Abstract
The present study evaluated the antibacterial activity and the synergy of the sanguisorbigenin (SGB) from the dried root of Sanguisorba officinalis L. combined with β‐lactam antibiotics against methicillin‐resistant Staphylococcus aureus. A total of six strains of reference strain and clinical isolates were used to determine the antibacterial activity using a broth microdilution assay, and the synergistic effects were determined using a checkerboard assay. To analyse the mechanism of synergy, we conducted the level of penicillin‐binding protein 2a by western blot. In addition, quantitative RT‐PCR was performed to analyse the mecA gene expression. The minimal inhibitory concentration values of SGB against six strains of S. aureus were in the range of 12·5–50 μg ml−1, and there were synergy, or partial synergy effects when SGB was combined with antibiotics. Furthermore, when treated with SGB, the level of penicillin‐binding protein 2a and the expression of the mecA gene was reduced significantly. In conclusion, this study demonstrated that SGB is a potential natural antibacterial agent against methicillin‐resistant S. aureus that represents a considerable burden on the healthcare system worldwide, and may an exceptionally modulator of β‐lactam antibiotics.
Keywords: mecA, methicillin‐resistant Staphylococcus aureus, penicillin‐binding protein 2a, sanguisorbigenin, synergy, β‐lactam antibiotics
Significance and Impact of the Study: Methicillin‐resistant Staphylococcus aureus is the pathogen represents a considerable burden on the healthcare system worldwide, and resistance to nearly all β‐lactam antibiotics. For the first time, we evaluated the antibacterial activity and the synergy of the sanguisorbigenin (SGB) combined with β‐lactam antibiotics against methicillin‐resistant S. aureus. The result indicated that SGB has a potent antibacterial effect and could effectively reverse the sense of β‐lactam antibiotics by inhibiting mecA expression and decrease the PBP2a expression. This study has a certain reference value for the development of novel antibacterial agents and antibiotic modulators.
Introduction
Staphylococcus aureus, a major bacterial pathogen of human beings, can acquire resistance to most antibiotics (Quadri et al. 2020). With the development of this ability, ‘superbug’ has appeared. For instance, the appearance of methicillin‐resistant Staphylococcus aureus (MRSA) was due to the clinical use of methicillin (Lakhundi and Zhang 2018). MRSA is a ubiquitous bacterium that causes a wide range of infections, from superficial skin infections to severe, even cause severe life‐threatening illnesses such as sepsis and endocarditis (Thapaliya et al. 2017). It represents a considerable burden on the healthcare system globally (Song et al. 2018). In recent years, the resistance of conventional anti‐MRSA antibiotics like vancomycin, teicoplanin, and linezolid have been found in the clinic(He et al. 2016). The emergence of novel antibacterial substances is required to combat and prevent antibiotic‐resistant bacterial infections. Resistance is usually achieved by obtaining a mecA gene encoding penicillin‐binding protein (PBP2a), which has a low affinity for β‐lactamase (Peacock and Paterson 2015). The resistant loci, such as blaR1 and blaZ, which is categorized as a class A β‐lactamase (Lakhundi and Zhang 2018), and mecR1 is divergently transcribed regulatory genes from the mecA promoter (He et al. 2016).
Sanguisorbigenin (SGB) is a natural compound isolated from the root of the perennial herbage, Sanguisorba officinalis L., family Rosaceae. Genus Sanguisorba mainly distributed among Asia, Europe, and North America (Meng et al. 2018). The common name ‘Sanguisorba’ has evolved from the Latin ‘Sanguis’—blood, and ‘sorbet’‐absorb, which its popular herbal use as an astringent bleeding (Pawlaczyk‐Graja et al. 2016). Sanguisorba has been used as an herbal medicine for centuries (Kim et al. 2018). Thousands of years, it is used for all kinds of bleeding due to various blood‐heat and scald. Besides, recent researches investigated that extracts from sanguisorba could be used to treat many types of allergic skin diseases (Yang et al. 2016), and have the activity of anti‐inflammatory, antiviral (Wu et al. 2018), anti‐cancer (Jang et al. 2018) and antibacterial (Su et al. 2019). Therefore, SGB may have inhibitory activity to MRSA and the underlying mechanism of the action of SGB against the MRSA has not yet been reported. In this study, we investigated the antibacterial effect of SGB and the antibacterial synergism of SGB combined with β‐lactam antibiotics against MRSA at the molecular level.
