Abstract
Background
Accurate and robust pathological image analysis for colorectal cancer (CRC) diagnosis is time-consuming and knowledge-intensive, but is essential for CRC patients’ treatment. The current heavy workload of pathologists in clinics/hospitals may easily lead to unconscious misdiagnosis of CRC based on daily image analyses.
Methods
Based on a state-of-the-art transfer-learned deep convolutional neural network in artificial intelligence (AI), we proposed a novel patch aggregation strategy for clinic CRC diagnosis using weakly labeled pathological whole-slide image (WSI) patches. This approach was trained and validated using an unprecedented and enormously large number of 170,099 patches, > 14,680 WSIs, from > 9631 subjects that covered diverse and representative clinical cases from multi-independent-sources across China, the USA, and Germany.
Results
Our innovative AI tool consistently and nearly perfectly agreed with (average Kappa statistic 0.896) and even often better than most of the experienced expert pathologists when tested in diagnosing CRC WSIs from multicenters. The average area under the receiver operating characteristics curve (AUC) of AI was greater than that of the pathologists (0.988 vs 0.970) and achieved the best performance among the application of other AI methods to CRC diagnosis. Our AI-generated heatmap highlights the image regions of cancer tissue/cells.
Conclusions
This first-ever generalizable AI system can handle large amounts of WSIs consistently and robustly without potential bias due to fatigue commonly experienced by clinical pathologists. It will drastically alleviate the heavy clinical burden of daily pathology diagnosis and improve the treatment for CRC patients. This tool is generalizable to other cancer diagnosis based on image recognition.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-021-01942-5.
Keywords: Colorectal cancer, Histopathology image, Deep learning, Cancer diagnosis
Background
Colorectal cancer (CRC) is the third leading cancer by incidence (6.1%) but second for mortality (9.2%) worldwide [1]. The global burden of CRC is expected to increase 60% by 2030, in terms of new cases and deaths [2]. The accurate and prompt detection of CRC is essential to improve treatment effectiveness and survivorship. The current diagnosis of CRC requires an extensive visual examination by highly specialized pathologists. Diagnoses are made using digital whole-slide images (WSIs) of the hematoxylin and eosin (H&E)-stained specimens obtained from formalin-fixed paraffin-embedded (FFPE) or frozen tissues. The challenges for the WSI analysis include very large image size (> 10,000 × 10,000 pixels), histological variations in size, shape, texture, and staining of nuclei, making the diagnosis complicated and time-consuming [3]. In most modern pathology departments, the average consultative workload increases by ~ 5–10% annually [4]. The current trends indicate a shortage of pathologists around the world, including USA [5] and low- to middle-income countries [6]. This results in overworked pathologists, which can lead to higher chances of deficiencies in their routine work and dysfunctions of the pathology laboratories with more laboratory errors [4]. While the demands of colon specimen examination in gastroenterology clinics are high, the training time of pathologists is long (> 10 years) [7]. It is thus imperative to develop reliable tools for pathological image analysis and CRC detection that can improve clinical efficiency and efficacy without unintended human bias during diagnosis.
State-of-the-art artificial intelligence (AI) approaches, such as deep learning (DL), are very powerful in classification and prediction. There have been many successful applications of DL, specifically convolutional neural network (CNN), in WSI analysis for lung [8, 9], breast [10, 11], prostate [12–14], and skin [15, 16] cancers. Most of the existing CNN for the CRC WSI analysis focused on the pathology work after cancer determination, including grade classification [17], tumor cell detection and classification [18–20], and survivorship prediction [21–23]. Although they resulted in reasonably high accuracy, their study sample sizes are limited and do not fully represent the numerous histologic variants of CRC that have been defined. These variants include tubular, mucinous, signet ring cell, and others [24]. These limitations inflate prediction error when applied to different independent samples. Meanwhile, most of the current DL models were developed from single data source without thorough validation using independent data. They only calculated the accuracy of patches without diagnosing WSIs or the patients. Their general applicability for CRC WSI diagnosis in various clinical settings, which may involve heterogeneous platforms and image properties, remains unclear. A DL approach generalizable to daily pathological CRC diagnosis that relieves clinical burden of pathologists and improves diagnostic accuracy is yet to be developed [25].
Here, we developed a novel automated AI approach centered on weakly labeled supervised DL for the very first general clinical application of CRC diagnosis. This AI approach uses Inception-v3 CNN architecture [26] with weights initialized from transfer learning. Weakly labeled supervised learning is advantageous in training massive and diverse datasets without exact labelling at object levels (e.g., small cancer cells) [12]. Transfer learning is a highly effective and efficient DL technique for image analysis that can utilize previously learned knowledge on general images for medical image analyses [27]. Our work is based on WSIs from multiple independent hospitals/sources in China (8554 patients), USA (1077 patients), and Germany (> 111 slides). This study has high practical value for improving the effectiveness and efficiency of CRC diagnosis and thus treatment. It highlights the general significance and utility of the application of AI to image analyses of other types of cancers.
Methods
Colorectal cancer whole-slide image dataset
We collected 14,234 CRC WSIs from fourteen independent sources (Table 1). All data were de-identified. The largest image set was from 6876 patients admitted between 2010 and 2018 in Xiangya Hospital (XH), Central South University (CSU, Changsha, China). XH is the largest hospital in Hunan Province and was established in 1906 with a close affiliation with Yale University [28]. The other independent sources were The Cancer Genome Atlas (TCGA) of the USA (https://portal.gdc.cancer.gov/) [29], the National Centre for Tumor Diseases (NCT) biobank and the University Medical Center Mannheim (UMM) pathology archive (NCT-UMM) of Germany (https://zenodo.org/record/1214456#.XgaR00dTm00, [22]), Adicon Clinical Laboratories (ACL), INC, and eleven hospitals in China (detailed in Table 1). The hospitals involved are located in the major metropolitan areas of China serving > 139 million population, including those most prestigious hospitals in pathology in China: XH, Fudan University Shanghai Cancer Center (FUS), Chinese PLA General Hospital (CGH), Southwest Hospital (SWH), and The First Affiliated Hospital Air Force Medical University (AMU); other state-level esteemed hospitals: Sun Yat-Sen University Cancer Center (SYU), Nanjing Drum Tower Hospital (NJD), Guangdong Provincial People’s Hospital (GPH), Hunan Provincial People’s Hospital (HPH), and The Third Xiangya Hospital of CSU (TXH); and a regional reputable Pingkuang Collaborative Hospital (PCH). All WSIs were from FFPE tissues, except parts (~ 75%) of TCGA WSIs were from frozen tissues [30]. The process of collection, quality control, and digitalization of the WSIs is described in Supplementary-Text 1.a (see Additional file 1).
