Abstract
Background
Tumors often develop resistance to surveillance by endogenous immune cells, which include natural killer (NK) cells. Ex vivo activated and/or expanded NK cells demonstrate cytotoxicity against various tumor cells and are promising therapeutics for adoptive cancer immunotherapy. Genetic modification can further enhance NK effector cell activity or activation sensitization. Here, we evaluated the effect of the genetic deletion of ubiquitin ligase Casitas B-lineage lymphoma pro-oncogene-b (CBLB), a negative regulator of lymphocyte activity, on placental CD34+ cell-derived NK (PNK) cell cytotoxicity against tumor cells.
Methods
Using CRISPR/Cas9 technology, CBLB was knocked out in placenta-derived CD34+ hematopoietic stem cells, followed by differentiation into PNK cells. Cell expansion, phenotype and cytotoxicity against tumor cells were characterized in vitro. The antitumor efficacy of CBLB knockout (KO) PNK cells was tested in an acute myeloid leukemia (HL-60) tumor model in NOD-scid IL2R gammanull (NSG) mice. PNK cell persistence, biodistribution, proliferation, phenotype and antitumor activity were evaluated.
Results
94% of CBLB KO efficacy was achieved using CRISPR/Cas9 gene editing technology. CBLB KO placental CD34+ cells differentiated into PNK cells with high cell yield and >90% purity determined by CD56+ CD3− cell identity. Ablation of CBLB did not impact cell proliferation, NK cell differentiation or phenotypical characteristics of PNK cells. When compared with the unmodified PNK control, CBLB KO PNK cells exhibited higher cytotoxicity against a range of liquid and solid tumor cell lines in vitro. On infusion into busulfan-conditioned NSG mice, CBLB KO PNK cells showed in vivo proliferation and maturation as evidenced by increased expression of CD16, killer Ig-like receptors and NKG2A over 3 weeks. Additionally, CBLB KO PNK cells showed greater antitumor activity in a disseminated HL60-luciferase mouse model compared with unmodified PNK cells.
Conclusion
CBLB ablation increased PNK cell effector function and proliferative capacity compared with non-modified PNK cells. These data suggest that targeting CBLB may offer therapeutic advantages via enhancing antitumor activities of NK cell therapies.
Keywords: immunotherapy, killer cells, natural, cell engineering
Introduction
Recent years have shown major advancements in the development and successful application of immunotherapies for cancer. Natural killer (NK) cells are key effector cells in cancer immunosurveillance and viral infections that use an array of receptors to distinguish between malignant and healthy cells, and mediate target cell killing. NK cells recognize universal stress ligands on tumor cells and do not elicit graft-versus-host disease,1 making them an attractive modality for developing off-the-shelf allogeneic cell therapies for treating cancer. Ex vivo expansion, activation, and novel genetic modification techniques, such as the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), can increase the antitumor activity of NK cells by overcoming resistance of tumor cells, increasing cytolytic activity, or promoting persistence and homing to tumor tissue.1 2
Antitumor efficacy of NK cells varies against different tumors, and it is driven by variability in the expression of ligands for NK activation or inhibition.3 Significant efforts to enhance anti-tumor responses of NK cells center around shifting the balance of signaling in favor of targeted activation, such as expression of chimeric antigen receptors (CARs) against various tumor antigens: CD19, CD20, CS1, CD138 and Her2.4–14 Alternatively, blockade of inhibitory receptors can lower the threshold for activation. For example, blocking Ly49 with an antibody was shown to improve antitumor function of murine NK cells.15 16 In a human NK cell line NK-92, blocking the inhibitory receptors KIR3DL1 and KIR2DL1 improved antibody-dependent cellular cytotoxicity in combination with rituximab against CD20-expressing B-cell lymphoma.17 Therefore, lowering the threshold for NK cell activation is a promising approach to enhance their antitumor function.
Activation and inhibitory signaling in lymphocytes is fine-tuned through intracellular checkpoints, such as E3 ubiquitin ligases. Members of the Casitas B-lineage lymphoma pro-oncogene ligase family negatively regulate the activation of various lymphocyte subsets through targeted ubiquitination of intracellular signaling molecules, including protein tyrosine kinases.18 Ubiquitination of proteins triggers their localization to lysosomes or proteasomes for degradation, thus negatively regulating the initiation and progression of immune responses.19 20 Casitas B-lineage lymphoma pro-oncogene-b (CBLB) interacts with its specific targets via phosphotyrosine-containing sequence motifs that are generated on activated protein tyrosine kinases that mediate activating signal transduction.21 22 CBLB broadly regulates T-cell function23 24 through the inhibition of the PI3K pathway,25 reduction of the linker for activation of T cells (LAT) phosphorylation and activation of PLC-γ1,26 negative regulation of Vav1 phosphorylation,23 and induces partial resistance to transforming growth factor beta-mediated suppression.27 Similarly, CBLB is a negative regulator of NK cell function. In NK cells, CBLB is activated and stabilized through inhibitory receptor signaling and reduces NK cell degranulation and cytotoxicity by targeted degradation of the adaptor protein LAT, which is involved in the recruitment of PLC-γ1/2 to the immunological synapse between an NK cell and a target cell.28 29
CBLB is induced in primary human NK cells by interleukin (IL)-15/IL-2 stimulation, interaction with K562 target cells30 or by signaling via the inhibitory receptor KIR2D1.28 Downregulation of CBLB in primary human NK cells significantly increased granzyme B (GZMB) and perforin expression, interferon gamma production and cytotoxicity against leukemia cell lines.30 Knockdown of CBLB in a human NK cell line NKL showed higher cytotoxicity against the Jurkat cell line and improved in vivo anticancer activity, effectively mitigating lung metastasis in a murine melanoma model through the release of an inhibitory effect on TAM receptors.21 Cblb knockout (KO) rendered mouse NK cells less susceptible to programmed death-ligand 1 (PD-L1)-mediated inhibition31 and enhanced responsiveness to cytokine stimulation.32 Thus, the deletion of CBLB is an attractive strategy to lower the threshold for activation and enhance the antitumor function of NK cells.
We have developed a good manufacturing practice procedure for generating human placental CD34+ cell-derived NK (PNK) cells with substantial cytolytic activity against several human tumor cell lines.33 PNK-007, a freshly formulated drug product, showed a high safety profile in phase I studies in patients with acute myeloid leukemia (AML) and multiple myeloma (MM)(Giarritta et al, manuscript in preparation)34–37. We hypothesize that the anti-tumor function of PNK cells can be further augmented by lowering their activation threshold by genetic deletion of negative regulators. Here, we demonstrate an effective use of CRISPR/Cas9 technology to genetically modify placenta-derived CD34+ hematopoietic stem cells and show that knocking out CBLB enhances the antitumor activity of PNK cells against several tumor cell lines and primary tumor cells. These data suggest that the deletion of negative regulators, such as CBLB in PNK cells, is a promising approach for developing more efficacious NK cell therapies for cancer.
