Abstract
An optimal clinical specimen for accurate detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) by minimizing the usage of consumables and reduce hazard exposure to healthcare workers is an urgent priority. The diagnostic performance of SARS‐CoV‐2 detection between healthcare worker‐collected nasopharyngeal and oropharyngeal (NP + OP) swabs and patient performed self‐collected random saliva was assessed. Paired NP + OP swabs and random saliva were collected and processed within 48 h of specimen collection from two cohort studies which recruited 562 asymptomatic adult candidates. Real‐time reverse‐transcription polymerase chain reaction targeting Open reading frame 1a (ORF1a) and nucleocapsid (N) genes was performed and the results were compared. Overall, 65 of 562 (28.1%) candidates tested positive for COVID‐19 based on random saliva, NP + OP swabs, or both testing techniques. The detection rate of SARS‐CoV‐2 was higher in random saliva compared to NP + OP testing (92.3%; 60/65 vs. 73.8%; 48/65; p < .05). The estimated sensitivity and specificity of random saliva were higher than NP + OP swabs (95.0; 99.9 vs. 72.2; 99.4). The C t values of ORF1a and N genes were significantly lower in random saliva compared to NP + OP swabs specimens. Our findings demonstrate that random saliva is an alternative diagnostic specimen for the detection of SARS‐CoV‐2. Self‐collected random oropharyngeal saliva is a valuable specimen that provides accurate SARS‐CoV‐2 surveillance testing of a community.
Keywords: COVID‐19, nasopharyngeal and oropharyngeal swabs, pandemic, random saliva, SARS‐CoV‐2, surveillance testing
1. INTRODUCTION
An outbreak of coronavirus disease (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has been spreading worldwide since late December 2019. 1 The number of infected SARS‐CoV‐2 cases worldwide has been rising dramatically despite various control measures. Globally, as of October 17, 2020, there have been 39,196,256 laboratory‐confirmed cases of COVID‐19, including 1,101,298 deaths reported to the World Health Organization (WHO). 2
Detection of SARS‐CoV‐2 in a population is an urgent priority for the prevention and containment of disease outbreaks in the community. This is due to the nature of illness characteristics where a large portion of the infected population is asymptomatic. 3 , 4 Collecting optimal clinical specimens for detection of SARS‐CoV‐2 is central in controlling the pandemic. 5
SAR‐CoV‐2 has been detected in various clinical specimens. 6 , 7 The current standard SARS‐CoV‐2 testing for surveillance as recommended by WHO is solely depending on nasopharyngeal (NP) and oropharyngeal (OP) swabs. 8 However, these sampling techniques have been proved to show relatively poor sensitivity and inconsistent. Moreover, these sampling techniques are known for their invasiveness and being patient unfriendly, which may limit compliance for repeat testing and expose healthcare workers to the detrimental virus or other pathogens. 7 , 9 , 10
Saliva has been reported as a better alternative to NP + OP swabs for detection of SARS‐CoV‐2. 11 , 12 , 13 , 14 In our previous study, saliva has been proved to have a better detection rate of SARS‐CoV‐2 among COVID‐19 patients. Other than that, it is less invasive, patient‐friendly, negates direct interaction of healthcare worker‐patient, and cost‐efficient by easing the supply demands on swabs, personal protective equipment (PPE), and manpower. 15 However, there are possible limitations in the previous study as study subjects were COVID‐19 confirmed cases.
Therefore, in our current study, we evaluated SARS‐CoV‐2 detection in paired NP + OP swabs and random saliva samples collected from asymptomatic persons under surveillance of the mass screening population.
2. METHODS
We conducted a large‐scale two cohort screening study to assess the comparability between nasopharyngeal and oropharyngeal (NP + OP) swabs and self‐collected random deep throat saliva for detection of SARS‐CoV‐2 via real‐time reverse‐transcription polymerase chain reaction (qRT‐PCR) among asymptomatic individuals.
2.1. Study design and population
A cross‐sectional two cohort diagnostic study was conducted on candidates who were undergoing surveillance testing for SARS‐CoV‐2 in Pokok Sena detention center at Kota Setar District, Kedah between October 3rd till October 10th, 2020 after detection of a new COVID‐19 cluster in this center 16 and among asymptomatic travelers arriving at Kuala Lumpur International Airport (KLIA) throughout the month of September 2020. The inclusion criteria for candidate selection were: (i) those above 18 years old, (ii) able to obey commands, and (iii) asymptomatic. Assent and written informed consent were obtained from the study candidates.
