea, Roseburia and Allobaculum were dependent on Arcolor1260 with CAR/PXR ablation; Gemmiger and Akkermansia were dependent only with CAR ablation; Odoribacter and Anaerotruncus were dependent only with PXR ablation; while the rest were dependent with both CAR and PXR ablation.
Fig. 4.
Effects of Aroclor1260, CAR and PXR on bacterial abundance at the genus level. The relative abundances of identified bacteria at the genus level, namely (A) Blautia, (B) Dorea, (C) Roseburia, (D) Ruminococcus, (E) Butyricimonas, (F) Lactobacillus, (G) Bifidobacterium and (H) Akkermansia were plotted for the different groups. Values are mean ± SEM, p < 0.05, a- Δ due to Aroclor1260 exposure within genotype, b, b*- Δ between WT and Car−/− without or with Aroclor1260 exposure, c, c*- Δ between WT and Pxr−/− without or with Aroclor1260 exposure, d, d*- Δ between Car and Pxr ablation without or with Aroclor1260 exposure.
3.4. Effects of Aroclor1260, CAR and PXR on bacterial abundance at the species level
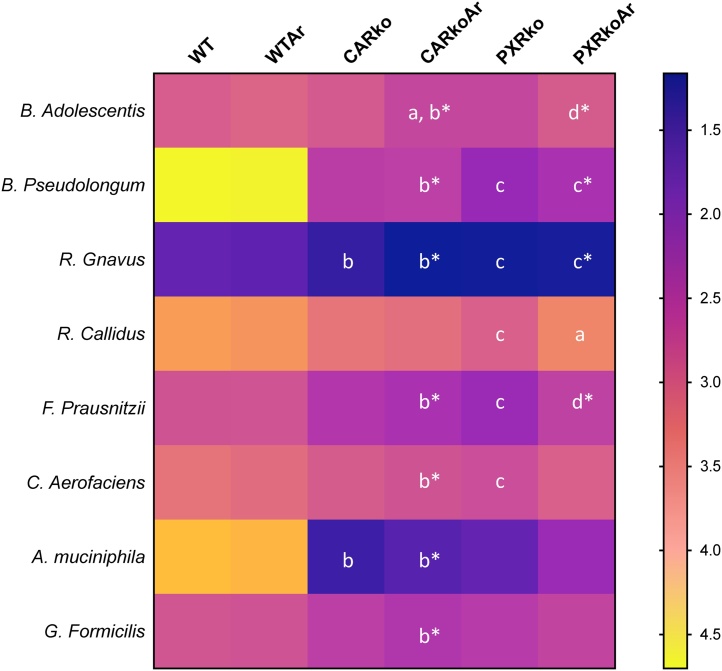
At the species level, 25 bacteria were identified and 8 were significantly different between groups. Mean abundance levels for the significantly different bacteria are illustrated in Fig. 5 and individual bar graphs for each bacteria are provided in Supplemental Fig. 3. Similar to bacterial abundance at the genus level, CAR or PXR ablation modified abundance at this level compared to WT mice, while Aroclor1260 had subtler effects (Fig. 5). CAR knockout mice exposed to Aroclor1260 had higher abundance of B. adolescentis compared to their unexposed counterparts; while B. Pseudolongum was higher in PXR knockout mice, irrespective of exposure, and also in exposed, CAR knockouts. Both knockout groups, with or without exposure, had higher abundance of R. Gnavus, whereas only PXR knockouts had increased R. Callidus abundance; however, Aroclor1260 exposure in PXR knockout mice decreased R. Callidus. Exposed, CAR knockout and unexposed PXR knockout mice both demonstrated higher abundance levels of F. Prausnitzii and C. Aerofaciens. Interestingly, both CAR knockout groups have increased abundance of A. Muciniphila, while only exposed, CAR knockout mice had increased G. Formicilis. To summarize, the changes in bacterial abundance for R. Callidus was dependent on Aroclor 1260 with PXR ablation; A. Muciniphila and G. Formicilis only with CAR ablation, while the rest were dependent with both CAR and PXR ablation.
Fig. 5.
Effects of Aroclor1260, CAR and PXR on bacterial abundance at the species level. A heatmap depicting the relative abundances of identified bacteria at the species level, namely B. Adolescentis, B. Pseudolongum, R. Gnavus, R. Callidus, F. Prausnitzii, C. Aerofaciens, A. Muciniphila and G. Formicilis for the different groups. Values are presented as -log (mean relative abundance), thereby lower values are reflective of higher relative abundance and vice versa. p < 0.05, a- Δ due to Aroclor1260 exposure within genotype, b, b*- Δ between WT and Car-/- without or with Aroclor1260 exposure, c, c*- Δ between WT and Pxr-/- without or with Aroclor1260 exposure, d, d*- Δ between Car and Pxr ablation without or with Aroclor1260 exposure.
