Summary
NIN (NODULE INCEPTION) is a transcription factor that plays a key role during root nodule initiation. However, its role in later nodule developmental stages is unclear.
Both NIN mRNA and protein accumulated at the highest level in the proximal part of the infection zone in Medicago truncatula nodules. Two nin weak allele mutants, nin‐13/16, form a rather normal nodule infection zone, whereas a fixation zone is not formed. Instead, a zone with defence responses and premature senescence occurred and symbiosome development gets arrested.
Mutations in nin‐13/16 resulted in a truncated NIN lacking the conserved PB1 domain. However, this did not cause the nodule phenotype as nin mutants expressing NINΔPB1 formed wild‐type‐like nodule. The phenotype is likely to be caused by reduced NIN mRNA levels in the cytoplasm. Transcriptome analyses of nin‐16 nodules showed that expression levels of defence/senescence‐related genes are markedly increased, whereas the levels of defence suppressing genes are reduced. Although defence/senescence seems well suppressed in the infection zone, the transcriptome is already markedly changed in the proximal part of infection zone.
In addition to its function in infection and nodule organogenesis, NIN also plays a major role at the transition from infection to fixation zone in establishing a functional symbiosis.
Keywords: defence, early senescence, Medicago truncatula, NIN (NODULE INCEPTION), nodule development, symbiosome
Introduction
Legumes have the ability to establish a root nodule symbiosis with bacteria belonging to different genera collectively named rhizobia. Rhizobia are hosted inside specialised nodule cells where they are able to reduce atmospheric nitrogen into ammonia. Root nodule formation involves two main processes, infection and nodule organogenesis (Oldroyd & Downie, 2008). Infection starts in root hairs with the formation of tube‐like structures called infection threads by which bacteria enter root cells (Oldroyd et al., 2011). Nodule organogenesis begins with mitotic reactivation of fully differentiated root cells leading to the formation of a nodule primordium (Xiao et al., 2014). NIN (NODULE INCEPTION) is a key transcription factor that is essential for both processes (Schauser et al., 1999; Marsh et al., 2007). Weak alleles of nin have been identified that allow the formation of nodules (Pislariu et al., 2012; Veerappan et al., 2016). These nodules are small and do not fix nitrogen, suggesting that NIN also plays a role at later stages of nodule development. However, it is not clear in which nodule developmental process NIN is involved. Here we studied the role of NIN during later nodule developmental stages in Medicago truncatula (Medicago).
At the start of nodulation, NIN is transcriptionally induced upon perception of rhizobia secreted lipo‐chito‐oligosaccharides, called Nod factors (NF). NF activate a signalling pathway that is shared with the more ancient arbuscular mycorrhizal symbiosis. NIN is the first induced transcription factor that distinguishes the rhizobium activated responses from that of arbuscular mycorrhizae (Oldroyd, 2013). NIN is not only essential for nodulation in legumes, but also in actinorhizal(‐like) plants such as Casuarina and Parasponia (Clavijo et al., 2015; Bu et al., 2020). In both legumes as well as actinorhizal plants, NIN is nodule specifically expressed (Schauser et al., 1999; Marsh et al., 2007; Clavijo et al., 2015; Bu et al., 2020). During nodule initiation in legumes, NIN is induced in the epidermis, where it is required for infection thread formation (Vernié et al., 2015). In Medicago, NIN is also induced in the pericycle and this marks the start of nodule primordium formation, after which the expression of NIN extends to the dividing cortical cells (Liu et al., 2019b).
Medicago forms indeterminate nodules that have a zonation representing successive developmental stages (Vasse et al., 1990). At the apex there is a meristem that continuously adds cells to nodule tissues. Proximal to the meristem is the infection zone, where cells are penetrated by infection threads from which rhizobia are released. During release, bacteria become surrounded by a plant‐derived membrane, these organelle‐like structures are called symbiosomes (Roth & Stacey, 1989). Subsequently, symbiosomes divide and gradually enlarge. Bacterial differentiation involves endoreduplication and this is controlled by numerous antimicrobial peptides called nodule‐specific cysteine‐rich (NCR) peptides (Van de Velde et al., 2010; Vinardell et al., 2013; Alunni & Gourion, 2016). NCRs are defensin‐like peptides that are targeted to symbiosomes (Mergaert et al., 2003; Van de Velde et al., 2010). Although the infected cells become massively infected by rhizobia, defence responses are well suppressed. Several host genes have been identified that contribute to this suppression, including Symbiotic Cysteine‐rich Receptor Kinase (SymCRK), Regulator of Symbiosome Differentiation (RSD), Defective in Nitrogen Fixation 2 (DNF2) and NODULES WITH ACTIVATED DEFENCE 1 (NAD1) (Bourcy et al., 2013; Sinharoy et al., 2013; Berrabah et al., 2014; Wang et al., 2016; Domonkos et al., 2017). Mutations in these genes activate strong defence responses in nodules leading to necrotic cell death and/or premature senescence. Proximal to infection zone is the fixation zone, where rhizobia fully differentiate and start to fix nitrogen. The switch from infection to the distal part of the fixation zone (interzone II/III according to Vasse et al., 1990) occurs rapidly, and is marked by changes in cell morphology as well as gene expression. For example, the rhizobial nif genes, encoding components of nitrogenase are induced, amyloplasts accumulate, bacteroids markedly enlarge and become radially aligned (Gavrin et al., 2014).
In older nodules, a senescence zone is formed at the basal part of the nodule, where both host cells and rhizobia are degraded (Van De Velde et al., 2006). A key feature of nodule senescence is the increase in proteolytic activities (Pierre et al., 2014). Cysteine protease genes are highly induced in senescent nodules (Perez Guerra et al., 2010; Pierre et al., 2014). Knockdown of cysteine proteinase genes delays nodule senescence, while their ectopic expression induces premature senescence (Pierre et al., 2014). This underlines a crucial role of cysteine proteases in nodule senescence.
NIN is the first gene of the NIN‐like protein (NLPs) family that was identified (Schauser et al., 2005). Two domains are highly conserved in this gene family: a DNA‐binding RWP‐RK domain and a Phox and Bem1 (PB1) domain involved in protein–protein interaction (Schauser et al., 2005). A first clue that NIN also functions at later nodule developmental stages was gained from two weak nin alleles: NF10547 (nin‐16) and NF0440 (Pislariu et al., 2012; Veerappan et al., 2016). Both mutants form nitrogen fixing deficient (Fix−) nodules. Here, we analysed these mutants in detail to uncover the role of NIN in later nodule developmental stages.
Materials and Methods
Plant material and growth conditions
Medicago (Medicago truncatula) ecotypes Jemalong A17 and R108, and two nin mutants (nin‐13 and nin‐16) were used in this study. nin‐16 was provided by Rebecca Dickstein (University of North Texas); nin‐13 was obtained from the Noble Research Institute (https://medicago‐mutant.noble.org/mutant/). nin‐16 were backcrossed with R108, and nin‐13 and nin‐16 were crossed (F1 hybrid) according to Chabaud et al. (2006). Seven nin‐13 and nin‐16 hybrid F1 plants were genotyped by PCR (primers listed in Supporting Information Table S1). The obtained PCR fragments were sequenced to confirm Tnt1 insertion positions. Seed sterilisation, germination and Agrobacterium msu440 mediated hairy root transformation was performed according to Limpens et al. (2004). Medicago plants were grown in perlite saturated with low nitrate (0.25 mM Ca(NO3)2) containing Färhaeus (Fä) medium (Catoira et al., 2000) at 21°C and a 16 h : 8 h, light : dark regime. After 1 wk of growth, plants were inoculated with Sinorhizobium meliloti 2011 wild‐type or constitutively expressing green fluorescent protein (GFP) or carrying the PronifH:GFP reporter (OD600 = 0.1, 2 ml per plant).
RNA isolation and qRT‐PCR
RNA was isolated from 2‐wk‐old nodules using the EZNA Plant RNA mini kit (Omega Bio‐tek, Norcross, GA, USA). Here, 1 µg RNA was used for cDNA synthesis with the iScript cDNA synthesis kit (Bio‐Rad). Real‐time qPCR was performed in 10 µl reactions using SYBR Green Supermix (Bio‐Rad) and a CFX real‐time system (Bio‐Rad). Primers are listed in Table S1. Gene expression was normalised using ACTIN2 as a reference gene.
