Abstract
The number of disease risk genes and loci identified through human genetic studies far outstrips our capacity to systematically study their functions. We applied a scalable genetic screening approach, in vivo Perturb-Seq, to functionally evaluate 35 autism spectrum disorder/neurodevelopmental delay (ASD/ND) de novo loss-of-function risk genes. Using CRISPR-Cas9, we introduced frameshift mutations in these risk genes in pools, within the developing mouse brain in utero, followed by single-cell RNA sequencing of perturbed cells in the postnatal brain. We identified cell type-specific and evolutionarily conserved gene modules from both neuronal and glial cell classes. Recurrent gene modules and cell types are affected across this cohort of perturbations, representing key cellular effects across sets of ASD/ND risk genes. In vivo Perturb-Seq allows us to investigate how diverse mutations affect cell types and states in the developing organism.
One sentence summary
An in utero Perturb-Seq genetic screen reveals recurrent and cell type-specific gene modules affected across a panel of psychiatric disorder risk gene perturbations during brain development.
Human genetic studies have uncovered associations between genetic variants in tens of thousands of loci and complex human diseases (1–3). In particular, analysis of trio-based whole-exome sequencing (WES) has implicated a large number of de novo loss-of-function variants contributing to risk of neurodevelopmental pathologies, including autism spectrum disorders and neurodevelopmental delay (ASD/ND) (4, 5). Compared to common variants identified by Genome-Wide Association Studies (GWAS), such de novo risk variants often have large effect sizes, are highly penetrant, and occur within a gene’s coding region, thus providing a crucial entry point for disease modeling and mechanistic studies.
However, a major challenge remains in identifying the point of action of each of these many risk genes. For example, ASD/ND comprises a broad collection of neurodevelopmental disorders with highly heterogeneous genetic contributions, including hundreds of highly penetrant de novo risk variant genes (6). Moreover, there is substantial diversity in the function of the gene products that these genes encode, precluding a clear a priori prediction of the underlying brain cell types, developmental processes, and molecular pathways affected during neurodevelopment (7). Few of these risk genes have been studied in animal or cellular models, and thus their function during brain development remains poorly defined. Due to the labor-intensive and time-consuming nature of generating and analyzing individual knockout animal models for functional investigation, it is crucial to develop phenotyping methods that are scalable, general-purpose, high-resolution, and high-content, to identify tissue- and cell-type specific effects of genetic perturbations in vivo.
To address these challenges, we developed in vivo Perturb-Seq, a scalable genetic screen, to investigate the function of large sets of genes at single-cell resolution in complex tissue in vivo. We applied in vivo Perturb-Seq in utero in mice to study the effect of a panel of ASD/ND risk genes on brain development.
In vivo Perturb-Seq to assess the function of ASD/ND risk genes
We chose ASD/ND candidate genes from a recently published WES study of 11,986 cases with 6,430 ASD/ND probands (8) (table S1). We initially prioritized 38 candidate genes (of which 35 were retained in the final analysis, table S1) that harbor de novo variants specific to ASD/ND patients within the broader class of neurodevelopmental disability (fig. S1A, table S1). These ASD/ND risk genes are expressed in human brain tissue, as assessed by the BrainSpan bulk RNA-seq dataset (9); some are highly expressed at embryonic stages, and others highly expressed from early postnatal to adult stages (fig. S1B). Based on mouse cortical single-cell RNA sequencing (scRNA-seq) data, the orthologs of these ASD/ND risk genes are expressed in diverse cell types (fig. S2) (E18.5 data from the 10x Genomics public dataset (10); P7 data from this work). Thus, these ASD/ND genes could, in principle, act in many different cell types and temporal frames, requiring scalable methods to test gene function across a range of cell types and developmental events.
For in vivo Perturb-Seq, we used Cas9-mediated genome editing (11–13) in a pooled approach to introduce mutations in each of the ASD/ND risk genes within progenitor cells of the mouse developing forebrain in utero, followed by scRNA-seq at P7 to read out both a barcode identifying the perturbation and the expression profile of the perturbed cells (Fig. 1A). Specifically, we used a transgenic mouse line that constitutively expresses Cas9 (14), and delivered pools of gRNAs targeting the different risk genes by lentiviral infection into the lateral ventricles of the developing embryo in utero. Each lentiviral vector contained two different gRNAs targeting the 5’-end coding exons of one ASD/ND gene (to enhance knockout efficiency), and a blue fluorescent protein (BFP) reporter with a unique barcode corresponding to the perturbation identity (11–13). To minimize vector recombination, we packaged each lentivirus separately and then pooled viruses at equal titers.
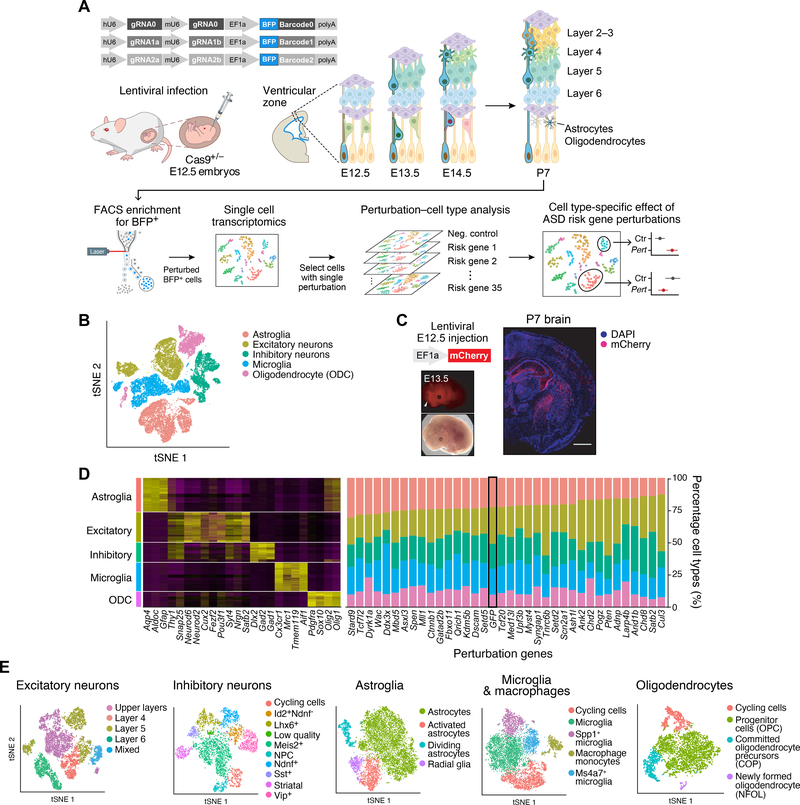
Figure 1. In vivo Perturb-Seq to investigate functions of a panel of ASD/ND risk genes harboring de novo variants.