3.5. Aroclor1260 altered ileal gene expression in WT and CAR knockout mice
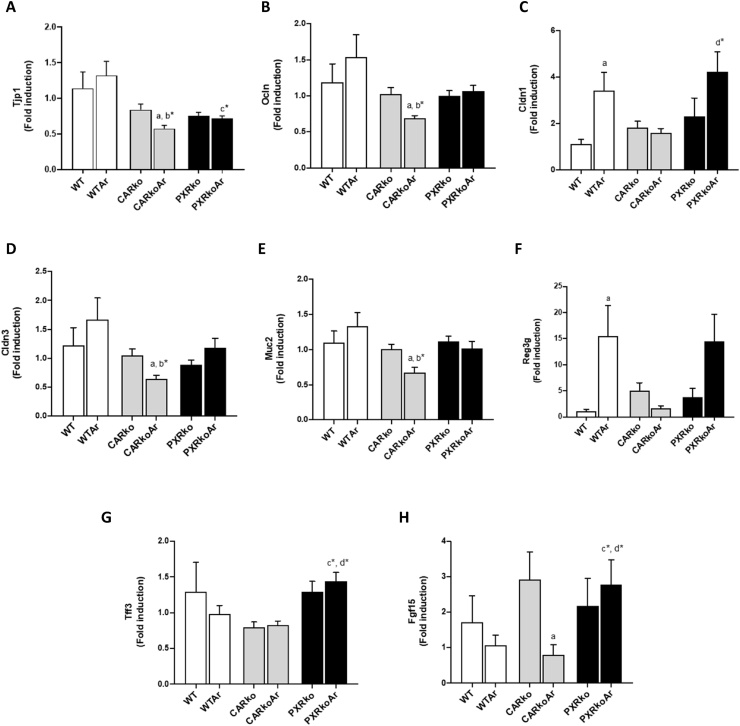
To gain a better understanding on how Aroclor1260 impacted gut function in WT and knockout mice, and how gut properties correlated with the altered microbiome, ileal markers of gut permeability and function were assessed. The ileum was chosen for the present study because it is considered to be the most immunologically active part of the gut and for its responsiveness to changes in barrier function [24]. Ileal mRNA levels of tight junction protein 1 (Tjp1), the gene encoding zonula occludens-1, a scaffolding protein linking tight junction transmembrane proteins including claudins and occludin to the actin cytoskeleton, was measured. Compared to exposed, WT mice, both knockout mice exposed to Aroclor1260 had lower ileal Tjp1 mRNA levels with the lowest seen in exposed, CAR knockouts (Fig. 6A). In fact, Aroclor1260 also decreased gene expression of the tight junction protein Occludin (Ocln) and adhesion molecule VE-cadherin (Cdh5) in CAR knockouts (Fig. 6B and Supplemental Fig. 4). With regards to claudins, there were no changes in claudin 2 (Cldn2) gene expression (Supplemental Fig. 4), while Aroclor1260 increased ileal expression of Cldn1 in WT mice and decreased Cldn3 in CAR knockouts (Fig. 6C & D). Exposed, CAR knockout mice also showed decreased gene expression for the mucus gel forming protein, mucin2 (Muc2) while there were no changes for Muc1 (Fig. 6E and Supplemental Fig. 4). Additionally, gene expression of intestinal antimicrobial and healthy mucosal markers including regenerating islet-derived protein 3-gamma (Reg3g), cathelicidin anti-microbial peptide (Camp) and trefoil factor 3 (Tff3) were assessed. Surprisingly, Aroclor1260 increased Reg3g mRNA levels in WT mice (Fig. 6F) while exposed, PXR knockouts had increased mRNA levels for Camp and Tff3 (Fig. 6G and Supplemental Fig. 4). Lastly, Aroclor1260 decreased gene expression levels of fibroblast growth factor 15 (Fgf15), implicated in maintenance of bile acid homeostasis and liver repair [25] in CAR knockout mice while exposed, PXR knockout mice had increased Fgf15 mRNA levels (Fig. 6H).
Fig. 6.
Aroclor1260 and CAR/PXR ablation altered ileal gene expression. Ileal mRNA levels for genes encoding proteins involved in maintenance of barrier integrity and intestinal inflammation/function including (A) Tjp1, (B) Ocln, (C) Cldn1, (D) Cldn3, (E) Muc2, (F) Reg3g, (G) Tff3 and (H) Fgf15 were measured using RT-PCR. Values are mean ± SEM, p < 0.05, a- Δ due to Aroclor1260 exposure within genotype, b, b*- Δ between WT and Car-/- without or with Aroclor1260 exposure, c, c*- Δ between WT and Pxr-/- without or with Aroclor1260 exposure, d, d*- Δ between Car and Pxr ablation without or with Aroclor1260 exposure.
