Highlight
-
•
An ICA was developed for P. aeruginosa detection.
-
•
The ICA strip showed a limit of detection of 2.41 × 104 CFU/mL.
-
•
The ICA could be applied to detect P. aeruginosa in water and food samples.
Keywords: Immunochromatographic assay, Pseudomonas aeruginosa, Monoclonal antibodies
Abstract
Pseudomonas aeruginosa (P. aeruginosa) is the common infection-causing bacterial pathogen. Conventional methods for the detection of P. aeruginosa are time-consuming, and therefore, a more rapid analytical method is required. Here, monoclonal antibodies (Mabs) against P. aeruginosa (CICC 10419) were prepared and based on paired Mabs, an immunochromatographic assay (ICA) was developed. The ICA strip showed a limit of detection of 2.41 × 104 CFU/mL and the linear range of detection was 3.13 × 104-1.0 × 106 CFU/mL. No cross-reactivity was observed when other common Gram-negative and Gram-positive bacteria were used. The analytical performance of the ICA strip indicated that the developed ICA had good specificity and stability. Moreover, the feasibility of the ICA strip was verified by detecting P. aeruginosa (CICC 10419) in spiked water and food samples. The ICA strip could detect samples contaminated with a low-level of P. aeruginosa (CICC 10419) after 8 h enrichment.
1. Introduction
Various types of infections triggered by Pseudomonas aeruginosa (P. aeruginosa) are increasingly becoming a major health concern worldwide (Castañeda-Montes et al., 2018, Croughs et al., 2018, Tacconelli et al., 2018). P. aeruginosa is an opportunist pathogen that can cause a number of infections, including sepsis, respiratory infection, endocarditis, urinary tract infection, and central nervous system infection, especially in patients suffering from severe burns, or who are immunocompromised due to cancer or cystic fibrosis (Jia et al., 2017, Juan et al., 2017, Alatraktchi et al., 2020). The spread of P. aeruginosa is caused by patient to patient contact via contaminated objects or by digestion of contaminated foods and water. More importantly, P. aeruginosa is a ubiquitous bacterium that is commonly found in most moist environments, such as soil, water and some foods (Cuttelod et al., 2011). In addition, in accordance with the stipulation from the European Communities and the Codex Alimentarius Commission (Tang et al., 2017aa, Tang et al., 2017bb), P. aeruginosa should be absent from all water and foods. Therefore, to ensure food safety and protect human health, a rapid, specific and sensitive method for the detection of the P. aeruginosa is urgently required.
A “gold standard” culture-based traditional detection method for P. aeruginosa, is in operation. However, this method cannot achieve the desired sensitivity needed and the method involves a series of tedious processes including sample preparation, enrichment, bacterial culture, and target bacteria identification. The whole process is inherently labor-intensive and time-consuming and therefore, is not suitable for certain situations and can cause a delay in detection of up to 2–3 days. Numerous efforts have been devoted toward development of new methods for P. aeruginosa detection. Several nucleic acid-based methods have been widely used, including the polymerase chain reaction (PCR) (Kunze et al., 2016, Tang et al., 2014), real-time PCR (Diawara et al., 2016), multiplex PCR (Aghamollaei et al., 2015), and loop-mediated isothermal amplification (LAMP) (Goto et al., 2010). Nucleic acid -based methods involve complicated steps such as DNA extraction, PCR amplification, and gel electrophoresis. Although it is known that PCR methods can provide sensitive results for pathogen detection, it requires expensive equipment and a highly well-trained technician, which makes it unsuitable for on-site detection. Biosensors have been widely recognized as a very promising analytical tool for pathogen detection because of their high sensitivity and specificity (Alhogail et al., 2019, Sarabaegi and Roushani, 2019). At the core of biosensor construction is a signal transducer and bio-recognition element coupled to the signal transducer, providing a corresponding analytical signal in response to the interaction between the bio-recognition element and the target. Optical biosensors, colorimetric biosensors, and electrochemical sensors have all been reported to be able to detect P. aeruginosa (Chen et al., 2019, Gao et al., 2018, Simoska et al., 2019, Zhang et al., 2019). Fast, real-time and accurate analysis of results from food samples can be achieved, but are highly dependent on sophisticated instruments and analysis systems. Additionally, biosensor methods also face the problem of high cost, susceptibility to the food matrix, and poor reproducibility.
As an alternative, immunological detection methods based on antigen–antibody interactions have been widely applied for the detection of pathogens due to their high sensitivity, specificity, and simplicity (Bever et al., 2019, Li et al., 2019). They have several advantages such as high-throughput detection, the use of relatively inexpensive equipment, and accuracy of the test results. Enzyme-linked immunosorbent assays (ELISA) based on the sandwich format is most frequently applied to microorganism detection, where a pair of antibodies (labeled capture Mab and immobilized detection Mab) is required to identify different antigen binding sites. Similarly, immunochromatographic assays (ICA) have been widely used to detect pathogens also (Shan et al., 2015, Wang et al., 2016). Compared to the sandwich ELISA, ICAs eliminate the complicated wash procedures and results can be attained within 10–20 min with the naked eye. Furthermore, quantitative results can be obtained by using a portable scanning reader, which makes this method suitable for on-site detection. Furthermore, recently, a portable one-step ICA for the rapid, on-site detection of P. aeruginosa in clinical samples has been developed (Wang et al., 2011), with a limit of detection (LOD) of 5.0 × 105 CFU/mL. However, there remain some limitations when using the ICA method such as its relatively low sensitivity. Therefore, in this study, we prepared a paired Mab against P. aeruginosa with higher sensitivity and specificity, and an ICA based on these paired Mabs was developed to detect P. aeruginosa in water and food samples.
2. Materials and methods
2.1. Materials and apparatus
Yeast extract powder and tryptone were purchased from Thermo Fisher Scientific (Shanghai, China). Agar powder was purchase from Beijing Solarbio Science &Tech Co., Ltd. (Beijing, China). Freund’s complete adjuvant (FCA) and Freund’s incomplete adjuvant (FIA), bovine serum albumin (BSA), horseradish peroxidase (HRP), HRP-labeled goat anti-mouse IgG and 3,3′,5,5′-Tetramethylbenzidine (TMB) were obtained from Sigma Aldrich (Shanghai, China). Sodium periodate, ethylene glycol and sodium cyanoborohydride were purchased from J&K Scientific Co., Ltd. (Shanghai, China). Cell culture media were purchased from Life Technologies Corporation (Shanghai, China). Other solvents and chemicals were purchased from the National Pharmaceutical Group Chemical Reagent Co., Ltd. (Shanghai, China) and were analytical reagent grade. All solutions were prepared with ultrapure water (Milli-Q ultrapure system, Millipore Co., Ltd. (Bedford, MA, USA). The SP2/0 myeloma cell line was purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). A commercial mouse Mab isotyping ELISA kit was purchased from Luoyang bai aotong experimental materials center (Luoyang, China). Sterilized homogenize bags were purchased from ELMEX Co., Ltd. (Shanghai, China). A stomacher machine (BagMixer 400) was obtained from Interscience (Saint Nom, France). Tap water samples were collected in Wuxi and stored at 4 °C until their analysis. Three different food items: orange fruit, milk, and beef were purchased from a local supermarket (Wuxi, China). The orange juice was obtained by crushing the orange fruit.