Constructs
NIN promotor (ProNIN3C‐5kb) and ProNIN3C‐5kb:NIN construct are described in Liu et al. (2019b). To construct ProNIN3C‐5kb:GFP‐NIN, ProNIN3C‐5kb:NIN‐GFP and ProNIN3C‐5kb:NINΔPB1, MultiSite Gateway (Thermo Fisher Scientific, Waltham, MA, USA) was used. pENTR‐4,1 and pENTR‐1,2 contained the 3C and 5 kb regions of the NIN promoter, respectively, pENTR‐2,3 was used to introduce coding sequences and 35S terminator (35Ster). NIN coding sequence was amplified (full‐length NIN or truncated NIN without the PB1 domain, the 3′‐UTR region of NIN was not included) by PCR using Medicago A17 nodule cDNA as template, Phusion high‐fidelity DNA polymerase (Finnzymes) and specific primers listed in Table S1. To make an N‐ or C‐terminus GFP fusion with NIN, pENTR‐TOPO‐NIN together with pENTR‐4,1‐35S:GFP and pENTR‐2,3‐35Ster or pENTR‐2,3‐GFP‐35Ster were recombined into Gateway binary vector pKGW‐RR‐MGW using LR Clonase II plus (Invitrogen) to generate Pro35S:GFP‐NIN‐35Ster or Pro35S:GFP‐NIN‐GFP‐35Ster. To fuse NINΔPB1 to 35S terminator, pENTR‐TOPO‐NINΔPB1 together with pENTR‐4,1‐35S:GFP and pENTR‐2,3‐35Ster were recombined into a pKGW‐RR‐MGW to generate Pro35S:GFP‐NINΔPB1‐35Ster. These constructs were used to amplify GFP‐NIN‐35Ster/NIN‐GFP‐35Ster/NINΔPB1‐35Ster by PCR using the forward primers with attB2 (GGGGACAGCTTTCTTGTACAAAGTGGAA) and reverse primers with attB3 (GGGGACAACTTTGTATAATAAAGTTGC) (Table S1). By the BP reaction these PCR fragments were introduced into pENTR‐2,3. Finally, the MultiSite Gateway reaction was performed to assemble pENTR‐2,3‐GFP‐NIN‐35Ster/NIN‐GFP35Ster/NINΔPB1‐35Ster with NIN promoter regions in pENTR‐4,1 and pENTR‐1,2 (TOPO) into the Gateway binary vector pKGW‐RR‐MGW.
Microscopy
Embedding of plant tissue in Technovit 7100 (Heraeus Kulzer), sectioning and staining were performed according to Xiao et al., 2014. Sections were analysed with DM5500B microscope equipped with a DFC425C camera (Leica). Bright‐field images were taken under a stereo macroscope (M165 FC, Leica). Hand‐sectioned nodules were examined using a Leica SP8 confocal microscope, excitation wavelengths of 488 nm and 543 nm were used for GFP/SYTO 9 and propidium iodide/DsRED, respectively. The accumulation of phenolic compounds was visualised by potassium permanganate staining (Bourcy et al., 2013). Live and dead staining of bacteria was performed as described by Haag et al. (2011)
Bacterial length measurement
R108 and nin‐13/16 were inoculated with Sinorhizobium meliloti 2011 constitutively expressing GFP. At 2 wk after inoculation, photographs of bacteria were taken using a Leica SP8 confocal microscope. The length of bacteria was measured using imagej software (Schneider et al., 2012).
RNA in situ hybridisation
RNA in situ hybridisation was conducted using Invitrogen ViewRNA ISH Tissue 1‐Plex Assay kits (Thermo Fisher Scientific) according to manufacturer's user guide and optimised for Medicago root and nodules sections (Kulikova et al., 2018). RNA ISH probe sets were designed and synthesised at Thermo Fisher Scientific. Catalogue numbers are VF1‐20312 for NIN (Medtr5g099060), VP2W7MP for CP2 (Medtr5g022560), VP7DPDU for SymCRK (Medtr3g079850), VPRWEMC for NAD1 (Medtr7g022640) and VF‐20311 for NF‐YA1 (Medtr1g056530). Each probe set was tested on tissue where it should not be present, in this case on noninoculated roots. As a negative control for each hybridisation procedure any probe set was omitted. In both cases no hybridisation signals were detected. The in situ images were taken with an AU5500B microscope equipped with a DFC425c camera (Leica).
RNA sequencing analyses
R108 and nin‐16 nodules were harvested 2 wk post inoculation. Three biological replicates were prepared for RNA sequencing (RNA‐seq). Total RNAs were isolated using the Qiagen RNeasy mini kit in accordance with the manufacturer's protocol. Isolated RNA samples were quality checked using an Agilent 2100 Bioanalyzer. Subsequently, an RNA‐seq library was constructed and sequenced on an BGISEQ‐500 platform with paired‐end reads at BGI (Hong Kong). About 4 Gb cleaned reads were obtained for each sample. Sequence reads were mapped to the M. truncatula genome v.5.0 (Pecrix et al., 2018) using the CLC genomics workbench 10.0.1 (Qiagen). Length fraction and similarity fraction were set to 0.9 during mapping and only uniquely mapped reads were considered in the analysis. All other parameters were set as default. Transcripts per million (TPM) (Wagner et al., 2012) and differential expression analyses were performed using the CLC genomic workbench 10.0.1. The criteria of fold change ≥ 4, false discovery rate (FDR) P‐value ≤ 0.01 and average TPM ≥ 5 were used to identify the differentially expressed genes.
Results
NIN is highly expressed in the nodule infection zone
According to the M. truncatula gene expression atlas, NIN is expressed in root nodules (Fig. S1). To determine in which stage of nodule development NIN could play a role, we performed RNA in situ hybridisation using a NIN‐specific probe set (Materials and Methods). This showed that in 2‐wk‐old nodules, NIN was expressed highest in the infected cells of the infection zone and the expression level was very low in the noninfected cells (Fig. 1a–c). NIN mRNA was also detected in the nodule vasculature (Fig. 1a). In the meristem and distal part of the infection zone the expression of NIN was very low (Fig. 1b), whereas in the proximal cell layers of the infection zone, expression of NIN was highest (Fig. 1a,b). At the transition from infection to fixation zone NIN expression level suddenly dropped (Fig. 1a–c). This switch coincides with amyloplast accumulation at the periphery of the infected cells (Fig. 1c) (Gavrin et al., 2014). A very low level of NIN expression was maintained throughout the fixation zone (Fig. 1a–c). The results of RNA in situ hybridisation are consistent with laser capture transcriptome data (Table S2) (Roux et al., 2014). This expression pattern suggests that NIN plays a role in processes that occur in the infection zone and/or at the transition from infection to fixation zone.
Fig. 1.
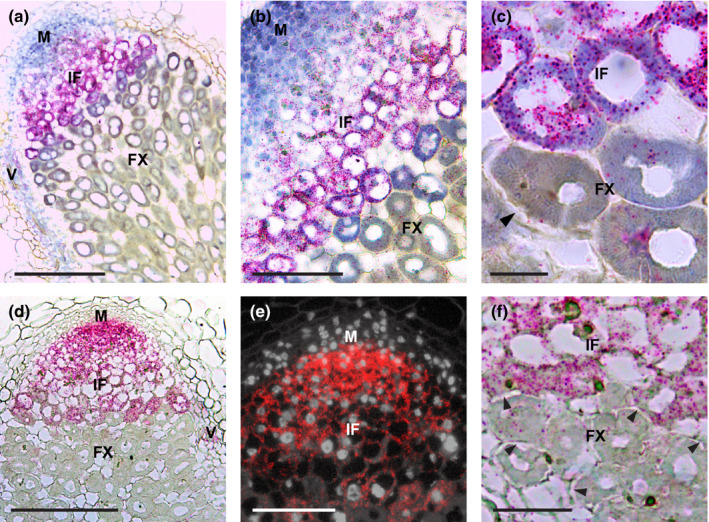
NIN and NF‐YA1 expression pattern in Medicago A17 nodule. An overview (a, d) and close‐up pictures (b, c, e, f) of RNA in situ localisation of NIN (a–c) or NF‐YA1 (d–f) in a nodule at 2 wk post inoculation (wpi). Hybridisation signals are visible as red dots. Arrowheads indicate amyloplast deposition. M, meristem; IF, infection zone; FX, fixation zone; V, vasculature. Bars: (a, d) 200 µm; (b, e) 100 µm; (c) 20 µm; (f) 50 µm.
NIN is nuclear located and is present at highest level in the proximal part of the infection zone
Studies on some NLPs have shown that they are located in the cytoplasm under low nitrate conditions and are translocated to the nucleus when the concentration of nitrate is high (Marchive et al., 2013; Cao et al., 2017; Lin et al., 2018). To determine in which cells NIN is located in nuclei, where it can induce expression of its target genes, we studied its subcellular localisation in nodules. The functional promoter of NIN (Liu et al., 2019b) was used to express NIN‐tagged with GFP at its C‐ or N‐terminus (ProNIN3C‐5kb:NIN‐GFP or ProNIN3C‐5kb:GFP‐NIN) in the nin knockout mutant nin‐1 (Marsh et al., 2007). At 4 wk after inoculation, pink nodules were formed in both cases (Fig. 2a,f), showing that both GFP fusions are biologically functional. Sections of these nodules showed that both C‐ and N‐terminus NIN‐GFP fusions localised in the nuclei in the meristem, infection zone, fixation zone and nodule vasculature (Fig. 2b–e,g–j). So, in all cells where NIN is expressed, NIN is located in the nucleus. NIN occurred at a very low levels in the distal part of the infection zone and at markedly higher levels in the proximal cell layers of infection zone (Fig. 2b,g). At the transition to the fixation zone, the NIN‐GFP/GFP‐NIN protein level dropped markedly, similar to the NIN mRNA (Fig. 2b,d,g,i).
Fig. 2.
