Abstract
Adjuvants are key immunostimulatory components in vaccine formulations, which improve the immune response to the co‐administered antigen. The saponin natural product QS‐21 is one of the most promising immunoadjuvants in the development of vaccines against cancer and infectious diseases but suffers from limitations that have hampered its widespread human use. Previous structure–activity relationship studies have identified simplified saponin variants with truncated carbohydrate chains, but have not focused on the influence of the linear oligosaccharide domain of QS‐21 in adjuvant activity. Herein, an expeditious 15‐step synthesis of new linear trisaccharide variants of simplified QS‐21‐derived adjuvants is reported, in which the complex terminal xylose‐rhamnose moiety has been replaced with commercially available, simpler lactose and cellobiose disaccharides in a β‐anomeric configuration. In vivo immunological evaluation of the synthetic saponins showed attenuated antibody responses, highlighting the negative impact of such carbohydrate modifications on adjuvant activity, which could be associated with higher saponin conformational flexibility.
Keywords: carbohydrates, computational chemistry, conformational analysis, in vivo immunological evaluation, synthetic saponin adjuvants, vaccines
Shorter route to vaccine adjuvants: These multidisciplinary studies have provided streamlined access to new terminal disaccharide variants of truncated, QS‐21 inspired saponin vaccine adjuvants through an expeditious synthetic route, and have identified a critical role for the native rhamnose‐xylose moiety in adjuvant activity in connection with saponin conformation.
Introduction
Molecular subunit vaccines based on homogeneous antigens are emerging as a more precise and safer vaccination approach against a variety of human diseases. [1] They provide a successful alternative to classical vaccines based on whole inactivated pathogens, which in addition to their safety issues, generate less targeted immune responses owing to their crude and unpredictable processing. [2] The molecularly defined subunit structures comprise the minimal antigenic epitope required to elicit an immune response; however, they lack some “danger signals” naturally present in the native pathogen, which makes them poorly immunogenic. To overcome their low immunogenicity, adjuvants have become crucial components to increase the effectiveness of such subunit vaccines. [3]
Adjuvants are immunostimulatory substances that modulate and enhance the immune response towards a co‐administered antigen, providing key benefits in modern vaccine formulations. [4] Notably, they enable dose sparing of rare, expensive, and otherwise impotent antigens, which is particularly important in short supply situations (e.g., during an epidemic), and allow risk groups such as elderly, children, and immunocompromised patients to respond better to the vaccine. [5] Despite their essential roles, only a handful of adjuvants are sufficiently potent and exhibit acceptable toxicity to be used in human vaccines. Alum, a mixture of aluminium salts approved first alone in the 1930s and as part of the AS04 adjuvant system in 2009, has relatively low potency but is still considered the golden standard against which other candidates are compared. [6] Emulsion‐based adjuvants containing squalene (MF59, AS03) have been incorporated in several human influenza vaccines, albeit with some side effects. [7]
QS‐21 is a saponin natural product adjuvant composed of four structural domains, with a central quillaic acid triterpene core flanked on either side by oligosaccharides and a terminal acyl chain along its periphery. The C3 hydroxyl group of the triterpene is linked to a branched trisaccharide, whereas the C28 carboxylic acid is attached to a linear tetrasaccharide, which is further elaborated at its bridging fucose unit with a glycosylated acyl side chain. QS‐21 has been investigated in a number of vaccine clinical trials for cancer, infectious diseases, and neurodegenerative disorders, and is a component of the recently licensed AS01 adjuvant system in vaccines against malaria and shingles. [8] Despite its potency and promise, its wider use and commercial advancement in human vaccines as a stand‐alone adjuvant is limited by several factors. These include scarcity [9] and heterogeneity [10] of the natural product, further emphasized by the isomeric composition of QS‐21 at the fourth, terminal sugar (≈65:35 apiose/xylose, 1 a/1 b) within the linear tetrasaccharide (Figure 1 a). [11] Moreover, QS‐21 is chemically unstable and undergoes spontaneous hydrolysis of the acyl chain esters at physiological pH and ambient temperature, [12] complicating formulation and storage protocols and thwarting vaccine deployment in the developing world. Finally, QS‐21 shows dose‐limiting toxicity associated to hydrolyzed hemolytic byproducts, with local erythema and swelling as well as systemic flu‐like symptoms. [13] These liabilities in advancing QS‐21 further are also exacerbated by the fact that the mechanism of action of QS‐21 is not fully understood, [14] which hampers rational development of more effective saponin adjuvants.
Figure 1.
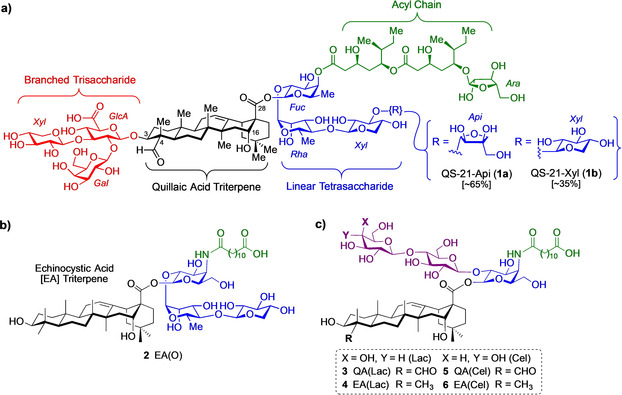
(a) Structure of the saponin natural product immunoadjuvant QS‐21 (65:35 mixture of 1 a/1 b) with its four structural domains. (b) Chemical structure of most recent echinocystic acid (EA) lead analog 2 EA(O). (c) Chemical structure of quillaic acid (QA) and echinocystic acid (EA) containing, terminal disaccharide lactose (Lac) and cellobiose (Cel) variants 3–6 studied herein.