Table 1.
Usage of datasets from multicenter data source
| Data source | Dataset usage | Sample preparation | Examination type Radical surgery/colonoscopy |
Population* | CRC | Non-CRC | Total | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Subjects | Slides | Subjects | Slides | Subjects | Slides | |||||
| Xiangya Hospital (XH) | A | FFPE | 100% / 0% | Changsha, China | 614 | 614 | 228 | 228 | 842 | 842 |
| NCT-UMM (NCT-CRC-HE-100 K) | B | FFPE | NA | Germany | NA | NA | NA | NA | NA | 86 |
| NCT-UMM (CRC-VAL-HE-7 K) | B | FFPE | NA | Germany | NA | NA | NA | NA | NA | 25 |
| XH | C | FFPE | 80% / 20% | Changsha, China | 3990 | 7871 | 1849 | 2132 | 5839 | 10,003 |
| XH | D | FFPE | 89% / 11% | Changsha, China | 98 | 99 | 97 | 114 | 195 | 213 |
| Pingkuang Collaborative Hospital (PCH) | C & D | FFPE | 60% / 40% | Jiangxi, China | 50 | 50 | 46 | 46 | 96 | 96 |
| The Third Xiangya Hospital of CSU (TXH) | C & D | FFPE | 61% / 39% | Changsha, China | 48 | 70 | 48 | 65 | 96 | 135 |
| Hunan Provincial People’s Hospital (HPH) | C & D | FFPE | 61% / 39% | Changsha, China | 49 | 50 | 49 | 49 | 98 | 99 |
| ACL | C & D | FFPE | 22% / 78% | Changsha, China | 100 | 100 | 107 | 107 | 207 | 207 |
| Fudan University Shanghai Cancer Center (FUS) | C & D | FFPE | 97% / 3% | Shanghai, China | 100 | 100 | 98 | 98 | 198 | 198 |
| Guangdong Provincial People’s Hospital (GPH) | C & D | FFPE | 77% / 23% | Guangzhou, China | 100 | 100 | 85 | 85 | 185 | 185 |
| Nanjing Drum Tower Hospital (NJD) | C & D | FFPE | 96% / 4% | Nanjing, China | 100 | 100 | 97 | 97 | 197 | 197 |
| Southwest Hospital (SWH) | C & D | FFPE | 93% / 7% | Chongqing, China | 99 | 99 | 100 | 100 | 199 | 199 |
| The First Affiliated Hospital Air Force Medical University (AMU) | C & D | FFPE | 95% / 5% | Xi’an, China | 101 | 101 | 104 | 104 | 205 | 205 |
| Sun Yat-Sen University Cancer Center (SYU) | C & D | FFPE | 100% / 0% | Guangzhou, China | 91 | 91 | 6 | 6 | 97 | 97 |
| Chinese PLA General Hospital (CGH) | C | FFPE | NA | Beijing, China | 0 | 0 | 100 | 100 | 100 | 100 |
| TCGA (TCGA-Frozen) | C | Frozen | 100% / 0% | U.S. | 631 | 1214 | 110 | 133 | 631** | 1347 |
| TCGA (TCGA-FFPE) | C | FFPE | 100% / 0% | U.S. | 441 | 441 | 5 | 5 | 446 | 446 |
| Total | 6612 | 11,100 | 3129 | 3469 | 9631 | 14,680 | ||||
*Location map available in Supplementary Text 1.a (see Additional file 1). **For the TCGA –Frozen data only, the non-CRC slides were made with normal intestinal tissues on part of the CRC slides
We formed four datasets (Table 1). Dataset-A includes slides from only XH and was used for patch-level training and testing (Table 2). We carefully selected WSIs to include all common tumor histological subtypes. Using incomplete information of cancer cells/tissues (e.g., location, shape, and demarcation), pathologists weakly labeled the patches from WSIs as either containing or not cancer cells/tissues. Two weakly labeled patches were provided as illustrative comparative examples with two fully labeled patches serving as contrasts (see Additional file 1: Supplementary-Figure 1). Patches from the same patient were all put into the same data set (either training or testing) so that the training and testing data sets are independent. To ensure an appropriate and comprehensive representation of cancer and normal tissue characteristics, we included an average of 49 patches per tumor sample and 144 patches per healthy sample. The number of patches containing a large proportion of cancer cells and the number of patches containing only a few cancer cells were approximately balanced so that the patches used for training were representative of cases seen in practice.
Table 2.
Dataset-A (training and testing) and Dataset-B (external validation) for patch-level analysis
| Dataset | Cancer | Non-cancer | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Subjects | Slides | Patches | Subjects | Slides | Patches | Subjects | Slides | Patches | |
| Training | 406 | 406 | 19,940 | 153 | 153 | 22,715 | 559 | 559 | 42,655 |
| Testing | 208 | 208 | 10,116 | 75 | 75 | 10,148 | 283 | 283 | 20,264 |
| Validation | NA | NA | 15,550 | NA | NA | 91,630 | NA | 111 | 107,180* |
| Total | > 614 | > 614 | 45,606 | > 228 | > 228 | 124,493 | > 842 | 953 | 170,099 |
* There are two datasets used for validation. The number is the sum of the two datasets
Patch-level performance was further validated using Dataset-B, which contained 107,180 patches downloaded from NCT-UMM. There were two independent subsets: 100,000 image patches of 86 hematoxylin and eosin stain (HE) slides of human cancer tissue (NCT-CRC-HE-100K) and 7180 image patches of 25 slides of CRC tissue (CRC-VAL-HE-7K) [22]. The overall split for patch-level training, testing, and external validation was about 2:1:5. All images are 224 × 224 pixels at 0.5 μm per pixel. More description can be found at https://zenodo.org/record/1214456#.XV2cJeg3lhF. The patches were rescaled to default input size before they are fed to the networks for testing.
Dataset-C was used for patient-level validation and is composed of slides from XH, the other hospitals, ACL, and frozen and FFPE samples of TCGA. Given the high imbalance of cancer and non-cancer slides in SYU and CGH (Table 1), they were combined in Dataset-C. In Dataset-C, the area occupied by cancer cells varied in images from different centers. Most (~ 72%) of the slides from the ten hospitals and ACL contained 10–50% cancer cells by area (see Additional file 1: Supplementary-Figure 2).
Dataset-D was used for the Human-AI contest and contained approximately equal number of slides from XH, the other hospitals, and ACL. There is an average of ~ 5045 patches on each slide, and more than 20% of the slides contain < 1000 patches. Supplementary-Text 1.b summarized the allocation of slides in the different datasets (see Additional file 1).
