Abstract
Background
Patients with cancer are particularly susceptible to SARS-CoV-2 infection. The systemic inflammatory response is a pathogenic mechanism shared by cancer progression and COVID-19. We investigated systemic inflammation as a driver of severity and mortality from COVID-19, evaluating the prognostic role of commonly used inflammatory indices in SARS-CoV-2-infected patients with cancer accrued to the OnCovid study.
Methods
In a multicenter cohort of SARS-CoV-2-infected patients with cancer in Europe, we evaluated dynamic changes in neutrophil:lymphocyte ratio (NLR); platelet:lymphocyte ratio (PLR); Prognostic Nutritional Index (PNI), renamed the OnCovid Inflammatory Score (OIS); modified Glasgow Prognostic Score (mGPS); and Prognostic Index (PI) in relation to oncological and COVID-19 infection features, testing their prognostic potential in independent training (n=529) and validation (n=542) sets.
Results
We evaluated 1071 eligible patients, of which 625 (58.3%) were men, and 420 were patients with malignancy in advanced stage (39.2%), most commonly genitourinary (n=216, 20.2%). 844 (78.8%) had ≥1 comorbidity and 754 (70.4%) had ≥1 COVID-19 complication. NLR, OIS, and mGPS worsened at COVID-19 diagnosis compared with pre-COVID-19 measurement (p<0.01), recovering in survivors to pre-COVID-19 levels. Patients in poorer risk categories for each index except the PLR exhibited higher mortality rates (p<0.001) and shorter median overall survival in the training and validation sets (p<0.01). Multivariable analyses revealed the OIS to be most independently predictive of survival (validation set HR 2.48, 95% CI 1.47 to 4.20, p=0.001; adjusted concordance index score 0.611).
Conclusions
Systemic inflammation is a validated prognostic domain in SARS-CoV-2-infected patients with cancer and can be used as a bedside predictor of adverse outcome. Lymphocytopenia and hypoalbuminemia as computed by the OIS are independently predictive of severe COVID-19, supporting their use for risk stratification. Reversal of the COVID-19-induced proinflammatory state is a putative therapeutic strategy in patients with cancer.
Keywords: inflammation, inflammation mediators, COVID-19
Introduction
SARS-CoV-2 is the novel Betacoronavirus first identified in Wuhan, China, that has resulted in over 104 million cases and more than two million deaths globally as of February 3, 2021.1 2 COVID-19, the resulting disease from SARS-CoV-2 infection, is characterized by a myriad of symptoms and systemic complications. Defining symptoms according to WHO guidelines include fever, cough, and dyspnea, alongside a number of non-specific constitutional symptoms.3 Complications are similarly varied and system-wide: complicated COVID-19 involves acute respiratory distress syndrome (ARDS), coagulopathy, septic shock, and acute kidney and cardiac injuries.1
Evolving clinical experience in the management of SARS-CoV-2 has confirmed that lethality from COVID-19 is related not only to the cytopathic effect of the virus but also to the host’s response to infection, which leads to end-organ damage and mortality through ignition of a sustained systemic inflammatory response.4 Patients with severe COVID-19 display heightened blood levels of interleukin (IL)-6, IL-2, IL-7, IL-10, and chemokines such as monocyte chemoattractant protein-1, implicating the innate immune response in COVID-19 progression.4 Assessment of inflammatory status at the time of COVID-19 diagnosis may lend itself as a bedside prognostic marker and novel immunological domain for therapeutic targets.
Systemic inflammation is in fact a unifying mechanism between cancer progression and COVID-19. The acute-phase response (APR) is a series of coordinated immune and metabolic changes that the host orchestrates in response to potentially harmful insults. The host’s innate immune response is central to the coordination of the APR, which is linked to both infectious and non-infectious noxae, including cancer.5 Activation of an APR is reflected in the acute derangement of hematological and biochemical parameters, including reactive leukocytosis, peripheral blood neutrophilia, lymphopenia, elevated C-reactive protein (CRP) levels, and hypoalbuminemia.5 6 A number of these laboratory parameters have been individually linked to worse prognosis from COVID-19.4 Inflammation-based indices, including the neutrophil:lymphocyte ratio (NLR),7 platelet:lymphocyte ratio (PLR),8 Prognostic Nutritional Index (PNI),9 10 modified Glasgow Prognostic Score (mGPS),11 and Prognostic Index (PI),12 have been derived from optimal integration of acute-phase reactants and have been consistently shown to predict for adverse prognosis independent of treatment, stage, and type of cancer. Evaluation of these indices in the context of SARS-CoV-2-infected patients with cancer may elucidate parameters of the APR that are linked to the progression of SARS-CoV-2 infection in its most lethal forms. We took special interest in the PI, which we renamed the OnCovid Inflammatory Score (OIS) in the context of COVID-19, as its calculation considers lymphopenia and hypoalbuminemia, two parameters individually shown to predict for COVID-19 prognosis.4
In a previous study, we had shown that a significant proportion of patients with cancer develop severe COVID-19 and that mortality is strongly related to host rather than oncological factors, including number of comorbidities and advancing age.13 14 However, the mechanisms underlying excess mortality observed in patients with cancer are not fully understood. In this study, we sought to determine whether the presence of a systemic inflammatory response at the time of COVID-19 diagnosis may help identify patients with severe SARS-CoV-2 infection and predict outcome in this population. To pursue this aim, we independently validated the OIS and a panel of inflammation-based indices in a large repository of patients with COVID-19 and cancer as part of the OnCovid study.
Materials and methods
Study design, patient demographics, and data collection
From the OnCovid repository, a retrospective registry of SARS-CoV-2-infected patients with cancer in Europe,14 we aimed to evaluate a panel of inflammation-based indices that are linked with the survival of patients with cancer for their prognostic role in the context of COVID-19 diagnosis.7–9 11 12 For the purpose of this study, the combination of hypoalbuminemia and lymphocytopenia (albumin concentration (g/L)+5total lymphocyte count (109/L)), previously qualified as a nutritional predictor of outcome (PNI), was redefined as the OIS in view of its qualification as an inflammatory predictor in the context of COVID-19 infection. Between February 27 and June 23, 2020, 1318 patients were consecutively referred from 23 academic centers in the UK (n=539), Spain (n=380), Italy (n=374), Belgium (n=19), and Germany (n=6, (online supplemental table 1)). Eligibility criteria for OnCovid included (1) SARS-CoV-2 infection confirmed by nasopharyngeal swab and subsequent reverse transcriptase PCR (RT-PCR)15 and (2) history of solid or hematological malignancy at any timepoint before or during COVID-19 disease course. Both patients with active malignancies and patients in cancer remission were included in the study. Patients were 18 years of age, and anonymized electronic medical record data were entered into the Research Electronic Data Capture (Vanderbilt University) tool hosted by the Medical Statistics Unit in Novara, Italy,16 17 which coordinated database access and curation.
