Abstract
COVID-19 has proven to be a deadly virus, and unfortunately, it triggered a worldwide pandemic. Its detection for further treatment poses a severe threat to researchers, scientists, health professionals, and administrators worldwide. One of the daunting tasks during the pandemic for doctors in radiology is the use of chest X-ray or CT images for COVID-19 diagnosis. Time is required to inspect each report manually. While a CT scan is the better standard, an X-ray is still useful because it is cheaper, faster, and more widely used. To diagnose COVID-19, this paper proposes to use a deep learning-based improved Snapshot Ensemble technique for efficient COVID-19 chest X-ray classification. In addition, the proposed method takes advantage of the transfer learning technique using the ResNet-50 model, which is a pre-trained model. The proposed model uses the publicly accessible COVID-19 chest X-ray dataset consisting of 2905 images, which include COVID-19, viral pneumonia, and normal chest X-ray images. For performance evaluation, the model applied the metrics such as AU-ROC, AU-PR, and Jaccard Index. Furthermore, it also obtained a multi-class micro-average of 97% specificity, 95% f1-score, and 95% classification accuracy. The obtained results demonstrate that the performance of the proposed method outperformed those of several existing methods. This method appears to be a suitable and efficient approach for COVID-19 chest X-ray classification.
Keywords: COVID-19, Chest X-ray, Deep learning, Classification
Introduction
The world will remember 2020 as a catastrophic year for humanity. Pneumonia of unknown etiology, which was identified in Wuhan, China in December 2019 [26] with its earliest death reported on 10th January 2020, has become a pandemic [51] and is rapidly gulping the entire world under its net. The World Health Organization (WHO) named it COVID-19 (Corona Virus Disease-2019), and the virus is also known as SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) [37]. According to the Johns Hopkins Bloomberg School of Public Health, globally confirmed coronavirus cases reached 20,306,856, with 741,723 deaths recorded as of 12th August 2020 [28]. Owing to the coronavirus flare-up, the Research and Development wings of various research communities are effectively participating in identifying an effective compelling symptomatic system and vaccination for its treatment [59]. Because the cure is under discovery, it is essential to take sufficient precautionary measures and maximize testing. Owing to the scarcity of test kits for this confirmatory test, the search for alternatives is ongoing. In general, coronavirus side effects range from the usual cold to fever, cough, brevity of breath, intense respiratory issues and multi-organ failure, and death [51]. These are challenging tasks for master clinicians at each medical clinic owing to the limited number of radiologists. Therefore, simple, accurate, and quick models can help conquer this issue and provide convenient help to patients.
Furthermore, the rapid rise of the COVID-19 epidemic has increased the need for expertise and expanded enthusiasm for creating automated discovery systems that depend on artificial intelligence (AI) methods. AI approaches can help acquire accurate results and are useful at eliminating hindrances such as lack of accessible real-time reverse transcription-polymerase chain reaction (RT- PCR) test kits and waiting time of test outcomes [6]. According to Ref. [51], acute respiratory problems are the primary symptoms of COVID-19 that can be detected using chest X-ray (CXR) images. Chest computed tomography (CT) scans can recognize the infection when the symptoms are mild [53]. The use of this information can overcome the constraints of different tools such as the absence of diagnostic kits. While CT scan is a better standard, X-ray is still valuable because it is less expensive, faster, and more widely used [45, 47]. The advantage of using X-ray images is the accessibility of imaging systems at most health centers and laboratories, even in rural areas. In the absence of common side effects, such as fever, the use of X-ray images of the chest has a relatively decent capacity to recognize the illness [63].
In previous studies, several machine learning algorithms have been used to automatically classify digitized chest images [11, 31, 35]. Although the application of machine learning techniques for automatic diagnosis is useful in the clinical field, if there are enough annotated images, deep learning approaches are superior compared to classical machine learning methods [3, 54]. Deep learning allows developing end-to-end models to accomplish guaranteed results using input information without the need for manual feature extraction [30, 32]. Various deep learning approaches have been effectively applied to numerous issues, including skin cancer classification [9, 19], breast cancer identification [7, 10], brain disease classification [56], pneumonia detection using X-ray images of the chest [44], and lung segmentation [14, 52].
Various ensemble learning methods have been proposed to improve the performance of deep learning neural networks. This improvement can be achieved by combining the predictions from multiple models [20]. Ensemble learning combines the predictions from multiple neural network models to reduce predictions’ variance and generalization error. Recently, many ensemble approaches have shown their efficient performance in many fields including ‘classification of rockburst intensity’ [62], ‘motor imagery classification’ [33], ‘cervical histopathology image classification’ [61], and ‘detection of misleading information on COVID-19’ [12]. By observing the advantages of deep learning applications shown in the clinical field, this study proposes a deep learning-based improved Snapshot Ensemble technique for the efficient classification of COVID-19 CXR images.
Contributions
Below is the list of technical contributions of this study.
This study proposes a deep learning-based improved Snapshot Ensemble technique for COVID-19 CXR classification.
A popular Convolutional Neural Network (CNN) architecture (ResNet50), which is a pre-trained network, is applied by the transfer learning approach.
Data augmentation is implemented to deal with a relatively small number of samples, which prevents the model from over-fitting, to provide efficient performance.
Snapshot Ensemble technique is implemented, which allows using an ensemble of multiple neural networks at no additional training cost.
An improved Snapshot Ensemble algorithm is proposed to enhance model training and accuracy.
The obtained results are evaluated using popular metrics such as AU- ROC, AU- PR, precision, recall, specificity, accuracy, f1-score, and Jaccard Index.
The proposed model is compared with baseline methods to show the efficiency of COVID-19 CXR classification.
Roadmap
The rest of this paper is arranged as follows. Section 2 describes the related work. Section 3 discusses the preliminaries of the proposed method including data augmentation, convolution neural network, and Snapshot Ensemble technique. Section 4 describes the proposed improved Snapshot Ensemble technique. Moreover, dataset details and experimental results are reported in Section 5. Section 6 presents the discussion of experimental results. Finally, Section 7 presents the conclusions of this study.
Related work
Although this study is not constrained to clinical or biotechnology fields, it includes specialists from different fields (e.g., from AI and data science) to prevent and control the pandemic by providing their specialized perspectives and potential solutions. In previous studies, several methods have been proposed to detect, cure, and predict COVID-19. Different analysis approaches offer models to predict the pandemic’s evolution in specific geographical areas, countries, or create a global model. The models allow us to predict virus behavior, which is used to make future response plans. Hernandez-Matamoros et al. in [24] have analyzed the spread of COVID-19 using an Auto-Regressive Integrated Moving Average (ARIMA) model to show the spread of the pandemic through 6 geographic regions (continents). The model created a relationship between the countries and predicted the spread of the virus, behavior, and geographic region cases. Koushlendra Kumar Singh et al. in [50] have reported a Kalman filter-based short-term prediction model for forecasting COVID-19 using the popular machine learning techniques such as Random Forest and Pearson Correlation. However, the proposed approach is not used to predict the geographic region cases; instead, it is used to classify CXR images.
Among other diagnosis methods, medical images are essential [2, 13, 17, 18, 60]. Recently, CNN has become one of the most mainstream and successful methodologies that uses numerous radiology images to detect COVID-19. CNN’s initial advantage is its capacity to automatically learn features from domain-specific images, unlike the classical machine learning methods. The mainstream system for preparing CNN architecture is to transfer learned knowledge from a pre-trained system that satisfied one undertaking into another assignment [39]. The transfer of knowledge via the fine-tuning mechanism showed outstanding performance in X-ray image classification [3, 15, 49].
