Abstract
Background
The effect of mannose‐binding lectin (MBL) gene polymorphisms on susceptibility of rheumatoid arthritis (RA) were evaluated in ethnically different populations, whereas the results were always inconsistent.
Materials and methods
Fourteen articles involving 36 datasets were recruited to evaluate the association between MBL gene polymorphisms and rheumatoid arthritis in a meta‐analysis. The random or fixed effect models were used to evaluate the pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs).
Results
Stratified analysis by ethnicities was conducted and the result revealed that rs1800450 (T vs C, OR = 1.32, 95% CI: 1.04‐1.67, P < .05) and MBL‐A/O (T vs C, OR = 1.20, 95% CI: 1.08‐1.34, P < .001) were strongly associated with RA in Brazilian populations. In addition, the significant relationship between rs11003125 (T vs C, OR = 1.16, 95% CI: 1.06‐1.26, P < .05) with RA were also observed in East Asian populations. Meanwhile, the inverse associations between rs5030737 with RA in East Asians and rs1800450 with RA in Indians were acquired. However, no association between any MBL polymorphism with RA susceptibility was confirmed in Caucasians.
Conclusions
The structural polymorphisms in exon 1 of MBL gene may significantly contribute to susceptibility and development of RA in Brazilian and Indian populations, whereas the functional polymorphisms in the promoter region were more likely to associate with RA in East Asians.
Keywords: mannose‐binding lectin, meta‐analysis, polymorphism, rheumatoid arthritis
1. INTRODUCTION
Rheumatoid arthritis (RA) is an autoimmune disease characterized by persistent synovitis, systemic inflammation and presence of autoantibodies. 1 RA affects 0.5% ~2% of the world population and the prevalence rate was 0.2%~0.4% in China. 2 , 3 At present, RA is one of the major diseases that seriously destructs life quality of patients and leads to the loss of labor capacity. 4 Although the pathogenesis of RA has not been fully clarified, it is generally believed that environmental, genetic and autoimmune factors may play a vital role in the onset of RA and genetic factors account for about 60% of RA susceptibility. 5
Mannose‐binding lectin (MBL) is a calcium‐dependent collagen lectin secreted by hepatocytes and plays an important role in innate immunity by opsonizing mannose‐ and N‐acetylglucosamine‐rich microorganisms and activating macrophages and complements. 6 The mutation of MBL gene can decrease the level of plasma MBL which may relate to immune deficiency. 7 The MBL gene is mapped on the long arm of chromosome 10q11.2‐10q21 and contains 4 exons. 8 Exon 1 has three functional single nucleotide polymorphisms (SNPs) at codons 54 (allele B, rs1800450), 57 (allele C, rs1800541), and 52 (allele D, rs5030737) in the structural part of the gene. 9 In addition, there are SNPs at positions −550 (allele L, rs11003125) and −221 (allele X, rs7096206) in the promoter region. 10 These variant alleles were associated with lowing serum MBL levels. Previous studies found that MBL gene variants can contribute to the susceptibility of acquired immune deficiency syndrome (AIDS), tuberculosis, systemic lupus erythematosus (SLE), Crohn's disease, RA and other infectious diseases. 11 , 12 , 13 , 14 , 15 However, the associations between MBL gene polymorphisms and RA were studied in various countries and ethnicities, but the results were inconsistent. 8 , 16 , 17 Tsutsumi et al. 18 found that codon 52 and 57 mutations of MBL gene were absent or extremely rare in Japanese; homozygous codon 54 mutation of the MBL gene was significantly increased in patients with autoimmune disorders. In addition, promotor regions of MBL gene suggested that individuals with absent or extremely low serum MBL were at risk of having autoimmune disorders. 19 Moreover, the B variant of the MBL2 gene may be associated with protection from RA in an Indian cohort and the promoter polymorphism rs1800450 seemed to have some roles in disease progression. 8 However, Stanworth et al. 20 declared that no evidence was found to support the association of MBL allele with protection from RA in Caucasian populations and genotype frequencies were similar in case and control groups. Similarly, the negative results were confirmed in a Japanese population. 18
Although many studies have been conducted so far to investigate the relationship between MBL gene polymorphisms and susceptibility of RA, the results were inconsistent. This discrepancy may be attributed to small sample sizes, low statistical power, different genetic background, or clinical heterogeneity. Meanwhile, inclusion of data that did not satisfy the requirement of meta‐analysis will produce a spurious association. Therefore, in order to reduce the limitations of a single study and to overcome the possible random errors, a large‐scale meta‐analysis involving multifarious ethnicities and multiple polymorphisms was performed in this study.
2. MATERIALS AND METHODS
2.1. Identification of eligible studies
To analyze the roles of MBL gene polymorphisms in susceptibility of RA, all published literature before June 2020 that researched the relationship between MBL gene polymorphisms and RA risk were included. The electronic databases were used including PubMed databases (National Center for Biotechnology, National Library of Medicine), CNKI (China National Knowledge Infrastructure), and Web of Science to retrieve articles by using the keywords “MBL gene”, “codon 54 (allele B, rs1800450)”, “codon 57 (allele C, rs1800541)”, “codon 52 (allele D, rs5030737)”, “‐550 (allele L, rs11003125)”, “‐221 (allele X, rs7096206)”, “polymorphism” connected to “RA”, “rheumatoid arthritis” without language restrictions. Finally, we extracted data from the published articles, not including meetings or any conference abstracts. All of studies were conducted with case–control or nested case–control design. The diagnosis of RA was according to the American College of Rheumatology (ACR) criteria and proper genotyping methods in most of the studies. 21 , 22 , 23
Three functional single nucleotide polymorphisms (SNP) in codons 54 (allele B), 57 (allele C), and 52 (allele D) were associated with changes in the structure and functional deficiency of protein. In codon 54, an A to G substitution alters an aspartic acid to a glycine at the protein level. In codon 57 there is a G to A substitution (glycine to glutamic acid), and in codon 52 a C to T substitution leads to a change from arginine to cysteine. Altogether, the presence of any variant alleles above has been collectively labeled O, while the simultaneous absence of variants at the 3 positions has been called allele A, the wild‐type allele. 24
2.2. Selection criteria and data extraction
Such major criteria must be followed for included studies: (a) original papers containing complete data; (b) case–control or cohort studies that assessed the association of MBL gene polymorphisms with RA; (c) sufficient data to calculate the odds ratio (OR) or P value; (d) relevant RA outcomes were angiographically confirmed according to the ACR criteria; 25 (e) the genotype distribution in the control group for each individual study should follow Hardy–Weinberg equilibrium (HWE). 26 , 27 The primary reasons for excluded studies: (a) case report, review or meta‐analysis articles; (b) deviation from the major selection criteria; (c) overlapping or that supplied inadequate data; (d) repeated publications or the same authors employed similar data in different papers, the data was only used once.
The study data were extracted based on standard protocols. 28 Disagreement was settled by a consensus between all authors. Where essential information was not presented in articles, every effort was made to contact the authors. All procedures conformed to the guidelines for meta‐analysis of observational studies in epidemiology. 29 The following information were extracted independently by individuals in our study: first author, year of publication, ethnicity, study design, types of RA, HWE status among controls, sample size of cases and controls, number of genotypes and allele frequency.
2.3. Statistical analysis
We calculated the allele frequency for each study in allele counting method; the HWE was tested by using the Chi‐square test. We employed pooled ORs and 95% confidence intervals (CIs) to evaluate the strength of association between polymorphisms and RA for every eligible study.