jitc-2020-002277supp001.pdf (34.1KB, pdf)
Inflammation-based indices
Eligible patients (n=1071) were stratified on the basis of a selected panel of five biomarkers of systemic inflammation (NLR, PLR, OIS, mGPS, and PI) at three timepoints: at the last oncological follow-up prior to SARS-CoV-2 infection (pre-COVID-19), at COVID-19 diagnosis, and, in surviving patients, at the first oncological follow-up after SARS-CoV-2 infection with a documented negative SARS-CoV-2 swab. Calculations for each inflammation-based index are shown in online supplemental table 2. Patients were stratified into good versus poor risk subgroups on the basis of predefined categories for the mGPS and PI, depending on whether they had an abnormal CRP and/or albumin (for mGPS) or white cell count (for PI) at the point of biomarker measurement.11 12 For inflammatory biomarkers characterized by a continuous distribution, good and poor risk groups were identified by the median value of the distribution.
jitc-2020-002277supp002.pdf (21.4KB, pdf)
Definitions and study endpoints
Features of severe COVID-19 such as ARDS and septic shock were defined according to WHO guidelines.3 Furthermore, all comorbid conditions, complications, and key COVID-19 symptoms were determined from WHO guidelines.3 Resolution of SARS-CoV-2 infection was confirmed via negative RT-PCR result following nasopharyngeal swab for surviving patients. Active malignancy was defined by presence of oncological disease according to clinical criteria for the corresponding tumor type. Patients categorized as undergoing active cancer therapy had received anti-cancer therapy within 4 weeks of COVID-19 diagnosis, and patients with no treatment history for cancer were categorized as treatment naïve. To evaluate the prognostic value of inflammation-based indices, we randomly divided eligible patients into training and validation sets. Primary study outcome was mortality. Overall survival (OS) was determined from the date of positive SARS-CoV-2 RT-PCR to the date of death or final follow-up.
Statistical analyses
Data following a normal distribution are presented as mean with SD, and data differing from normal distribution are presented as median with IQR. The Mann-Whitney U test and the Wilcoxon signed-rank test were used as appropriate to compare the medians of non-normally distributed continuous data. Categorical variables, shown as absolute counts or percentages, were compared using Pearson’s χ2 test. Kaplan-Meier estimates with a log-rank test were employed for univariable survival plot analysis. Univariable HRs were calculated through logistic Cox regression, and multivariable HRs were calculated through Cox regression with conditional backward elimination using a removal value of 0.10 and entry value of 0.05. Harrell’s concordance index (C-index) was used to evaluate the predictive ability of inflammation-based indices and the multivariable model. 150 bootstrap samples were used to calculate the 95% CI for each C-index, estimate the optimism due to potential overfitting, and provide optimism-adjusted C-index values with corresponding CIs. A two-tailed p value of 0.05 was considered the threshold for statistical significance. Dataset management and statistical analyses were performed using Statistical Product and Service Solutions V.24 for Apple Macintosh OSX (IBM, Armonk, New York, USA). Figures were created in Prism V.8 for Apple Macintosh OSX (GraphPad, San Diego, California, USA).
Patient involvement
Patients were involved via retrospective analysis of anonymized patient chart data. Patients did not participate in the planning of this study, and this study involved no interventions.
Results
Inclusion criteria, training and validation set assortment, and demographics
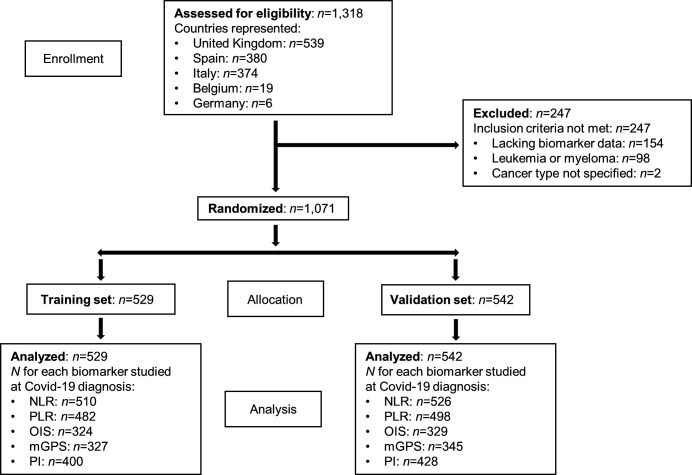
A total of 1318 patients ≥18 years of age with a history of active malignancy or in cancer remission and possessing a positive COVID-19 diagnosis were consecutively entered into the main OnCovid registry. In total, 247 patients were excluded, leaving a final dataset of 1071 eligible patients (figure 1). Reasons for exclusion were (1) lack of laboratory data to calculate at least one biomarker of interest (n=154), (2) leukemia or myeloma diagnosis (n=98), and/or (3) lack of specification of cancer type (n=2). Patients with leukemia and myeloma were excluded due to the potential confounding effect of the neoplastic derangement on peripheral blood values used to compute bone marrow-derived inflammation indices.18–20 From this final study population (n=1071), patients were randomly allocated into a training set (n=529) or a validation set (n=542) matched for age, comorbid burden, number of COVID-19 complications, and proportion of patients with active malignancy (online supplemental table 3). Out of 1071 eligible patients, the majority were male (n=625, 58.3%) with a mean age of 68.2 years (SD 13.4, range 21–99 years). Patient distribution across participating academic centers is shown in online supplemental table 1. Genitourinary cancers comprised the most common primary tumor site (n=216, 20.2%), and the majority of patients had evidence of active malignancy (n=687, 64.1%). At COVID-19 diagnosis, 519 (48.5%) patients had localized or locoregional disease; 420 patients (39.2%) had metastatic/advanced disease; and 516 (48.2%) patients were receiving anticancer therapy. Most patients had at least one comorbidity (n=844, 78.8%), most commonly hypertension (n=496, 46.3%) or cardiovascular disease (n=257, 24.0%).
Figure 1.