Results and discussion
Antibacterial activity of SGB and synergy effect
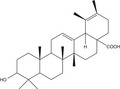
Of late, the emergence of resistant bacteria has become increasingly common (Thapaliya et al. 2017). As such, novel studies of antibiotics are urgently required. This review focuses on the study of the antibacterial activity of SGB and the mechanism of the synergistic effect in combination with β‐lactam antibiotics. SGB, a triterpene compound (Fig. 1), is the main chemical constituents isolated from S. officinalis L. The researchers of triterpenes have mainly focused on the antioxidant, anti‐inflammatory, and anti‐tumour activities in recent years (Wu et al. 2018). Moreover, the anti‐MRSA activity of SGB has not been reported. For the first time, we measured the antibacterial activity of SGB and the synergistic effect. The MIC values of ampicillin and oxacillin against the six S. strains were 15·6–62·5 μg ml−1 and 125–1000 μg ml−1 (Table 1). The MIC values of SGB as low as 12·5–50 μg ml−1, the values were lower than the MIC values of two antibiotics. The results showed that SGB exhibited a high antibacterial activity against MRSA. The natural product preparation has the characteristics of high economic efficiency and high bioavailability (Xiao et al. 2017), and are potential clues for drug development (Dias et al. 2012). Therefore, SGB is a potential natural antibacterial substance, and may be valuable for the development of anti‐MRSA substances. Synergistic effects were tested by using an SGB combination with two β‐lactam antibiotics, the data indicated that the MIC of ampicillin and oxacillin reduced by 2‐ to 6‐fold and 2‐ to 8‐fold in the presence of SGB respectively (Table 1). The results showed that the SGB combination with two antibiotics all have the synergy or partial synergy effect. It provides evidence for the SGB‐induced reversal effect on β‐lactam antibiotic resistance of MRSA and may be helpful to reduce the use of antibiotics.
Figure 1.
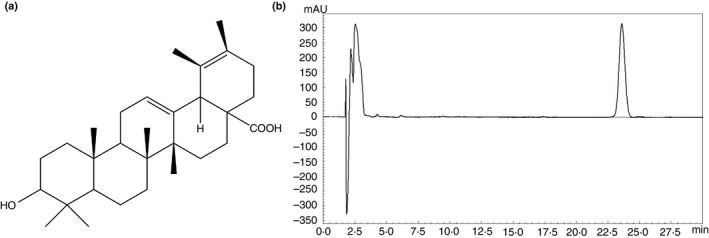
(a) The chemical structure of sanguisorbigenin (SGB). (b) HPLC Chromatogram of sanguisorbigenin. HPLC conditions: Kinetex XB‐C18 column (100 × 4·6 mm × 2·6 µm); mobile phase, water (A) and acetonitrile (B) in gradient mode (0–2 min, 29–31% B; 2–13 min, 31–35% B; 13–15 min, 35–40% B; 15–23 min, 40–44% B; 23–25 min, 44–46% B; 25–31 min, 46–49% B; and 31–38 min, 49–55% B); flow rate, 1·0 ml min−1; column temperature, 30°C; detection wavelength, 210 nm.
Table 1.