To address the scarcity and heterogeneity of QS‐21 from the bark extract, Gin and co‐workers developed synthetic routes to QS‐21, procuring synthetic access to both isomers of the natural product (QS‐21‐Api, 1 a [15] and QS‐21 Xyl, 1 b [16] ; Figure 1 a) in pure form by total synthesis, albeit in an excessively long route totaling 76 steps. This chemical technology was subsequently applied in a more efficient and streamlined semisynthetic strategy that shortened the synthesis of QS‐21 to 56 total steps and also enabled the development of improved synthetic analogs of QS‐21 through structure–activity relationship studies. [17] The chemical instability issue was solved by the Gin group by incorporating amide linkages in the acyl chain instead of the labile ester groups, providing stable amide‐derived variants that retained adjuvant activity even with a simplified, linear side chain. [18] Wang et al. synthesized other chemically stable, adjuvant‐active QS‐21 analogs that lacked the original acyl chain but incorporated different linear side chains at the branched trisaccharide glucuronic acid through amidation.[ 19 , 20 ] Systematic truncation of the linear tetrasaccharide by Gin and co‐workers revealed that the fourth apiose/xylose unit was dispensable for adjuvant activity, identifying a potent linear trisaccharide analog accessed in 24 steps, albeit with considerable toxicity. [21] Although this work highlighted the importance of maintaining at least a trisaccharide moiety for optimal activity, the impact of the carbohydrate residues within this domain on adjuvant activity was not investigated, hampering the identification of more streamlined linear oligosaccharide saponin variants. Subsequent studies with central glycosidic ester linkage variants showed that this region is quite sensitive to structural modification, highlighting a β‐thioester variant with potent adjuvant activity but dose‐limiting toxicity. [22] Truncation of the left‐hand branched trisaccharide and molecular editing at the C4 and C16 triterpene positions provided simplified variants, which decoupled adjuvant activity from toxicity, [23] albeit in a 22‐step synthesis as exemplified by our most recent lead analog 2 EA(O) [24] (Figure 1 b).
Herein, we report new structure– and conformation–activity investigations into an unexplored structural feature of QS‐21, the right‐hand trisaccharide moiety previously identified as optimal for adjuvant activity. To probe specific structural requirements within this region, we implemented expeditious, flexible syntheses of new carbohydrate variants (3–6) incorporating modified terminal disaccharides (Figure 1 c). These variants were designed to increase the ease of synthesis by strategic selection of easily scalable sugar precursors, and could be prepared in fewer steps than our previously reported lead compound 2 EA(O) (as few as 15 total steps). [25] Through a combination of in vivo immunological evaluation and computational conformational analysis, we then investigated the effect of the terminal disaccharide motif in adjuvant activity and saponin conformation. These carbohydrate truncated variants induced lower antibody responses than the parent analog 2, indicative of weak adjuvant activity, and showed higher conformational flexibility in molecular dynamics simulations.
Results and Discussion
Chemical synthesis
In our earlier efforts, the adjuvant‐active saponin 2 EA(O), which lacks the branched trisaccharide but retains the original, truncated linear trisaccharide, was prepared in 22 total steps. [25] In particular, 16 of the total steps were already devoted to the synthesis of the linear trisaccharide itself, with each of the selectively protected monosaccharide building blocks requiring 2–6 steps. Therefore, in considering further specific molecular modifications of the QS saponin scaffold, our goal was to not only gain insight into the importance of the linear trisaccharide substructure for adjuvant activity, but also provide expedient access to saponin variants that can be prepared efficiently through abbreviated, expeditious syntheses.
With this double purpose in mind, we synthesized four saponin variants (3–6) glycosylated with two different, commercially available terminal disaccharides [lactose (Lac) and cellobiose (Cel); Figure 1 c], each of which incorporated the quillaic acid (QA) and echinocystic acid (EA) cores as key triterpene moieties present in previously identified adjuvant‐active saponins.[ 23 , 24 ] The rational selection of commercially available lactose and cellobiose avoided the lengthy synthesis of the original rhamnosylxylose disaccharide, enabling a more concise, scalable route that could be amenable to large‐scale production, while also probing the role of this moiety in adjuvant activity. Notably, instead of using the standard convergent approach employed for the synthesis of the parent saponin 2 EA(O), involving en bloc glycosylation of the triterpene with a pre‐assembled trisaccharide, these new carbohydrate variants were synthesized by following a streamlined, divergent strategy in which the bridging monosaccharide can be varied independently of the terminal disaccharide unit. [26] Thus, key steps in the overall route towards the target saponins involve initial coupling of the 4‐azidogalactose monosaccharide to the triterpene (QA, EA), followed by installation of the commercially available lactose or cellobiose disaccharides.
The synthetic sequence to prepare these variants was initiated with TES‐protected QA and EA triterpenes 7 and 8, respectively, [23] which were glycosylated at their C28 carboxylic acid with conveniently protected 4‐azidogalactosyl imidate 9 [21] under Schmidt coupling conditions to provide the corresponding glycosyl esters 10 and 11 (β anomers only) in excellent yields (Scheme 1).
Scheme 1.
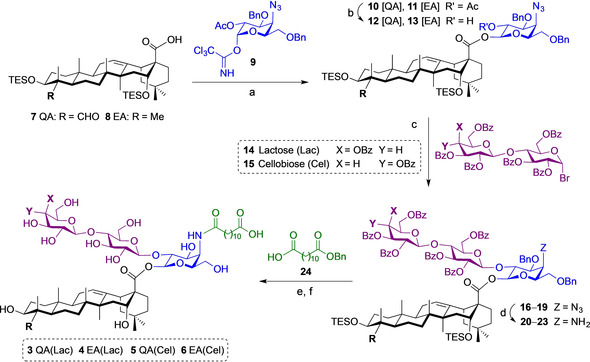
Expeditious 15‐step synthesis of lactose and cellobiose variants 3–6 via a flexible synthetic strategy. Reagents and conditions: (a) BF3 ⋅Et2O, 4 Å molecular sieves (MS), CH2Cl2, −78 °C to −45 °C, 0.5 h, 98 % for 10 [QA], 87 % for 11 [EA]; (b) NaOMe, MeOH, CH2Cl2, H2O, 35 °C, 22 h, 87 % for 12 [QA], 93 % for 13 [EA]; (c) 14, AgOTf, TTBPy, 4 Å MS, CH2Cl2/toluene (1:2), r.t., 1.5 h, 54 % for 16 [QA(Lac)], 72 % for 17 [EA(Lac)], 63 % for 18 [QA(Cel)], 84 % for 19 [EA(Cel)]; (d) PhSeH, Et3N/THF/toluene (10:3:3), 40 °C, 4 h, 83 % for 20 [QA(Lac)], 94 % for 21 [EA(Lac)], 99 % for 22 [QA(Cel)], 95 % for 23 [EA(Cel)]; (e) 24, EtOCOCl, Et3N, THF, 0 °C to r.t., 4 h, 85 % for [QA(Lac)], 90 % for [EA(Lac)], 89 % for [QA(Cel)], 91 % for [EA(Cel)]; (f) i. Pd/C, H2 (1 atm), EtOH/THF 1:1, r.t., 16 h, ii. TFA/H2O 3:1, 0 °C, 1 h, iii. NaOMe, MeOH, H2O, r.t., 6 h, 55 % overall for 3 [QA(Lac)], 57 % overall for 4 [EA(Lac)], 34 % overall for 5 [QA(Cel)], 49 % overall for 6 [EA(Cel)]. QA=quillaic acid; EA=echinocystic acid; Lac=lactose, Cel=cellobiose.