Patient disposition. mGPS, modified Glasgow Prognostic Score; NLR, neutrophil:lymphocyte ratio; OIS, OnCovid Inflammatory Score; PI, Prognostic Index; PLR, platelet:lymphocyte ratio.
jitc-2020-002277supp003.pdf (35.4KB, pdf)
Presenting SARS-CoV-2 symptoms included fever (n=660, 61.6%) and cough (n=542, 50.6%). Of the 957 (89.4%) patients hospitalized, 124 (13.0%) were escalated to intensive or subintensive care. The majority of patients required supplemental oxygen therapy (n=627, 58.5%), and a subset required mechanical ventilation (n=115, 10.7%). Most patients received at least one COVID-19-specific drug treatment (n=708, 66.1%), with the majority receiving an antimalarial or antiviral agent (n=622, 58.1%). A total of 754 (70.4%) patients developed COVID-19 complications, most frequently acute respiratory failure (n=627, 58.5%). At the time of database censoring (June 23, 2020), 375 (35.0%) patients were deceased and 680 (63.5%) were discharged from the hospital or were in-hospital survivors (online supplemental table 4 highlights COVID-19 complications and complete patient demographics).
jitc-2020-002277supp004.pdf (31.7KB, pdf)
Systemic inflammatory response identifies patients with adverse outcomes
In eligible patients (n=1071), median NLR, PLR, and OIS at COVID-19 diagnosis were 6.1 (IQR 8.7), 270.1 (IQR 260.6), and 38.5 (IQR 10.5) respectively. In total, 526 (49.1%) patients had NLR6, 491 (45.8%) had PLR270, and 384 (35.9%) had OIS40. mGPS was available in 672 (62.7%) patients at COVID-19 diagnosis, with 503 (47.0%) patients being defined as intermediate (mGPS 1, n=196, 18.3%) or poor risk (mGPS 2, n=307, 28.7%). A total of 828 (77.3%) patients were evaluable for PI, with 686 (64.1%) classifying as intermediate (PI 1, n=546, 51.0%) or poor risk (PI 2, n=140, 13.1%) subgroups.
As shown in online supplemental figure 1, OIS values were significantly lower in patients with active malignancy (median 38.1, IQR 11.2), a factor known to predict for outcome from COVID-19,13 compared with patients in remission (median 39.5, IQR 9.6, p<0.05). Patients with active malignancy were also more likely to classify in poorer mGPS risk groups (p<0.05). A comparison of inflammation scores across tumor types is shown in online supplemental table 5.
jitc-2020-002277supp009.pdf (63.6KB, pdf)
jitc-2020-002277supp005.pdf (17.2KB, pdf)
Median NLR, PLR, and OIS at COVID-19 diagnosis were studied in relation to patients’ mortality along with the distribution of the mGPS and PI. Mortality from COVID-19 was associated with categorization into the poorer risk group for each index (p0.05, figure 2A).
Figure 2.
Relationship between inflammatory markers and COVID-19 outcomes and marker values at timepoints. (A) Median inflammatory index values or distributions at COVID-19 diagnosis in living versus deceased patients for the NLR (n=661 alive, n=360 dead; p0.0001), PLR (n=628 alive, 338 dead; p0.05), OIS (n=413 alive, n=230 dead; p0.0001), mGPS (n=430 alive, n=229 dead; p0.0001), and PI (n=525 alive, n=295 dead; p0.0001). (B) Median values and distributions across timepoints for the NLR (n=55), PLR (n=50), OIS (n=23), mGPS (n=143 pre-COVID-19, n=672 at diagnosis, n=32 post-COVID-19), and PI (n=163 pre-COVID-19, n=828 at diagnosis, n=36 post-COVID-19). *P0.05, **P0.01, ***P0.001, ****P0.0001. Error bars represent 95% CIs from the median. mGPS, modified Glasgow Prognostic Score; NLR, neutrophil:lymphocyte ratio; ns, not significant; OIS, OnCovid Inflammatory Score; PI, Prognostic Index; PLR, platelet:lymphocyte ratio.
To evaluate the relative contribution of active SARS-CoV-2 infection over underlying malignancy in determining the systemic inflammatory response, we evaluated dynamic changes across the pre-COVID-19, at, and post-COVID-19 diagnosis timepoints in a subset of SARS-CoV-2 survivors (n=65, 6.1%). All samples taken at post-COVID-19 diagnosis were defined as measurements taken during the first outpatient oncological follow-up after COVID-19 with a negative SARS-CoV-2 swab at the moment of blood sampling. Patients had data for all indices (NLR, n=55; PLR, n=50; OIS, n=23; mGPS, n=8; and PI, n=13). The mean interval between pre-COVID-19 and at COVID-19 diagnosis timepoints was 270 days (SD±855), and the mean interval between COVID-19 diagnosis and post-COVID-19 diagnosis timepoints was 39 days (SD±24).
As shown in figure 2B, NLR values were significantly higher at COVID-19 diagnosis (median 6.1, IQR 8.7) compared with pre-COVID-19 (median 2.9, IQR 3.1, p0.001) and post-COVID-19 timepoints (median 2.6, IQR 2.8, p0.01), whereas pre-COVID and post-COVID-19 values did not differ (p=0.82). Similar dynamic changes were described for the OIS, which appeared lower at COVID-19 diagnosis (median 38.5, IQR 10.5) compared with pre-COVID-19 (median 47.0, IQR 9.6, p0.001) and post-COVID-19 timepoints (47.8, IQR 10.0, p0.0001), with no difference noted between pre-COVID-19 and post-COVID-19 measurements (p=0.58). mGPS and PI distribution also varied across timepoints (figure 2B).
Next, we sought to determine the relationship between patients’ inflammatory scores at COVID-19 diagnosis and previously established predictors of mortality or disease severity.13 21–24 As shown in online supplemental table 6, patients in the poor risk groups for all inflammatory indices developed a greater number of COVID-19 complications. Furthermore, patients in poor risk groups for all indices except the PLR were older and had a higher comorbid burden. Patients in poor risk categories for the NLR, mGPS, and PI were less likely to be undergoing anticancer therapy at COVID-19 diagnosis and were more often male (online supplemental table 6).
jitc-2020-002277supp006.pdf (41.4KB, pdf)
We also investigated the association between inflammatory markers and laboratory variables known to predict for increased mortality from COVID-19.21–24 As shown in online supplemental figure 2, an NLR of ≥6 was associated with increased lactate dehydrogenase (LDH, p0.0001), D-dimer (p0.01), troponin (p<0.001), and ferritin (p<0.01) levels compared with an NLR<6. An OIS40 was associated with heightened LDH (p0.001), D-dimer (p0.001), and ferritin (p0.001) levels. Poor risk mGPS and PI categories also exhibited an association with clinical variables.
jitc-2020-002277supp010.pdf (119KB, pdf)
OIS predicts for decreased OS and greater risk of severe COVID-19 in patients with cancer
To accurately evaluate the prognostic performance of inflammation-based indices, we randomly assigned the 1071 eligible patients into a training (n=529) and validation set (n=542, figure 1). Both datasets were matched for key characteristics that are known to influence prognosis from COVID-19, including age ≥65 years, number of comorbidities, number of COVID-19 complications, and presence of active malignancy (online supplemental table 3).13 Table 1 presents complete demographics of both the training and validation sets. Unadjusted mortality rates were similar across datasets (36.1% in the training set and 35.0% in the validation set, p=0.16).