The bacterial strains used in this study are as follows: P. aeruginosa (CICC 10419), P. aeruginosa (CICC 21625), P. aeruginosa (CICC 10351), P. aeruginosa (ATCC 27853), P. aeruginosa (ATCC 15442), P. aeruginosa (ATCC 25619), P. aeruginosa (CICC 10299), Escherichia coli (E. coli) O157:H7 (CICC 21530), Vibrio parahaemolyticus (V. parahaemolyticus) (CICC 21617), Campylobacter jejuni (C. jejuni) (CICC 22936), Shigella flexneri (S. flexneri) (CICC 10865), Staphylococcus aureus (S. aureus) (ATCC 29213), Listeria monocytogenes (L. monocytogenes) (ATCC 19115), Enterobacter sakazakii (E.sakazakii) (ATCC 29544), Salmonella typhimurium (S. typhimurium) (ATCC 13311).
The polyvinylchloride (PVC) backing cards, sample pads (glass-fiber membrane, CB-SB08) and absorption pads (SX18) were purchased from JieYi Biotechnology Co., Ltd. (Shanghai, China) and the nitrocellulose (NC) membranes were acquired from Whatman-Xinhua Filter Paper Co., Ltd. (Hangzhou, China). A CM 4000 guillotine cutting module was obtained from Gene Co., Ltd. (Shanghai, China). The Airjet Quanti 3000™ dispenser was obtained from Xinqidian Gene Technology Co., Ltd. (Beijing, China). A hand-held strip scan reader was supplied by Huaan Magnech Bio-Tech Co., Ltd. (Beijing, China) and Protein-G affinity chromatography (Cell Signaling Technology, Shanghai, China) was used to purify the antibodies. Constant temperature shaker (THZ-420) was purchased from Shanghai Jinghong Experimental Equipment Co., Ltd. (Shanghai, China).
2.2. Bacterial strains culture.
P. aeruginosa was grown in 100 mL of LB broth at 37 °C overnight under constant agitation at a setting of 180 rpm and then, P. aeruginosa was heat-inactivated in the water bath at 100 °C for 15 mins. Bacteria were harvested from 100 mL of overnight culture by centrifugation at 5000 × g for 20 min at 4 °C, washed with sterile phosphate-buffered saline (PBS), resuspended in 5 mL of PBS, and finally stored at −20 °C for use in the enzyme immunoassay as the immune-antigens. Strains used in the sandwich ELISA and ICA strip to determine the LOD were prepared and stored at −20 °C. For the solid medium, 15 g/L of agar was added and standard LB agar plate counting was used to evaluate the concentration of bacteria. The number of colony forming units (CFU) was determined and 10-fold serial dilutions of bacterial cells were plated on LB agar plates and cultured overnight at 37 °C.
2.3. Immunization, Mab production and purification.
The immunization schedule was similar to that of Zhou et al. (2020). Female mice (6–8 weeks old) were firstly immunized by subcutaneous injection of 1.0 × 108 CFU/mL of heat-inactivated P. aeruginosa (CICC 10419) emulsified in an equal volume of FCA. After the initial immunization, three sequential booster injections were performed at approximately three-week intervals, with half the dose of the first immunization emulsified in FIA. Mouse serum was then evaluated by a noncompetitive indirect enzyme-linked immunosorbent assay (NCI-ELISA), the coating concentration of P. aeruginosa (CICC 10419) and other tested strains were 1.0 × 108 CFU/mL, 3.0 × 107 CFU/mL and 1.0 × 107 CFU/mL, respectively. The mouse with the highest serum titer against P. aeruginosa (CICC 10419) was selected as the spleen donor. The fusion of splenocytes and SP2/0 myeloma cells was performed as published previously (Ye et al., 2018). The culture supernatant from the hybridoma cells was screened first using the coated P. aeruginosa (CICC 10419) 96-well plates, the positive wells with the absorbance higher than 1.5 at 450 nm were selected. Then the wells selected were determined using the plates coating other test bacteria, the positive wells that did not cross-react with other test bacteria for P. aeruginosa (CICC 10419) were selected. The positive clones were selected at the same procedure and separated by the three rounds of limiting dilution. Subsequently, mass-produced in paraffin primed BALB/c mice. The Mabs were ultimately isolated and purified by protein G agarose affinity chromatography. Ten stable Mabs against P. aeruginosa (CICC 10419) were obtained. Isotypes of the Mabs were determined with a commercial mouse Mab isotyping ELISA kit. All animal studies in this work were performed in compliance with the institutional ethical guidelines for Care and Use of Laboratory Animals of Jiangnan University and were approved by the Animal Ethics Committee of Jiangsu province.
2.4. Conjugation of the Mab to HRP
The HRP-antibody conjugation reaction was performed as previously reported, with slight modifications (Kuang et al., 2013). Briefly, 0.2 mL of 10 mg/mL HRP and 0.2 mL of 0.06 M NaIO4 were reacted at 4 °C for 30 min, producing the generation of aldehyde groups by oxidation of the hydroxyl groups on HRP. Then, 0.2 mL of 0.16 M glycol was added to the mixture to eliminate the excess NaIO4 at room temperature. Next, 2 mg of purified Mab was added and the pH was adjusted to 9 by the addition of 0.05 M carbonate buffer, in which aldehyde groups can be linked to the amino groups of the antibody to produce the corresponding Schiff bases. After stirring for 20 h at 4 °C, the stable HRP-antibody conjugate was formed by adding 90 μL of 5 mg/mL NaBH4 and precipitated by adding an equal volume of saturated ammonium sulfate solution. After centrifugation for 10 min at 5000 × g, the pellet was resuspended with 0.01 M PBS (pH 7.4). The HRP-antibody conjugation was dialyzed against 0.01 M PBS for 36 h at 4 °C and then added to an equal volume of glycerol, and stored at −20 °C for long-term storage until use. All the reactions, including the dialysis should be protected from light and the HRP-antibody conjugation was characterized by direct ELISA. The purified Mabs and the HRP-Mab conjugations were then paired with each other in a sandwich ELISA format.
2.5. Sandwich ELISA
The anti-P. aeruginosa (CICC 10419) Mab diluted with coating buffer (100 μL/well), was added to a 96-well plate and incubated at 4 ℃ overnight. After incubation, the plate was washed three times with washing buffer and blocking buffer (220 μL/well) was added and incubated for 2 h at 37 ℃ to avoid non-specific binding. After washing, 100 μL of sample was added to each well and incubated for 1 h at 37 ℃. After washing the plate, HRP-labeled anti-P. aeruginosa (CICC 10419) Mab was added and incubated for a further 1 h at 37 °C. After another wash, 100 μL of TMB substrate solution was added, and reacted with the labeled Mab at 37 °C for 15 min in the dark. The reaction was stopped by the addition of 2 M sulfuric acid (50 μL/well), and the absorbance was measured at 450 nm with a microplate reader and all measurements were performed in triplicate.
2.6. Pairwise interaction analysis
A checkerboard method was designed using sandwich ELISA format for pairwise interaction analysis. Briefly, plates were coated with ten Mabs diluted to a concentration of 4 μg/mL as the capture antibody (generated from 10 cell lines). After blocking, 100 μL of 1.0 × 107 CFU/mL P. aeruginosa or 0.01 M PBS were added as positive and negative controls, respectively. Then, 100 μL of 2 μg/mL of HRP-labeled Mabs used as detection antibodies were added. The sandwich ELISA procedure was the same as described previously. The highest OD 450 nm ratio of the positive and negative controls (P/N value) was considered as the optimal combination.
