Abstract
Breast cancer is one of the most prevalent types of malignant tumors in the world, resulting in a high incidence of death. The development of new molecules and technologies aiming to apply more effective and safer therapy strategies has been intensively explored to overcome this situation. The association of nanoparticles with known antitumor compounds (including plant-derived molecules such as curcumin) has been considered an effective approach to enhance tumor growth suppression and reduce adverse effects. Therefore, the objective of this systematic review was to summarize published data regarding evaluations about efficacy and toxicity of curcumin nanoparticles (Cur-NPs) in in vivo models of breast cancer. The search was carried out in the databases: CINAHL, Cochrane, LILACS, Embase, FSTA, MEDLINE, ProQuest, BSV regional portal, PubMed, ScienceDirect, Scopus, and Web of Science. Studies that evaluated tumor growth in in vivo models of breast cancer and showed outcomes related to Cur-NP treatment (without association with other antitumor molecules) were included. Of the 528 initially gathered studies, 26 met the inclusion criteria. These studies showed that a wide variety of NP platforms have been used to deliver curcumin (e.g., micelles, polymeric, lipid-based, metallic). Attachment of poly(ethylene glycol) chains (PEG) and active targeting moieties were also evaluated. Cur-NPs significantly reduced tumor volume/weight, inhibited cancer cell proliferation, and increased tumor apoptosis and necrosis. Decreases in cancer stem cell population and angiogenesis were also reported. All the studies that evaluated toxicity considered Cur-NP treatment to be safe regarding hematological/biochemical markers, damage to major organs, and/or weight loss. These effects were observed in different in vivo models of breast cancer (e.g., estrogen receptor-positive, triple-negative, chemically induced) showing better outcomes when compared to treatments with free curcumin or negative controls. This systematic review supports the proposal that Cur-NP is an effective and safe therapeutic approach in in vivo models of breast cancer, reinforcing the currently available evidence that it should be further analyzed in clinical trials for breast cancer treatments.
Keywords: breast cancer, nanoparticle, curcumin, apoptosis, antitumor, toxicity, in vivo, systematic review
Graphical Abstract.
Introduction
Among women, breast cancer is the most prevalent cancer worldwide, with 2.1 million cases reported in 2018 (with an annual increase of 3.1%) and more than 620,000 deaths per year (1, 2). Breast cancer is divided into five major intrinsic molecular subtypes: luminal A-like (60–70%), luminal B-like human epidermal growth factor receptor-type 2 negative (HER2–) (10–20%), HER2-enriched (non-luminal) and luminal B-like HER2+ (13–15%), and triple-negative (10–15%) (1). Early diagnosis of breast cancer raises the chances of total recuperation of patients, and the treatment concept is decided based on several criteria such as subtype and grade (3). Chemotherapy and endocrine therapy are typical systemic therapies in non-metastatic breast cancer. They can be associated with local therapy like surgery and radiation (3). In most cases, metastatic breast cancer remains incurable and therapy aims to prolong life and alleviate symptoms (3). However, these conventional treatments present some limitations, like resistance to chemotherapy or endocrine therapy, and some adverse effects (4). Thus, alternative treatments must be investigated to improve the recovery of breast cancer patients, reduce adverse effects, and circumvent therapy resistance.
Natural products are considered as promising alternatives for the development of new antitumor drugs (5, 6). Curcumin, or diferuloylmethane, is a yellow pigment extracted from the rhizomes of Curcuma longa Linn, also known as turmeric. It is the most abundant polyphenol and curcuminoid present in the root of this plant (7). Usually used in culinary and traditional medicine, curcumin also interests conventional medicine by demonstrating antioxidant and anti-inflammatory activities (7). Its anti-cancer effect was reported for the first time in 1985, by Kuttan and co-workers, in cells and animal models of lymphoma (8). In breast cancer, curcumin inhibits cell proliferation, induces apoptosis (5) and acts as a potent antiangiogenic, anti-invasive, and anti-metastatic agent in vitro and in vivo (9). Curcumin also demonstrated the capacity to reverse chemotherapeutic resistance in doxorubicin-resistant breast cancer cells (MCF-7/DOX and MDA-MB-231/DOX) (10). Moreover, curcumin led to the downregulation of aldehyde dehydrogenase-1, and p-glycoprotein-mediated multidrug resistance gene expression, increasing sensitivity of breast cancer cells (MCF-7) to paclitaxel (11). However, the therapeutic application of curcumin is limited due to its poor water solubility and low bioavailability. Thus, some studies have investigated the use of nanoparticles (NPs) to deliver curcumin to breast cancer cells and enhance its bioavailability and efficacy (12).
Nanotechnology is a strong alternative tool to improve application of hydrophobic molecules. The use of NPs increases the stability and bioavailability of antitumor compounds, reduces therapeutic doses, and minimizes possible adverse effects (13). Several types of NPs can be used as drug delivery systems, such as polymer NPs, liposomes, nanoemulsions, metal NPs, micelles, solid lipid NPs, dendrimers, nanospheres, and nanocapsules (14). These NPs can also be associated with other molecules like aptamers, antibodies, or polymers as active targeting moieties. This surface modification can improve the specificity of NPs to tumor cells, facilitate their interaction, and, consequently, increase antitumor effects (13).
At present, there are several clinical trials in which curcumin has been evaluated, mainly after oral administration regimens, in breast cancer patients (e.g., NCT03980509, NCT01042938, NCT03847623, NCT03865992, NCT01740323, NCT01975363, NCT02556632, NCT01246973, NCT03482401) (15). So far, only one clinical trial using intravenous administration of a curcumin water-soluble formulation (CUC-1®) in combination with paclitaxel in breast cancer patients has been registered (NCT03072992) (15).
A substantial number of studies have been published describing the activity of Cur-NPs in in vivo models of breast cancer. Nevertheless, to the best of our knowledge, a systematic review on this subject has not been published yet. Therefore, considering the importance of in vivo studies and their clinical translation, the aim of this systematic review was to summarize published data regarding evaluations about efficacy and toxicity of Cur-NPs in in vivo models of breast cancer, as well as showing evidence for the potential of this therapeutic approach for clinical trial investigations. Although several works describing interesting data regarding the combination of curcumin with chemotherapeutic drugs have been published (16, 17), the present systematic review was performed to cover studies evaluating curcumin as the active antitumor agent, associated with NPs, to understand better the effects on breast tumor progression of curcumin itself and the advantages/limitations of using NPs as its dug carrier.
Materials and Methods
Protocol and Registration
The present study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (18). The protocol for this systematic review was registered in the International Prospective Register of Systematic Reviews (PROSPERO) (19) with registration number: CRD42020209159.
Eligibility Criteria
Inclusion Criteria
This systematic review based the inclusion criteria on the PICOS (Population, Intervention, Comparison, Outcome, and Study Design) approach (20). We considered studies which evaluated the efficacy and toxicity (O) of Cur-NPs (I) compared with free curcumin and/or negative control (C) on in vivo models of breast cancer in mice or rats (P).
Exclusion Criteria
Studies were excluded for the following reasons: i) reviews, letters, personal opinions, book chapters, and conference abstracts; ii) in vitro studies and clinical trials; iii) use of only free curcumin or curcumin derivatives; iv) other types of cancer; v) use of Cur-NPs associated with other antitumor compounds; vi) full paper copy not available; vii) low quality.
Information Sources and Search Strategy
Individual search strategies were designed for each of the following bibliographic databases: CINAHL, Cochrane, LILACS, Embase, FSTA, MEDLINE, ProQuest, BSV regional portal, PubMed, ScienceDirect, Scopus, and Web of Science ( Table S1 ). The search on databases was performed on August 10 and 11, 2020, with no time restriction. Duplicated references were removed by reference manager software (Mendeley®). There were no restrictions on language and period of publication.
Study Selection
The articles were selected in two phases: screening of titles and abstracts (phase 1) and full text reading (phase 2). In phase 1, two authors (A.S.O. and V.R.P.S) reviewed titles and abstracts of all references identified in the electronic databases and selected articles that seemed to meet the inclusion criteria. In phase 2, six pairs of authors (A.S.O and V.R.P.S.; L.R.A and G.J.F; W.O.P and A.C.P; J.V.O and M.P.G; M.S and G.F.G; P.M.C. and G.A.J.) were formed to independently analyze the full text of articles selected in phase 1 and exclude studies that did not meet the inclusion criteria ( Table S2 ). A third author was consulted if disagreements between the two initial evaluators were not solved by consensus. Extraction of relevant data was done in all included studies to identify animal model, intervention (treatment regimen, dose, route, and NP platform) and outcomes (antitumoral activity and toxicity analysis).
Risks of Bias and Quality in Individual Studies
The quality of the articles included was estimated by applying the 10-question form from SYRCLE’s RoB Toll ( Table S3 ) (21), to analyze the risk of selection, performance, detection, attrition, and other bias. The items were answered in each study by two reviewers individually and the disagreements were resolved by a third reviewer. YES answers indicated low risk of bias, NO indicated high risk of bias, and UNCLEAR indicated it was not possible to assign bias. As a secondary analysis, the quality of the studies was also assessed through 15 questions related to the methodology ( Table S4 ) [based and adapted from the ARRIVE Guideline (22)], and measurement of the outcomes, which were raised by the reviewers and applied in order to classify the studies according to the percentage of YES responses to the criteria raised, being considered as high quality studies with > 70%, moderate quality with 50–69%, and low quality < 49%.
Results
Study Selection
Research on breast cancer has significantly increased. This can be observed, for example, when using the search string “(TITLE-ABS-KEY ((“Breast Cancer” OR “Breast Neoplasm” OR “Mammary Cancer” OR “Malignant Neoplasm of Breast” OR (“Mammary Carcinoma” AND Human) OR “Breast Carcinoma” OR “Cancer of Breast”)))”, on September 27, 2020, in the scientific database Scopus (23), for example, 530,186 document results were found. Of these, some of the regions/countries that published the most were: United States > China > United Kingdom > Germany > Italy > France > Japan > Canada > Australia > India, where the United States published 33.53% and China 8.32% of the studies.