Table 1.
Patient demographics of training and validation sets: features of patients from both the training (n=529) and validation (n=542) sets
| Characteristics | Training set (total, n=529) |
Validation set (total, n=542) |
| Age (years), mean±SD | 67.9±13.3 | 68.5±13.5 |
| Age 65 years, n (%) | 311 (58.8) | 334 (61.6) |
| Sex, n (%) | ||
| Male | 296 (56.0) | 329 (60.7) |
| Female | 231 (43.7) | 212 (39.1) |
| Information unavailable | 2 (0.3) | 1 (0.2) |
| Smoking history, n (%) | ||
| Never smoker | 205 (38.8) | 207 (38.2) |
| Current/former smoker | 228 (43.1) | 234 (43.2) |
| Unknown | 96 (18.1) | 101 (18.6) |
| Cancer type, n (%) | ||
| Head and neck | 22 (4.2) | 12 (2.2) |
| Lung and thoracic | 67 (12.7) | 87 (16.1) |
| Gastroesophageal | 30 (5.7) | 22 (4.1) |
| Hepatobiliary | 33 (6.2) | 22 (4.1) |
| Duodenal and lower GI tract | 61 (11.5) | 71 (13.1) |
| Breast | 85 (16.1) | 92 (17.0) |
| Gynecological | 35 (6.6) | 22 (4.1) |
| Genitourinary | 108 (20.4) | 108 (19.9) |
| Skin | 17 (3.2) | 26 (4.8) |
| Lymphoma | 49 (9.3) | 38 (7.0) |
| Other | 22 (4.1) | 42 (7.6) |
| Tumor stage, n (%) | ||
| Localized | 173 (32.7) | 187 (34.5) |
| Locoregional | 72 (13.6) | 87 (16.1) |
| Metastatic | 223 (42.2) | 197 (36.3) |
| Information unavailable | 61 (11.5) | 71 (13.1) |
| Tumor status at COVID-19 diagnosis, n (%) | ||
| Active malignancy | 353 (66.7) | 334 (61.6) |
| Remission | 150 (28.4) | 182 (33.6) |
| Information unavailable | 26 (4.9) | 26 (4.8) |
| Ongoing anticancer therapy at COVID-19 diagnosis, n (%) | 264 (49.9) | 252 (46.5) |
| Prior radical therapies, n (%) | 285 (53.9) | 306 (56.5) |
| Surgery | 244 (46.1) | 266 (49.0) |
| Adjuvant/neoadjuvant chemotherapy | 171 (32.3) | 148 (27.3) |
| Prior palliative systemic therapy, n (%) | 147 (27.8) | 130 (24.0) |
| Chemotherapy | 97 (18.3) | 87 (16.1) |
| Immunotherapy | 21 (4.0) | 25 (4.6) |
| Endocrine therapy | 29 (5.5) | 24 (4.4) |
| Targeted therapy | 22 (4.2) | 23 (4.2) |
| Prior curative systemic therapy, n (%) | 32 (6.0) | 30 (5.5) |
| Prior radiotherapy, n (%) | 150 (28.4) | 169 (31.2) |
| Prior lines of palliative therapy, n (%) | ||
| 1 | 75 (14.2) | 60 (11.1) |
| 2 | 30 (5.7) | 29 (5.4) |
| 3 | 33 (6.2) | 28 (5.2) |
| Comorbidities, n (%) | 425 (80.3) | 419 (77.3) |
| Hypertension | 251 (47.4) | 245 (45.2) |
| Diabetes | 115 (21.7) | 123 (22.7) |
| Cardiovascular disease | 128 (24.2) | 129 (23.8) |
| Chronic pulmonary disease | 80 (15.1) | 80 (14.8) |
| Chronic kidney disease | 62 (11.7) | 63 (11.6) |
| Cerebrovascular disease | 37 (7.0) | 41 (7.6) |
| Dementia | 27 (5.1) | 36 (6.6) |
| Peripheral vascular disease | 19 (3.6) | 21 (3.9) |
| Liver impairment | 11 (2.1) | 10 (1.8) |
| Immunosuppression | 16 (3.0) | 29 (5.4) |
| Steroid therapy in progress | 23 (4.3) | 27 (5.0) |
| Other | 164 (31.0) | 144 (26.6) |
| Number of comorbidities (%) | ||
| 0 | 104 (19.7) | 123 (22.7) |
| 1 | 149 (28.2) | 149 (27.5) |
| 2 | 129 (24.4) | 118 (21.8) |
| 3 | 147 (27.7) | 152 (28.0) |
| COVID-19 symptoms at diagnosis, n (%) | 497 (94.0) | 514 (94.8) |
| Fever | 317 (59.9) | 343 (63.3) |
| Cough | 257 (48.6) | 285 (52.6) |
| Dyspnea | 206 (38.9) | 232 (42.8) |
| Fatigue | 132 (25.0) | 125 (23.1) |
| Myalgia | 60 (11.3) | 57 (10.5) |
| Diarrhea | 50 (9.5) | 76 (14.0) |
| Coryzal symptoms | 23 (4.3) | 26 (4.8) |
| Nausea or vomiting | 33 (6.2) | 47 (8.7) |
| Sore throat | 18 (3.4) | 9 (1.7) |
| Headache | 21 (4.0) | 21 (3.9) |
| Dysgeusia | 14 (2.6) | 21 (3.9) |
| Anosmia | 13 (2.5) | 16 (3.0) |
| Other (ie, confusion and delirium) | 119 (22.5) | 124 (22.9) |
| Number of symptoms at diagnosis (%) | ||
| 0 | 32 (6.0) | 28 (5.2) |
| 1 | 125 (23.6) | 104 (19.2) |
| 2 | 151 (28.5) | 149 (27.5) |
| 3 | 221 (41.9) | 261 (48.2) |
| Hospitalization rate, n (%) | 477 (90.2) | 480 (88.6) |
| Admission to intensive or subintensive care unit, n (%) | 59/477 (12.4) | 65/480 (13.5) |
| COVID-19-specific drug treatments, n (%) | 349 (66.0) | 359 (66.2) |
| Antibiotics | 294 (55.6) | 301 (55.5) |
| Hydroxychloroquine or chloroquine | 199 (37.6) | 193 (35.6) |
| Lopinavir/ritonavir | 86 (16.3) | 86 (15.9) |
| Systemic corticosteroids | 45 (8.5) | 46 (8.5) |
| Remdesivir | 5 (0.9) | 7 (1.3) |
| Tocilizumab | 18 (3.4) | 28 (5.2) |
| Others | 57 (10.8) | 71 (13.1) |
| COVID-19-specific oxygen interventions, n (%) | 309 (58.4) | 323 (59.6) |
| Oxygen therapy | 307 (58.0) | 320 (59.0) |
| Mechanical ventilation | 44 (8.3) | 71 (13.1) |
| COVID-19 complications, n (%) | 372 (70.3) | 382 (70.5) |
| Acute respiratory failure | 307 (58.0) | 320 (59.0) |
| ARDS | 61 (11.5) | 70 (12.9) |
| Acute kidney injury | 42 (7.9) | 44 (8.1) |
| Secondary infection | 49 (9.3) | 37 (6.8) |
| Sepsis | 29 (5.5) | 23 (4.2) |
| Septic shock | 22 (4.2) | 23 (4.2) |
| Acute cardiac injury | 12 (2.3) | 12 (2.2) |
| Acute liver injury | 4 (0.8) | 6 (1.1) |
| Others (ie, DIC) | 33 (6.2) | 28 (5.2) |
| Number of complications (%) | ||
| 0 | 157 (29.7) | 160 (29.5) |
| 1 | 190 (35.9) | 204 (37.6) |
| 2 | 101 (19.1) | 93 (17.2) |
| 3 | 42 (7.9) | 47 (8.7) |
| Information unavailable | 39 (7.4) | 38 (7.0) |
ARDS, acute respiratory distress syndrome; DIC, disseminated intravascular coagulation; GI, gastrointestinal.