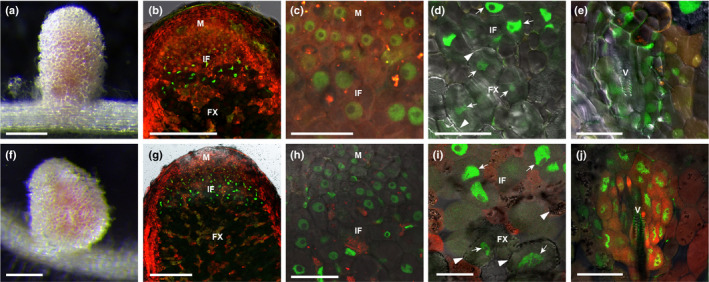
NIN protein is nuclear located in Medicago nodules. Pink nodules formed on nin‐1 roots transformed with ProNIN:NIN‐GFP (a) and ProNIN:GFP‐NIN (f) constructs at 6 wk post inoculation (wpi). Confocal images show that both NIN‐GFP (b–e) and GFP‐NIN (g–j) are located in the nucleus of cells in the meristem (c, h), infection zone (b–d, g–i), fixation zone (d, i) and vasculature (e, j). Transgenic roots were identified by DsRED fluorescent (red colour). Arrowheads indicate amyloplast deposition. Arrows indicate nucleus. M, meristem; IF, infection zone; FX, fixation zone; V, vasculature. Bars: (a, f) 2 mm; (b, g) 250 µm; (c–e, h–j) 40 µm.
The expression level of NF‐YA1 does not correlate with the level of NIN
NF‐YA1 has been shown to be a direct target of NIN in Lotus (Lotus japonicus) (Soyano et al., 2013). To test whether the level of NIN correlates with the level of NF‐YA1 expression in Medicago nodules, we determined the expression pattern of NF‐YA1 using RNA in situ hybridisation. This showed that NF‐YA1 was expressed in the entire infection zone as found for NIN (Fig. 1d). However, differently from NIN, the expression level of NF‐YA1 was high in the proximal part but not detectable in the distal part of the meristem (Fig. 1e). NF‐YA1 was also highly expressed in the distal part of infection zone, after which it decreased (Fig. 1d,e). At the transition to the fixation zone, its expression level markedly dropped as for NIN (Fig. 1f). Like NIN, NF‐YA1 was also expressed in the nodule vasculature (Fig. 1d). So the spatial expression patterns of NIN and NF‐YA1 were similar, but the expression level of NF‐YA1 did not correlate with that of NIN. This difference indicates the complexity of regulation of NF‐YA1 by NIN and suggests that additional transcription factors are involved.
nin‐13 and nin‐16 nodules showed premature senescence, defence responses and defects in symbiosome differentiation
To study the function of NIN in nodules, we analysed two weak alleles of nin (NF10547 and NF0440) that have been reported to form nonfunctional nodules (Pislariu et al., 2012; Veerappan et al., 2016). NF10547 was named nin‐16, and we named NF0440 nin‐13. nin‐16 is a monogenic recessive mutant (Veerappan et al., 2016), with a Tnt1 insertion in the last exon, between the DNA‐binding domain (RWP‐RK) and protein–protein interaction domain (PB1) (Fig. 3a). nin‐13 also has a Tnt1 insertion between the RWP‐RK and PB1 domains, but in nin‐13 it is located 22 bp upstream of that in nin‐16 (Fig. 3a). At 2 wk after inoculation, nin‐16 had formed white nodules indicating that they were not functional (Fig. 3e). This was confirmed by inoculation with Sinorhizobium meliloti 2011 containing PronifH:GFP. PronifH:GFP was highly expressed in the fixation zone of wild‐type nodules, but not in nin‐16 nodules (Fig. 3f,g). The number of nodules formed on nin‐16 was similar to that of wild‐type (R108) (Fig. 3b), indicating that nodule initiation was not affected. Semithin sections of nin‐16 nodules showed that an apical meristem was formed (Fig. 3k), infection threads penetrated nodule cells and rhizobia were released and had divided (Fig. 3m). However, before the rhizobia were fully elongated, nodule development was arrested and premature senescence was induced (Fig. 3o). This was also supported by live/dead staining of rhizobia that showed many dead bacteria in infected cells (Fig. S2). nin‐13 displayed the same nodule phenotype as nin‐16 (Fig. S3). F1 plants obtained by crossing these two mutants also had the same nodule phenotype, confirming that these mutants were allelic (Fig. S4). In the subsequent studies we mainly focused on nin‐16.
Fig. 3.
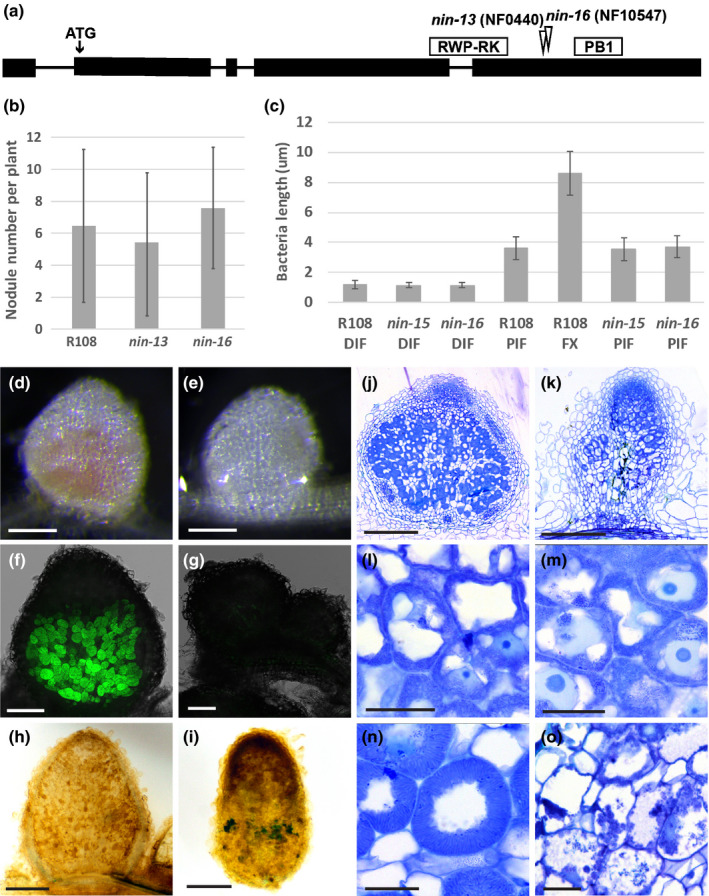
Phenotype of Medicago nin‐13 and nin‐16 mutants. (a) Schematic representation of the Tnt1 insertion site in nin‐13 and nin‐16 alleles. Exons of NIN are shown as black boxes, and introns are represented by lines. (b) The number of nodules formed on R108 (wild‐type), nin‐13 and nin‐16 are similar. Nodule numbers per plant (n = 19) were counted at 2 wk post inoculation (wpi). Data are means ± SD. (c) Rhizobia in nin‐13/16 were able to elongate to the size that in wild‐type plants reached before the transition to the fixation zone. Bacteria length are measured at 2 wpi. n 70, data are means ± SD. DIF, distal part of infection zone; PIF, proximal part of infection zone; FX, fixation zone. At 2 wpi with Sinorhizobium meliloti 2011, wild‐type nodules were pink (d) whereas nin‐16 nodules remained white (e) that indicate the absence of leghaemoglobin. Formation of nonfunctional nodules on nin‐16 was further confirmed by inoculation with Sinorhizobium meliloti 2011 carrying the PronifH::GFP reporter construct (f, g). nifH was highly induced in R108 (f) but not induced in nin‐16 nodules (g). Semithin sections stained with toluidine blue show that in comparison with wild‐type nodules (j, l, n), an apical meristem was formed on nin‐16 mutant nodule (k), rhizobia were released and divided (m), however, their development was arrested, and premature senescence was induced (o). Potassium permanganate/methylene blue staining shows accumulation of phenolic compound in nin‐16 nodule (i), but not in wild‐type nodule (h). nin‐13 shows similar phenotype as nin‐16, figures shown in Supporting Information Fig. S3. Bars: (d, e) 2 mm; (f, g) 200 µm; (h, i) 500 µm; (j, k) 300 µm; (l–o) 30 µm.
To determine when rhizobial development was blocked in the nin‐16 mutant nodules, we measured the length of the rhizobia (Fig. 3c). In both wild‐type and mutant nodules the rhizobia in the infection thread, or just released from it, were about 1 μm long. In the most proximal part of infection zone of the wild‐type nodules, the symbiosomes were about 3.5 μm in length. After the transition to the fixation zone, the fully differentiated rhizobia were about 8 μm long. Rhizobia in nin‐13/16 nodules could reach a length of c. 3.5 μm (Fig. 3c). This in combination with the lack of nif expression, indicated that the block of differentiation occurred at the stage when, in the wild‐type, the transition from infection to fixation zone takes place.
The block in rhizobia differentiation might be due to premature senescence and a not sufficiently suppressed defence response. The accumulation of phenolic compounds is commonly used to identify a defence response in nodules (Bourcy et al., 2013; Berrabah et al., 2015, 2018; Wang et al., 2016; Domonkos et al., 2017). Potassium permanganate/methylene blue staining showed that phenolic compounds indeed accumulated in nin‐16 nodules (Fig. 3i). This means that a defence response was induced in mutant nodules.
