Abstract
Objective
MicroRNA‐34a‐5p (miR‐34a‐5p) expression is elevated in the synovial fluid of patients with late‐stage knee osteoarthritis (OA); however, its exact role and therapeutic potential in OA remain to be fully elucidated. This study was undertaken to examine the role of miR‐34a‐5p in OA pathogenesis.
Methods
Expression of miR‐34a‐5p was determined in joint tissues and human plasma (n = 71). Experiments using miR‐34a‐5p mimic or antisense oligonucleotide (ASO) treatment were performed in human OA chondrocytes, fibroblast‐like synoviocytes (FLS) (n = 7–9), and mouse OA models, including destabilization of the medial meniscus (DMM; n = 22) and the accelerated, more severe model of mice fed a high‐fat diet and subjected to DMM (n = 11). Wild‐type (WT) mice (n = 9) and miR‐34a–knockout (KO) mice (n = 11) were subjected to DMM. Results were expressed as the mean ± SEM and analyzed by t‐test or analysis of variance, with appropriate post hoc tests. P values less than 0.05 were considered significant. RNA sequencing was performed on WT and KO mouse chondrocytes.
Results
Expression of miR‐34a‐5p was significantly increased in the plasma, cartilage, and synovium of patients with late‐stage OA and in the cartilage and synovium of mice subjected to DMM. Plasma miR‐34a‐5p expression was significantly increased in obese patients with late‐stage OA, and in the plasma and knee joints of mice fed a high‐fat diet. In human OA chondrocytes and FLS, miR‐34a‐5p mimic increased key OA pathology markers, while miR‐34a‐5p ASO improved cellular gene expression. Intraarticular miR‐34a‐5p mimic injection induced an OA‐like phenotype. Conversely, miR‐34a‐5p ASO injection imparted cartilage‐protective effects in the DMM and high‐fat diet/DMM models. The miR‐34a–KO mice exhibited protection against DMM‐induced cartilage damage. RNA sequencing of WT and KO chondrocytes revealed a putative miR‐34a‐5p signaling network.
Conclusion
Our findings provide comprehensive evidence of the role and therapeutic potential of miR‐34a‐5p in OA.
INTRODUCTION
Despite its being the most common type of arthritis, the pathogenesis of osteoarthritis (OA) is not fully understood due to its complex etiology. Consequently, no disease‐modifying therapies have been effective to date. Obesity is the most prominent modifiable risk factor for OA development (1). A combination of mechanical and molecular mechanisms drives progressive articular cartilage loss. Molecular mechanisms contributing to OA include increased expression of extracellular matrix (ECM) catabolic enzymes (e.g., matrix metalloproteinase 13 [MMP‐13] and ADAMTS‐5), inflammatory cytokines (e.g., interleukin‐1β [IL‐1β], IL‐6, and tumor necrosis factor [TNF]), and mediators of apoptosis (e.g., caspase 3 and its substrate, poly(ADP‐ribose) polymerase [PARP]), in addition to reduced expression of anabolic genes (e.g., COL2A1 and ACAN) and modification of homeostatic processes, including autophagy (2, 3, 4, 5, 6, 7, 8, 9). These changes result in cartilage degradation, synovial inflammation, fibrosis, and subchondral bone sclerosis, which contribute to joint pain, stiffness, and reduced mobility (10, 11).
Concerted efforts have been dedicated to understanding the genetic and epigenetic mechanisms underlying OA (12). The expression of a wide array of genes that regulate joint structure and function is controlled by microRNAs (miRNAs) (13). MiRNAs are short noncoding RNA segments that bind specific complementary sequences in the 3′‐untranslated region of target messenger RNAs (mRNAs) to negatively regulate gene expression (14). Differential miRNA expression patterns in OA patients compared to healthy individuals highlight the importance of miRNAs in OA pathophysiology (15, 16, 17, 18, 19). Previous studies have identified critical roles for miRNAs in chondrogenesis, ECM regulation, inflammatory cytokine production, and other biologic processes that govern normal joint function and maintain homeostasis (19, 20, 21, 22, 23). For instance, miR‐26a expression is reported to alleviate both synovial inflammation and cartilage injury in surgically induced OA in rats (24).