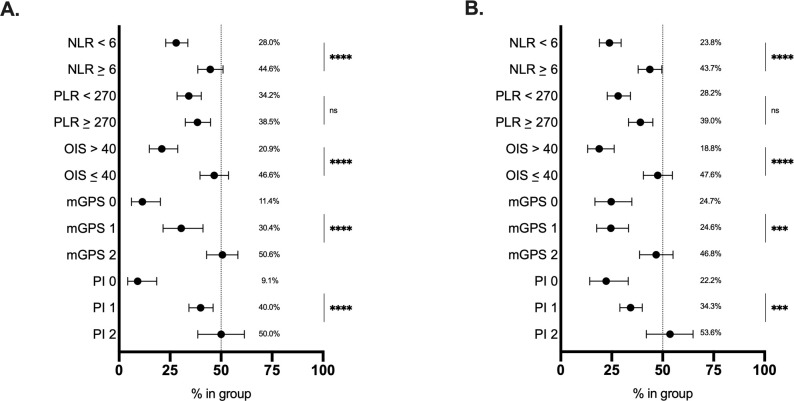
In the training set, all tested inflammatory indices but the PLR were associated with unadjusted mortality rates (p<0.0001), as shown in figure 3A. Analysis of unadjusted mortality in the validation set confirmed these findings (figure 3B). Patients with NLR of 6, OIS of 40, and poor risk mGPS and PI values had shorter median OS (p0.001, figure 4 and online supplemental table 7). Additionally, as shown in table 2, patients in poor risk groups for all inflammatory indices except the PLR (p=0.55) presented univariable HRs above 1.00 (p≤0.001). This was confirmed in the validation set analysis (p<0.01; figure 5, table 2 and online supplemental table 7).
Figure 3.
Relationship between inflammatory markers and unadjusted mortality. Inflammatory marker values at COVID-19 diagnosis divided into good and poor risk groups plotted against unadjusted mortality rates for the (A) training set and (B) validation set. Error bars represent lower and upper limits. ***P<0.001, ****P<0.0001. mGPS, modified Glasgow Prognostic Score; NLR, neutrophil:lymphocyte ratio; ns, not significant; OIS, OnCovid Inflammatory Score; PI, Prognostic Index; PLR, platelet:lymphocyte ratio.
Figure 4.
Univariable survival analysis of inflammatory markers (training set). Kaplan-Meier estimates for univariable survival analysis are shown for the NLR (n=447, p0.001), OIS (n=273, p0.0001), mGPS (n=278, p0.0001), and PI (n=361, p0.0001) at COVID-19 diagnosis with significance calculated following log-rank methodology. ***P0.001, ****P0.0001. mGPS, modified Glasgow Prognostic Score; NLR, neutrophil:lymphocyte ratio; OIS, OnCovid Inflammatory Score; PI, Prognostic Index.
Table 2.
Univariable and multivariable HRs for prognostic factors
| Training | n=529 | Validation | n=542 | |||||
| Prognostic factor | Univariable HR (95% CI) | P value | Multivariable HR (95% CI) | P value | Univariable HR (95% CI) | P value | Multivariable HR (95% CI) | P value |
| NLR <6/≥6 |
1.68 (1.25 to 2.27) | 0.001 | 0.97 (0.60 to 1.55) | 0.882 | 2.07 (1.50 to 2.86) | <0.0001 | 0.75 (0.45 to 1.26) | 0.275 |
| PLR <270/≥270 |
1.10 (0.81 to 1.45) | 0.550 | 0.76 (0.49 to 1.19) | 0.234 | 1.55 (1.12 to 2.13) | 0.008 | 0.94 (0.52 to 1.70) | 0.842 |
| OIS >40/≤40 |
2.78 (1.78 to 4.34) | <0.0001 | 1.97 (1.19 to 3.26) | 0.008 | 2.83 (1.80 to 4.45) | <0.0001 | 2.48 (1.47 to 4.20) | 0.001 |
| mGPS 0/2 |
5.12 (2.57 to 10.22) | <0.0001 | 2.35 (0.77 to 7.14) | 0.132 | 2.18 (1.32 to 3.60) | 0.002 | 1.37 (0.18 to 10.78) | 0.764 |
| PI 0/2 |
5.61 (2.36 to 13.34) | <0.0001 | 1.41 (0.13 to 15.76) | 0.781 | 2.71 (1.49 to 4.90) | 0.001 | 1.76 (0.74 to 4.17) | 0.200 |
| Sex (Female/male) |
1.38 (1.03 to 1.862) | 0.032 | 0.69 (0.45 to 1.07) | 0.097 | 1.40 (1.02 to 1.92) | 0.039 | 0.86 (0.51 to 1.45) | 0.576 |
| Age <65/≥65 |
2.22 (1.60 to 3.06) | <0.0001 | 1.47 (0.93 to 2.33) | 0.103 | 2.56 (1.78 to 3.67) | <0.0001 | 2.80 (1.50 to 5.22) | 0.001 |
| Comorbid burden 0/≥2 |
1.86 (1.24 to 2.80) | 0.003 | 1.28 (0.71 to 2.30) | 0.412 | 2.40 (1.53 to 3.76) | <0.0001 | 2.19 (0.89 to 5.39) | 0.090 |
| Malignancy status Remission/active |
1.55 (1.09 to 2.21) | 0.015 | 1.60 (0.95 to 2.72) | 0.080 | 1.20 (0.86 to 1.65) | 0.284 | 1.16 (0.69 to 1.95) | 0.575 |
| Anticancer therapy at COVID-19 diagnosis No/yes |
0.59 (0.44 to 0.79) | <0.001 | 0.65 (0.41 to 1.02) | 0.063 | 0.72 (0.53 to 0.97) | 0.032 | 0.94 (0.54 to 1.63) | 0.813 |
Prognostic factors for both the training (n=529) and validation (n=542) are shown with their corresponding univariable and multivariable HRs with 95% CIs and p values. Univariable HRs were calculated through logistic Cox regression, and multivariable HRs were calculated through Cox regression with conditional backward elimination. Statistical signifiance indicated via bold and italics. HR, hazard ratio; NLR, neutrophil:lymphocyte ratio; PLR, platelet:lymphocyte ratio; OIS, OnCovid Inflammatory Score; mGPS, modified Glasgow Prognostic Score; PI, Prognostic Index.