PB1 domain of NIN is not essential for nodule development
The Tnt1 insertion in both nin‐13 and nin‐16 will result in a truncated NIN protein lacking the protein–protein interaction domain (PB1). To test whether the nin‐13/16 phenotype is caused by the absence of the PB1 domain, we introduced a construct encoding a truncated NIN (deletion starts at the position of the Tnt1 insertion in nin‐16, including the PB1 domain) (ProNIN3C‐5kb:NINΔPB1) into the knockout mutant nin‐1 by hairy root transformation. Surprisingly, pink nodules were formed on transgenic roots 4 wk after inoculation (Fig. 4a). Next, we inoculated transgenic roots with rhizobia containing PronifH:GFP, and nifH was highly induced in the formed nodules (Fig. 4c,d). Semithin sections of these nodules revealed that 23 out of 67 (34.3%) were wild‐type‐like (Fig. 4b), comparable with 12 out of 38 (31.6%) wild‐type‐like nodules formed on nin‐1 roots transformed with full‐length NIN (ProNIN3C‐5kb:NIN). This suggests that the PB1 domain of NIN is not essential for the formation of functional nodules.
Fig. 4.
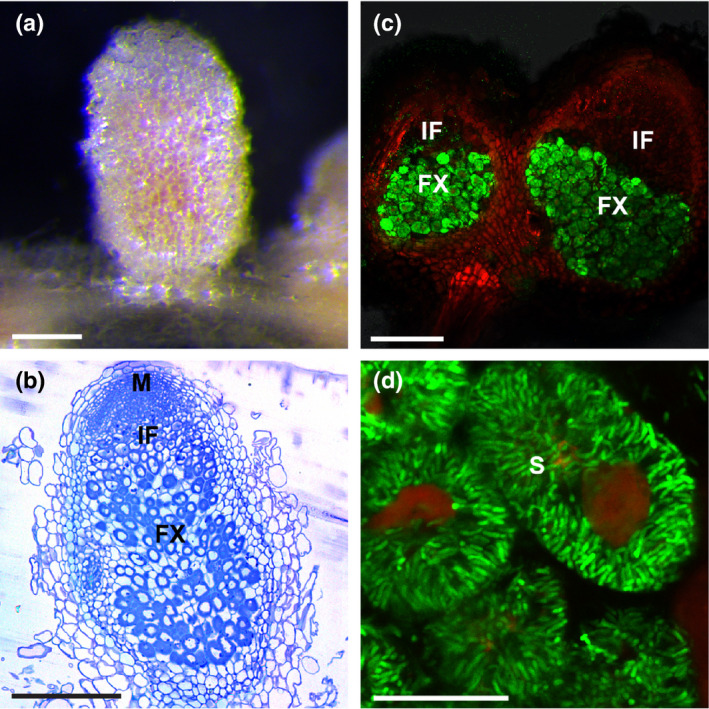
Wild‐type‐like pink nodules formed on Medicago nin knockout mutant (nin‐1) roots transformed with truncated NINΔPB1. Pink nodules formed on nin‐1 roots transformed with ProNIN3C‐5kb:NINΔPB1 at 4 wk post inoculation (wpi) (a). Semithin sections of the nodules stained with toluidine blue display normal zonation (b). Confocal image of nodules formed on the ProNIN3C‐5kb:NINΔPB1 transgenic nin‐1 roots at 4 wpi with a rhizobial strain carrying nifH:GFP showed that nifH is highly expressed in the fixation zone (c). Close‐up of nodule cells in fixation zone, showing well developed symbiosomes (d). M, meristem; IF, infection zone; FX, fixation zone; S, symbiosome. Bars: (a) 2 mm; (b) 300 µm; (c) 250 µm; (d) 30 µm.
Reduced NIN mRNA level in the cytoplasm of nin‐13/16 nodule cells
The transition from infection to fixation zone does not occur in nin‐13/16. As NIN accumulates at a high level at this transition site, we analysed the NIN expression pattern in nin‐13/16 nodules by RNA in situ hybridisation. As found in wild‐type (R108) nodules (Fig. 5a), NIN was highly expressed in the infection zone of the mutant nodules (Fig. 5b,c), however there was a marked difference between nin‐13/16 and wild‐type nodules. NIN RNA in nin‐13/16 nodules accumulated to higher levels in nuclei compared with in the cytoplasm, which is not the case in wild‐type nodules (Fig. 5d,e,f). This indicated that the transport of NIN mRNA from the nucleus to the cytoplasm was hampered. By contrast, transport of NF‐YA1 RNA was not affected (Fig. S5), suggesting that the inhibition of mRNA transport was most likely to be specific for NIN in nin‐13/16. By quantitative real‐time PCR (qRT‐PCR) we showed that NIN‐Tnt1 fusion transcripts were formed in nin‐13/16 nodules (Fig. S6). In nin‐13, about 80% of NIN transcripts contained Tnt1 sequences, and about 60% of NIN transcripts contained Tnt1 sequences in nin‐16 (Fig. S6). Moreover, nin‐16 RNA sequencing data (see below) revealed that some reads were mapped to the intron and promoter region of NIN (Fig. S7). The frequency of aberrantly spliced NIN transcripts in nin‐16 was between 30.24% and 64.80%. This is markedly higher than in R108, in which this frequency was between 1.39% and 4.37% (Fig. S7). This finding indicated that the Tnt1 insertion affected NIN transcription and splicing. As the export of unspliced and incompletely spliced transcripts from nuclei is prevented (Schmid & Jensen, 2008), we assumed that this might have caused the reduced transport of NIN RNA to the cytoplasm. By using NIN‐specific primers (upstream of Tnt1 insertion), we showed that the expression level of NIN in wild‐type and mutant nodules was similar (Fig. 5g,h). As a substantial part of NIN mRNA had accumulated in nuclei of nin‐13/16, the level of NIN mRNA that can be translated (in the cytoplasm) was reduced, this might have caused the phenotype of nin‐13/16 nodules.
Fig. 5.
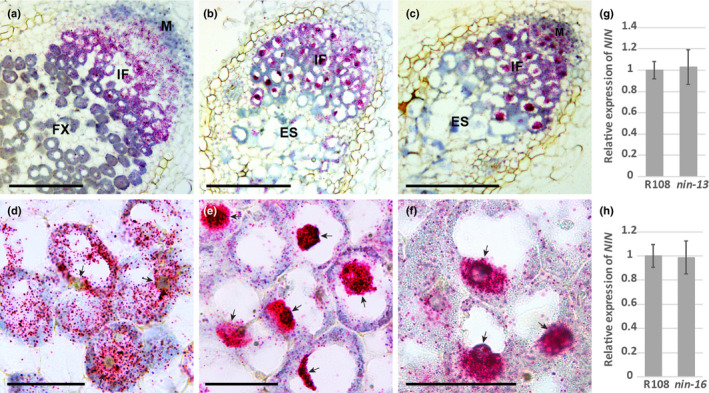
NIN mRNA level reduced in the cytoplasm of nodule cells of Medicago nin‐16. RNA in situ localisation of NIN shows that the substantial part of NIN mRNA accumulates in nuclei of nin‐13 (b, e) and nin‐16 nodule cells (c, f), compared with wild‐type (R108) nodule section (a, d). Hybridisation signals are visible as red dots (a–f). Arrows indicate the nuclei (d–f). M, meristem; IF, infection zone; FX, fixation zone; ES, early senescence zone. Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) shows that NIN expression level in wild‐type (R108), nin‐13 and nin‐16 nodules were similar (g, h). Data are means ± SD of three biological replicates. Bars: (a–c) 250 µm; (d–f) 40 µm.
To test this hypothesis, we introduced ProNIN3C‐5kb:NINΔPB1 into nin‐16 by hairy root transformation. Semithin sections of the nodules formed on transgenic roots showed that symbiosome development was restored (symbiosomes reached the length about 8 μm) and the defence/premature senescence phenotype was markedly reduced (Fig. S8). This result supports the conclusion that the nin‐13/16 nodule phenotype is caused by a reduced NIN mRNA level, but not the absence of the PB1 domain of NIN.
nin‐16 nodule transcriptome switched from symbiosis to defence/senescence‐related processes
To determine which NIN‐controlled processes were disturbed in nin‐13/16 mutants, we compared the transcriptomes of 2‐wk‐old nin‐16 and wild‐type nodules; 31 841 gene transcripts were detected in these nodules (Table S2). We identified 2744 genes that were differentially expressed in nin‐16 nodules compared with wild‐type (Table S3) of which 1281 (46.7%) genes were upregulated and 1463 (53.3%) genes were downregulated in nin‐16 (Table S3).
The histology of the nin‐13/16 nodules indicated that the infection zone was not much affected, whereas a fixation zone was not formed. Consistent with this finding is that only a small fraction of the genes that are especially expressed in the meristem or the distal part of the infection zone of wild‐type nodule (according to Roux et al., 2014) is differentially expressed in nin‐16. By contrast, a markedly higher percentage of genes that are especially expressed in fixation zone of wild‐type nodules is differentially expressed (Table 1). However, for the genes especially expressed in the proximal part of the infection zone of wild‐type nodules, the percentage of differentially expressed genes in nin‐16 was also high (Table 1). The few differentially expressed genes that are especially expressed in the distal part of the infection zone were mainly upregulated (Table 1). Several of these upregulated genes are related to the defence response (Table S4). By contrast, differentially expressed genes that are especially expressed in the proximal part of the infection zone or the distal part of the fixation zone (interzone II/III according to (Roux et al., 2014)) in wild‐type nodules, were mainly downregulated (Table 1). Most of these genes were related to symbiotic functions such as leghaemoglobin and NCR genes (Table S4). For the genes especially expressed in the proximal part of the fixation zone (zone III, according to Roux et al., 2014), for which the expression level was changed, similar numbers were upregulated and downregulated, respectively (Table 1). The upregulated genes were mainly related to senescence and defence and downregulated genes included symbiotic genes (Table S4). We do not know in which part of the nin‐13/16 nodules the upregulated genes are expressed and this could be different from their expression in wild‐type nodules. For example, the defence‐related genes that are expressed in the meristem of wild‐type nodules might be induced in the early senescence zone of nin‐13/16 nodules.