Hemdan et al. [23] have utilized a deep learning model called COVIDX-Net to analyze COVID-19 chest X-ray images containing 7 CNN models. Wang et al. [58] have proposed a deep learning CNN model for COVID-19 recognition (COVID-Net) using chest X-rays, which obtained a 92.4% accuracy when classifying COVID-19, non-COVID pneumonia, and normal classes. Apostolopoulos et al. [5] have built a deep transfer learning model using 224 positive COVID-19 images. This model achieved a 98.75% accuracy for binary class and a 93.48% progress rate for multi-class data. Ali Narin et al. [36] have achieved a 98% COVID-19 identification accuracy using CXR images combined with the ResNet50 model.
Recently, Sethy and Behera [48] have proposed a CNN-based model that relies on different ImageNet pre-prepared models to extract high-level features. Those features were fed into SVM as a machine learning classifier to distinguish COVID-19 CXR images. The abovementioned study shows that the combination of the ResNet50 model and SVM-classifier produced useful results. The abovementioned study suggested that transfer learning can separate critical biomarkers that are identified with the COVID-19 disease. Harsh Panwar et al. [40] have developed nCOVnet (i.e., a fast screening method for the detection of COVID-19 by analyzing X-rays), which is a deep neural network method. Rodolfo M. Pereira et al. have developed an RYDLS-20 model [41] using a resampling method. The model used a CXR image database and obtained a 0.65 f1-score.
Turker Tuncer et al. [57] have applied an automated Residual Exemplar Local Binary Pattern and iterative ReliefF-based method for COVID-19 lung X-ray image classification. A modified deep CNN model has also been proposed by Mohammad Rahimzadeh and Abolfazl Attar [43] for detecting COVID-19 and pneumonia in CXR images by concatenating Xception and ResNet50V2 methods. Asmaa Abbas et al. [1] have proposed another deep CNN model called DeTraC (Decompose, Transfer, and Compose) to classify COVID-19 CXR images and deal with random images by investigating class boundaries. Tulin Ozturk et al. have proposed an automatic COVID-19 detection method (DarkCovidNet) for CXR images using the DarkNet model with a transfer learning approach. Recently, Perumal et al. [42] have presented a transfer learning model to accelerate the prediction process and assist medical professionals in identifying COVID-19 using CT scan and CXR images. However, the tests used to identify COVID-19 are not sufficiently fast. The proposed approach overcomes the limitation of a long testing period using an automated deep learning-based technique. The proposed approach allows for obtaining results in less time, especially during the initial stages of virus development.
Preliminaries
This section describes details of the methods used for distinguishing COVID-19 from CXR images.
Data augmentation
The data imbalance problem makes the model more or less biased towards certain classes [4]. The proposed method uses the data augmentation approach to solve class imbalance, which artificially adds images to fewer categories to equal those of the largest class. The proposed approach randomly chose and copied the images belonging to the class with fewer samples to create duplicate images while resampling. However, because deep neural networks perform better with a large amount of data, data augmentation helps create images that depict its class’s features at every possible angle. Data augmentation ensures that the trained model can predict a class with higher precision at any angle the image is obtained. Different techniques used for data augmentation are as follows:
Randomly rotate images in the range (0 to 180∘)
Randomly zoom image
Randomly shift images horizontally
Randomly shift images vertically
Randomly flip the images horizontally
Randomly flip the images vertically
CNN
In general, deep learning approaches uncover the dataset’s highlights, such as images and videos that are hidden in the original data. Among these deep learning techniques, CNNs are widely used for medical image classification [21]. CNNs are feed-forward Artificial Neural Networks (ANN) [25] with alternating convolutional and sub-sampling layers. Profound 2D-CNN has many hidden layers and parameters. It can learn intricate patterns, given that it is trained on a gigantic size of visual database with ground-truth labels. Further, it is a modern architecture that processes high volumes of information with higher accuracy and relatively low computational expense compared to other classification algorithms owing to the efficiency in handling extensive data. Using different filters to identify specific features in images, CNN uses a unique way of image classification. Furthermore, the deep learning model’s relevant filters grasp the more in-depth features and convert them into predetermined features using pooling layers.
Transfer learning
The transfer learning approach is faster and simple to apply without the requirement for an enormous annotated dataset for training. Accordingly, numerous analysts tend to apply this strategy, particularly in medical imaging. This approach can be accomplished using the following important situations:
“Shallow- tuning” which adapts only the last classification layer to adapt to the new task and freezes the parameters of remaining layers without training;
“Deep- tuning” aims to retrain all of the parameters of the pre-trained model from the end-to-end approach;
“Fine- tuning” intends to continuously train more layers by tuning the learning parameters to achieve a considerable performance boost.
Transfer knowledge via the fine-tuning approach demonstrated exceptional performance in X-ray image classification [49].
Cyclic learning rate scheduling
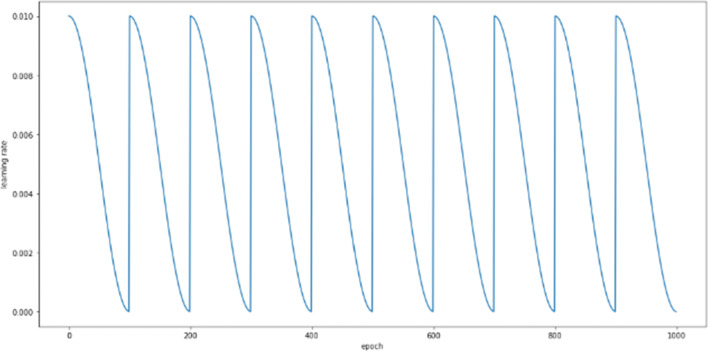
To improve results and make the model converge at a global minimum instead of a local minimum, the learning rate should be increased periodically instead of exponentially to determine the optimal learning rate. Cyclic learning rate scheduling makes this possible by cyclically changing the learning rate, which helps the model escape several global minima. In addition, this eliminates the necessity to find an optimal maximum learning rate manually. The utilized cyclic learning rate approach is shown in Fig. 1.
Fig. 1.
Visualization of the cyclic learning rate, where the X-axis shows the number of epochs, and the Y-axis shows the learning rate
Snapshot ensemble technique
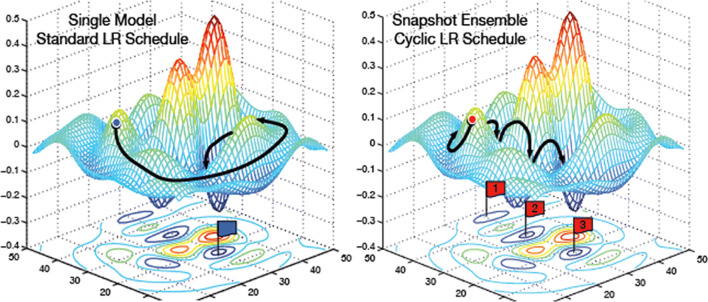
Ensemble models of neural networks are known to be substantially robust and accurate than individual networks. However, training multiple deep networks for model averaging is computationally expensive. Therefore, ‘Snapshot Ensembling’ has been proposed to ensemble multiple neural networks at no additional training cost with consistent lower error rates [27]. By adopting cyclic learning rate scheduling, Snapshot Ensembling has confirmed its compatibility with diverse network architectures and learning tasks. The proposed approach used this technique to periodically save model parameters during training. When the model converges to local minima during a cycle, these parameters are saved, and the learning rate increases to apply another model. This approach allows us to gather an ensemble of models in a single training cycle. Figure 2 shows the illustration of Stochastic Gradient Descent (SGD) optimization (with a typical learning rate schedule) and the illustration of Snapshot Ensemble [27].