Materials and methods
Placenta CD34+ cell isolation and culture
Placental CD34+ cells were acquired from healthy donors under fully informed consent. With donor eligibility documentation, tissues were qualified using a series of tests including serology and bacteriology (Lifebank USA). Blood was isolated from healthy donor tissues and processed by red blood cell depletion using Hetastarch (Hospira). The resulting cells were then magnetically labeled using Direct CD34 Progenitor Cell Isolation Kit (Miltenyi Biotec). CD34+ cells were positively selected using AutoMACS Cell Separator following manufacturer’s protocol. Placental CD34+ cells were then cryopreserved in CryoStor CS10 (Biolife Solutions) and stored in liquid nitrogen before use.
For PNK culture, placental CD34+ cells were thawed and cultivated following a three-stage process in the presence of cytokines, including thrombopoietin, SCF, Flt3 ligand, IL-7, IL-15 and IL-2 (Thermo Fisher Scientific), for 35 days to generate PNK cells. Nucleofection of CRISPR reagents was performed at day 5-7 of culture. Cell count and passage were performed every 2–3 days and cell expansion was recorded. At the end of the culture, cell phenotype was evaluated by flow cytometry to confirm that the cells expressed typical NK receptors and cytolytic markers.
Immunophenotypical characterization
The phenotype of PNK cells was analyzed by multicolor flow cytometry. First, PNK cells were washed and stained with fluorochrome-conjugated antibodies diluted in staining buffer (10% fetal bovine serum in phosphate-buffered saline (PBS)) according to the manufacturer’s instructions. CD244 (clone: 2B4)—FITC (BD Biosciences), CD226 (DNAM-1) (clone: DX11)—PE (Miltenyi Biotec), CD94 (clone: HP-3D9)—PerCP-Cy5.5 (BD Biosciences), CD314 (NKG2D) (clone: 1D11)—APC (Miltenyi Biotec), CD56 (clone: NCAM16.2)—Pe-Cy7 (BD Biosciences), CD3 (clone: SK7)—APC-H7 (BD Biosciences), CD14 (clone: MφP9)—APC-H7 (BD Biosciences), CD19 (clone: HIB19)—APC-Cy7 (BD Biosciences), CD69 (clone: FN50)—AF700 (BD Biosciences), NKp46 (CD335) (clone: 9E2)—BV650 (BD Biosciences), TIGIT (clone: A15153G)—BV605 (Biolegend), CD336 (NKp44) (clone: p44-8)—BUV395 (BD Biosciences), CD337 (NKp30) (clone: p30-15)—BV421 (BD Biosciences), CD11a (clone: HI111)—BV711 (BD Biosciences), CD16 (clone: 3G8)—BV786 (BD Biosciences), CD158a (clone: HP-3E4)—PE (BD Biosciences), CD158e1/e2 (clone: Z27.3.7)—PE (Beckman Coulter), CD158b1/b2, j (clone: GL183)—PE (Beckman Coulter), CD159a (NKG2A) (clone: 3D12HLA-E)—APC (Thermo Fisher Scientific), GZMB (clone: X40)—AF700 (BD Biosciences), perforin (clone: dG9)—BV421 (BD Biosciences), TIM-3 (CD366) (clone: F38-2E2)—BV605 (Biolegend). Dead cells were labelled with the Live/Dead Fixable Aqua Stain (Thermo Fisher Scientific) and gated out. For intracellular staining, surface-stained cells were permeabilized using the Fixation/Permeabilization Solution Kit (BD Biosciences) according to manufacturer’s recommendations and stained with fluorochrome-conjugated antibodies recognizing intracellular molecules. Stained cells were acquired on BD Fortessa X20 flow cytometer (BD Biosciences) and data were analyzed using FlowJo software.
Gene editing by CRISPR/Cas9
CBLB guide RNAs (gRNA) were designed using an online design tool from the Zhang lab at Massachusetts Institute of Technology (http://crispr.mit.edu/). The gRNA sequence of TAATCTGGTGGACCTCATGAAGG was selected. gRNA with 2′-O-methyl 3′phosphorothioate modifications incorporated in the three terminal nucleotides at both the 5′ and 3′ ends were custom-ordered from TriLink Biotechnologies. Cas9 messenger RNA (mRNA), which is capped using antireverse cap analog (Cap 0), polyadenylated and modified with 5-methylcytidine and pseudouridine (5meC, Ψ), was produced by TriLink.
Transfection of PNK cells
At days 5–7 of culture, transfection with Cas9 mRNA and CBLB gRNA (TriLink BioTechnologies) was conducted using the Lonza Nucleofector and the Human CD34+ Cell Nucleofector Kit (Lonza) according to manufacturer’s protocol. Briefly, 15 µg of Cas9 mRNA and 15 µg of CBLB gRNA were transfected into 2×106 cells in a 100 µL volume. After transfection, cells were transferred to PNK culture media at a density of 200,000/mL. For the control group, cells were either not transfected or transfected with only 15 µg of Cas9 mRNA.
Tracking of indels by decomposition (TIDE) assay
Gene editing efficiency was determined by TIDE analysis at days 12–14 and at the end of the PNK culture. Briefly, cells were lysed with Quick Extract buffer (Thermo Fisher Scientific). CBLB gene specific primers 5′-TGATAGCCTAGGACTGTTTGAGAGAA-3′ (forward) 5′-CCAGGCTCACAGCATCTGATAAC-3′ (reverse) were used to amplify CBLB genomic region by PCR. PCR samples were sequenced using the Sanger method and analyzed using the TIDE web tool (https://tide.nki.nl/).38
Western blot
To determine the efficiency of CBLB deletion at the protein expression level, mature PNK cells were lysed in radioimmunoprecipitation assay (RIPA) Lysis and Extraction Buffer (Thermo Fisher Scientific) including 1× Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Clarified cell lysates were separated on Novex Tris-Glycine Mini Gels (Thermo Fisher Scientific) by electrophoresis, and the total protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific). The membrane was blocked with Superblock T20 Blocking Buffer (Thermo Fisher Scientific) and incubated overnight at 4°C with rabbit anti-CBLB (Cell Signaling Technologies, clone: D3C12) diluted at 1:500 and mouse anti-β-Actin (Cell Signaling Technologies, clone: 8H10D10) diluted at 1:2000 in blocking buffer. After washing, the membrane was incubated for 1 hour at room temperature with IRDye 680RD donkey anti-mouse (LI-COR Biosciences) and IRDye 800CW donkey anti-rabbit (LI-COR Biosciences) at 1:15 000 in blocking buffer. The membrane was imaged using LiCOR CLx Imager (LI-COR Biosciences).