Bold and italicized values are statistically significant (P<0.05).
mGPS, modified Glasgow prognostic score; NLR, neutrophil:lymphocyte ratio; OIS, OnCovid inflammatory score; PI, prognostic index; PLR, platelet:lymphocyte ratio.
Figure 5.
Univariable survival analysis of inflammatory markers (validation set). Kaplan-Meier estimates for univariable survival analysis are shown for the NLR (n=452, p0.0001), OIS (n=272, p0.0001), mGPS (n=291, p0.001), and PI (n=384, p0.01) at COVID-19 diagnosis with significance calculated following log-rank methodology. **P0.01, ***P0.001, ****P0.0001. mGPS, modified Glasgow Prognostic Score; NLR, neutrophil:lymphocyte ratio; OIS, OnCovid Inflammatory Score; PI, Prognostic Index.
jitc-2020-002277supp007.pdf (23.5KB, pdf)
In multivariable Cox regression models, the inflammatory indices were evaluated for their independent prognostic ability against established prognostic factors for COVID-19.13 21–24 In the training set analysis, the OIS was the only factor emerging as significantly prognostic with patients in the poor risk group (OIS≤40) presenting a multivariable HR of 1.97 (95% CI 1.19 to 3.26, p=0.008; table 2). The independent prognostic value of the OIS also emerged in multivariable analyses of the validation set (OIS≤40, HR 2.48, 95% CI 1.47 to 4.20, p=0.001; table 2), confirming its external validity as a prognostic factor for COVID-19. Analysis of inflammatory indices for prognostic ability was further evaluated through calculation of Harrell’s C-index (online supplemental table 8). The mGPS (optimism-adjusted C-index 0.634, 95% CI 0.595 to 0.680) and OIS (adjusted C-index 0.603, 95% CI 0.555 to 0.646) emerged as most predictive in the training set. OIS was most predictive in validation set analysis (adjusted C-index 0.611, 95% CI 0.564 to 0.668) followed by the mGPS (adjusted C-index 0.596, 95% CI 0.548 to 0.651; online supplemental table 8).
jitc-2020-002277supp008.pdf (27.6KB, pdf)
Discussion
Previous work has emphasized the contribution of comorbid burden, advanced age, and a diagnosis of hematological malignancy as risk factors for mortality from COVID-19; however, the molecular and immunological mechanisms that lead to a more adverse COVID-19 course in patients with cancer are largely unknown.13 25
In our study, we demonstrate that the presence of a proinflammatory diathesis as measured by the NLR, OIS, mGPS and PI is generally associated with worsening levels of established biomarkers of COVID-19 severity such as higher circulating D-dimer, troponin, ferritin and LDH concentration and less strongly related to the presence of measurable or active malignancy. These findings, paired with evidence of dynamic changes of inflammatory markers, strengthen the link between systemic inflammation as a SARS-CoV-2-driven disease mechanism linked with adverse clinical outcome.
Interestingly, activation of a systemic inflammatory response appears unevenly distributed across tumor sites. Patients with breast cancer, for instance, had lower mortality in our study (20.9%) and were more likely to cluster within better risk categories according to inflammatory prognostic indices. The relationship between inflammatory scores and mortality in selected tumors such as breast cancer is highly provocative and warrants further research into whether certain tumor types are associated with a more favorable immune response to COVID-19.
Irrespective of tumor-specific considerations, analysis of survival confirms the NLR, OIS, mGPS, and PI as tumor-agnostic predictors of OS in both the training and validation sets, strengthening the external validity of the APR as a driver of mortality from COVID-19. In our study, the PLR did not emerge as a predictor of survival, suggesting reactive thrombocytosis as a suboptimal marker to identify severe COVID-19. This is perhaps unsurprising given that thrombocytopenia, rather than thrombocytosis, has been implicated in severe COVID-19.26 27
Our study shows that the systemic inflammatory response in patients with cancer diagnosed with COVID-19 is influenced by advanced age and comorbid burden. A number of studies have reproducibly shown these features to predict for adverse clinical course in SARS-CoV-2-infected patients irrespective of a diagnosis of cancer.13 21–24 Our data contribute to shed light on this relationship by highlighting that these categories at risk are more likely to mount an exaggerated innate response to SARS-CoV-2 as evidenced by significantly worse derangement biomarkers of systemic inflammation in these subgroups, tracing an important link between the extent of such proinflammatory response and outcome in these high-risk categories.
Because the panel of proinflammatory markers we studied relies on diverse and often non-redundant acute-phase reactants (hypoalbuminemia and neutrophilia) an important aim of our study was to establish the prognostic accuracy of each score in predicting patients’ OS.
We therefore tested the inflammatory indices in multivariable models including age, comorbid burden, and sex. We confirmed the combination of hypoalbuminemia and lymphopenia as computed by the OIS as a strong, independent predictor of survival, leading us to optimally select proinflammatory features that best scale with outcome in the context of COVID-19 and cancer. Both lymphopenia and hypoalbuminemia have previously been identified as independent prognostic factors in patients affected by COVID-19 without cancer.27 28 Our study shows that combination of these traits optimally scales with the prognosis of SARS-CoV-2-infected patients with cancer, an approach that was not considered in previous studies.
Taken together, our results qualify the OIS in SARS-CoV-2-infected patients with cancer as a readily available, inexpensive, and validated predictor of outcome in this patient population characterized by high risk of mortality following infection with SARS-CoV-2. The independence of and tumor-agnostic value of the OIS yields potential promise as a source of therapeutic targets to modulate innate immunity and prevent end-organ damage and COVID-19-related mortality in patients with cancer.
Our study acknowledges a number of limitations. The retrospective study design is a limitation shared by a number of studies published in this area29–31 and contributes to heterogeneity in treatment decisions, including escalation to intensive/high-dependence care and missing outcome data. Furthermore, non-cancer controls would have helped us understand whether the impact of proinflammatory scores is similar in patients who do not have cancer. Additionally, our study included only patients with confirmed SARS-CoV-2 infection determined by RT-PCR, leaving us no information on immune dysfunction in asymptomatic carriers. Finally, the 270-day gap between pre-COVID-19 and at COVID-19 diagnosis inflammatory index calculations for patients with pre-COVID-19, at, and post-COVID-19 diagnosis information available is a study limitation as is the smaller size of this 65-patient cohort.