Fig. 2.
Illustration of SGD optimization using typical learning rate schedule (left), and the illustration of Snapshot Ensembling (right) [27]
Proposed method
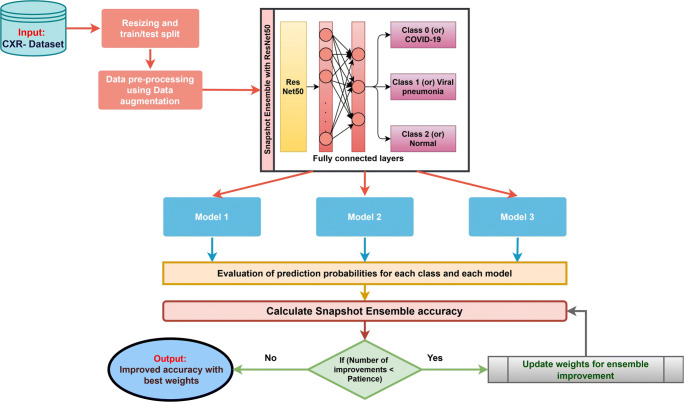
This section describes the proposed model creation and improved Snapshot Ensemble technique. Figure 3 visualizes the workflow of the proposed method.
Fig. 3.
Workflow of the proposed method
Model creation
The idea behind the transfer learning approach is to use the CNN model, which has been already trained on the ImageNet [46] data. The model is then applied to the lower layers of the proposed model to capture the generic features. In the proposed approach, the higher layers are fine-tuned to our specific domain and redefine the last layer that outputs three values that correspond to three different classes. The proposed approach uses ResNet50 [22] pre-trained architecture owing to its better results. These results were obtained while experimenting with five epochs on different pre-trained architectures. The proposed approach also uses the ‘Adam’ (adaptive moment estimation) optimizer [29] with weight decay to reduce overfitting and obtain the best validation accuracy upon training the data. Adam is one of the latest algorithms in the family of optimizers for model training. It combines two powerful optimizers: RMSProp (Root Mean Square Propagation) and AdaGrad (Adaptive Gradient). Unlike other optimizers, as training proceeds, it uses a different learning rate for every parameter in the network and then adjusts it along with the parameter.
The proposed model is built in Python using Keras Sequential API. In this API, we have to attach one layer to the model at a time. First, the ResNet50 architecture (a pre-trained architecture, which is used to capture generic features) is added to the model. Then, a dropout layer is added, which is a new regularization method. The dropout layer randomly ignores a few nodes from each training sample and makes the model learn features in a distributed way. In addition, it improves the generalization and reduces overfitting. A dense layer with 128 nodes is added, which is a part of a fully connected layer where different features from ResNet50 are converted to provide an output from 128 nodes. Then, a dropout layer is added, followed by the dense output layer with three nodes, which correspond to 3 different CXR types; the net output provides the probability of each class. These layers utilize commonly used ReLU (Rectified Linear Unit) [16] as an activation function, which adds nonlinearity to the model. Furthermore, the Snapshot Ensemble technique is added to the abovementioned model. Table 1 shows the proposed model summary.
Table 1.
Proposed model summary
| Layer (type) | Output shape | Param # |
|---|---|---|
| ResNet50 (Model) | (None, 2048) | 23587712 |
| dropout_1 (Dropout) | (None, 2048) | 0 |
| dense_1 (Dense) | (None, 128) | 262272 |
| dropout_2 (Dropout) | (None, 128) | 0 |
| dense_2 (Dense) | (None, 3) | 387 |
| Total param # : 23,850,371 | ||
| Trainable param # : 9,193,987 | ||
| Non-trainable param # : 14,656,384 | ||
The core of Snapshot Ensembling is an optimization process that visits few local minima before converging to the ultimate solution. It gradually saves snapshots at each local minimum and averages their predictions to quickly obtain the global minimum. Nevertheless, in the regular model, we have to travel for a long time to reach a global minimum. Thus, this ensemble model helps us to reach the global minimum in fewer epochs. To converge to multiple local minima, Snapshot Ensemble follows a Cyclic Cosine Annealing schedule [34] as a cyclic learning rate schedule. This method splits the training process into C cycles; each cycle starts with a large learning rate, annealed to a lower learning rate. The learning rate of α for the iteration t is calculated as follows:
| 1 |
Here, α0 represents the initial learning rate; T represents the total number of training iterations, and C represents the number of cycles.
Improved snapshot ensemble technique
Improved Snapshot Ensemble technique proposes to consider the weighted average instead of taking the average of probabilities of all models. To choose the weights for different models, random weight initialization is considered. After checking the improvements, new weights are added to the best weights. If there is no improvement, the number of improvements counter increases. We continue this process until the number of improvements counter reaches a specific limit; here, the limit is named patience. Thus, the final accuracy and final improved weights can be obtained. Algorithm 1 represents the pseudo-code of the improved Snapshot Ensemble algorithm.

Improved snapshot ensemble calculation
The proposed Snapshot Ensemble model uses three models. During model execution, three snapshots of the model have been saved. The snapshots produce three different weight probabilities for each model evaluation. An ensemble probability is calculated by taking the ‘average’ of all three probabilities. Further, the weights of the resultant class with the highest probability are used to obtain the ensemble model accuracy. Example illustrations of ‘Snapshot Ensemble model’ and ‘improved Snapshot Ensemble model’ calculations are shown in Tables 2 and 3, respectively. The details of Tables 2 and 3 are as follows:
C1,...Cn:represents the number of classes
M1,...Mm:represents the number of models
Pa:represents the predicted class probabilities using the Snapshot Ensemble model and
Pw:represents the predicted class probabilities using the improved Snapshot Ensemble model and
p11,...pn1:represents the prediction probabilities of model ‘M1’ for ‘n’ classes
Similarly, p1m,...pnm:represents the prediction probabilities of model ‘Mm’ for ‘n’ classes
p11,...p1m:represents the prediction probabilities of class ‘C1’ for ‘m’ number of models
Similarly, pn1,...pnm:represents the prediction probabilities of class ‘Cn’ for ‘m’ number of models
w1,...wm: represents the random weights initialized to ‘m’ number of models for ‘n’ number of classes
Table 2.
Illustration of the Snapshot Ensemble model prediction
| M1 | ..... | Mm | Pa | |
|---|---|---|---|---|
| C1 | p11 | ..... | p1m | =Average(p11,...., p1m) |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| Cn | pn1 | ..... | pnm | =Average(pn1,...., pnm) |
Table 3.
Illustration of the improved Snapshot Ensemble model prediction
| M1 | ..... | Mm | Pw | |
|---|---|---|---|---|
| C1 | p11w1 | ..... | p1mwm | =Average(p11w1,...., p1mwm) |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| . | . | ..... | . | . |
| Cn | pn1w1 | ..... | pnmwm | =Average(pn1w1,...., pnmwm) |
Experimental evaluation
This section describes the evaluation metrics, dataset details, and experimental procedure, along with obtained results and comparisons. Test executions are performed using Python and Keras Sequential API.