Table 1.
Analysis of genes specifically expressed in different Medicago wild‐type nodule developmental zones (Roux et al., 2014) differentially expressed in nin‐16.
| Genes 70% expressed in zone: | Meristem | Distal infection zone | Proximal infection zone | Distal fixation zone (interzone) | Proximal fixation zone (zone III) |
|---|---|---|---|---|---|
| Total number of genes | 1901 | 370 | 191 | 665 | 536 |
| Number of genes differentially expressed in nin‐16 | 126 | 24 | 51 | 184 | 82 |
| Percentage of genes of differentially expressed in nin‐16 | 6.63% | 6.49% | 26.70% | 27.67% | 15.30% |
| Number of genes up regulated in nin‐16 | 101 | 21 | 1 | 2 | 35 |
| Percentage of genes up regulated in nin‐16 | 80.16% | 87.50% | 1.96% | 1.09% | 42.68% |
| Number of genes down regulated in nin‐16 | 25 | 3 | 50 | 182 | 47 |
| Percentage of genes down regulated in nin‐16 | 19.84% | 12.50% | 98.04% | 98.91% | 57.32% |
Darker shade indicates higher percentage of genes differentially expressed in nin‐16. Higher percentage of genes up regulated in nin‐16 shown in darker red colour; higher percentage of genes downregulated in nin‐16 shown in darker green colour.
Consistent with the fact that the early nodule developmental stages were not much affected, the expression levels of genes required for intracellular infection were not significantly changed. This group included Ethylene response factor (ERF), transcription factor required for nodulation 1 (ERN1) and direct targets of NIN, NF‐YA1 and nodulation pectate lyase (NPL) (Fig. 6b) (Xie et al., 2012; Soyano et al., 2013; Laporte et al., 2014). Release of rhizobia from infection threads occurred in the distal part of the infection zone. The symbiotic exocytosis pathway‐related genes Vamp721d, Vamp721e and SYP13II are important for release (Ivanov et al., 2012; Huisman et al., 2016). Their level of expression in nin‐16 nodules was not significantly changed (Fig. 6b), which was consistent with the occurrence of release.
Fig. 6.
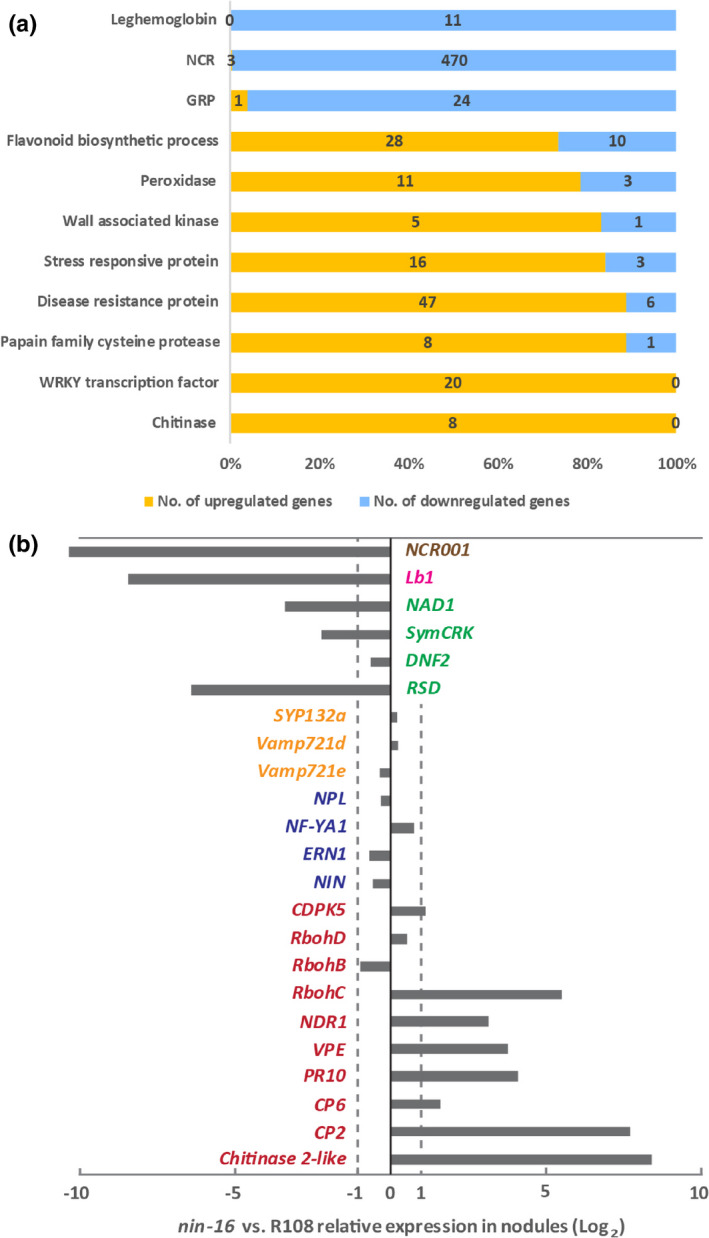
Medicago nin‐16 transcriptome is consistent with the reduced suppression of defence and early senescence. Transcriptional regulation of gene families/metabolism pathways in nodules (a). The classification of each gene in the families/metabolism pathways is based on the protein function indicated in the Mt 5.0 annotation or gene ontology terms. NCR, Nodule Cysteine‐Rich peptide; GRP, Nodule‐specific Glycine Rich Peptide. Analysis of representative genes from the transcriptome data (b). Genes required for primordium initiation and infection formation (blue colour), including NIN (Nodule inception) itself, Ethylene response factor required for nodulation 1 (ERN1), Nuclear transcription factor Y subunit A‐1 (NF‐YA1) and nodulation pectate lyase (NPL). The symbiotic exocytosis pathway‐related genes (yellow colour) required for rhizobial release, including VAMP721d and VAMP721e that are members of the vesicle‐associated membrane protein 721 family, and SYNTAXIN OF PLANTS 132 (SYP132). Nodule‐specific cysteine‐rich (NCR) peptides (brown colour) and leghaemoglobin (LB) (pink colour), are known to be required for bacteria differentiation and nitrogen fixation. Genes related to defence and senescence (red colour), including calcium‐dependent protein kinase (CDPK), respiratory burst oxidase homologs (Rboh), cysteine proteinases (CPs), nonrace‐specific disease resistance 1 (NDR1), vacuolar processing enzyme (VPE), pathogenesis‐related protein 10 (PR10) and Chitinase genes. See detailed information about selected genes in Supporting Information Table S1.
In nin‐16 nodules, rhizobia elongated and reached a size that in wild‐type nodules is characteristic for rhizobia in the most proximal part of the infection zone (Fig. 3c). The elongation of rhizobia was initiated in nin‐16 nodules (Fig. 3c) and this was consistent with an unaffected or even upregulated expression level of some NCR genes that were expressed in the infection zone (Fig. 6a; Table S2). The block in further differentiation might be caused by the lack of fixation zone formation, where other NCR genes are expressed. The accumulation of phenolic compounds and the induction of premature senescence suggested that defence/senescence was not sufficiently suppressed in nin‐13/16 nodules. Consistent with this, there were many defence/senescence‐related genes that were significantly upregulated in nin‐16 nodules (Fig. 6a; Table S5) including well kno wn marker genes such as PR10 (16.9‐fold upregulated) and NDR1 (8.7‐fold upregulated) (Fig. 6b; Table S3). The expression of flavonoid biosynthesis genes also supported the occurrence of a defence response. Among 38 differentially expressed genes, 28 were upregulated. Most of these genes had putative chalcone synthase activity or chalcone isomerase activity that is involved in the formation of flavonoid scaffolds. By contrast, all the 10 downregulated genes had putative glucosyltransferase activity that was related to the diversity of flavonoids (Fig. 6a; Table S5) (Saito et al., 2013). Cysteine proteases (CPs) and vacuolar processing enzyme (VPE) have been shown to play a crucial role in the onset of nodule senescence and their expression can also be the consequence of plant defence responses (Sheokand & Brewin, 2003; Grudkowska & Zagdańska, 2004; Hara‐Nishimura et al., 2005; Wang et al., 2016). In nin‐16 nodules, CP2 and VPE are 202.6‐fold and 13.6‐fold upregulated, respectively (Fig. 6b; Table S3), and they were among the most highly expressed genes (Table S2). Moreover, genes involved in suppression of plant defence in nodules such as DNF2, SymCRK, NAD1 and RSD were 1.6‐, 4.7‐, 10.6‐ and 83.9‐fold downregulated in nin‐16 (Fig. 6b; Table S2). Taken together, these data supported the conclusion that nin‐16 transcriptome switched from symbiosis to defence/senescence‐related processes.
Defence/senescence is suppressed in the infection zone of nin‐13/16 nodules
Many plant defence/senescence‐related genes were highly induced in nin‐16 nodules. To determine whether the induction of these genes had already occurred in the infection zone, we used CP2 as a marker gene as it was the highest expressed defence/senescence‐related gene in nin‐16. In situ hybridisation showed that, in 2‐wk‐old mutant nodules, CP2 was extremely highly expressed in the infected cells of the premature senescence zone, but not in the infection zone or other nodule tissue (Fig. 7b,c). This suggested that the defence and early senescence response was well suppressed in the nodule infection zone, but that suppression was not sufficient at the stage when transition to fixation zone would occur.