In vitro cytotoxicity assay
RPMI8266, U266, ARH77, HL-60, K562, KG1 and U251 MG cell lines were purchased from American Type Culture Collection (ATCC) and cultured following ATCC recommendations. For cytotoxicity against RPMI8226, U266, ARH77, K562, KG1 and HL-60, target cells were washed with PBS and labeled with PKH26 (Sigma-Aldrich) according to manufacturer’s protocol. CBLB KO PNK or control cells were cultured together with PKH26-labeled target cells in 96-well U-bottom plates at different effector-to-target (E:T) ratios for 4 hours.39 TO-PRO-3 (Thermo Fisher Scientific) was added at the end of the 4-hour culture at 0.25 µM concentration for 20 min, followed by flow cytometric analysis. Cytotoxicity was expressed as the percentage of dead cells (PKH26+ TO-PRO-3+) within the total PKH26+ target tumor cells. Lytic units (LU20/106 cells) were calculated as follows: % of specific lysis (Y axis) was plotted in function of E:T ratio (log scale X axis) and linear regression calculated. E:T value for 20% cytotoxicity (E:T20) was derived from the trendline. Lytic units were calculated based on the equation LU20/106 cells=106×((E:T20)×Twell)−1, where Twell is target cell number per well.40 Cytotoxicity against solid tumor cells (U251) was measured using the xCELLigence RTCA platform (ACEA Bioscience). U251 were cultured in 96-well electronic microtiter plates overnight, followed by addition of PNK cells at an E:T ratio of 2.5:1. Cell index, indicating the impedance of electron flow caused by adherent cells, was recorded real time. Percentage of cytolysis was calculated using the formula provided by ACEA Bioscience: (cell index of no effector–cell index of effector)/cell index of no effector×100.
Animal studies
Animal studies were conducted by a contract research organization accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, and studies were approved by the Institutional Animal Care and Use Committee. The studies were performed on female NOD-scid IL2R gammanull (NSG) mice, 8 weeks old, approximately 18–22 g at study initiation.
In vivo biodistribution and persistence
In vivo distribution and persistence of CBLB KO PNK cells were evaluated relative to control cells transduced with Cas9 mRNA (control) in NSG mice. Specifically, NSG mice were pretreated with 30 mg/kg busulfan (Alfa Aesar) at day −1. The next day, 1×107 CBLB KO PNK or control cells were intravenously injected via the tail vein. Recombinant human (rh)IL-15 (Thermo Fisher Scientific) was given via intraperitoneal injection at 0.5 µg/mouse/day every 2 days from day 0 to day 12. Animals (n=4–5) from each group were sacrificed at days 7, 14, and 21. At each time point, peripheral blood, bone marrow, lungs, liver and spleen were harvested. Lungs, liver and spleen were dissociated into single-cell suspensions using the gentleMACS Dissociator (Miltenyi Biotec) according to manufacturer’s instructions. Single-cell suspensions from all other tissues were filtered through 100 µm nylon filters and centrifuged. Red blood cells were lysed using ACK lysis buffer (Gibco). By flow cytometry, PNK cells were subsequently quantified by the amount of human CD45+ cells in peripheral blood, bone marrow, lungs, liver, and spleen samples.
In vivo proliferation and maturation
To evaluate cell proliferation in vivo, PNK (CBLB KO or control) cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) dye at day 35 of culture and infused into NSG mice (n=4). Cell labeling was performed using CellTrace CFSE Cell Proliferation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. CFSE labeled PNK cells were intravenously administered in busulfan preconditioned NSG mice at a dose of 1×107 cells/mouse. rhIL-15 was given intraperitoneally at 0.5 µg/mouse/day every 2 days from days 0 to 12. After 3, 7, 14 and 21 days of infusion, mouse liver was harvested and dissociated into single cell suspension as described previously. CFSE was analyzed by flow cytometry to characterize PNK cell proliferation in vivo. Additionally, in a separate study with the same design, unlabeled PNK cells were infused into busulfan preconditioned mice (n=5). On days 3, 7, 14 and 21 after infusion, cells were harvested from the peripheral blood, bone marrow, lungs, liver and spleen as described previously, and were analyzed for cell proliferation marker Ki67 (clone: B56)—PE (BD Biosciences) by flow cytometry. Cells harvested from the mouse livers were analyzed for human NK cell maturation markers CD11a, CD16, killer Ig-like receptors (KIRs), and NKG2A by flow cytometry.
In vivo antitumor efficacy
Antitumor activity of PNK cells was evaluated in a Xenograft AML model in NSG mice. To establish the model, HL-60 cells were transduced with a lentiviral construct (Amsbio) to express green fluorescence protein and luciferase. Cells were sorted for the top 2.5% expressors using FACSAria flow cytometer (BD) and were used for the animal study. Specifically, 1×105 HL-60 cells expressing luciferase were inoculated intravenously to NSG mice at day 0 after busulfan preconditioning (day −1). At days 2 and 7, 1×107 CBLB KO (n=7) or control (n=7) PNK cells were injected intravenously and PBS was used as the non-treatment control. rhIL-15 was given intraperitoneally at 0.5. g/mouse/day every 2 days from days 2 to 20. Tumor growth was measured by in vivo bioluminescence imaging twice weekly starting from day 2. Mouse body weight was measured every 4 days until study termination at day 20.
Statistical analysis
Experimental data were graphed and analyzed with Prism. All data were presented as mean±SD. For in vitro expansion and phenotype data, paired t-test was performed to determine the difference between CBLB KO and control groups. For in vitro cytotoxicity data and in vivo data, two-way analysis of variance was first performed, and then a pairwise comparison was made. Log-rank test was used to compare survival curves between groups. A p value of ≤0.05 is considered significant. An asterisk indicates significance: *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001; no asterisk indicates no statistical significance.