Synergy effects of SGB combination of antibiotics
| Staphylococcus aureus strains | MIC (μg ml−1) | FICI | Interpretation | MIC (μg ml−1) | FICI | Interpretation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Agent | Alone | Combination | Agent | Alone | Combination | |||||
| ATCC 33591 | SGB | 12·5 | 6·25 | 0·63 | Partial S. | SGB | 12·5 | 6·3 | 0·75 | Partial S. |
| AMP | 62·5 | 7·8 | OXA | 125 | 31·3 | |||||
| CCARM 3090 | SGB | 25 | 6·3 | 0·5 | Synergy | SGB | 25 | 6·3 | 0·38 | Synergy |
| AMP | 31·3 | 3·9 | OXA | 125 | 15·6 | |||||
| CCARM 3091 | SGB | 25 | 6·3 | 0·5 | Synergy | SGB | 25 | 6·3 | 0·5 | Synergy |
| AMP | 31·3 | 7·8 | OXA | 1, 000 | 250 | |||||
| CCARM 3095 | SGB | 25 | 12·3 | 0·63 | Partial S. | SGB | 25 | 6·3 | 0·5 | Synergy |
| AMP | 15·6 | 7·8 | OXA | 250 | 62·5 | |||||
| CCARM 3102 | SGB | 25 | 12·5 | 0·63 | Partial S. | SGB | 25 | 6·3 | 0·75 | Partial S. |
| AMP | 15·6 | 7·8 | OXA | 250 | 125 | |||||
| DPS‐1 | SGB | 50 | 25 | 0·63 | Partial S. | SGB | 50 | 25 | 0·75 | Partial S. |
| AMP | 31·3 | 3·9 | OXA | 500 | 125 | |||||
SGB, sanguisorbigenin; AMP, ampicillin; OXA, oxacillin; MIC; minimal inhibitory concentration; S. aureus, Staphylococcus aureus; FICI, fractional inhibitory concentration index; Partial S., partial synergy. Index interpretation: <0·5, synergy; 0·5–0·75, partial synergy; 0·75–1, additive effect; 1–4, no effect; and >4, antagonism. Values represent the average of three independent experiments.
Reverse transcription and qRT‐PCR
The resistance mechanisms to β‐lactam antibiotics are associated with the presence of a penicillin‐binding protein (PBP2) encoded by the mecA gene in MRSA (Moosavian et al. 2020). The affinity of most β‐lactam reduced due to the overexpression of mecA encoded PBP2a (Hui et al. 2020). However, only when MRSA encounters β‐lactam, it produces the mechanism of resistance to β‐lactam antibiotics, which is regulated by the transmembrane sensor/signal transducer proteins blaR1 and mecR1 (Belluzo et al. 2019). We speculated that SGB may damage the mecA gene and inhibit PBP2a growth. To this end, we performed qRT‐PCR to test the blaR1, mecA, mecR1, and blaZ gene. When exposed to 1/2 MIC of SGB, the transcriptional levels of blaR1 is almost undetectable, the transcriptional levels of mecA and blaZ were reduced by 5‐ and 4‐fold respectively (Fig. 2). The expression of blaR1, mecA and blaZ was significantly inhibited in MRSA in a dose‐dependent manner when it was treated with sub‐MIC (1·56–6·25 μg ml−1) concentrations of SGB. As expected, SGB remarkably reduced the gene expression of mecA. Moreover the gene expression of blaZ and blaR1 that participate in the regulation of mecA also remarkably reduced. However, a weak influence on the reverse transcription of mecR1 was measured in S. aureus, which is likely due to SGB does not suppress mecA expression in MRSA through regulating mecR1.
Figure 2.
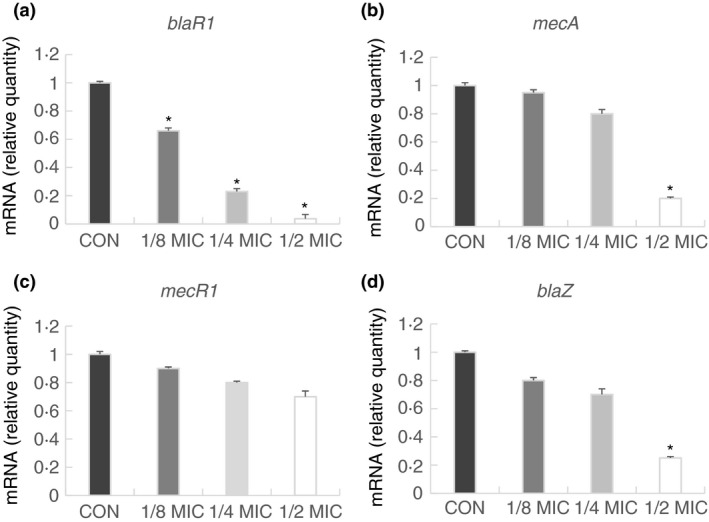
Relative gene expression of blaR1, mecA, mecR1 and blaZ in Staphylococcus aureus (ATCC 33591) after growth at various concentrations of sanguisorbigenin. The relative gene expression of blaR1 (a), mecA (b), mecR1 (c) and blaZ (d) was reduced in a dose‐dependent manner. Values represent the mean and standard error of three independent experiments. *represents P < 0·05.