After the slides were digitalized, the visual verification of the cancer diagnosis labels was performed with high stringency and accuracy. Dataset-A and Dataset-C included more than 10,000 slides, which were independently reviewed by two senior and seasoned pathologists with initial and second read. When their diagnoses were consistent with the previous clinical diagnosis conclusion, the slides were then included in the dataset. If the two experts disagreed with each other or with the previous clinical diagnosis, the slides were excluded. The labels of slides from TCGA were obtained from the original TCGA database. The labels of Dataset-B were from the NCT-UMM. The binary labels of Dataset-D for the Human-AI contest were more strictly checked. Three highly experienced senior pathologists independently reviewed the pathological images without knowing the previous clinical diagnosis. If a consensus was reached, the slides were included; otherwise, two other independent pathologists would join the review. After a discussion among the five pathologists, the sample was included only if they reached an agreement; otherwise, it was excluded.
Study design and pipeline
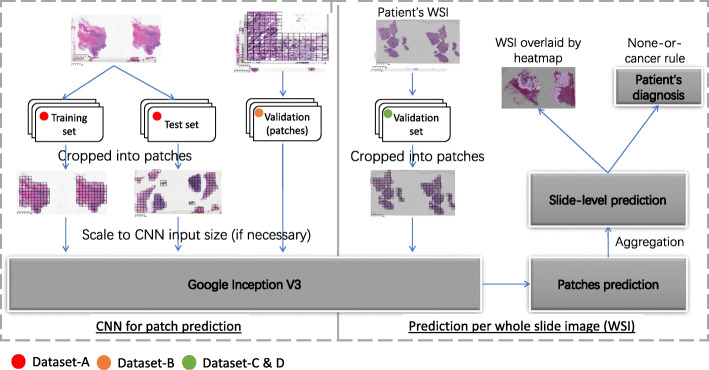
Our approach to predict patient cancerous status involved two major steps: DL prediction for local patches and patch-level results aggregation for patient-level diagnosis (Fig. 1). The WSIs after preprocessing served as the input for patch-level prediction. A deep-learning model was constructed to analyze the patches. The patch-level prediction was then aggregated by a novel patch-cluster-based approach to provide slide and patient-level diagnosis. The performance of patch-level prediction and the way of aggregation would determine to a large extent the accuracy of patient-level diagnosis. Our empirical results showed that a patch-level sensitivity of ~ 95% and specificity of ~ 99% was sufficient to achieve a high predictive power and control the false positive rate (FPR) at the patient-level using our proposed aggregation approach (see Additional file 1: Supplementary-Text 1.c). In addition, the heatmap and activation map were generated to show the informative area on the slide. The details for each step are illustrated as follows.
Fig. 1.
Study pipeline and dataset usage
Image preprocessing for patch-level training
There were 3 steps in the image preprocessing. First, we tiled each WSI at × 20 magnification with non-overlapping 300 × 300 pixel patches, which can be easily transformed to the required input size of most CNN architectures (such as the 299 × 299 input size required by Inception-v3 [26], see Additional file 1: Supplementary-Table 1). The use of a smaller patch size compared with other studies with patches of 512 × 512 pixels would make the boundaries of cancer regions more accurate [19]. Second, we removed non-informative background patches according to two criteria: the maximum difference among the 3 color channel values of the patch was less than 20, or the brightness of more than 50% of the patch surface was less than 220 in grayscale [8]. Combining these two criteria, we removed background patches and kept as many tissue patches as possible. Third, regular image augmentation procedures were applied, such as random flipping and random adjustment of the saturation, brightness, contrast, and hue. The color of each pixel was centered by the mean of each image and its range was converted/normalized from [0, 255] to [− 1, 1].
Patch-level training by deep learning
Our DL model used Inception-v3 as the CNN architecture to classify cancerous and normal patches. The Inception network uses different kernel sizes and is specifically powerful in learning diagnostic information in pathological image from differing scales. This architecture has achieved near human expert performance in the analyses of other cancer types [8, 15, 31, 32]. There are a few Inception architectures performed well on the ImageNet dataset [33] and WSIs analysis [33], such as the Inception-v1 [34], Inception-v3 [26], and Inception-v4 [35]. We chose Inception-v3 based on extensive comparison of their patch-level and patient-level performance in testing sets, which showed that the complexity and multiscale modules in Inception-v3 made it more appropriate to recognize the histopathology WSIs (see Additional file 1: Supplementary-Text 1.d) [26, 34–39]. During the study, we also tested some most recent algorithms, such as DenseNet [37] and ResNeXt [39]. Inception-v3 still performs best at the patch-level CRC classification.
We initialized the CNN by transfer learning with pre-trained weights from ImageNet [26], which were optimized to capture the structures in general images [27]. With transfer learning, our model can recognize pivotal image features for CRC diagnosis most efficiently. The 300 × 300 pixel patches were resized to a size of 299 × 299 pixels. Accordingly, the patches in the testing sets were rescaled to 299 × 299 pixels (0.37 μm/pixels) before they were fed to the network. The network was deeply fine-tuned by following training steps. Given the possible high false positive rate after aggregating the patch-level results, the optimal set of hyper-parameters was randomly searched with an objective of reaching > 95% sensitivity and > 99% specificity. We showed that, with this objective at the patch level, the error rate at the patient level was well controlled (see Additional file 1: Supplementary-Text 1.c). The network was finalized after 150,000 epochs of fine-tuning the parameters at all layers using the RMSProp [40] optimizer with a weight decay of 0.00004, a momentum value of 0.9, and RMSProp decay set to 0.9. The initial learning rate was 0.01 and was exponentially decayed with epochs to the final learning rate of 0.0001. The optimized result was achieved when the batch size was 64. The training and testing procedures were implemented in a Linux server with an NVIDIA P100 GPU. We used Python v2.7.15 and Tensorflow v1.8.0 for data preprocessing and CNN model training and testing.
Patient diagnosis and false positive control
Considering the high false positive rate (FPR) accumulated from multiple patch-level predictions, we proposed a novel patch-cluster-based aggregation method for slide-level prediction based on the fact that the tumor cells tend to gather together (especially at × 20 magnification). Motivated by the clustering inference of fMRI [41], we predicted the WSI as cancer positive if there were several positive patches topologically connected as a cluster on the slide (defined by the cluster size), such as four patches as a square. Otherwise, we predicted the slide as negative. We tested various cluster sizes and chose a cluster size of four as the result of an empirically observed best balance of sensitivity and FPR in the testing dataset (see Additional file 1: Supplementary-Text 1.e). For a patient who had one or multiple slides, denoted by S = {s1, s2, …, sl}, we provided the patient-level diagnosis D(S) combining the results from all of the patient’s slides: D(S) = D(s1) ∪ D(s2) ∪ … ∪ D(sl), where D(sl)= 1 or 0 indicated a positive or negative classification of the lth slide respectively. The patient will be diagnosed as having cancer as long as one of the slides indicates diagnosis.