Candidates’ sociodemographic and symptoms at the time of sampling were collected. As a standard protocol, NP and OP swabs from candidates were collected using sterile flocked swabs and placed in a sterile tube containing viral transport medium (VTM). Before that, candidates were briefed to provide self‐collected random deep throat/oropharyngeal saliva by clearing the throat into a sterile collection container. Briefly, candidates were instructed to avoid food and water for 1 h during the collection of 2 ml of deep throat saliva. The time span taken between random saliva and NP + OP swabs was approximately 2–3 h. Samples were stored at 4–8°C by packing in a polystyrene box filled with ice cubes and the cold chain was maintained throughout the transport to the central laboratory. Samples collected from Pokok Sena district were transited at Makmal Kesihatan Awam Ipoh (MKAI) by keeping them in a cold room for approximately 1–2 h before transferred to a central laboratory, within 6–9 h. However, samples from Kuala Lumpur International Airport, KLIA were transferred to the central laboratory directly within 3–4 h. Samples were processed and analyzed within 48 h upon arrival at the research laboratory (Institute for Medical Research).
2.2. SARS‐COV‐2 detection via RT‐PCR assay
On arrival at the research lab, all clinical specimens were inactivated at 65°C for 15 min in the Biosafety cabinet. Congealed or viscous saliva specimens were diluted in 500 μl of normal saline. Total nucleic acid extraction of 200 μl of VTM containing both NP and OP swabs or 200 μl of saliva was performed using the Magna Pure 96 system with the Magna Pure 96 DNA and Viral NA Small Volume extraction kit (Roche Diagnostic GmBH). The extracted RNA was then eluted in 50 μl of elution buffer. A known positive sample from our routine screening was used as a substitute for external positive control in every batch of extraction to ensure the quality of RNA extraction. For SARS‐CoV‐2 RNA detection, 5 μl of RNA template was tested using one‐step RT‐PCR of a DiaPlexQ™ Novel Coronavirus (2019‐nCoV) Detection Kit (Solgent Co., Ltd.), which has been certified for in vitro diagnostic product use by International Medical Device Regulators Forums (IMDRF) jurisdiction. This kit uses probes and primers targeting the nucleocapsid (N) gene and open reading frame 1a (ORF1a) gene of SARS‐CoV‐2 and PCRC gene as an internal control (IC). As we lacked a reliable quantified positive control in our laboratory, we adopted the limit of detection (LOD) as described in the kit instruction (C t: 38.1 ± 1.3 of NP and 36.2 ± 0.98 of sputum/saliva for 40 viral copies/ml). Samples were classified as positive for SARS‐CoV‐2 when either one or both gene primer‐probe sets were detected at a C t value of <40 as per kit's instruction.
2.3. Data processing and analysis
Data were analyzed for normality and descriptive statistics were described as a number (%) for categorical variables and mean ± standard deviation (SD) or median (interquartile range [IQR]) for continuous variables. McNemar's test was used to compare the detection rate for two sampling methods in terms of the number of patients. The kappa statistic was used to assess the interrater reliability between two sampling methods. C t values of concordance results were compared using paired t‐test. The correlation of concordance results was assessed by using the Pearson correlation coefficient. A p < .05 was considered statistically significant. All statistical analysis was performed using Jamovi software Version 1.2.22.0. 17
2.4. The accuracy of the diagnostic test
Assuming the current standard test (NP + OP swabs) is of unknown accuracy, invasive, and specialized personnel dependent, a Bayesian Latent Class Model was used to estimate the true sensitivity and specificity of each diagnostic method. Two tests in the two populations model were used. This statistical model was analyzed by using an open‐access web‐based application “Modelling for Infectious disease Center, MICE.” 18 Noninformative prior distribution is used for all parameters (beta distribution (0.5,0.5)) and probable ranges of specificity of all diagnostic tests in the model are between 0.4 and 1. Positive results of either test specimen (NP + OP swab) or random saliva was assumed as a perfect gold standard.
3. RESULTS
3.1. Candidates demographics
Of the 660 candidates screened, 584 (88.5%) candidates consented to the study. However, only 562 (85.2%) candidates were included in the analysis (Figure 1). The reasons for exclusion were symptomatic candidates (n = 22, 3.3%) and underage candidates (n = 76, 11.5%). Paired samples of NP + OP swabs and saliva were collected from 562 candidates. One hundred‐seventy (30.2%) candidates were women. The median (IQR) age was 33 (25–42) years. All candidates were asymptomatic at the time of sampling. The demographic characteristics of the studied candidates are shown (Table 1).