When filtering these results with the search string “(TITLE-ABS-KEY ((“Breast Cancer” OR “Breast Neoplasm” OR “Mammary Cancer” OR “Malignant Neoplasm of Breast” OR (“Mammary Carcinoma” AND human) OR “Breast Carcinoma” OR “Cancer of Breast”) AND (curcumin OR “Turmeric Yellow” OR (yellow AND turmeric) OR diferuloylmethane)) AND TITLE-ABS KEY ((nanoparticles OR nanoparticle OR nanogels OR “Nanocomposite Gels” OR “Nanocomposite Gel” OR nanocapsule OR nanocapsules OR nanoemulsion OR micelle OR micelles OR liposome OR liposomal))),” in order to find studies that have specifically investigated curcumin associated with NPs, 402 documents were obtained ( Figure 1 ). The countries/regions that most published on “breast cancer AND curcumin AND nanotechnology” were India, United States, China, Iran, Italy, among others, according to the Scopus database.
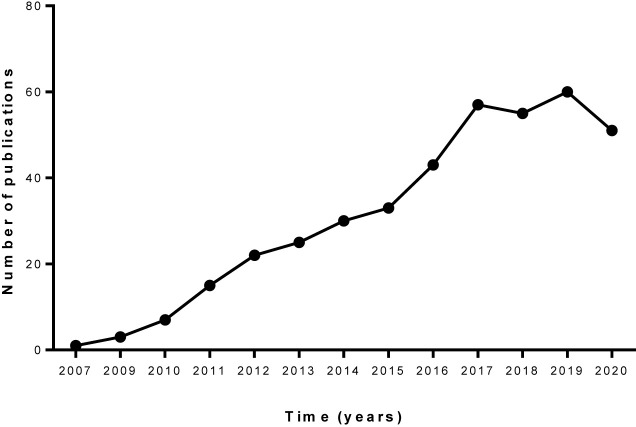
Figure 1.
Number of publications listed in Scopus database with MeSH terms for curcumin nanoparticles and breast cancer. Data obtained on September 27, 2020.
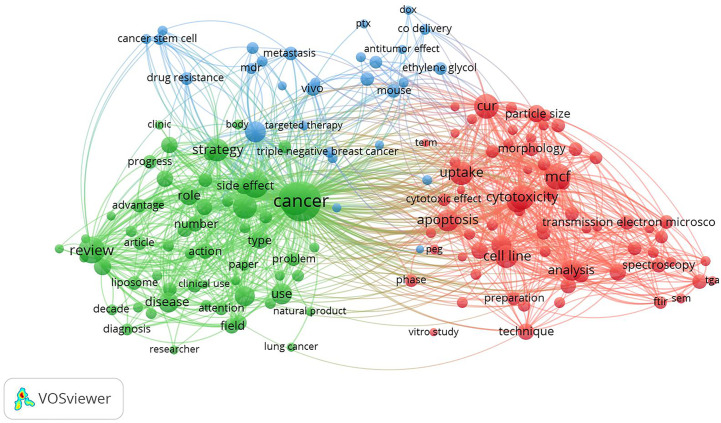
When conducting a bibliometric study of the results obtained in the last search in the Scopus database, using the VOSviewer 1.6.15 program (24), 159 terms were obtained, when establishing at least 10 co-occurrences and “binary count” (presence or absence in each study) configurations. Figure 2 shows the interrelation of three clusters among the most recurrent terms used, where therapy, cytotoxicity, anticancer activity, and apoptosis were among the terms of greatest interest to the scientific community regarding the evaluated topic.
Figure 2.
Bibliometric study of publications listed in Scopus database, with MeSH terms for curcumin nanoparticles and breast cancer, using the VOSviewer program (24), in “title and abstract”, with at least 10 co-occurrences and binary count configurations. Data obtained on September 27, 2020.
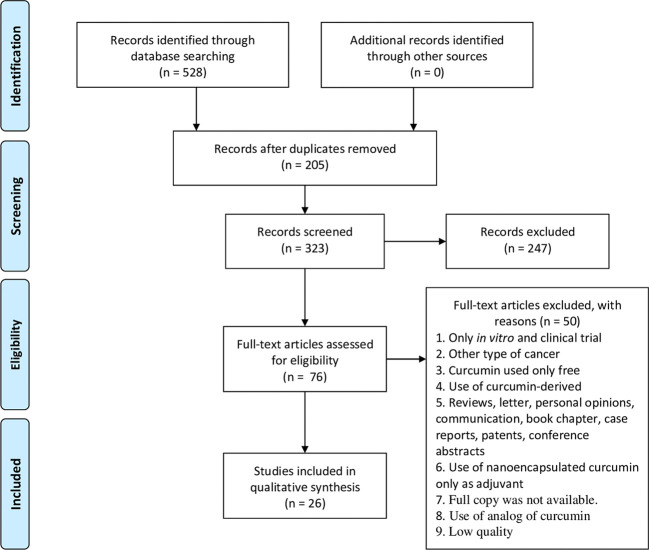
Data presented in Figures 1 and 2 were obtained using only the Scopus database in order to give a general overview of the topics discussed herein. However, in the present systematic review, a total of 528 studies were identified from different databases (57 from ScienceDirect; 35 from LILACS; 113 from Embase; 52 from MEDLINE; 60 from Portal Regional da BSV; 55 from PubMed; 41 from Web of Science; 112 from Scopus; 1 from CINAHL, 1 from FSTA, and 1 from ProQuest) ( Table S1 ). After duplicate removal, 320 studies remained and an evaluation of “title and abstract” resulted in the exclusion of 244 studies. The remaining 76 articles were analyzed by full-text review. This process led to exclusion of 50 articles according to exclusion criteria ( Table S2 ). In the end, 26 articles were maintained and included in this systematic review ( Table 1 ) (25–29, 31–50). A flowchart detailing this process is shown in Figure 3 .
Table 1.
Summary of descriptive characteristics of the included studies.
| Study | Population | Intervention | Outcomes | |||
|---|---|---|---|---|---|---|
| Authors, year (reference)/country | Animal model a | Treatment regimen b | Dose; route | Nanostructure platform | Antitumor activity | Toxicity analysis |
| Shukla et al. (25)/India | Balb/c mice/n = 3/ | Ten days from tumor inoculation; | 100 mg/kg; | Lipid based CPC-SNEDDS NPs (Phospholipid, castor oil, Tween 80, PEG 400); | 1) Cur-NP ↓TV (58.9%); free Cur ↓TV (29.5%); p<0.001 | (ND) |
| 4T1/mouse/ | Daily for 28 days | Oral | HD: 83.27 nm/PDI: 0.151; | |||
| (1 x 106 cells)/ | - Free Cur vehicle: | ZP: −16 mV/EE: 29.1% | ||||
| subcutaneously on back skin | gum acacia (1%, w/v). | |||||
| Chen et al. (26)/China | Balb/c nude mice/n = 5/ | TV of 200 mm3 | 5 mg/kg; | Micelles NPs [POCA4C6 (phosphorylated calixarene) micelles—PM]; | 1) Cur-NP ↓TV (~60%) and ↓ TW (~80%); free Cur: ↓TV (~34%) and ↓ TW (~60%); p<0.05 | No damage in major organs; |
| BT-549/human/ | Fourteen days at every 2 days | Intratumoral | HD: 3.86 nm/PDI: 0.125; | 2) Cur-NP ↑TNC and ↑TAP | No WL; hematological indices: ~ control | |
| (2 x 106 cells)/ | - Free Cur vehicle: (NM) | ZP: −25.18 mV/EE: 95.4% | 3) Cur-NP ↓CD44+ CD133+ cancer stem cells | |||
| Subcutaneously on the upper right thigh | p<0.05 | |||||
| Mahalunkar et al. (27)/India, Germany, and Norway | Balb/c mice/n = 6/ | First day of treatment: (NM) | 10 mg/kg; | Metal gold NPs (CurAu-PVP) with folic acid (FA) (HAuCl4 and PVP polymer). | 1) Cur-NP-FA ↓TV (~51%); free Cur: no ↓TV; p<0.006 | (ND) |
| 4T1/mouse/ | Twice a week for 2 weeks | Intratumoral | HD: 358.7 nm/PDI: 0.6 | 2) Cur-NP-FA ↓TW (~44%); free Cur: no TV↓; p<0.05 | ||
| (1 x 105 cells)/ | - Free Cur vehicle: (NM) | ZP: −12.5 mV/EE: (NM) | ||||
| Mammary fat pad | ||||||
| Alizadeh et al. (28)/Iran | Balb/c mice/n = 8/ | 14 days after tumor induction; | Dose: (NM); | Micelles/polymersomes NPs (PNP) [monomethoxy-PEG (mPEG 2000), oleic acid (OA)] | 1) Cur-NP ↓TV (~80%); p<0.05 | 31.25 mg/Kg of PNP-CUR: no damage in major organs; |
| Transplantation of spontaneous mouse mammary tumor/ | Daily for 24 days | Intraperitoneal | HD: 99.44 nm/PDI: 0.182; | 2) Cur-NP ↑TAP; ↓ANG (CD31); ↓PROL (Ki-67); p<0.05 | Hematological and biochemical indices: ~ control | |
| Pieces < 0.3 cm3/ | ZP: −29.3 mV mV/EE: 64% | p<0.05 | ||||
| Subcutaneous on the left flank | ||||||
| Jung et al. (29)/Republic of Korea | Balb/c nude mice/n = 4/ | TV of 50 mm3; | 10 mg/kg; | Micelle NPs (DSPE-PEG micelle with or without EGFR specific targeting—EGF-Cur-NP) | 1) Cur-NP-EGFR ↓TV (~59.1%); Cur-NP no ↓TV; p<0.05 | No WL |
| MDA-MB-468/human/ | Three times a week; total of eight injections | Intraperitoneal | Cur-NP and EGF-Cur-NP: HD: 248.9 and 229.3 nm/PDI: 0.170 and 0.200, respectively | p<0.05 | ||
| (5 x 106 cells)/ | Cur-NP and EGF-Cur-NP: ZP: −3.6 and −1.73 mV, respectively/EE: (NM) | |||||
| Right shoulder | ||||||
| Wang et al. (30)/China | Nuke mice/n = (NM)/ | Two months after tumor induction; | 1 × 10−3 M; | Polymeric NPs (MPEG-PCL); | 1) Cur-NP ↓TV (~82%); free Cur:↓TV (~49%); p<0.01 | (ND) |
| MDA-MB-231/human/ | Daily for 2 weeks | Intravenous | HD: 139 nm/PDI: (NM); | 2) Cur-NP ↑TAP | ||
| (1.5 x 106 cells)/ | - Free Cur vehicle: (NM) | ZP and EE: (NM) | ||||
| Subcutaneous | ||||||
| Laha et al. (31)/India and USA | Balb/c mice/n =6/ | Ten days after tumor induction; | 2 mg/kg (*unclear); | Metal organic frameworks NPs (IRMOF-3) with or without folic acid (FA) [(Zn (NO3)2; NH2-H2BDC] | 1) Cur-NP-FA ↓TV (~61%); Cur-NP↓TV (~44%); p<0.05 | Biochemical markers for liver and kidney: ~ control |
| 4T1/mouse/ | Every 5 days for 4 times | Route of administration: (NM) | HD: 371,7 nm/PDI: 0.397 | 2) Cur-NP ↓TW (~74%); Cur-NP-FA ↓TW (~85%); p<0.05 | p<0.05 | |
| (NM)/ | ZP: −10.9 mV/EE: 98% | 3) Cur-NP and Cur-NP-FA ↓ tumor cell density | ||||
| Mammary fat pad | ||||||
| Vakilinezhad et al. (32)/Iran | Sprague Dawley rats/n = 6/ | Four months after tumor induction; | 2.5 mg; | Polymeric NPs (PLGA-PVA) | 1) Cur-NP ↓TV (~20%); Free Cur: ↓TV (~16%); p<0.05 | (ND) |
| Chemically induced mammary tumors (MNU) | Once a week for 4 weeks | Intravenous | HD: 92.4 nm/PDI: 0.150 | |||
| - Free Cur vehicle: aqueous suspension | ZP: −5.12 mV/EE: 89.4% | |||||
| Yuan et al. (33)/China | Balb/c nude mice/n = 6/ | TV of 100 mm3; | 2.5 mg/kg; | Polymeric NPs (mPEG- PLGA-Pglu) | 1) Cur-NP ↓TV (~28.0%); p<0.05 | No damage in major organs; |
| MCF-7/human/ | Every other day 4 times; total 18 d | Intravenous | HD: 228.5 nm/PDI: (NM) | 2) Cur-NP ↓TW (~22.5%); p<0.05 | No WL | |
| (3 x 106 cells)/ | ZP: −23.8 mV/EE: 76.9% | 3) Cur-NP ↓breast cancer stem cells (~62%); p<0.05 | p<0.05 | |||
| Right flank | ||||||
| Sahne et al. (34)/Iran | Balb/c mice/n = 4/ | TV of 50–100 mm3; | 4 mg/kg; | Graphene oxide NPs (GO NPs with CMC, PVP, PEG, FA); | 1) Cur-NP-FA↓TV (~86%); p<0.05 | No damage in major organs; |
| 4T1/mouse/ | Daily for a total 3 weeks | Intravenous | HD: ~60 nm/PDI: (NM) | 2) Cur-NP-FA ↓TW (~76%); p<0.05 | No WL | |
| (NM)/ | ZP: −48 mV/EE: 94% | 3) Cur-NP-FA ↑ ST and ↓ metastasis; p<0.05 | p<0.05 | |||
| Subcutaneous on the flank | 4) Cur-NP-FA: ↑TNC; ↓ cell density; ↓ANG (CD31, CD34); ↑ pro-inflammatory response in the tumor microenvironment; p<0.05 | |||||
| Ji et al. (35)/China | Balb/c mice/n = 5/ | First day of treatment: (NM); | 5 mg/kg; | Nanocrystals NPs with or without HA | 1) Cur-NP-HA ↓TV (~86%); Cur-NP↓TV (~39%); free Cur: ↓TV (~21%); p<0.05 | No damage in major organs; no WL; |
| 4T1/mouse/ | Every 2 days for a total 10 days | Intravenous | Cur-NP and HA-Cur-NP: HD: 101.4 and 161.9 nm/PDI: ~0.330 and 0.250, respectively | 2) Cur-NP-HA ↓TW (~75%); Cur-NP↓TW (~37.5%); Free Cur: ↓TW (~25%); p<0.05 |
No hemolysis (<5%); | |
| (1 x 106 cells)/ | - Free Cur vehicle: (NM) | HA-Cur-NP: ZP: −25.0 mV, respectively/EE: (NM) | 3) ↑ ST: Cur-NP-HA > Cur-NP > Free Cur; p<0.05 | Hematological and biochemical indices: ~ healthy control | ||
| Subcutaneous on the right flank | p<0.05 | |||||
| He et al. (36)/China | Balb/c mice/n = 6/ | TV of 100 mm3 | 5mg/kg; | Polymeric micellar NPs [amphiphilic diblock copolymer— mPEG-b-PLG (Se)-TP]; | 1) Cur-NP ↓TV (~65%); Free Cur: ↓TV (~49%); p<0.05 | No damage in major organs; no WL |
| 4T1/mouse/ | Every 4 days, for 4 times; total 21 days | Intravenous | HD: 136 nm/PDI: 0.071 | 2) Cur-NP ↓TW (~82%); free Cur: ↓TW (~62%); p<0.05 | p<0.05 | |
| (1 x 106 cells)/ | - Free Cur vehicle: (NM) | ZP: (NM)/EE: ~ 68% | 3) Cur-NP ↑TNC and ↑TAP; ↓ANG (CD31); ↓PROL (Ki-67); p<0.05 | |||
| Subcutaneous on the right back | ||||||
| Jin et al. (37)/China and USA | Balb/c nude mice/n = 5/ | 7 days after tumor induction; | 5 mg/kg; | Polymeric NPs with or without EGFR-targeting peptides (GE11) (PLGA-PEG); | 1) Cur-NP-GE11 and Cur-NP ↓TV (~80%); free Cur: no TV↓; p<0.05 | Inflammatory cytokine levels: ~ healthy mice |
| MCF-7/human/ | Every 24 h for 20 times | Intravenous | HD: 210 nm/PDI: 0.112; | 2) Cur-NP-GE11↓TW (~56%); Cur-NP ↓TW (~48%); free Cur: no TW↓; p<0.05 | p<0.05 | |
| (1 x 107 cells)/ | ZP: −22 mV/EE: 92.3 | 3) Cur-NP-GE11and Cur-NP ↑TAP | ||||
| Subcutaneous on the dorsal flank | - Free Cur vehicle: (NM) | |||||
| Abd-Ellatef et al. (38)/Italy and Egypt | Balb/c mice/n = 8/ | TV of 50 mm3; | 5 mg/kg; | Solid lipid nanoparticles (SLN) with or without chitosan (CS) coating (cholesterol; trilaurin, butyl lactate, Epikuron®200, Cremophor®RH60, sodium taurocholate, Pluronic® F68); | 1) Cur-NP-CS and Cur-NP ↓TV (~35%); Free Cur: no TV↓; p<0.01 | Biochemical indices: ~ control |
| JC/mouse/ | Thrice (on day 1st, 7th, 14th) | Intravenous | HD: < 200 nm/PDI: (NM) | p<0.05 | ||
| (1 x 107 cells)/ | - Free Cur vehicle: 10% v/v DMSO suspension |
ZP: (NM)/EE: 70–75% | ||||
| Mammary fat pad | ||||||
| Li et al. (39)/China | Balb/c mice/n = 4/ | Tumor diameter of 4 mm; | 8 mg/kg; | Mesoporous silica nanoparticles with hyaluronan (MSN-HA) or polyethyleneimine-folic acid (MSN-PEI-FA). | 1) Cur-NP-PEI-HA ↓TV (~50%); Free Cur: no TV↓; p<0.01 | No damage in major organs; no WL; |
| MDA-MB-231/human/ | Every 3 days for a total of six times | Intravenous | HD: < 300 nm/PDI: (NM)/ | 2) Cur-NP-PEI-HA ↓TW (~70%); free Cur: no TW↓; p<0.01 | Hemolysis (<5%); | |
| (1 x 107 cells)/ | - Free Cur vehicle: (NM) | ZP: ~ −20 mV (MSN-HA); ~ +40 mV (MSN-PEI-FA) |
Biochemical indices: ~ healthy control | |||
| Subcutaneous | p<0.05 | |||||