Removal of the C2 acetyl group was effected by using a sodium methoxide in methanol solution under carefully monitored conditions to prevent deprotection of the triterpene hydroxyl groups, affording the divergent intermediates 12 and 13. This triterpene‐monosaccharide building block was then used as a late‐stage diversification point for elongation with different commercially available disaccharides to access the corresponding new linear trisaccharide substructures. Among these, we selected lactose (β‐d‐galactopyranosyl‐(1→4)‐d‐glucopyranose) as one of the most common, inexpensive disaccharides with immunomodulatory functions [27] and cellulose‐derived cellobiose (β‐d‐glucopyranosyl‐(1→4)‐d‐glucopyranose) as a potential danger‐associated molecular pattern in immunity. [28] In contrast to the original rhamnosylxlyose disaccharide, which required a seven‐step synthesis, these alternative disaccharide surrogates were easily converted, upon HBr treatment (HBr solution, 33 wt % in acetic acid), into the hepta‐O‐benzoylated bromides 14 and 15. [29] These convenient sugar donors were subsequently coupled to the respective triterpene‐monosaccharide alcohol intermediates 12 and 13 through a silver triflate‐promoted Koenigs–Knorr glycosylation in the presence of 2,4,6‐tri‐tert‐butylpyridine (TTBPy) to provide the corresponding triterpene‐conjugated trisaccharides 16–19 with complete β‐selectivity owing to the C2‐benzoyl anchimeric assistance.
The azide groups in 16–19 were reduced by using benzeneselenol (PhSeH, prepared in situ) under basic conditions (Et3N) to afford the corresponding amines 20–23, onto which the carboxylic acid acyl chain was then appended. Thus, N‐acylation with dodecanedioic acid monobenzyl ester 24 was accomplished by initial carboxylate activation with ethyl chloroformate in the presence of Et3N, providing the resulting fully protected saponins. Finally, a three‐step global deprotection sequence involving hydrogenolytic removal of the O‐benzyl groups (H2, Pd/C), followed by acidic desilylation (TFA/H2O at 0 °C) of the triterpene hydroxyl groups and subsequent NaOMe‐mediated Zemplén saponification of the lactose/cellobiose benzoate esters provided the terminal disaccharide variants 3–6 in 15 total steps [25] and good overall yields.
Immunological evaluation
With the synthetic saponin targets in hand, we next sought to evaluate their adjuvant activity in comparison to QS‐21 and the parent echinocystic acid‐linear trisaccharide lead compound 2 EA(O). In this preliminary in vivo study, cohorts of five mice (C57BL/6, five animals per group, seven groups in total) were immunized subcutaneously (100 μL injections) three times every two weeks (days 0, 14, and 28) with the model antigen ovalbumin (OVA; 20 μg dose/mouse), either alone as a no adjuvant control group, or in combination with the saponin variant of interest (50 μg/mouse), dissolved in phosphate‐buffered saline (PBS). As positive controls, one group of mice was vaccinated with OVA plus QS‐21 (20 μg/mouse) and another group was co‐administered with OVA and our previous lead compound 2 EA(O) (50 μg/mouse) for comparison. Mice were bled the day before (day 27) and two weeks after the third vaccination (day 41), and the presence of antibodies was examined by enzyme‐linked immunosorbent (ELISA) assays to determine antibody responses against OVA. Serially diluted pre‐ and post‐treatment mouse sera were added to OVA‐coated ELISA plates, which were then washed. Specific antibody detection was carried out by using goat anti‐mouse immunoglobulin G (IgG) conjugated with horseradish peroxidase, followed by measurement of the absorbance of the sample at 450 nm. Thus, total anti‐OVA IgG antibody titers were analyzed by indirect ELISA at day 27 and day 41 after the first immunization, following the previously reported method. [22] The results obtained are shown in Figure 2.
Figure 2.
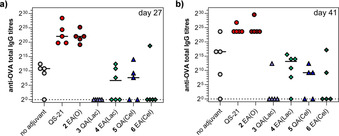
Immunological evaluation in mice of saponin variants 3–6 with OVA antigen. Antibody endpoint titers of total anti‐OVA IgG on (a) day 27 and (b) day 41 after first immunization. Data points correspond to individual mice (five animals per group) and horizontal bars indicate median titers.
Compared with the parent trisaccharide variant 2 EA(O), all four of the novel terminal disaccharide variants 3–6 showed lower antibody responses, indicative of attenuated adjuvant activity (Figure 2). Notably, echinocystic acid lactose variant 4 EA(Lac) showed generally higher and more consistent antibody titers compared with most of the other terminal disaccharide variants, followed closely by 5 QA(Cel) with slightly lower antibody levels on day 41. Moreover, the analysis of splenic T cell activation levels at the time of sacrifice (day 41) confirmed that 4, and to a lesser extent 5, induced the highest activation of CD4+ T cells, which was comparable to lead compound 2 EA(O) and significantly higher than the rest of synthetic variants (data not shown). Overall, the diminished antibody responses induced by these variants suggests the importance of the identity of the original linear trisaccharide for adjuvant activity.