2.7. Sensitivity and cross-reactivity analysis
The concentration of capture antibody and detection antibody were optimized and as previously, a checkerboard method was designed. Plates were coated with capture Mab (8, 4, 2 and 1 μg/mL) and after blocking, P. aeruginosa (CICC 10419) was diluted with PBS to 1.0 × 107 CFU/mL and added along with 0.01 M PBS as a negative control. Then, the detection antibody (4, 2, 1 and 0.5 μg/mL) was added and after color development and termination the highest P/N value corresponding to the optimal concentrations of capture antibody and detection antibody were calculated. Next under the predetermined optimal conditions, P. aeruginosa (CICC 10419) was serially diluted with PBS to seven different concentrations (1.0 × 107 CFU/mL, 3.33 × 106 CFU/mL, 1.11 × 106 CFU/mL, 3.70 × 105 CFU/mL, 1.23 × 105 CFU/mL, 4.12 × 104 CFU/mL, 1.37 × 104 CFU/mL) and added to the plate well, and again, 0.01 M PBS was used as a negative control. Then, a calibration curve for P. aeruginosa (CICC 10419) was established. In addition, a total of seven P. aeruginosa, and eight other bacterial strains from various sources were used to evaluate the specificity of our method in this study. All the tested strains were diluted to 1.0 × 108 CFU/mL in 0.01 M PBS.
2.8. Lateral-flow ICA strip development
2.8.1. Gold nanoparticle (GNP)-labeled Mab preparation
The GNP-labeled Mab conjugation was obtained according to the method developed within our laboratory (Luo et al., 2019). Colloidal GNPs were prepared using the sodium citrate reduction method (Wang, Guo, Liu, Kuang, & Xu, 2018). Transmission electron microscopic (TEM) image of the GNPs was obtained with a transmission electron microscope (JEOL JEM-2100) operating at an acceleration voltage of 200 kV. Then, the pH of the GNP solution was adjusted to 8.2 by the addition of 0.1 M K2CO3. After this, 100 μL of Mab was added into the 10 mL of GNP solution and stirred continuously for 1 h at room temperature. Next, 1 mL of 0.5% (w/v) BSA was added to block the GNP surface for 2 h, to avoid non-specific adsorption. The solution was then centrifuged for 30 min at 7000 × g and the precipitate was resuspended in 5 mL of 0.02 M PBS containing 0.04% NaN3, 0.1% Tween, 0.1% PEG 6000, 1% mannitol, 2% sorbitol, 5% sucrose. The GNP-labeled Mab conjugation was used as the detection antibody.
2.8.2. Construction of the lateral-flow ICA strip
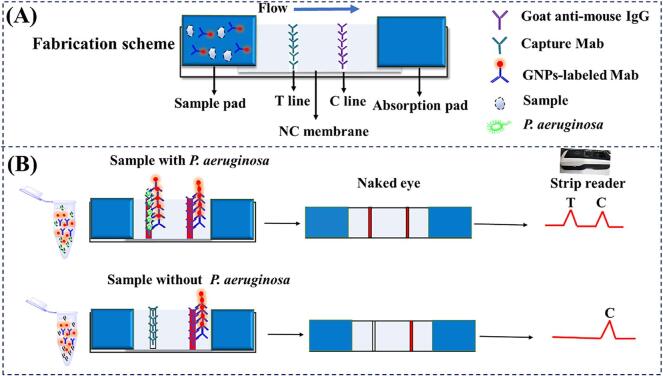
The lateral-flow ICA strip was constructed in a sandwich format as previously reported. As shown in Fig. 1A, the components of the strip were composed of a sample pad, an NC membrane with test (T line) and control lines (C line), an absorbent pad, and a bottom pad. The T and C lines were formed by spraying anti-P. aeruginosa (CICC 10419) Mab (the capture antibody) and goat anti-mouse IgG on to them. The NC membrane was dried at 37 °C for 6 h. To assemble the lateral-flow ICA strips, the NC membrane was pasted on to the center of the bottom pad and was overlapping with the sample and absorption pads on each side. The assembled pad was cut into 4.0 mm wide strips using a guillotine and stored in a desiccator at room temperature until use.
Fig. 1.
Schematic diagrams of the ICA strip for P. aeruginosa detection. (A) The fabrication scheme of the ICA strip; (B) The principle of the ICA strip for P. aeruginosa detection.
2.8.3. The principle of the lateral-flow ICA strip.
The lateral-flow ICA strip prepared in this work is shown in Fig. 1B. For analysis, 150 μL of the standard or samples are first reacted with 50 μL of GNP-labeled Mab for 5 min. When the strip with the sample pad is inserted into the solution which moves from the sample pad to the absorbent pad by capillary action. In the presence of target bacteria (positive sample), the GNP-labeled Mab firstly reacts with the target bacteria, and then is captured by the anti-P. aeruginosa (CICC 10419) Mab on the T line. Excess GNP-labeled Mab is captured by the goat anti-mouse IgG antibody at the C line and eventually a visible red band at both the T line and C line are formed. The color intensity increases as the bacterial concentration increases. In the absence of target bacteria (negative sample), the GNP-labeled Mab only binds to the goat anti-mouse IgG antibody at the C line and therefore, only a red C line is observed. The C line should always be observed in both positive and negative samples indicating that the ICA strip has worked properly. This entire lateral-flow ICA strip procedure takes approximately 5–10 min.
2.8.4. ICA strip performance
The immobilized Mab on the T line, the labeled Mab, and the running buffer solution were firstly optimized to give the ICA strip optimal performance. P. aeruginosa (CICC 10419) was serially diluted with PBS to give six different concentrations (1.0 × 106, 5.0 × 105, 2.5 × 105, 1.25 × 105, 6.25 × 104 and 3.13 × 104 CFU/mL). Under optimal conditions, a range of P. aeruginosa (CICC 10419) concentrations and PBS solutions were tested. To evaluate the specificity of the ICA strip, five P. aeruginosa strains and eight foodborne pathogenic bacteria were tested at concentration of 2.0 × 108 CFU/mL, P. aeruginosa (CICC 21625) and P. aeruginosa (CICC 10351) at concentration of 2.0 × 109 CFU/mL were tested with the ICA strips. In addition, the ICA strips were stored at 4 °C for one, two, three, and four months. Then the strips were used to detect PBS and the P. aeruginosa (CICC 10419) at concentrations of 1.0 × 106 CFU/mL to assess its stability.