3.6. Correlation between changes in bacterial abundance and inflammation/energy metabolism
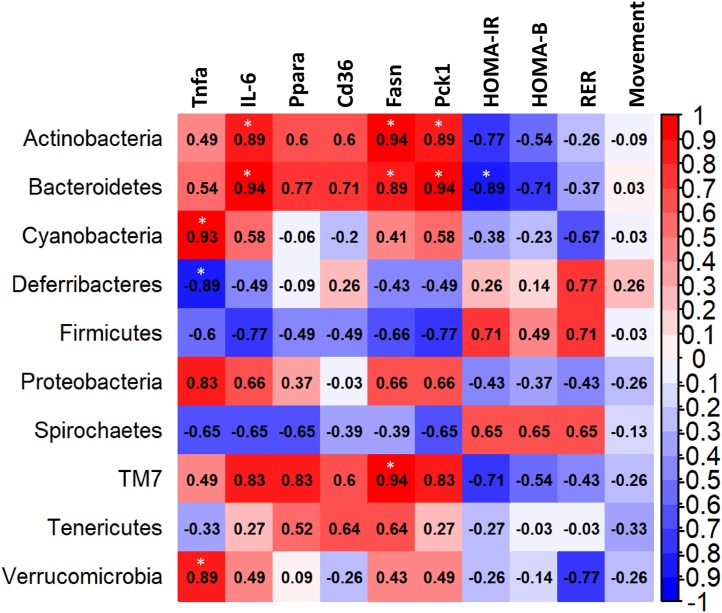
In our previous study, toxicity endpoints pertinent to NAFLD and TASH were evaluated including hepatic inflammation and disrupted energy homeostasis such as altered hepatic gene expression, glucose metabolism and overall energy expenditure [13]. To better understand how microbiome remodeling correlated with these metabolic phenotypic observations, a Spearman Correlation analysis was performed on bacterial abundance (phyla level) and selected phenotypic endpoints (Fig. 7). With regards to hepatic inflammation, increased abundance of Verrucomicrobia and Cyanobacteria were significantly correlated with increased hepatic tumor necrosis factor (Tnfa) expression while Actinobacteria and Bacteroidetes were positively correlated with hepatic interleukin IL-6 expression. Deferribacteres was negatively correlated with hepatic Tnfa expression. In terms of gene expression related to lipid and glucose metabolism genes, Bacteroidetes and Actinobacteria showed strong positive correlations with gene expression of Fasn (fatty acid synthase, fatty acid synthesis) and Pck1 (phosphoenolpyruvate carboxykinase, gluconeogenesis) while TM7 was positively correlated with hepatic Fasn and showed a trend (p = 0.058) for positive correlations with hepatic expression of IL-6, Pck1 and Ppara (peroxisome-proliferator activated receptor alpha, fatty acid oxidation). In contrast, Bacteroidetes showed a negative correlation with homeostasis model assessment for insulin resistance (HOMA-IR). There were no significant correlations for gene expression of Cd36 (lipid uptake) nor for pancreatic beta cell function (HOMA-B), respiratory exchange rate (RER) and movement.
Fig. 7.
Correlation between inflammatory/metabolic endpoints and bacterial abundance at the phyla level. Spearman Correlation analysis was performed on bacterial abundance at the phyla level and selected phenotypic endpoints for inflammation (hepatic Tnfa and IL-6 mRNA levels), lipid and glucose metabolism (hepatic Ppara, CD36, Fasn and Pck1 mRNA levels), insulin resistance (HOMA-IR and HOMA-B) and energy expenditure (RER and movement). * p < 0.05.
4. Discussion
PCBs’ mechanisms of toxicity in liver disease have been investigated extensively [10]. While major contributors to these mechanistic pathways included activation of hepatic receptors such as the AhR, CAR and PXR; there are also more novel mechanisms recently reported including epigenetic modifications, phospho-signaling disruption and microbiome alterations [10,11]. Our previous study illustrated that the commercial PCB mixture, Aroclor1260, exacerbated NAFLD endpoints, particularly in the presence of diet-induced obesity, in part through CAR and PXR activation, since the mixture was composed mainly of non-coplanar congeners [8]. While it has been proposed that PCBs act as a “second hit” that worsen the transition of diet-induced steatosis to steatohepatitis; more recently, extensive investigations on PCB-induced alterations on the hepatic proteome suggested that PCBs can also act as a “first hit” by compromising normal liver function and rendering it susceptible to hepatic insults caused by HFD feeding [11,12]. Nonetheless, the net effects included a myriad of inflammatory and metabolic complications pertaining to NAFLD/TASH and driven by multiple mechanistic pathways including CAR and PXR activation. To understand the role of CAR and PXR in PCB-associated NAFLD and TASH observed in diet-induced obese mice, a thorough study was carried out to identify key changes caused by Aroclor1260 exposure in CAR and PXR knockout mice [13]. Interestingly, key findings from the study revealed that while CAR and PXR activation play pivotal roles in driving PCB-mediated perturbations in inflammation, energy metabolism, endocrine disruption and behavior, primarily though receptor cross-talk; absence of these receptors did not necessarily protect mice from NAFLD/TASH caused by co-exposure to PCBs and HFD feeding [13]. In fact, CAR and PXR ablation resulted in higher basal inflammation and deranged lipid metabolism, while PCB-exposed CAR and PXR knockout mice exhibited dissociations between obesity, steatosis, insulin resistance and inflammation, thereby implicating complexities of PCB-nuclear receptor interactions and existence of other “off-targets” that potentially impact overall effects. Based on reported findings [[17], [18], [19]], we postulated that one of these “off-targets” effects that could provide insight into pathways pertinent to PCB-nuclear receptor interactions in NAFLD/TASH are effects on gut microbiota and the accompanying gut-liver axis.