Conformational analysis
To seek a molecular rationale for the attenuation in adjuvant activity observed for these saponin variants, which could be associated to the potential role of the terminal disaccharide in saponin conformation, we next performed an in silico conformational analysis on the synthetic terminal disaccharide variants 3–6 and the parent lead compound 2 EA(O) using all‐atom molecular dynamics (MD) simulations in explicit water (see the Supporting Information for complete details). Strikingly, the β‐linked terminal disaccharide variant 4 EA(Lac) showed a higher conformational flexibility than the more potent parent structure 2 EA(O), in which the terminal rhamnose‐xylose disaccharide is α‐linked to the bridging monosaccharide (Figure S1 in the Supporting Information). The distinct conformational preferences observed among the variants suggest that the chemical identity of the terminal disaccharide dictates the preferred global shape of the corresponding saponins. These observations point to a potential correlation between saponin conformational flexibility (i.e., preorganization into bioactive conformations) and attenuation in adjuvant activity, as observed for the synthetic variants 3–6 studied herein. Moreover, our investigations show that the α‐glycosidic linkage at the rhamnose anomeric position in 2 EA(O), which is also found in potent saponin adjuvants, [30] seems to be crucial to attain conformational rigidity correlating with immunostimulatory activity. This relationship between higher structural rigidity and increased adjuvant potency is in line with our previous studies with related saponin molecules.[ 22 , 24 ]
Conclusion
The amphiphilic properties and overall conformation of QS‐21 and other saponins, which is associated to their combined lipophilic and hydrophilic substructures, have been proposed to be more important for adjuvant activity than the contribution of the saponin individual functional groups. [31] Notably, the QS‐21 carbohydrate moieties have been suggested to play a role in saponin adjuvant activity by binding to cell surface lectins. [32] Herein, we have investigated the importance of the terminal disaccharide within the truncated right‐hand carbohydrate of QS‐21 by exploiting a flexible, 15‐step synthetic sequence, which streamlined the original convergent route by seven steps and provided an expeditious, facile access to saponin variants (3–6) with modified sugars at the terminus of the linear trisaccharide. Thus, we have combined the immunological evaluation and conformational analysis of these synthetic variants to study the impact of the terminal disaccharide modification in adjuvant activity in connection with saponin conformation. The obtained immunological results suggest that the original rhamnose‐xylose moiety within this domain is important for adjuvant activity, as their replacement with more easily accessible and scalable disaccharide surrogates led to diminished antibody responses. Notably, the molecular dynamics simulations yielded distinct conformational preferences for the four rather flexible terminal disaccharide variants (3–6) compared with the more structurally rigid, adjuvant‐active parent structure 2 EA(O), providing insights into the implications of such modification on saponin conformation, especially in terms of the higher conformational flexibility associated with attenuated adjuvant activity. Thus, these multidisciplinary studies suggest that saponin adjuvant activity is more closely connected to the specific substructures and three‐dimensional shape of the molecule rather than simply to their hydrophilic–hydrophobic balance, as has also been recently reported by Wang and co‐workers. [33] This is consistent with a mechanism of action involving interaction with discrete molecular receptor(s), whereby higher conformational flexibility would play against productive interactions necessary for proper target binding. Structure–activity and conformational studies of this kind may guide the rational design of synthetically accessible yet active saponin vaccine adjuvants in the near future. Going forward, the flexible synthetic design exploited herein leaves the door open for the potential development and rapid assessment of additional terminal disaccharide variants that may exhibit increased adjuvant activity, coupled with the expeditious access and scalability enabled by this route, which could facilitate future clinical deployment.
Experimental Section
General experimental procedure for synthesis of terminal disaccharide saponin variants
The selectively protected triterpenes (7, 8) [23] were monoglycosylated (BF3 ⋅OEt2, CH2Cl2) with 4‐azidogalactosyl trichloroacetimidate donor 9 [26] to give β‐glycosyl esters 10 and 11. Subsequent deacetylation (NaOMe, MeOH/CH2Cl2/H2O) gave the triterpene‐azidogalactose alcohols 12 and 13, as glycosyl acceptor building blocks for diversification. Koenigs–Knorr glycosylation (AgOTf, TTBPy, CH2Cl2/toluene) with perbenzoylated lactosyl and cellobiosyl bromides 14 and 15 [29] provided the triterpene‐trisaccharide azides 16–19. Subsequent reduction (PhSeH, Et3N/THF/toluene) to the corresponding amines 20–23 followed by N‐acylation with dodecanedioic acid monobenzyl ester (24) (EtOCOCl, Et3N, THF) gave the fully protected saponins S1–S4 (see the Supporting Information). Final global deprotection by hydrogenolysis (H2, Pd/C, EtOH/THF) and acid hydrolysis (TFA/H2O) followed by Zemplén de‐O‐benzoylation provided the corresponding terminal disaccharide saponin variants 3–6. See the Supporting Information for complete details.