| Kundu et al. (40)/India | Swiss albine mice/n = 6/ | Ten days after induction; | 10 mg/kg; | Metal NPs [Zinc oxide nanoparticles (ZnO) with PBA]; | 1) Cur-NP↓TV (~77%); free Cur: ↓TV (~66%); p<0.05 | No damage in the liver and kidney; |
| Ehrlish ascites carcinoma cells/ | Alternate days for 14 days. | Intravenous | HD: 413.63 nm/PDI: (NM) | 2) Cur-NP ↓TW (~72%); free Cur: ↓TW (~50%); p<0.05 | Biochemical markers: ~control; ↓ tumor-induced splenomegaly | |
| (1.0 x 107/ml) | - Free Cur vehicle: (NM) | ZP: −16.4 mV/EE: 27% | 3) Cur-NP and free Cur: ↑TAP; p<0.05 | p<0.05 | ||
| Left flank | ||||||
| Lv et al. (41)/China | Kunming (mice)/n = 6/ | TV of 300 mm3; | 10 mg/kg; | Polymeric NPs (PEG-PCDA) with or without biotin; | 1) Cur-NP ↓TV (~69%); Cur-NP-biotin ↓TV (~79%); free Cur: ↓TV (~32%); p<0.05 | no WL |
| EMT6/mouse/ | Daily for 9 days; total 14 days | Intravenous | PEG-PCDA and biotin-PEG-PCDA: HD: 94.2 and 125.1 nm/PDI: 0.170 and 0.08, respectively | 2) Cur-NP ↓TW (~70%); Cur-NP-biotin ↓TW (~85%); free Cur: ↓TW (~25%); p<0.05 | p<0.05 | |
| (1.0 x 107/ml) | - Free Cur vehicle: cremophor EL: dehydrated alcohol (1:1, v/v) and diluted with physiological saline | PEG-PCDA and biotin-PEG-PCDA: ZP: −9.56 and −12.86 mV/EE: (NM) | 3) Cur-NP and Cur-NP-biotin: ↑TNC and ↑TAP; ↓ANG (CD31; VEGF; COX-2); ↓PROL (Ki-67); p<0.05 | |||
| Subcutaneously | ||||||
| Yang et al. (42)/China | Balb/c nude mice/n = 5 | TV of 200 mm3 | 10 mg/kg; | Micelle NPs (triblock copolymer PPBV); | 1) Cur-NP ↓TV (~58.5%, day 12); ↓TV (~28.9%, day 20); p<0.05 | No WL |
| MCF-7/human/ | Every other day for 5 times; total 20 days | Intravenous | HD: 6.7 nm/PDI: 0.117 | 2) Cur-NP ↓TW (~22%, day 20); p<0.05 | p<0.05 | |
| (1 x 107 cells)/ | ZP: −1.42 mV/EE: 68.5% | |||||
| Subcutaneous on the flank | - Free Cur vehicle: (NM) | |||||
| Yang et al. (43)/China | Balb/c nude mice/n = 5/ | TV of 200 mm3 | 15 mg/kg; | Hybrid NPs [PLGA NPs coated with a modified hialuronic acid (HA-hybrid)] | 1) Cur-NP-HA ↓TV (~43.8%, day 12); ↓TV (~24%, day 20); p<0.05 | No WL |
| MCF-7/human/ | Every other day for 5 times; total 20 days | Intravenous | HD: 350 nm/PDI: (NM) | 2) Cur-NP-HA ↓TW (~22%, day 20); p<0.05 | p<0.05 | |
| (1 x 107 cells)/ | ZP: −22 mV/EE: 32% | 3) Cur-NP-HA ↓tumor cell density; p<0.05 | ||||
| Subcutaneous on the flank | - Free Cur vehicle: (NM) | |||||
| Greish et al. (44)/Bahrain | Balb/c mice/n = 5/ | TV of 100 mm3; | 10 and 20 mg/kg; | Micelles (curcumin–metal complex and SMA) | 1) Cur-NP-10mg/Kg ↓TV (~61%); Cur-NP-20mg/Kg ↓TV (~92%); p<0.05 |
(ND) |
| 4T1/mouse/ | frequency of treatment: unclear; total 10 days | Intravenous | HD: 248 nm/PDI: 0.274; | |||
| (1 x 106 cells)/ | ZP: −11 mV/EE: 80% | |||||
| Bilaterally on the flanks | ||||||
| Mukerjee et al. (45)/USA | Balb/c nude mice/n = 8/ | TV of 70 mm3; | 20 mg/kg; | Polymeric NPs [PLGA/PVA with or without antibody targeting (AnxA2)] | 1) Cur-NP- AnxA2 ↓TV (~44.0%); Cur-NP ↓TV (~33.5%); p<0.05 | No WL |
| MCF10CA1a/human/ | Thrice week for a total 30 days | Intravenous | Cur-NP and AnxA2-Cur-NP: HD: 150 and 157 nm/PDI: ~0.240 and 0.200, respectively | 2) Cur-NP- AnxA2 ↓TW (~53.0%); Cur-NP ↓TW (~30%); p<0.05 | p<0.05 | |
| (3 x 106 cells)/ | Cur-NP and AnxA2-Cur-NP: ZP: −27.5 and −28.5 mV, respectively/EE: 89.2% | 3) Cur-NP- AnxA2 and Cur-NP: ↓ANG; ↓ NFkβ; ↓PROL (Ki-67); p<0.05 | ||||
| Flank | ||||||
| Mukhopadhyay et al. (46)/India | Balb/c nude mice/n = 5/ | 8 days after induction; | 20 mg/kg | Polymeric NPs [PLGA/PVA with or without folate (F)] | 1) Cur-NP-F ↓TV (~90%); Cur-NP↓TV (~75%); p<0.05 | No damage in major organs |
| MDA-MB-231/human/ | Thrice week for a total 21 days | Route of administration: unclear | HD: 170 nm/PDI: 0.186; | 2) Cur-NP-F ↓TW (~92%); Cur-NP↓TW (~61.5%); p<0.05 | p<0.05 | |
| (5 x 106 cells)/ | ZP: −28.2 mV/EE: 68.6% | 3) Cur-NP-F and Cur-NP ↓ cell density | ||||
| Right flank | ||||||
| Yu et al. (47)/China | Balb/c nude mice/n = 5/ | TV of 100–400 mm3; | 40 mg/kg | Micelles NPs (MPEG-PLA with or without PAE) | 1) Cur-NP-PAE ↓TV (~65.6%); Cur-NP ↓TV (~47.1%); p<0.05 | no WL |
| MCF-7/human/ | Every other day for 5 times for a total 24 days | Intravenous | HD: 128.4 nm to 171.0 nm/ PDI: 0.118 to 0.134 |
2) Cur-NP-PAE ↓TW (~76%); Cur-NP ↓TW (~53%); p<0.05 |
p<0.05 | |
| (3 x 106 cells)/ | ZP: −2.0 to +4.0 mV/ EE: 96.5 to 98.8% |
|||||
| Subcutaneously right flank | ||||||
| Huang et al. (48)/China | Balb/c mice/n = 5/ | TV of 40–50 mm3/ | 50 mg/kg | Polymeric NPs (HA-CHEMS); pH-sensitive | 1) Cur-NP ↓TV (~38%); p<0.05 | ↓ Damage in major organs |
| 4T1/mouse/ | Every 2 days for a total of 5 times | Intravenous | HD: 144 nm/PDI: (NM); | 2) Cur-NP ↑ ST; | no WL | |
| (NM)/ | ZP: −21.25 mV/EE: (NM) | 3) Cur-NP ↑TNC and ↑TAP | p<0.05 | |||
| Flank | ||||||
| Shiri et al. (49)/Iran | Balb/c mice/n = 9/ | Third day after tumor induction/ | 40 mg/kg or 80 mg/kg | Dendrosome NPs (DNC) [composition: not mentioned (patent number: 71753)] | 1) NP-40mg/Kg ↓TV (~72%); NP-80mg/Kg ↓TV (~76%); p<0.05 | no WL |
| 4T1/mouse/ | daily for 35 consecutive days | Route of administration: (NM) | HD; PD; ZP; EEI: (NM) | 2) NP-40mg/Kg ↓TV (~61%); NP-80mg/Kg ↓TV (~64%); p<0.05 | No change in food intake and behavior | |
| (1 x 106 cells)/ | 3) NP ↑ ratio of M1/M2 macrophages | p<0.05 | ||||
| left flank | ||||||
| Lin et al. (50)/China | Balb/c nude mice/n = 6/ | First day of treatment: (NM)/ | Dose: (NM)/ | Lipid based NPs (NLC) with or without folate (FA) coating (PEG-DSPE, soya lecithin, castor oil, Tween 80, Precirol ATO-5); | 1) Cur-NP-FA ↓TV (~83%); Cur-NP ↓TV (~66%); free Cur: ↓TV (~31%); | No WL |
| MCF-7/human/ | once every 3 days for 15 days | Intravenous | HD: 126.8 nm/PDI: 0.16 | p<0.05 | ||
| (NM)/ | ZP: +12.6 mV/EE: 82.7% | |||||
| Subcutaneous in the right armpit | - Free Cur vehicle: (NM) | |||||
Animal type/replicates/cell type injected/source/cell concentration/local of cell insertion;
First day of treatment (or tumor volume)/treatment frequence and total experiment time; ANG, angiogenesis; CHEMS, cholesteryl hemisuccinate; CMC, carboxymethyl cellulose; CPC-SNEDDS, curcumin-phospholipid complex self-nanoemulsifying drug delivery systems; Cur, curcumin; d, days; DSPE, distearoyl phosphatidyl- ethanolamine; EGFR, epidermal growth factor receptor; FA, folic acid; HA, hyaluronic acid; MNU, N-methyl nitroso urea; mPEG-b-PLG (Se)-TP, poly-(ethylene glycol)-b-poly(L-glutamic acid)-two-photon AIE fluorophores [mPEG-b-PLG (Se)-TP] amphiphilic copolymer with selenide group (Se) conjugated and a two-photon AIE fluorogen (TP) on the terminal group of PLG segments.; MPEG-PCL, methoxypoly(ethylene glycol)-polycaprolactone; NF-κB, nuclear factor kappa b; NH2-H2BDC, 2-amino terephthalic acid; (ND), not determined; (NM), not mentioned; NPs, nanoparticles; PAE, poly (b-aminoester; PBA, phenyl boronic acid; PCDA, pentacosadiynoic acid; PEG, poly(ethylene glycol); PGlu, poly L-glutamic acid; PLA, polylactic-co-glycolic acid; PPBV, mPEG-PBLA-PVIm triblock copolymer; PROL, tumor proliferation; PVA, polyvinyl alcohol; PVP, polyvinylpyrrolidone; SMA, poly(styrene)-co-maleic acid; ST, survival time; TAP, tumor apoptosis; TNC, tumor necrosis; TV, tumor volume; TW, tumor weight; WL, weight loss.
Figure 3.
Flow diagram of literature search and selection criteria adapted from PRISMA (18).
Characteristics of the Included Studies
All included studies are research articles that evaluated antitumoral activity of Cur-NPs in in vivo models of breast cancer. The main characteristics of the studies are summarized in Table 1 .
The studies were conducted in several countries: China (n = 13); India (n = 6); Bahrain (n = 1); Iran (n = 4); Italy (n = 1); US (n = 1); Republic of Korea (n = 1), and all of them were published from 2014 to 2020 in the English language.
Cur-NPs used in the included studies were mainly described by hydrodynamic diameter (HD) (n=26), polydispersity index (PdI) (n=17), and zeta potential (n=21) through dynamic light scattering and electrophoretic mobility. Some studies assessed NPs’ size and/or morphology by transmission electron microscopy (TEM) and/or scanning electron microscopy (SEM). Curcumin encapsulation efficiency (EE%) was evaluated mostly by high-performance liquid chromatography (HPLC) (n=18).
All in vivo studies followed the progression of tumor volume during the experimental period by measuring tumor small/large diameters or width/length and calculating the final volumes with mathematical formulas. Studies also assessed tumor weight (n=16); survival time (n=3); tumoral stem cells through flow cytometry (n=2); ratio of M1/M2 macrophages through RT-PCR (n=1); apoptosis (n=8), necrosis (n=5), proliferation (n=4), angiogenesis (n=5), cell density (n=4), inflammatory response in the tumor (n=1), and metastasis (n=1) through classical histology (HE) and immunohistochemistry. Regarding toxicity analysis, 21 studies used at least one parameter of evaluation. Studies assessed weight loss (n=15); damage of major organs through classical histology (HE) (n=9); food intake/behavior (n=1); inflammatory cytokine levels (n=1) through ELISA; hemolysis (n=2) through absorbance; and hematological (n=3) and biochemical parameters (n=6) through animal blood counter and commercial kits.
Quality and Risk of Bias in Individual Studies
When analyzed by questions based on ARRIVE guidelines, as seen in Table 2 , 23 studies were graded as of high quality and 3 as of moderate quality. Most studies clearly reported the encapsulation methods of curcumin (n=24) and investigated characteristics of NPs (n=23). Animal models were considered adequate in all studies and ethical committee approval was clearly reported in 18 studies. Six studies did not clearly report ethical approval and two other articles did not mention this criterion. Furthermore, animal conditions during the experiment were not clearly described in 13 studies. The study design of anticancer activity was well executed in most studies and some articles clearly mentioned time of treatment (n=25), route of administration (n=22), dose of curcumin (n=25), and presence of control groups (n=25). Nevertheless, only 15 studies investigated anticancer activity of free curcumin to compare with the Cur-NP effect. Additionally, a toxicity assay was performed in 21 studies. Lastly, statistical models were considered as unclear in all studies due to lack of information.
Table 2.
Overall quality of the selected studies. Detailed description of the evaluated parameters is found in Table S4 .
| Critical analysis of the articles included | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Methodology | Toxicity | Outcomes | Score | Classification | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | % | |||
| Abd-Ellatef et al. (38) | 2020 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
93 | High quality |
| Alizadeh et al. (28) | 2015 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
80 | High quality |
| Chen et al. (26) | 2017 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
93 | High quality |
| Greish et al. (44) | 2018 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
73 | High quality |
| He et al. (36) | 2019 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
| Huang et al. (48) | 2020 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
66 | Moderate quality |
| Ji et al. (35) | 2020 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
| Jin et al. (37) | 2017 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
| Jung et al. (29) | 2018 |