CAR and PXR play significant roles in endobiotic metabolic processes such as lipid and bile acid metabolism, albeit being fundamentally known as drug and xenobiotic receptors and are considered potential therapeutic targets for NAFLD and non-alcoholic steatohepatitis (NASH) [14,26]. Presumably, CAR is an anti-obesity nuclear receptor due to its ability to improve insulin resistance and alleviate hepatic steatosis in rodents [27,28]. In contrast, PXR is thought to be an obesity-promoting receptor due to the obesogenic effects resulting from PXR activation in rodents, although there are conflicting reports [[29], [30], [31]]. Notably, studies such as ours have unveiled that the consequential effects brought upon by activation of these ligand-activated transcription factors are far more complex and involved receptor-receptor interactions, epigenetic modifications and alterations in transcriptional machinery. Newer studies concomitant to CAR and PXR function with respect to the general microbiome and their impact on the gut-liver axis have also illustrated the significance of these receptors in normal gut function including intestinal repair from inflammatory bowel diseases (IBD) [[32], [33], [34]]. The interconnectedness of PXR’s anti-inflammatory action and the gut microbiome is exemplified by additional studies, demonstrating PXR activation counteracting Toll-like receptor 4 and nuclear factor-kappa B (NF-kB) pathways to mitigate intestinal inflammation and bacterial translocation through the “leaky gut” [[35], [36], [37], [38]]. Conversely, the commensal microbiota has also been shown to influence CAR and PXR expression and activity and consequently impact host xenobiotic and endobiotic metabolism [39]. Based on these studies, it can be postulated that perhaps the absence of beneficial CAR and PXR in the knockout mice, in part, resulted in higher basal inflammation in our previously reported study [13].
Indeed, the findings from the current study, using our previously reported diet-induced obesity model, reiterated the critical role of these nuclear receptors in regulating the microbiome and vice versa. In addition, this is the first study to explore the contribution of Aroclor1260 in altering gut microbiome in the presence of HFD. The results clearly indicated that CAR and/or PXR ablation, regardless of exposure status, increased bacterial alpha diversity which is a sign of microbiome wellness and health. In contrast, Aroclor1260 did not affect alpha diversity significantly. While CAR and PXR ablation drastically changed the bacterial composition (beta diversity) from WT mice, Aroclor1260 exposure also altered bacterial composition, consistent with reported findings that both dioxin-like and non-dioxin-like PCBs can impact microbial diversity [10]. However, this effect was observed only in WT and CAR knockout mice but not in PXR knockouts. A plausible explanation for this could be that HFD feeding and PXR ablation may have already altered the microbiome significantly that the Aroclor1260 effects appeared somewhat diminished. In the current study, Firmicutes were the most predominant bacterial phyla observed in the mouse gut microbiota, concordant with other findings [18]. The Firmicutes/Bacteroidetes ratio, which is considered as an obesity indicator and positively correlated with body mass index in humans over time [40,41], was calculated. Knocking out CAR or PXR resulted in decreased Firmicutes/Bacteroidetes ratio, which is somewhat surprising given that there were no changes in body weight gain between groups [13]. However, an inverse relationship has also been reported for the Firmicutes/Bacteroidetes ratio and NASH, with NASH patients exhibiting decreased Firmicutes and higher Bacteroidetes abundance [16,42,43]. This is potentially applicable to our current study given that both knockout groups manifested basal hepatic inflammation and NASH endpoints. Notably, our findings showed that Aroclor1260 increased Bacteroidetes abundance in WT mice and these diet-induced obese mice have been shown to exhibit NASH features [8,13]. Actinobacteria abundance was higher in CAR and PXR knockouts which reflected their role in regulating this particular phylum, given that pharmacological activation of PXR and CAR led to decreased abundance of the bile acid metabolizing Actinobacteria class in conventional mice, resulting in decreased secondary bile acids in the liver [15]. Additionally, correlation findings between microbiome remodeling and mouse phenotype revealed that both Bacteroidetes and Actinobacteria were positively correlated with hepatic inflammation and dysregulated energy metabolism in the current study. This is somewhat intriguing and reinforces the contradictory reports on the role of Bacteroidetes in NAFLD and NASH [44], while it is also important to note that CAR and PXR ablation may also influence these correlations.