(A) Schematics of the in vivo Perturb-Seq platform, which introduces mutations in individual genes in utero at E12.5, followed by transcriptomic profiling of the cellular progeny of these perturbed cells at P7 via single-cell RNA sequencing (scRNA-seq). (B) tSNE of five major cell populations identified in the Perturb-Seq cells. (C) In vivo Perturb-Seq lentiviral vector carrying an mCherry reporter drives detectable expression within 24h, and can sparsely infect brain cells across many brain regions. Scale bar is 1000μm. (D) Cell-type analysis of in vivo Perturb-Seq of ASD/ND de novo risk genes. Canonical marker genes were used to identify major cell clusters (left), and cell-type distribution in each perturbation group (right). Negative control (GFP) is highlighted by a black rectangle. (E) tSNEs showing the subclusters of each of the five major cell types, identified by re-clustering each cell type separately.
We injected a pool of lentiviruses with equal gRNA representation into the ventricles of the developing forebrain at E12.5 (Fig. 1A). In this approach, lentiviral injection leads to infection of neural progenitors lining the lateral ventricle of the developing forebrain, including progenitors of the neocortex and the ganglionic eminences. Since lentiviral vectors integrate into the genome, the progeny of the infected progenitors are labeled by BFP and carry a perturbation barcode corresponding to the target ASD/ND gene.
Both immunohistochemical analysis and scRNA-seq of BFP+ cells at P7 showed that the Perturb-Seq vectors were expressed across a variety of neuronal and glial cell types in the cortex (Figs. 1B–C and S3A–B). While microglia originate mostly from outside the targeted germinal zones, we detected lentiviral vector expression in cortical microglia, indicated by the presence of BFP as well as perturbation barcode expression, across multiple individual experiments (fig. S4E–F). While it is unclear how microglia are labeled in our experimental procedure, it is possible that the in utero injection could have led to either local lesions that recruited and expanded the number of microglia along the injection tract, or that microglia were labeled within the parenchyma along the same tract. Overall, our approach allowed us to examine the effects of each perturbation across a wide range of cell types from distinct brain regions (i.e., cortical projection neurons, interneurons, astroglia, oligodendroglia, etc.), and, importantly, under sparse labeling conditions where less than 0.1% of cells in the cortex were perturbed, and thus development of individual perturbed cells is highly unlikely to be affected by perturbed neighbors (fig. S3A–C).
In vivo Perturb-Seq targets diverse cell types without affecting overall cell type composition
We performed the experiment with 18 different cohorts of pregnant mice, for a total of 163 embryos, each subjected to the entire pool of perturbations. We micro-dissected and dissociated cortical tissues separately at P7, FACS-enriched the perturbed cells by selecting for BFP expression, and used droplet-based scRNA-seq to obtain each cell’s expression profile along with its perturbation barcode. The cell survival rate after FACS was 78%, and we confirmed a 40–70% frameshift insertion/deletion for each gRNA target among the infected cells (fig. S3D–E).
This multiplexed experimental design allowed us to test the cell-autonomous effect of all perturbations against the effect of a negative control construct targeting the endogenous GFP in the Rosa26 locus, thus controlling for effects related to viral infection, among other confounders. To minimize batch-dependent variation, the control construct was included in the same pool as the perturbation vectors (fig. S3F). After quality control, we retained for further analysis a total of 46,770 neocortical cells across 17 high-quality experimental batches. We partitioned the cells into major cell classes using Louvain clustering (15) and annotated them by known marker gene expression (16, 17) (Fig. 1D).
We focused on five broad cell populations from this cortical dataset for downstream analysis: cortical projection neurons (8,450 cells), cortical inhibitory neurons (5,532 cells), astrocytes (9,526 cells), oligodendrocytes (4,279 cells), and microglia/macrophages (8,070 cells) (thus excluding vascular, endothelial, and contaminant hippocampal and striatal cells). We further filtered out some remaining low-quality cells in these five major cell classes, retaining 35,857 high-quality cells (median of 2,436 detected genes per cell overall, and median of 4,084 genes in the projection neuron cluster, as expected from their large size and known high RNA content (fig. S3G)). We subclustered each of the five major cell types separately and annotated biologically meaningful subclusters (Figs. 1E and S6).
From inspecting the perturbation barcodes from the lentiviral constructs, 92% (33,231 cells) of the cells in these five major cell classes had at least one perturbation read assigned to them, and 50% had barcodes for a single gene (fig. S4A–C, 18,044 cells), reflecting the low multiplicity of infection (fig. S4D). As it is rare for multiple ASD/ND loss-of-function risk gene mutations to co-occur in patients, we focused on the 18,044 cells that carried a single perturbation. We found a median of 338 cells per perturbation: after excluding perturbations with <70 perturbed cells, we retained 35 ASD/ND risk gene perturbations. BFP from the lentiviral vector was robustly detected as one of the most highly expressed genes in all retained cells (fig. S4E). The BFP detection rate in each cell type correlated with the average number of genes detected (fig. S4F), further supporting the reliability of the readout.
ASD/ND risk gene perturbations had a very modest effect on the presence and proportions of these five major cell types relative to the negative control (targeting the GFP gene). Only loss of Dyrk1a had a significant effect on cell type composition, increasing the proportion of oligodendrocytes and reducing the proportion of microglia/macrophages [FDR-corrected P<0.05 using Poisson regression (18)] (Figs. 1D and S5).
Co-varying gene modules associate with cell states
To assess whether ASD/ND genetic perturbations caused molecular changes and alterations in cell states, we first sought to define gene modules that co-vary within each of the five broad cell classes. As previous work has shown (11–13, 19), focusing on gene modules instead of individual genes provides more statistical power to detect biologically-meaningful perturbation effects using fewer cells than would be required for single gene-level analysis, and can capture diversity both within and across cell types.
We first tested if the expression of known Gene Ontology (GO) gene sets (20) was affected, by calculating a gene-set expression score for each cell and fitting a linear regression model to this score. After correcting for multiple hypothesis testing, no GO terms were significantly altered by any perturbation (Table S8). However, this approach is limited by the large number of tests performed (one test per GO term per cell type per perturbation, for a total of 510,265 tests), as well as the limited number of GO terms relevant to the developing cortex.