Figure 1.
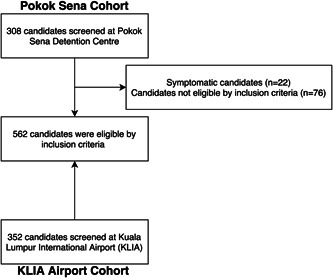
Screened and eligible candidates for the study
Table 1.
Characteristic of studied candidates
| Characteristic | Results | p value |
|---|---|---|
| Age (years), median (IQR) | ||
| Overall, n = 562 | 33 (25–42) | <.05 |
| COVID‐19, n = 65 | 31 (26–41) | |
| Non‐COVID‐19, n = 497 | 33 (29–42) | |
| Gender, male, n (%) | ||
| Overall | 392 (69.8%) | <.05 |
| COVID‐19 | 59 (15.1%) | |
| Non‐COVID‐19 | 333 (84.9%) |
Abbreviations: COVID‐19, coronavirus disease 2019; IQR, interquartile range.
3.2. Comparison of SARS‐CoV‐2 detection between NP + OP swabs and random deep throat saliva
Of 562 eligible candidates, 11.6% (65 of 562) of candidates tested positive for SARS‐CoV‐2 either by NP + OP swab, random saliva, or both. There was an overall significant substantial agreement between the two sampling methods (96.1%; 65 of 562, κ coefficient 0.78, 95% CI 0.69–0.87, p < .05). Among candidates with concordant results, 66.2% (43 of 65) had the virus detected in both random saliva and NP + OP swabs. However, 33.8% (22 of 65) candidates had discordant results between random saliva and NP + OP swabs by which 26.2% (17 of 65) candidates had the virus detected in random saliva but not in NP + OP swabs and 7.7% (5 of 65) candidates had the virus detected in NP + OP swabs but not in random saliva. The detection rate of SARS‐CoV‐2 virus in random saliva was significantly higher than of NP + OP swab (92.3%; 60/65 vs. 73.8%; 48/65; p < .05). The PCRC gene (IC) was detectable in all specimens. Data of the overall study are available in Table S3.
3.3. Viral load comparison between NP + OP swabs and random deep throat saliva
The median (IQR) C t values of N and ORF1a genes were 30.6 ± 7.06 and 30.0 ± 6.86, respectively in NP + OP swabs, and 29.2 ± 7.87 and 28.5 ± 7.70, respectively, in random saliva. Among candidates with concordant results, there was a significant average difference between C t values of N and ORF1a genes of random saliva and NP + OP swabs (t N‐gene = 5.07, p < .05; t ORF1a‐gene = 5.39, p < .05). On average, C t values of N and ORF1a genes of saliva were 3.44 (95% CI, 2.07–4.81) and 3.45 (95% CI, 2.16–4.73) lower than NP + OP swabs. Both C t values of random saliva and NP + OP swabs were moderately and positively correlated (r N‐gene = 0.80, p < .05; r ORF1a ‐gene = 0.82, p < .05). Data on concordant samples available in Tables S5 and S6.
3.4. The accuracy of diagnostic tests by Bayesian latent class model (LCM)
The sensitivity of NP + OP swab and random saliva were 72.2% (95% credible interval, 60.0–82.6) and 95.0% (95% credible interval, 83.8–100), respectively. Meanwhile, the specificity of NP + OP swab and random saliva was 99.4% (95% credible interval, 98.2–100) and 99.9% (95% credible interval, 98.9–100), respectively. The prevalence estimated by Bayesian LCM of the Pokok Sena outbreak population and KLIA traveler population were 28.0 (95% CI, 22.1–34.7) and 1.2 (95% CI, 0.4–2.9), respectively (Table S4). Table 2 describes the statistical and Bayesian latent class model findings.
Table 2.