Despite the acknowledged limitations, our study is the largest to attempt a robust and standardized evaluation of novel biomarkers of outcome in SARS-CoV-2-infected patients with cancer. The geographical diversity of our patient population and the choice of external validation limit the risk of overfitting of our survival estimates and facilitate a broader generalizability of our results.
In conclusion, we have demonstrated that the systemic inflammatory response can be used not only to facilitate an individualized risk assessment of patients’ mortality but also to better understand the disease biology of SARS-CoV-2 infection. Our work ties a dysfunctional proinflammatory response to poorer outcomes from COVID-19 in cancer and validates the combination of lymphopenia and hypoalbuminemia as measured by the OIS as a tumor-agnostic predictor of mortality following adjustment for key clinicopathological features, including comorbid burden and age. In the clinic, patients presenting with an OIS below 40 at COVID-19 diagnosis should be considered at higher risk of adverse outcome by treating physicians. Future study of the OIS in patients without cancer is warranted to validate its utility as a predictor of outcome in the general population. Our findings provide a clinical rationale for therapeutic targeting of the inflammatory response in SARS-CoV-2-infected patients with cancer.
Acknowledgments
We acknowledge the following OnCovid study collaborators: Meritxell Mollà (Department of Radiation Oncology, Hospital Clinic, Barcelona, Spain); Anna Carbó, Isabel García-Fructoso, and Anna Sureda (Medical Oncology, Catalan Institute of Oncology, Girona, Spain); Riccardo Bruna (Department of Translational Medicine, Università del Piemonte Orientale, Vercelli, Italy); Sarah Benafif, Myria Galazi, and Palma Dileo (Cancer Division, University College London Hospitals, London, UK); Rachel Würstlein (Gynecology and Obstetrics Clinic, Breast Center, University of Munich and CCCLMU); Marta Betti (Infrastruttura Ricerca Formazione Innovazione, Azienda Ospedaliera SS Antonio e Biagio e Cesare Arrigo, Alessandria, Italy); Salvatore Provenzano (Medical Oncology Unit, Cancer Medicine Department, Instituto Nazionale dei Tumori, Milan, Italy); Alessia Dalla Pria (National Centre for HIV Malignancies, Chelsea and Westminster Hospital, London, UK); Francesca D’Avanzo (Oncology, Ospedale Maggiore della Carità, Novara, Italy); Judith Swallow, Maria Martinez, and Joanne S Evans (Division of Surgery and Cancer, Imperial College London); Rachel Sharkey (Medical Oncology, Chelsea and Westminster Hospital, London, UK); Clara Maluquer (Department of Medical Oncology, ICO L’Hospitalet, L’Hospitalet de Llobregat, Barcelona, Spain); Gianluca Gaidano (Hematology, Division of Hematology, Amedeo Avogadro University of Eastern Piedmont); and Macarena Izuzquiza and Alba Cabirta (Hematology, Vall d’Hebron University Hospital, Barcelona, Spain).
Footnotes
Twitter: @DettorreGino
Correction notice: This paper has been updated to amend author affiliations.
Contributors: Full access to all data in the study and responsibility for data integrity and analysis, drafting of the manuscript and statistical analysis: GMD. Study concept and design, analysis and interpretation of data: GMD and DJP. Acquisition of data: DJP, AZ, JAC, MB, CS, RS, AB, JB, RM, ES, FB, DG, SG, GR, ML, AM, NH, BB, RB, DO, AC, RB, SB, AM, RW, CC, NC, CT, OM, VT, MB, SP, VF, CAC, FD, JSE, NS, EF, MG, IGF, AJXL, TND, AP, DGI, RR, PD, RS, YNSW, DF, JMH, AS, CM, IRC, LR, LC, MI, AC, MF, AS, AP, JT, and AG. Obtainment of funding and study supervision: DJP. Manuscript revision and input: all authors.
Funding: This work was supported by grant funding from the Wellcome Trust Strategic Fund (PS3416 to DJP) and acknowledges infrastructural support by the Cancer Research UK Imperial Centre and the Imperial NIHR BRC. AG is supported by the AIRC IG (grant number 14230), Associazione Italiana per la Ricerca sul Cancro Foundation, Milan, Italy.
Competing interests: DJP received lecture fees from ViiV Healthcare and Bayer Healthcare and travel expenses from BMS and Bayer Healthcare; consulting fees for Mina Therapeutics, EISAI, Roche, and Astra Zeneca; research funding (to institution) from MSD and BMS. AP has declared personal honoraria from Pfizer, Novartis, Roche, MSD Oncology, Eli Lilly, and Daiichi Sankyo; travel, accommodations, and expenses paid by Daiichi Sankyo; research funding from Roche and Novartis; and consulting/advisory role for NanoString Technologies, Amgen, Roche, Novartis, Pfizer and Bristol-Myers Squibb. TND has declared consulting/advisory role for Amgen, Bayer, AstraZeneca, BMS, Boehringer Ingelheim, Eli Lilly, MSD, Novartis, Otsuka, Pfizer, Roche, and Takeda; speakers fees from AstraZeneca, MSD, Roche, Takeda; and travel, accommodations and expenses paid by AstraZenca, BMS, Boehringer Ingelheim, Lilly, MSD, Otsuka, Roche, and Takeda. JB has declared consulting/advisory role for MSD and Astra Zeneca. PPS has declared consulting/advisory role for Sanofi and Abbvie. AP has declared consulting/advisory role for Takeda and Sanofi. MP has declared consulting/advisory role for Gilead and Bayer. AG has declared consulting/advisory role for Roche, MSD, Eli Lilly, Pierre Fabre, EISAI, and Daichii Sankyo; speakers bureau for Eisai, Novartis, Eli Lilly, Roche, Teva, Gentili, Pfizer, Astra Zeneca, Celgene, and Daichii Sankyo; research funds: EISAI, Eli Lilly, and Roche. LR reports receiving consulting fees from Amgen, ArQule, AstraZeneca, Basilea, Bayer, Celgene, Eisai, Exelixis, Hengrui, Incyte, Ipsen, Lilly, MSD, Nerviano Medical Sciences, Roche, and Sanofi; lectures fees from AbbVie, Amgen, Eisai, Gilead, Incyte, Ipsen, Lilly, Roche, and Sanofi; travel fees from Ipsen; and institutional research funding from Agios, ARMO BioSciences, AstraZeneca, BeiGene, Eisai, Exelixis, Fibrogen, Incyte, Ipsen, Lilly, MSD, and Roche. All remaining authors have declared no conflicts of interest.