We therefore sought to identify gene modules de novo in our data using two approaches: Weighted Gene Correlation Network Analysis (WGCNA), which identifies “modules” of genes with correlated expression, and structural topic modeling (STM), which attempts to reduce the dimensionality of the gene expression matrix and returns “topics” corresponding to the components of this representation (Figs. 2A and S6–8, Tables S2–3) (21, 22). We performed these analyses for each of the five major cell clusters separately, to better identify effects associated with specific cell types; our nomenclature for the modules incorporates the cell cluster analysis it is derived from (e.g., PN1 represents a module identified by analysis of projection neurons). Each of these analyses used the full set of perturbations, in order to identify effects shared across multiple perturbations. We focused our subsequent analysis on the 14 modules identified by WGCNA, because they were highly correlated with one or more topics returned by STM (fig. S7).
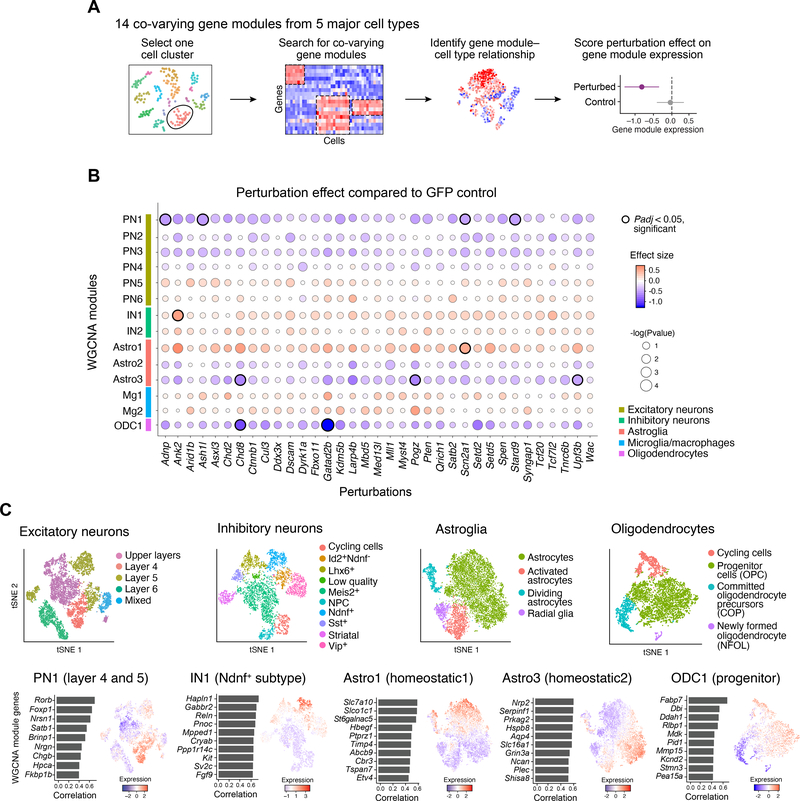
Figure 2. In vivo Perturb-Seq reveals cell-type specific effects of ASD/ND risk gene perturbations.
(A) Schematic illustration of the Perturb-Seq analysis pipeline. (B) ASD/ND risk gene perturbation effects in different WGCNA gene modules compared to GFP controls. Dot color corresponds to effect size, dot size corresponds to negative base 10 log(P-value). Module gene lists are presented in Table S2. P-values were calculated from linear modeling, Padj was calculated by Benjamini & Hochberg FDR correction. (C) The four cell types and five gene modules that were altered by ASD risk gene perturbations. Top row: subcluster tSNE of each cell class (repeated from Fig. 1E for ease of comparison). Bottom row: feature plots of gene module expression scores and the top correlated genes within each module across the relevant cell class.
The 14 WGCNA modules comprised two broad categories. Some reflect common biological processes and were present across multiple cell subsets (e.g., cell cycle, differentiation, maturation). For example, module PN2 is associated with genes involved in neurite development and varied across cells in multiple projection neuron subclusters (fig. S6A). Others represent cell type-specific features unique to only some subsets (e.g., subcluster-specific features of a neuronal sub-type). For example, module PN1 is a module associated with two defined subclusters of projection neurons of Layer 4 and Layer 5 (fig. S6A).
ASD/ND gene perturbations affect cell states in multiple cell classes
As the WGCNA analysis is expected to recover gene modules associated with many kinds of variation across the data, we next tested the association of each risk gene perturbation with the 14 individual WGCNA gene modules. We estimated the effect size of each perturbation on each gene module by fitting a joint linear regression model, estimating how module gene expression in cells from each perturbation group deviated from the GFP control cells (Fig. 2A–B). To ensure that no single perturbation or batch dominated the linear model, we down-sampled the cells in each cell category such that no perturbation had more than two times the median number of cells over all perturbations. This linear regression analysis was performed on mean-centered and standard deviation-scaled module scores, so effect sizes can be interpreted in terms of standard deviations from the population mean (Fig. 2B). Our modeling approach assumes that module expression in individual cells is independent after conditioning on the experimental batch, and that noise is normally distributed. To evaluate the effects of these assumptions, we also compared alternative approaches, including a linear mixed model-based approach and a permutation-based approach (fig. S9, table S9).
Perturbations in 9 ASD/ND genes (Adnp, Ank2, Ash1l, Chd8, Gatad2b, Pogz, Scn2a1, Stard9, and Upf3b) had significant effects across 5 modules (Fig. 2B, highlighted circles, compared to the GFP control, FDR corrected P<0.05, Table S4): a module associated with projection neurons of Layer 4 and 5 (PN1, affected by perturbations in Adnp, Ash1l, Scn2a1, and Stard9); modules representing two distinct homeostatic signatures in astrocytes (Astro1 affected by perturbation of Scn2a1, and Astro3 affected by perturbations of Chd8, Pogz, and Upf3b); a module associated with oligodendrocyte progenitor cells (ODC1, Chd8 and Gatad2b); and a module associated with Ndnf+ interneurons (IN1, Ank2) (Fig. 2C, fig. S6).
Notably, the oligodendrocyte progenitor module (ODC1) also had a significant amount of its variation across the oligodendrocyte cell cluster explained by the perturbation state overall (van der Waerden test, a non-parametric alternative to ANOVA analysis, FDR corrected P<0.05) (fig. S5C), suggesting that this module represents convergent effects across different perturbed genes. Collectively, the data indicate that a selected group of perturbations was able to affect recurrent gene modules with cell-type specificity, and point to some convergent effects across diverse ASD/ND risk genes.