Human-AI contest
Six pathologists (A-F) with varying experience of 1 to 18 clinical practice years joined the contest (see Additional file 1: Supplementary-Table 2). The pathologists independently provided a diagnosis specifying cancer or non-cancer for each patient after reading the WSIs in Dataset-D. The pathologists did not participate in the data collection or labeling. An independent analyst blindly summarized and compared the accuracy and speed of AI and human experts in performing diagnosis.
Statistical analysis and visualization
We assessed the performance of the AI and pathologists in terms of sensitivity, specificity, and accuracy () for the diagnosis. The receiver operating characteristic (ROC) curve that plotted the sensitivity versus the FPR and the corresponding area under the ROC curve (AUC) were computed. The AUCs of AI and each of the pathologists in multiple datasets were compared by the paired Wilcoxon signed-rank test. We examined the pairwise agreements among AI and pathologists by Cohen’s Kappa statistic (K). The statistical analyses were done in R v3.5 (Vienna, Austria), using packages caret, ggplot2, pROC, and psych among others. Statistical significance level was set at an alpha level of 0.05.
To locate the CRC region in the WSI, we visualized the WSI as a heatmap based on the confidence score of each patch. Brighter regions indicate higher confidence that the classifier would consider the region cancer positive. The heatmap was generated by Python (https://www.python.org/) and overlaid with the original WSI by gimp (https://www.gimp.org/).
Results
Highest accuracies in patch-level prediction by our model
We divided the 842 WSIs from Dataset-A (Table 1) into 62,919 non-overlapping patches (Table 2) to construct the CNN for patch-level prediction based on fine-tuning of Inception-v3. An average of ~ 75 patches per WSI were included to ensure an appropriate and comprehensive representation of cancer and normal tissue characteristics. Three major CRC histological subtypes were involved for the training and testing, including 74.76% tubular, 24.59% mucinous, and 0.65% signet ring cell patches, roughly reflecting their clinical incidences [42]. In the training, 19,940 (46.75%) patches had cancer, and 22,715 (53.25%) patches were normal. Using another independent set of 10,116 (49.92%) cancer and 10,148 (50.08%) non-cancer patches, the AI for patch-level prediction achieved a testing accuracy of 98.11% and an AUC of 99.83%. The AUC outperformed that of all the previous AI studies for CRC diagnosis and prediction (79.2–99.4%) and even for the majority of other types of cancer (82.9–99.9%, see Additional file 1: Supplementary-Tables 3, [8, 12, 17, 19, 22, 43–48]). The specificity was 99.22% and the sensitivity 96.99%, both outstanding. In the external validation Dataset-B, our model yielded an accuracy and AUC of 96.07% and 98.32% in NCT-CRC-HE-100 K, and 94.76% and 98.45% in CRC-VAL-HE-7 K, which matched the performance from in-house data and outplayed the patch-level validation analysis in other AI studies (AUC 69.3–95.0%, see Additional file 1: Supplementary-Table 3). The patch-level testing and validation result was summarized in Table 3.
Table 3.
Patch-level (Dataset-A and Dataset-B) and patient-level (Dataset-C and Dataset-D) performance summary
| Source | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|
| Dataset-A (patch-level testing) | ||||
| XH | 96.99% | 99.22% | 98.11% | 99.83% |
| Dataset-B (patch-level validation) | ||||
| NCT-CRC-HE-100 K | 92.03% | 96.74% | 96.07% | 98.32% |
| CRC-VAL-HE-7 K | 94.24% | 94.87% | 94.76% | 98.45% |
| Dataset-C (patient-level validation) | ||||
| XH | 98.80% | 99.51% | 99.02% | 99.16% |
| TCGA-Frozen | 94.04% | 88.06% | 93.44% | 91.05% |
| TCGA-FFPE | 97.96% | 100.00% | 97.98% | 98.98% |
| SYU-CGH | 98.90% | 92.45% | 95.43% | 95.68% |
| Dataset-D (patient-level Human-AI contest) | ||||
| XH | 97.96% | 100% | 98.97% | 98.99% |
| SYU | 98.90% | 100% | 98.97% | 99.45% |
| Dataset-C and Dataset-D (patient-level validation and Human-AI contest) | ||||
| PCH | 96.00% | 97.83% | 96.88% | 97.91% |
| TXH | 100% | 97.92% | 98.96% | 99.20% |
| HPH | 97.96% | 97.96% | 97.96% | 98.98% |
| FUS | 100% | 97.96% | 98.99% | 99.99% |
| GPH | 100% | 97.65% | 98.91% | 99.15% |
| NJD | 92.93% | 97.94% | 95.41% | 95.84% |
| SWH | 98.99% | 97.00% | 97.99% | 99.42% |
| AMU | 97% | 97.06% | 97.04% | 98.37% |
| ACL | 100% | 97.20% | 98.55% | 99.83% |
Diagnosis of CRC at patient level using DL-predicted patches
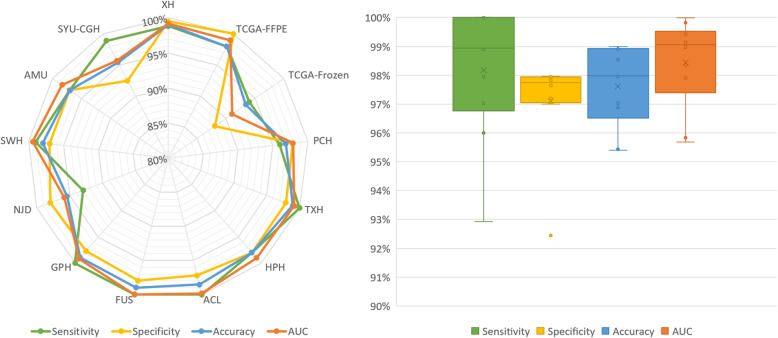
Our AI approach was tested for patient diagnosis with 13,514 slides from 8594 patients (Dataset-C). In the largest subset (5839 patients) from XH, our approach produced an accuracy of 99.02% and an AUC of 99.16% (Fig. 2, Table 3). In other independent multicenter datasets, our approach consistently performed very well. For the FFPE slides from other hospitals, TCGA-FFPE, and ACL, the AI approach yielded an average AUC and accuracy higher than 97.65% (Fig. 2). For frozen slides TCGA-Frozen, the AI accuracy and AUC were 93.44% and 91.05% respectively (Fig. 2). Our AUC values (ranging from 91.05 to 99.16%) were higher than that of other AI-based approaches for independent datasets (ranging from 83.3 to 94.1%). Of note, because the majority of those earlier AI approaches were tested on datasets of much smaller sample sizes (see Additional file 1: Supplementary-Table 3), their performances may be over-estimated. The limited number of negative slides in TCGA may result in an imbalanced classification problem that needs further investigation, which is beyond the scope of this study. The results on TCGA-Frozen slides showed that our method did learn the histological morphology of cancer and normal tissues for cancer diagnosis, which is preserved in both the FFPE and frozen samples, even though our method was developed based on the FFPE samples. Table 3 summarized the complete patient-level result.