Results
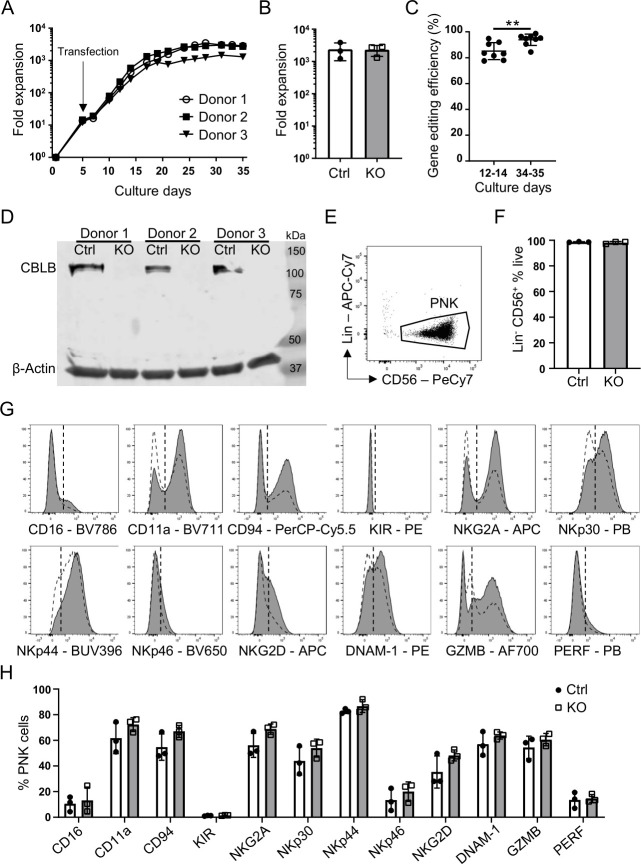
As CBLB acts as an important gatekeeper limiting NK cell cytotoxicity in various tumor models,21 we used CRISPR/Cas9 technology to generate CBLB KO PNK cells to enhance their cytotoxic potential. PNK cells are generated from placenta-derived CD34+ stem cells following a 35-day culture process. To generate CBLB KO PNK cells, placenta-derived CD34+ cells were transfected with CBLB guide RNAs and Cas9 mRNA on day 5 of culture. Control cells only received Cas9 mRNA. To evaluate the possible effect of transfection on CD34+ cell culture, we monitored their expansion over the 35-day culture process (figure 1A). The fold expansion of three independent gene modification experiments was analyzed. Both CBLB KO and control cells resulted in an average of >2000-fold expansion by the end of the 35-day culture process with no difference between treatment conditions (figure 1B). A growth arrest of 2–3 days was observed after electroporation at day 5, likely due to the transfection event. The cells quickly recovered, demonstrating a fast and steady expansion from days 7 to 25, and thereafter reached a plateau until harvest on day 35. The data suggest that CBLB deletion does not affect CD34+ cell proliferation and differentiation towards PNK cells. Gene editing efficiency was evaluated on culture days 12–14 and 34–35 using the TIDE assay.38 A high CBLB editing efficiency was achieved: 85% (range 78%–95%) on days 12–14 and 94% (range 84%–99%) on days 34–35 (figure 1C). Successful CBLB deletion was confirmed by a lack of CBLB protein detected by Western blot in gene-edited PNK cells on day 35 of culture (figure 1D). A strong protein band was detected at 125 kDa only in non-modified control cells corresponding to the expected size of the CBLB protein. The data show that complete ablation of CBLB protein in PNK cells was achieved with high efficiency by genetic modification of CD34+ stem cells using CRISPR/Cas9 technology.
Figure 1.
Efficient CRISPR/Cas9-mediated CBLB gene editing and conserved NK cell identified in human PNK cells. (A) representative growth curves of CBLB KO PNK cell cultures. Cas9 mRNA and CBLB gRNA were introduced by transfection on day 5 (n=3). (B) Quantification of Ctrl or KO PNK cell expansion at day 35 of PNK culture, mean±SD (n=3). (C) CBLB gene editing efficiency using tide analysis summarizing eight individual experiments after transfection and on days 34–35 of PNK cell culture. Gene editing efficiency is determined as the frequency of nucleotide additions or deletions, mean±SD (n=8). (D) Western blot analysis of CBLB protein in day 35 Ctrl and KO PNK cells. β-actin serves as a loading control (n=3). (E) Representative dot plot of PNK cell gating. Indicated population was first gated of live single cells (not shown). Lins were CD3, CD14 and CD19. (F) Quantification of Lin− CD56+ cells in Ctrl and KO PNK cell cultures, mean±SD (n=3). (G) Representative histograms demonstrating the expression of indicated NK cell markers in Ctrl (white dashed) and KO PNK (gray) cells on day 35 of culture. KIR=CD158e1/e2 and CD158b1/b2, j. Vertical dotted line marks isotype control. (H) Quantification of the expression of indicated NK cell markers on Ctrl and KO cells, mean±SD (n=3). **P<0.01. CBLB, Casitas B-lineage lymphoma pro-oncogene-b; Ctrl, control; KIR, killer Ig-like receptor; KO, knockout; Lin, lineage marker; NK, natural killer; PNK, placental CD34+ cell-derived NK; PERF, perforin.
To confirm that CRISPR/Cas9-mediated gene modification did not affect the differentiation of PNK cells, we used flow cytometry to immunophenotype CBLB KO PNK cells and donor-matched control cells harvested on day 35. CBLB gene modification did not affect PNK cell purity, indicated by >95% of CD56+ cells that lacked T cell, B cell or monocyte markers CD3, CD19 or CD14, respectively (figure 1E, F). We next performed comprehensive characterization of CBLB KO PNK cells to eliminate any effect of CBLB deletion on their receptor profile and activation potential. Similarly to control cells, CBLB KO PNK cells demonstrated high CD56, minimal CD16, and no killer Ig-like receptor (KIR) expression, consistent with a phenotype for NK cells differentiated from hematopoietic stem cell precursors (figure 1G, H).41 NK cell proteins that mediate target recognition (NKp30, NKp44, NKp46, NKG2D, DNAM-1, NKG2A, and CD94), formation of the immunological synapse (CD11a) and cytotoxic response (GZMB and perforin) were expressed on both control and CBLB KO PNK cells. There was no significant change in the expression of any of these markers comparing CBLB KO to control PNK cells, suggesting that CBLB ablation did not affect the differentiation and maturation of PNK cells during cultivation.
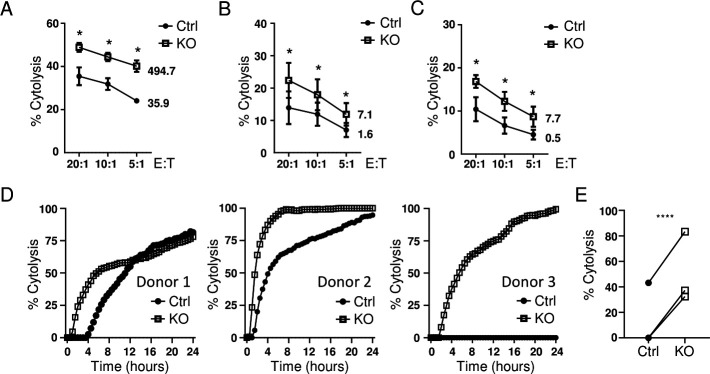
We have previously demonstrated that PNK cells exhibit cytotoxicity against a wide range of tumor cell lines.33 To evaluate the effect of CBLB ablation on PNK cell effector functions, we first tested their cytotoxicity in vitro. The cytolytic activity of CBLB KO PNK cells was evaluated in a flow cytometry-based cytotoxicity assay against myeloma cell lines RPMI8226 and U266, and plasma cell leukemia cell line ARH77 (figure 2A–C). Significantly higher cytotoxicity of CBLB KO PNK cells was observed against tumor cell lines at every E:T ratio tested compared with non-modified control cells from the same donor. We also used an impedance-based cytotoxicity assay to analyze the cytotoxicity of CBLB-deficient PNK cells against the U-251 MG glioblastoma cell line. CBLB KO cells showed improved relative killing activity over control cells during a 24-hour period (figure 2D, E). CBLB deletion improved overall cytotoxicity of PNK cells against liquid and solid tumor cell lines in vitro.