Fig. 7.
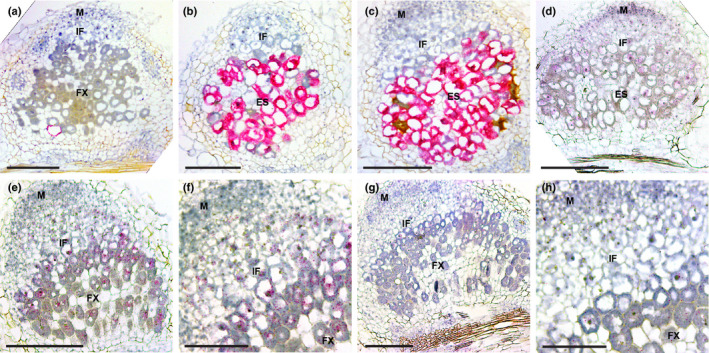
Expression pattern of defence/senescence response and suppression genes. RNA in situ localisation of defence/senescence response gene CP2 in Medicago R108 (a), nin‐13 (b) and nin‐16 (c) nodules at 2 wk post inoculation (wpi). Overview (e, g) and close‐up (f, h) pictures of expression patterns of defence suppression genes SymCRK (e, f) and NAD1 (g, h) in 2 wpi R108 nodules. (d) Expression pattern of SymCRK in 2 wpi nin‐16 nodule. Hybridisation signals are visible as red dots. M, meristem; IF, infection zone; FX, fixation zone; ES, early senescence zone. Bars: (a–e, g) 200 µm; (f, h) 100 µm.
symcrk and nad1 mutants form nodules in which defence responses are induced and symbiosome differentiation becomes arrested (Berrabah et al., 2014b; Wang et al., 2016; Domonkos et al., 2017; Yu et al., 2018). This is similar to the nodule phenotype of nin‐13/16. rsd and dnf2 mutants also have a similar nodule phenotype (Bourcy et al., 2013; Sinharoy et al., 2013), however the expression level of RSD was very low and the DNF2 expression level in nin‐16 was not significantly changed (Table S2). To determine when plant defence suppressing genes were activated during nodule development, we checked the expression pattern of SymCRK and NAD1. RNA in situ hybridisation showed that both SymCRK and NAD1 were expressed in the infected cells of the infection zone and that their expression levels increased from the distal to the proximal part (Fig 7e–h) and was similar to the NIN expression pattern (Figs 1a, 5a). However, SymCRK and NAD1 remained expressed at a relatively high level in the fixation zone (Fig. 7e–h), whereas NIN expression markedly dropped at the transition from infection to fixation zone (Figs 1a–c, 5a). To determine where in the nodule the expression of defence suppressing genes was affected, we studied SymCRK expression in nin‐16 nodules. RNA in situ hybridisation showed that SymCRK was expressed in the infection zone (Fig. 7d). In the senescing cells, SymCRK mRNA was hardly detectable.
These results indicated that the expression level of defence suppressing genes in the infection zone was sufficient to suppress the defence response and early senescence, whereas at the transition to fixation zone, the suppression was no longer sufficient.
Discussion
Here we showed that NIN plays, in addition to its previously identified role in infection and nodule organogenesis, a key role in the late steps of nodule development leading to a functional symbiosis. nin‐13/16 nodules displayed a particularly normal infection zone, whereas a fixation zone was not formed. Instead, a zone with defence responses and premature senescence occurred. The phenotype is most likely to be caused by reduced NIN mRNA levels in the cytoplasm. Consistent with this phenotype, transcriptome analyses of nin‐16 nodules showed that defence/senescence‐related genes were highly induced and defence suppressing genes were reduced. However, although defence/senescence seemed well suppressed in the infection zone, the transcriptome is most likely already markedly changed in the proximal part of the nodule infection zone. NIN mRNA and protein accumulated at the highest level in the proximal part of the infection zone. At the transition to fixation zone, this level suddenly dropped to a much lower level, suggesting that NIN is involved in the switch from infection to fixation zone.
In both nin‐13 and nin‐16, Tnt1 is inserted upstream of the region that encodes the PB1 domain, which is located at the carboxy‐terminus of NIN. Therefore, we expected that loss of this domain would have caused the mutant nodule phenotype. However, by introducing truncated NINΔPB1 into a nin knockout mutant (nin‐1), we showed that the PB1 domain was not necessary for the formation of wild‐type‐like nodules in Medicago A17 (Fig. 4). The importance of the PB1 domain is underlined by being conserved in the NLP family, including NIN. The PB1 domain has been shown to be essential for the formation of homodimers and heterodimers of Arabidopsis NLP6 and NLP7 (Guan et al., 2017). In addition, Medicago NLP1 is required for suppression of nodulation by nitrate, and this suppression probably involves a physical interaction of NIN and NLP1 by their PB1 domains (Lin et al., 2018). Therefore, the PB1 domain of NIN may be involved in regulating its activity in response to, for example, exogenous nitrate, whereas it is not essential for intrinsic nodule development.
The nin‐13/16 nodule phenotype is probably caused by reduced NIN mRNA levels in the cytoplasm. In situ hybridisation showed that a substantial part of NIN RNA accumulated in nin‐13/16 nuclei. Furthermore, by introducing truncated NIN into nin‐16, symbiosome development was restored and the defence and early senescence phenotype was markedly reduced. This confirmed that the mutant nodule phenotype was caused by reduced NIN mRNA level, but not the absence of the PB1 domain of NIN. The NIN RNA transcribed from the nin‐16 allele was altered by the Tnt1 insertion, therefore, it might be possible that this improper nature of the NIN transcripts caused (partial) retainment of it in nuclei. Nuclear retention of transcripts might also occur in other mutants with the Tnt1 insertion. Although the qPCR data showed that NIN‐Tnt1 chimeric sequences were present, they did not prove that full‐length NIN‐Tnt1 chimeric sequences were formed, as promoter and terminator regions existed in the identical 5′ and 3′ long terminal repeats (LTR) of Tnt1 (Grandbastien et al., 1989; Grandbastien, 2015). It has been shown that some LTR regulatory regions can induce transcription in an opposite direction (Grandbastien et al., 1989; Grandbastien, 2015). This could explain the reads mapped to the promoter region of NIN in nin‐16 as a readout transcript of LTR.
In Pislariu et al. (2012), more mutants with a Tnt1 insertion in NIN were described. However, except in nin‐13 (NF0440), they all showed a Nod− phenotype, including two mutants (NF0825 and NF2700) that had the Tnt1 insertion at a position that was downstream of that in nin‐13/16. However, it cannot yet be concluded that the truncated NIN proteins that are slightly longer than in nin‐13 and nin‐16 are not functional. In both nin‐13 and nin‐16, NIN mRNA is retained in the nucleus. It seems probable that the Tnt1 insertions in NF0825 and NF2700 also cause such NIN mRNA retention. If this is stronger than in nin‐13/16, this could explain the Nod− phenotype of these mutants. It is also possible that other Tnt1 insertions in these mutants caused the Nod− phenotype, as whether the nodule phenotype co‐segregates with Tnt1 insertion in NIN has not yet been studied.
Several NLPs are located in the cytoplasm under low nitrate conditions and are translocated to the nucleus under high nitrate conditions (Marchive et al., 2013; Cao et al., 2017; Lin et al., 2018). Here we showed that NIN was located in nuclei in all nodule cells where it is expressed. For the nitrate‐triggered translocation to nuclei Ser205 of AtNLP7 becomes phosphorylated. This serine is conserved in several NLPs, but is lacking in NIN (Liu et al., 2017), so NIN might have lost the information required for retention in the cytoplasm.
In Medicago, NIN is expressed in the infection zone and meristem. This is consistent with findings of a previous study on pea (Pisum sativum) (Borisov et al., 2003). Both NIN mRNA and NIN protein had reached their highest level in the proximal part of the infection zone in Medicago nodules, and this suddenly dropped at the transition to fixation zone. However, it is not clear whether in pea the decrease in NIN expression also occurred at the switch to interzone/distal fixation zone or somewhat later (Borisov et al., 2003). The sudden change in the NIN levels at transition from the infection to fixation zone coincided with several other rapid changes such as the accumulation of starch, the induction of bacterial nif genes and the radial alignment of the symbiosomes (Gavrin et al., 2014). Rapid changes in protein/mRNA activity/levels often involve negative and positive feedback mechanisms. For example, in the Arabidopsis stem cell niche, an asymmetric cell division initiates the formation of root endodermis and cortex. This involves the SHORT ROOT (SHR)/SCARECROW (SCR) complex. This complex induces the expression of cyclinD6,1 that inactivates RETINOBLASTOMA‐RELATED, the repressor of SCR activity. Through this positive feedback loop, the SHR–SCR complex activity increases and triggers asymmetrical division. This was followed by a negative feedback involving degradation of the SHR–SCR complex (Cruz‐Ramírez et al., 2012), therefore the accumulation of NIN to a high level, which is followed by a sudden drop, was consistent with a role in a molecular switch. Whether and how NIN is involved in a molecular circuit controlling the switch from infection to fixation zone remains to be studied.