Kappa coefficient (κ), agreement (%), McNemar test, prevalence, sensitivities, and specificities estimated by assuming either NP + OP swabs or random saliva as a perfect gold standard
| Cohort study | Pokok Sena detention centre (n = 210) | Airport travelers (n = 352) |
|---|---|---|
| Total positive case, (n) | 59 | 6 |
| Kappa coefficient, κ (p) | 0.75 (p < .05) | 0.66, (p < .05) |
| Agreement, (%) | 91.0 | 99.1 |
| McNemar test, (p) | <.05 | .564 |
| Prevalence by Bayesian LCM, (95% credible interval) | 28.0 (22.1–34.7) | 1.2 (0.4–2.9) |
| Nasopharygneal and oropharyngeal swabs (NP + OP swabs) (95% credible interval): | ||
| (i) Sensitivity | 72.2 (60.0–82.6) | |
| (ii) Specificity | 99.4 (98.2–100) | |
| (iii) PPV | 93.9 (82.6–99.7) | |
| (iv) NPV | 96.6 (94.6–98.0) | |
| Random saliva (95% credible interval): | ||
| (i) Sensitivity | 95.0 (83.8–100) | |
| (ii) Specificity | 99.9 (98.9–100) | |
| (iii) PPV | 98.9 (91.0–100) | |
| (iv) NPV | 99.4 (97.8–100) | |
4. DISCUSSION
Rapid detection of asymptomatic carriers is central in controlling and mitigating the global pandemic. Collecting an optimal clinical specimen that offers a high positivity rate and minimizes the risk of viral transmission to healthcare workers as well as a cost‐effective method in resource‐limited settings are indeed in demand in the current pandemic. 5 , 19 This study assessed the detection rate of SARS‐CoV‐2 by qRT‐PCR using NP + OP swabs as recommended by the interim guideline and random saliva specimens in the surveillance of asymptomatic candidates. 20
Our results demonstrated the value of testing random saliva as a noninvasive and easily obtained specimen for detection of SARS‐CoV‐2 in mass screening. Overall, our results showed a high detection rate of the virus in random saliva with comparable performance to the current standard technique of collecting NP + OP swabs. A significant substantial agreement and κ coefficient were observed between these two sampling methods suggesting high concordance between results (p < .05), as shown in earlier studies. 14 , 21 This finding differs from our previous study 15 which focused on asymptomatic positive COVID‐19 cases and had lower concordance due to the study design.
Previous studies comparing the viral load between NP + OP and saliva specimens report paradoxical results. A few studies have demonstrated that viral load in saliva is higher in comparison to NP + OP swabs, 11 , 15 while others have reported equivalent viral load between these two specimens. 12 , 13 Nonetheless, a few studies have demonstrated that saliva is less sensitive in comparison to NP + OP sampling. 22 , 23 Our findings among the concordance showed that on average the viral load in saliva is higher than the standard sampling technique. In addition to that, our results were not affected by changing early morning saliva to random saliva as compared to our previous study. 15 However, we presume the discrepancy between studies performed in other countries is possibly due to distinct sampling techniques, detection kit, study population, and prevalence of the disease.
Nevertheless, 1/3 (32.2%) of the studied candidates showed discordant results for detection of SARS‐CoV‐2. Of which 17 of the studied candidates had their random saliva test positive for SARS‐CoV‐2 while their NP + OPS were negative. These could be due to the property of saliva which acts as inhibitor for RNA decomposition. 24 It is also possible that nasopharyngeal debris (including virus RNA) drained into the oropharynx and mixed with saliva or the salivary gland, the tongue and oropharyngeal mucosa being an entry point for the virus to replicate as described by Azzi et al. 25 On the other hand, we had five candidates who were positive for SARS‐CoV‐2 by NP + OP but tested negative for saliva. Of these five NP + OP specimens, the average C t value of ORF1a and N genes were 31.9 ± 6.03 and 32.5 ± 7.23, respectively. However, the C t value of these genes in random saliva were out of range for the kit's interpretation. This could further be explained by dilution effect of normal saline to the saliva as the samples were congeal and viscous during extraction process. We presume the C t value of these samples as described in our study are possibly affected by dilutional effect, which may add up to false‐negative results.