Single perturbation of Ank2 confirms Perturb-Seq effect on an interneuron gene expression module
In our multiplex in vivo Perturb-Seq results, Ank2 perturbation led to increased expression of an interneuron module (IN1) (FDR corrected P<0.05, fig. S10). This module was strongly correlated with a subcluster of inhibitory interneurons expressing Ndnf (fig. S10C, D), and contains genes such as Kcnq5 (a voltage-gated potassium channel) and Gabbr2 (GABA receptor subunit) (fig. S6B and table S2). To validate our finding from the pooled Perturb-Seq experiment, we performed a single perturbation targeting either Ank2 or GFP (control), followed by scRNA-seq of neocortical cells at P7, resulting in 2,943 and 1,716 high-quality cells, respectively.
The individual perturbation experiment confirmed the results from the pooled Perturb-Seq screen. Ank2-perturbed cells were present across all cell types and overall proportions of cells were not significantly changed (fig. S10B). Within the Ndnf+ interneurons, Ank2 perturbation led to upregulation of the IN1 module (FDR corrected P<0.05, fig. S10E), confirming the Perturb-Seq result. This finding indicates that multiplexing perturbations in the pooled approach does not significantly distort the results observed for an individually perturbed gene.
Ank2 encodes an ankyrin protein and is expressed broadly in excitatory and inhibitory neurons as well as glial cells in the brain (23). Studies examining Ank2 loss-of-function suggest that it is involved in axonal morphology, connectivity, and calcium signaling in excitatory neurons (24–27). Our Perturb-Seq data suggests a role of Ank2 in the Ndnf+ interneuron subtype during cortical development, in addition to its known roles in excitatory neurons.
The ASD/ND risk genes Chd8 and Gatad2b alter gene modules in oligodendrocyte progenitors
In our Perturb-Seq experiment, Chd8 and Gatad2b perturbations significantly decreased the expression of the ODC1 module in the oligodendrocyte cluster (Fig. 3A–D, FDR corrected P<0.05; see alternative measurement of effect size fig. S11A, estimated by log transcripts per million (TPM) gene expression differences). The ODC1 module is highly expressed in cycling cells and oligodendrocyte precursor cells (OPC), and lowly expressed in committed oligodendrocyte progenitor cells (COP) and newly formed oligodendrocytes (NFOL), suggesting that this module is linked to oligodendrocyte maturation (Fig. 3A), and therefore that perturbation in Chd8 and Gatad2b might accelerate oligodendrocyte maturation. This is consistent with recent reports that Chd8 loss-of-function potentiates an impaired OPC development phenotype caused by deletion of Chd7 (28).
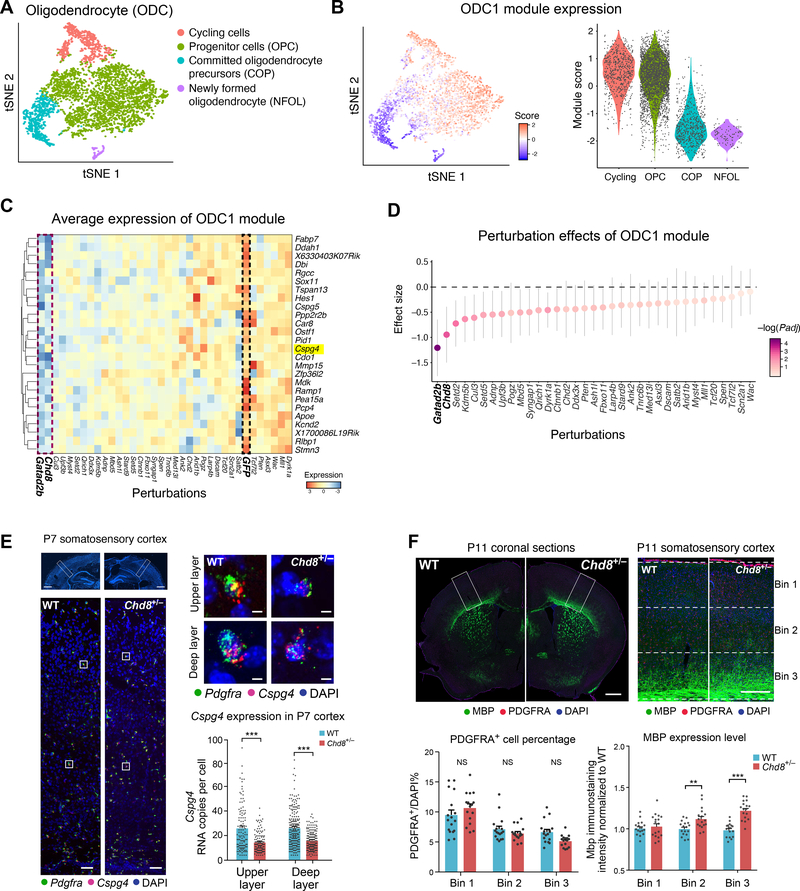
Figure 3. Perturbation effect in oligodendrocytes and validation in the Chd8+/− mouse model.
(A) tSNE of oligodendrocyte subtypes from the Perturb-Seq data. (B) The ODC1 gene module expression score in each cell (left) and in each subcluster (right). (C) Average expression of genes in the ODC1 gene module (by row) in each perturbation group (by column), scaled by row. (D) Effect size of each perturbation on the ODC1 gene module compared to the control group. Note that the perturbation effects of the different genes present a continuous gradient. Error bars represent 95% confidence intervals. (E) In situ hybridization for Cspg4, a gene in module ODC1 that is a known marker of oligodendrocyte precursor cells (OPC), in the somatosensory cortex of P7 Chd8+/− and wild-type littermates. The bottom images of represent the higher magnifications of top images, and the right images represent higher magnifications for each cell. Right: quantification of Cspg4 expression in P7 cortex of Chd8+/− and wild-type littermates. Each dot represents the gene expression value from one cell; error bars represent standard error of the mean (n=3 animals per genotype). Scale bar is 1000μm (left bottom panel), 100μm (left top panel), and 10μm (right panel), respectively. (F) Immunohistochemistry for PDGFRA and MBP (markers for immature OPC and mature oligodendrocytes, respectively), PDGFRA+ cell counts, and distribution of MBP expression, in the somatosensory cortex of P11 Chd8+/− animals and wild-type littermates. Scale bar is 1000μm (left panel) and 250μm (right panel), respectively.