The methodology of Cochran's Q‐statistic was used to evaluate the heterogeneity, which is similar to the previous study in our lab. 22 , 23 If the P value in heterogeneity test was higher than 0.1, the fixed effect model was used. Moreover, the random effect model was used. We used the following formula to quantify the effect of heterogeneity: I 2 = 100% × (Q − dƒ)/Q. 30 The proportion of between‐study variability attributable to heterogeneity was indicated by I 2 value, and I 2 values of 25%, 50% and 75% were considered to be of low, moderate and high heterogeneity, respectively. If study groups revealed no heterogeneity, the similar results were produced in fixed and random effects models and, otherwise the random effects model usually produced wider CIs than the fixed effects model. 31 In this meta‐analysis, P value of less than .05 was considered as statistically significant.
In order to get exacting search results, we evaluated possible publication bias by Egger's linear regression test. If P value <.05 the statistical publication bias was considered. Moreover, the Begg's test also used a funnel plot to evaluate the publication bias. 32 For sensitivity analysis, we removed 1 study orderly from the total and tested residual studies. 33 All standard methods in this meta‐analysis were conducted in a previous study by us. Statistical analysis was carried out using the software program STATA15.0 (Stata Corporation).
3. RESULTS
3.1. Studies included in the meta‐analysis
In this meta‐analysis, totally 318 relevant articles were searched. After reading titles and abstracts, we excluded irrelevant studies, leaving 105 articles for further reading. Then, we excluded 54 articles, because of no data, insufficient data, repeated date, family‐based studies and not referring to RA. Thus, 51 articles met the study inclusion criteria. Lastly, 15 articles that included insufficient data, 5 articles in which the control populations deviated from HWE and 18 reviews or meta‐analysis researches about MBL gene polymorphisms were excluded. 25 , 44 After filtering, 13 eligible studies involving 36 data sets were finally included. 8 , 53 Eventually, 13 studies provided 5972 cases and 6663 controls: codon 54 (allele B, rs1800450), 1472 patients and 1554 controls; codon 57 (allele C, rs1800541), 240 patients and 264 controls; codon 52 (allele D, rs5030737), 520 patients and 642 controls; −550 (allele L, rs11003125), 785 patients and 841 controls; −221 (allele X, rs7096206), 1097 patients and 1151 controls; MBL‐A/O, 1858 patients and 2211 controls were pooled to evaluate the relationship between SNPs of MBL and RA in the meta‐analysis (Table 1). The flowchart of selecting articles is presented in Figure 1.
TABLE 1.
The basic information of included studies in this meta‐analysis
| Study | Y | Ethnicity | Polymorphisms | Sample size | Genotypes | Allele frequencies (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Case | Control | Case | ||||||||||||
| Control | Case | CC | CT | TT | CC | CT | TT | C | T | C | T | ||||
| Bhawna et al. 8 | 2005 | Indian | rs5030737 | 119 | 120 | 104 | 15 | 0 | 100 | 20 | 0 | 93.7 | 6.3 | 91.7 | 8.3 |
| Bhawna et al. 8 | 2005 | Indian | rs5030737 | 145 | 120 | 132 | 13 | 0 | 100 | 20 | 0 | 95.5 | 4.5 | 91.7 | 8.3 |
| Hou et al. 47 | 2013 | Chinese | rs5030737 | 378 | 280 | 362 | 15 | 0 | 278 | 2 | 0 | 97.9 | 2.1 | 99.6 | 0.4 |
| Stanworth et al. 20 | 1999 | Caucasian | rs1800450 | 114 | 182 | 84 | 28 | 2 | 142 | 35 | 5 | 86.0 | 14.0 | 87.6 | 12.4 |
| Ip et al. 46 | 2000 | Chinese | rs1800450 | 196 | 211 | 153 | 42 | 1 | 143 | 64 | 4 | 88.8 | 11.2 | 82.9 | 17.1 |
| Horiuchi et al. 18 | 2000 | Japanese | rs1800450 | 105 | 59 | 66 | 29 | 10 | 42 | 13 | 4 | 76.7 | 23.3 | 82.2 | 17.8 |
| Tsutsumi et al. 53 | 2001 | Japanese | rs1800450 | 129 | 95 | 88 | 39 | 2 | 58 | 32 | 5 | 83.3 | 16.7 | 77.9 | 22.1 |
| Bhawna et al. 8 | 2005 | Indian | rs1800450 | 119 | 120 | 72 | 47 | 0 | 106 | 14 | 0 | 80.3 | 19.7 | 94.2 | 5.8 |
| Bhawna et al. 8 | 2005 | Indian | rs1800450 | 145 | 120 | 94 | 49 | 2 | 106 | 14 | 0 | 81.7 | 18.3 | 94.2 | 5.8 |
| Min et al. 49 | 2006 | Chinese | rs1800450 | 48 | 93 | 36 | 12 | 0 | 66 | 24 | 3 | 87.5 | 12.5 | 83.9 | 16.1 |
| Hou et al. 47 | 2013 | Chinese | rs1800450 | 378 | 280 | 231 | 138 | 10 | 195 | 76 | 9 | 79.2 | 20.8 | 83.2 | 16.8 |
| Isabela et al. 16 | 2014 | Brazilian | rs1800450 | 200 | 156 | 148 | 45 | 7 | 96 | 55 | 5 | 85.3 | 14.7 | 79.2 | 20.8 |
| Isabela et al. 16 | 2014 | Brazilian | rs1800450 | 120 | 156 | 79 | 41 | 0 | 96 | 55 | 5 | 82.9 | 17.1 | 79.2 | 20.8 |
| Bhawna et al. 8 | 2005 | Indian | rs1800451 | 119 | 120 | 111 | 8 | 0 | 110 | 10 | 0 | 96.6 | 3.4 | 95.8 | 4.2 |