In previous studies on nin‐13/16 (Pislariu et al., 2012; Veerappan et al., 2016), the accumulation of phenolic compounds was not described. The level of accumulation of phenolic compounds in these two mutants seems lower in comparison with some other mutants (Bourcy et al., 2013; Berrabah et al., 2014b; Wang et al., 2016; Domonkos et al., 2017), therefore, it might have been overlooked in previous studies. Furthermore, it has been shown that in some mutants, for example dnf2 (Berrabah et al., 2014a), the accumulation of phenolic compounds is conditional. Therefore, it cannot be excluded that less phenolic compounds had accumulated in nin‐13/16 nodules due to different growth conditions. nin‐13 previously has been reported as Nod− at 21 d post inoculation with rhizobia (Pislariu et al., 2012). The reason for this difference is not clear, as in all our experiments the nodule number formed on the mutant was similar to that of the wild‐type (R108).
Although nin‐13/16 has been reported previously, the phenotypic characterisation was not detailed (Pislariu et al., 2012; Veerappan et al., 2016). Here we analysed the nin‐13/16 nodule phenotype explicitly. Both mutant nodules made an infection zone that at the cytological level was rather normal. Rhizobia were released from infection threads, they multiplied and the symbiosomes started to elongate, reaching the size that is obtained in the proximal part of the infection zone in WT nodules. By contrast, processes that occurred at the transition to fixation zone did not take place. For example, nif genes were not induced, symbiosomes did not further elongate and amyloplasts did not accumulate at the periphery of the infected cells. This phenotype is in part consistent with the transcriptome data. Genes that were especially expressed in the fixation zone were downregulated (except genes related to senescence and the defence response). By contrast, expression levels of genes especially active in the distal infection zone were in most cases not affected, except defence‐related genes, which were upregulated. It is not known where these upregulated genes are expressed in the nin‐16 nodules. Furthermore, the expression levels of many genes that were especially expressed in the proximal part of the infection zone were already affected, the genes related to symbiotic functions were downregulated.
Symbiosome development became arrested in nin‐13/16 nodules and defence and early senescence were induced. The defence suppressing genes SymCRK and NAD1 were markedly downregulated in nin‐16 nodules. Loss‐of‐function mutations in these genes caused a similar symbiosome phenotype as found for nin‐16 (Berrabah et al., 2014b; Wang et al., 2016; Domonkos et al., 2017). Therefore, it is possible that symbiosome development was blocked due to the defence/senescence‐related responses, which were not sufficiently suppressed. In addition, other studies have shown that arrest of symbiosome development did not induce defence responses (Berrabah et al., 2015), therefore defence is more likely to be a cause than a result of blocking symbiosome development.
Laser capture transcriptome data suggested that NF‐YA1 is highly expressed in the nodule meristem and distal part of infection zone (Roux et al., 2014), Our in situ hybridisation data showed that NF‐YA1 was not expressed in the distal part of the meristem, but was expressed at a high level in the proximal part of the meristem and the distal part of the infection zone, where NIN is only present at a very low level. This finding supports the idea that NF‐YA1 expression can be regulated not only by NIN but also by other transcription factor(s).
SymCRK might also be a direct target of NIN as indicated using chromatin immunoprecipitation sequencing (ChIP‐seq) analyses on Lotus roots ectopically expressing NIN (Soyano et al., 2014; Liu et al., 2019a). In the infection zone, NIN is expressed at a very high level, so it could control SymCRK expression. If NIN also directly controls SymCRK expression in the fixation zone, then a very low level of NIN might still be sufficient, considering that a low level of NIN is sufficient to induce a high level of NF‐YA1 expression. Alternatively, it has been shown that members of the NLP family can also bind to the promoter of NIN target gene CLE‐RS2 (Nishida et al., 2018). Moreover, NLPs are highly expressed in Medicago nodules, especially MtNLP1/2/3 (Lin et al., 2018), therefore it is possible that they take over the function of NIN to maintain expression of this defence suppression gene in the fixation zone.
So, in addition to its function in nodule organogenesis and infection, NIN has a major function in establishing a functional symbiosis in Medicago nodules. It will be interesting to see whether this function is shared with other legumes or even actinorhizal plants. Such studies in an evolutionary context can provide first insight into when this additional function of NIN evolved or whether it is related to a function of the ancestor of NIN that was recruited when nodulation evolved.
Author contributions
JL, OK and TB designed the research. JL, MR and OK performed experiments. JL, MR, TZ, JK, OK and TB analysed data. JL, OK and TB wrote the manuscript.
Supporting information
Fig. S1 Expression profile of Medicago NIN/Medtr5g099060 (probe id. Mtr.28094.1.S1.st) based on M. truncatula Gene Atlas in various tissues.
Fig. S2 Live/dead staining shows prematurely death of rhizobia in Medicago nin‐16 nodules.
Fig. S3 Medicago nin‐13 mutant nodules show the same phenotype as nin‐16.
Fig. S4 F1 plants obtained by crossing Medicago nin‐13 and nin‐16 showed the same nodule phenotype as the parental plants.
Fig. S5 NF‐YA1 expression pattern in Medicago nin‐16 nodule.
Fig. S6 Tnt1 was transcribed in Medicago nin‐13 and nin‐16 mutant nodules.
Fig. S7 NIN RNA transcribed from the Medicago nin‐16 allele was altered by Tnt1 insertion.
Fig. S8 Complementation of Medicago nin‐16 nodule phenotype with ProNIN3C‐5kb:NINΔPB1 and ProNIN3C‐5kb:NIN.
Table S1 Primers used in this study.
Table S2 Genes with transcripts detected in Medicago nin‐16 or R108 (wild‐type).
Table S3 Genes differentially expressed in Medicago nin‐16.
Table S4 Genes specifically expressed in different Medicago wild‐type nodule developmental zones differentially expressed in nin‐16.
Table S5 Differentially expressed gene families/metabolism pathways in Medicago nin‐16.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We would like to thank Rebecca Dickstein (University of North Texas) for providing NF10547 (nin‐16) seeds. This research was supported by the European Research Council (ERC‐2011‐AdG‐294790), and the China Scholarship Council (201506300062 to JL).
References
- Alunni B, Gourion B. 2016. Terminal bacteroid differentiation in the legume−rhizobium symbiosis: nodule‐specific cysteine‐rich peptides and beyond. New Phytologist 211: 411–417. [DOI] [PubMed] [Google Scholar]
- Berrabah F, Balliau T, Aït‐Salem EH, George J, Zivy M, Ratet P, Gourion B. 2018. Control of the ethylene signaling pathway prevents plant defenses during intracellular accommodation of the rhizobia. New Phytologist 219: 310–323. [DOI] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Cayrel A, Eschstruth A, Mondy S, Ratet P, Gourion B. 2014a. Growth conditions determine the DNF2 requirement for symbiosis. PLoS ONE 9: e91866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B et al. 2014b. A nonRD receptor‐like kinase prevents nodule early senescence and defense‐like reactions during symbiosis. New Phytologist 203: 1305–1314. [DOI] [PubMed] [Google Scholar]
- Berrabah F, Ratet P, Gourion B. 2015. Multiple steps control immunity during the intracellular accommodation of rhizobia. Journal of Experimental Botany 66: 1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N et al. 2003. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus . Plant Physiology 131: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P. 2013. Medicago truncatula DNF2 is a PI‐PLC‐XD‐containing protein required for bacteroid persistence and prevention of nodule early senescence and defense‐like reactions. New Phytologist 197: 1250–1261. [DOI] [PubMed] [Google Scholar]
- Bu F, Rutten L, Roswanjaya YP, Kulikova O, Rodriguez‐Franco M, Ott T, Bisseling T, van Zeijl A, Geurts R. 2020. Mutant analysis in the nonlegume Parasponia andersonii identifies NIN and NF‐YA1 transcription factors as a core genetic network in nitrogen‐fixing nodule symbioses. New Phytologist 226: 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Qi S, Sun M, Li Z, Yang Y, Crawford NM, Wang Y. 2017. Overexpression of the maize ZmNLP6 and ZmNLP8 can complement the arabidopsis nitrate regulatory mutant nlp7 by restoring nitrate signaling and assimilation. Frontiers in Plant Science 8: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, De Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J. 2000. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Lichtenzveig J, Ellwood S, Pfaff T, Journet P. 2006. Vernalization, crossings and testing for pollen viability. In: Mathesius U, Journet E‐P, eds. Medicago truncatula Handbook. Ardmore, OK, USA: The Samuel Roberts Noble Foundation, 1–13. [Google Scholar]
- Clavijo F, Diedhiou I, Vaissayre V, Brottier L, Acolatse J, Moukouanga D, Crabos A, Auguy F, Franche C, Gherbi H et al. 2015. The Casuarina NIN gene is transcriptionally activated throughout Frankia root infection as well as in response to bacterial diffusible signals. New Phytologist 208: 887–903. [DOI] [PubMed] [Google Scholar]
- Cruz‐Ramírez A, Díaz‐Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P et al. 2012. A bistable circuit involving SCARECROW‐RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos Á, Kovács S, Gombár A, Kiss E, Horváth B, Kováts GZ, Farkas A, Tóth MT, Ayaydin F, Bóka K et al. 2017. NAD1 controls defense‐like responses in Medicago truncatula symbiotic nitrogen fixing nodules following rhizobial colonization in a BacA‐independent manner. Genes 8: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrin A, Kaiser BN, Geiger D, Tyerman SD, Wen Z, Bisseling T, Fedorova EE. 2014. Adjustment of host cells for accommodation of symbiotic bacteria: vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in medicago. Plant Cell 26: 3809–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien MA. 2015. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms 1849: 403–416. [DOI] [PubMed] [Google Scholar]