We further investigated and validated this result by examining oligodendrocyte development in a Chd8 germline heterozygous mutant model (as homozygous mutation is embryonic lethal (29)), using several orthogonal methods. First, we used in situ hybridization for two canonical OPC markers known to be involved in fate specification, Cspg4 (a member of the ODC1 module) and Pdgfra. Both were downregulated in P7 Chd8+/− cortex (Figs. 3E and S11B–D), consistent with our in vivo Perturb-Seq results. Second, we used immunohistochemistry to examine a later developmental time point, P11. OPC cell number (e.g., PDGFRA+ cells) did not show significant differences between the WT and Chd8+/− littermates, also consistent with in vivo Perturb-Seq; however, cells positive for the MBP protein, a marker of myelinating oligodendrocytes, were increased in number and displayed elevated MBP levels in the Chd8+/− mutant (FDR corrected P<0.05, nonparametric ANOVA test) (Fig. 3F). In combination with the Perturb-Seq result showing reduction in the signature of oligodendrocyte progenitors and of the progenitor-expressed ODC1 module in Chd8-perturbed cells, this suggests that Chd8 perturbation may result in acceleration of the progressive increase in MBP levels that occurs postnatally. These data further demonstrate that in vivo Perturb-Seq has the power to identify cell type-specific molecular changes similar to those observed in a single-gene, germline-modified mouse model.
Perturb-Seq gene modules are conserved between human and mouse
To establish whether the perturbed gene modules identified in the mouse cerebral cortex are conserved in human cells, we examined the expression of each module across multiple scRNA-seq datasets from human tissues: adult human cortex (30), ASD donor cortex with matched controls (31), fetal human cortex (32), and 3 month and 6 month-old human brain organoids (33) (Fig. 4A–B). In the fetal brain and the 3-month brain organoid samples, glial cell types were sparsely represented due to the early developmental stages of the samples (fig. S12A). We identified human genes that had 1:1 orthologs to the mouse genes in each module, and asked whether the modules were conserved, using two metrics: whether the orthologous genes were also expressed in the corresponding cell type in the human datasets, and whether the expression of the genes in each module co-varied across single cells (as estimated by correlation), reflecting the degree of “modularity” of these mouse gene programs in humans.
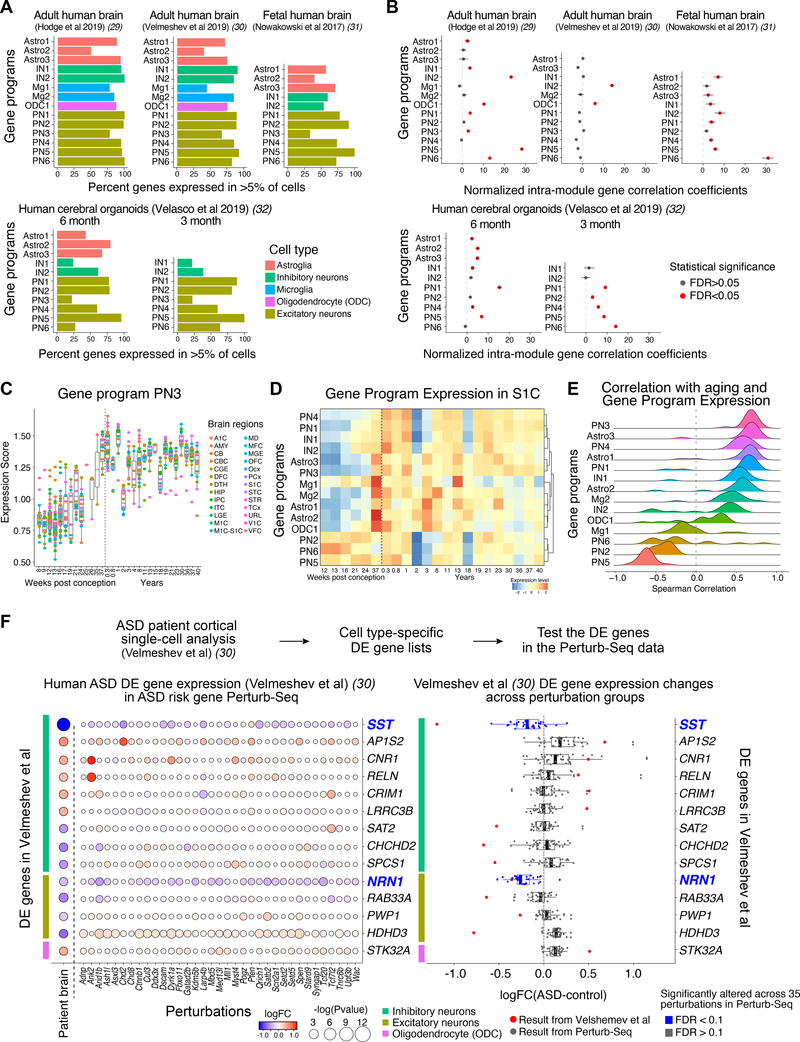
Figure 4. Cell-type specific gene modules from Perturb-Seq are conserved in developing human brains.
(A) Percent of genes with a human orthologue expressed in >5% of cells of the associated cell type in scRNA-seq datasets from the human brain or human brain organoids. (B) Normalized average pairwise correlation of gene expression within each gene module in the human brain or human brain organoids. Correlation values were normalized to the mean correlation from the background distribution, and divided by the standard deviation of the background distribution. Correlations are shown for modules with at least 4 genes after filtering out genes expressed in less than 5% of cells. Bars represent 95% confidence intervals. Red color represents statistical significance (FDR<0.05). (C) Expression of module PN3 over developmental time in human brain tissues across regions (BrainSpan data) (9). (D) Expression of each module over developmental time in human primary somatosensory cortex S1C (BrainSpan) (9). (E) Distribution of the Spearman correlation of module expression with age in human brain data over various brain regions (BrainSpan) (9). (F) Differential gene expression analysis of human prefrontal cortical samples from ASD donors and controls. Left: Expression of differentially expressed (DE) genes across cell types (color bars) from Velmeshev et al (31) (rows) in the Perturb-Seq data across a panel of ASD/ND risk genes (columns). Right: DE gene expression changes in Perturb-Seq data (black dots; each dot represents an ASD/ND risk gene perturbation) compared to DE values for the 14 genes found to be DE in ASD patients in the Velmeshev et al dataset (31) (FDR<0.2) (red dots). The two highlighted genes, SST and NRN1, showed decreased expression in the Perturb-Seq data (FDR<0.1), consistent with the ASD patient dataset.
The expression of each module was largely conserved in all human datasets, with different modules showing distinct levels of conservation of expression in each dataset (Fig. 4A). Some modules like PN1, PN2, and PN5 displayed high levels of conservation of expression (with at least 75% of the genes in these modules being expressed by at least 5% of cells in the corresponding associated cell type) across all datasets. The proportions of the genes expressed in the corresponding cell types in human tissues were generally lower than in mouse tissues (fig. S12B).