Evaluation metrics
After the implementation of the proposed approach, the proposed model performance is evaluated using popular evaluation metrics such as Area Under the Curve (AUC) - Receiver Operating Characteristic (ROC), Area Under Precision-Recall curve (AU-PR), Specificity (or) True Negative Rate (TNR), Precision (or) Positive Predictive Value (PPV), Recall (or) Sensitivity (or) True Positive Rate (TPR), f1-score (or) F-measure, accuracy, and Jaccard Index. ROC is a 2-dimensional graph that plots between TPR and False Positive Rate (FPR). Similarly, it may be characterized as an exchange between sensitivity and specificity. The ROC curve contains TPR on the Y-axis and FPR on the X-axis. AUC-ROC is most suitable when both classes maintain approximately the same number of samples. In the case of data imbalance, majority samples have a higher impact on the curve than minority samples, which causes a biased result. However, AUC-PR is mostly used for class imbalance problems because it does not consider false positives and false negatives, which produces unbiased results without sample influence. Medical studies require higher AUC results.
The Jaccard Index, which is also known as the Jaccard similarity coefficient, is a statistic that is used to understand similarities between sample sets. The mathematical representation of the Jaccard Index is as follows:
| 2 |
where, M and F represent the sample sets (if M and F are both empty, define J(M, F)), and 0 ≤ J(M, F) ≤ 1.
Similarly, the formulae used to evaluate the performance of the model (e.g., TNR, PPV, TPR, f1-score, and accuracy) are given as follows:
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
Where TP, FP, TN, and FN represent True Positive, False Positive, True Negative, and False Negative in independent datasets, respectively.
Dataset details
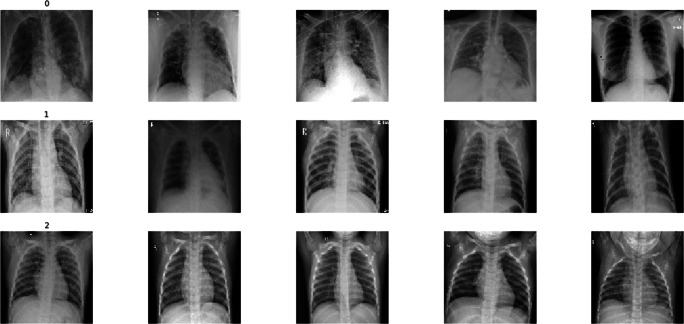
This study uses the collection of 2905 X-ray images from the COVID-19 CXR dataset. The image dataset is a publicly accessible COVID-19 CXR dataset [8], which is derived from the COVID-19 Radiography Database.1 It contains 219 COVID-19, 1345 viral pneumonia, and 1341 normal CXR images. Table 4 shows the details of the dataset. Figure 4 shows sample images from the X-ray dataset containing COVID-19, viral pneumonia, and normal types.
Table 4.
COVID-19 CXR dataset details
| X-ray type | Number of samples |
|---|---|
| COVID-19 | 219 |
| Viral Pneumonia | 1345 |
| Normal | 1341 |
| Total | 2905 |
Fig. 4.
Visualization of few X-ray samples [Note: From top to bottom, rows with labels 0, 1, and 2 represent the class names such as COVID-19, viral pneumonia, and normal samples, respectively]
Resizing of images and normalization
Keeping the same-size ratio does not result in the loss of information in the image. Because all original images in the dataset have different sizes, a considerable computation time is required to train the data. Therefore, all images in the dataset are resized to the dimension (75 × 100 × 3).
Because each input image pixel value ranges from 0 to 255, and the neural networks do not support the format, the following normalization method is applied to all images.
| 8 |
Here, NI represents the normalized image; X represents the original image pixel value; μ represents the mean of all corresponding pixel values, and σ represents the standard deviation of all corresponding pixel values.
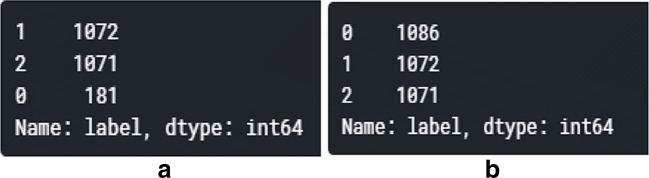
After normalization, each color code format value changed to the range of -2 to 2, which is preferred by neural networks. The obtained data were further divided into training and testing sets, i.e., 80% and 20%, respectively. The training dataset is divided into training and validation parts at the 80:20 ratio. After analyzing the data, the datasets were determined to be imbalanced. Then, according to Section 3.1, data augmentation was applied only to the final training data. Figure 5 shows the number of training instances of each category obtained after data preprocessing.
Fig. 5.
Details representing before and after data balancing using data augmentation for the CXR dataset [Note: Labels 0, 1, and 2 represent the class names such as COVID-19, viral pneumonia, and normal samples, respectively]. a Before data balancing. b After data balancing
Model training and testing
Testing is necessary to measure the classification accuracy of the testing data. The proposed model’s test accuracy is obtained using different combinations of the epochs and number of ensemble models. The proposed model is trained using an exponential learning rate reducer to improve the test accuracy. Apart from this, to reduce the computation time, the proposed model is trained with combinations of a different number of models and number of epochs so that the number of models × Number of epochs per model = 30, which makes the total number of epochs for the entire model to be 30. After reviewing the abovementioned combinations, the following optimal values are fixed: the number of models = 3, the number of epochs per model = 10, batch size = 10, and the maximum learning rate remains 0.001.
Furthermore, to determine the efficiency of the proposed model, performance evaluation is made for the following models, such as the proposed model (i.e., ResNet50 + improved Snapshot Ensemble + data balance), ResNet50 with data balance, and ResNet50 without data balance, on the CXR dataset. Table 5 shows the performance details of the proposed model on the CXR dataset. The ResNet50 model’s performance with data balance on the CXR dataset is shown in Table 6. Similarly, Table 7 shows the performance of the ResNet50 model without data balance on the CXR dataset. These results show that the proposed model can achieve an overall accuracy of 95% and specificity of 97% for the multi-class CXR dataset. Moreover, the micro average of precision, recall, f1-score, and Jaccard similarity is determined to be 95%, 95%, 95%, and 91%, respectively. Whereas the ResNet50 model, with data balance, achieved an overall accuracy of 92% for three classes of the same dataset. The micro average of precision, recall, f1-score, and Jaccard similarity obtained are 92%, 92%, 92%, and 86%, respectively.
Table 5.
Test accuracy of the proposed model on the COVID-19 CXR dataset
| X-ray | Support | Accu- | Jaccard | Precision | Recall | f1- | Specifi- |
|---|---|---|---|---|---|---|---|
| type | racy | Index | score | city | |||
| COVID-19 | 38 | 1 | 0.97 | 0.97 | 1 | 0.99 | 1 |
| Viral- | 273 | 0.90 | 0.90 | 1 | 0.90 | 0.95 | 0.99 |
| pneumonia | |||||||
| Normal | 270 | 0.99 | 0.91 | 0.91 | 1 | 0.95 | 0.92 |
| Sum | 581 | ||||||
| Average | 0.95 | 0.91 | 0.95 | 0.95 | 0.95 | 0.97 |
Table 6.