| Bhawna et al. 8 | 2005 | Indian | rs1800451 | 145 | 120 | 129 | 16 | 0 | 110 | 10 | 0 | 94.5 | 5.5 | 95.8 | 4.2 |
| Ip et al. 46 | 2000 | Chinese | rs7096206 | 174 | 115 | 119 | 50 | 5 | 68 | 41 | 6 | 82.8 | 17.2 | 76.5 | 23.5 |
| Bhawna et al. 8 | 2005 | Indian | rs7096206 | 119 | 120 | 70 | 43 | 6 | 60 | 50 | 10 | 76.9 | 23.1 | 70.8 | 29.2 |
| Bhawna et al. 8 | 2005 | Indian | rs7096206 | 90 | 120 | 46 | 32 | 12 | 60 | 50 | 10 | 68.9 | 31.1 | 70.8 | 29.2 |
| Min et al. 49 | 2006 | Chinese | rs7096206 | 48 | 50 | 38 | 9 | 1 | 33 | 15 | 2 | 88.5 | 11.5 | 81.0 | 19.0 |
| Isabela et al. 16 | 2014 | Brazilian | rs7096206 | 200 | 156 | 130 | 58 | 12 | 109 | 38 | 9 | 79.5 | 20.5 | 82.1 | 18.9 |
| Isabela et al. 16 | 2014 | Brazilian | rs7096206 | 120 | 156 | 91 | 28 | 1 | 109 | 38 | 9 | 87.5 | 12.5 | 82.1 | 17.9 |
| Hou et al. 17 | 2020 | Chinese | rs7096206 | 400 | 380 | 232 | 160 | 8 | 230 | 143 | 7 | 77.9 | 22.1 | 79.3 | 20.7 |
| Ip et al. 46 | 2000 | Chinese | rs11003125 | 174 | 115 | 48 | 87 | 39 | 20 | 56 | 39 | 52.6 | 47.4 | 41.7 | 58.3 |
| Bhawna et al 8 | 2005 | Indian | rs11003125 | 119 | 120 | 16 | 54 | 49 | 15 | 54 | 51 | 36.1 | 63.9 | 35.0 | 65.0 |
| Bhawna et al. 8 | 2005 | Indian | rs11003125 | 100 | 120 | 17 | 42 | 41 | 15 | 54 | 51 | 38.0 | 62.0 | 35.0 | 65.0 |
| Min et al. 49 | 2006 | Chinese | rs11003125 | 48 | 50 | 17 | 23 | 8 | 15 | 25 | 10 | 59.4 | 40.6 | 54.0 | 46.0 |
| Hou et al. 17 | 2020 | Chinese | rs11003125 | 400 | 380 | 111 | 225 | 64 | 100 | 181 | 99 | 56.0 | 44.0 | 50.2 | 49.8 |
| Jacobsen et al. 51 | 2000 | Caucasian | MBL‐A/O | 250 | 68 | 157 | 86 | 7 | 35 | 28 | 5 | 80.0 | 20.0 | 72.1 | 27.9 |
| Koert et al. 45 | 2008 | Caucasian | MBL‐A/O | 194 | 218 | 120 | 65 | 9 | 128 | 81 | 9 | 78.6 | 21.4 | 77.3 | 22.7 |
| Fernanda et al. 19 | 2012 | Brazilian | MBL‐A/O | 345 | 322 | 207 | 120 | 18 | 171 | 131 | 20 | 77.4 | 22.6 | 73.4 | 26.6 |
| Fernanda et al. 19 | 2012 | Brazilian | MBL‐A/O | 244 | 300 | 148 | 83 | 13 | 160 | 123 | 17 | 77.7 | 22.3 | 73.8 | 26.2 |
| Fernanda et al. 19 | 2012 | Brazilian | MBL‐A/O | 101 | 22 | 59 | 37 | 5 | 11 | 8 | 3 | 76.7 | 23.3 | 68.2 | 31.8 |
| Isabela et al. 16 | 2014 | Brazilian | MBL‐A/O | 200 | 156 | 119 | 73 | 8 | 75 | 73 | 8 | 77.8 | 22.2 | 71.4 | 28.6 |
| Isabela et al. 16 | 2014 | Brazilian | MBL‐A/O | 120 | 156 | 62 | 58 | 0 | 75 | 73 | 8 | 75.8 | 24.2 | 71.4 | 28.6 |
| Malthe et al. 48 | 2014 | Caucasian | MBL‐A/O | 383 | 301 | 159 | 193 | 31 | 130 | 143 | 28 | 66.7 | 33.3 | 66.9 | 33.1 |
| Malthe et al. 48 | 2014 | Caucasian | MBL‐YA/O | 374 | 315 | 150 | 193 | 31 | 144 | 143 | 28 | 65.9 | 34.1 | 68.4 | 31.6 |
C, represent wild‐type allele; T, represent minor allele; MBL‐A/O, the presence of any of rs5030737, rs1800450, rs1800451 has been collectively labeled O, while the simultaneous absence of variants at the 3 positions has been called allele A, the wild‐type allele; MBL‐YA/O, the MBL‐A/O and presence of rs7096206.
FIGURE 1.
The process of the articles selected in this meta‐analysis
3.2. Meta‐analysis results
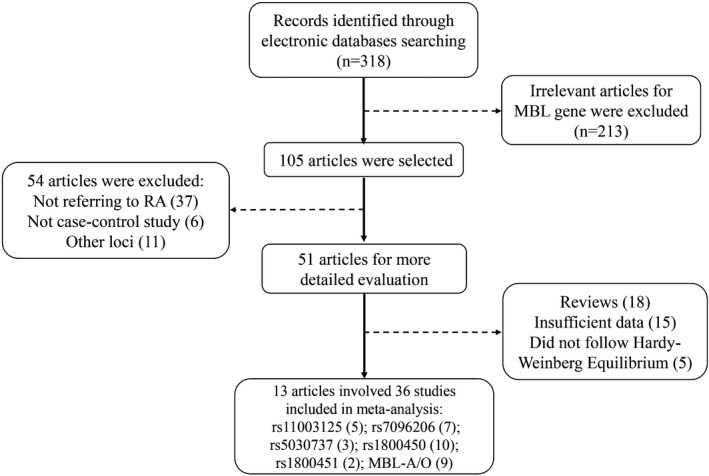
In this meta‐analysis we recruited allele model, dominant gene model and recessive gene model to confirm the association between 5 MBL SNPs with RA in multiple ethnicities. The results of stratification by ethnicity revealed the heterogeneity had disappeared (P > .01, I 2 < 30%; Figure 2A‐D).
FIGURE 2.
Forest plot for the meta‐analysis of allele model (T vs C). A, MBL gene polymorphisms and RA in Brazilians. B, MBL gene polymorphisms and RA in Caucasians. C, MBL gene polymorphisms and RA in East Asians. D, MBL gene polymorphisms and RA in Indians
3.3. Mannose‐binding lectin SNPs and RA in Brazilians
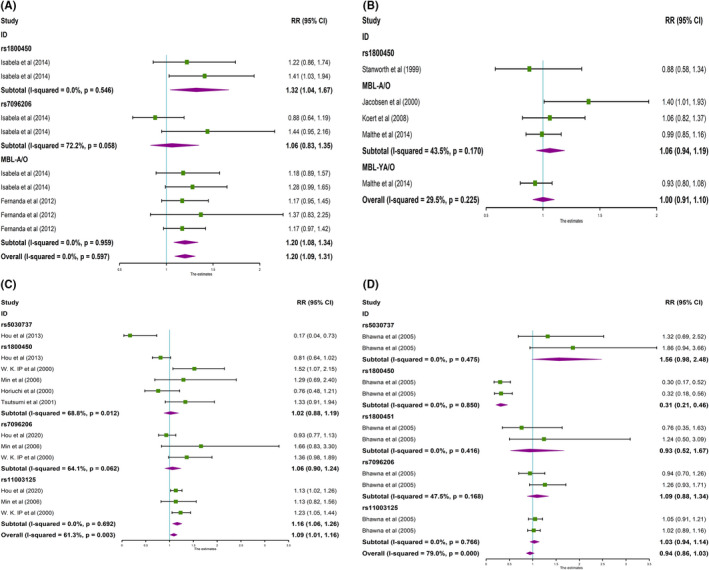
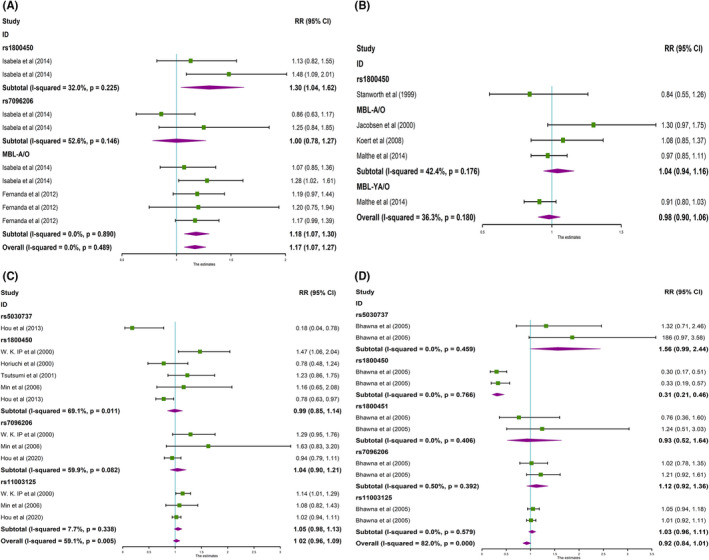
The results found that rs1800450 (T vs C, OR = 1.32, 95% CI: 1.04‐1.67, P OR < .05) and MBL‐A/O (T vs C, OR = 1.20, 95% CI: 1.08‐1.34, P OR < .001) were strongly associated with RA in a Brazilian population (Table 2, Figure 2A). Meanwhile, the overall study displayed the same significant association (T vs C, OR = 1.20, 95% CI: 1.09‐1.31, P OR < .001), and no heterogeneity (P H = .597) (Table 2, Figure 2A). In addition, rs1800450 (TT + TC vs CC, OR = 1.30, 95% CI: 1.04‐1.62, P OR < .05) and MBL‐A/O (TT + TC vs CC, OR = 1.18, 95% CI: 1.07‐1.31, P OR < .05) were strongly related to RA in the dominant model (Table 2, Figure 3A), whereas, the association was weak in the recessive gene model (Table 2, Figure 4A). However, pooled associations of rs1800450 and MBL‐A/O not only in the dominant model (TT + TC vs CC, OR = 1.17, 95% CI: 1.07‐1.27, P OR < .0001) were strong (Table 2, Figure 3A), but also in the recessive model (TT vs TC + CC, OR = 1.44, 95%CI: 1.04‐1.99, P OR < .05) (Table 2, Figure 4A).