Figure 2.
CBLB KO PNK cells demonstrate enhanced cytotoxicity against hematological cancer and solid tumor lines in vitro. The efficacy of tumor cell cytolysis was analyzed in CBLB KO and their corresponding Ctrl cells in a 4-hour in vitro cytotoxicity assay against RPMI8266 (A), ARH77 (B) and U266 (C) cell lines using E:T ratios of 20:1, 10:1 and 5:1. The proportion of dead target cells was quantified by flow cytometry. The figures next to the curves indicate lytic units (LU20/106 cells), mean±SD (n=3). (D, E) Cytolysis of Ctrl and KO PNK cells against of an adherent cancer cell line U-251 MG from three donors at an E:T ratio of 2.5:1 analyzed using an impedance-based RTCA platform. Cytolysis against U-251 MG over 24 hours (D) and at a 4-hour time point (E) (n=3). *P<0.05, ****P<0.0001. CBLB, Casitas B-lineage lymphoma pro-oncogene-b; Ctrl, control; E:T, effector-to-target; KO, knockout; PNK, placental CD34+ cell-derived NK; RTCA, real time cytolytic assay.
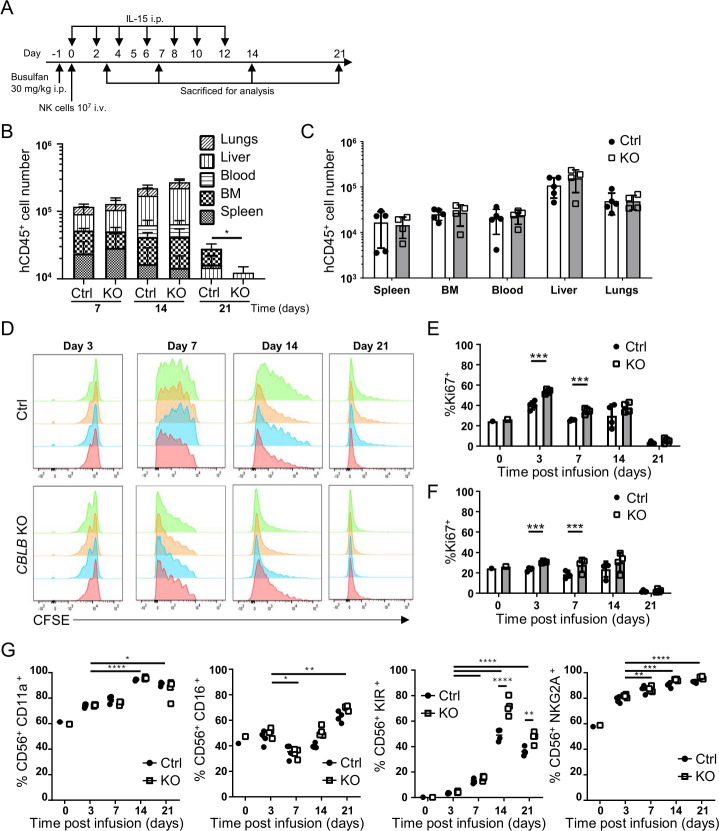
Clinical studies have suggested that persistence and expansion of donor NK cells are required to show efficacy of NK cell therapy.42 Therefore, we first examined the persistence and biodistribution of CBLB KO and control cells in lymphocyte-deficient NSG mice. Ten million CBLB KO or control cells were injected into busulfan-treated (day −1) NSG mice via the tail vein (figure 3A). rhIL-15 was given intraperitoneally every 2 days until day 12. Animals were sacrificed at different time points to analyze PNK cell persistence using anti-hCD45 staining and flow cytometry. Infused PNK cells, characterized by hCD45+ expression, were detected in all tissues examined, including peripheral blood, bone marrow, spleen, lungs and liver of mice on days 7, 14 and 21 (figure 3B, C). hCD45+ cell number in blood and the tissues analyzed increased from days 7 to 14, but significantly dropped by day 21 (figure 3B). Importantly, hCD45+ cell counts were comparable in animals receiving either CBLB KO or control PNK cells for the first 2 weeks. Figure 3C shows a representative PNK cell distribution in all tissues examined (day 14 time point), with the highest cell number found in the liver and the lungs. Control and CBLB KO PNK cells were found to have similar biodistribution in mice with comparable numbers for all organs analyzed. These data demonstrate that CBLB deficiency does not affect the biodistribution and persistence of PNK cells in vivo.
Figure 3.
Biodistribution and persistence of CBLB KO PNK cells in vivo. (A) Schematic of experimental procedures for in vivo biodistribution and proliferation studies. CBLB KO or Ctrl PNK cells were injected intravenously into NSG mice 1 day after intraperitoneal busulfan treatment. rhIL-15 was administered intraperitoneally every 2 days until day 12. Animals were sacrificed on days 3, 7, 14 and 21 for analysis of PNK cells using hCD45 staining (n=4–5). (B) Quantification of hCD45+ cell numbers quantified in blood, BM, liver, lungs and spleen on days 7, 14 and 21 after infusion of Ctrl or CBLB KO PNK cells. (C) Quantification of hCD45+ cells in indicated organs on day 14 after PNK cell infusion, mean±SD (n=5). (D) CFSE-labeled Ctrl or CBLB KO PNK cells were injected intravenously in NSG mice, and animals were sacrificed on days 3, 7, 14 or 21 to analyze CFSE signal on PNK cells. Each histogram represents data from one mouse (n=4). (E, F) Proportion of PNK cells isolated from the liver (E) or the lungs (F), expressing the proliferation marker Ki67. (G) Quantification of indicated NK cell surface receptors on Ctrl or CBLB KO PNK cells after harvested from mice. Day 0 sample represents the preinfusion phenotype. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. BM, bone marrow; CBLB, Casitas B-lineage lymphoma pro-oncogene-b; CFSE, carboxyfluorescein succinimidyl ester; Ctrl, control; h, human; IL, interleukin; KO, knockout; NK, natural killer; NSG, NOD-scid IL2R gammanull; PNK, placental CD34+ cell-derived NK.