Previous studies reported the paradoxical sensitivity of saliva over nasopharyngeal swab for detection of SARS‐CoV‐2. 26 , 27 These problems were due to disease misclassification by the imperfect gold standard. However, the airport cohort was inadequate to confirm random saliva as a better alternative than NP + OP swabs due to lower prevalence among KLIA travelers. In contrast, the sensitivity and specificity values estimated by our study are unbiased because they are based on the true status of candidates predicted by the Bayesian LCM. The true sensitivity of random saliva for detecting SARS‐CoV‐2 estimated by Bayesian LCM was 95.0% which is higher than 72.2% estimated for NP + OP swabs. In addition, the Bayesian LCM also estimated that the true specificity of random saliva was higher than NP + OP swabs similarly to Isao et al. 21
Other than sharing diagnostic benefits, self‐collected random saliva has significant advantages over NP + OP swabs mainly in the setting of mass screening. Self‐collected random saliva is noninvasive which may enhance recruitment of individuals for community surveillance, requires no trained personnel for collection, able to scale down nosocomial hazards by negating the need for direct healthcare worker‐patient interaction and cost effective by easing the supply demands on swabs, PPE, and manpower. Overall, in the long run, as the current pandemic, saliva abates the demands on swabs and PPE, 28 reduces plastic pollution as a result of COVID‐19 pandemic 29 and reduces the burnout rate of healthcare workers which ensures optimal patient care. 30
4.1. Limitation of this study
In this study, we only recruited adult candidates, therefore, further evaluation should be conducted in the pediatric population. Secondly, the spectrum of the disease ranges from asymptomatic to severely ill patients but our study only focused on homogenously composed asymptomatic carriers. This is due to our priority of identifying asymptomatic carriers that is in parallel with the prevention and containment of disease outbreaks in the communities. Thirdly, the random saliva collected were not screened microscopically to evaluate the quality of saliva. Lastly, the C t value in this study portrays a trend in viral load but not the viral copies per ml as we lacked a reliable quantified positive control in our laboratory.
5. SUMMARY AND CONCLUSION
Random deep throat/oropharyngeal saliva is an alternative for nasopharyngeal and oropharyngeal swab for detection of SARS‐CoV‐2 via qRT‐PCR assay as it has a better sensitivity and detection rate. Self‐collected random deep throat saliva provides accurate results for the diagnosis of COVID‐19 and could enable surveillance testing.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHORS CONTRIBUTIONS
Mohan Rao, Rohaidah Hashim, and Norazah Ahmad designed the study. Mawaddah Ghazali, Anizan A. Manaf, Harishah Talib, Nurul A. Abdullah, Hanisah Ahmad, S. L. Aren, and Shareh A. S. Ali collected the samples from the field. Fairuz A. Rashid, Fashihah S. A. H. Sabri, Nur Nadia Jamil, Valentinus Seradja processed the samples. Rozainanee Zain, Ravindran Thayan, Fairuz Amran, Tahir Aris analyzed the data. Mohan Rao, Rohaidah Hashim, Norazah Ahmad wrote the paper with input from all authors.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
We are grateful to the Institute for Medical Research, Kuala Lumpur, Kuala Lumpur International Airport Health Office, and Kota Setar District Health Office for their support and facilities. The authors would like to thank the Director‐General of Health Malaysia for allowing us to publish our findings. This study obtained approval from the National Health Institute Human Research Ethics Committee, Ministry of Health Malaysia (KKM/NIHSEC/P20‐1935). Although this study did not involve any types of intervention in diagnosis and treatment, written informed consent was deemed necessary. It involves the use of anonymized candidate medical data and only socio‐demographic data were obtained. Therefore, there was no breach of privacy or confidentiality of the studied candidates. This study only reviewed human candidates and no animals were involved in any aspect of the study. This study was supported by a grant from the Ministry of Health Malaysia Research Grant (NMRR‐20‐1821‐56223).
Rao M, Rashid FA, Sabri FSAH, et al. COVID‐19 screening test by using random oropharyngeal saliva. J Med Virol. 2021;93:2461–2466. 10.1002/jmv.26773
DATA AVAILABILITY STATEMENT
Raw data of this study are available upon request.
REFERENCES
- 1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020; 395(10223):470‐473. 10.1016/S0140-6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization . WHO coronavirus disease (COVID‐19) dashboard. Covid‐19 Dashboard. 2020:1‐1. https://covid19.who.int/. Accessed June 3, 2020. [Google Scholar]
- 3. Chen Q, Zheng Z, Zhang C, et al. Clinical characteristics of 145 patients with corona virus disease 2019 (COVID‐19) in Taizhou, Zhejiang, China. Infection. 2020;1:3. 10.1007/s15010-020-01432-5. Accessed June 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Quilty BJ, Clifford S, Flasche S, Eggo RM. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019‐nCoV). Eurosurveillance. Vol. 25. European Centre for Disease Prevention and Control (ECDC); 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization . Guidelines for the collection of clinical specimens during field investigation of outbreaks WHO/CDS/CSR/EDC/2000.4. World Health Organ. 2000:1‐51. http://www.who.int/emc Accessed June 22, 2020. [Google Scholar]
- 6. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. J Am Med Assoc. 2020; 323(18):2‐3. https://jamanetwork.com/journals/jama/fullarticle/2762997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. WHO Use of Laboratory Methods for SARS Diagnosis. http://www.who.int/csr/sars/labmethods/en/. Accessed October 18, 2003.