PXR and CAR ablation had a massive impact on bacterial abundance across different genera and species and also influenced Aroclor1260 effects on these taxa. Aroclor1260 increased Ruminococcus abundance in WT mice which is intriguing, given that elevated Ruminococcus abundance was reported in NASH patients and associated with increased liver fibrosis [45]. This positively corresponded with our recent proteomics findings that Aroclor1260 exposure increased pro-fibrotic proteins in the liver, thereby making it susceptible to fibrosis development [12]. Interestingly, CAR and PXR knockouts had higher levels of R. Gnavus, a known pro-inflammatory microbe often associated with inflammatory diseases including Crohn’s disease [46], and lower abundance of the short-chain fatty acid producers, Lactobacillus, which is beneficial in IBD and previously shown to increase with CAR activation [47], further corroborating the protective role of these nuclear receptors. The butyrate-producing Butyricimonas was increased by Aroclor1260 which was somewhat counterintuitive since butyrate aids in the maintenance of a healthy gut and reduces inflammation; however, it was almost undetected in the knockout groups. Additionally, CAR and PXR knockouts also had higher abundance of the bile acid metabolizing Bifidobacterium, implicating changes in bile acid metabolism in these groups. The Aroclor1260-exposed, CAR knockouts and unexposed, PXR knockouts also had higher levels of identified genera of the Lachnospiraceae family including Blautia and Dorea which have reportedly been identified as microbiota signatures of NAFLD and NASH progression [48,49]. Interestingly, assessment of ileal gene expression suggested that Aroclor1260 decreased tight junction markers and Fgf15 expression in CAR knockouts, in spite of this group having higher abundance levels of microbes beneficial in intestinal integrity and liver injury including B. adolescentis and A. muciniphila [50,51]. A plausible explanation for this could be that the impact on ileal gene expression preceded gut-microbiome alterations and perhaps prolonging the duration of exposure will result in microbiome effects that are fairly consistent with the observed intestinal findings.
Although the current study brings to light the importance of the gut microbiome when assessing Aroclor1260 toxicity and the role of CAR and PXR in influencing PCB-mediated toxicity on the gut-liver axis, it is not without limitations. While PCB-driven effects on bile acid metabolism through CAR and PXR activation have been implicated [17], changes on hepatic and circulating levels of bile acids were not accounted for in the present study. Moreover, the study utilized only male mice and sexually dimorphic effects on the liver have been reported with PCB exposures [52]. Importantly, the study did not use littermate controls, or mice that were heterozygous for the null alleles but from the same breeding, and this may have attributed to the stark changes in microbiome composition between WT and knockout groups. In addition, the animal experiment was performed only once and similar studies are needed to replicate such findings. Finally, a more thorough evaluation on PCB-mediated intestinal effects including ileal pathology and assessment of intestinal CAR and PXR gene battery are needed. Therefore, future studies will include investigating alterations in bile acid homeostasis in relation to changes in gut microbiome, evaluating the role of sexual dimorphism in influencing the observed gut-liver axis perturbations, utilizing humanized CAR and PXR mouse models and multi-pollutant exposures to further understand the importance of these receptors in NAFLD/NASH caused by environmental chemical exposures in humans. Such studies will help address some of the real-life effects observed with toxicant exposures [53]. In addition, future studies are also aimed to identify and investigate bacterial metabolites that may target CAR and PXR and modulate their functions.
Altogether, the data from the current study suggested that CAR and PXR ablation played a critical role in modifying microbial composition in the presence of HFD, while concurrently dictating Aroclor1260 effects on both gut microbial composition and intestinal integrity. Even though knocking out these nuclear receptors promoted microbial diversity and richness; this however did not protect the animals from developing NASH caused by co-exposures to PCBs and HFD. Additionally, abundance of microbes reportedly associated with decreased intestinal inflammation and injury and known to be beneficial in IBD were altered in CAR and PXR knockout mice, providing further evidence that these receptors play a therapeutic role in inflammatory diseases. Furthermore, it can be argued that because Aroclor1260 activated CAR and PXR, the microbiota profiles observed in the current study with Aroclor1260 exposure would be similar to those triggered by pharmacological CAR and PXR agonists. The results also provided a strong argument that these nuclear receptors, particularly CAR, may potentially play a protective role in Aroclor1260-induced toxicity pertaining to the gut-liver axis and that PCB-mediated CAR activation by itself may not necessarily be a mechanism of PCB-induced hepatotoxicity as previously proposed. Moreover, this study was performed in diet-induced obese mice and it is well-known that high fat consumption can impact the gut microflora significantly [54], thus this needs to be accounted for when interpreting the observed results. In conclusion, the study demonstrated the significance of the gut-liver axis as a potential target for toxicant-induced liver injury and TASH as seen by Aroclor1260 effects in WT mice, and underscores the role of xenobiotic hepatic receptors such as CAR and PXR in influencing the degree of toxicity as observed by distinct changes in gut microbiome in the knockout mice.
Author statement
Banrida Wahlang: Conceptualization, Project Administration, Methodology, Investigation, Formal Analysis, Visualization, Writing - original draft.