2.9. Detection of P. Aeruginosa in water and food samples
To determine whether the developed ICA strip can be applied for P. aeruginosa detection from contaminated real samples, tap water, orange juice, milk and beef samples were spiked with different amounts of P. aeruginosa (CICC 10419). Each sample was verified to be free of any bacteria by plate counting. P. aeruginosa was grown in LB overnight, centrifuged at 5000 × g for 20 min at 4 °C and resuspended in PBS. And then diluted to the target inoculation number. For liquid samples, 1 mL of P. aeruginosa (CICC 10419) at known concentrations (0–109 CFU/mL) were separately added to 9 mL of the sample solution. The target inoculation number were 106, 5 × 105, 2.5 × 105, 1.25 × 105, 6.25 × 104, 3.13 × 104 CFU/mL in tap water sample, while the target inoculation number were 1.0 × 108, 2.0 × 107, 4.0 × 106, 8.0 × 105, 1.6 × 105, and 3.2 × 104 CFU/mL in orange juice and milk samples. For beef samples, 10 g of beef samples was put into a sterilized homogenize bag. And then, 1 mL of the diluted bacteria solutions was added. The beef samples were homogenized with 90 mL of PBS in a stomacher machine for 1 min. The solution was poured into a sterile tube and centrifuged 5000 × g for 10 min. and the sediment was suspended in 10 mL and tested by the ICA strip. Non-inoculated samples were used as negative controls. For bacterium enrichment, 10 mL (10 g) of samples were inoculated with a fewer number of P. aeruginosa (CICC 10419). and then 90 mL of LB broth was added. The samples inoculated were incubated in LB broth at 37 °C for 6 h, 8 h and 10 h, respectively. After enrichment, the liquid sample solutions were tested directly. The beef sample was put into a stomacher machine, centrifuged and concentrated as above-mentioned. Finally, 150 μL samples were reacted with 50 μL of GNP-labeled Mab for 5 min. then the solution was applied to the sample pad of the ICA strip. The solution migrated towards the absorption pad and the signal intensity on the T line was recorded and all tests were performed in triplicate.
3. Results and discussion
3.1. Pairwise analysis
To obtain the best pair combination for the sandwich ELISA for P. aeruginosa (CICC 10419) identification, a checkboard method was executed. As shown in Table 1, 100 combinations were obtained using purified Mab as a capture antibody and HRP-labeled Mab as a detection antibody in the development of the sandwich ELISA. 1.0 × 107 CFU/mL of P. aeruginosa (CICC 10419) was added as positive control (100 µL/well), and 100 µL of standard diluted solution (0.01 M PBS, pH 7.4) was added as negative control. The highest P/N value was obtained using Mab 3C7 as the capture antibody and HRP-labeled Mab 2H10 as the detection antibody, respectively. Therefore, this combination was used for further experiments.
Table 1.
The P/N value of pairwise interaction analysis using sandwich ELISAa.
| Detection Mab |
Capture Mab |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1H12 | 2B10 | 2H10 | 3C7 | 4C10 | 5C12 | 7H3 | 8G12 | 9C2 | 10D10 | |
| 1H12-HRP | 6.52 | 5.90 | 9.52 | 6.60 | 6.43 | 6.09 | 7.82 | 5.61 | 8.18 | 6.06 |
| 2B10-HRP | 5.78 | 5.52 | 8.37 | 5.52 | 5.73 | 5.62 | 5.85 | 4.51 | 6.06 | 5.07 |
| 2H10-HRP | 7.44 | 8.76 | 5.06 | 14.75 | 12.65 | 11.78 | 10.42 | 7.29 | 7.03 | 11.76 |
| 3C7-HRP | 5.25 | 3.44 | 8.07 | 5.20 | 4.71 | 3.92 | 5.31 | 4.84 | 6.89 | 4.13 |
| 4C10-HRP | 5.33 | 4.94 | 13.27 | 7.75 | 7.67 | 5.63 | 7.65 | 7.49 | 9.87 | 5.00 |
| 5C12-HRP | 8.58 | 5.86 | 9.43 | 8.10 | 8.32 | 7.49 | 8.08 | 10.47 | 10.19 | 7.60 |
| 7H3-HRP | 2.41 | 2.29 | 2.83 | 3.49 | 3.14 | 2.69 | 2.26 | 1.99 | 2.19 | 2.28 |
| 8G12-HRP | 6.66 | 3.07 | 5.61 | 5.73 | 5.58 | 6.70 | 5.45 | 3.11 | 6.81 | 4.41 |
| 9C2-HRP | 3.15 | 2.31 | 2.17 | 5.47 | 4.97 | 4.03 | 3.06 | 3.90 | 3.42 | 4.05 |
| 10D10-HRP | 9.20 | 6.51 | 11.05 | 6.98 | 5.86 | 8.90 | 9.05 | 8.83 | 10.77 | 7.42 |
P/N value was the ratio of OD450 value of the positive to the negative control.
3.2. Sensitivity and cross-reactivity analysis
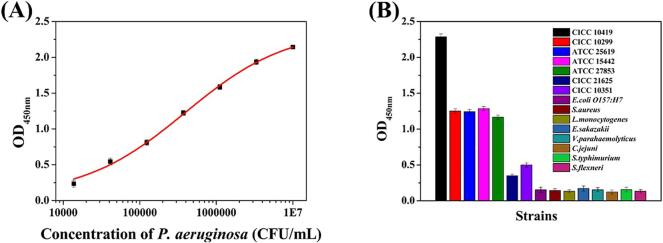
To evaluate the sensitivity of the sandwich ELISA, the concentrations of Mab 3C7 and HRP-labeled Mab 2H10 were optimized. As shown in Table S1, this combination gave the highest P/N. This represented a concentration of Mab 3C7 of 4 μg/mL and HRP-labeled Mab 2H10 at 1 μg/mL. Under the optimal concentrations, a calibration curve for the optimized sandwich ELISA was developed. As shown in Fig. 2A, the detection limit (P/N ≥ 2.1) of the sandwich ELISA was 1.37 × 104 CFU/mL and the detection ranged from 1.37 × 104 CFU/mL to 1.0 × 107 CFU/mL. To characterize the specificity of the paired Mab, the binding ability of the paired Mab towards fifteen pathogens was analyzed. The results (Fig. 2B) showed that the paired Mab has a relatively lower cross-reactivity with P. aeruginosa (CICC 21625) and P. aeruginosa (CICC 10351), and has a high affinity with P. aeruginosa (ATCC 27853), P. aeruginosa (ATCC 15442), P. aeruginosa (ATCC 25619), P. aeruginosa (CICC 10299). This may be attributed to the complexity of bacteria surface antigens. There are chemical and antigenically variable antigens such as lipopolysaccharide (LPS) O-antigens and flagella. Additionally, P. aeruginosa expresses some highly conserved antigens such as out membrane protein. Also, no cross-reactivity was observed with E. coli O157: H7, S. aureus, L. monocytogenes, E. sakazakii, V. parahaemolyticus, C. jejuni, S. typhimurium, or S. flexneri.
Fig. 2.
Sandwich ELISA development. (A) The calibration curve of the optimized sandwich ELISA for P. aeruginosa (CICC 10419); (B) Specificity of the anti-P. aeruginosa (CICC 10419) Mab towards 15 types of foodborne pathogens.
3.3. ICA strip optimization
GNPs have extensive applications for the detection of microbes ((Tang et al., 2017aa, Tang et al., 2017bb)), mycotoxins (Liu et al., 2018), veterinary drugs (Wang et al., 2019) and pesticides (Facure et al., 2017). The size distribution and structure of the synthesized GNPs were characterized by TEM, and the UV–Vis absorption spectrum was recorded. As shown in Fig. S1, the GNPs were homogeneous, and had an average size of 20 nm with a maximum absorption wavelength of 523 nm. These results confirmed that the GNPs were prepared successfully.