Ethics approval and consent to participate: The United Kingdom Health Research Authority (20/HRA/1608) granted approval in addition to the corresponding ethics bodies of each participating center. Prospective informed consent was waived due to the anonymized nature of data access and retrospective study design.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
References
- 1. Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19). JAMA 2020;324:782. 10.1001/jama.2020.12839 [DOI] [PubMed] [Google Scholar]
- 2. Center for Systems Science and Engineering . Coronavirus COVID-19 global cases, 2019. Available: https://coronavirus.jhu.edu/map.html
- 3. Organization WH . Clinical management of COVID-19: interim guidance. V 1.5. 1.5 ED 2020.
- 4. Tay MZ, Poh CM, Rénia L, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020;20:363–74. 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moses AGW, Maingay J, Sangster K, et al. Pro-Inflammatory cytokine release by peripheral blood mononuclear cells from patients with advanced pancreatic cancer: relationship to acute phase response and survival. Oncol Rep 2009;21:1091–5. 10.3892/or_00000328 [DOI] [PubMed] [Google Scholar]
- 6. Baumann H, Gauldie J. The acute phase response. Immunol Today 1994;15:74–80. 10.1016/0167-5699(94)90137-6 [DOI] [PubMed] [Google Scholar]
- 7. Guthrie GJK, Charles KA, Roxburgh CSD, et al. The systemic inflammation-based neutrophil-lymphocyte ratio: experience in patients with cancer. Crit Rev Oncol Hematol 2013;88:218–30. 10.1016/j.critrevonc.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 8. Kwon H-C, Kim SH, Oh SY, et al. Clinical significance of preoperative neutrophil-lymphocyte versus platelet-lymphocyte ratio in patients with operable colorectal cancer. Biomarkers 2012;17:216–22. 10.3109/1354750X.2012.656705 [DOI] [PubMed] [Google Scholar]
- 9. Sun K, Chen S, Xu J, et al. The prognostic significance of the prognostic nutritional index in cancer: a systematic review and meta-analysis. J Cancer Res Clin Oncol 2014;140:1537–49. 10.1007/s00432-014-1714-3 [DOI] [PubMed] [Google Scholar]
- 10. Sanghera C, Teh JJ, Pinato DJ. The systemic inflammatory response as a source of biomarkers and therapeutic targets in hepatocellular carcinoma. Liver Int 2019;39:2008–23. 10.1111/liv.14220 [DOI] [PubMed] [Google Scholar]
- 11. Proctor MJ, Morrison DS, Talwar D, et al. An inflammation-based prognostic score (mGPS) predicts cancer survival independent of tumour site: a Glasgow inflammation outcome study. Br J Cancer 2011;104:726–34. 10.1038/sj.bjc.6606087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kato A, Tsuji T, Sakao Y, et al. A comparison of systemic inflammation-based prognostic scores in patients on regular hemodialysis. Nephron Extra 2013;3:91–100. 10.1159/000355148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pinato DJ, Zambelli A, Aguilar-Company J, et al. Clinical portrait of the SARS-CoV-2 epidemic in European cancer patients. Cancer Discov 2020. 10.1158/2159-8290.CD-20-0773. [Epub ahead of print: 31 Jul 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pinato DJ, Lee AJX, Biello F, et al. Presenting features and early mortality from SARS-CoV-2 infection in cancer patients during the initial stage of the COVID-19 pandemic in Europe. Cancers 2020;12. 10.3390/cancers12071841. [Epub ahead of print: 08 07 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)-a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harris PA, Taylor R, Minor BL, et al. The REDCap Consortium: building an international community of software platform partners. J Biomed Inform 2019;95:103208. 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Küppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer 2009;9:15–27. 10.1038/nrc2542 [DOI] [PubMed] [Google Scholar]
- 19. Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med 1999;341:1051–62. 10.1056/NEJM199909303411407 [DOI] [PubMed] [Google Scholar]
- 20. Barlogie B, Shaughnessy J, Tricot G, et al. Treatment of multiple myeloma. Blood 2004;103:20–32. 10.1182/blood-2003-04-1045 [DOI] [PubMed] [Google Scholar]
- 21. Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol 2020;146:110–8. 10.1016/j.jaci.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lippi G, Favaloro EJ. D-Dimer is associated with severity of coronavirus disease 2019: a pooled analysis. Thromb Haemost 2020;120:876–8. 10.1055/s-0040-1709650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippi G, Lavie CJ, Sanchis-Gomar F. Cardiac troponin I in patients with coronavirus disease 2019 (COVID-19): evidence from a meta-analysis. Prog Cardiovasc Dis 2020;63:390–1. 10.1016/j.pcad.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gómez-Pastora J, Weigand M, Kim J, et al. Hyperferritinemia in critically ill COVID-19 patients - Is ferritin the product of inflammation or a pathogenic mediator? Clin Chim Acta 2020;509:249–51. 10.1016/j.cca.2020.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dai M, Liu D, Liu M, et al. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov 2020;10:783–91. 10.1158/2159-8290.CD-20-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta 2020;506:145–8. 10.1016/j.cca.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther 2020;5:33. 10.1038/s41392-020-0148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Wang X, Jia X, et al. Risk factors for disease severity, unimprovement, and mortality in COVID-19 patients in Wuhan, China. Clin Microbiol Infect 2020;26:767–72. 10.1016/j.cmi.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. UK Coronavirus Cancer Monitoring Project team . The UK coronavirus cancer monitoring project: protecting patients with cancer in the era of COVID-19. Lancet Oncol 2020;21:622–4. 10.1016/S1470-2045(20)30230-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang L, Zhu F, Xie L, et al. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann Oncol 2020;31:894–901. 10.1016/j.annonc.2020.03.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whisenant JG, Trama A, Torri V, et al. TERAVOLT: thoracic cancers international COVID-19 collaboration. Cancer Cell 2020;37:742–5. 10.1016/j.ccell.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002277supp001.pdf (34.1KB, pdf)
jitc-2020-002277supp002.pdf (21.4KB, pdf)
jitc-2020-002277supp003.pdf (35.4KB, pdf)
jitc-2020-002277supp004.pdf (31.7KB, pdf)
jitc-2020-002277supp009.pdf (63.6KB, pdf)
jitc-2020-002277supp005.pdf (17.2KB, pdf)
jitc-2020-002277supp006.pdf (41.4KB, pdf)
jitc-2020-002277supp010.pdf (119KB, pdf)
jitc-2020-002277supp007.pdf (23.5KB, pdf)
jitc-2020-002277supp008.pdf (27.6KB, pdf)
Data Availability Statement
Data are available upon reasonable request.