|

|

|

|

|

|

|

|

|

|

|
- | - |

|

|
86 | High quality |
| Kundu et al. (40) | 2019 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
80 | High quality |
| Laha et al. (31) | 2019 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
73 | High quality |
| Li et al. (39) | 2018 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
| Lin et al. (50) | 2016 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
80 | High quality |
| Lv et al. (41) | 2015 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
60 | Moderate quality |
| Mahalunkar et al. (27) | 2019 |

|

|

|

|

|

|

|

|

|

|

|
- | - |

|

|
86 | High quality |
| Murkerjee et al. (45) | 2016 |

|

|

|

|

|

|

|

|

|

|

|
- | - |

|

|
73 | High quality |
| Mukhopadhyay et al. (46) | 2020 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
80 | High quality |
| Sahne et al. (34) | 2019 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
93 | High quality |
| Shiri et al. (49) | 2015 |

|

|

|

|

|

|

|

|

|

|

|
- | - |

|

|
53 | Moderate quality |
| Shukla et al. (25) | 2017 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
| Vaklinezhad et al. (32) | 2019 |

|

|

|

|

|

|

|

|

|

|

|
- | - |

|

|
73 | High quality |
| Wang et al. (30) | 2018 |

|

|

|

|

|

|

|

|

|

|

|
- | - |

|

|
86 | High quality |
| Yang et al. (42) | 2017a |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
| Yang et al. (43) | 2017b |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
80 | High quality |
| Yu et al. (47) | 2014 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
80 | High quality |
| Yuan et al. (33) | 2018 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
86 | High quality |
 Yes
Yes  No
No  Unclear - Not evaluated.
Unclear - Not evaluated.
Risk of bias assessment based on SYRCLEs’ RoB guidelines of all included studies is summarized in Table 3 . Criteria were considered unclear when they were not clearly reported or gave incomplete information. Most studies did not clearly describe information on allocation, randomization, and blinding, which is required for quality assessment. Animals were not randomly housed during the experiment in two studies. Additionally, three studies did not adequately address incomplete outcome data. In parallel, the experimental groups were considered similar in 18 studies, and 24 studies showed low risk of selection bias of reported outcomes.
Table 3.
Risk of bias in individual studies (SYRCLE’s Rob toll criteria). YES answers indicated low risk of bias, NO indicated high risk of bias, and UNCLEAR indicated it was not possible to assign bias. Detailed description of the evaluated parameters is found in Table S3 .
| SYRCLE’s Risk of Bias | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Selection | Performance | Detection | Attrition | Reporting | Other | ||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Abd-Ellatef et al. (38) | 2020 |