We further calculated whether the co-variation of expression of the genes in each module (their “modularity”) was also comparable in humans. To do so, for each module and each dataset we calculated the average pairwise expression correlation coefficient between the genes in a given module, and compared it to a module-specific null-distribution based on random gene sets with similar expression levels, to calculate both a P-value for the correlation of our modules, and a normalized correlation coefficient. 8 out of 14 modules showed greater intra-module correlation than a comparable random gene set in the adult human brain dataset from Hodge et al (30) (Fig. 4B). Correlation also increased with the age of the human samples across brain regions of the BrainSpan dataset (9) (Figs. 4C–E and S13). As a control, we used the same approach to calculate the expression and modularity of each gene module in non-associated cell types. We found that the modularity was decreased in non-associated cell types (fig. S12D–E), reflected by both the proportion of comparisons with significant correlation and by the strength of the significant correlations, suggesting that our modules reflect cell type-specific effects.
Altogether, our results suggest that expression and modularity of most gene modules in the mouse are conserved in human brain tissue, pointing at potential shared functions, and suggesting that processes identified as affected in our Perturb-Seq experiments are relevant to biological processes that may be developmentally regulated in the human brain.
Mouse Perturb-Seq results are correlated with expression changes in ASD patient brain tissues
Finally, we explored whether the effects observed in mouse Perturb-Seq may be similar to changes observed in postmortem brains of ASD patients. To this end, we compared our data to a single-nucleus RNA-seq (snRNA-seq) dataset of postmortem ASD brain samples (31), and bulk RNA-seq of postmortem psychiatric disorder brain samples from the PsychEncode project (34).
Using a dataset of snRNA-seq profiles from 15 ASD donors and 16 controls (31), we defined differentially expressed (DE) genes in each cell type using a statistically conservative pseudobulk-based analysis with DESeq2 (35, 36), correcting for age, sex, and patient-to-patient variability. We identified genes that were differentially expressed between patients and controls in at least one of three major cell types (inhibitory neurons, excitatory neurons, or oligodendrocytes) with FDR < 0.2, and selected those that have 1:1 orthologs in mice, resulting in 14 genes (Fig. 4F, Table S10).
We then compared these 14 genes to our Perturb-Seq data and asked if these ASD-patient DE genes were also affected by the 35 ASD risk gene perturbations in our dataset. We aggregated the effects of all 35 perturbations, and asked whether the aggregated gene expression changes agreed more strongly with the gene expression changes in the ASD patient data than would be expected by chance. For each ASD patient DE gene, we took its mouse orthologue and calculated the median fold change of expression (logFC) over all perturbations in the Perturb-Seq data. We then compared this logFC with the corresponding logFC in the ASD patient data, and generated an agreement score for each gene, defined as a high median logFC and a similar direction of change as in the human data. We then binned genes by their expression, and compared each ASD patient DE gene to others in the same bin to extract p-values (with FDR correction). From this analysis, we identified two genes, SST in interneurons and NRN1 in excitatory neurons, both of which showed decreased expression in ASD patients and were likewise significantly decreased in expression across our panel of perturbations (FDR < 0.1), albeit with different effect sizes (Fig. 4F, Table S10). This indicates that despite the different developmental stages, high clinical heterogeneity in ASD, and patient genetic diversity, similar genes and cell types can be identified as affected in both our analyses and in studies of human patient tissue.
We also analyzed the 14 gene modules reported in the PsychEncode study of 700 bulk RNA-seq samples of human cortex from a panel of psychiatric disorders (34). 6 of the 14 modules previously reported to be altered in the ASD patients in the PsychEncode analysis were also significantly affected across 8 of our ASD/ND risk gene perturbations (fig. S14). Although these analyses are limited by the relatively few available datasets of ASD patient brain samples, they suggest that our Perturb-Seq experiments can identify gene program abnormalities seen in human ASD patients.
Discussion
In vivo Perturb-Seq can serve as a scalable tool for systems genetic studies of large gene panels to reveal their cell-intrinsic functions at single-cell resolution in complex tissues. In this work, we demonstrated the application of in vivo Perturb-Seq to ASD/ND risk genes in the developing brain; this method can be applied across diverse diseases and tissues.
ASD/ND affects brain function profoundly, but its cellular and molecular substrates are not yet defined. The large number of highly penetrant de novo risk genes implicated through human genetic studies offers an entry point to identify the cell types, developmental events, and mechanisms underlying ASD/ND. However, this requires scalable methods to define the function of risk-associated genes with cell-type specificity. Using Perturb-Seq to functionally test large gene sets in the developing embryo, we observed gene expression changes linked to ASD/ND genes in different cell types and processes. Within the power of the analysis that can be achieved with the number of cells that can be reasonably sequenced, we find that some recurrent modules are affected across more than one ASD/ND risk gene perturbation. It is likely that this represents an underestimation of the number of convergent modules across perturbations which might be revealed by larger-scale experiments using greater numbers of cells.
We were particularly interested in validating the observed effects of Ank2 perturbation, because of its known roles in the brain. Ank2 encodes an ankyrin protein and is expressed broadly in excitatory and inhibitory neurons as well as glial cells in the brain (23). Ankyrin homologs interact with ion channels in many neuronal types, and Ankyrin-G has been shown to stabilize GABAergic synapses (37). The roles of Ank2 in the brain have largely been studied in the context of excitatory neurons. Ank2 loss-of-function results in hypoplasia of the corpus callosum and pyramidal tract, and ultimately optic nerve degeneration (24), suggesting that it is required in the maintenance of premyelinated axons in excitatory neurons in early neurodevelopment. Ank2 mutants showed misregulation of intracellular calcium homeostasis and calcium channel expression in excitatory neurons (25, 26), as well as increased axonal branching and ectopic connectivity (27). Our Perturb-Seq data suggests an additional role of Ank2 in the Ndnf+ interneuron subtype, along with its known roles in excitatory neurons.
In addition to neurons, oligodendrocytes and astrocytes were also affected by several perturbations. Oligodendrocytes modulate and consolidate neural circuit refinement, and abnormal maturation of oligodendrocytes may be linked to long-lasting changes in neural wiring and brain function (38). One of the risk genes, Chd8, encodes a protein that binds directly to β-catenin to recruit histone proteins and negatively regulates the Wnt signaling pathway, which plays a crucial role in neuronal progenitor proliferation and differentiation in the forebrain (39–42). Our results showed that Chd8 modulates gene modules for oligodendrocyte differentiation and maturation, consistent with previously reported ChIP-Seq results showing that CHD8 interacts directly with OPC maturation genes at perinatal stages of development (28, 43).