MicroRNA‐34a‐5p (miR‐34a‐5p) is a p53‐regulated tumor suppressor miRNA that modulates biologic functions such as p53‐induced cell cycle arrest, apoptosis, senescence, and proliferation (25, 26, 27). We previously showed that miR‐34a‐5p expression is significantly increased in the synovial fluid of patients with late‐stage OA (Kellgren/Lawrence [K/L] grade 3 or 4) compared to patients with K/L grade 1 or 2 radiographic knee OA (28). Its expression is also increased in chondrocytes from late‐stage OA cartilage compared to healthy cartilage and has been linked to increased apoptosis and reduced proliferation (29, 30). Zhang et al showed that rats receiving a miR‐34a antagomir injection followed by surgical OA induction have reduced chondrocyte death and cartilage degeneration (31), indicating a prophylactic effect. However, the overall contribution of miR‐34a‐5p to OA pathophysiology, including its function, signaling, and therapeutic potential, remains to be fully elucidated.
In this study, we examined the role and therapeutic potential of targeting miR‐34a‐5p in knee OA pathogenesis using human OA biospecimens, in vivo and in vitro models subjected to miR‐34a‐5p mimic and miR‐34a‐5p antisense oligonucleotide (ASO) treatments, and miR‐34a–knockout (KO) mice.
MATERIALS AND METHODS
OA patient recruitment
Blood, cartilage, and synovial tissue were obtained from patients with a K/L grade of 3 or 4 who were scheduled to undergo total knee replacement (TKR), using Research Ethics Board (REB)–approved protocols (07‐0383‐BE and 14‐7592‐AE). Informed consent was obtained from all patients. Plasma was obtained from healthy individuals without musculoskeletal injury, pain that persisted at least 1 month, or pain present on at least half the days of the month prior to providing consent (REB 07‐0383‐BE). Synovial tissue was obtained from patients with early radiographic knee OA (K/L grade 0 or 1) undergoing joint arthroscopy (REB 16‐5969‐AE). Biospecimens used in this study were not donor‐matched (Supplementary Table 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). The sample numbers for individual biospecimen groups are detailed in the respective text below.
Expression of miR‐34a‐5p in plasma and joint tissues
Expression of miR‐34a‐5p in human plasma (n = 37 patients undergoing TKR; n = 34 healthy controls) was determined by quantitative reverse transcriptase–polymerase chain reaction (qRT‐PCR). To account for OA‐related changes in the circulatory levels of miR‐34a‐5p as practically as possible, samples from patients who self‐reported various preexisting conditions were excluded (see Supplementary Methods, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). Expression of miR‐34a‐5p was also determined in human articular cartilage (n = 11 patients with OA [undergoing TKR]; n = 8 normal cadaver controls) and synovial tissue (n = 7 patients with late radiographic OA [K/L grade 3 or 4]; n = 7 patients with early radiographic OA [K/L grade 0 or 1]) by qRT‐PCR. To determine the localization of miR‐34a‐5p in joint tissues, in situ hybridization (ISH) was performed on tissue sections of human articular cartilage with no evidence of degeneration (n = 4) and articular cartilage obtained from TKR patients (n = 4), and on knee joints of 10‐week‐old C57BL/6J mice subjected to destabilization of the medial meniscus (DMM) or sham surgery (n = 4 per group). Additional details are provided in Supplementary Methods.
Treatment of chondrocytes and fibroblast‐like synoviocytes (FLS) with miR‐34a‐5p mimic or miR‐34a‐5p ASO
OA chondrocytes and FLS from each individual patient sample (n = 7–9 independent TKR patient samples) were equally seeded in a 12‐well plate (60,000 cells/well) and transfected with 100 nM miR‐34a‐5p mimic, miR‐34a‐5p ASO, or control oligonucleotide for 24 hours. Relative expression levels of key OA phenotypic markers (Supplementary Table 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract) were investigated by qRT‐PCR, as detailed in Supplementary Methods, and analyzed in a pairwise manner to account for this non‐independence.
Intraarticular injection of miR‐34a‐5p mimic in C57BL/6J mice
Ten‐week‐old male C57BL/6J mice were injected intraarticularly with 5 μg of in vivo–ready miRCURY LNA miR‐34a‐5p mimic or control mimic into the right or left knee, respectively (see Supplementary Methods for details). Joints were harvested 8 weeks post‐injection (n = 8 per group). Histopathologic analysis, Osteoarthritis Research Society International (OARSI) scoring, and immunohistochemistry were performed as described in Supplementary Methods.