NSG mice infused with PNK cells were treated with rhIL-15 to promote PNK cell proliferation and persistence. We evaluated the proliferation of CBLB KO and control cells by administering CFSE-labeled PNK cells to NSG mice according to the protocol described in figure 3A. hCD45+ cells harvested from the mouse liver were analyzed for the intensity of CFSE signal by flow cytometry on different days postinjection. CFSE intensity decreased from days 3 to 21 in both the control and CBLB KO PNK cells, indicating cell proliferation in vivo (figure 3D). Additionally, reduced and rapidly decreasing CFSE signal, indicating increased proliferation, was detected in CBLB KO cells compared with control PNK cells (figure 3D). We also analyzed the proliferation marker Ki67 expression on CBLB KO and control PNK cells in different tissues (figure 3E, F and online supplemental figure 1). An increase of in vivo proliferation of CBLB KO PNK cells was detected in the liver and the lungs on days 3 and 7 (figure 3E, F). PNK cell numbers in tissues (figure 3B) and Ki67 expression (figure 3E, F) correlated with the rhIL-15 infusion period (figure 3A), demonstrating a significant role of IL-15 on NK cell proliferation and persistence.
jitc-2020-001975supp001.pdf (184.6KB, pdf)
In vitro cultured NK cells further mature in vivo and acquire a phenotype resembling peripheral blood NK cells.41 To characterize PNK cell phenotype after infusion to NSG mice, the expression of surface markers representing the differentiation and maturation of NK cells, including CD11a, CD16, NKG2A and KIR, were analyzed on hCD45+ cells in the liver (figure 3G). In both CBLB KO and control cells, all four markers increased from days 3 to 21 after infusion compared with preinfusion cells, suggesting that PNK cells underwent further maturation in vivo and that CBLB ablation did not impair this process. CBLB KO cells, however, showed significantly higher KIR expression on days 14 and 21 compared with control cells, suggesting that the ablation of CBLB resulted in enhanced PNK differentiation in vivo.
Next, we evaluated if the in vivo infused PNK cells maintained their cytolytic potential and analyzed the cytotoxicity of CBLB KO and control PNK cells ex vivo against myeloid leukemia cell lines K562, HL-60 and KG1. The results confirmed that CBLB KO cells isolated from mice demonstrated enhanced cytotoxicity against all three cell lines compared with control cells (online supplemental figure 2).
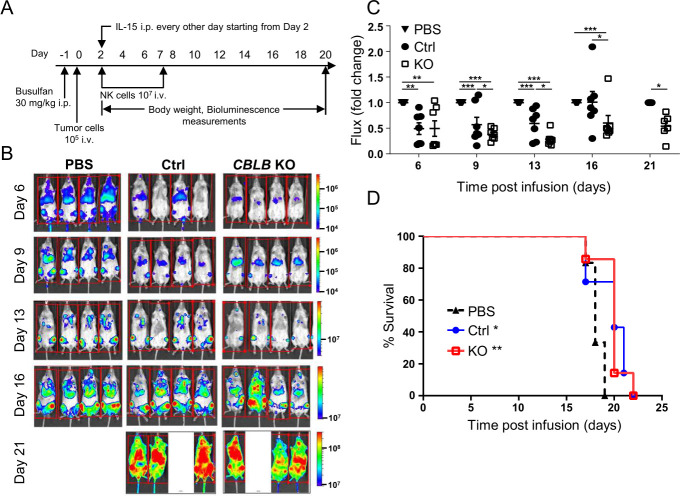
To analyze whether elevated in vitro cytotoxicity and higher in vivo proliferation and maturation of CBLB KO PNK cells translated into enhanced in vivo antitumor efficacy, we established a human AML cell line HL-60 xenograft in the NSG mouse (figure 4A). HL-60 cells expressing luciferase were injected at 1×105 intravenously into NSG mice (day 0), followed by 1×107 CBLB KO or control PNK cells on days 2 and 7. Non-treated control mice were injected with PBS. Both CBLB KO and control PNK cell-treated mice showed significantly less tumor burden between days 6 and 16 compared with PBS-treated control animals that were all sacrificed between days 17 and 19 due to aggressive progression of the tumor. CBLB KO PNK cells showed significantly improved control of the tumor compared with control PNK cells from days 9 to 21 (figure 4B, C). Although there was significant reduction in tumor burden in mice receiving CBLB KO cells compared with control cells, this difference did not translate into a survival benefit. However, animals that received control or CBLB KO PNK cells lived longer than PBS-treated control mice (figure 4D).
Figure 4.
CBLB KO PNK cells reduce tumor burden in vivo. (A) Schematic of experimental procedures for the evaluation of antitumor activity of Ctrl or CBLB KO PNK cells in vivo using HL-60 xenograft mouse model (n=6–7). One day after busulfan treatment, 1×105 HL-60 cells expressing luciferase were injected intravenously to NSG mice and NK cells were injected on days 2 and 7 post-tumor cell injection to three randomized animal groups (PBS, Ctrl PNK or CBLB KO PNK cells). Each animal received 1×107 cells. rhIL-15 was administered intraperitoneally every 2 days until day 20. (B) Representative bioluminescence imaging of each group shows the tumor burden at days 6, 9, 13, 16 and 20. All animals were sacrificed at day 21. (C) Tumor burden of each group normalized to PBS Ctrl group as tumor growth fold change. (D) Survival plot of the HL-60 xenograft mouse administered with PBS, Ctrl or CBLB KO PNK cells. Asterisks (*) indicate significant difference between Ctrl and KO groups compared with the PBS group. *P<0.05, **P<0.01, ***P<0.001. CBLB, Casitas B-lineage lymphoma pro-oncogene-b; Ctrl, control; IL, interleukin; KO, knockout; NK, natural killer; NSG, NOD-scid IL2R gammanull; PBS, phosphate-buffered saline; PNK, placental CD34+ cell-derived NK; rh, recombinant human.
Discussion
Novel immunotherapies for cancer are rapidly changing the way tumors are treated in the clinic. Major advancements have recently been made in the development and successful application of monoclonal antibodies and engineered immune cells. While the introduction of tumor-specific CAR T cells for autologous cell therapy has demonstrated striking results against hematological malignancies, CAR T cell therapies still face many challenges, which include high cost, a long manufacturing process and issues associated with autologous approaches. NK cells are cytotoxic innate immune cells that use an array of universal receptors, which recognize markers of oncogenic transformation on malignant cells and therefore require no prior sensitization for target identification. The safety and clinical effectiveness of adoptive NK cell therapy have been shown in multiple clinical studies using ex vivo activated and/or expanded NK cells derived from various sources.43 For these properties, NK cells are actively investigated as an attractive alternative to CAR T cells in multiple ongoing clinical trials targeting hematological or solid tumors.44 Ex vivo activation, expansion, and genetic modification can further enhance NK cell cytotoxic potential and overcome resistance of tumor cells to NK cell recognition and killing.
In this study, we established a process to produce genetically modified NK cells with improved cytotoxic function using CRISPR/Cas9 gene editing technology. By knocking out CBLB gene in CD34+ hemopoietic stem cells derived from full-term postpartum human placenta, we significantly improved the cytotoxic function of placental CD34+ cell-derived NK (PNK) cells in vitro and in vivo. It is widely recognized that the E3 ligase CBLB is a central player in the negative regulation of lymphocyte function, including NK cells, and was therefore selected for deletion in this proof-of-concept study.18 Our data demonstrate that CBLB is a negative regulator of PNK cell function and that genetic manipulation of placental CD34+ cells is a feasible approach to enhance antitumor cytotoxicity for immunotherapy.