Test accuracy of the ResNet50 model with data balance on the COVID-19 CXR dataset
| X-ray | Support | Accu- | Jaccard | Precision | Recall | f1- | Specifi- |
|---|---|---|---|---|---|---|---|
| type | racy | Index | score | city | |||
| COVID-19 | 38 | 0.97 | 0.86 | 0.90 | 0.95 | 0.92 | 0.99 |
| Viral- | 273 | 0.86 | 0.85 | 0.99 | 0.86 | 0.92 | 0.99 |
| pneumonia | |||||||
| Normal | 270 | 0.95 | 0.86 | 0.88 | 0.99 | 0.93 | 0.88 |
| Sum | 581 | ||||||
| Average | 0.92 | 0.86 | 0.92 | 0.92 | 0.92 | 0.96 |
Table 7.
Test accuracy of the ResNet50 model without data balance on the COVID-19 CXR dataset
| X-ray | Support | Accu- | Jaccard | Precision | Recall | f1- | Specifi- |
|---|---|---|---|---|---|---|---|
| type | racy | Index | score | city | |||
| COVID-19 | 38 | 0.97 | 0.90 | 0.93 | 0.97 | 0.95 | 0.99 |
| Viral- | 273 | 0.84 | 0.83 | 0.99 | 0.84 | 0.91 | 0.99 |
| pneumonia | |||||||
| Normal | 270 | 0.98 | 0.86 | 0.87 | 0.99 | 0.92 | 0.87 |
| Sum | 581 | ||||||
| Average | 0.91 | 0.85 | 0.92 | 0.92 | 0.92 | 0.96 |
Similarly, the ResNet50 model without data balance achieved an overall accuracy of 91% for the same dataset. In addition, the micro average of precision, recall, f1-score, and Jaccard similarity is determined to be 92%, 92%, 92%, and 85%, respectively. Tables 5, 6, and 7 show that the proposed model exhibits an efficient performance compared with ResNet50 with data balance and ResNet50 without data balance. In addition, by observing the proposed model’s class results, the COVID-19 class has achieved a 100% accuracy, 100% specificity, and 99% f1-score. Moreover, the ensemble accuracy for three models, along with the improved accuracy using the proposed algorithm, are shown in Table 8. The proposed algorithm acquired a multi-class micro average accuracy of 95.18%. Therefore, the proposed model shows the potential to assist in COVID-19 treatment and decision making at critical stages.
Table 8.
Improved accuracy with the best weights using the proposed approach
| Model (or) Improvement | Accuracy (%) (or) Weight |
|---|---|
| Model-1 | 94.15 |
| Model-2 | 93.80 |
| Model-3 | 94.66 |
| Snapshot Ensemble | 94.84 |
| Improvement-1 | 95.01 |
| Improvement-2 | 95.18 |
| Best weight for model-1 | 0.020691 |
| Best weight for model-2 | 0.54205966 |
| Best weight for model-3 | 0.43724934 |
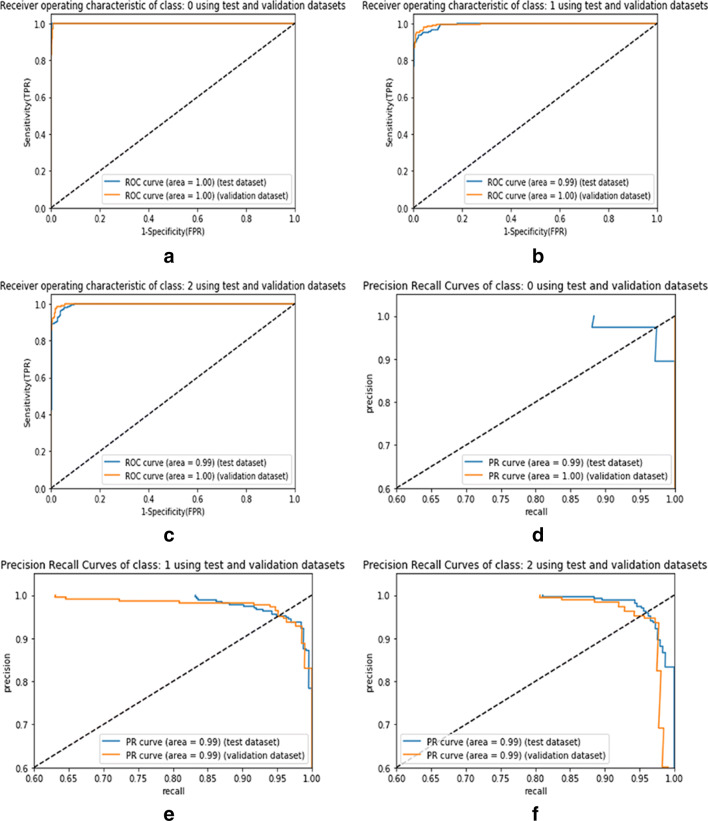
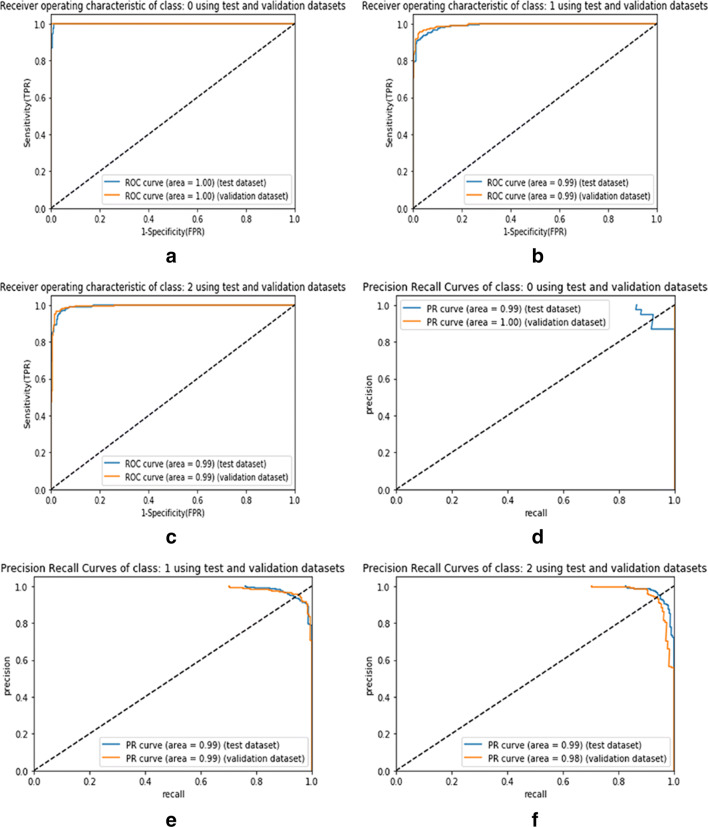
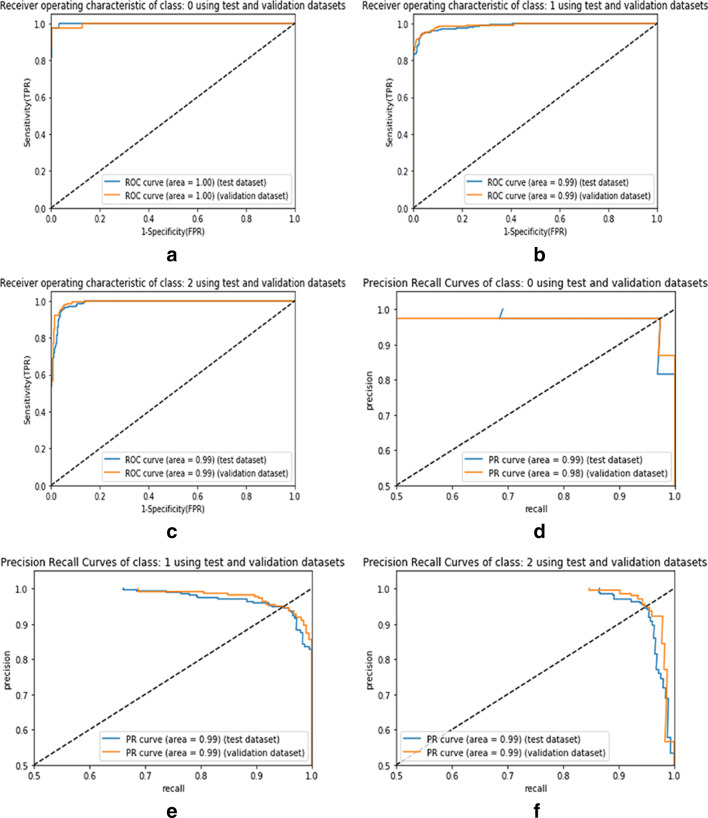
In Fig. 6, the AU-ROC and AU-PR curves represent the performance analysis of the proposed model for individual classes such as COVID-19, pneumonia, and normal CXR data. Similarly, in Fig. 7, the AU-ROC and AU-PR curves are plotted for the ResNet50 model with data balance. In Fig. 8, the AU-ROC and AU-PR curves are plotted for the ResNet50 model without data balance. Moreover, the proposed model obtained AU-ROC values of 1.00 for COVID-19, 0.99 for viral pneumonia, and 0.99 for normal data. Similarly, it obtained the AU-PR values of 0.99 for the COVID-19 class, 0.99 for viral pneumonia, and 0.99 for the normal class. These AU-ROC and AU-PR curves show the strength of the proposed approach in dealing with different types of image data.