Nicholas C. Alexander II: Investigation, Formal Analysis, Writing - review & editing.
Xiaohong Li: Formal Analysis, Software, Visualization, Writing - review & editing.
Eric C. Rouchka: Formal Analysis, Resources, Software, Writing - review & editing.
Irina A. Kirpich: Writing - review & editing.
Matthew C. Cave: Funding acquisition, Supervision, Writing - review & editing.
Funding information
This work was supported in part by the National Institute of Environmental Health Sciences [R35ES028373, P42ES023716, P30ES030283, R01ES032189, R21ES031510, T32ES011564, T35ES014559]; the National Institute of General Medical Sciences [P20GM113226, P20GM103436] and the National Institute on Alcohol Abuse and Alcoholism [R01AA024102, P50AA024337]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgements
The authors would like to acknowledge the Kentucky Biomedical Research Infrastructure Network (KBRIN) and the University of Louisville Genomics Core Facility for assistance with the project.
Edited by Dr. A.M. Tsatsaka
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.toxrep.2021.03.010.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Cave M., Appana S., Patel M., Falkner K.C., McClain C.J., Brock G. Polychlorinated biphenyls, lead, and mercury are associated with liver disease in American adults: NHANES 2003-2004. Environ. Health Perspect. 2010;118:1735–1742. doi: 10.1289/ehp.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clair H.B., Pinkston C.M., Rai S.N., Pavuk M., Dutton N.D., Brock G.N., Prough R.A., Falkner K.C., McClain C.J., Cave M.C. Liver disease in a residential cohort with elevated polychlorinated biphenyl exposures. Toxicol. Sci. 2018;164:39–49. doi: 10.1093/toxsci/kfy076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ATSDR . U.S. Department of Health and Human Services, Public Health Service; Atlanta, GA: 2000. Toxicological Profile for Polychlorinated Biphenyls (PCBs) Agency for Toxic Substances and Disease Registry (ATSDR) [Google Scholar]
- 4.Schecter A., Colacino J., Haffner D., Patel K., Opel M., Papke O., Birnbaum L. Perfluorinated compounds, polychlorinated biphenyls, and organochlorine pesticide contamination in composite food samples from Dallas, Texas, USA. Environ. Health Perspect. 2010;118:796–802. doi: 10.1289/ehp.0901347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., Dong S., Wang P., Su X., Fu J. Polychlorinated biphenyls are still alarming persistent organic pollutants in marine-origin animal feed (fishmeal) Chemosphere. 2019;233:355–362. doi: 10.1016/j.chemosphere.2019.05.250. [DOI] [PubMed] [Google Scholar]
- 6.Safe S. Toxicology, structure-function relationship, and human and environmental health impacts of polychlorinated biphenyls: progress and problems. Environ. Health Perspect. 1993;100:259–268. doi: 10.1289/ehp.93100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahlang B., Falkner K.C., Clair H.B., Al-Eryani L., Prough R.A., States J.C., Coslo D.M., Omiecinski C.J., Cave M.C. Human receptor activation by aroclor 1260, a polychlorinated biphenyl mixture. Toxicol. Sci. 2014;140:283–297. doi: 10.1093/toxsci/kfu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahlang B., Song M., Beier J.I., Cameron Falkner K., Al-Eryani L., Clair H.B., Prough R.A., Osborne T.S., Malarkey D.E., States J.C., Cave M.C. Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 2014;279:380–390. doi: 10.1016/j.taap.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahlang B., Beier J.I., Clair H.B., Bellis-Jones H.J., Falkner K.C., McClain C.J., Cave M.C. Toxicant-associated steatohepatitis. Toxicol. Pathol. 2013;41:343–360. doi: 10.1177/0192623312468517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahlang B., Hardesty J.E., Jin J., Falkner K.C., Cave M.C. Polychlorinated biphenyls and nonalcoholic fatty liver disease. Curr. Opin. Toxicol. 2019;14:21–28. doi: 10.1016/j.cotox.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlang B., Jin J., Beier J.I., Hardesty J.E., Daly E.F., Schnegelberger R.D., Falkner K.C., Prough R.A., Kirpich I.A., Cave M.C. Mechanisms of environmental contributions to fatty liver disease. Curr. Environ. Health Rep. 2019;6:80–94. doi: 10.1007/s40572-019-00232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardesty J.E., Wahlang B., Falkner K.C., Shi H., Jin J., Zhou Y., Wilkey D.W., Merchant M.L., Watson C.T., Feng W., Morris A.J., Hennig B., Prough R.A., Cave M.C. Proteomic analysis reveals novel mechanisms by which polychlorinated biphenyls compromise the liver promoting diet-induced steatohepatitis. J. Proteome Res. 2019;18:1582–1594. doi: 10.1021/acs.jproteome.8b00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahlang B., Prough R.A., Falkner K.C., Hardesty J.E., Song M., Clair H.B., Clark B.J., States J.C., Arteel G.E., Cave M.C. Polychlorinated biphenyl-xenobiotic nuclear receptor interactions regulate energy metabolism, behavior, and inflammation in non-alcoholic-Steatohepatitis. Toxicol. Sci. 2016;149:396–410. doi: 10.1093/toxsci/kfv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cave M.C., Clair H.B., Hardesty J.E., Falkner K.C., Feng W., Clark B.J., Sidey J., Shi H., Aqel B.A., McClain C.J., Prough R.A. Nuclear receptors and nonalcoholic fatty liver disease. Biochim. Biophys. Acta. 2016;1859:1083–1099. doi: 10.1016/j.bbagrm.