TABLE 2.
The association between MBL polymorphisms and RA risk in meta‐analysis
| Sub‐group analysis | No. of data sets | No. of cases/controls | Allele model (T vs C) | Dominant model (CC vs TT + CT) | Recessive model (TT vs CC + CT) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | P OR | P H | OR (95% CI) | P OR | P H | OR (95% CI) | P OR | P H | ||
| Brazilian | ||||||||||||
| rs1800450 | 2 | 754 | 740 | 1.32 (1.04‐1.67)∗ | <.05 | .546 | 1.30 (1.04‐1.62)∗ | <.05 | .225 | 1.55 (0.59‐4.12) | .377 | .141 |
| rs7096206 | 2 | 736 | 752 | 1.06 (0.83‐1.35) | .666 | .058 | 1.00 (0.78‐1.27) | .969 | .146 | 1.54 (0.74‐3.20) | .248 | .070 |
| MBL‐A/O | 5 | 2432 | 2479 | 1.20 (1.08‐1.34)** | <.001 | .959 | 1.18 (1.07‐1.31)∗ | <.05 | .890 | 1.39 (0.95‐2.05) | .093 | .379 |
| Overall | 9 | 3922 | 3971 | 1.20 (1.09‐1.31)*** | <.0001 | .597 | 1.17 (1.07‐1.27) *** | <.0001 | .489 | 1.44 (1.04‐1.99)∗ | <.05 | .298 |
| Caucasian | ||||||||||||
| rs1800450 | 1 | 409 | 260 | 0.88 (0.58‐1.34) | .556 | NA | 0.84 (0.55‐1.26) | .391 | NA | 1.57 (0.31‐7.94) | .588 | NA |
| MBL‐A/O | 3 | 1510 | 2092 | 1.06 (0.94‐1.20) | .357 | .170 | 1.04 (0.94‐1.16) | .463 | .176 | 1.20 (0.81‐1.78) | .372 | .310 |
| MBL‐YA/O | 1 | 829 | 1003 | 0.93 (0.80‐1.08) | .326 | NA | 0.91 (0.80‐1.03) | .141 | NA | 1.07 (0.66‐1.75) | .779 | NA |
| Overall | 5 | 2748 | 3355 | 1.00 (0.91‐1.10) | .973 | .225 | 0.98 (0.90‐1.06) | .592 | .180 | 1.16 (0.86‐1.57) | .335 | .623 |
| East Asian | ||||||||||||
| rs5030737 | 1 | 562 | 772 | 0.17 (0.04‐0.73) * | <.05 | NA | 0.18 (0.04‐0.78) * | <.05 | NA | NA | ||
| rs1800450 | 5 | 1735 | 2017 | 1.02 (0.88‐1.19) | .769 | .012 | 0.99 (0.85‐1.14) | .872 | .011 | 1.44 (0.81‐2.56) | .214 | .426 |
| rs7096206 | 3 | 1320 | 1492 | 1.06 (0.90‐1.24) | .495 | .062 | 1.04 (0.90‐1.21) | .567 | .082 | 1.28 (0.63‐2.61) | .499 | .647 |
| rs11003125 | 3 | 1649 | 1800 | 1.16 (1.06‐1.26) * | <.05 | .692 | 1.05 (0.98‐1.13) | .133 | .338 | 1.56 (1.25‐1.94) | <.0001 | .783 |
| Overall | 12 | 5266 | 6081 | 1.09 (1.02‐1.17)* | <.05 | .003 | 1.02 (0.96‐1.09) | .531 | .005 | 1.52 (1.24‐1.84) | <.0001 | .851 |
| Indian | ||||||||||||
| rs5030737 | 2 | 520 | 556 | 1.56 (0.98‐2.48) | .062 | .475 | 1.56 (1.00‐2.44) | .053 | .459 | NA | NA | NA |
| rs1800450 | 2 | 508 | 628 | 0.31 (0.21‐0.46)*** | <.0001 | .850 | 0.31 (0.21‐0.46)*** | <.0001 | .766 | 0.24 (0.01‐4.98) | .357 | NA |
| rs1800451 | 2 | 500 | 552 | 0.93 (0.52‐1.67) | .802 | .416 | 0.93 (0.52‐1.64) | .798 | .406 | NA | NA | NA |
| rs7096206 | 2 | 620 | 529 | 1.09 (0.88‐1.34) | .434 | .168 | 1.12 (0.92‐1.36) | .268 | .392 | 0.94 (0.52‐1.71) | .836 | .130 |
| rs11003125 | 2 | 792 | 714 | 1.03 (0.94‐1.14) | .524 | .766 | 1.03 (0.96‐1.11) | .415 | .579 | 1.03 (0.83‐1.28) | .760 | .985 |
| Overall | 10 | 2940 | 2979 | 1.07 (0.98‐1.18)*** | .184 | <.0001 | 1.09 (1.00‐1.19) | .085 | <.0001 | 1.00 (0.82‐1.23) | .981 | .510 |
Abbreviations: C, represent wild‐type allele; 95%CI, 95% confidence interval; OR, odd ratio; P OR, P value for the test of association; P H, P value for heterogeneity analysis; T, represent minor allele; NA, none.
P OR < .05.
P OR < .001.
P OR < .0001.
FIGURE 3.
Forest plot for the meta‐analysis of allele model dominant model (CC vs TT + CT). A, MBL gene polymorphisms and RA in Brazilians. B, MBL gene polymorphisms and RA in Caucasians. C,, MBL gene polymorphisms and RA in East Asians. D, MBL gene polymorphisms and RA in Indians
FIGURE 4.