Expression of PBP2a protein in MRSA treated with SGB
We speculated that SGB inhibited mecA at both transcriptional and translational expression levels. Western blotting was performed to detect the protein level of PBP2a in MRSA. PBP2a was exposed to subinhibitory concentrations (1·56–6·25 μg ml−1) of SGB. When exposed to subinhibitory concentrations (lanes 2–4) of SGB, all the PBP2a levels were reduced markedly, and when exposed to 6·25 μg ml−1 of SGB (Lane 2), the PBP2a levels were hard to trace (Fig. 3). The reduced PBP2a expression level in MRSA after the addition of SGB corresponded to the low expression of mecA transcription after primer extension. The results confirm our speculated that SGB could damage the mecA gene, and then inhibit PBP2a growth, leading to the reduction of MRSA resistance.
Figure 3.
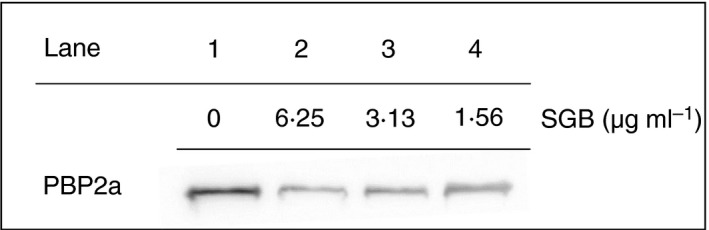
Expression of PBP2a in Staphylococcus aureus (ATCC 33591) cultures grown in the presence of various concentrations of sanguisorbigenin. The PBP2a production was reduced significantly after exposure to S. aureus strains with 6·25 µg ml−1 SGB (Lane 2), 3·13 µg ml−1 SGB (Lane 3) and 1·56 µg ml−1 SGB (Lane 4). CON, is control S. aureus strain, which without the drug (Lane 1).
The results of the present study suggest that SGB has a potent antibacterial effect and could effectively reverse the sense of antibiotics by inhibiting the mecA expression and decrease PBP2a expression. According to previous reports, ethanol extract of S. officinalis significantly inhibited MRSA biofilm formation in an ica‐dependent manner (Chen et al. 2015). Hence, we speculated that SGB could inhibit MRSA biofilm, which improves the efficiency of antibiotics entering cells, then improves the sensitivity of antibiotics. Further studies of biofilm inhibition assay would be conducted to verify the mechanism of SGB in combination with conventional antibiotics against MRSA, and multiresistant strains would be used in the experiment. The triterpene glycosides isolated from S. officinalis L. has been reported that exhibited the markedly cytotoxic potential with low IC50 values against tumour cell lines (Hu et al. 2015). Therefore, the cytotoxicity assay of SGB would be tested to evaluate whether it is suitable for clinical application.
Materials and methods
Plant materials
The dried roots of S. officinalis L. was collected in October 2016 from Shaoyang, Hunan province, China. A voucher specimen (No. 20161125) was identified by one of the authors (X. Q. L) and deposited in the School of Pharmacy, Hunan University of Chinese Medicine.