|

|

|

|

|

|

|

|

|

|
| Alizadeh et al. (28) | 2015 |

|

|

|

|

|

|

|

|

|

|
| Chen et al. (26) | 2017 |

|

|

|

|

|

|

|

|

|

|
| Greish et al. (44) | 2018 |

|

|

|

|

|

|

|

|

|

|
| He et al. (36) | 2019 |

|

|

|

|

|

|

|

|

|

|
| Huang et al. (48) | 2020 |

|

|

|

|

|

|

|

|

|

|
| Ji et al. (35) | 2020 |

|

|

|

|

|

|

|

|

|

|
| Jin et al. (37) | 2017 |

|

|

|

|

|

|

|

|

|

|
| Jung et al. (29) | 2018 |

|

|

|

|

|

|

|

|

|

|
| Kundu et al. (40) | 2019 |

|

|

|

|

|

|

|

|

|

|
| Laha et al. (31) | 2019 |

|

|

|

|

|

|

|

|

|

|
| Li et al. (39) | 2018 |

|

|

|

|

|

|

|

|

|

|
| Lin et al. (50) | 2016 |

|

|

|

|

|

|

|

|

|

|
| Lv et al. (41) | 2015 |

|

|

|

|

|

|

|

|

|

|
| Mahalunkar et al. (27) | 2019 |

|

|

|

|

|

|

|

|

|

|
| Murkerjee et al. (45) | 2016 |

|

|

|

|

|

|

|

|

|

|
| Mukhopadhyay et al. (46) | 2020 |

|

|

|

|

|

|

|

|

|

|
| Sahne et al. (34) | 2019 |

|

|

|

|

|

|

|

|

|

|
| Shiri et al. (49) | 2015 |

|

|

|

|

|

|

|

|

|

|
| Shukla et al. (25) | 2017 |

|

|

|

|

|

|

|

|

|

|
| Vaklinezhad et al. (32) | 2019 |

|

|

|

|

|

|

|

|

|

|
| Wang et al. (30) | 2018 |

|

|

|

|

|

|

|

|

|

|
| Yang et al. (42) | 2017a |

|

|

|

|

|

|

|

|

|

|
| Yang et al. (43) | 2017b |

|

|

|

|

|

|

|

|

|

|
| Yu et al. (47) | 2014 |

|

|

|

|

|

|

|

|

|

|
| Yuan et al. (33) | 2018 |

|

|

|

|

|

|

|

|

|

|
 Yes
Yes  No
No  Unclear - Not evaluated.
Unclear - Not evaluated.
Synthesis of Results
A wide variety of NP types were used in the included studies. Polymer NPs were mostly used for curcumin delivery (n = 9), followed by micelles (n = 6), lipid-based NPs (n=3), metal NPs (n = 2), hybrid NPs (n=2), dendrosomal NP (n =1), nanocrystal (n=1), graphene oxide NP (n=1), and mesoporous silica NP (n=1). Poly(ethylene glycol) chains (PEG) were present in NP composition in 11 studies. Nine studies evaluated NPs associated with targeting moieties such as folic acid (n=5), hyaluronic acid (n=1), EGF peptides (n=2), and AnxA2 (n=1). With HD ranging from 101.4 to 371.7 nm, most Cur-NPs presented negative Zeta potential (−48 to +40 mV) (n=22) and EE% from 32 to 98% ( Table 1 ).
Concerning experimental design, studies were heterogeneous regarding animal model, route of administration, duration of the experiment, and dose of treatment. Ten studies adopted nude mice as animal models when tumor was induced with human cells and another 16 used mice with a native immune system for tumor induction with murine cells. One study used rats with MNU-chemically induced mammary tumors (n=1). The main human cell lines used were MCF-7 (n=6), MDA-MB-231 (n=3), MDA-MD-468 (n=1), MCF10CA1a (n=1), and BT-549 (n=1). The murine cell lines used were 4T1 (n=9), EMT6 (n=1), JC (n=1), and Ehrlich ascites carcinoma cells (n=1). One study used the transplantation of spontaneous mouse mammary tumor pieces (n=1) as the breast cancer model. Implantation of tumor cells into the mammary fat comprised three studies, while the others adopted subcutaneous implantation into the flank (n=14) or armpit (n=1).
The first day of treatment was described according to days after induction (n=9), ranging from 3 days to 4 months, or tumor volume (n=13) in the range of 40 to 400 mm3. The curcumin doses used in the treatments varied between 2 and 100 mg/Kg and were administered daily (n=8), every other day (every 2 days) (n=7), three times a week (n=6); twice a week or less (n=4). Intravenous administration was the main route of administration used in the included studies (n=18). Few studies used intraperitoneal (n=2), intratumoral (n=2), or oral administration route (n=1); and one did not clearly mention this information (n=1).
Discussion
Summary of Evidence
The structure of curcumin has chemical groups that allow interactions of diverse chemical natures (e.g., covalent, non-covalent, hydrophobic, and hydrogen bonds) with molecules involved in the different pathways of breast carcinogenesis (9, 16). It has been reported that curcumin inhibits cell proliferation, tumor invasion, and angiogenesis. As an anti-proliferative agent, curcumin induces cell cycle arrest and p53-dependent apoptosis. It also alters signaling protein expression, such as Ras, protein kinase B (Akt), and phosphatidylinositol-3-kinase (PI3K) (51). Additionally, the use of curcumin has been described as a potential strategy for inhibiting EZH2 (enhancer of zeste homolog-2), a histone modifier protein subunit involved in tumor growth, metastatic potential, and in the regulation of drug resistance. In breast cancer, it has been reported that curcumin is able to inhibit the proliferation of human breast cancer MDA-MB-435 cells in correlation with the downregulation of EZH2 expression (52, 53).
Curcumin has demonstrated anti-invasive effects through downregulation of matrix metalloproteinase (MMP-2) and upregulation of tissue inhibitor of metalloproteinase (TIMP-1) in MDA-MB-231 breast cancer cells (54). Interestingly, emerging evidence indicates that the chemopreventive and chemotherapeutic properties of curcumin are closely linked to the modulation of miRNAs involved in tumorigenesis and metastasis signaling pathways, e.g., hedgehog, notch-1, PI3K/Akt/mTOR, Wnt/β-catenin, IGF, VEGF, and TGF-β/smad3 pathways (55, 56). Gallardo et al. (57) demonstrated that curcumin prevents the migration and invasion of breast cancer cells (MCF-10F and MDA-MB-231) by targeting miR-34a as a regulator of Rho-A and other genes involved in epithelial-mesenchymal transition, such as Axl, Slug, and CD24 (57). Curcumin can also prevent angiogenesis by inhibiting vascular endothelial growth factor (VEGF) (58, 59) and suppressing angiogenic cytokine interleukin-6 (60). All of the mentioned mechanisms cited above along with anti-inflammatory action and inhibition of cell growth factors, support confirmation of the wide activity of curcumin in the regulation of tumor growth by acting in different cancer hallmarks (9).
However, the hydrophobic property of curcumin limits its applications and demonstrates less impressive success in clinical trials (14). Additionally, free curcumin can undergo biomodifications and may be mostly excreted in feces or in bile in animal models (61). Therefore, the use of drug delivery systems, such as NP platforms, is an alternative to improve drug bioavailability, administer lower doses, increase time of circulation, and enhance its biological activity. The natural product-based nanomedicine field for cancer treatment has increased and demonstrated great potential (6, 14). The present systematic review reports the effects of Cur-NPs on antitumoral activity and toxicity of in in vivo models of breast cancer.
Analysis of the included studies showed that curcumin evaluated in in vivo models of breast cancer has been loaded in a wide variety of NP platforms, including different compositions, sizes, and zeta potential. In fact, advantages were evidenced in all the included studies that compared tumor volume reduction after Cur-NP treatment with curcumin treatments in its free form and/or with negative controls ( Table 1 ). For instance, a volume reduction of ~21% was observed in tumors of animals treated with free curcumin, while a significant reduction of ~86% was observed in animals treated with curcumin nanocrystals coated with hyaluronic acid (35).
NPs are able to accumulate into solid tumors (e.g., breast cancer). The classical concept states that NPs extravasate the tumor’s vascular barrier through gaps between endothelial cells (owing to irregular angiogenic growth) and are retained in the tumor mass due to poor local lymphatic drainage—a passive process known as the enhanced permeability and retention (EPR) effect (62, 63) Nevertheless, this pathway has been currently under debate, and updated data show evidence that it may not be the dominant mechanism of NPs’ extravasation into solid tumors (64, 65). Other mechanisms of NPs’ tumor accumulation have been investigated, such as the trans-endothelial pathway, which is a metabolically active process that requires endothelial cells to rearrange their structure to present vesicles that can uptake NPs and further deliver them to tumor cells nearby (64).
Improvements in tumor NP accumulation and favorable interaction between NPs and cancer cells can be obtained by tailoring the surface of the NP with moieties able to confer prolonged blood-circulation time (e.g., PEG) and specific active targeting (e.g., ligands with affinity to molecules overexpressed in tumor cells) (66). The main active targeting moieties found in the included studies were folic acid (FA) and hyaluronic acid (HA). FA shows affinity to folate receptors, which are tumor-associated proteins overexpressed in more than 40% of human tumors, including breast cancer (67). A metal organic framework of FA-Cur-NPs significantly improved curcumin antitumor efficacy (~61%), while non-modified NP accounted for ~44% for tumor volume reduction (31). Similarly, attachment of HA, a natural polysaccharide consisting of repeating disaccharide units, to the surface of NPs, has been investigated since it binds to the cell surface molecule CD44, a surface protein widely expressed in breast cancer (68). Curcumin associated with HA-mesoporous silica NPs showed a significant ~70% reduction in tumor weight, while no significant effect was observed for free curcumin (39). Other modifications of the curcumin-NP surface with ligands specific to different tumor surface biomarkers for breast cancer have also been explored (29, 37, 45).
Once in the tumor site, NPs can be internalized by tumor cells and/or release their cargo in the tumor microenvironment. It is known that the tumor microenvironment is acidic (69), and this pathological characteristic of cancer can be used as a strategy for the controlled release of NPs responsive to acidic pH (40). This strategy prevents cargo release to non-target tissues and aids in the mitigation of possible adverse effects. Kundu and co-workers (40) designed their study based on this approach by using pH-sensitive NPs. They observed that the release of curcumin from the nanohybrid zinc oxide NPs was improved in decreased pH and resulted in an increased accumulation of curcumin in tumor tissue and a significative tumor volume reduction (~77%). In addition, no biochemical modifications or structural damage were observed in the liver and kidneys (40). Huang and co-workers (48) encapsulated curcumin in pH-sensitive polymeric NPs and showed a significant reduction in tumor volume followed by increased survival time ( Table 1 ) (48). Internalization of NPs can be mediated or not by active targeting ligands (depending on the mechanism triggered), and it occurs mainly through endocytosis pathways where the main mechanisms comprise a) clathrin-mediated endocytosis; b) caveolae-mediated endocytosis, for NPs up to 200 nm; c) macropinocytosis; and d) other clathrin and caveola-independent endocytosis for NPs with sizes between 250 nm and 3 µm (70, 71). Once inside the cells, NPs can interact with specific organelles and/or release their cargo to reach potential targets, such as the ones involved in cell death/survival and cell proliferation pathways (66).