Forest plot for the meta‐analysis of recessive model (TT vs CC + CT). A, MBL gene polymorphisms and RA in Brazilians. B, MBL gene polymorphisms and RA in Caucasians. C, MBL gene polymorphisms and RA in East Asians. D, MBL gene polymorphisms and RA in Indians
3.4. MBL SNPs and RA in East Asians
The significant relationship between rs11003125 (T vs C, OR = 1.16, 95% CI: 1.06‐1.26, P OR < .05) with RA susceptibility was observed in East Asian populations (Table 2, Figure 2C). Meanwhile, significant association was found in the recessive gene model (TT vs TC + CC, OR = 1.56, 95% CI: 1.25‐1.94, P OR < .0001) (Table 2, Figure 4C). The rs5030737 (T vs C, OR = 0.17, 95% CI: 0.04‐0.73, P OR < .05) was reversely associated with RA in East Asians (Table 2, Figure 2C), and the reverse association was maintained in the dominant model (TT + TC vs CC, OR = 0.18, 95% CI: 0.04‐0.78, P OR < .05) (Table 2, Figure 3C). However, pooled associations of rs11003125 and rs5030737 were observed, but the heterogeneity (P H < .01, I 2 > 30%) was also found.
3.5. MBL SNPs and RA in Indians
In this stratification, the heterogeneity was resolved. The rs1800450 (T vs C, OR = 0.31, 95% CI: 0.21‐0.46, P OR < .0001) was reversely associated with RA in an Indian population (Table 2, Figure 2D). Meanwhile, the reverse association was maintained in the dominant model (TT + TC vs CC, OR = 0.31, 95% CI: 0.21‐0.46, P OR < .0001) (Table 2, Figure 3D).
3.6. MBL SNPs and RA in Caucasians
In this meta‐analysis, 5 studies involved rs1800450 and pooled MBL‐A/O polymorphisms to research the association with RA in Caucasians. However, the results showed that no association between any MBL polymorphism with RA susceptibility was confirmed in Caucasian (P OR > .05) (Table 2, Figures 2, 3 and 4B).
3.7. Comparing allele frequency of MBL SNPs to the 1000 genome phase 3 population
We compared allele frequencies of different ethnicities in our meta‐analysis to 1000 genome allele frequencies in Table 3. In view of the sample size and population, the allelic frequencies of MBL polymorphisms in this meta‐analysis were consistent with the allelic frequencies in the 1000 Genome Project East Asian ancestry and Caucasians. However, there was distinction between the allele frequencies in Indians and the 1000 Genomes Project. Meanwhile, allele frequency of rs7096206 was inconsistent in any ethnicity compared to the 1000 Genomes Project.
TABLE 3.
The allele frequency comparison between the meta‐analysis and 1000 Genomes Project
| Polymorphisms | Populations | Meta‐analysis (alleles frequencies) | 1000 genomes (alleles frequencies) | ||||
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
| C | T | C | T | C | T | ||
| rs5030737 | Brazilian | NA | NA | NA | NA | 1.00 | 0.00 |
| Caucasian | NA | NA | NA | NA | 0.94 | 0.06 | |
| East Asian | 1.00 | 0.00 | 0.98 | 0.02 | 1.00 | 0.00 | |
| Indian | 0.92 | 0.08 | 0.95 | 0.05 | 0.97 | 0.03 | |
| All | 0.96 | 0.04 | 0.97 | 0.03 | 0.97 | 0.03 | |
| rs1800450 | Brazilian | 0.79 | 0.21 | 0.84 | 0.16 | 0.85 | 0.15 |
| Caucasian | 0.88 | 0.12 | 0.86 | 0.14 | 0.86 | 0.14 | |
| East Asian | 0.82 | 0.18 | 0.82 | 0.18 | 0.85 | 0.15 | |
| Indian | 0.94 | 0.06 | 0.81 | 0.19 | 0.78 | 0.22 | |
| All | 0.84 | 0.16 | 0.83 | 0.17 | 0.88 | 0.12 | |
| rs1800451 | Brazilian | NA | NA | NA | NA | 0.97 | 0.03 |
| Caucasian | NA | NA | NA | NA | 0.99 | 0.01 | |
| East Asian | NA | NA | NA | NA | 1.00 | 0.00 | |
| Indian | 0.96 | 0.04 | 0.95 | 0.05 | 0.98 | 0.02 | |
| All | 0.96 | 0.04 | 0.95 | 0.05 | 0.92 | 0.08 | |
| rs11003125 | Brazilian | NA | NA | NA | NA | 0.47 | 0.53 |
| Caucasian | NA | NA | NA | NA | 0.61 | 0.39 | |
| East Asian | 0.49 | 0.51 | 0.55 | 0.45 | 0.55 | 0.45 | |
| Indian | 0.35 | 0.65 | 0.37 | 0.63 | 0.60 | 0.40 | |
| All | 0.45 | 0.55 | 0.51 | 0.49 | 0.69 | 0.31 | |
| rs7096206 | Brazilian | 0.82 | 0.18 | 0.83 | 0.18 | 0.13 | 0.87 |
| Caucasian | NA | NA | NA | NA | 0.22 | 0.78 | |
| East Asian | 0.79 | 0.21 | 0.80 | 0.20 | 0.19 | 0.81 | |
| Indian | 0.71 | 0.29 | 0.73 | 0.27 | 0.13 | 0.87 | |
| All | 0.78 | 0.22 | 0.80 | 0.20 | 0.20 | 0.80 | |
| MBL‐A/O | Brazilian | 0.73 | 0.27 | 0.77 | 0.23 | NA | NA |
| Caucasian | 0.70 | 0.30 | 0.71 | 0.29 | NA | NA | |
| All | 0.72 | 0.28 | 0.74 | 0.26 | NA | NA | |
Abbreviations: C, represent wild‐type allele; T, represent minor allele.
3.8. Publication bias and sensitivity analysis
Begg's funnel plot and Egger's test were performed to estimate publication bias (Figure 5A‐D). No evidence of publication bias for MBL gene polymorphisms under the allele genetic model was found in any ethnicity. In addition, no significant difference was found in the Egger's test, suggesting no obvious bias of publication in the present meta‐analysis. We also conducted sensitivity analysis to assess the influence of individual studies on the pooled ORs. We found the pooled OR was not substantially altered, when a single study involved in the meta‐analysis was deleted each time (Figure 6A‐D).
FIGURE 5.
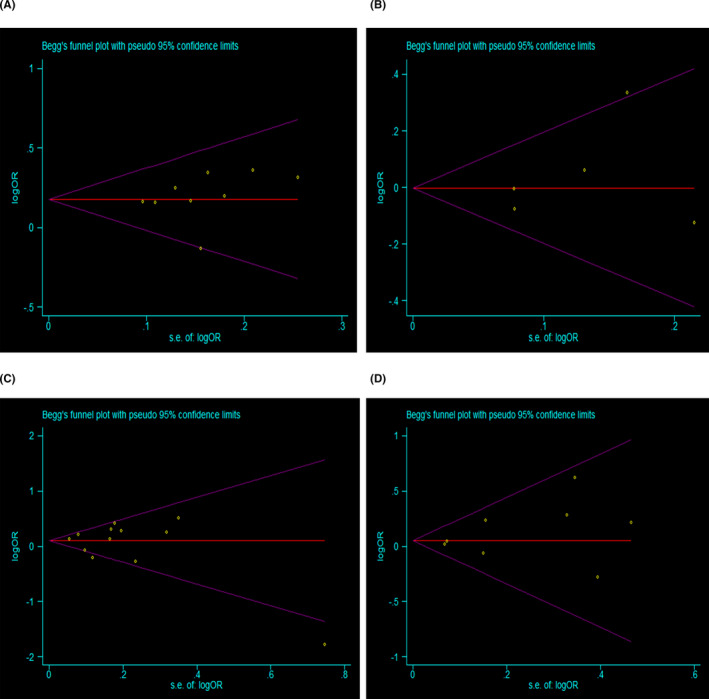
Begg's funnel plot of publication bias in the meta‐analysis of the association of MBL gene polymorphisms with RA risk. A, MBL gene polymorphisms and RA in Brazilian. B, MBL gene polymorphisms and RA in Caucasian. C, MBL gene polymorphisms and RA in East Asian. D, MBL gene polymorphisms and RA in Indian
FIGURE 6.