CBLB gene modification of CD34+ cells did not result in any obvious developmental or phenotypical defects in PNK cells, as we achieved greater than 95% of the cells expressing CD56 and other NK cell prototypic markers at a comparable level to non-modified cells. This agrees with a previous report that CBLB KO mice develop NK cells normally.21 In vivo, PNK cell numbers, their proliferation and maturation correlated with rhIL-15 treatment period, highlighting the key role of IL-15 cytokine on NK cell persistence and homeostasis. We observed increased proliferation of CBLB KO cells compared with unmodified PNK cells in vivo, similarly to what has been observed by others.21 The increased in vivo proliferation of CBLB KO PNK cells could partially explain the reduced tumor burden in mice treated with the CBLB KO cells.
Phenotypically, PNK cells, either unmodified or CBLB KO, are of CD56bright CD16- phenotype that is considered to be more immature compared with the CD56dim CD16+ NK cells in the blood.45 PNK also show lower expression of the inhibitory KIRs but increased expression of the activating NKp30, NKp44 and NKp46 receptors, similar to reports by others for CD34+ cell-derived NK cells.46 In vivo, PNK cells demonstrated rapid maturation, illustrated by the upregulation of CD16, CD11a and KIRs, a phenotype that is associated with increased cytotoxicity.45
CBLB ablation in PNK cells increased cytotoxicity against tumor cells both in in vitro and in vivo models. This is in accordance with recent studies from Lu et al demonstrating significantly increased cytotoxicity of primary human NK cells on knockdown of CBLB.30 Increased cytotoxicity of CBLB KO PNK over control cells appears to be comparable to previously reported murine CBLB KO NK cells over their wild-type counterparts,21 suggesting a conserved role of CBLB in the regulation of NK cell cytotoxicity in mice and men. The in vivo study showed that PNK cells significantly reduced tumor (HL-60) burden in mice and that CBLB deficiency further enhanced PNK cytotoxicity and reduced tumor burden compared with control cells. Although there was no survival benefit when using CBLB KO PNK cells versus control cells, it is possible that the decrease in tumor burden mediated by CBLB KO in PNK cells was not sufficient to confer a survival benefit in this aggressive tumor model using the HL-60 cancer cell line that leads to 100% of lethality in mice within 3 weeks.
In conclusion, we have established a process to generate clinical-grade PNK cells for immunotherapy of various cancers or infectious disease, and we demonstrated the feasibility of using genetic modification by CRISPR/Cas9 technology to enhance their antitumor function. CBLB, a negative regulator of NK cell function, was selected as a target for deletion. While a high KO efficiency was achieved, CBLB deletion did not affect CD34+ cell proliferation or differentiation into PNK cells. Furthermore, CBLB deletion enhanced proliferative capacity and effector function of PNK cells in vivo. Altogether, the data suggest that genetic modification is a promising approach to further enhance NK cell persistence and antitumor potential for the purpose of offering therapeutic advantages in cancer immunotherapy.
Acknowledgments
We thank Tianjian Li, Chuan Wang, Mini Bharathan and Vladimir Jankovic for their scientific contributions to the article.
Footnotes
XG and TM contributed equally.
Contributors: XG, QY, SS, SH, RH and XZ conceptualized the study. RH and XZ acquired funding. XG, TM, QY, SS, SH, WvdT, RH and XZ led the study and provided supervision. XG, TM, SS, HR, QY, SH, AD and JG conducted experiments and analyzed the data. XG, QY and SS prepared the first draft of the manuscript. TM, QY, TM, XG, SS and SH wrote the manuscript. All authors contributed to the review and editing of the manuscript and approved the final version.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: No data are available. All data relevant to the study are included in the article or uploaded as supplementary information. Data are available upon reasonable request.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Shimasaki N, Jain A, Campana D. Nk cells for cancer immunotherapy. Nat Rev Drug Discov 2020;19:200–18. 10.1038/s41573-019-0052-1 [DOI] [PubMed] [Google Scholar]
- 2.Carlsten M, Childs RW. Genetic manipulation of NK cells for cancer immunotherapy: techniques and clinical implications. Front Immunol 2015;6:266. 10.3389/fimmu.2015.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivier E, Tomasello E, Baratin M. Functions of natural killer cells. Nat Immunol 2008. 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- 4.Romanski A, Uherek C, Bug G, et al. CD19-CAR engineered NK-92 cells are sufficient to overcome NK cell resistance in B-cell malignancies. J Cell Mol Med 2016;20:1287–94. 10.1111/jcmm.12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oelsner S, Waldmann A, Billmeier A, et al. Genetically engineered CAR NK cells display selective cytotoxicity against FLT3-positive B-ALL and inhibit in vivo leukemia growth. Int J Cancer 2019;145:1935–45. 10.1002/ijc.32269 [DOI] [PubMed] [Google Scholar]
- 6.Liu E, Tong Y, Dotti G, et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 2018;32:520–31. 10.1038/leu.2017.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Liu Q, Zhong M, et al. 2B4 costimulatory domain enhancing cytotoxic ability of anti-CD5 chimeric antigen receptor engineered natural killer cells against T cell malignancies. J Hematol Oncol 2019;12:49. 10.1186/s13045-019-0732-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimasaki N, Fujisaki H, Cho D, et al. A clinically adaptable method to enhance the cytotoxicity of natural killer cells against B-cell malignancies. Cytotherapy 2012;14:830–40. 10.3109/14653249.2012.671519 [DOI] [PubMed] [Google Scholar]
- 9.Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood 2005;106:376–83. 10.1182/blood-2004-12-4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu Y, Hochberg J, Yahr A, et al. Targeting CD20+ aggressive B-cell non-Hodgkin lymphoma by anti-CD20 CAR mRNA-modified expanded natural killer cells in vitro and in NSG mice. Cancer Immunol Res 2015;3:333–44. 10.1158/2326-6066.CIR-14-0114 [DOI] [PubMed] [Google Scholar]
- 11.Jiang H, Zhang W, Shang P, et al. Transfection of chimeric anti-CD138 gene enhances natural killer cell activation and killing of multiple myeloma cells. Mol Oncol 2014;8:297–310. 10.1016/j.molonc.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu J, Deng Y, Benson DM, et al. CS1-specific chimeric antigen receptor (CAR)-engineered natural killer cells enhance in vitro and in vivo antitumor activity against human multiple myeloma. Leukemia 2014;28:917–27. 10.1038/leu.2013.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esser R, Müller T, Stefes D, et al. Nk cells engineered to express a GD2 -specific antigen receptor display built-in ADCC-like activity against tumour cells of neuroectodermal origin. J Cell Mol Med 2012;16:569–81. 10.1111/j.1582-4934.2011.01343.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genßler S, Burger MC, Zhang C, et al. Dual targeting of glioblastoma with chimeric antigen receptor-engineered natural killer cells overcomes heterogeneity of target antigen expression and enhances antitumor activity and survival. Oncoimmunology 2016;5:e1119354. 10.1080/2162402X.2015.1119354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koh CY, Blazar BR, George T, et al. Augmentation of antitumor effects by NK cell inhibitory receptor blockade in vitro and in vivo. Blood 2001;97:3132–7. 10.1182/blood.V97.10.3132 [DOI] [PubMed] [Google Scholar]