Extraction and isolation
The air‐dried roots of S. officinalis L. (3 kg) was extracted with 70% aqueous MeOH (100 ml × 3) at room temperature and filtered. The filtrate was concentrated under reduced pressure using a rotavapor to acquire the MeOH extract (0·6 kg). The crude extract was suspended in distilled H2O and successively solvent‐partitioned with petroleum ether, EtOAc and n‐BuOH respectively. The EtOAc fraction was separated over macro‐porous resin column chromatography (CC) and eluted with an EtOH‐H2O gradient system (0–95%) to yield fractions A–E. Fraction D (75% EtOH, 18·0 g) was subjected to silica gel CC (CHCl3‐MeOH‐H2O, 25 : 1 : 0 to 1 : 1 : 0·2) to afford subfractions D1‐D15. Fractions D3 (100 mg) was chromatographed on silica gel H (CHCl3‐MeOH‐H2O, 15 : 1 : 0 to 6 : 1 : 0·1) to afford six subfractions D3·1–D3·6. Subfraction D3·3 (88·0 mg) was purified by ODS column chromatography (MeOH‐H2O, 70–100%) to obtain SGB (18·0 mg). The structures of compounds were identified by analysing the signals of spectral data (mass spectrometry, 1H‐and 13C‐nuclear magnetic resonance) and the spectral data were compared with those reported previously (WADA et al. 1964).
High‐performance liquid chromatography (HPLC)
The compound used in this study was checked by high‐performance liquid chromatography (HPLC) and was >98% pure. HPLC analysis of SGB was performed on Agilent 1200 HPLC equipment with a Kinetex XB‐C18 analytical column (100 × 4·6 mm × 2·6 µm; Phenomenex, Inc., Torrance, CA) at 30°C. The mobile phase of an experiment where water (A) and Acetonitrile (B) following a gradient profile of 0–2 min, 29–31% by 2–13 min, 31–35% by; 13–15 min, 35–40% by; 15–23 min, 40–44% by; 23–25 min, 44–46% by; 25–31 min, 46–49% by; and 31–38 min, 49–55% by with a flow rate of 1·0 ml min−1 and a wavelength of 210 nm. The purity, value was found to be >98% using a peak area normalization method. The purity value was obtained by calculating the percentage of the SGB peak area to that of the total peaks in the HPLC chromatogram.
Bacterial strains and medium
A total of six strains MRSA used in this study, A reference strain (ATCC 33591) was purchased from the American Type Culture Collection (ATCC; Manassas, VA); one clinical isolate of MRSA (DPS‐1) was collected from the patient at Wonkwang University Hospital (Jeonbuk, Korea) (Kong et al. 2016); The other four strains, CCARM 3090, CCARM 3091, CCARM 3095, CCARM 3102, were provided by the Culture Collection of Antimicrobial Resistant Microbes (National Research Resource Bank, Seoul, Korea). Mueller‐Hinton agar (MHA) and Muller‐Hinton broth (MHB) as the culture medium for bacterial at 37˚C.
Reagents
Mueller‐Hinton agar, MHB, and skim milk were obtained from Difco Laboratories (Baltimore, MD). Ampicillin and oxacillin were obtained from Sigma‐Aldrich Co. (St. Louis, MO). SMART™ bacterial protein extraction solution was purchased from Intron Bio Technology, Inc. (Seongnam, Korea). The chemiluminescent ECL assay kit was purchased from ATTO Corp. (Tokyo, Japan). E.Z.N.A. Bacterial RNA Kit was purchased from Omega Bio‐Tek (Norcross, GA). The sequences of primers used in this study were purchased from Bioneer (Daejeon, Korea) (Table 2). QuantiTect Reverse Transcription Kit was purchased from (Dusseldorf, Germany). Power SYBR Green PCR Master Mix was purchased from Life Technologies LTD (Warrington, UK).
Table 2.