Fig. 2.
Patient-level testing performance on twelve independent datasets from Dataset-C. Left: the radar map of the sensitivity, specificity, accuracy, and AUC in each dataset from Dataset-C. Right: the boxplot showing the distribution of sensitivity, specificity, accuracy, and AUC in datasets excluding XH and TCGA. The horizontal bar in the box indicates the median, while the cross indicates the mean. Circles represent data points
Contest with six human experts
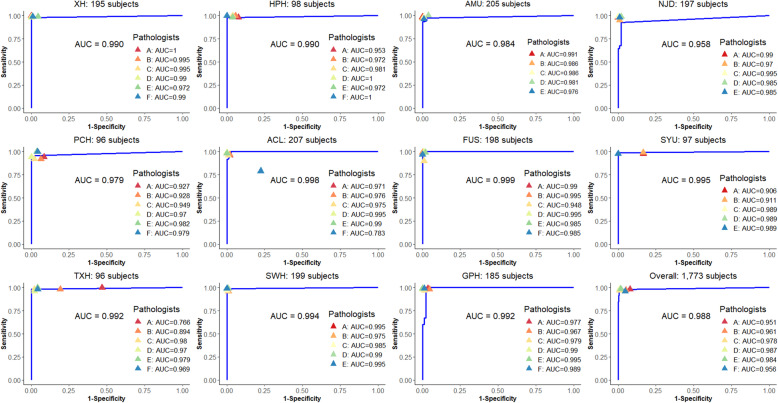
The performance of our AI approach was consistently comparable to the pathologists in diagnosing 1831 WSIs from independent centers (Dataset-D, Fig. 3). The AI resulted in an average accuracy and AUC of 98.06% (95% confidence interval [CI] 97.36 to 98.75%) and 98.83% (95% CI 98.15 to 99.51%), which both ranked top three out of the seven competitors (AI plus the six pathologists) and were greater than the average of the pathologists (accuracy 97.14% (95% CI 96.12 to 98.15%) and AUC 96.95% (95% CI 95.74 to 98.16%)). The paired Wilcoxon signed-rank test of AUCs in multicenter datasets found there were no significant differences between AI and each of the pathologists. The AI yielded the highest sensitivity (98.16%) relative to the average (97.47%) of the pathologists (see Additional file 1: Supplementary-Table 4). The pathologists (D and E) who slightly outperformed the AI have 7 and 12 years of clinical experience respectively, while the AI outperformed the other 4 pathologists with 1, 3, 5, and 18 years of experience respectively. Cohen’s Kappa statistic (K) showed an excellent agreement (K ≥ 0.858, average 0.896) between AI and every pathologist (see Additional file 1: Supplementary-Table 5). Our approach is thus proven generalizable to provide diagnosis support for potential CRC subjects like an independent pathologist, which can drastically relieve the heavy clinical burden and training cost of professional pathologists. Details of the Human-AI contest are given in Supplementary-Tables 4 & 5 (see Additional file 1)
Fig. 3.
ROC analysis of AI and pathologists in the Human-AI contest using Dataset-D. The blue line is the estimated ROC curve for AI. The colored triangles indicate the sensitivity and specificity achieved by the six pathologists
The pathologists were all informed to compete with our AI and with each other; hence, their performances were achieved under their best possible conditions with very best effort, which represented their highest skill with least error. However, with heavy workload in clinic, their performance in terms of accuracy and speed will not be as stable as that of AI. The current study of AI in cancer diagnosis using WSI has shown that AI can accurately diagnose in ~ 20 s [8] or less (~ 13 s in our case). With evolved DL techniques and advanced computing hardware, the AI can constantly improve and provide steady, swift, and accurate first diagnosis for CRC or other cancers.
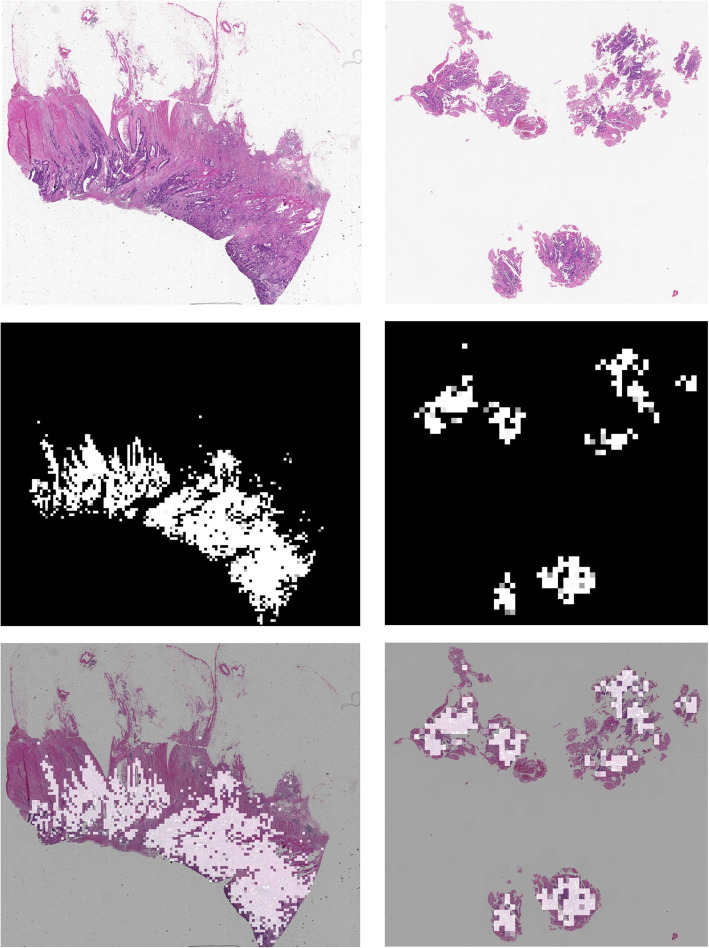
Slide-level heatmap
Our approach offers an additional distinct feature: heatmap for highlighting potential cancer regions (as patches) in WSI. In Fig. 4, we presented two WSIs, which were overlaid with the predicted heatmap. For both radical surgery WSI and colonoscopy WSI, the true cancerous region was highly overlapped with highlighted patches obtained by AI, which was also verified by pathologists. See more examples in Supplementary-Figure 3 (see Additional file 1). In addition, to visualize informative regions utilized by DL for the CRC detection, we provided the activation maps in Supplementary-Figure 4 (see Additional file 1).