Fig. 6.
AU-ROC and AU-PR curves representing the proposed model for the individual classes such as COVID-19, viral pneumonia, and normal. Figure 6a to c visualize the AU-ROC curves, and Fig. 6d to f visualize the AU-PR curves. [Note: Labels 0, 1, and 2 represent the class names such as COVID-19, viral pneumonia, and normal samples, respectively]
Fig. 7.
AU-ROC and AU-PR curves representing the ResNet50 model with balanced data for the individual classes such as COVID-19, viral pneumonia, and normal. Figure 7a to c visualize the AU-ROC curves, and Fig. 7d to f visualize the AU-PR curves. [Note: Labels 0, 1, and 2 represent the class names such as COVID-19, viral pneumonia, and normal samples, respectively]
Fig. 8.
AU-ROC and AU-PR curves representing the ResNet50 model without data balance for the individual classes such as COVID-19, viral pneumonia, and normal. Figure 8a to c visualize the AU-ROC curves, and Fig. 8d to f visualize the AU-PR curves. [Note: Labels 0, 1, and 2 represent the class names such as COVID-19, viral pneumonia, and normal samples, respectively]
Comparison
Table 9 provides a detailed comparison of the proposed model with recent baseline models to demonstrate the effectiveness of the proposed model. All comparisons are made only for the multi-class data. The majority of investigations referenced to compare with the proposed work used the COVID-19 CXR data, which was acquired from different openly accessible sources. The proposed model utilized a total of 2905 CXR images [219 COVID-19 (+), 1345 Viral Pneumonia, and 1341 Normal]. Although many proposed models in the literature showed efficient results while classifying CXR images, the proposed model obtained a multi-class micro-average of 97.16% specificity, 95.23% precision, 95.63% recall, 95.42% f1-score, and 95.18% classification accuracy.
Table 9.
Comparison of the proposed model with baseline models
| Method | Image | Number of | Accuracy | Precision | Recall | f1-score | Specifi- |
|---|---|---|---|---|---|---|---|
| name | type | cases | (%) | (%) | (%) | (%) | city (%) |
| ResNet+ Location | Chest CT | 219 COVID-19 (+) | 86.70 | 81.30 | 86.70 | 83.90 | 93.10 |
| Attention [60] | 224 Viral pneumonia | ||||||
| 175 Healthy | |||||||
| DarkCovidNet [38] | CXR | 125 COVID-19 (+) | 87.02 | 89.96 | 85.35 | 87.37 | 92.18 |
| 500 Pneumonia | |||||||
| 500 No-FIndings | |||||||
| Xception and | CXR | 180 COVID-19 (+) | 91.40 | 35.27 | 80.53 | 49.05 | 99.56 |
| ResNet50V2 [43] | 6054 Pneumonia | ||||||
| 8851 Normal | |||||||
| ResNet50 (applied | CXR | 219 COVID-19 (+) | 92.25 | 92.35 | 92.47 | 92.41 | 96.12 |
| in this work) | 1345 Viral Pneumonia | ||||||
| 1341 Normal | |||||||
| DeTraC [1] | CXR | 105 COVID-19 (+) | 93.10 | - | 100 | - | 85.18 |
| 11 SARS | |||||||
| 80 Normal | |||||||
| COVID-Net [58] | CXR | 358 COVID-19 (+) | 93.30 | 98.90 | 91.00 | 94.78 | 95.67 |
| 5538 COVID-19 (-) | |||||||
| 8066 Healthy | |||||||
| VGG-19 [5] | CXR | 224 COVID-19 (+) | 93.48 | 93.27 | 92.85 | 93.06 | 98.75 |
| 700 Pneumonia | |||||||
| 504 Healthy | |||||||
| ResNet18 [55] | CXR | 423 COVID-19 (+) | 95.02 | 93.88 | 93.61 | 93.74 | 92.41 |
| 144 MERS | |||||||
| 134 SARS | |||||||
| RYDLS-20 [41] | CXR | 1000 Normal | 95.10 | 57.61 | 82.76 | 65.00 | 99.30 |
| 90 COVID-19 (+) | |||||||
| 10 MERS | |||||||
| 11 SARS | |||||||
| 10 Varicella | |||||||
| 12 Streptococcus | |||||||
| 11 Pneumocystis | |||||||
| Proposed | CXR | 219 COVID-19 (+) | 95.18 | 95.23 | 95.63 | 95.42 | 97.16 |
| model | 1345 Viral Pneumonia | ||||||
| 1341 Normal | |||||||
Discussion
Because the scarcity of COVID-19 test kits necessitated the need for automated discovery systems that depend on AI methods, the proposed improved Snapshot Ensemble technique utilizes ResNet50 (which is a transfer learning approach) to provide a quick alternative to aid the diagnosis process. Many models described in the literature utilized the advantage of transfer learning technique and ResNet models to achieve better performance results. For example, Xiaowei Xu et al. in [60] obtained an 86.70% accuracy using the combination of ResNet18 model and location-attention mechanism for early screening and distinguishing COVID-19 from influenza-A viral pneumonia (IAVP) and healthy CT images. Further, a deep learning DarkCovidNet [38] model was utilized to detect COVID-19. This approach utilized 1125 CXR images consisting of 125 COVID-19 positive, 500 pneumonia, and 500 no-findings samples to develop the model. This approach showed an accuracy of 87.02%.