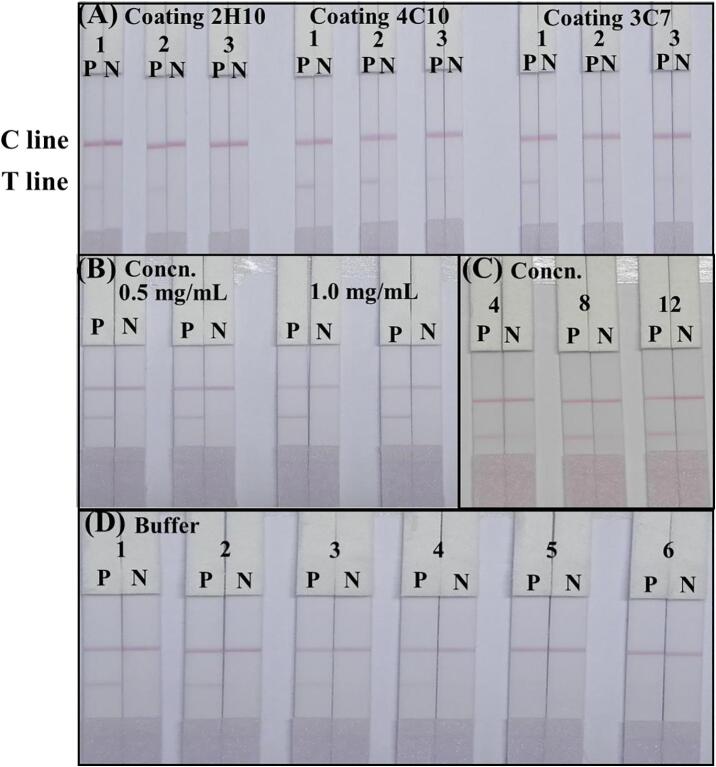
The ICA strip was developed based on the sandwich format, in which the immobilized Mab on the T line and the GNP-labeled Mab recognize the target and generated a signal. The sensitivity of the ICA strip was strongly affected by the Mab on the T line and the GNP-labeled Mab. Three types of immobilized Mabs (3C7, 4C10 and 2H10) and three types of GNP-labeled Mab (3C7, 4C10 and 2H10) were compared using 1.0 × 106 CFU/mL of P. aeruginosa (CICC 10419) culture and PBS. As shown in Fig. 3A, the immobilized Mab 3C7 on the T line and the GNP-labeled Mab 2H10 showed a red band on the T line. Similarly, the concentration of immobilized Mab on the T line was optimized. Concentrations of 0.5 mg/mL and 1.0 mg/mL of Mab 3C7 immobilized on the T line were compared using 1.0 × 106 CFU/mL of P. aeruginosa (CICC 10419) and PBS and the intensity of the color at the T line increases with an increase in the concentration of Mab. The concentration of Mab at 1.0 mg/mL gave a deeper color on the T line (Fig. 3B). Also, the GNP-labeled antibody binding is related to Mab concentration, where the concentration of the GNP-labeled Mab influences the visibility of the ICA strip. Different concentrations of Mab (4, 8 and 12 µg/mL) were used for the preparation of the GNP-labeled Mab conjugation. We found that the optimal concentration of Mab was 12 µg/mL, giving a deeper color generation on the T line (Fig. 3C). In addition, the running buffer solution dramatically affected the migration of GNP-labeled Mab. Therefore, six different types of running buffer solution were used. As shown in Fig. 3D, the color of the visual band in the ICA strip using basic buffer containing 1% Triton X-100 as running buffer was significantly brighter than those using other buffers. Based on these results, a basic buffer containing 1% Triton X-100 was selected as the optimal running buffer.
Fig. 3.
The optimization of the ICA strip. (A) Optimization the Mab sprayed on the T line and GNP-labeled Mab. 1, 2 and 3 were GNP-labeled Mab 2H10, 4C10 and 3C7, respectively. (B) Optimization the concentration of immobilized Mab on the T line at 0.5 and 1.0 mg/mL, respectively. (C) Optimization the concentration of GNP-labeled Mab at 4, 8 and 12 µg/mL, respectively. (D) Optimization running buffer. 1–6 represent the basic buffer, basic buffers containing 1% Triton X-100, 1% PEG, 1% PVA, 1% BSA, and 1% casein, respectively. P: 1.0 × 106 CFU/mL of P. aeruginosa; N: PBS.
3.4. ICA strip performance
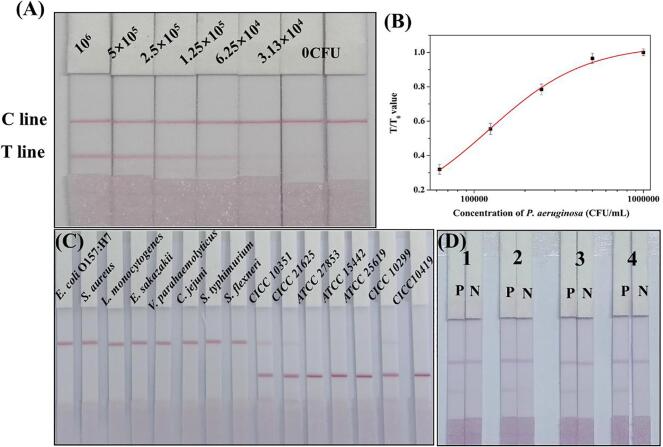
To evaluate the sensitivity of the ICA strip, P. aeruginosa (CICC 10419) with concentrations ranging from 0 to 106 CFU/mL were analyzed using our ICA strip under optimized conditions, respectively. The evaluation was performed by the naked eyes and a strip reader. As shown in Fig. 4A, the color of the T line on the ICA strip continuously deepened with the concentrations of P. aeruginosa (CICC 10419) increased. A weaker color on the T line was generated at the P. aeruginosa (CICC 10419) concentration of 6.25 × 104 CFU/mL than the negative control. The visual LOD of the ICA strip was defined as the lowest concentration of P. aeruginosa (CICC 10419) that generated a weaker T line compared with that of negative samples. And the cut-off value was defined as the threshold concentration that contributed to the color on the T line disappears completely. Therefore, the visual LOD was 6.25 × 104 CFU/mL. Meanwhile, a colorless T line was observed at the P. aeruginosa (CICC 10419) concentrations of 3.13 × 104 CFU/mL. Consequently, the cut-off value of the ICA strip was 3.13 × 104 CFU/mL. For quantitative analysis, the color density of the T and C lines were determined using a strip reader. A calibration curve was established by plotting T/T0 values (the ratio of the color density of T line for the positive sample to that of the negative sample) (y-axis) against corresponding concentration of P. aeruginosa (CICC 10419) (x-axis). As shown in Fig. 4B, the calculated LOD of the ICA strip was 2.41 × 104 CFU/mL from the calibration curve.
Fig. 4.
Analytical performance of the ICA strip. (A) The image for P. aeruginosa (CICC 10419) detection with ICA strip. (B) The calibration curve for P. aeruginosa (CICC 10419) detection. (C) The assessment of the specificity of the ICA strip. (D) The stability evaluation of ICA strips after storing at 4 °C for one month, two months, three months and four months, respectively. P: 1.0 × 106 CFU/mL of P. aeruginosa; N: PBS.