Fig. 4.
Heatmap produced by AI. Top row: WSI from radical surgery (left) and colonoscopy (right); middle row: AI predicted heatmap corresponding to the first row, with white coloration indicating predicted cancer region; bottom row: heatmap overlaid on the corresponding WSI
Discussion
We collected high-quality, comprehensive, and multiple independent human WSI datasets for training, testing, and external validation of our AI-based approach focusing on pathological diagnosis of CRC under common clinical settings. We mimicked the clinical procedure of WSI analysis, including the image digitalization, slide review, and expert consultations of the disputed slides. Different from other studies [21], we did not apply any manual selection of slides or the area of interest when building the study dataset. Given the complex histologic variants of CRC, we randomly selected training patches from three most commonly seen subtypes roughly proportional to their incidences. The number of patches from images with large and small cancer tissue area was balanced and well represented in patch-level analysis. The collected images were labeled by agreement of at least two senior experts in CRC pathology (see Additional file 1: Supplementary-Text 1.b). The testing dataset from different locations in China, USA, and Germany served as a representative pool for validation and generalization. Our dataset well represents the slides seen in clinics. Consequently, the trained AI model is robust and generalizable to analyze images of different production protocols and image quality.
For a fast-growing area, we are aware of that several new CNN architectures have been proposed after the completion of the study of the present paper, such as the DenseNet [37], Squeeze-and-Excitation network [38], and ResNeXt [39]. We did some exploratory analysis by comparing the ResNet152V2, DenseNet201, and NASNetLarge relative to the Inception-v3 in classifying patches. DenseNet201 produced similar evaluation metrics as Inception-v3, while the other two architectures yielded less accuracy and AUC than Inception-v3. Although these new models have been shown to increase the prediction accuracy on ImageNet dataset compared to Inception-v3, the complexity (depth and number of parameters) and the multiscale modules in Inception-v3 may be appropriate to recognize the CRC WSIs. The performance of the new architectures on pathology images analysis and cancer diagnosis deserves more focused dedicated research for more detailed technical comparison. Moreover, we identify other techniques that may extend the current study, such as the semi- and unsupervised learning [49, 50], which can learn from more WSIs with and without labels efficiently, and the multiscale decision aggregation and data augmentation [51], which can work in the presence of limited data. Given the highly accurate performance already achieved in the current approach presented, we can investigate if and how these new techniques might attain the current prediction performance with less data collection and labelling effort in future studies.
There are several histological types that were too rare (less than 0.5% in incidence [52]) to be included, such as medullary, micropapillary, and serrated. Our AI approach performed only slightly less satisfactory in frozen samples than in FFPE samples. With WSIs from rare types and more frozen samples available for training in the future, we expect our approach can be constantly improved to be more generalizable.
Most of the previous studies obtained the patient’s diagnosis by integrating the patch-level recognition results, since it is not feasible to process the large-size WSI directly. This strategy is difficult to control the accumulated false positive rate (FPR) from multiple predictions based on individual patches. Recently, Coudray et al. used the proportion of positive patches or the average probability of all patches as the prediction criterion for the WSI [8]. Although their results were verified in three independent datasets (all with small sample sizes (340 slides)), their aggregation method may not be valid for those images with only a small area of cancer tissues where it will yield false negative findings for cancer patients. Instead, we proposed a novel aggregation strategy for patch-based WSI or patient-level prediction, which is intuitive and can easily balance the sensitivity and specificity. Specifically, we aggregated information from the cluster of patches that are topologically connected on the slide to determine the cancer status. In practice, setting the cluster size to four is most likely to exceed the average accuracy of pathologists, while cluster size of two can be used for pathological screening with an average sensitivity of ~ 99.78% and an average specificity of ~ 72.29% according to our test data (see Additional file 1: Supplementary-Text 1.e).
Conclusions
In summary, we developed a novel AI-based histopathological image classification approach for CRC diagnosis using deep learning, which achieved the best performance with the largest number of sample sizes and data sources in the field so far. Our approach was able to quickly and accurately distinguish CRC cases from healthy or inflammatory cases and was comparable to or even superior to pathologists in the testing of large-scale multicenter data. To the best of our knowledge, this is the first AI study for a reliable, generalized, and robust auxiliary tool for daily clinical pathology diagnosis of CRC initial screening. Our approach may also be adapted and applied to the histological analysis of other cancer types via the code available upon request.
Supplementary Information
Additional file 1: Supplementary-Text 1.a Collection and digitalization of the WSIs. Supplementary-Text 1.b Dataset-A, B, C and D. Supplementary-Text 1.c Patch-level performance and patient-level accuracy. Supplementary-Text 1.d Comparison of different architectures at patch-level. Supplementary-Text 1.e Comparison of different cluster sizes for aggregation of patch-level results. Supplementary-Table 1. Input patch size for common CNN. Supplementary-Table 2 Pathologist info. Supplementary-Table 3 List of AUCs of AI applied in CRC and other cancer types. Supplementary-Table 4 Overall performance of AI and pathologists in Human-AI contest. Supplementary-Table 5 Cohen’s Kappa coefficient for agreement among human experts and AI. Supplementary-Figure 1 Weakly-labeled and fully-labeled CRC patches. Supplementary-Figure 2.The distribution of cancerous area in multiple independent WSI datasets measured by the proportion of patches (P) containing cancer cells on the WSI. Supplementary-Figure 3 Heatmap produced by AI. Supplementary-Figure 4 Activation map produced by AI.
Acknowledgements
Part of the computing for this project was performed at the OU Supercomputing Center for Education & Research (OSCER) at the University of Oklahoma (OU). The authors would like to appreciate the assistance of Lance Ford, The University of Oklahoma Health Sciences Center, in editing the manuscript. We thank the editor and the three anonymous reviewers whose comments/suggestions helped improve and clarify this manuscript.