Similarly, Rahimzadeh and Attar [43] achieved a 91.40% accuracy using the concatenation of Xception and ResNet50V2 networks. Abbas et al. [1] obtained 93.10% accuracy and 100% recall using the DeTraC model. Furthermore, the COVID-Net [58] model achieved a 93.30% progress rate for COVID-19 detection using radiography pictures. Moreover, Apostolopoulos and Mpesiana developed a transfer learning approach using VGG19 and MobileNet v2 model [5] for a similar reason as COVID-Net. For this experiment, 224 COVID-19 positive, 700 cases of pneumonia, and 504 ordinary radiology pictures were used, and a 93.48% accuracy was achieved.
Furthermore, the studies presented in [55] and [41] have shown excellent performance by achieving greater than 95% accuracy. Most of these prior studies experienced data scarcity during the building of the models. The proposed model is developed to deal with the challenging COVID-19 problem by exploiting data augmentation for the class imbalance problem. Although many existing methods achieved excellent results, the proposed model demonstrated efficient results by achieving a high f1-score of 95.42% compared to other similar methods applied to CXR images. The proposed method demonstrated its robustness in coping with the limited availability of training data and irregularities in the data distribution. More importantly, the proposed improved Snapshot Ensemble algorithm provides a generic solution to improve the model’s efficiency. Moreover, the utilized metrics (e.g., micro average accuracy, precision, recall, f1-score, specificity, Jaccard similarity, AU-ROC, and AU-PR) showed excellent results to support the efficient performance of the proposed model.
The proposed method’s significant advantages are as follows. X-ray images are considered owing to the readily available disease diagnosis methods. It is an efficient approach to assist technicians with diagnosing to get fast predictions. CT scan is an expensive and not readily available procedure because this equipment is usually located in big hospitals. It is essential to collect balanced data for better predictions. Here, data augmentation and class balancing are essential in model performance, as has been previously observed. Moreover, another important benefit of the proposed method is that it does not depend on the disease stage; it can be applied even at an early stage.
In addition to classic image processing techniques, pre-defined generative models can be used to improve the model’s performance. The proposed approach used the ReLU activation function, which is the most commonly used activation function. However, it is recommended to try the available advanced activation functions. In the future, the proposed work can be addressed using various sources of data for COVID-19 diagnosis to compare the outputs with the current CXR image outcomes. It is also possible to get local CXR images of COVID-19 patients and evaluate them using the proposed model. After the evaluation, the proposed model can be deployed at local health centers.
Conclusion
In this study, a deep learning-based improved Snapshot Ensemble technique is proposed to efficiently classify COVID-19 CXR images. The proposed model takes advantage of the popular transfer learning approach (e.g., ResNet50) for efficient deep feature extraction. The proposed model enhanced the existing Snapshot Ensemble technique by providing an improved Snapshot Ensemble algorithm. The proposed model demonstrated efficient performance in classifying COVID-19, viral pneumonia, and normal CXR images. The model can obtain high micro average multi-class accuracy of 95% (with 97% specificity and 95% f1-score). The model can also obtain AU-ROC and AU-PR values of 1.00 and 0.99 for the COVID-19 class. Moreover, the model achieved a high f1-score compared to several modern methods. These results clearly show that this model can assist in COVID-19 treatment and decision making at critical stages.
Biographies
Samson Anosh Babu P
received his B.Tech. degree from Department of Information Technology at RVR & JC College of Engineering, Guntur, India, in 2009 and M.Tech. degree from Department of Computer Science and Engineering at National Institute of Technology Rourkela, India in 2012. Further, he worked as an Adhoc lecturer in the Department of Computer Science and Engineering at National Institute of Technology Calicut, India, from 2012 to 2013. He is currently a Full-time Research Scholar in the Department of Computer Science and Engineering at Indian Institute of Technology (Indian School of Mines) Dhanbad, India. His research interest includes Machine Learning techniques for Biological data analysis.

Chandra Sekhara Rao Annavarapu
is currently working as an Assistant Professor in the Department of Computer Science and Engineering at Indian Institute of Technology (Indian School of Mines) Dhanbad, India. He received his PhD from Computer Science and Engineering department, Indian Institute of Technology (Indian School of Mines) Dhanabd, India. He has more than 15 years of teaching experience and published many research papers in reputed journals and conferences. His research interests include Data and Knowledge-based System, Machine Learning, Evolutionary Algorithms, and Bioinformatics.

Funding
This study was not funded by any grants.
Declarations
Conflict of Interests
The authors declare that they have no conflicts of interest.
Ethical Approval
We further confirm that any aspect of the work covered in this manuscript has not involved human patients.
Footnotes
This article belongs to the Topical Collection: Artificial Intelligence Applications for COVID-19, Detection, Control, Prediction, and Diagnosis
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Samson Anosh Babu P, Email: samson.enosh1@gmail.com.
Chandra Sekhara Rao Annavarapu, Email: acsrao@iitism.ac.in.
References
- 1.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2021;51:854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alimadadi A, Aryal S, Manandhar I, Munroe PB, Joe B, Cheng X. Artificial intelligence and machine learning to fight COVID-19. Physiol Genomics. 2020;52(4):200–202. doi: 10.1152/physiolgenomics.00029.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou A, Storkey A, Edwards H (2017) Data augmentation generative adversarial networks. arXiv:1711.04340
- 5.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caobelli F. Artificial intelligence in medical imaging: game over for radiologists? Eur J Radiol. 2020;126:108940–108940. doi: 10.1016/j.ejrad.2020.108940. [DOI] [PubMed] [Google Scholar]
- 7.Celik Y, Talo M, Yildirim O, Karabatak M, Acharya UR. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit Lett. 2020;133:232–239. doi: 10.1016/j.patrec.2020.03.011. [DOI] [Google Scholar]
- 8.Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Emadi NA, Reaz MBI, Islam MT. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 9.Codella NCF, Nguyen Q, Pankanti S, Gutman DA, Helba B, Halpern AC, Smith JR. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Develop. 2017;61(4/5):5:1–5:15. doi: 10.1147/JRD.2017.2708299. [DOI] [Google Scholar]
- 10.Cruz-Roa A, Basavanhally A, González F, Gilmore H, Feldman M, Ganesan S, Shih N, Tomaszewski J, Madabhushi A (2014) Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In: Gurcan MN, Madabhushi A (eds) Medical imaging 2014: digital pathology. 10.1117/12.2043872, vol 9041. International society for optics and photonics, SPIE, pp 1–15
- 11.Dandıl E, Çakiroğlu M, Ekşi Z, Özkan M, Kurt ÖK, Canan A (2014) Artificial neural network-based classification system for lung nodules on computed tomography scans. In: 2014 6th international conference of soft computing and pattern recognition (SoCPaR). 10.1109/SOCPAR.2014.7008037, pp 382–386
- 12.Elhadad MK, Li KF, Gebali F (2020) An ensemble deep learning technique to detect COVID-19 misleading information. In: International conference on network-based information systems. Springer, pp 163–175
- 13.Fanelli D, Piazza F. Analysis and forecast of COVID-19 spreading in China, Italy and France. Chaos, Solitons & Fractals. 2020;134:109761. doi: 10.1016/j.chaos.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaál G, Maga B, Lukács A (2020) Attention U-net based adversarial architectures for chest X-ray lung segmentation. arXiv:2003.10304