Curcumin can modulate multiple apoptosis signaling pathways. The predominant apoptotic mechanism—extrinsic (receptor-mediated) or intrinsic (mitochondrial) – differs between cell types, differentiation stages, or curcumin concentrations. Increase of Bax/Bcl-2 ratio, activation of caspase-3, inhibition of telomerase, DNA fragmentation, and induction of redox signaling are some of the apoptotic mechanisms activated by curcumin in distinct breast cancer cells (72–74). Cell cycle arrest by free curcumin has also been described and is potentially associated with its antiproliferative effects (74). Regarding the antitumor mechanisms reported in the included studies, Cur-NPs were shown to induce at least tumor apoptosis, necrosis, and/or cell proliferation blockage in in vivo breast cancer models (26, 28, 30, 34, 36, 37, 40, 41, 45, 48).
Angiogenesis involves the development of new blood vessels from pre-existing vessels and plays an important role in tumor growth, maintenance, and metastasis (26, 30). Free curcumin has been described presenting anti-angiogenesis effects by inhibiting or modulating many pro-angiogenesis factors such as vascular endothelial growth factor (VEGF), matrix metalloproteinases (MMPs), and basic fibroblast growth factor (bFGF) in in vitro and in vivo studies (26, 30, 58). Similar effects were reported in the included studies when curcumin was associated with micelles, graphene oxide, or polymeric NPs and administered in in vivo breast cancer models (28, 34, 36, 41, 45).
It is known that a population of cancer stem cells (CSCs) is present within the tumor microenvironment. These cells are able to activate self-sustaining and self-renewal mechanisms, giving rise to heterogeneous cancer cells that comprise the tumor (75). CSCs are also known to present a high expression of P-glycoprotein, a well-known protein involved in multidrug resistance (MDR), making them less susceptible to antitumor therapies (76). Interestingly, free curcumin has been described affecting CSCs with no toxicity to normal stem cells. The mechanisms involve modulation of P-glycoprotein (77); suppression of the release of cytokines such as interleukin (IL)-6, IL-8, and IL-1, which stimulate CSCs; among others (75). Cur-NPs of the studies evaluated herein seems to maintain this property since studies with curcumin associated with micelles and polymeric NPs have shown a significant reduction in the proportion of CSCs present in in vivo breast cancer models (26, 33) ( Table 1 ).
Metastasis is the process where cells from the primary tumor spread to distant sites and give rise to a secondary tumor. Advanced breast cancer includes both stage (IV) of metastatic breast cancer and inoperable locally advanced breast cancer, which has not spread to distant organs. The most common site affected by breast cancer cells are the axillary lymph nodes, lungs, liver, and bones (1, 78). In the present review, Cur-NPs showed significant effects against tumor metastasis in in vivo breast cancer models. For instance, curcumin associated with graphene oxide NPs reduced the regions of metastasis in a triple negative breast cancer model (34).
Different routes of administration were adopted among the studies evaluated herein. The oral administration route is preferred over other routes for drugs and NPs due to advantages such as ease of ingestion, good patient compliance, and pain avoidance. The main limitation of this route is the knowledge of the real dose absorbed (79). Only one included study explored oral administration with curcumin entrapped in a lipid-based NP (25). Interestingly, a remarkable tumor volume reduction of ~60% was achieved; nevertheless, the dose of curcumin administered was the highest among all the other studies (100 mg/Kg), probably due to variations in NP absorption through this route.
Other routes of administration can be applied to avoid the gastrointestinal tract and potential degradation (80). Intratumoral administration is an interesting option for breast cancer therapy. NPs can be administered with a minimal invasive procedure with a regular biopsy needle, for example, right in the tumor site, increasing the lifetime of drugs in contact with malignant target cells, reducing adverse effects on healthy tissues, and bypassing liver metabolism (81). Two of the included studies used this route to administer micelle NPs to triple negative breast cancer models and showed similar outcomes in terms of tumor volume reduction (51–60%) (26, 27).
The intraperitoneal (IP) route is commonly used in rodents and consists of an injection of pharmacological drugs into the peritoneal cavity. Quick and minimally stressful for animals, the IP route permits safe administration of a large volume of drug and it is more appropriate when the intravenous route is challenging. The IP route is an entry portal for blood circulation through the capillary system (80). Two of the included studies reported the success of Cur-micelle NP administration via IP route with a reduction of tumor volume of ~80 and ~59.1%, respectively (28, 29).
The intravenous (IV) route of administration enables the rapid circulation of an administered drug in the bloodstream. Although approximately 70% of the included studies adopted this route of administration, precise efficacy comparisons regarding composition and other characteristics (e.g., HD, PDI, PZ) of NPs are limited due to variations in cell lines, number of cells used for induction, moment of the first treatment, and dose/treatment regimens. However, some comparisons involving dose concentrations and presence of active targeting were possible when analyzing studies that evaluated more than one experimental variable in the same experimental design. In general, it was observed that all the types of NPs used led to improved outcomes in terms of tumor volume reduction in models of estrogen receptor (ER) positive, chemically induced, and triple negative breast cancer ( Table 1 ).
MCF-7 is one of the human cell lines most commonly used for breast cancer research, since it expresses substantial levels of estrogen receptor (ER) mimicking the majority of breast cancers diagnosed nowadays (82). Analyzing MCF-7 models of the included studies, it was observed that the influence of active targeting in improved efficacy outcomes depends on the moiety used. Lin and co-workers (50) reported significant improvements in tumor volume reduction when attaching FA, as the targeting moiety, to lipid-based NPs (~83%) when compared to non-targeted NPs (~66%) (50). Similar results were observed in MDA-MB-231 in vivo models (triple negative breast cancer), where the presence of folate, as an active targeting moiety, showed improved tumor volume reduction (~90%) when compared to non-targeted NPs (~75%) (46). On the other hand, no significant improvements were observed when peptide moieties with affinity to EGFR were used in polymeric NPs when compared with non-targeted NPs (37). Another interesting aspect that seems to improve efficacy outcomes in MCF-7 models is the design of pH-sensitive NPs. Yu and co-workers (47) reported improved reduction of tumor volume in animals treated with pH-sensitive micelles (mPEG-PLA with PAE) (~65.6%) when compared to the ones treated with non-pH-sensitive micelles (~47.1%) (47).
Both MCF-7 and MDA-MB-231 conventional in vivo models are induced in immunocompromised mice due to the human origin of these cell lines (83). These models, also known as xenograft, lack relevance when the study aims to evaluate/associate the outcomes with a functional immune system. In this case, syngeneic models, where cells of the same genetic background (murine) are implanted into a mouse with a native immune system, are recommended (83). Syngeneic breast cancer models usually use the 4T1 cell line as a representative model that mimics triple negative breast cancers (84). Analyzing 4T1 models used in the included studies, it was observed that the outcome of tumor volume reduction showed a tendency to respond in a dose-dependent manner. Greish and co-workers (44) showed that a 20mg/Kg dose led to improved tumor volume reduction (~92%) when compared to a 10 mg/ml dose (~61%) (44). Nevertheless, when comparing high doses, such as 40 and 80 mg/Kg, no significant improvements were observed between them (49). The presence of the active targeting moieties FA or HA in the NPs showed improved efficacy, as reported by Laha and co-workers (31) and Ji and co-workers (31, 35).
Analyzing only the studies of Sahne and co-workers (34), Ji and co-workers (35) and He and co-workers (36), all with breast cancer induction with 4T1 cells (106) in Balb/c mice, and with the treatments performed intravenously and in very similar doses, 4 or 5 mg/kg, but with a difference in the treatment schedule ( Table 1 ), (in the work of Sahne and co-workers, the treatment was daily, for 21 days; in the work of Ji and co-workers, the treatment was every two days for 10 days; and in the work of He and co-workers, it was every four days in 21 total days), it can be observed that, interestingly, the treatments with the Cur-NPs, either by FA-GO-NP or HA-Cur-NP, promoted a similar percentage of reduction in tumor volume (approximately 86%, Table 1 ). Assessing NPs, HA-Cur-NP is a nanocrystal that has a 162 nm HD, while FA-GO-NP is a graphene oxide NP that has a 60 nm HD. In the treatment aspect, treatment every 2 days for only 10 days of HA-Cur-NP had the same antitumor efficiency as daily treatment for 21 days of FA-GO-NP. Therefore, it is a shorter and less aggressive therapeutic regimen, presenting similar efficiency (34–36).
The elimination of NPs occurs in organs and tissue systems after i.v. injection by two main clearance systems: reticuloendothelial system (RES) or mononuclear phagocyte system (MPS) and by renal and hepatic systems. Properties of NPs, including core type, surface chemistry, size, shape, degradability, and surface charge influence the process of clearance (85). The MPS is based on phagocytosis (mostly for NPs between 50 and 200 nm) or pinocytosis, and degraded NPs are excreted into the blood circulation, decreasing the injected dose (85, 86). Renal and hepatic systems are the main clearance organs of NPs less than 100 nm through glomerular filtration and tubular secretion in the kidney. NPs that are not cleared by the kidney can be processed in the liver due to the presence of a large number of Kupffer cells that can sequester foreign bodies, and the very permeable sinusoidal endothelial cells that enhance liver uptake and retention of NPs (85).
Among the included studies that evaluated toxicity effects, none of the Cur-NPs provoked toxicity, considering biochemical markers, hematological changes, damage to major organs, and weight loss ( Table 1 ). It is worth pointing out that the majority of studies (~80%) analyzing the efficacy of Cur-NPs also evaluated at least one toxicity outcome, showing that research into NPs is not only interested in treatment efficacy but also considers safety issues. Nevertheless, ~33% of such studies used only body weight as a parameter for toxicity analysis. Thus, it is important to pursue deeper investigations beyond these parameters in order to understand better the safety of the treatment and enable its clinical translation.
Limitations
Some limitations were encountered during the elaboration of this systematic review. First, there was high heterogeneity regarding NP type, characteristics of NPs, animal models, period of administration, and intervention concentrations which made meta‐analysis unfeasible. Furthermore, one study was excluded in phase 2 because its full copy could not be obtained. Moreover, most SYRCLE’ RoB criteria were unclearly reported in most included studies therefore, limiting the evaluation of study quality.
Conclusion