Abbreviations
- AI
Artificial intelligence
- AMU
The First Affiliated Hospital Air Force Medical University
- AUC
Average area under the receiver operating characteristics curve
- CNN
Convolutional neural network
- CGH
Chinese PLA General Hospital
- CRC
Colorectal cancer
- DL
Deep learning
- FFPE
Formalin-fixed paraffin-embedded
- FPR
False positive rate
- FUS
Fudan University Shanghai Cancer Center
- GPH
Guangdong Provincial People’s Hospital
- HPH
Hunan Provincial People’s Hospital
- H&E
Hematoxylin and eosin
- NJD
Nanjing Drum Tower Hospital
- PCH
Pingkuang Collaborative Hospital
- SWH
Southwest Hospital
- SYU
Sun Yat-Sen University Cancer Center
- TXH
The Third Xiangya Hospital of CSU
- WSI
Whole-slide image
- XH
Xiangya Hospital
Authors’ contributions
Conceptualization, H.W.D., H.M.X., K.S.W., G.Y., C.X., and X.H.M.; Methodology, G.Y., K.S.W., X.H.M., C.X., C.Y.Z., J.L., W.Z., K.Z., Y.P.W., S.C.W., Z.C.H., J.H., H.M.X., and H.W.D.; Investigation, H.M.X., K.S.W., G.Y., C.X., X.H.M., and H.W.D.; Writing—original draft, C.X., X.H.M., G. Y, and K.S.W.; Writing—review and editing, H.W.D., H.M.X., Y.P.W., C.Y.Z., J.L., W.Z., K.Z., C.W., and H.S.; Funding acquisition, H.M.X., H.W.D., and K.S.W.; Resources, H.M.X., K.S.W., G.Y., and H.W.D.; Data curation, K.S.W., J.Z., C.Z., Z.D., L.S., R.L., S.S., X.Z., Q.L., J.L., J.W., K.M., J.Q., Z.H., P.T., J.D., X.Q., B.Y.L., W.D.S., R.P.Q., J.T.Y., L.Y.H., Y.X., Z.C.Y., Z.L., H.R., C.L., W.G., Y.L., H.X., Y.G., J.P.Y., D.H., Z.S., X.F., L.C., X.Y., Z.L., and C.Y.; Supervision, H.M.X. and H.W.D. The author(s) read and approved the final manuscript.
Funding
H.S. and H.W.D. were partially supported by grants from National Institutes of Health (R01AR059781, P20GM109036, R01MH107354, R01MH104680, R01GM109068, R01AR069055, U19AG055373, R01DK115679), the Edward G. Schlieder Endowment and the Drs. W. C. Tsai and P. T. Kung Professorship in Biostatistics from Tulane University. H.M.X. was partially supported by the National Key Research and Development Plan of China (2017YFC1001103, 2016YFC1201805), National Natural Science Foundation of China (#81471453), and Jiangwang Educational Endowment. K.S.W. was partially supported by the National Natural Science Foundation of China (#81972490) and the Natural Science Foundation of Hunan Province (#2015JJ2150). Z.C.Y. was partially supported by the National Natural Science Foundation of China (#81673491). K.Z. was partially supported by grants from National Institutes of Health (2U54MD007595).
Availability of data and materials
The datasets analyzed during the current study are not publicly available due to limited computing/storage resources but are available from the corresponding author on reasonable request. The source code of our approach is available at GitHub: https://github.com/csu-bme/DeepPathology-CRC.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
H.W. Deng is Lead Contact
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
K.S. Wang, G. Yu, C. Xu, X.H. Meng is Equal first authors
Contributor Information
H. M. Xiao, Email: hmxiao@csu.edu.cn
H. W. Deng, Email: hdeng2@tulane.edu
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maung R. Pathologists’ workload and patient safety. Diagnostic Histopathol. 2016;22(8):283–287. doi: 10.1016/j.mpdhp.2016.07.004. [DOI] [Google Scholar]
- 5.Metter DM, Colgan TJ, Leung ST, Timmons CF, Park JY. Trends in the US and Canadian pathologist workforces from 2007 to 2017. JAMA Netw Open. 2019;2(5):e194337. doi: 10.1001/jamanetworkopen.2019.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayed S, Lukande R, Fleming KA. Providing pathology support in low-income countries. J Glob Oncol. 2015;1(1):3–6. doi: 10.1200/JGO.2015.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black-Schaffer WS, Morrow JS, Prystowsky MB, Steinberg JJ. Training pathology residents to practice 21st century medicine: a proposal. Acad Pathol. 2016;3:2374289516665393. doi: 10.1177/2374289516665393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–67. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hua K-L, Hsu C-H, Hidayati SC, Cheng W-H, Chen Y-J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther. 2015;8:2015–2022. doi: 10.2147/OTT.S80733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, Gonzalez F, Larsen AB, Vestergaard JS, Dahl AB, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20(1):237–248. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Ehteshami Bejnordi B, Veta M. Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J, the CC, Hermsen M, Manson QF et al: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campanella G, Hanna MG, Geneslaw L, Miraflor A. Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ: Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–9. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulten W, Pinckaers H, van Boven H, Vink R, de Bel T, van Ginneken B, van der Laak J, Hulsbergen-van de Kaa C, Litjens G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 2020;21(2):233–241. doi: 10.1016/S1470-2045(19)30739-9. [DOI] [PubMed] [Google Scholar]
- 14.Strom P, Kartasalo K, Olsson H. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study (vol 21, pg 222, 2020) Lancet Oncol. 2020;21(2):E70. doi: 10.1016/S1470-2045(19)30738-7. [DOI] [PubMed] [Google Scholar]
- 15.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu L, Chen H, Dou Q, Qin J, Heng PA. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging. 2017;36(4):994–1004. doi: 10.1109/TMI.2016.2642839. [DOI] [PubMed] [Google Scholar]
- 17.Sari CT, Gunduz-Demir C. Unsupervised feature extraction via deep learning for histopathological classification of colon tissue images. IEEE Trans Med Imaging. 2019;38(5):1139–1149. doi: 10.1109/TMI.2018.2879369. [DOI] [PubMed] [Google Scholar]
- 18.Sirinukunwattana K, Ahmed Raza SE, Yee-Wah T, Snead DR, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine Colon Cancer histology images. IEEE Trans Med Imaging. 2016;35(5):1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 19.Haj-Hassan H, Chaddad A, Harkouss Y, Desrosiers C, Toews M, Tanougast C. Classifications of multispectral colorectal cancer tissues using convolution neural network. J Pathol Inform. 2017;8:1. doi: 10.4103/jpi.jpi_47_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaddad A, Tanougast C. Texture analysis of abnormal cell images for predicting the continuum of colorectal Cancer. Anal Cell Pathol (Amst) 2017;2017:8428102. doi: 10.1155/2017/8428102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, Walliander M, Lundin M, Haglund C, Lundin J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8(1):3395. doi: 10.1038/s41598-018-21758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis C-A, Gaiser T, Marx A, Valous NA, Ferber D, et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. Plos Med. 2019;16(1):e1002730. doi: 10.1371/journal.pmed.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestol K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395(10221):350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 24.Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3(3):153–173. doi: 10.3978/j.issn.2078-6891.2012.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Algorithms May Assist Expert Pathologists in Prostate Cancer Diagnosis. Cancer Discov 2020, 10(3):OF1. [DOI] [PubMed]
- 26.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR): 27–30 June 2016 2016. 2016. Rethinking the Inception Architecture for Computer Vision; pp. 2818–2826. [Google Scholar]
- 27.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Li LJ, Lu GX. How medical ethical principles are applied in treatment with artificial insemination by donors (AID) in Hunan, China: effective practice at the Reproductive and Genetic Hospital of CITIC-Xiangya. J Med Ethics. 2005;31(6):333–337. doi: 10.1136/jme.2004.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velazquez Vega JE, Brat DJ, Cooper LAD. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A. 2018;115(13):E2970–E2979. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Accuracy of a deep learning algorithm for detection of diabetic retinopathy. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 32.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine. 2018;27:317–328. doi: 10.1016/j.ebiom.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR): 7–12 June 2015 2015. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 35.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Thirty-First AAAI Conference on Artificial Intelligence: 2017. 2017. Inception-v4, inception-resnet and the impact of residual connections on learning. [Google Scholar]
- 36.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556 2014.