To assess the specificity of the ICA strip, thirteen bacterial strains at a concentration of 2.0 × 108 CFU/mL, P. aeruginosa (CICC 21625) and P. aeruginosa (CICC 10351) at concentration of 2.0 × 109 CFU/mL were tested with the ICA strips. The results are displayed in Fig. 4C, a bright red band on the T line was generated when testing P. aeruginosa (CICC 10419), P. aeruginosa (ATCC 27853), P. aeruginosa (ATCC 15442), P. aeruginosa (ATCC 25619), P. aeruginosa (CICC 10299). The ICA strip based on the paired mAb that reflects a relatively weak affinity to the other two P. aeruginosa could detect the two P. aeruginosa at the concentration of 2.0 × 109 CFU/mL. Thus, high sensitivity antibody preparation is necessary. A colorless T line was observed when the other species were tested. These results indicate that the ICA strip reacted strongly toward P. aeruginosa (CICC 10419), P. aeruginosa (ATCC 27853), P. aeruginosa (ATCC 15442), P. aeruginosa (ATCC 25619), P. aeruginosa (CICC 10299), which could be applied for the detection of P. aeruginosa.
The stability of the ICA strip was evaluated by testing strips that had been stored at 4 °C for one, two, three, and four months and the results (Fig. 4D) were consistent with initial data (Fig. 4A), indicating that these strips can be stored at 4 °C for at least four months, with no loss of performance.
3.5. Application in water and food samples
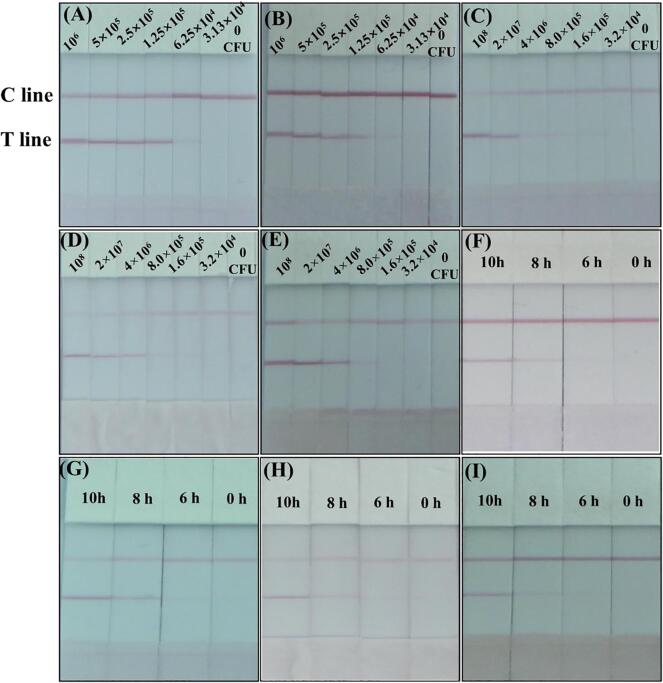
Tap water, orange juice, milk, and beef samples were used to validate the feasibility of the ICA strip. No bacteria were detected in the blank tap water, orange juice, milk, and beef samples using the colony counting method. The tap water, orange juice, milk, and beef samples were spiked with varying concentrations of P. aeruginosa (CICC 10419) separately and tested. Since the matrix of real samples is complex, the color intensities of the ICA after spiking with P. aeruginosa (CICC 10419) in PBS and real samples were compared. As shown in Fig. 5A and B, a faint red color was observed at 6.25 × 104 CFU/mL in PBS and tap water samples. The cut-off values were all obtained at 3.13 × 104 CFU/mL. The results indicated that the ICA strip was valid for P. aeruginosa (CICC 10419) detection in tap water samples with little matrix effects on the results. For orange juice, milk, beef samples, a weaker color on the T line was generated at 8.0 × 105 CFU/mL. Therefore, the cut-off value for P. aeruginosa (CICC 10419) detection were 1.6 × 105 CFU/mL in orange juice, milk and beef samples. It was found that the cut-off values increased due to the influence of the food matrix, but P. aeruginosa (CICC 10419) could be detected in real samples by the ICA strip.
Fig. 5.
Analysis of samples by ICA strip. (A), (B), (C), (D) and (E) Detection of P. aeruginosa (CICC 10419) spiked in PBS, tap water, orange juice, milk, and beef samples, respectively. (F), (G), (H) and (I) Detection of the tap water, orange juice, milk, and beef samples of P. aeruginosa (CICC 10419) after 6 h, 8 h and 10 h enrichment. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Analyzing low levels of pathogen that exist below the LOD in real samples is still challenging. Therefore, bacteria enrichment is indispensable and effective for the detection. The real samples were contaminated with P. aeruginosa (CICC 10419) 10 CFU/mL or CFU/g. The number of bacteria was measured after 6 h, 8 h, 10 h enrichment in LB broth. After 6 h, a colorless on the T line was observed. The number of bacteria was lower than the minimum number of bacteria required for the test. While a faint color was generated on the T line after enrichment of 8 h, indicating that the bacteria number reach the lowest threshold for the test. It is concluded that 10 CFU/mL or 10 CFU/g of contamination can be detected after 8 h enrichment. The ICA test process was conducted and the results were obtained within 15 min using the ICA strip after enrichment. Considering the sample pretreatment, bacteria enrichment, and detection, positive results were obtained in 9 h. The combination of the bacteria enrichment with the use of ICA strip can detect the presence of P. aeruginosa (CICC 10419) quickly compared with the culture-based detection method. The inoculation and experiments with water and food samples suggest that the proposed ICA strip can be applied to detect rapidly P. aeruginosa from contaminated water and food samples.
Recently, efforts have been devoted to the rapid and sensitive detection of P. aeruginosa using various approaches. Different recognition elements are used, including monoclonal antibody (Ellairaja et al., 2017), DNA aptamers (Zhong, Gao, Chen, & Jia, 2020), fluorescent organic nanoparticles (Kaur, Raj, Kaur, & Singh, 2015). These elements enhance the sensitivity for P. aeruginosa detection and reduce the assay time. Our future studies will focus on screening for high sensitivity antibodies, using new formats and labels, combining other methods to improve detection sensitivity and decrease the time of ICA for P. aeruginosa detection.
4. Conclusions
In the present study, Mabs against P. aeruginosa (CICC 10419) were successfully prepared and based on the paired Mab method, an ICA for the detection of P. aeruginosa (CICC 10419) in water and food samples was established. We also studied the factors that affect the performance of the ICA strip, including the immobilized Mab on the T line, the GNP-labeled Mab and the running buffer. Under optimal conditions, the developed sandwich ICA exhibited a linear range of 3.13 × 104-1.0 × 106 CFU/mL and a lower limit of detection of 2.41 × 104 CFU/mL. In addition, the specificity of the ICA strip was evaluated and no cross-reactivity was observed with other common Gram-negative and Gram-positive bacteria. Furthermore, the ICA strip can be applied to detect P. aeruginosa (CICC 10419) in contaminated water and food samples. After enrichment. water and food samples with low-level contamination were detected by the ICA strip within 15 min. Positive results were obtained in 9 h. This successful, rapid detection of P. aeruginosa in water and food samples indicates the feasibility of the ICA for practical applications. Noticeably, this research would provide a good basis for future work on improvements, probably using mAbs directed against highly conserved outer membrane proteins of the species.