Lactosyl‐(4‐(11‐carboxyundecanoamido)‐4‐deoxygalactosyl) quillaic acid ester [3 QA(Lac)]: Fully deprotected quillaic acid lactose variant 3 was obtained as a white solid following the corresponding general experimental procedures (see the Supporting Information for complete details). HPLC: t R=18.41 min, λ max=196 nm; 1H NMR (600 MHz, [D4]MeOH): δ=9.31 (s, 1 H, H‐23 [CHO] QA), 5.33 (d, J=8.0 Hz, 1 H, H‐1 N‐Gal), 5.31 (t, J=3.6 Hz, 1 H, H‐12 QA), 4.75 (d, J=7.9 Hz, 1 H, H‐1′ Glc), 4.73 (t, J=2.9 Hz, 1 H, H‐16 QA), 4.36–4.33 (m, 2 H, H‐1′′ Gal, H‐4 N‐Gal), 4.07 (dd, J=9.8, 8.0 Hz, 1 H, H‐2 N‐Gal), 4.03–3.96 (m, 2 H, H‐3 N‐Gal, H‐6a“ Glc), 3.89 (dd, J=12.0, 4.8 Hz, 1 H, H‐6b” Glc), 3.84 (d, J=3.2 Hz, 1 H, H‐4′′ Gal), 3.80–3.75 (m, 2 H, H‐3 QA, H‐6a′′ Gal), 3.74–3.70 (m, 2 H, H‐6b′′ Gal, H‐5 N‐Gal), 3.62–3.58 (m, 1 H, H‐5′′ Gal), 3.58–3.46 (m, 5 H, H‐2′′ and H‐3′′ Gal, H‐3′ and H‐4′ Glc, H‐6a N‐Gal), 3.46–3.40 (m, 2 H, H‐6b N‐Gal, H‐5′ Glc), 3.24 (dd, J=8.8, 8.1 Hz, 1 H, H‐2′ Glc), 2.96 (dd, J=14.4, 4.3 Hz, 1 H, H‐18 QA), 2.37–2.23 (m, 5 H, ‐CH 2(a)CONH and ‐CH 2(a′)CO2H ac, H‐19a QA), 2.00–1.91 (m, 4 H, H‐11a,b QA, H‐22a and H‐21a QA), 1.79–1.67 (m, 6 H, H‐9, H‐15a, H‐22b and H‐1a QA, ‐CH 2(b′)CH2CO2H ac), 1.65–1.50 (m, 6 H, H‐2a,b QA, H‐7a and H‐6a QA, ‐CH 2(b)CH2CONH ac), 1.39 (s, 3 H, CH3 C‐27 QA), 1.37–1.28 (m, 15 H, ‐[CH 2(c)]6‐ ×6 ac, H‐15b, H‐7b and H‐5 QA), 1.17–1.10 (m, 2 H, H‐21b and H‐1b QA), 1.09–1.04 (m, 1 H, H‐19b QA), 1.02 (s, 3 H, CH3 C‐24 QA), 1.00 (s, 3 H, CH3 C‐25 QA), 0.95 (s, 3 H, CH3 C‐30 QA), 0.94–0.90 (m, 1 H, H‐6b QA), 0.89 (s, 3 H, CH3 C‐29 QA), 0.78 ppm (s, 3 H, CH3 C‐26 QA); 13C NMR (151 MHz, [D4]MeOH): δ=208.9 (CHO QA), 178.3 (CONH ac), 177.8 (CO2H ac), 177.1 (CO2 [C‐28] QA), 145.0 (C‐13 QA), 123.1 (C‐12 QA), 105.3 (C‐1′′ Gal), 104.1 (C‐1′ Glc), 94.6 (C‐1 N‐Gal), 81.6 (C‐4′ Glc), 77.1 (C‐5′′ Gal), 76.84 (C‐5 N‐Gal), 76.81 (C‐2 N‐Gal), 76.7 (C‐5′ Glc), 76.6 (C‐3′ Glc), 75.4 (C‐2′ Glc), 74.9 (C‐3′′ Gal), 74.4 (C‐16 QA), 73.7 (C‐3 N‐Gal), 72.9 (C‐3 QA), 72.5 (C‐3′′ Gal), 70.2 (C‐4′′ Gal), 62.5 (C‐6′ Glc), 62.4 (C‐6′′ Gal), 62.0 (C‐6 N‐Gal), 56.8 (C‐4 QA), 52.0 (C‐4 N‐Gal), 49.9 (C‐17 QA), 48.9 (C‐5 QA), 48.1 (C‐9 QA), 47.8 (C‐19 QA), 42.6 (C‐14 QA), 42.0 (C‐18 QA), 41.2 (C‐8 QA), 39.5 (C‐1 QA), 37.0 (C‐10 QA), 36.7 (CH2CONH ac, C‐15 QA), 36.4 (C‐21 QA), 35.0 (CH2CO2H ac), 33.3 (CH3 C‐29 QA), 31.5 (C‐22 QA), 31.3 (C‐20 QA), 30.6, 30.5, 30.45, 30.40, 30.3, 30.2 (‐[CH2(c)]6‐ ×6 ac), 27.3 (CH3 C‐27 QA), 27.2 (C‐2 QA), 27.0 (CH2(b′)CH2CO2H ac), 26.1 (CH2(b)CH2CONH ac), 25.0 (CH3 C‐30 QA), 24.5 (C‐11 QA), 21.8 (C‐6 QA), 17.8 (CH3 C‐26 QA), 16.3 (CH3 C‐25 QA), 9.4 ppm (CH3 C‐24 QA); HRMS (MALDI) m/z: calcd for [C60H97NO22Na]+ [M+Na]+: 1206.6394; found: 1206.6341.
Lactosyl‐(4‐(11‐carboxyundecanoamido)‐4‐deoxygalactosyl) echinocystic acid ester [4 EA(Lac)]: Fully deprotected echinocystic acid lactose variant 4 was obtained as a white solid following the corresponding general experimental procedures (see the Supporting Information for complete details). HPLC: t R=19.31 min, λ max=196 nm; 1H NMR (600 MHz, [D4]MeOH): δ=5.32 (d, J=8.0 Hz, 1 H, H‐1 N‐Gal), 5.30 (t, J=3.5 Hz, 1 H, H‐12 EA), 4.74 (d, J=7.9 Hz, 1 H, H‐1′ Glc), 4.74–4.71 (m, 1 H, H‐16 EA), 4.38 (d, J=7.7 Hz, 1 H, H‐1′′ Gal), 4.36 (dd, J=4.8, 1.5 Hz, 1 H, H‐4 N‐Gal), 4.08 (dd, J=9.8, 8.0 Hz, 1 H, H‐2 N‐Gal), 4.04–3.97 (m, 2 H, H‐3 N‐Gal, H‐6a“ Glc), 3.91 (dd, J=12.0, 4.7 Hz, 1 H, H‐6b” Glc), 3.83 (d, J=3.3 Hz, 1 H, H‐4′′ Gal), 3.79 (dd, J=11.3, 7.4 Hz, 1 H, H‐6a′′ Gal), 3.74–3.69 (m, 2 H, H‐6b′′ Gal, H‐5 N‐Gal), 3.63–3.60 (m, 1 H, H‐5′′ Gal), 3.59–3.48 (m, 5 H, H‐2′′ Gal, H‐3′ Glc, H‐6a N‐Gal, H‐4′ Glc, H‐3′′ Gal), 3.46–3.40 (m, 2 H, H‐6b N‐Gal, H‐5′ Glc), 3.28–3.24 (m, 1 H, H‐2′ Glc), 3.15 (dd, J=11.6, 4.7 Hz, 1 H, H‐3 EA), 2.95 (dd, J=14.5, 4.3 Hz, 1 H, H‐18 EA), 2.38–2.21 (m, 5 H, CH 2(a)CONH and CH 2(a′)CO2H ac, H‐19a EA), 2.00–1.87 (m, 4 H, H‐22a, H‐11ab, H‐21a EA), 1.79–1.72 (m, 2 H, H‐22b, H‐15a EA), 1.68–1.50 (m, 10 H, H‐9 and H‐1a EA, CH 2(b)CH2CONH ac, CH 2(b′)CH2CO2Bn ac, H‐2ab, H‐6a and H‐7a EA), 1.47–1.40 (m, 1 H, H‐6b EA), 1.39–1.29 (m, 17 H, including 1.36 [s, 3 H, CH3 C‐27 EA], H‐7b and H‐15b EA, ‐[CH2(c)]6‐ ×6 ac), 1.16–1.12 (m, 1 H, H‐21b EA), 1.08–1.00 (m, 2 H, H‐19b and H‐1b EA), 0.98 (s, 3 H, CH3 C‐23 EA), 0.96 (s, 3 H, CH3 C‐25 EA), 0.95 (s, 3 H, CH3 C‐30 EA), 0.89 (s, 3 H, CH3 C‐29 EA), 0.78 (s, 6 H, CH3 C‐26 and CH3 C‐24 EA), 0.77–0.73 ppm (m, 1 H, H‐5 EA); 13C NMR (151 MHz, [D4]MeOH): δ=178.3 (CONH ac), 177.7 (CO2H ac), 177.1 (CO [C‐28] EA), 144.8 (C‐13 EA), 123.4 (C‐12 EA), 105.3 (C‐1′′ Gal), 104.2 (C‐1′ Glc), 94.6 (C‐1 N‐Gal), 81.4 (C‐4′ Glc), 79.8 (C‐3 EA), 77.1 (C‐5′′ Gal), 76.9 (C‐2 N‐Gal), 76.85 (C‐5 N‐Gal), 76.79 (C‐5′ Glc), 76.6 (C‐3′ Glc), 75.4 (C‐2′ Glc), 74.9 (C‐3′′ Gal), 74.5 (C‐16 EA), 73.6 (C‐3 N‐Gal), 72.6 (C‐2′′ Gal), 70.3 (C‐4′′ Gal), 62.5 (C‐6′ Glc), 62.4 (C‐6′′ Gal), 61.9 (C‐6 N‐Gal), 56.9 (C‐5 EA), 52.0 (C‐4 N‐Gal), 50.0 (C‐17 EA), 48.2 (C‐9 EA), 47.8 (C‐19 EA), 42.6 (C‐14 EA), 42.1 (C‐18 EA), 40.9 (C‐8 EA), 40.0 (C‐1 EA), 39.9 (C‐4 EA), 38.2 (C‐10 EA), 36.8 (CH2(a)CONH ac), 36.7 (C‐15 EA), 36.4 (C‐21 EA), 35.0 (CH2(a′)CO2H ac), 34.4 (C‐7 EA), 33.3 (CH3 C‐29 EA), 31.30 (C‐22 EA), 31.27 (C‐20 EA), 30.58, 30.56, 30.46, 30.41, 30.3, 30.2 (‐[CH2(c)]6‐ ×6 ac), 28.8 (CH3 C‐23 EA), 27.9 (C‐2 EA), 27.3 (CH3 C‐27 EA), 27.2 (CH2(b′)CH2CO2H ac), 26.1 (CH2(b)CH2CONH ac), 25.1 (CH3 C‐30 EA), 24.5 (C‐11 EA), 19.6 (C‐6 EA), 17.9 (CH3 C‐26 EA), 16.3 (CH3 C‐24 EA), 16.1 ppm (CH3 C‐25 EA); HRMS (MALDI) m/z: calcd for [C60H99NO21Na]+ [M+Na]+: 1192.6602; found: 1192.6682.