- 37.Huang G, Liu Z, Lvd M, Weinberger KQ. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR): 21–26 July 2017 2017. 2017. Densely Connected Convolutional Networks; pp. 2261–2269. [Google Scholar]
- 38.Hu J, Shen L, Sun G. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition: 18–23 June 2018 2018. 2018. Squeeze-and-Excitation Networks; pp. 7132–7141. [Google Scholar]
- 39.Veit A, Alldrin N, Chechik G, Krasin I, Gupta A, Belongie S. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR): 21–26 July 2017 2017. 2017. Learning from Noisy Large-Scale Datasets with Minimal Supervision; pp. 6575–6583. [Google Scholar]
- 40.GSN H, Swersky K. Lecture 6a Overview of Mini-Batch Gradient Descent. Lecture Notes Distributed in CSC321 of University of Toronto. 2014. [Google Scholar]
- 41.Heller R, Stanley D, Yekutieli D, Rubin N, Benjamini Y. Cluster-based analysis of FMRI data. Neuroimage. 2006;33(2):599–608. doi: 10.1016/j.neuroimage.2006.04.233. [DOI] [PubMed] [Google Scholar]
- 42.Liu T. Diagnostic pathology. 3. Beijing: People’s Medical Publishing House; 2013. [Google Scholar]
- 43.Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, Chang EI. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics. 2017;18(1):281. doi: 10.1186/s12859-017-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kainz P, Pfeiffer M, Urschler M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ. 2017;5:e3874. doi: 10.7717/peerj.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponzio F, Macii E, Ficarra E, Di Cataldo S. Colorectal Cancer classification using deep convolutional networks - an experimental study. 2018. [Google Scholar]
- 46.Cruz-Roa A, Basavanhally A, González F, Gilmore H, Feldman M, Ganesan S, Shih N, Tomaszewski J, Madabhushi A: Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In: Medical Imaging 2014: Digital Pathology: 2014: International Society for Optics and Photonics; 2014:904103.
- 47.Araujo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, Polonia A, Campilho A. Classification of breast cancer histology images using convolutional neural networks. PLoS One. 2017;12(6):e0177544. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jannesari M, Habibzadeh M, Aboulkheyr H, Khosravi P, Elemento O, Totonchi M, Hajirasouliha I. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM): 3–6 Dec. 2018 2018. 2018. Breast cancer histopathological image classification: a deep learning approach; pp. 2405–2412. [Google Scholar]
- 49.Feng YQ, Zhang L, Mo J. Deep manifold preserving autoencoder for classifying breast cancer histopathological images. Ieee Acm T Comput Bi. 2020;17(1):91–101. doi: 10.1109/TCBB.2018.2858763. [DOI] [PubMed] [Google Scholar]
- 50.Alirezazadeh P, Hejrati B, Monsef-Esfahani A, Fathi A. Representation learning-based unsupervised domain adaptation for classification of breast cancer histopathology images. Biocybern Biomed Eng. 2018;38(3):671–683. doi: 10.1016/j.bbe.2018.04.008. [DOI] [Google Scholar]
- 51.Karimi D, Nir G, Fazli L, Black PC, Goldenberg L, Salcudean SE. Deep learning-based Gleason grading of prostate cancer from histopathology images-role of multiscale decision aggregation and data augmentation. IEEE J Biomed Health Inform. 2020;24(5):1413–1426. doi: 10.1109/JBHI.2019.2944643. [DOI] [PubMed] [Google Scholar]
- 52.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. 4. Lyon: International Agency for Research on Cancer; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary-Text 1.a Collection and digitalization of the WSIs. Supplementary-Text 1.b Dataset-A, B, C and D. Supplementary-Text 1.c Patch-level performance and patient-level accuracy. Supplementary-Text 1.d Comparison of different architectures at patch-level. Supplementary-Text 1.e Comparison of different cluster sizes for aggregation of patch-level results. Supplementary-Table 1. Input patch size for common CNN. Supplementary-Table 2 Pathologist info. Supplementary-Table 3 List of AUCs of AI applied in CRC and other cancer types. Supplementary-Table 4 Overall performance of AI and pathologists in Human-AI contest. Supplementary-Table 5 Cohen’s Kappa coefficient for agreement among human experts and AI. Supplementary-Figure 1 Weakly-labeled and fully-labeled CRC patches. Supplementary-Figure 2.The distribution of cancerous area in multiple independent WSI datasets measured by the proportion of patches (P) containing cancer cells on the WSI. Supplementary-Figure 3 Heatmap produced by AI. Supplementary-Figure 4 Activation map produced by AI.
Data Availability Statement
The datasets analyzed during the current study are not publicly available due to limited computing/storage resources but are available from the corresponding author on reasonable request. The source code of our approach is available at GitHub: https://github.com/csu-bme/DeepPathology-CRC.