5. Disclosure statement
No potential conflict of interest was reported by the authors.
6. Compliance with ethical standards
Funding
This work was funded by the Key Programs from MOST (2018YFC1602906).
Ethical approval
This article does not contain any studies with human subjects. All animal studies were carried out under the guidance of the animal welfare committee of Jiangnan University.
Author contributions
Xu C. and Kuang H. designed the experiments. Zeng L. and Wang Z. performed the experiments. Guo L., Xu X., Ding H., and Song S. participated in data analysis. Xu C. and Xu L. drafted the manuscript, and all authors read and approved the manuscript prior to submission.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2021.100117.
Contributor Information
Liguang Xu, Email: xlg@jiangnan.edu.cn.
Chuanlai Xu, Email: xcl@jiangnan.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aghamollaei H., Moghaddam M.M., Kooshki H., Heiat M., Mirnejad R., Barzi N.S. Detection of Pseudomonas aeruginosa by a triplex polymerase chain reaction assay based on lasI/R and gyrB genes. Journal of Infection and Public Health. 2015;8(4):314–322. doi: 10.1016/j.jiph.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Alatraktchi F.A., Dimaki M., Støvring N., Johansen H.K., Molin S., Svendsen W.E. Nanograss sensor for selective detection of Pseudomonas aeruginosa by pyocyanin identification in airway samples. Analytical Biochemistry. 2020;593:113586. doi: 10.1016/j.ab.2020.113586. [DOI] [PubMed] [Google Scholar]
- Alhogail S., Suaifan G.A.R.Y., Bikker F.J., Kaman W.E., Weber K., Cialla-May D., Popp J., Zourob M.M. Rapid Colorimetric Detection of Pseudomonas aeruginosa in Clinical Isolates Using a Magnetic Nanoparticle Biosensor. ACS Omega. 2019;4(26):21684–21688. doi: 10.1021/acsomega.9b02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever, C. S., Scotcher, M., Cheng, L. W., Hnasko, R. M., & Stanker, L. H. (2019). Development and Characterization of Monoclonal Antibodies to Botulinum Neurotoxin Type E. Toxins, 11(7). Doi: 10.3390/toxins11070407. [DOI] [PMC free article] [PubMed]
- Castañeda-Montes F.J., Avitia M., Sepúlveda-Robles O., Cruz-Sánchez V., Kameyama L., Guarneros G., Escalante A.E. Population structure of Pseudomonas aeruginosa through a MLST approach and antibiotic resistance profiling of a Mexican clinical collection. Infection, Genetics and Evolution. 2018;65:43–54. doi: 10.1016/j.meegid.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Chen Y., Ji W., Yan K., Gao J., Zhang J. Fuel cell-based self-powered electrochemical sensors for biochemical detection. Nano Energy. 2019;61:173–193. doi: 10.1016/j.nanoen.2019.04.056. [DOI] [Google Scholar]
- Croughs P.D., Klaassen C.H.W., van Rosmalen J., Maghdid D.M., Boers S.A., Hays J.P., Goessens W.H.F. Unexpected mechanisms of resistance in Dutch Pseudomonas aeruginosa isolates collected during 14 years of surveillance. International Journal of Antimicrobial Agents. 2018;52(3):407–410. doi: 10.1016/j.ijantimicag.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Cuttelod M., Senn L., Terletskiy V., Nahimana I., Petignat C., Eggimann P., Bille J., Prod'hom G., Zanetti G., Blanc D.S. Molecular epidemiology of Pseudomonas aeruginosa in intensive care units over a 10-year period (1998-2007) Clinical Microbiology and Infection. 2011;17(1):57–62. doi: 10.1111/j.1469-0691.2010.03164.x. [DOI] [PubMed] [Google Scholar]
- Diawara I., Katfy K., Zerouali K., Belabbes H., Elmdaghri N. A duplex real-time PCR for the detection of Streptococcus pneumoniae and Neisseria meningitidis in cerebrospinal fluid. Journal of Infection in Developing Countries. 2016;10(1):53–61. doi: 10.3855/jidc.5647. [DOI] [PubMed] [Google Scholar]
- Ellairaja S., Krithiga N., Ponmariappan S., Vasantha V.S. Novel Pyrimidine Tagged Silver Nanoparticle Based Fluorescent Immunoassay for the Detection of Pseudomonas aeruginosa. Journal of Agriculture and Food Chemistry. 2017;65(8):1802–1812. doi: 10.1021/acs.jafc.6b04790.s001. [DOI] [PubMed] [Google Scholar]
- Facure M.H.M., Mercante L.A., Mattoso L.H.C., Correa D.S. Detection of trace levels of organophosphate pesticides using an electronic tongue based on graphene hybrid nanocomposites. Talanta. 2017;167:59–66. doi: 10.1016/j.talanta.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Gao R., Zhong Z., Gao X., Jia L.i. Graphene Oxide Quantum Dots Assisted Construction of Fluorescent Aptasensor for Rapid Detection of Pseudomonas aeruginosa in Food Samples. Journal of Agriculture and Food Chemistry. 2018;66(41):10898–10905. doi: 10.1021/acs.jafc.8b02164.s001. [DOI] [PubMed] [Google Scholar]
- Goto M., Shimada K., Sato A., Takahashi E., Fukasawa T., Takahashi T., Ohka S., Taniguchi T., Honda E., Nomoto A., Ogura A., Kirikae T., Hanaki K.-I. Rapid detection of Pseudomonas aeruginosa in mouse feces by colorimetric loop-mediated isothermal amplification. Journal of Microbiological Methods. 2010;81(3):247–252. doi: 10.1016/j.mimet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Jia F., Xu L., Yan W., Wu W., Yu Q., Tian X., Dai R., Li X. A magnetic relaxation switch aptasensor for the rapid detection of Pseudomonas aeruginosa using superparamagnetic nanoparticles. Microchimica Acta. 2017;184(5):1539–1545. doi: 10.1007/s00604-017-2142-2. [DOI] [Google Scholar]
- Li J., Liu Q., Wan Y., Wu X., Yang Y., Zhao R., Chen E., Cheng X., Du M. Rapid detection of trace Salmonella in milk and chicken by immunomagnetic separation in combination with a chemiluminescence microparticle immunoassay. Analytical and Bioanalytical Chemistry. 2019;411(23):6067–6080. doi: 10.1007/s00216-019-01991-z. [DOI] [PubMed] [Google Scholar]
- Juan, C., Pena, C., & Oliver, A. (2017). Host and Pathogen Biomarkers for Severe Pseudomonas aeruginosa Infections. Journal of Infectious Diseases, 215, S44-S51. https://doi.org/10.1093/infdis/jiw299. [DOI] [PubMed]
- Kaur G., Raj T., Kaur N., Singh N. Pyrimidine-based functional fluorescent organic nanoparticle probe for detection of Pseudomonas aeruginosa. Organic & Biomolecular Chemistry. 2015;13(16):4673–4679. doi: 10.1039/C5OB00206K. [DOI] [PubMed] [Google Scholar]