Cellobiosyl‐(4‐(11‐carboxyundecanoamido)‐4‐deoxygalactosyl) quillaic acid ester [5 QA(Cel)]: Fully deprotected quillaic acid cellobiose variant 5 was obtained as a white solid following the corresponding general experimental procedures (see the Supporting Information for complete details). HPLC: t R=15.70 min, λ max=195.52 nm; 1H NMR (600 MHz, [D4]MeOH): δ=9.31 (s, 1 H, H‐23 CHO QA), 7.97 (d, J=12.2 Hz, 1 H, NH), 5.33 (d, J=8.0 Hz, 1 H, H‐1 N‐Gal), 5.31 (t, J=3.6 Hz, 1 H, H‐12 QA), 4.75 (d, J=7.9 Hz, 1 H, H‐1′ Glc), 4.72 (t, J=3.1 Hz, 1 H, H‐16 QA), 4.40 (d, J=7.9 Hz, 1 H, H‐1′′ Glc), 4.35 (dd, J=4.8, 1.4 Hz, 1 H, H‐4 N‐Gal), 4.07 (dd, J=9.8, 8.0 Hz, 1 H, H‐2 N‐Gal), 4.03–3.96 (m, 2 H, H‐3 N‐Gal, H‐6′ Glc), 3.93–3.85 (m, 2 H, H‐6′ Glc, H‐6′′ Glc), 3.79–3.75 (m, 1 H, H‐3 QA), 3.74–3.66 (m, 2 H, H‐5 N‐Gal, H‐6′′ Glc), 3.55–3.32 (m, 8 H, H‐6ab N‐Gal, H‐3′, H‐4′ and H‐5′ Glc, H‐3′′, H‐4′′ and H‐5′′ Glc), 3.25–3.21 (m, 2 H, H‐2′ Glc, H‐2′′ Glc), 2.96 (dd, J=14.3, 4.2 Hz, 1 H, H‐18 QA), 2.37–2.27 (m, 2 H, CH 2(a)CONH ac), 2.26–2.23 (m, 2 H, CH 2(a′)CO2H ac, H‐19a QA), 2.00–1.89 (m, 4 H, H‐11ab, H‐22a, H‐21a QA), 1.79–1.66 (m, 6 H, H‐22b, H‐15b, H‐1b and H‐9 QA, CH 2(b′)CH2CO2H ac), 1.65–1.50 (m, 6 H, H‐6a, H‐7a and H‐2ab QA, CH 2(b)CH2CONH ac), 1.39 (s, 3 H, CH3 C‐27 QA), 1.38–1.28 (m, 15 H, H‐5, H‐15b and H‐7b QA, ‐[CH2(c)]6‐×6 ac), 1.17–1.10 (m, 2 H, H‐21b and H‐1b QA), 1.07 (dd, J=12.9, 2.9 Hz, 1 H, H‐19b QA), 1.02 (s, 3 H, CH3 C‐24 QA), 1.00 (s, 3 H, CH3 C‐25 QA), 0.95 (s, 3 H, CH3 C‐30 QA), 0.94–0.90 (m, 1 H, H‐6b QA), 0.89 (s, 3 H, CH3 C‐29 QA), 0.78 ppm (s, 3 H, CH3 C‐26 QA); 13C NMR (201 MHz, [D4]MeOH): δ=208.7 (CHO QA), 178.3 (CONH ac), 178.2 (CO2H ac), 177.1 (CO2 C‐28 QA), 145.0 (C‐13 QA), 123.1 (C‐12 QA), 104.8 (C‐1′′ Glc), 104.1 (C‐1′ Glc), 94.6 (C‐1 N‐Gal), 81.5 (C‐4′ Glc), 78.2 (C‐4′′ Glc), 77.9 (C‐3′′ Glc), 76.8 (C‐5 N‐Gal), 76.7 (C‐2 N‐Gal, C‐5′ Glc), 76.6 (C‐3′ Glc), 75.5 (C‐2′ Glc), 74.9 (C‐2′′ Glc), 74.3 (C‐16 QA), 73.7 (C‐3 N‐Gal), 72.9 (C‐3 QA), 71.3 (C‐5′′ Glc), 62.45 (C‐6′ Glc), 62.36 (C‐6′′ Glc), 61.9 (C‐6 N‐Gal), 56.8 (C‐4 QA), 52.1 (C‐4 N‐Gal), 49.9 (C‐17 QA), 48.7 (C‐5 QA), 48.1 (C‐9 QA), 47.8 (C‐19 QA), 42.6 (C‐14 QA), 42.0 (C‐18 QA), 41.2 (C‐8 QA), 39.5 (C‐1 QA), 37.0 (C‐10 QA), 36.75 (CH2(a)CONH ac), 36.72 (C‐15 QA), 36.4 (CH2(a′)CO2H ac, C‐21 QA), 33.6 (C‐7 QA), 33.3 (CH3 C‐29 QA), 31.35 (C‐20 QA), 31.28 (C‐22 QA), 30.8, 30.6, 30.4, 30.3 (‐[CH2(c)]6‐ ×6 ac), 27.3 (CH3 C‐27 QA), 27.2 (C‐2 QA), 27.0 (CH2(b′)CH2CO2H ac), 26.6 (CH2(b)CH2CONH ac), 25.1 (CH3 C‐30 QA), 24.5 (C‐11 QA), 21.8 (C‐6 QA), 17.9 (CH3 C‐26 QA), 16.3 (CH3 C‐25 QA), 9.4 ppm (CH3 C‐24 QA); HRMS (MALDI) m/z: calcd for [C60H97NO22Na]+ [M+Na]+: 1206.6394; found: 1206.6393.