This systematic review evidences that the use of NPs as drug delivery systems for curcumin is a promising approach for the treatment of breast cancer. The results show significant tumor volume reduction in all breast cancer models, which could be attributed to increased apoptosis and necrosis rate, reduction of tumor cell proliferation and impairment of angiogenesis, and even reduction of the population of stem cancer cells, which might also be correlated with improved survival times. All of these improved outcomes are also related to no or low adverse effects in terms of body weight, histopathology of major organs (e.g., liver, kidneys, lungs, spleen), or alterations in hematological/biochemical parameters.
Variations in NP structure should be considered according to the type of breast tumor as well as the route of administration and dose schedule. In addition, the cost-effective and large-scale manufacturing of the proposed NP platforms is also of considerable importance to enable a real translation of these remarkable technologies from the bench to the bedside.
Although Cur-NPs’ association with other therapeutic approaches is not within the scope of the present work, it is recommended that systematic evaluations of outcomes regarding efficacy and toxicity of Cur-NPs when associated with other plant-derived molecules or currently prescribed therapies (e.g., chemotherapy, radiotherapy) should be further considered. Altogether, this systematic review supports the proposal that Cur-NPs provide an effective and safe therapeutic approach in in vivo models of breast cancer, reinforcing the currently available evidence that their usage should be further analyzed in clinical trials for breast cancer treatments.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author Contributions
AO, VS, GL, and GJ conceived the idea and prepared, edited, and finalized the manuscript. The articles were selected in two phases: screening of titles and abstracts (phase 1), and full-text reading (phase 2). In phase 1, two authors (AO and VS) reviewed titles and abstracts of all references identified in the electronic databases and selected articles that seemed to meet the inclusion criteria. In phase 2, six pairs of authors (AO and VS; LA and GF; WP and AP; JO and MG; MS and GG; PC and GJ) were formed to independently analyze the full text of articles selected in phase 1 and exclude studies that did not meet the inclusion criteria. LA and PC revised the manuscript. GG prepared the graphical abstract. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES—Finance Code 001), Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq), Instituto Nacional de Ciência e Tecnologia em Nanobiotecnologia (INCT Nanobiotecnologia), and Fundação de Amparo à Pesquisa do Distrito Federal (FAP-DF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.612903/full#supplementary-material
References
- 1. Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers (2019) 5:1–2. 10.1038/s41572-019-0111-2 [DOI] [PubMed] [Google Scholar]
- 2. WHO . Breast cancer. (2020). Available at: https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en.
- 3. Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA - J Am Med Assoc (2019) 321(3):288–300. 10.1001/jama.2018.19323 [DOI] [PubMed] [Google Scholar]
- 4. Falagan-Lotsch P, Grzincic EM, Murphy CJ. New advances in nanotechnology-based diagnosis and therapeutics for breast cancer: An assessment of active-targeting inorganic nanoplatforms. Bioconjug Chem (2017) 28(1):135–52. 10.1021/acs.bioconjchem.6b00591 [DOI] [PubMed] [Google Scholar]
- 5. Hu S, Xu Y, Meng L, Huang L, Sun H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp Ther Med (2018) 16(2):1266–72. 10.3892/etm.2018.6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kashyap D, Tuli HS, Yerer MB, Sharma A, Sak K, Srivastava S, et al. Natural product-based nanoformulations for cancer therapy: Opportunities and challenges. Semin Cancer Biol (2019). Academic Press. 10.1016/j.semcancer.2019.08.014 [DOI] [PubMed] [Google Scholar]
- 7. Sharifi-Rad J, Rayess Y EL, Rizk AA, Sadaka C, Zgheib R, Zam W, et al. Turmeric and Its Major Compound Curcumin on Health: Bioactive Effects and Safety Profiles for Food, Pharmaceutical, Biotechnological and Medicinal Applications. Front Pharmacol (2020) 11:1021. 10.3389/fphar.2020.01021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuttan R, Bhanumathy P, Nirmala K, George MC. Potential anticancer activity of turmeric (Curcuma longa). Cancer Lett (1985) 29(2):197–202. 10.1016/0304-3835(85)90159-4 [DOI] [PubMed] [Google Scholar]
- 9. Banik U, Parasuraman S, Adhikary AK, Othman NH. Curcumin: The spicy modulator of breast carcinogenesis. J Exp Clin Cancer Res (2017) 36(1):1–16. 10.1186/s13046-017-0566-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wen C, Fu L, Huang J, Dai Y, Wang B, Xu G, et al. Curcumin reverses doxorubicin resistance via inhibition the efflux function of ABCB4 in doxorubicin−resistant breast cancer cells. Mol Med Rep (2019) 19(6):5162–8. 10.3892/mmr.2019.10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Attia YM, El-Kersh DM, Ammar RA, Adel A, Khalil A, Walid H, et al. Inhibition of aldehyde dehydrogenase-1 and p-glycoprotein-mediated multidrug resistance by curcumin and vitamin D3 increases sensitivity to paclitaxel in breast cancer. Chem Biol Interact (2020) 315:108865. 10.1016/j.cbi.2019.108865 [DOI] [PubMed] [Google Scholar]
- 12. Javadi M, Khadem Haghighian H, Goodarzy S, Abbasi M, Nassiri-Asl M. Effect of curcumin nanomicelle on the clinical symptoms of patients with rheumatoid arthritis: A randomized, double-blind, controlled trial. Int J Rheum Dis (2019) 22(10):1857–62. 10.1111/1756-185X.13688 [DOI] [PubMed] [Google Scholar]
- 13. Kalyane D, Raval N, Maheshwari R, Tambe V, Kalia K, Tekade RK. Employment of enhanced permeability and retention effect (EPR): Nanoparticle-based precision tools for targeting of therapeutic and diagnostic agent in cancer. Mater Sci Eng C (2019) 98::1252–76. 10.1016/j.msec.2019.01.066 [DOI] [PubMed] [Google Scholar]
- 14. Watkins R, Wu L, Zhang C, Davis RM, Xu B. Natural product-based nanomedicine: Recent advances and issues. Int J Nanomedicine (2015) 10:6055–74. Dove Medical Press Ltd. 10.2147/IJN.S92162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Home . ClinicalTrials.gov. (2020). Available at: https://www.clinicaltrials.gov.
- 16. Giordano A, Tommonaro G. Curcumin and cancer. Nutrients (2019) 11:2376. MDPI AG. 10.3390/nu11102376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Momtazi-Borojeni AA, Ghasemi F, Hesari A, Majeed M, Caraglia M, Sahebkar A. Anti-Cancer and Radio-Sensitizing Effects of Curcumin in Nasopharyngeal Carcinoma. Curr Pharm Des (2018) 24(19):2121–8. 10.2174/1381612824666180522105202 [DOI] [PubMed] [Google Scholar]
- 18. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PloS Med (2009) 6(7):e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. PROSPERO . (2020). https://www.crd.york.ac.uk/prospero/.
- 20. Methley AM, Campbell S, Chew-Graham C, McNally R, Cheraghi-Sohi S. PICO. PICOS and SPIDER: A comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC Health Serv Res (2014) 14(1):2. 10.1186/s12913-014-0579-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hooijmans CR, Rovers MM, De Vries RBM, Leenaars M, Ritskes-Hoitinga M, Langendam MW. SYRCLE’s risk of bias tool for animal studies. BMC Med Res Methodol (2014) 14(1):1–9. 10.1186/1471-2288-14-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PloS Biol (2020) 18(7):e3000410. 10.1371/journal.pbio.3000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scopus preview . Scopus - Welcome to Scopus. (2020). Available at: https://www.scopus.com/home.uri.
- 24. VOSviewer . Features - Highlights. (2020). Available at: https://www.vosviewer.com/features/highlights.
- 25. Shukla M, Jaiswal S, Sharma A, Srivastava PK, Arya A, Dwivedi AK, et al. A combination of complexation and self-nanoemulsifying drug delivery system for enhancing oral bioavailability and anticancer efficacy of curcumin. Drug Dev Ind Pharm [Internet] (2017) 43(5):847–61. 10.1080/03639045.2016.1239732 [DOI] [PubMed] [Google Scholar]
- 26. Chen W, Li L, Zhang X, Liang Y, Pu Z, Wang L, et al. Curcumin: A calixarene derivative micelle potentiates anti-breast cancer stem cells effects in xenografted, triple-negative breast cancer mouse models. Drug Deliv (2017) 24(1):1470–81. 10.1080/10717544.2017.1381198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahalunkar S, Yadav AS, Gorain M, Pawar V, Braathen R, Weiss S, et al. Functional design of pH-responsive folate-targeted polymer-coated gold nanoparticles for drug delivery and in vivo therapy in breast cancer. Int J Nanomed (2019) 14:8285–302. 10.2147/IJN.S215142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alizadeh AM, Sadeghizadeh M, Najafi F, Ardestani SK, Erfani-Moghadam V, Khaniki M, et al. Encapsulation of curcumin in diblock copolymer micelles for cancer therapy. BioMed Res Int (2015) 2015:2–7. 10.1155/2015/824746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jung K-HKH, Lee JHJH, Park JWJW, Kim DHDH, Moon SHS-H, Cho YSYS, et al. Targeted therapy of triple negative MDA-MB-468 breast cancer with curcumin delivered by epidermal growth factor-conjugated phospholipid nanoparticles. Oncol Lett (2018) 15(6):9093–100. 10.3892/ol.2018.8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Luo Z, Wang Z, You M, Xie S, Peng Y, et al. Effect of curcumin-loaded nanoparticles on mitochondrial dysfunctions of breast cancer cells. J Nanopart Res (2018) 20(10):2–6. 10.1007/s11051-018-4382-4 [DOI] [Google Scholar]
- 31. Laha D, Pal K, Chowdhuri AR, Parida PK, Sahu SK, Jana K, et al. Fabrication of curcumin-loaded folic acid-tagged metal organic framework for triple negative breast cancer therapy in in vitro and in vivo systems. New J Chem (2019) 43(1):217–29. 10.1039/C8NJ03350A [DOI] [Google Scholar]
- 32. Vakilinezhad MA, Amini A, Dara T, Alipour S. Methotrexate and Curcumin co-encapsulated PLGA nanoparticles as a potential breast cancer therapeutic system: In vitro and in vivo evaluation. Colloids Surf B Biointerfaces (2019) 184:110515. 10.1016/j.colsurfb.2019.110515 [DOI] [PubMed] [Google Scholar]
- 33. Yuan JD, ZhuGe DL, Tong MQ, Lin MT, Xu XF, Tang X, et al. pH-sensitive polymeric nanoparticles of mPEG-PLGA-PGlu with hybrid core for simultaneous encapsulation of curcumin and doxorubicin to kill the heterogeneous tumour cells in breast cancer. Artif Cells Nanomed Biotechnol (2018) 46(sup1):302–13. 10.1080/21691401.2017.1423495 [DOI] [PubMed] [Google Scholar]
- 34. Sahne F, Mohammadi M, Najafpour GD. Single-Layer Assembly of Multifunctional Carboxymethylcellulose on Graphene Oxide Nanoparticles for Improving in Vivo Curcumin Delivery into Tumor Cells. ACS Biomater Sci Eng (2019) 5(5):2595–609. 10.1021/acsbiomaterials.8b01628 [DOI] [PubMed] [Google Scholar]
- 35. Ji P, Wang L, Chen Y, Wang S, Wu Z, Qi X. Hyaluronic acid hydrophilic surface rehabilitating curcumin nanocrystals for targeted breast cancer treatment with prolonged biodistribution. Biomater Sci (2020) 8(1):462–72. 10.1039/C9BM01605H [DOI] [PubMed] [Google Scholar]
- 36. He H, Zhuang W, Ma B, Su X, Yu T, Hu J, et al. Oxidation-Responsive and Aggregation-Induced Emission Polymeric Micelles with Two-Photon Excitation for Cancer Therapy and Bioimaging. ACS Biomater Sci Eng (2019) 5(5):2577–86. 10.1021/acsbiomaterials.9b00212 [DOI] [PubMed] [Google Scholar]
- 37. Jin H, Pi J, Zhao Y, Jiang J, Li T, Zeng X, et al. EGFR-targeting PLGA-PEG nanoparticles as a curcumin delivery system for breast cancer therapy. Nanoscale (2017) 9(42):16365–74. 10.1039/C7NR06898K [DOI] [PubMed] [Google Scholar]
- 38. Abd-Ellatef GEF, Gazzano E, Chirio D, Hamed AR, Belisario DC, Zuddas C, et al. Curcumin-loaded solid lipid nanoparticles bypass p-glycoprotein mediated doxorubicin resistance in triple negative breast cancer cells. Pharmaceutics (2020) 12(2):1–20. 10.3390/pharmaceutics12020096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li N, Wang Z, Zhang Y, Zhang K, Xie J, Liu Y, et al. Curcumin-loaded redox-responsive mesoporous silica nanoparticles for targeted breast cancer therapy. Artif Cells Nanomed Biotechnol (2018) 46(sup2):921–35. 10.1080/21691401.2018.1473412 [DOI] [PubMed] [Google Scholar]