Sensitivity analysis to assess the stability of the meta‐analysis. A, MBL gene polymorphisms and RA in Brazilian. B, MBL gene polymorphisms and RA in Caucasian. C, MBL gene polymorphisms and RA in East Asian. D, MBL gene polymorphisms and RA in Indian
4. DISCUSSION
The comprehensive meta‐analysis confirmed that the biological roles of 5 loci in different ethnicities were distinct. It was verified that the structural polymorphisms in exon 1 of MBL gene may significantly contribute to susceptibility and development of RA in Brazilian and Indian populations, whereas the functional polymorphisms in the promoter region were more likely to associate with RA in East Asians.
MBL was structurally and functionally similar to C1q, and shared the same phagocytic receptor on phagocytes, platelets, and endothelial cells. 54 MBL plays a key role in the innate immune system by activating complements and macrophages, and by inducing opsonization. MBL mediates lectin‐dependent activation of the complement pathway, and resembles C1q in terms of structure and function. 55 Low serum levels of MBL may result in impaired opsonization of complement‐containing immune complexes. 56 The activation of MBL variants could contribute to damage tissue and consequently to disease severity. Inversely, deficiencies of complement proteins may enhance autoimmunity. 15 Considering that, the lectin pathway is involved in the clearance of pathogens and apoptotic bodies that may act as potential autoimmune initiators, deficiencies of components could enhance susceptibility and severity of some rheumatic disorders. 36 , 57 , 58 The functional MBL exon 1 codon 54 (allele B), codon 57 (allele C), and codon 52 (allele D) variants cause structural changes of the MBL basic unit, producing a lower molecular weight protein and reduced serum MBL levels. 16 Besides the exon 1 variant alleles, SNPs at promoter −550 (allele L) and −221 (allele X) have been associated with low serum MBL levels. 48
A low MBL level caused by MBL variant alleles has been associated with human immunodeficiency virus and hepatitis C virus infections, and with SLE. 13 , 15 , 59 Since MBL2 variants are the major determinants of MBL circulating levels, various studies demonstrated that variations on MBL serum levels seem to influence RA development and prognosis in different ways. 60 Although MBL2 low‐producing polymorphisms were associated with increased susceptibility to RA, disease progression and clinical manifestations, the B variant was reported to confer protection against RA in an Indian population. 8 High producing genotype YA/YA conferred an increased risk of myocardial infarction and death in RA patients with ischemic heart disease. 61 , 62 Similarly, high producing MBL2 genotypes enhanced the risk of cardiovascular disease in patients with rheumatic fever. 54 , 63 Nevertheless, no association between RA and MBL2 polymorphisms was reported by others. 64
In addition, a meta‐analysis was conducted with 8 researches by others and this found \ a significant association between the MBL D allele and RA in the overall population (OR = 1.708, P = .023). 42 An association was also found between the MBL L allele and RA in the overall group (OR = 1.936, P = .005), as well as between the MBL X allele and RA in the overall group (OR = 1.582, P = .001). Their meta‐analysis demonstrated an association between the MBL D, L, and X alleles and the risk of RA. However, the mixed ethnic population and limited sample size may make their results unreliable, or serious deviation from the real situation. Moreover, Stefanie et al. 35 also conducted a meta‐analysis and the results showed that MBL2 low‐producing OO and XX genotypes do not confer higher risk to RA, even when data were analyzed according to the cohort's ethnicity. Due to the diversity of MBL2 alleles and divergent concepts about high and low‐producing genotypes, they analyzed first only exon 1 polymorphisms and classified the data according to the presence of AA, AO and OO genotypes. Of course, so far, some research had reported that there was no association found between rs1800450 and RA, which was contradictory with our findings. 43 It is normal that such distinct consequences were obtained in separated studies. RA is considered to be a common multifactor autoimmune disease due to its complicated pathogenesis. It was validated that body mass index (BMI) and smoking will significantly contribute to susceptibility and development of RA. In addition, the gender difference was the key role in RA morbidity. However, lack of BMI level in participants might lead to inconsistent results. These phenomena and discrepancies need further investigation on the basis of large sample size. Moreover, the concentration of MBL may be regulated by other mechanisms than by variants on the MBL2 gene; additional studies including both polymorphisms and functional assays could give a better insight into the relationship between MBL and RA.
Although we revealed some new discoveries in this study, there were still several limitations which should be taken into consideration. In our study, the overall sample size is large, but the size of each study is relatively small; the smallest sample is 50 cases and 48 controls, and we need numerous data to validate the relationship between MBL SNPs and RA for further study in Caucasian populations. Second, in stratification analysis, the number of studies included in each ethnicity was unbalanced, some just for one study. Additionally, we are unable to analyze the actual impact of immanent factors on RA because of the incomplete data. Meanwhile, how the interaction of genes with environmental factors and genes with dietary models relate to the risk of RA is unclear. Further efforts should be put on investigating the association of the functional mutations in the MBL gene with RA, and the interactions of potential gene‐gene and gene‐environment factors should be comprehensively analyzed.
5. CONCLUSIONS
We conducted a meta‐analysis to evaluate the effects of MBL polymorphisms (rs1800450, rs1800541, rs5030737, rs11003125, rs7096206) on the risk of RA. The structural polymorphisms in exon 1 of MBL gene may significantly contribute to susceptibility and development of RA in Brazilian and Indian populations, whereas the functional polymorphisms in the promoter region were more likely to associate with RA in East Asians. Meanwhile, the reverse association between rs5030737 with RA in East Asians was displayed. However, the polymorphisms in exon 1 of MBL gene lacked the connection with RA.
CONFLICT OF INTERESTS
The authors declare they have no competing interests.
AUTHOR CONTRIBUTIONS
KQL and JJX made substantial contributions to the conception; JJX designed the work; GC, ZY and MCQ interpreted data; WTT for the main data analysis; XBZ created new software used in the work. LZ and YMZ drafted the work or substantively revised it. All authors reviewed and approved the final manuscript.