Primers used in qRT‐PCR
| Primer | Sequence (5′–3′) |
|---|---|
| 16S | F: ACTCCTACGGGAGGCAGCAG |
| R: ATTACCGCGGCTGCTGG | |
| mecA | F: CAATGCCAAAATCTCAGGTAAAGTG |
| R: AACCATCGTTACGGATTGCTTC | |
| mecR1 | F: GTGCTCGTCTCCACGTTAATTCCA |
| R: GACTAACCGAAGAAGTCGTGTCAG | |
| blaR1 | F: CACTATTCTCAGAATGACTTGGT |
| R: TGCATAATTCTCTTACTGTCATG | |
| blaZ | F: GCTTTAAAAGAACTTATTGAGGCTTC |
| R: CCACCGATYTCKTTTATAATTT |
Susceptibility testing of SGB with antibiotics
The MIC value of SGB, either alone or in combination with antibiotics (ampicillin and oxacillin), was determined using the microdilution and checkerboard assays. The strains were first sub‐cultured twice on MHA at 37°C for 24 h. Serial dilutions of SGB with antibiotics were mixed in MHB. The MRSA inocula were adjusted to 0·5 McFarland standard (1·5 × 108 colony‐forming units (CFU) per well) in MHB. The bacterial concentration of the final inoculum was 1·5 × 105 CFU per well. Each MIC value was determined after a 24 h incubation period at 37°C and defined as the lowest concentration of SGB, either alone or in combination with antibiotics, which inhibited bacterial growth. The interaction between the two drugs is usually quantified by determining the fractional inhibitory concentration index (FICI). The FICI was used to analyse the interaction between SGB and antibiotic at a 1 : 1 ratio, and was calculated as follows:
where [A] and [B] denote the MICs of SGB and antibiotic respectively and MICA and MICB represent the MICs of SGB and antibiotic respectively when the two drugs are used in combination. The FICI values obtained were interpreted as follows: values ≤0. 5 indicated synergy, 0·5 <values ≤0·75 indicated partial synergy, 0·75 ≤ values ≤1 indicated additive effect, 1 < values ≤4 indicated no interaction, and values >4 indicated antagonism (Mun et al. 2013).
Reverse transcription and quantitative RT‐PCR (qRT‐PCR)
ATCC 33591 was cultured in MHB overnight until the OD600 value reached 0·9, and then SGB with sub inhibition concentration (1/8 MIC, 1/4 MIC and 1/2 MIC) was added for 0·5 h. A control was included without SGB. First, we extracted RNA according to the protocol of E.Z.N.A. Bacterial RNA Kit. Second, then reverse transcribed RNA into cDNA according to the manufacturer's instructions of QuantiTect Reverse Transcription Kit, and then the PCR was set up was as follows: 10 μl of 2× SYBR premix, 2 μl sample cDNA and 1 μl of each primer (10 μmol l−1), and deionized water to a total volume of 20 μl. The sequences of primer are presented in Table 2. The PCR was run using the StepOnePlus real‐time PCR system (Applied Biosystems, Courtaboeuf, France) in the third part (Mun et al. 2016).
Western blotting analysis
The MRSA (ATCC 33591) culture was grown to an OD600 value of 0·9 in MHB and treated with sub‐inhibitory concentrations (1·56–6·25 μg ml−1) of SGB, and a control was included without SGB in the western blotting analysis. Cell protein extracts were harvested after 30 min of incubation and suspended in the bacterial protein extract (iNtRON Biotechnology, Applied Biosystems) containing Tris‐HCI (pH 7·5). The procedures were performed according to the manufacturer's description. Protein concentrations were measured using a Bio‐Rad protein assay reagent (Bio‐Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. The aliquots of equal protein were analysed via sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE). The electrophoresed gels were transferred to Amersham HybondTM‐P membranes (GE Healthcare, Piscataway, NJ) for the western blotting analysis. The membranes were blocked using 5% skim milk in Tris‐buffered saline with Tween‐20 buffer (150 mmol l−1, NaCl, 20 mmol l−1 Tris‐HCl and 0·05% Tween‐20, pH 7·4). After blocking, the membranes were probed with monoclonal mouse anti‐PBP2a primary antibody (diluted 1 : 1000; DiNonA, Seoul, Korea) and re‐probed with anti‐mouse IgG secondary antibody (diluted 1 : 2000; Enzo Life Sciences, Ann Arbor, MI). The membranes were then treated with ECL™ Prime western blotting detection reagent (GE Healthcare Life Sciences, Incheon, Korea), and the bands were visualized using an ImageQuant LAS‐4000 mini chemical luminescent imager (GE Healthcare Life Sciences) (Zhou et al. 2017).