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dempsey J.L., Wang D., Siginir G., Fei Q., Raftery D., Gu H., Yue Cui J. Pharmacological activation of PXR and CAR downregulates distinct bile acid-metabolizing intestinal Bacteria and alters bile acid homeostasis. Toxicol. Sci. 2019;168:40–60. doi: 10.1093/toxsci/kfy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker S.S., Baker R.D. Gut microbiota and liver injury (II): chronic liver injury. Adv. Exp. Med. Biol. 2020;1238:39–54. doi: 10.1007/978-981-15-2385-4_4. [DOI] [PubMed] [Google Scholar]
- 17.Cheng S.L., Li X., Lehmler H.J., Phillips B., Shen D., Cui J.Y. Gut microbiota modulates interactions between polychlorinated biphenyls and bile acid homeostasis. Toxicol. Sci. 2018;166:269–287. doi: 10.1093/toxsci/kfy208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi Y., Lin Y., Zhu H., Huang Q., Ye G., Dong S. PCBs-high-fat diet interactions as mediators of gut microbiota dysbiosis and abdominal fat accumulation in female mice. Environ. Pollut. 2018;239:332–341. doi: 10.1016/j.envpol.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Chi Y., Lin Y., Lu Y., Huang Q., Ye G., Dong S. Gut microbiota dysbiosis correlates with a low-dose PCB126-induced dyslipidemia and non-alcoholic fatty liver disease. Sci. Total Environ. 2019;653:274–282. doi: 10.1016/j.scitotenv.2018.10.387. [DOI] [PubMed] [Google Scholar]
- 20.Kuczynski J., Stombaugh J., Walters W.A., Gonzalez A., Caporaso J.G., Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinf. 2011 doi: 10.1002/0471250953.bi1007s36. Chapter 10, Unit 10 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goecks J., Nekrutenko A., Taylor J., Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A., Nielsen J., Ley R.E., Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Wang Y.D., Chen W.D., Wang X., Lou G., Liu N., Lin M., Forman B.M., Huang W. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology. 2012;56:2336–2343. doi: 10.1002/hep.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs C.D., Traussnigg S.A., Trauner M. Nuclear receptor modulation for the treatment of nonalcoholic fatty liver disease. Semin. Liver Dis. 2016;36:69–86. doi: 10.1055/s-0036-1571296. [DOI] [PubMed] [Google Scholar]
- 27.Gao J., He J., Zhai Y., Wada T., Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J. Biol. Chem. 2009;284:25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J., Chen B., Lu J., Xie W. Deciphering the roles of the constitutive androstane receptor in energy metabolism. Acta Pharmacol. Sin. 2015;36:62–70. doi: 10.1038/aps.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y., Liu D. Activation of pregnane X receptor by pregnenolone 16 alpha-carbonitrile prevents high-fat diet-induced obesity in AKR/J mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spruiell K., Richardson R.M., Cullen J.M., Awumey E.M., Gonzalez F.J., Gyamfi M.A. Role of pregnane X receptor in obesity and glucose homeostasis in male mice. J. Biol. Chem. 2014;289:3244–3261. doi: 10.1074/jbc.M113.494575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L.Y., Xu J.Y., Shi Z., Englert N.A., Zhang S.Y. Pregnane X receptor (PXR) deficiency improves high fat diet-induced obesity via induction of fibroblast growth factor 15 (FGF15) expression. Biochem. Pharmacol. 2017;142:194–203. doi: 10.1016/j.bcp.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Terc J., Hansen A., Alston L., Hirota S.A. Pregnane X receptor agonists enhance intestinal epithelial wound healing and repair of the intestinal barrier following the induction of experimental colitis. Eur. J. Pharm. Sci. 2014;55:12–19. doi: 10.1016/j.ejps.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Garg A., Zhao A., Erickson S.L., Mukherjee S., Lau A.J., Alston L., Chang T.K., Mani S., Hirota S.A. Pregnane X receptor activation attenuates inflammation-associated intestinal epithelial barrier dysfunction by inhibiting cytokine-induced myosin light-chain kinase expression and c-Jun N-terminal kinase 1/2 activation. J. Pharmacol. Exp. Ther. 2016;359:91–101. doi: 10.1124/jpet.116.234096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson G.M., Flannigan K.L., Erickson S.L., Vicentini F.A., Zamponi A., Hirota C.L., Alston L., Altier C., Ghosh S., Rioux K.P., Mani S., Chang T.K., Hirota S.A. Constitutive androstane receptor regulates the intestinal mucosal response to injury. Br. J. Pharmacol. 