- 40. Kundu M, Sadhukhan P, Ghosh N, Chatterjee S, Manna P, Das J, et al. pH-responsive and targeted delivery of curcumin via phenylboronic acid-functionalized ZnO nanoparticles for breast cancer therapy. J Adv Res (2019) 18:161–72. 10.1016/j.jare.2019.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lv L, Guo Y, Shen Y, Liu J, Zhang W, Zhou D, et al. Intracellularly Degradable, Self-Assembled Amphiphilic Block Copolycurcumin Nanoparticles for Efficient In Vivo Cancer Chemotherapy. Adv Healthc Mater (2015) 4(10):1496–501. 10.1002/adhm.201500075 [DOI] [PubMed] [Google Scholar]
- 42. Yang Z, Sun N, Cheng R, Zhao C, Liu J, Tian Z. Hybrid nanoparticles coated with hyaluronic acid lipoid for targeted co-delivery of paclitaxel and curcumin to synergistically eliminate breast cancer stem cells. J Mater Chem B (2017) 5(33):6762–75. 10.1039/C7TB01510K [DOI] [PubMed] [Google Scholar]
- 43. Yang Z, Sun N, Cheng R, Zhao C, Liu Z, Li X, et al. pH multistage responsive micellar system with charge-switch and PEG layer detachment for co-delivery of paclitaxel and curcumin to synergistically eliminate breast cancer stem cells. Biomaterials (2017) 147:53–67. 10.1016/j.biomaterials.2017.09.013 [DOI] [PubMed] [Google Scholar]
- 44. Greish K, Pittalà V, Taurin S, Taha S, Bahman F, Mathur A, et al. Curcumin–copper complex nanoparticles for the management of triple-negative breast cancer. Nanomaterials (2018) 8(11):2–8. 10.3390/nano8110884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mukerjee A, Ranjan AP, Vishwanatha JK. Targeted Nanocurcumin Therapy Using Annexin A2 Anitbody Improves Tumor Accumulation and Therapeutic Efficacy Against Highly Metastatic Breast Cancer. J Biomed Nanotechnol (2016) 12: (7):1374–92. 10.1166/jbn.2016.2240 [DOI] [PubMed] [Google Scholar]
- 46. Mukhopadhyay R, Sen R, Paul B, Kazi J, Ganguly S, Debnath MC. Gemcitabine Co-Encapsulated with Curcumin in Folate Decorated PLGA Nanoparticles; a Novel Approach to Treat Breast Adenocarcinoma. Pharm Res (2020) 37(3):10–14. 10.1007/s11095-020-2758-5 [DOI] [PubMed] [Google Scholar]
- 47. Yu Y, Zhang X, Qiu L. The anti-tumor efficacy of curcumin when delivered by size/charge-changing multistage polymeric micelles based on amphiphilic poly(β-amino ester) derivates. Biomaterials (2014) 35(10):3–10. 10.1016/j.biomaterials.2013.12.096 [DOI] [PubMed] [Google Scholar]
- 48. Huang C, Chen F, Zhang L, Yang Y, Yang X, Pan W. 99mTc radiolabeled HA/TPGS-based curcumin-loaded nanoparticle for breast cancer synergistic theranostics: Design, in vitro and in vivo evaluation. Int J Nanomedicine (2020) 15:2987–98. 10.2147/IJN.S242490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shiri S, Alizadeh AM, Baradaran B, Farhanghi B, Shanehbandi D, Khodayari S, et al. Dendrosomal curcumin suppresses metastatic breast cancer in mice by changing M1/M2 macrophage balance in the tumor microenvironment. Asian Pac J Cancer Prev (2015) 16(9):3917–22. 10.7314/APJCP.2015.16.9.3917 [DOI] [PubMed] [Google Scholar]
- 50. Lin M, Teng L, Wang Y, Zhang J, Sun X. Curcumin-guided nanotherapy: a lipid-based nanomedicine for targeted drug delivery in breast cancer therapy. Drug Deliv (2016) 23(4):1420–5. 10.3109/10717544.2015.1066902 [DOI] [PubMed] [Google Scholar]
- 51. Song X, Zhang M, Dai E, Luo Y. Molecular targets of curcumin in breast cancer (Review). Mol Med Rep (2019) 19(1):23–9. 10.3892/mmr.2018.9665 [DOI] [PubMed] [Google Scholar]
- 52. Shahabipour F, Caraglia M, Majeed M, Derosa G, Maffioli P, Sahebkar A. Naturally occurring anti-cancer agents targeting EZH2. Cancer Lett (2017) 400:325–35. 10.1016/j.canlet.2017.03.020 [DOI] [PubMed] [Google Scholar]
- 53. Hua WF, Fu YS, Liao YJ, Xia WJ, Chen YC, Zeng YX, et al. Curcumin induces down-regulation of EZH2 expression through the MAPK pathway in MDA-MB-435 human breast cancer cells. Eur J Pharmacol (2010) 637(1–3):16–21. 10.1016/j.ejphar.2010.03.051 [DOI] [PubMed] [Google Scholar]
- 54. Shao ZM, Shen ZZ, Liu CH, Sartippour MR, Go VL, Heber D, et al. Curcumin exerts multiple suppressive effects on human breast carcinoma cells. Int J Cancer (2002) 98(2):234–40. 10.1002/ijc.10183 [DOI] [PubMed] [Google Scholar]
- 55. Simental-Mendía LE, Caraglia M, Majeed M, Sahebkar A. Impact of curcumin on the regulation of microRNAs in colorectal cancer. Expert Rev Gastroenterol Hepatol (2017) 11:99–101. 10.1080/17474124.2017.1268528 [DOI] [PubMed] [Google Scholar]
- 56. Li J, Wei H, Liu Y, Li Q, Guo H, Guo Y, et al. Curcumin Inhibits Hepatocellular Carcinoma via Regulating miR-21/TIMP3 Axis. Evidence-Based Complement Altern Med (2020) 2020:4–8. 10.1155/2020/2892917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gallardo M, Kemmerling U, Aguayo F, Bleak TC, Muñoz JP, Calaf GM. Curcumin rescues breast cells from epithelial−mesenchymal transition and invasion induced by anti−miR−34a. Int J Oncol (2020) 56(2):480–93. 10.3892/ijo.2019.4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saberi-Karimian M, Katsiki N, Caraglia M, Boccellino M, Majeed M, Sahebkar A. Vascular endothelial growth factor: An important molecular target of curcumin. Crit Rev Food Sci Nutr (2019) 59:299–312. 10.1080/10408398.2017.1366892 [DOI] [PubMed] [Google Scholar]
- 59. Ferreira L, Arbab A, Jardim-Perassi B, Borin T, Varma N, Iskander A, et al. Effect of Curcumin on Pro-angiogenic Factors in the Xenograft Model of Breast Cancer. Anticancer Agents Med Chem (2015) 15 (10):1285–96. 10.2174/1871520615666150520093644 [DOI] [PubMed] [Google Scholar]
- 60. Ghandadi M, Sahebkar A. Curcumin: An Effective Inhibitor of Interleukin-6. Curr Pharm Des (2016) 23: (6):921–31. 10.2174/1381612822666161006151605 [DOI] [PubMed] [Google Scholar]
- 61. Basnet P, Skalko-Basnet N. Curcumin: An anti-inflammatory molecule from a curry spice on the path to cancer treatment. Molecules (2011) 16(6):4567–98. 10.3390/molecules16064567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maeda H. Toward a full understanding of the EPR effect in primary and metastatic tumors as well as issues related to its heterogeneity. Adv Drug Deliv Rev (2015) 91:3–6. 10.1016/j.addr.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 63. Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater (2016) 1:16014. 10.1038/natrevmats.2016.14 [DOI] [Google Scholar]
- 64. Sindhwani S, Syed AM, Ngai J, Kingston BR, Maiorino L, Rothschild J, et al. The entry of nanoparticles into solid tumours. Nat Mater. Nat Mater (2020) 19(5):566–75. 10.1038/s41563-019-0566-2 [DOI] [PubMed] [Google Scholar]
- 65. Shi Y, van der Meel R, Chen X, Lammers T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics (2020) 10:7921–4. 10.7150/thno.49577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gmeiner WH, Ghosh S. Nanotechnology for cancer. Nanotechnol Rev (2014) 3: (2):111–22. 10.1515/ntrev-2013-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jordan Price R, Lillycrop KA, Burdge GC. Folic acid induces cell type-specific changes in the transcriptome of breast cancer cell lines: A proof-of-concept study. J Nutr Sci (2016) 5:1–8. 10.1017/jns.2016.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tan S, Yamashita A, Gao SJ, Kurisawa M. Hyaluronic acid hydrogels with defined crosslink density for the efficient enrichment of breast cancer stem cells. Acta Biomater (2019) 94:320–9. 10.1016/j.actbio.2019.05.040 [DOI] [PubMed] [Google Scholar]
- 69. Anemone A, Consolino L, Arena F, Capozza M, Longo DL. Imaging tumor acidosis: a survey of the available techniques for mapping in vivo tumor pH. Cancer Metastasis Rev (2019) 38:25–49. 10.1007/s10555-019-09782-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Controlled Release (2010) 145:182–95. 10.1016/j.jconrel.2010.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Manzanares D, Ceña V. Endocytosis: The nanoparticle and submicron nanocompounds gateway into the cell MDPI AG. Pharmaceutics (2020) 12:6. 10.3390/pharmaceutics12040371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Karunagaran D, Rashmi R, Kumar T. Induction of Apoptosis by Curcumin and Its Implications for Cancer Therapy. Curr Cancer Drug Targets (2005) 5(2):117–29. 10.2174/1568009053202081 [DOI] [PubMed] [Google Scholar]
- 73. Reuter S, Eifes S, Dicato M, Aggarwal BB, Diederich M. Modulation of anti-apoptotic and survival pathways by curcumin as a strategy to induce apoptosis in cancer cells. Biochem Pharmacol (2008) 76:4. 10.1016/j.bcp.2008.07.031 [DOI] [PubMed] [Google Scholar]
- 74. Liczbiński P, Michałowicz J, Bukowska B. Molecular mechanism of curcumin action in signaling pathways: Review of the latest research. Phytother Res (2020) 34:1992–2005. 10.1002/ptr.6663 [DOI] [PubMed] [Google Scholar]
- 75. Sordillo PP, Helson L. Curcumin and cancer stem cells: Curcumin has asymmetrical effects on cancer and normal stem cells. Anticancer Res (2015) 35(2):599–614. [PubMed] [Google Scholar]
- 76. Cho Y, Kim YK. Cancer Stem Cells as a Potential Target to Overcome Multidrug Resistance. Front Oncol (2020) 10:764. 10.3389/fonc.2020.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lopes-Rodrigues V, Sousa E, Vasconcelos MH. Curcumin as a modulator of P-glycoprotein in cancer: Challenges and perspectives. Pharmaceuticals (2016) 9:4–5. 10.3390/ph9040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cao H, Dan Z, He X, Zhang Z, Yu H, Yin Q, et al. Liposomes Coated with Isolated Macrophage Membrane Can Target Lung Metastasis of Breast Cancer. ACS Nano (2016) 10(8):7738–48. 10.1021/acsnano.6b03148 [DOI] [PubMed] [Google Scholar]
- 79. Zhang J, Xie Z, Zhang N, Zhong J. Nanosuspension drug delivery system: preparation, characterization, postproduction processing, dosage form, and application. In: . Nanostructures for Drug Delivery (2017). p. 413–43. [Google Scholar]
- 80. Al Shoyaib A, Archie SR, Karamyan VT. Intraperitoneal Route of Drug Administration: Should it Be Used in Experimental Animal Studies? Pharm Res (2020) 37(1):1. 10.1007/s11095-019-2745-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kollenda SA, Klose J, Knuschke T, Sokolova V, Schmitz J, Staniszewska M, et al. In vivo biodistribution of calcium phosphate nanoparticles after intravascular, intramuscular, intratumoral, and soft tissue administration in mice investigated by small animal PET/CT. Acta Biomater (2020) 109:244–53. 10.1016/j.actbio.2020.03.031 [DOI] [PubMed] [Google Scholar]
- 82. Lee AV, Oesterreich S, Davidson NE. MCF-7 Cells - Changing the Course of Breast Cancer Research and Care for 45 Years. J Natl Cancer Inst (2015) 107:1–2. 10.1093/jnci/djv073 [DOI] [PubMed] [Google Scholar]
- 83. Holen I, Speirs V, Morrissey B, Blyth K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis Model Mech (2017) 10:359–71. 10.1242/dmm.028274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Paschall AV, Liu K. An Orthotopic mouse model of spontaneous breast cancer metastasis. J Vis Exp (2016) 2016(114):2–5. 10.3791/54040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wei Y, Quan L, Zhou C, Zhan Q. Factors relating to the biodistribution & clearance of nanoparticles & their effects on in vivo application. Nanomedicine (2018) 13(12):1495–512. 10.2217/nnm-2018-0040 [DOI] [PubMed] [Google Scholar]
- 86. Shreffler JW, Pullan JE, Dailey KM, Mallik S, Brooks AE. Overcoming hurdles in nanoparticle clinical translation: The influence of experimental design and surface modification. Int J Mol Sci (2019) 20(23):1–25. 10.3390/ijms20236056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.