Statistical analysis
Analyses were performed in triplicate and data were presented as the mean ± standard deviation. The results were statistically analysed using an independent Scheffe'st test (SPSS software ver. 22.0; IBM SPSS, Armonk, NY). A P‐value of <0·05 was considered statistically significant.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgements
The present study was supported by Wonkwang University in 2019.
Contributor Information
O.‐H. Kang, Email: kangokhwa@daum.net.
D.‐Y. Kwon, Email: sssimi@wku.ac.kr.
References
- Belluzo, B.S. , Abriata, L.A. and Giannini, E. (2019) An experiment‐informed signal transduction model for the role of the Staphylococcus aureus MecR1 protein in β‐lactam resistance. Sci Rep 9, 19558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Shang, F. , Meng, Y. , Li, L. , Cui, Y. , Zhang, M. , Qi, K. and Xue, T. (2015) Ethanol extract of Sanguisorba officinalis L. inhibits biofilm formation of methicillin‐resistant Staphylococcus aureus in an ica‐dependent manner. J Dairy Sci 98, 8486–8491. [DOI] [PubMed] [Google Scholar]
- Dias, D.A. , Urban, S. and Roessner, U. (2012) A historical overview of natural products in drug discovery. Metabolites 2, 303–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M. , Shao, L. , Liu, Q. , Li, J. , Lin, H. , Jing, L. , Li, M. and Chen, D. (2016) Mechanism of synergy between SIPI‐8294 and β‐lactam antibiotics against methicillin‐resistant Staphylococcus aureus . Lett Appl Microbiol 63, 3–10. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Song, Y. , Li, H. , Yang, B. , Mao, X. , Zhao, Y. and Shi, X. (2015) Cytotoxic triterpene glycosides from the roots of Sanguisorba officinalis . Arch Pharm Res 38, 984–990. [DOI] [PubMed] [Google Scholar]
- Hui, J. , Dong, P.T. , Liang, L. , Mandal, T. , Li, J. , Ulloa, E.R. , Zhan, Y. , Jusuf, S. et al. (2020) Photo‐disassembly of membrane microdomains revives conventional antibiotics against MRSA. Adv Sci (Weinh) 7, 1903117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, E. , Inn, K.S. , Jang, Y.P. et al. (2018) Phytotherapeutic activities of Sanguisorba officinalis and its chemical constituents: a review. Am J Chin Med 46, 299–318. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Oh, S. and Noh, H.B. (2018) In vitro antioxidant and anti‐propionibacterium acnes activities of cold water, hot water, and methanol extracts, and their respective ethyl acetate fractions, from Sanguisorba officinalis L. roots. Molecules 23, e3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, R. , Kang, O.H. , Seo, Y.S. , Mun, S.H. , Zhou, T. , Shin, D.W. and Kwon, D.Y. (2016) The inhibition effect of Chlorpromazine against the β‐lactam resistance of MRSA. Asian Pac J Trop Med 9, 542–546. [DOI] [PubMed] [Google Scholar]
- Lakhundi, S. and Zhang, K. (2018) Methicillin‐resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31, 00020‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X.X. , Xian, Y.F. , Xiang, L. , Zhang, D. , Shi, Y.H. , Wu, M.L. , Dong, G.Q. , Ip, S.P. et al. (2018) Complete chloroplast genomes from sanguisorba: identity and variation among four species. Molecules 23, e2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosavian, M. , Dehkordi, P.B. and Hashemzadeh, M. (2020) Characterization of SCCmec, Spa types and multidrug resistant of methicillin‐resistant Staphylococcus aureus isolates in Ahvaz, Iran. Infect Drug Resist 13, 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun, S.H. , Joung, D.K. , Kim, Y.S. , Kang, O.H. , Kim, S.B. , Seo, Y.S. , Kim, Y.C. , Lee, D.S. et al. (2013) Synergistic antibacterial effect of curcumin against methicillin‐resistant Staphylococcus aureus . Phytomedicine 20, 714–718. [DOI] [PubMed] [Google Scholar]