- 15.Gao M, Bagci U, Lu L, Wu A, Buty M, Shin HC, Roth H, Papadakis GZ, Depeursinge A, Summers RM, Xu Z, Mollura DJ. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Computer methods in biomechanics and biomedical engineering. Imaging Visual. 2018;6(1):1–6. doi: 10.1080/21681163.2015.1124249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics, pp 315–323
- 17.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E (2020) Rapid AI development cycle for the coronavirus (COVID-19) pandemic:, Initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv:2003.05037
- 18.Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding COVID-19 from chest X-rays using deep learning on a small dataset. arXiv:2004.02060
- 19.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Derma. 2018;138(7):1529–1538. doi: 10.1016/j.jid.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 20.Hansen LK, Salamon P. Neural network ensembles. IEEE Trans Pattern Anal Machine Intell. 1990;12(10):993–1001. doi: 10.1109/34.58871. [DOI] [Google Scholar]
- 21.Haskins G, Kruger U, Yan P. Deep learning in medical image registration: a survey. Mach Vis Appl. 2020;31(1):8. doi: 10.1007/s00138-020-01060-x. [DOI] [Google Scholar]
- 22.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR). 10.1109/CVPR.2016.90, pp 770–778
- 23.Hemdan EED, Shouman MA, Karar ME (2020) COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv:2003.11055
- 24.Hernandez-Matamoros A, Fujita H, Hayashi T, Perez-Meana H. Forecasting of COVID19 per regions using ARIMA models and polynomial functions. Applied Soft Computing. 2020;96:106610. doi: 10.1016/j.asoc.2020.106610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopfield JJ. Artificial neural networks. IEEE Circ Devices Magazine. 1988;4(5):3–10. doi: 10.1109/101.8118. [DOI] [Google Scholar]
- 26.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang G, Li Y, Pleiss G, Liu Z, Hopcroft JE, Weinberger KQ (2017) Snapshot ensembles: train 1, get M for free. In: 5th international conference on learning representations, ICLR 2017, Toulon, France, April 24-26, 2017, Conference Track Proceedings. OpenReview.net. https://openreview.net/forum?id=BJYwwY9ll
- 28.JHU (2020) Johns Hopkins University coronavirus resource center. Accessed 12 Aug 2020. https://coronavirus.jhu.edu/
- 29.Kingma DP, Ba J (2015) Adam: a method for stochastic optimization. CoRR arXiv:1412.6980
- 30.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 31.Kuruvilla J, Gunavathi K. Lung cancer classification using neural networks for CT images. Comput Methods Programs Biomed. 2014;113(1):202–209. doi: 10.1016/j.cmpb.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 32.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 33.Lee BH, Jeong JH, Lee SW. Sessionnet: feature similarity-based weighted ensemble learning for motor imagery classification. IEEE Access. 2020;8:134524–134535. doi: 10.1109/ACCESS.2020.3011140. [DOI] [Google Scholar]
- 34.Loshchilov I, Hutter F (2016) SGDR: stochastic gradient descent with warm restarts. arXiv:1608.03983
- 35.Manikandan T, Bharathi N. Lung cancer detection using fuzzy auto-seed cluster means morphological segmentation and SVM classifier. J Med Syst. 2016;40(7):181. doi: 10.1007/s10916-016-0539-9. [DOI] [PubMed] [Google Scholar]
- 36.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv archivePrefix [DOI] [PMC free article] [PubMed]
- 37.Organization WH (2020) Naming the coronavirus disease (covid-19) and the virus that causes it. Accessed 02 Nov 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-ithttps://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-ithttps://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- 38.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 40.Panwar H, Gupta P, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos, Solitons & Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira RM, Bertolini D, Teixeira LO, Silla CN, Costa YM. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed. 2020;194:105532. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perumal V, Narayanan V, Rajasekar SJS. Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl Intell. 2021;51:341–358. doi: 10.1007/s10489-020-01831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform Med Unlocked. 2020;19:100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren MP, Ng AY (2017) CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv archivePrefix
- 45.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim JJ, Martin IBK, Anderson DJ, Kong C, Altes T, Bush A, Desai SR, Goldin O, Goo JM, Humbert M, Inoue Y, Kauczor HU, Luo F, Mazzone PJ, Prokop M, Remy-Jardin M, Richeldi L, Schaefer-Prokop CM, Tomiyama N, Wells AU, Leung AN. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the fleischner society. Radiology. 2020;296(1):172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, et al. Imagenet large scale visual recognition challenge. Int J Comput Vision. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 47.Self WH, Courtney DM, McNaughton CD, Wunderink RG, Kline JA. High discordance of chest X-ray and computed tomography for detection of pulmonary opacities in ED patients: implications for diagnosing pneumonia. Amer J Emerg Med. 2013;31(2):401–405. doi: 10.1016/j.ajem.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sethy PK, Behera SK. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. 2020;2020030300:2020. [Google Scholar]
- 49.Shin H, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh KK, Kumar S, Dixit P, Bajpai MK (2020) Kalman filter based short term prediction model for COVID-19 spread. Appl Intell [DOI] [PMC free article] [PubMed]
- 51.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Souza JC, Bandeira Diniz JO, Ferreira JL, França da Silva GL, Corrêa Silva A, de Paiva AC. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput Methods Programs Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Sun D, Li H, Lu XX, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World J Pediatr. 2020;16:251–259. doi: 10.1007/s12519-020-00354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun W, Zheng B, Qian W (2016) Computer aided lung cancer diagnosis with deep learning algorithms. In: Tourassi GD, III SGA (eds) Medical imaging 2016: computer-aided diagnosis. 10.1117/12.2216307, vol 9785. International Society for Optics and Photonics, SPIE, pp 241–248
- 55.Tahir A, Qiblawey Y, Khandakar A, Rahman T, Khurshid U, Musharavati F, Kiranyaz S, Chowdhury ME (2020) Coronavirus: comparing COVID-19. SARS and MERS in the eyes of AI. arXiv:2005.11524
- 56.Talo M, Yildirim O, Baloglu UB, Aydin G, Acharya UR. Convolutional neural networks for multi-class brain disease detection using MRI images. Computerized Medical Imaging and Graphics. 2019;78:101673. doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 57.Tuncer T, Dogan S, Ozyurt F. An automated residual exemplar local binary pattern and iterative ReliefF based COVID-19 detection method using chest X-ray image. Chemomet Intell Labor Syst. 2020;203:104054. doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Wong A (2020) COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. arXiv:2003.09871 [DOI] [PMC free article] [PubMed]
- 59.WHO (2020) World health organization- coronavirus disease (COVID-2019) R & D. Accessed 24 June 2020. https://www.who.int/teams/blueprint/covid-19
- 60.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J, Lang G, Li Y, Zhao H, Liu J, Xu K, Ruan L, Sheng J, Qiu Y, Wu W, Liang T, Li L (2020) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 10.1016/j.eng.2020.04.010. http://www.sciencedirect.com/science/article/pii/S2095809920301636 [DOI] [PMC free article] [PubMed]
- 61.Xue D, Zhou X, Li C, Yao Y, Rahaman MM, Zhang J, Chen H, Zhang J, Qi S, Sun H. An application of transfer learning and ensemble learning techniques for cervical histopathology image classification. IEEE Access. 2020;8:104603–104618. doi: 10.1109/ACCESS.2020.2999816. [DOI] [Google Scholar]
- 62.Zhang J, Wang Y, Sun Y, Li G. Strength of ensemble learning in multiclass classification of rockburst intensity. Int J Numer Anal Methods Geomech. 2020;44(13):1833–1853. doi: 10.1002/nag.3111. [DOI] [Google Scholar]
- 63.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296(2):E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]