- 9. To KK‐W, Tsang OT‐Y, Leung W‐S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469.Accessed October 18, 2020. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 11. Wyllie AL, Fournier J, Casanovas‐Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS‐CoV‐2. N Engl J Med. 2020;383:1283‐1286. http://www.ncbi.nlm.nih.gov/pubmed/32857487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chau NVV, Lam VT, Dung NT, et al. The natural history and transmission potential of asymptomatic SARS‐CoV‐2 infection. medRxiv. 2020. https://www.medrxiv.org/content/10.1101/2020.04.27.20082347v1 [Google Scholar]
- 13. Jamal AJ, Mozafarihashjin M, Coomes E, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin Infect Dis. 2020.2000‐xxxx 1‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pasomsub E, Watcharananan SP, Boonyawat K, et alSaliva sample as a non‐invasive specimen for the diagnosis of coronavirus disease 2019: a cross‐sectional study. Clin Microbiol Infect. 2020. http://www.clinicalmicrobiologyandinfection.com/article/S1198743X20302780/fulltext. Accessed June 23, 2020. [DOI] [PMC free article] [PubMed]
- 15. Rao M, Rashid FA, Sabri FSAH, et al. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS‐CoV‐2. Clin Infect Dis. 2020. https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa1156/5882012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. CARVALHO M. Covid‐19: New cluster identified in Kedah after death of man in emergency ward. The Star . 2020. https://www.thestar.com.my/news/nation/2020/09/30/covid-19-new-cluster-identified-in-kedah-after-death-of-man-in-emergency-ward. Accessed October 16, 2020.
- 17. The jamovi project. jamovi—Stats. Version 1.2; 2020. https://www.jamovi.org/. Accessed June 22, 2020.
- 18. Lim C, Wannapinij P, White L, et al. Using a web‐based application to define the accuracy of diagnostic tests when the gold standard is imperfect. PLOS One. 2013; 8(11). http://mice.tropmedres.ac [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS‐CoV‐2. J Infect. 2020;1‐6. 10.1016/j.jinf.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson ML. General principles of specimen collection and transport. Clin Infect Dis. 1996;22(5):766‐777. https://academic.oup.com/cid/article-abstract/22/5/766/360981 [DOI] [PubMed] [Google Scholar]
- 21. CDC . Interim Guidelines for Clinical Specimens for COVID‐19. Centers for Disease Control and Prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed June 22, 2020.
- 22. Yokota I, Peter, Shane Y , et al. Mass screening of asymptomatic persons for SARS‐CoV‐2 using saliva. 2020. https://www.academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa1388/5911780. Accessed October 20, 2020.
- 23. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non‐invasive specimen for detection of SARS‐CoV‐2. J Clin Microbiol. 2020;50. https://jcm.asm.org/content/early/2020/04/17/JCM.00776-20. Accessed June 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Becker D, Sandoval E, Amin A, et al. Saliva is less sensitive than nasopharyngeal swabs for COVID‐19 detection in the community setting. medRxiv. 2020:1‐15. https://www.medrxiv.org/content/10.1101/2020.05.11.20092338v2 [Google Scholar]
- 25. Tiwari M. Science behind human saliva. J Nat Sci Biol Med. 2011; 2:53‐58. 10.4103/0976-9668.82322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarode GS, Sarode SC, Sengupta N, et al. Clinical status determines the efficacy of salivary and nasopharyngeal samples for detection of SARS‐CoV‐2. Clin Oral Investig. 2020:1‐2. 10.1007/s00784-020-03630-9. Accessed November 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Czumbel LM, Kiss S, Farkas N, et al. Saliva as a candidate for COVID‐19 diagnostic testing: a meta‐analysis. Front Med. 2020;7:465. 10.3389/fmed.2020.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ranney ML, Griffeth V, Jha AK. Critical supply shortages—The need for ventilators and personal protective equipment during the COVID‐19 pandemic. New Eng J Med. 2020;382:E41. [DOI] [PubMed] [Google Scholar]
- 29. Patrício Silva AL, Prata JC, Walker TR, et al. Increased plastic pollution due to COVID‐19 pandemic: Challenges and recommendations. Chem Eng J. 2020;405. 10.1016/j.cej.2020.126683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khasne RW, Dhakulkar BS, Mahajan HC, Kulkarni AP. Burnout among healthcare workers during COVID‐19 pandemic in India: results of a questionnaire‐based survey. Indian J Crit Care Med. 2020;24(8):664‐671. https://creativecommons [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
Raw data of this study are available upon request.