2017;174:1857–1871. doi: 10.1111/bph.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah Y.M., Ma X., Morimura K., Kim I., Gonzalez F.J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1114–1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 36.Cheng J., Shah Y.M., Ma X., Pang X., Tanaka T., Kodama T., Krausz K.W., Gonzalez F.J. Therapeutic role of rifaximin in inflammatory bowel disease: clinical implication of human pregnane X receptor activation. J. Pharmacol. Exp. Ther. 2010;335:32–41. doi: 10.1124/jpet.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S., Fleet J.C., Kortagere S., Mukherjee P., Fasano A., Le Ven J., Nicholson J.K., Dumas M.E., Khanna K.M., Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohandas S., Vairappan B. Pregnane X receptor activation by its natural ligand Ginkgolide-A improves tight junction proteins expression and attenuates bacterial translocation in cirrhosis. Chem. Biol. Interact. 2020;315 doi: 10.1016/j.cbi.2019.108891. [DOI] [PubMed] [Google Scholar]
- 39.Collins S.L., Patterson A.D. The gut microbiome: an orchestrator of xenobiotic metabolism. Acta Pharm. Sin. B. 2020;10:19–32. doi: 10.1016/j.apsb.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 41.John G.K., Mullin G.E. The gut microbiome and obesity. Curr. Oncol. Rep. 2016;18:45. doi: 10.1007/s11912-016-0528-7. [DOI] [PubMed] [Google Scholar]
- 42.Zhu L., Baker S.S., Gill C., Liu W., Alkhouri R., Baker R.D., Gill S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 43.Sobhonslidsuk A., Chanprasertyothin S., Pongrujikorn T., Kaewduang P., Promson K., Petraksa S., Ongphiphadhanakul B. The association of gut microbiota with nonalcoholic steatohepatitis in thais. Biomed Res. Int. 2018;2018 doi: 10.1155/2018/9340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau E., Carvalho D., Freitas P. Gut microbiota: association with NAFLD and metabolic disturbances. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/979515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boursier J., Mueller O., Barret M., Machado M., Fizanne L., Araujo-Perez F., Guy C.D., Seed P.C., Rawls J.F., David L.A., Hunault G., Oberti F., Cales P., Diehl A.M. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henke M.T., Kenny D.J., Cassilly C.D., Vlamakis H., Xavier R.J., Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh R.K., Chang H.W., Yan D., Lee K.M., Ucmak D., Wong K., Abrouk M., Farahnik B., Nakamura M., Zhu T.H., Bhutani T., Liao W. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Chierico F., Nobili V., Vernocchi P., Russo A., De Stefanis C., Gnani D., Furlanello C., Zandona A., Paci P., Capuani G., Dallapiccola B., Miccheli A., Alisi A., Putignani L. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451–464. doi: 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]
- 49.Shen F., Zheng R.D., Sun X.Q., Ding W.J., Wang X.Y., Fan J.G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2017;16:375–381. doi: 10.1016/S1499-3872(17)60019-5. [DOI] [PubMed] [Google Scholar]
- 50.Wu W., Lv L., Shi D., Ye J., Fang D., Guo F., Li Y., He X., Li L. Protective effect of Akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front. Microbiol. 2017;8:1804. doi: 10.3389/fmicb.2017.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Lv L., Ye J., Fang D., Shi D., Wu W., Wang Q., Wu J., Yang L., Bian X., Jiang X., Jiang H., Yan R., Peng C., Li L. Bifidobacterium adolescentis CGMCC 15058 alleviates liver injury, enhances the intestinal barrier and modifies the gut microbiota in D-galactosamine-treated rats. Appl. Microbiol. Biotechnol. 2019;103:375–393. doi: 10.1007/s00253-018-9454-y. [DOI] [PubMed] [Google Scholar]
- 52.Wahlang B., Jin J., Hardesty J.E., Head K.Z., Shi H., Falkner K.C., Prough R.A., Klinge C.M., Cave M.C. Identifying sex differences arising from polychlorinated biphenyl exposures in toxicant-associated liver disease. Food Chem. Toxicol. 2019;129:64–76. doi: 10.1016/j.fct.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsiaoussis J., Antoniou M.N., Koliarakis I., Mesnage R., Vardavas C.I., Izotov B.N., Psaroulaki A., Tsatsakis A. Effects of single and combined toxic exposures on the gut microbiome: current knowledge and future directions. Toxicol. Lett. 2019;312:72–97. doi: 10.1016/j.toxlet.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Bibbo S., Ianiro G., Giorgio V., Scaldaferri F., Masucci L., Gasbarrini A., Cammarota G. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 2016;20:4742–4749. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.