Cellobiosyl‐(4‐(11‐carboxyundecanoamido)‐4‐deoxygalactosyl) echinocystic acid ester [6 EA(Cel)]: Fully deprotected echinocystic acid cellobiose variant 6 was obtained as a white solid following the corresponding general experimental procedures (see the Supporting Information for complete details). HPLC: t R=19.51 min, λ max=196 nm; 1H NMR (600 MHz, [D4]MeOH): δ=5.33 (d, J=8.0 Hz, 1 H, H‐1 N‐Gal), 5.30 (t, J=3.6 Hz, 1 H, H‐12 EA), 4.75 (d, J=7.9 Hz, 1 H, H‐1′ Glc), 4.71 (t, J=3.5 Hz, 1 H, H‐16 EA), 4.43 (d, J=7.9 Hz, 1 H, H‐1′′ Glc), 4.36 (d, J=4.7 Hz, 1 H, H‐4 N‐Gal), 4.08 (dd, J=9.8, 8.0 Hz, 1 H, H‐2 N‐Gal), 4.04–3.97 (m, 2 H, H‐3 N‐Gal, H‐6′a Glc), 3.94–3.87 (m, 2 H, H‐6b“ Glc, H‐6a′′ Glc), 3.75–3.65 (m, 2 H, H‐5 N‐Gal, H‐6b′′ Glc), 3.56–3.50 (m, 3 H, H‐4′ Glc, H‐6a N‐Gal, H‐3′ Glc), 3.47–3.41 (m, 2 H, H‐6b N‐Gal, H‐5′ Glc), 3.40–3.32 (m, 3 H, H‐3′′, H‐4′′ and H‐5′′ Glc), 3.29–3.22 (m, 2 H, H‐2′ Glc, H‐2′′ Glc), 3.15 (dd, J=11.4, 4.7 Hz, 1 H, H‐3 EA), 2.95 (dd, J=14.3, 3.6 Hz, 1 H, H‐18 EA), 2.39–2.19 (m, 5 H, CH 2(a)CONH and CH 2(a′)CO2H ac, H‐19 EA), 2.00–1.86 (m, 4 H, H‐22a, H‐21a, H‐11ab EA), 1.79–1.71 (m, 2 H, H‐15a, H‐22b EA), 1.70–1.49 (m, 10 H, H‐9 and H‐1a EA, CH 2(b′)CH2CO2H and CH2(b)CH 2CONH ac, H‐2ab, H‐6a and H‐7a EA), 1.47–1.27 (m, 18 H, including 1.35 [s, 3 H, CH3 C‐27], H‐6b, H‐7b and H‐15b EA, ‐[CH2(c)]6‐ ×6 ac]), 1.18–1.10 (m, 1 H, H‐21b EA), 1.06 (dd, J=12.8, 3.4 Hz, 1 H, H‐19b EA), 1.04–0.99 (m, 1 H, H‐1b EA), 0.98 (s, 3 H, CH3 C‐23 EA), 0.97–0.93 (m, 6 H, CH3 ×2 [C‐25 and C‐30 EA]), 0.89 (s, 3 H, CH3 C‐29 EA), 0.80–0.77 (m, 6 H, CH3 ×2 [C‐26 and C‐24 EA]), 0.77–0.73 ppm (m, 1 H, H‐5 EA); 13C NMR (151 MHz, [D4]MeOH): δ=178.3 (CONH ac), 177.8 (CO2H ac), 177.1 (CO2 [C‐28] EA), 144.7 (C‐13 EA), 123.4 (C‐12 EA), 104.8 (C‐1′′ Glc), 104.1 (C‐1′ Glc), 94.6 (C‐1 N‐Gal), 81.3 (C‐4′ Glc), 79.7 (C‐3 EA), 78.1 (C‐4′′ Glc), 77.9 (C‐3′′ Glc), 76.8 (C‐5 and C‐2 N‐Gal), 76.7 (C‐5′ Glc), 76.5 (C‐3′ Glc), 75.5 (C‐2′ Glc), 74.9 (C‐2′′ Glc), 74.4 (C‐16 EA), 73.7 (C‐3 N‐Gal), 71.2 (C‐5′′ Glc), 62.4 (C‐6′ Glc), 62.3 (C‐6′′ Glc), 61.9 (C‐6 N‐Gal), 56.9 (C‐5 EA), 52.0 (C‐4 N‐Gal), 50.1 (C‐17 EA), 48.2 (C‐9 EA), 47.7 (C‐19 EA), 42.6 (C‐14 EA), 42.1 (C‐18 EA), 40.9 (C‐8 EA), 40.0 (C‐1 EA), 39.9 (C‐4 EA), 38.2 (C‐10 EA), 36.8 (CH2CONH ac), 36.7 (C‐15 EA), 36.3 (C‐21 EA), 35.0 (CH2CO2H ac), 34.3 (C‐7 EA), 33.3 (CH3 C‐29 EA), 31.2 (C‐20 EA), 31.1 (C‐22 EA), 30.6, 30.5, 30.45, 30.39, 30.3, 30.2 (‐[CH2(c)]6‐ ×6 ac), 28.8 (CH3 C‐23 EA), 27.9 (C‐2 EA), 27.3 (CH3 C‐27 EA), 27.2 (CH2(b′)CH2CO2H ac), 26.1 (CH2(b)CH2CONH ac), 25.2 (CH3 C‐30 EA), 24.5 (C‐11 EA), 19.6 (C‐6 EA), 18.0 (CH3 C‐26 EA), 16.3 (CH3 C‐24 EA), 16.1 ppm (CH3 C‐25 EA); HRMS (MALDI) m/z: calcd for [C60H99NO21Na]+ [M+Na]+: 1192.6602; found: 1192.6643.