- Mun, S.H. , Kong, R. , Seo, Y.S. , Zhou, T. , Kang, O.H. , Shin, D.W. and Kwon, D.Y. (2016) Subinhibitory concentrations of punicalagin reduces expression of virulence‐related exoproteins by Staphylococcus aureus . FEMS Microbiol Lett 363, e253. [DOI] [PubMed] [Google Scholar]
- Pawlaczyk‐Graja, I. , Balicki, S. and Ziewiecki, R. (2016) Inhibitory effects of Sanguisorba officinalis root extract on HYBID (KIAA1199)‐mediated hyaluronan degradation and skin wrinkling. Int J Biol Macromol 41, 12–20. [DOI] [PubMed] [Google Scholar]
- Peacock, S.J. and Paterson, G.K. (2015) Mechanisms of methicillin resistance in Staphylococcus aureus . Annu Rev Biochem 84, 577–601. [DOI] [PubMed] [Google Scholar]
- Quadri, S.A. , Al‐Sultan, A.A. , Al‐Ramdan, A.M. , Badger‐Emeka, L. and Ali, S. (2020) Frequency of panton‐valentine leukocidin gene among clinical isolates of methicillin‐resistant Staphylococcus aureus in Eastern Province of Saudi Arabia. J Glob Infect Dis 12, 37–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. , Zeng, Q. , Xiang, Y. , Gao, L. , Gao, L. , Huang, J. , Huang, J. , Wu, K. et al. (2018) The antibacterial effect of topical ozone on the treatment of MRSA skin infection. Mol Med Rep 17, 2449–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, X.D. , Guo, R.H. , Yang, S.Y. , Kim, Y.H. and Kim, Y.R. (2019) Anti‐bacterial effects of components from Sanguisorba officinalis L. on Vibrio vulnificus and their soluble epoxide hydrolase inhibitory activity. Nat Prod Res 33, 3445–3449. [DOI] [PubMed] [Google Scholar]
- Thapaliya, D. , Hellwig, E.J. , Kadariya, J. , Rush, H. , Yee, C. , Oet, M. , Lohani, S. and Smith, T.C. (2017) Prevalence and characterization of Staphylococcus aureus and methicillin‐resistant Staphylococcus aureus on public recreational beaches in Northeast Ohi. Geohealth 1, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, H. , Nakata, H. and Hirata, Y. (1964) Structure of sanguisorbigenin, a triterpene obtaines from Sanguisorba officinalis L. Yakugaku Zasshi 84, 477–479. [PubMed] [Google Scholar]
- Wu, C. , Yao, M. , Li, W. , Cui, B. , Dong, H. , Ren, Y. , Yang, C. and Gan, C. (2018) Simultaneous determination and pharmacokinetics study of six triterpenes in rat plasma by UHPLC‐MS/MS after oral administration of Sanguisorba officinalis L. Molecules 23, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Zhao, Y. and Xie, Y. (2017) Design and implementation of fast allergy skin test detector for traditional Chinese medicine injections. Exp Ther Med 13, 1884–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.H. , Yoo, J.M. , Cho, W.K. and Ma, J.Y. (2016) Anti‐inflammatory effects of Sanguisorbae Radix water extract on the suppression of mast cell degranulation and STAT‐1/Jak‐2 activation in BMMCs and HaCaT keratinocytes. BMC Complement Altern Med 16, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T. , Li, Z. , Kang, O.H. , Mun, S.H. , Seo, Y.S. , Kong, R. , Shin, D.W. , Liu, X.Q. and et al. (2017) Antimicrobial activity and synergism of ursolic acid 3‐O‐α‐L‐arabinopyranoside with oxacillin against methicillin‐resistant Staphylococcus aureus . Int J Mol Med 40, 1285–1293. [DOI] [PubMed] [Google Scholar]