Although we focused on the perinatal neocortex in this study, in vivo Perturb-Seq can be applied to study gene functions systematically across other tissues and developmental ages, to reveal tissue-specific as well as broadly-distributed gene functions. This approach can uncover both the impact of individual disease-associated genes and of combinations of genes and the overall set of processes that they affect. Our findings underscore the importance of using single-cell profiles as a rich, comprehensive, and interpretable phenotypic readout. With advances in other single-cell profiling approaches (e.g., single-cell ATAC-seq (44), single-cell multi-omics (45), and spatial genomics (46, 47)), we expect in vivo Perturb-Seq to be coupled in the near future with diverse readouts to better define the function of disease-risk associated variants, from molecular mechanisms to non-cell autonomous effects in tissues. Spatial transcriptomics in particular should be well suited for combination with in vivo Perturb-Seq, and should help uncover non-cell autonomous effects. In vivo Perturb-Seq can enable discoveries of pathways and cell types affected in heterogenous genetic pathologies, directing downstream studies and informing the development of refined models for genetic disorders and mechanistic studies as we move from genetic variants to function.
Methods Summary
In vivo Perturb-Seq experiment
The backbone plasmid contains antiparallel cassettes of two gRNAs (Table S5) under mouse U6 and human U6 promoters, and the EF1a promoter to express puromycin, BFP, and a polyadenylated barcode unique to each perturbation. Cloning and lentiviral packaging of the 38 vectors were done individually.
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees (IACUC) of Harvard University and of the Broad Institute of MIT and Harvard. In utero lentiviral injection into the lateral ventricles was performed at E12.5 in Cas9 transgenic mice (14) (4–6 month old, Jax #026179), and each single-cell library was made by combining the BFP+ cells from 1–3 litters (4–20 animals) of P7 animals harvested on the same day. Tissue dissociation was performed with the Papain Dissociation kit (Worthington, #LK003152). The FACS-purified cells were sorted into cold Hibernate A/B27 medium and subjected to single-cell RNA sequencing library preparation. Our analysis comprises 17 independent libraries of Perturb-Seq cells.
Single-cell RNA sequencing libraries were created using the Chromium Single Cell 3’ Solution v2 kit (10x Genomics) following the manufacturer’s protocol. Each library was sequenced with Illumina NextSeq high-output 75-cycle kit with sequencing saturation above 70%. Dial-out PCR was performed to extract the perturbation barcode in each cell.
We identified perturbation barcodes by two complementary methods. We first used the dial-out sequences to create a cell-by-perturbation UMI count matrix by a modification of from the original Perturb-Seq work (12). In addition, we extracted barcode sequences from the 10x Genomics Cell Ranger bam file. Reads were then assigned to the perturbation they mapped best. Cell barcodes and UMIs were extracted, and a cell-by-perturbation UMI count matrix was created. We then only kept cells for which either i) the assigned 10x and dialout perturbations agree or ii) the cell was assigned to a perturbation by one method but not assigned to any perturbation in the other.
Perturb-Seq analysis
UMI count data was loaded into R and processed using the Seurat v 2.2 package (48). Clusters were assigned to cell types based on marker genes from the literature, mousebrain.org (16), and DropViz.com (23). We focused only on cells of 5 key types (projection neurons, inhibitory neurons, oligodendrocytes, microglia/macrophages, and astroglia) and removed the rest.
WGCNA and Structural topic modelling (STM) were performed for each cell cluster based on the published pipelines (21, 22). Linear regression was used to test the relationship between perturbations and WGCNA gene scores, correcting for batch and number of genes. To test for correlations between perturbations and topics, the theta matrix (the matrix containing proportions of topics per cell) was extracted from the STM matrix. For each topic, linear regression was used to test how the per-cell proportions for each topic related to perturbations (after setting GFP to be the reference perturbation), correcting for nGene and batch.
RNA in situ hybridization and immunohistochemistry
Multiplexed RNAscope fluorescent in situ hybridization and immunohistochemistry was performed on fixed-frozen tissue. Probes against the following mRNAs were used: Pdgfra, Cspg4, and Fezf2 (ACDBio). The antibodies and dilutions were: Mouse anti-NeuN antibody (mab377, 1:500; Millipore), Mouse anti-GS antibody (mab302, 1:500; Millipore), Goat anti-Pdgfra antibody (AF1062, 1:200; R&D System), Rabbit Iba1 antibody (019–19741, 1:400; Wako), Chicken anti-GFP antibody (ab16901, 1:500; Millipore), Mouse anti-Satb2 (ab51502, 1:50; Abcam), Rat anti-Ctip2 (ab18465, 1:100, Abcam), Rabbit anti-Sox6 (ab30455, 1:500; Abcam), Rat anti-Mbp (mab386, 1:100; Millipore). We double-blinded the staining, imaging, and quantifications.
Analysis of human single nucleus or single cell RNA-seq data
For each single cell/nucleus human dataset, the UMI count matrix and metadata were downloaded and processed with Seurat to create Seurat objects. Cell types were extracted from the metadata, and combined into more general cell types, namely: Microglia, Astroglia (including Radial Glia), Inhibitory neurons, Excitatory neurons, Oligodendrocytes, and other. For differential expression analysis for data from Velmeshev et al (31), we removed data from all individuals of <12 years of age and separated PFC and ACC regions. For each cell type in each region a pseudobulk profile was constructed and genes expressed in <5% of cells or with <10 reads were removed. DESeq2 v 1.20.0 (35) was then used to perform differential expression analysis between the ASD patients and the controls, correcting for sex and age. We then extracted all genes with 1:1 mouse orthologs (BioMart) and calculated FDR corrected P-values on these genes for both ACC and PFC. Only analysis on the PFC yielded significant hits, which are presented in Fig 4F.
To compare these results to the Perturb-Seq data, for each human DE gene, an agreement score was calculated by taking the absolute value of its mouse orthologues’ median logFC over all perturbations (calculated with Limma), and giving it a positive sign if its direction agreed with that of the human data, a negative sign otherwise. Finally, genes were binned by expression, and p-values were calculated for each gene by comparing the agreement scores to other genes in the same bin.
Detailed procedures for the experiments and data analyses are described in the supplementary materials.