- 16.Koh CY, Ortaldo JR, Blazar BR, et al. Nk-cell purging of leukemia: superior antitumor effects of NK cells H2 allogeneic to the tumor and augmentation with inhibitory receptor blockade. Blood 2003;102:4067–75. 10.1182/blood-2003-04-1367 [DOI] [PubMed] [Google Scholar]
- 17.Binyamin L, Alpaugh RK, Hughes TL, et al. Blocking NK cell inhibitory self-recognition promotes antibody-dependent cellular cytotoxicity in a model of anti-lymphoma therapy. J Immunol 2008;180:6392–401. 10.4049/jimmunol.180.9.6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutz-Nicoladoni C, Wolf D, Sopper S. Modulation of immune cell functions by the E3 ligase Cbl-b. Front Oncol 2015;5:58. 10.3389/fonc.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 2005;6:599–609. 10.1038/nrm1700 [DOI] [PubMed] [Google Scholar]
- 20.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature 2009;458:430–7. 10.1038/nature07959 [DOI] [PubMed] [Google Scholar]
- 21.Paolino M, Choidas A, Wallner S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 2014;507:508–12. 10.1038/nature12998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohapatra B, Ahmad G, Nadeau S, et al. Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta 2013;1833:122–39. 10.1016/j.bbamcr.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmaier K, Krawczyk C, Kozieradzki I, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 2000;403:211–6. 10.1038/35003228 [DOI] [PubMed] [Google Scholar]
- 24.Paolino M, Thien CBF, Gruber T, et al. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. J Immunol 2011;186:2138–47. 10.4049/jimmunol.1003390 [DOI] [PubMed] [Google Scholar]
- 25.Fang D, Liu YC. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat Immunol 2001;2:870–5. 10.1038/ni0901-870 [DOI] [PubMed] [Google Scholar]
- 26.Jeon M-S, Atfield A, Venuprasad K, et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity 2004;21:167–77. 10.1016/j.immuni.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 27.Wohlfert EA, Gorelik L, Mittler R, et al. Cutting edge: deficiency in the E3 ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. J Immunol 2006;176:1316–20. 10.4049/jimmunol.176.3.1316 [DOI] [PubMed] [Google Scholar]
- 28.Matalon O, Barda-Saad M. Cbl ubiquitin ligases mediate the inhibition of natural killer cell activity. Commun Integr Biol 2016;9:e1216739. 10.1080/19420889.2016.1216739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matalon O, Fried S, Ben-Shmuel A, et al. Dephosphorylation of the adaptor LAT and phospholipase C-γ by SHP-1 inhibits natural killer cell cytotoxicity. Sci Signal 2016;9:ra54. 10.1126/scisignal.aad6182 [DOI] [PubMed] [Google Scholar]
- 30.Lu T, Chen L, Mansour AG, et al. Cbl-B is upregulated and plays a negative role in activated human NK cells. J Immunol 2021;206:677–85. 10.4049/jimmunol.2000177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara M, Anstadt EJ, Clark RB. Cbl-B deficiency mediates resistance to programmed death-ligand 1/programmed death-1 regulation. Front Immunol 2017;8:42. 10.3389/fimmu.2017.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lametschwandtner G, Sachet M, Haslinger I, et al. Cbl-B silenced human NK cells respond stronger to cytokine stimulation. J Immunother Cancer 2015;3. 10.1186/2051-1426-3-S2-P230 [DOI] [Google Scholar]
- 33.Kang L, Voskinarian-Berse V, Law E, et al. Characterization and ex vivo expansion of human placenta-derived natural killer cells for cancer immunotherapy. Front Immunol 2013;4:101. 10.3389/fimmu.2013.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooley S, Hari P, McCloskey J. Abstract CT079: a phase I study of PNK-007, allogeneic, off the shelf NK cell in relapsed/refractory AML (NCT02781467) 2019. 10.1158/1538-7445.AM2019-CT079 [DOI]
- 35.Holstein SA, Cooley SA, Hari P. Abstract CT108: a phase I study of PNK-007, allogeneic, off the shelf NK cell, post autologous transplant in multiple myeloma (NCT02955550) 2019. 10.1158/1538-7445.AM2019-CT108 [DOI]
- 36.Koppisetti S, Balint C, Giarritta E, et al. PS1215 safety and tolerability of allogeneic, OFF-THE shelf placental natural killer cells (PNK 007) in phase 1 multiple myeloma and acute myeloid leukemia studies (NCT02955550 and NCT02781467). Hemasphere 2019;3:554. 10.1097/01.HS9.0000563144.13954.0d [DOI] [Google Scholar]
- 37.Holstein SA, Cooley S, Hari P, et al. Results of a phase I study of Pnk-007, allogeneic, off the shelf NK cell, post autologous transplant in multiple myeloma (NCT02955550). Blood 2019;134:4451. 10.1182/blood-2019-128467 [DOI] [Google Scholar]
- 38.Brinkman EK, Chen T, Amendola M, et al. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014;42:e168. 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee-MacAry AE, Ross EL, Davies D, et al. Development of a novel flow cytometric cell-mediated cytotoxicity assay using the fluorophores PKH-26 and TO-PRO-3 iodide. J Immunol Methods 2001;252:83–92. 10.1016/S0022-1759(01)00336-2 [DOI] [PubMed] [Google Scholar]
- 40.Mbitikon-Kobo F-M, Bonneville M, Sekaly R-P, et al. Ex vivo measurement of the cytotoxic capacity of human primary antigen-specific CD8 T cells. J Immunol Methods 2012;375:252–7. 10.1016/j.jim.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 41.Cichocki F, Grzywacz B, Miller JS. Human NK cell development: one road or many? Front Immunol 2019;10:2078. 10.3389/fimmu.2019.02078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geller MA, Miller JS. Use of allogeneic NK cells for cancer immunotherapy. Immunotherapy 2011;3:1445–59. 10.2217/imt.11.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin MH, Kim J, Lim SA, et al. Nk cell-based immunotherapies in cancer. Immune Netw 2020;20. 10.4110/in.2020.20.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu E, Marin D, Banerjee P, et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N Engl J Med 2020;382:545–53. 10.1056/NEJMoa1910607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol 2001;22:633–40. 10.1016/S1471-4906(01)02060-9 [DOI] [PubMed] [Google Scholar]
- 46.Yoon SR, Lee YS, Yang SH, et al. Generation of donor natural killer cells from CD34(+) progenitor cells and subsequent infusion after HLA-mismatched allogeneic hematopoietic cell transplantation: a feasibility study. Bone Marrow Transplant 2010;45:1038–46. 10.1038/bmt.2009.304 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001975supp001.pdf (184.6KB, pdf)