Xu J, Chen G, Yan Z, et al. Effect of mannose‐binding lectin gene polymorphisms on the risk of rheumatoid arthritis: Evidence from a meta‐analysis. Int J Rheum Dis. 2021;24:300–313. 10.1111/1756-185X.14060
Funding information
This study was supported by the National Natural Science Foundation of China (81871831), the Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars of China (LR17H070001). The funding agencies had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Calabresi E, Petrelli F, Bonifacio AF, et al. One year in review 2018: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol. 2018;36(2):175‐184. [PubMed] [Google Scholar]
- 2. Daniel A, Smolen JS. Diagnosis and management of rheumatoid arthritis. A review. JAMA. 2018;14(34):254‐261. [DOI] [PubMed] [Google Scholar]
- 3. Mizoguchi F, Slowikowski K, Wei K, et al. Functionally distinct disease‐associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9(1):789‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bae SC, Lee YH. Causal association between body mass index and risk of rheumatoid arthritis: a Mendelian randomization study. Eur J Clin Invest. 2019;49(4):126‐138. [DOI] [PubMed] [Google Scholar]
- 5. Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(20):257‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Auriti C, Prencipe G, Moriondo M, et al. Mannose‐binding lectin: biologic characteristics and role in the susceptibility to infections and ischemia‐reperfusion related injury in critically ill. J Immunol Res. 2017;8(6):11‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamamoto K, Okada Y, Suzuki A, et al. Genetics of rheumatoid arthritis in Asia‐present and future. Nat Rev Rheumatol. 2015;11(6):375‐379. [DOI] [PubMed] [Google Scholar]
- 8. Gupta B, Agrawal C, Raghav SK, et al. Association of mannose‐binding lectin gene (MBL2) polymorphisms with rheumatoid arthritis in an Indian cohort of case‐control samples. J Hum Genet. 2005;50(11):583‐591. [DOI] [PubMed] [Google Scholar]
- 9. Takahashi M, Tanii H, Sawada T, et al. Meta‐analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44(5):511‐516. [DOI] [PubMed] [Google Scholar]
- 10. Lee YH, Bae S‐C, Choi SJ, et al. Genome‐wide pathway analysis of genome‐wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39(12):10627‐10635. [DOI] [PubMed] [Google Scholar]
- 11. Liang Y, Ziming T, Bin Z, et al. Relationship between polymorphism of mannose binding lectin gene and susceptibility to Tuberculosis. Med Information. 2018;71(13):341‐349. [Google Scholar]
- 12. Laura C, Francis V, Frederic L, et al. Polymorphisms in the mannose‐binding lectin gene are associated with defective mannose‐binding lectin functional activity in Crohn's disease patients. Sci Rep. 2016;6(1):1641‐1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ji X, Gewurz H, Spear GT. Mannose binding lectin (MBL) and HIV. Mol Immunol. 2005;42(2):145‐152. [DOI] [PubMed] [Google Scholar]
- 14. Hamvas RMJ, Johnson M, Vlieger AM, et al. Role for mannose binding lectin in the prevention of Mycoplasma infection. Infect Immun. 2005;73(8):5238‐5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garred P, Voss A, Madsen HO, et al. Association of mannose‐binding lectin gene variation with disease severity and infections in a population‐based cohort of systemic lupus erythematosus patients. Genes Immun. 2001;2(8):442‐450. [DOI] [PubMed] [Google Scholar]
- 16. Goeldner I, Skare TL, Utiyama SR, et al. Mannose binding lectin and susceptibility to rheumatoid arthritis in Brazilian patients and their relatives. PLoS One. 2014;9(4):91‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hou L, Wang Y, Zhou Y. Study on the correlation between MBL gene single nucleotide polymorphism and rheumatoid arthritis. Sci Technol Vis. 2020;7(3):146‐149. [Google Scholar]
- 18. Horiuchi T, Tsukamoto H, Morita C, et al. Mannose binding lectin (MBL) gene mutation is not a risk factor for systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) in Japanese. Genes Immun. 2000;1(7):464‐466. [DOI] [PubMed] [Google Scholar]
- 19. Martiny FL, Veit TD, Brenol CV, et al. Mannose‐binding lectin gene polymorphisms in Brazilian patients with rheumatoid arthritis. J Rheumatol. 2012;39(1):6‐9. [DOI] [PubMed] [Google Scholar]
- 20. Stanworth SJ, Donn RP, Hassall A, et al. Absence of an association between mannose‐binding lectin polymorphism and rheumatoid arthritis. Br J Rheumatol. 1999;37(2):186‐188. [DOI] [PubMed] [Google Scholar]
- 21. Teng J, Ye J, Zhou Z, et al. A comparison of the performance of the 2019 European League Against Rheumatism/American College of Rheumatology criteria and the 2012 Systemic Lupus International Collaborating Clinics criteria with the 1997 American College of Rheumatology classification criteria for systemic lupus erythematous in new‐onset Chinese patients. Lupus. 2020;29(6):617‐624. [DOI] [PubMed] [Google Scholar]
- 22. Xu JJ, Liu KQ, Ying ZM, et al. Effect of CD14 polymorphisms on the risk of cardiovascular disease: evidence from a meta‐analysis. Lipids Health Dis. 2019;18(74):156‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bracken MB. Meta‐analysis requires independent observations and freedom from bias. Br J Clin Pharmacol. 2016;81(6):56‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van de Geijn FE, Hazes JMW, Geleijns K, et al. Mannose‐binding lectin polymorphisms are not associated with rheumatoid arthritis–confirmation in two large cohorts. Rheumatology (Oxford). 2008;47(5):1168‐1171. [DOI] [PubMed] [Google Scholar]
- 25. Hamvas RMJ, Turner MW. Mannose‐binding lectin: structure, function, genetics and disease associations. Rev Immunog. 2000;2(3):305‐322. [PubMed] [Google Scholar]
- 26. Lin L, Chu H. Quantifying publication bias in meta‐analysis. Biometrics. 2018;74(3):2‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meisner J, Albrechtsen A. Testing for Hardy‐Weinberg equilibrium in structured populations using genotype or low‐depth next generation sequencing data. Mol Ecol Resour. 2019;19(5):1144‐1152. [DOI] [PubMed] [Google Scholar]
- 28. Piepho HP, Madden LV, Roger J, et al. Estimating the variance for heterogeneity in arm‐based network meta‐analysis. Pharm Stat. 2018;17(3):33‐37. [DOI] [PubMed] [Google Scholar]
- 29. Steyerberg EW, Nieboer D, Debray TPA, et al. Assessment of heterogeneity in an individual participant data meta‐analysis of prediction models: an overview and illustration. Stat Med. 2019;38(22):234‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoneoka D, Henmi M. Clinical heterogeneity in random‐effect meta‐analysis: between‐study boundary estimate problem. Stat Med. 2019;38(21):4131‐4145. [DOI] [PubMed] [Google Scholar]
- 31. Downey LA, Guzzetta NA. A problem of too much heterogeneity. Anest Analg. 2020;130(6):1591‐1593. [DOI] [PubMed] [Google Scholar]
- 32. Pustejovsky JE, Rodgers MA. Testing for funnel plot asymmetry of standardized mean differences. Res Synth Methods. 2019;10(1):57‐71. [DOI] [PubMed] [Google Scholar]