Further experimental details are available in the Supporting Information.
Conflict of interest
A. F.‐T. is co‐inventor on patents and patent applications that include saponin molecules presented in this work.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
Funding from the ERC (ERC‐2016‐STG‐716878 to A. F.‐T.), Spanish Ministry of Science and Innovation (CTQ2017‐87530‐R, RYC‐2015‐17888 to A. F.‐T.; RTI2018‐096494‐B‐100 to J. A.; RTI2018‐099592‐B‐C22 to G. J.‐O.; Severo Ochoa Accreditation SEV‐2016‐0644 to CIC bioGUNE), and the Mizutani Foundation for Glycoscience (200077 to G. J.‐O.) is gratefully acknowledged. A. F.‐T. thanks Raquel Fernández for inspiration.
R. Fuentes, A. Ruiz-de-Angulo, N. Sacristán, C. D. Navo, G. Jiménez-Osés, J. Anguita, A. Fernández-Tejada, Chem. Eur. J. 2021, 27, 4731.
Dedicated to Prof. Jesús Jiménez‐Barbero on the occasion of his 60th birthday
References
- 1. Moyle P. M., Toth I., ChemMedChem 2013, 8, 360–376. [DOI] [PubMed] [Google Scholar]
- 2. Jones L. H., Nat. Chem. 2015, 7, 952–960. [DOI] [PubMed] [Google Scholar]
- 3. Sarkar I., Garg R., van Drunen Littel-van den Hurk S., Expert Rev. Vaccines 2019, 18, 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reed S. G., Orr M. T., Fox C. B., Nat. Med. 2013, 19, 1597–1608. [DOI] [PubMed] [Google Scholar]
- 5. Gellin B. G., Salisbury D. M., Vaccine 2015, 33, B44–B46. [DOI] [PubMed] [Google Scholar]
- 6. HogenEsch H., O'Hagan D. T., Fox C. B., NPJ Vaccines 2018, 3, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barker C. I. S., Snape M. D., Lancet Infect. Dis. 2014, 14, 227–238. [DOI] [PubMed] [Google Scholar]
- 8. Lacaille-Dubois M. A., Phytomedicine 2019, 60, 152905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kensil C. R., Patel U., Lennick M., Marciani D., J. Immunol. 1991, 146, 431–437. [PubMed] [Google Scholar]
- 10. Kamstrup S., San Martin R., Doberti A., Grande H., Dalsgaard K., Vaccine 2000, 18, 2244–2249. [DOI] [PubMed] [Google Scholar]
- 11. Soltysik S., Bedore D. A., Kensil C. R., Ann. N. Y. Acad. Sci. 1993, 690, 392–395. [DOI] [PubMed] [Google Scholar]
- 12. Cleland J. L., Kensil C. R., Lim A., Jacobsen N. E., Basa L., Spellman M., Wheeler D. A., Wu J. Y., Powell M. F., J. Pharm. Sci. 1996, 85, 22–28. [DOI] [PubMed] [Google Scholar]
- 13. Ragupathi G., Gardner J. R., Livingston P. O., Gin D. Y., Expert Rev. Vaccines 2011, 10, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marciani D. J., Trends Pharmacol. Sci. 2018, 39, 573–585. [DOI] [PubMed] [Google Scholar]
- 15. Kim Y., Wang P., Navarro-villalobos M., Rohde B. D., Gin D. Y., J. Am. Chem. Soc. 2006, 128, 11906–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng K., Adams M. M., Damani P., Livingston P. O., Ragupathi G., Gin D. Y., Angew. Chem. Int. Ed. 2008, 47, 6395–6398; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2008, 120, 6495–6498. [Google Scholar]
- 17. Fernández-Tejada A., Tan D. S., Gin D. Y., Acc. Chem. Res. 2016, 49, 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams M. M., Damani P., Perl N. R., Won A., Hong F., Livingston P. O., Ragupathi G., Gin D. Y., J. Am. Chem. Soc. 2010, 132, 1939–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang P., Dai Q., Thogaripally P., Zhang P., Michalek S. M., J. Org. Chem. 2013, 78, 11525–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang P., Devalankar D. A., Dai Q., Zhang P., Michalek S. M., J. Org. Chem. 2016, 81, 9560–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chea E. K., Fernández-Tejada A., Damani P., Adams M. M., Gardner J. R., Livingston P. O., Ragupathi G., Gin D. Y., J. Am. Chem. Soc. 2012, 134, 13448–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walkowicz W. E., Fernández-Tejada A., George C., Corzana F., Jiménez Barbero J., Ragupathi G., Tan D. S., Gin D. Y., Chem. Sci. 2016, 7, 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fernández-Tejada A., Chea E. K., George C., Pillarsetty N., Gardner J. R., Livingston P. O., Ragupathi G., Lewis J. S., Tan D. S., Gin D. Y., Nat. Chem. 2014, 6, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghirardello M., Ruiz-De-Angulo A., Sacristan N., Barriales D., Jiménez-Barbero J., Poveda A., Corzana F., Anguita J., Fernández-Tejada A., Chem. Commun. 2020, 56, 719–722. [DOI] [PubMed] [Google Scholar]
- 25.Step counts based on number of isolated, characterized intermediates.
- 26. Fernández-Tejada A., Tan D. S., Gin D. Y., Chem. Commun. 2015, 51, 13949–13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan L. L., Deng Y. Y., Wang R., Wu C., Li J., Niu W., Yang Q., Bhatia M., Gudmundsson G. H., Agerberth B., Diana J., Sun J., Front. Immunol. 2018, 9, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Souza C. A., Li S., Lin A. Z., Boutrot F., Grossmann G., Zipfel C., Somerville S. C., Plant Physiol. 2017, 173, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lichtenthaler F. W., Kaji E., Weprek S., J. Org. Chem. 1985, 50, 3505–3515. [Google Scholar]
- 30. Deng K., Adams M. M., Gin D. Y., J. Am. Chem. Soc. 2008, 130, 5860–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kenji O., Matsuda H., Murakami T., Katayama S., Ohgitani T., Yoshikawa M., Biol. Chem. 2000, 381, 67–74. [DOI] [PubMed] [Google Scholar]
- 32. Marciani D. J., Drug Discovery Today 2003, 8, 934–943. [DOI] [PubMed] [Google Scholar]
- 33. Skalamera D., Kim H., Zhang P., Michalek S. M., Wang P., J. Org. Chem. 2020, 85, 15837–15848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