Supplementary Material
Acknowledgements
We thank C. Dulac, W. Gilbert, M. Meselson and C.I. Bargmann for critical reading of our manuscript; L. Gaffney, A. Hupalowska, R. Macrae, J. Brown, S. Smith, as well as members of the Levin lab, Regev lab, Zhang lab, and Arlotta lab for technical and intellectual support.
Funding: This work is supported by the Stanley Center for Psychiatric Research at the Broad Institute, NIH grants (U01MH115727, R01MH096066, P50MH094271 Conte Center) (to P.A); NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation and Harvard William F. Milton Grant (to X.J.); Stanley Center for Psychiatric Research at the Broad Institute (to J.Z.L.); The Klarman Cell Observatory, HHMI and an NHGRI Center for Cell Circuits CEGS grant (to A.R.); NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201), HHMI, New York Stem Cell and Mathers Foundations, the Poitras Center for Affective Disorders Research at MIT, the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT, and J. and P. Poitras (to F.Z.); F.Z. is a New York Stem Cell Foundation–Robertson Investigator.
Footnotes
Competing interests: P.A. is a SAB member in System 1 Biosciences and Foresite Labs and is a co-founder of FL60. AR is a co-founder of and equity holder in Celsius Therapeutics, Equity holder in Immunitas. Until July 31, 2020, A.R. was an SAB member of ThermoFisher Scientific, Syros Pharmaceuticlas, Asimov, and Neogene Therapeutics. From August 2020, A.R. is an employee of Genetech. F.Z. is a co-founder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences. X.J., S.S., A.R., F.Z. and P.A. are co-inventors on in vivo Perturb-Seq and CRISPR inventions filed by the Broad Institute relating to the work in this manuscript.
Data and materials availability:
Data generated for this study are available through the Gene Expression Omnibus (accession GSE157977) as well as the Broad single cell portal (https://singlecell.broadinstitute.org/single_cell/study/SCP521). The analysis pipeline is deposited on GitHub repository (https://github.com/klarman-cell-observatory/ivPerturbSeq) and Zenodo (DOI: 10.5281/zenodo.4019534). All other data are available in the manuscript or the supplementary materials.
Reference
- 1.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH, Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22, 345–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia C Working Group of the Psychiatric Genomics, Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jostins L et al. , Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satterstrom FK et al. , Novel genes for autism implicate both excitatory and inhibitory cell lineages in risk. bioRxiv, 484113 (2018). [Google Scholar]
- 5.Sanders SJ et al. , De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JA, Penagarikano O, Belgard TG, Swarup V, Geschwind DH, The emerging picture of autism spectrum disorder: genetics and pathology. Annu Rev Pathol 10, 111–144 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Mullins C, Fishell G, Tsien RW, Unifying Views of Autism Spectrum Disorders: A Consideration of Autoregulatory Feedback Loops. Neuron 89, 1131–1156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satterstrom FK et al. , Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180, 568–584 e523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller JA et al. , Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Data of 9k brain cells from an E18 mouse, 10x Genomics: https://support.10xgenomics.com/single-cell-gene-expression/datasets/2.1.0/neuron_9k.
- 11.Adamson B et al. , A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882 e1821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixit A et al. , Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866 e1817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaitin DA et al. , Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896 e1815 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Platt RJ et al. , CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E, Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment 2008, P10008 (2008). [Google Scholar]
- 16.Zeisel A et al. , Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014 e1022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancinelli S, Lodato S, Decoding neuronal diversity in the developing cerebral cortex: from single cells to functional networks. Curr Opin Neurobiol 53, 146–155 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Haber AL et al. , A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan B et al. , Model-based understanding of single-cell CRISPR screening. Nat Commun 10, 2233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. The Gene Ontology, The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts M, Stewart B, Tingley D, stm: R Package for Structural Topic Models. Journal of Statistical Software. [Google Scholar]
- 22.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders A et al. , Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030 e1016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scotland P, Zhou D, Benveniste H, Bennett V, Nervous system defects of AnkyrinB (−/−) mice suggest functional overlap between the cell adhesion molecule L1 and 440-kD AnkyrinB in premyelinated axons. J Cell Biol 143, 1305–1315 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuvia S, Buhusi M, Davis L, Reedy M, Bennett V, Ankyrin-B is required for intracellular sorting of structurally diverse Ca2+ homeostasis proteins. J Cell Biol 147, 995–1008 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kline CF, Scott J, Curran J, Hund TJ, Mohler PJ, Ankyrin-B regulates Cav2.1 and Cav2.2 channel expression and targeting. J Biol Chem 289, 5285–5295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang R et al. , ANK2 autism mutation targeting giant ankyrin-B promotes axon branching and ectopic connectivity. Proc Natl Acad Sci U S A 116, 15262–15271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marie C et al. , Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc Natl Acad Sci U S A, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiyama M et al. , Early embryonic death in mice lacking the beta-catenin-binding protein Duplin. Mol Cell Biol 24, 8386–8394 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodge RD et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velmeshev D et al. , Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowakowski TJ et al. , Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velasco S et al. , Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandal MJ et al. , Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lun ATL, Marioni JC, Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics 18, 451–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng WC, Jenkins PM, Tanaka M, Mooney R, Bennett V, Giant ankyrin-G stabilizes somatodendritic GABAergic synapses through opposing endocytosis of GABAA receptors. Proc Natl Acad Sci U S A 112, 1214–1219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bercury KK, Macklin WB, Dynamics and mechanisms of CNS myelination. Dev Cell 32, 447–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt RJ et al. , Chd8 Mutation Leads to Autistic-like Behaviors and Impaired Striatal Circuits. Cell Rep 19, 335–350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katayama Y et al. , CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature 537, 675–679 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Durak O et al. , Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci 19, 1477–1488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakamoto I et al. , A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J Biol Chem 275, 32871–32878 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Zhao C et al. , Dual Requirement of CHD8 for Chromatin Landscape Establishment and Histone Methyltransferase Recruitment to Promote CNS Myelination and Repair. Dev Cell 45, 753–768 e758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubin AJ et al. , Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 176, 361–376 e317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bian S et al. , Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 362, 1060–1063 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Rodriques SG et al. , Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X et al. , Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doench JG et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joung J et al. , Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arlotta P et al. , Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie ME et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finak G et al. , MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biology 16, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study are available through the Gene Expression Omnibus (accession GSE157977) as well as the Broad single cell portal (https://singlecell.broadinstitute.org/single_cell/study/SCP521). The analysis pipeline is deposited on GitHub repository (https://github.com/klarman-cell-observatory/ivPerturbSeq) and Zenodo (DOI: 10.5281/zenodo.4019534). All other data are available in the manuscript or the supplementary materials.