- 33. Gjerdevik M, Heuch I. Improving the error rates of the Begg and Mazumdar test for publication bias in fixed effects meta‐analysis. BMC Med Res Methodol. 2014;14(109):2‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barton A, Platt H, Salway F, et al. Polymorphisms in the mannose binding lectin (MBL) gene are not associated with radiographic erosions in rheumatoid or inflammatory polyarthritis. J Rheumatol. 2004;31(3):442‐447. [PubMed] [Google Scholar]
- 35. Epp Boschmann S, Goeldner I, Tuon FF, et al. Mannose‐binding lectin polymorphisms and rheumatoid arthritis: a short review and meta‐analysis. Mol Immunol. 2016;69(2):77‐85. [DOI] [PubMed] [Google Scholar]
- 36. Freudenberg J, Lee AT, Siminovitch KA, et al. Locus category based analysis of a large genomewide association study of rheumatoid arthritis. Hum Mol Genet. 2010;19(19):3863‐3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maury CPJ, Aittoniemi J, Tiitinen S, Laiho K, Kaarela K, Hurme M. Variant mannose‐binding lectin 2 genotype is a risk factor for reactive systemic amyloidosis in rheumatoid arthritis. J Intern Med. 2007;262(4):466‐469. [DOI] [PubMed] [Google Scholar]
- 38. Jacobsen S, Garred P, Madsen H, et al. Mannose‐binding lectin gene polymorphisms are associated with disease activity and physical disability in untreated, anti‐cyclic citrullinated peptide‐positive patients with early rheumatoid arthritis. J Rheumatol. 2009;36(4):731‐735. [DOI] [PubMed] [Google Scholar]
- 39. Garred P, Madsen HO, Marquart H, et al. Two edged role of mannose binding lectin in rheumatoid arthritis: a cross sectional study. J Rheumatol. 2000;27(1):26‐34. [PubMed] [Google Scholar]
- 40. Gupta B, Raghav SK, Agrawal C, Chaturvedi VP, Das RH, Das HR. Anti‐MBL autoantibodies in patients with rheumatoid arthritis: prevalence and clinical significance. J Autoimmun. 2006;27(2):125‐133. [DOI] [PubMed] [Google Scholar]
- 41. Song GG, Bae S‐C, Seo YH, et al. Meta‐analysis of functional MBL polymorphisms. Z Rheumatol. 2014;73(7):657‐664. [DOI] [PubMed] [Google Scholar]
- 42. Wang H, Li S‐L, Zhu J, et al. The association of mannose‐binding lectin genetic polymorphisms with the risk of rheumatoid arthritis: a meta‐analysis. J Recept Signal Transduct Res. 2015;35(6):357‐362. [DOI] [PubMed] [Google Scholar]
- 43. Xie Q, Wang S‐C, Bian G, et al. Association of MIF−173G/C and MBL2 codon 54 gene polymorphisms with rheumatoid arthritis: a meta‐analysis. Hum Immunol. 2012;73(9):966‐971. [DOI] [PubMed] [Google Scholar]
- 44. Zhang C, Zhang C, Zhu J, et al. The association of mannose‐binding lectin genetic polymorphisms with the risk of rheumatoid arthritis: a meta‐analysis. J Recept Signal Transduct. 2015;35(4):357‐362. [DOI] [PubMed] [Google Scholar]
- 45. Dolman KM, Brouwer N, Frakking FN, et al. Mannose‐binding lectin deficiency is associated with early onset of polyarticular juvenile rheumatoid arthritis: a cohort study. Arthritis Res Ther. 2008;10(2):32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ip WK, Lau YL, Chan SY, et al. Mannose‐binding lectin and rheumatoid arthritis in southern Chinese. Arthritis Rheum. 2000;43(8):1679‐1687. [DOI] [PubMed] [Google Scholar]
- 47. Ling H, Donghua Y, Jinrui X. Study on the relationship between MBL gene mutation and rheumatoid arthritis in Ningxia. Modern. Prev Med. 2013;40(14):2710‐2713. [Google Scholar]
- 48. Malthe K, Morten F, Hans OM, et al. Smoking and polymorphisms of genes encoding mannose‐binding lectin and surfactant protein‐D in patients with rheumatoid arthritis. Rheumatol Int. 2014;34(3):373‐380. [DOI] [PubMed] [Google Scholar]
- 49. Kang M, Wang H. Peixuan C. Study of single nucleotide polymorohisms in th promoter of mannose‐binding lectin gene in patients with juvenile idiopathic arthritis. Chin J Eugenics Genet. 2006;5(4):21‐22. [Google Scholar]
- 50. Jacobsen S, Madsen HO, Klarlund M, et al. The influence of mannose binding lectin polymorphisms on disease outcome in early polyarthritis. TIRA Group. J Rheumatol. 2001;28(5):935‐942. [PubMed] [Google Scholar]
- 51. Jacobsen S, Madsen HO, Klarlund M, et al. The influence of mannose binding lectin polymorphisms on disease outcome in early polyarthritis. J Rheumatol. 2001;28(5):935‐942. [PubMed] [Google Scholar]
- 52. Saevarsdottir S, Vikingsdottir T, Vikingsson A, et al. Low mannose binding lectin predicts poor prognosis inpatients with early rheumatoid arthritis. A prospective study. J Rheumatol. 2001;28(4):728‐734. [PubMed] [Google Scholar]
- 53. Tsutsumi A, Sasaki K, Wakamiya N, et al. Mannose‐binding lectin gene: polymorphisms in Japanese patients with systemic lupus erythematosus, rheumatoid arthritis and Sjögren’s syndrome. Genes Immun. 2001;2(2):99‐104. [DOI] [PubMed] [Google Scholar]
- 54. Baspinar O, Balat A, Sever T, et al. Association of macrophage migration inhibitory factor and mannose‐binding lectin‐2 gene polymorphisms in acute rheumatic fever. Cardiol Young. 2013;23(4):486‐490. [DOI] [PubMed] [Google Scholar]
- 55. Coelho AVC, Brandão LAC, Guimarães RL, et al. Mannose binding lectin and mannose binding lectin‐associated serine protease‐2 genes polymorphisms in human T‐lymphotropic virus infection. J Med Virol. 2013;85(10):1829‐1835. [DOI] [PubMed] [Google Scholar]
- 56. Figueiredo GG, Cezar RD, Freire NM, et al. Mannose‐binding lectin gene (MBL2) polymorphisms related to the mannose‐binding lectin low levels are associated to dengue disease severity. Immuno genetics. 2016;77(7):571‐575. [DOI] [PubMed] [Google Scholar]
- 57. Freudenberg J, Lee HS, Han BG, et al. Genome‐wide association study of rheumatoid arthritis in Koreans: Population‐specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63(4):884‐893. [DOI] [PubMed] [Google Scholar]
- 58. Diogo D, Okada Y, Plenge RM. Genome‐wide association studies to advance our understanding of critical cell types and pathways in rheumatoid arthritis: recent findings and challenges. Curr Opin Rheumatol. 2014;26(1):85‐92. [DOI] [PubMed] [Google Scholar]
- 59. Azeem WAE, Faried AA, Mahmoud EA, Diab KA. Relationship between mannose‐binding lectin‐2 gene polymorphism and CD25 with hepatocellular carcinoma‐induced hepatitis‐C development. Menoufia Med J. 2017;30(4):1203‐1209. [Google Scholar]
- 60. Özerkan K, Oral B, Uncu G. Mannose‐binding lectin levels in endometriosis. Fertil Steril. 2009;94(2):775‐776. [DOI] [PubMed] [Google Scholar]
- 61. Troelsen LN, Garred P, Madsen HO, et al. Genetically determined high serum levels of mannose‐binding lectin and agalactosyl IgG are associated with ischemic heart disease in rheumatoid arthritis. Arthritis Rheum. 2007;56(1):21‐29. [DOI] [PubMed] [Google Scholar]
- 62. Troelsen LN, Garred P, Jacobsen S. Mortality and predictors of mortality in rheumatoid arthritis–a role for mannose‐binding lectin? J Rheumatol. 2010;37(3):536‐543. [DOI] [PubMed] [Google Scholar]
- 63. Gomaa MH, Ali SS, Fattouh AM, et al. MBL2 gene polymorphism rs1800450 and rheumatic fever with and without rheumatic heart disease: an Egyptian pilot study. Pediatr Rheumatol. 2018;16(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Best LG, Davidson M, North KE, et al. Prospective analysis of mannose‐binding lectin genotypes and coronary artery disease in American Indians: the Strong Heart Study. Circulation. 2004;109(4):471‐475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