- Grandbastien M‐A, Spielmann A, Caboche M. 1989. Tntl, a mobile retroviral‐like transposable element of tobacco isolated by plant cell genetics. Nature 337: 376–380. [DOI] [PubMed] [Google Scholar]
- Grudkowska M, Zagdańska B. 2004. Multifunctional role of plant cysteine proteinases. Acta Biochimica Polonica 51: 609–624. [PubMed] [Google Scholar]
- Guan P, Ripoll J‐J, Wang R, Vuong L, Bailey‐Steinitz LJ, Ye D, Crawford NM. 2017. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proceedings of the National Academy of Sciences, USA 114: 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A, Longhi R, Boncompagni E, Hérouart D, Dall’Angelo S et al. 2011. Protection of sinorhizobium against host cysteine‐rich antimicrobial peptides is critical for symbiosis. PLoS Biology 9: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara‐Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. 2005. Vacuolar processing enzyme: An executor of plant cell death. Current Opinion in Plant Biology 8: 404–408. [DOI] [PubMed] [Google Scholar]
- Huisman R, Hontelez J, Mysore KS, Wen J, Bisseling T, Limpens E. 2016. A symbiosis‐dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host‐microbe interface in symbiosis. New Phytologist 211: 1338–1351. [DOI] [PubMed] [Google Scholar]
- Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, Bisseling T. 2012. Rhizobium‐legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proceedings of the National Academy of Sciences, USA 109: 8316–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova O, Franken C, Bisseling T. 2018. In situ hybridization method for localization of mRNA molecules in medicago tissue sections. Methods in Molecular Biology 1822: 145–159. [DOI] [PubMed] [Google Scholar]
- Laporte P, Lepage A, Fournier J, Catrice O, Moreau S, Jardinaud M‐F, Mun J‐H, Larrainzar E, Cook DR, Gamas P et al. 2014. The CCAAT box‐binding transcription factor NF‐YA1 controls rhizobial infection. Journal of Experimental Botany 65: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. 2004. RNA interference in Agrobacterium rhizogenes‐transformed roots of Arabidopsis and Medicago truncatula . Journal of Experimental Botany 55: 983–992. [DOI] [PubMed] [Google Scholar]
- Lin JS, Li X, Luo ZL, Mysore KS, Wen J, Xie F. 2018. NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula . Nature Plants 4: 942–952. [DOI] [PubMed] [Google Scholar]
- Liu C, Breakspear A, Guan D, Cerri MR, Jackson K, Jiang S, Robson F, Radhakrishnan GV, Roy S, Bone C et al. 2019a. NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiology 179: 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rutten L, Limpens E, van der Molen T, van Velzen R, Chen R, Chen Y, Geurts R, Kohlen W, Kulikova O et al. 2019b. A remote cis‐regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula . Plant Cell 31: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Niu Y, Konishi M, Wu Y, Du H, Chung HS, Li L, Boudsocq M, Mccormack M, Maekawa S et al. 2017. Discovery of nitrate – CPK – NLP signalling in central nutrient – growth networks. Nature 545: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A. 2013. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature Communications 4: 1–9. [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GED. 2007. Medicago truncatula NIN is essential for rhizobial‐independent nodule organogenesis induced by autoactive calcium/calmodulin‐dependent protein kinase. Plant Physiology 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. 2003. A novel family in Medicago truncatula consisting of more than 300 nodule‐specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiology 132: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Tanaka S, Handa Y, Ito M, Sakamoto Y, Matsunaga S, Betsuyaku S, Miura K, Soyano T, Kawaguchi M et al. 2018. A NIN‐LIKE PROTEIN mediates nitrate‐induced control of root nodule symbiosis in Lotus japonicus . Nature Communications 9: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annual Review of Plant Biology 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume‐rhizobial symbiosis. Annual Review of Genetics 45: 119–144. [DOI] [PubMed] [Google Scholar]
- Pecrix Y, Staton SE, Sallet E, Lelandais‐Brière C, Moreau S, Carrère S, Blein T, Jardinaud M‐F, Latrasse D, Zouine M et al. 2018. Whole‐genome landscape of Medicago truncatula symbiotic genes. Nature Plants 4: 1017–1025. [DOI] [PubMed] [Google Scholar]
- Perez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M. 2010. Comparison of developmental and stress‐induced nodule senescence in Medicago truncatula . Plant Physiology 152: 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre O, Hopkins J, Combier M, Baldacci F, Engler G, Brouquisse R, Hérouart D, Boncompagni E. 2014. Involvement of papain and legumain proteinase in the senescence process of Medicago truncatula nodules. New Phytologist 202: 849–863. [DOI] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J, Cosson V, Muni RR, Wang M, Benedito VA, Andriankaja A, Cheng X, Jerez IT et al. 2012. A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiology 159: 1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth LE, Stacey G. 1989. Bacterium release into host cells of nitrogen‐fixing soybean nodules the symbiosome membrane comes from three sources. European Journal of Cell Biology 49: 13–23. [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carre S, Sallet E, Courcelle E, Moreau S et al. 2014. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser‐capture microdissection coupled to RNA sequencing. The Plant Journal 77: 817–837. [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura‐Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. 2013. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiology and Biochemistry 72: 21–34. [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schauser L, Wieloch W, Stougaard J. 2005. Evolution of NIN‐like proteins in Arabidopsis, rice, and Lotus japonicus . Journal of Molecular Evolution 60: 229–237. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. 2008. Quality control of mRNP in the nucleus. Chromosoma 117: 419–429. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheokand S, Brewin NJ. 2003. Cysteine proteases in nodulation and nitrogen fixation. Indian Journal of Experimental Biology 41: 1124–1132. [PubMed] [Google Scholar]
- Sinharoy S, Torres‐Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK. 2013. The C2H2 transcription factor REGULATOR OF SYMBIOSOME DIFFERENTIATION represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula . Plant Cell 25: 3584–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M. 2014. Nodule Inception creates a long‐distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proceedings of the National Academy of Sciences, USA 111: 14607–14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M. 2013. NODULE INCEPTION directly targets NF‐Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus . PLoS Genetics 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, De Billy F, Camut S, Truchet G. 1990. Correlation between ultrastructural differentiation of bacteriods and nitrogen fixation in alfalfa nodules. Journal of Bacteriology 172: 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerappan V, Jani M, Kadel K, Troiani T, Gale R, Mayes T, Shulaev E, Wen J, Mysore KS, Azad RK et al. 2016. Rapid identification of causative insertions underlying Medicago truncatula Tnt1 mutants defective in symbiotic nitrogen fixation from a forward genetic screen by whole genome sequencing. BMC Genomics 17: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Velde W, Guerra JCP, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. 2006. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula . Plant Physiology 141: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H et al. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 328: 25–28. [DOI] [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho‐Niebel F, Oldroyd GED. 2015. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinardell JM, Fedorova E, Cebolla A, Kevei Z, Horvath G, Kelemen Z, Tarayre S, Roudier F, Mergaert P, Kondorosi A et al. 2013. Endoreduplication mediated by the anaphase‐promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. The Plant Cell 23: 2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA‐seq data: RPKM measure is inconsistent among samples. Theory in Biosciences 131: 281–285. [DOI] [PubMed] [Google Scholar]
- Wang C, Yu H, Luo L, Duan L, Cai L, He X, Wen J, Mysore KS, Li G, Xiao A et al. 2016. NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula . New Phytologist 212: 176–191. [DOI] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T. 2014. Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GED, Downie JA. 2012. Legume pectate lyase required for root infection by rhizobia. Proceedings of the National Academy of Sciences, USA 109: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xiao A, Dong R, Fan Y, Zhang X, Liu C, Wang C, Zhu H, Duanmu D, Cao Y et al. 2018. Suppression of innate immunity mediated by the CDPK‐Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytologist 220: 425–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression profile of Medicago NIN/Medtr5g099060 (probe id. Mtr.28094.1.S1.st) based on M. truncatula Gene Atlas in various tissues.
Fig. S2 Live/dead staining shows prematurely death of rhizobia in Medicago nin‐16 nodules.
Fig. S3 Medicago nin‐13 mutant nodules show the same phenotype as nin‐16.
Fig. S4 F1 plants obtained by crossing Medicago nin‐13 and nin‐16 showed the same nodule phenotype as the parental plants.
Fig. S5 NF‐YA1 expression pattern in Medicago nin‐16 nodule.
Fig. S6 Tnt1 was transcribed in Medicago nin‐13 and nin‐16 mutant nodules.
Fig. S7 NIN RNA transcribed from the Medicago nin‐16 allele was altered by Tnt1 insertion.
Fig. S8 Complementation of Medicago nin‐16 nodule phenotype with ProNIN3C‐5kb:NINΔPB1 and ProNIN3C‐5kb:NIN.
Table S1 Primers used in this study.
Table S2 Genes with transcripts detected in Medicago nin‐16 or R108 (wild‐type).
Table S3 Genes differentially expressed in Medicago nin‐16.
Table S4 Genes specifically expressed in different Medicago wild‐type nodule developmental zones differentially expressed in nin‐16.
Table S5 Differentially expressed gene families/metabolism pathways in Medicago nin‐16.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