- Kuang H., Wang W., Xu L., Ma W., Liu L., Wang L., Xu C. Monoclonal antibody-based sandwich ELISA for the detection of staphylococcal enterotoxin A. International Journal of Environmental Research and Public Health. 2013;10(4):1598–1608. doi: 10.3390/ijerph10041598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze A., Dilcher M., Abd El Wahed A., Hufert F., Niessner R., Seidel M. On-Chip Isothermal Nucleic Acid Amplification on Flow-Based Chemiluminescence Microarray Analysis Platform for the Detection of Viruses and Bacteria. Analytical Chemistry. 2016;88(1):898–905. doi: 10.1021/acs.analchem.5b03540.s001. [DOI] [PubMed] [Google Scholar]
- Liu L., Xu L., Suryoprabowo S., Song S., Kuang H. Development of an immunochromatographic test strip for the detection of ochratoxin A in red wine. Food and Agricultural Immunology. 2018;29(1):434–444. doi: 10.1080/09540105.2017.1401043. [DOI] [Google Scholar]
- Luo P., Chen X., Xiao J., Zhao Y., Wang Z. Rapid detection of iprodione in cucumber and apple using an immunochromatographic strip test. Food and Agricultural Immunology. 2019;30(1):701–712. doi: 10.1080/09540105.2019.1625309. [DOI] [Google Scholar]
- Sarabaegi M., Roushani M. A nano-sized chitosan particle based electrochemical aptasensor for sensitive detection of P. aeruginosa. Analytical Methods. 2019;11(43):5591–5597. doi: 10.1039/C9AY01509D. [DOI] [Google Scholar]
- Shan S., Lai W., Xiong Y., Wei H., Xu H. Novel Strategies To Enhance Lateral Flow Immunoassay Sensitivity for Detecting Foodborne Pathogens. Journal of Agriculture and Food Chemistry. 2015;63(3):745–753. doi: 10.1021/jf5046415. [DOI] [PubMed] [Google Scholar]
- Simoska O., Sans M., Fitzpatrick M.D., Crittenden C.M., Eberlin L.S., Shear J.B., Stevenson K.J. Real-Time Electrochemical Detection of Pseudomonas aeruginosa Phenazine Metabolites Using Transparent Carbon Ultramicroelectrode Arrays. ACS Sens. 2019;4(1):170–179. doi: 10.1021/acssensors.8b01152.s001. [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., Ouellette M., Outterson K., Patel J., Cavaleri M., Cox E.M., Houchens C.R., Grayson M.L., Hansen P., Singh N., Theuretzbacher U., Magrini N., Aboderin A.O., Al-Abri S.S., Awang Jalil N., Benzonana N., Bhattacharya S., Brink A.J., Burkert F.R., Cars O., Cornaglia G., Dyar O.J., Friedrich A.W., Gales A.C., Gandra S., Giske C.G., Goff D.A., Goossens H., Gottlieb T., Guzman Blanco M., Hryniewicz W., Kattula D., Jinks T., Kanj S.S., Kerr L., Kieny M.-P., Kim Y.S., Kozlov R.S., Labarca J., Laxminarayan R., Leder K., Leibovici L., Levy-Hara G., Littman J., Malhotra-Kumar S., Manchanda V., Moja L., Ndoye B., Pan A., Paterson D.L., Paul M., Qiu H., Ramon-Pardo P., Rodríguez-Baño J., Sanguinetti M., Sengupta S., Sharland M., Si-Mehand M., Silver L.L., Song W., Steinbakk M., Thomsen J., Thwaites G.E., van der Meer J.WM., Van Kinh N., Vega S., Villegas M.V., Wechsler-Fördös A., Wertheim H.F.L., Wesangula E., Woodford N., Yilmaz F.O., Zorzet A. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. The Lancet Infectious Diseases. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- Tang R., Yang H., Gong Y., You MinLi, Liu Z., Choi J.R., Wen T., Qu Z., Mei Q., Xu F. A fully disposable and integrated paper-based device for nucleic acid extraction, amplification and detection. Lab on a Chip. 2017;17(7):1270–1279. doi: 10.1039/C6LC01586G. [DOI] [PubMed] [Google Scholar]
- Tang Y., Ali Z., Zou J., Jin G., Zhu J., Yang J., Dai J. Detection methods for Pseudomonas aeruginosa: History and future perspective. RSC Advances. 2017;7(82):51789–51800. doi: 10.1039/C7RA09064A. [DOI] [Google Scholar]
- Tang Y., Ali Z., Zou J., Yang K., Mou X., Li Z.…He N. Detection of Pseudomonas aeruginosa Based on Magnetic Enrichment and Nested PCR. Journal of Nanoscience and Nanotechnology. 2014;14(7):4886–4890. doi: 10.1166/jnn.2014.8707. [DOI] [PubMed] [Google Scholar]
- Wang, J., Katani, R., Li, L., Hegde, N., Roberts, E. L., Kapur, V., & DebRoy, C. (2016). Rapid Detection of Escherichia coli O157 and Shiga Toxins by Lateral Flow Immunoassays. Toxins, 8(4). https://doi.org/10.3390/toxins8040092. [DOI] [PMC free article] [PubMed]
- Wang Y., Dou H., Chen K., Zhang H., Hu C. Development of a colloidal gold-based immunochromatographic test strip for the rapid, on-site detection of Pseudomonas aeruginosa in clinical samples. Scandinavian Journal of Infectious Diseases. 2011;43(5):329–338. doi: 10.3109/00365548.2011.552519. [DOI] [PubMed] [Google Scholar]
- Wang Z., Guo L., Liu L., Kuang H., Xu C. Colloidal gold-based immunochromatographic strip assay for the rapid detection of three natural estrogens in milk. Food Chemistry. 2018;259:122–129. doi: 10.1016/j.foodchem.2018.03.087. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang J., Liu L., Wu X., Kuang H., Xu C., Xu L. A colorimetric paper-based sensor for toltrazuril and its metabolites in feed, chicken, and egg samples. Food Chemistry. 2019;276:707–713. doi: 10.1016/j.foodchem.2018.10.047. [DOI] [PubMed] [Google Scholar]
- Ye L., Wu X., Xu L., Zheng Q., Kuang H. Preparation of an anti-thiamethoxam monoclonal antibody for development of an indirect competitive enzyme-linked immunosorbent assay and a colloidal gold immunoassay. Food and Agricultural Immunology. 2018;29(1):1173–1183. doi: 10.1080/09540105.2018.1523373. [DOI] [Google Scholar]
- Zhang X., Xie G., Gou D., Luo P., Yao Y., Chen H. A novel enzyme-free electrochemical biosensor for rapid detection of Pseudomonas aeruginosa based on high catalytic Cu-ZrMOF and conductive Super P. Biosensors and Bioelectronics. 2019;142:111486. doi: 10.1016/j.bios.2019.111486. [DOI] [PubMed] [Google Scholar]
- Zhong Z., Gao R., Chen Q., Jia L.i. Dual-aptamers labeled polydopamine-polyethyleneimine copolymer dots assisted engineering a fluorescence biosensor for sensitive detection of Pseudomonas aeruginosa in food samples. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2020;224:117417. doi: 10.1016/j.saa.2019.117417. [DOI] [PubMed] [Google Scholar]
- Zhou S., Kuang H., Liu L. Development of an ic-ELISA and colloidal gold strip for the detection of the beta-blocker carazolol. Food and Agricultural Immunology. 2020;31(1):217–230. doi: 10.1080/09540105.2019.1710113. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.