Intraarticular injection of the miR‐34a‐5p inhibitor locked nucleic acid (LNA)–ASO in the DMM mouse model of knee OA
Ten‐week‐old male C57BL/6J mice were subjected to DMM of the right knee (n = 22). Mice were injected with 5 μg of in vivo–ready miRCURY LNA miR‐34a‐5p‐ASO (mmu‐miR‐34a‐5p [catalog no. Y104100981; Exiqon]; n = 11) or control oligonucleotide (catalog no. Y100199006 [Exiqon]; n = 11) at 2, 4, and 6 weeks after DMM surgery (a total of 3 injections). Mouse knee joints were collected 10 weeks after DMM surgery for histopathologic analysis, OARSI scoring, and immunohistochemistry (see Supplementary Methods for details).
Expression of miR‐34a‐5p in mice fed a high‐fat diet
Ten‐week‐old male C57BL/6J mice were fed a high‐fat diet (n = 19) or lean diet (n = 25) for 18 weeks, and miR‐34a‐5p expression was determined in mouse plasma and knee joints by qRT‐PCR and ISH, respectively, as detailed in Supplementary Methods.
Intraarticular injection of miR‐34a‐5p LNA‐ASO in mice fed a high‐fat diet and subjected to DMM surgery
Ten‐week‐old male C57BL/6J mice were fed a high‐fat diet for 9 weeks. At the end of the diet, the 19‐week‐old mice were subjected to DMM surgery. Mice were injected with miR‐34a‐5p LNA‐ASO (n = 6) or control oligonucleotide (n = 5) at 2, 3, and 4 weeks after DMM surgery (a total of 3 injections). Mouse knee joints were collected 5 weeks after DMM surgery for histologic analysis and OARSI scoring, as described in Supplementary Methods.
MiR‐34a–KO mice
Ten‐week‐old male homozygous miR‐34a–KO mice (n = 11) and wild‐type (WT) controls (n = 9) were subjected to DMM surgery followed by histopathologic assessment and OARSI scoring 10 weeks after DMM surgery. Additionally, articular chondrocytes isolated from 5‐week‐old female WT and miR‐34a–KO mouse cartilage (hip and knee) were subjected to qRT‐PCR (n = 9 WT and 7 KO mice) (Supplementary Table 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract), RNA sequencing (n = 3 WT and 3 KO mice), and computational analysis. See Supplementary Methods for details.
Statistical analysis
Data are presented as scatterplots or bar graphs with error bars (showing the mean ± SEM). Relative expression data and percent positivity were log‐transformed to fit a normal distribution prior to statistical analyses. ROUT outlier test (Q [maximum false discovery rate] =1%) was conducted on log‐transformed data to determine the presence of outliers using GraphPad Prism software, version 8. Statistical analysis was performed on log fold‐change values of miR‐34a‐5p mimic–treated or miR‐34a‐5p ASO–treated chondrocytes and FLS compared to paired control oligonucleotide–treated corresponding cells to test for differences in response to treatment using Student’s paired t‐test. ROUT outlier test was also conducted on log fold‐change values. To accurately display donor variability, as well as the response of individual patient cells to miR‐34a‐5p mimic or miR‐34a‐5p ASO compared to control oligonucleotide, relative gene expression data were graphed in raw scale. Statistical significance comparing 2 groups with parametric data was assessed by Student’s unpaired or paired 2‐tailed t‐tests. Statistical significance comparing 2 groups with nonparametric data was assessed by Mann‐Whitney U tests. Statistical analysis comparing multiple groups with parametric data was performed by two‐way analysis of variance followed by Tukey’s multiple comparisons post hoc tests. Correlation analysis was conducted using Pearson’s correlation coefficient on log‐transformed values. P values less than 0.05 were considered significant for all comparison tests. Additional experimental procedures are provided in Supplementary Methods.
RESULTS
Systemically and locally increased miR‐34a‐5p expression in patients with late‐stage radiographic knee OA
Expression of miR‐34a‐5p has been shown to be significantly increased in synovial fluid from patients with late‐stage radiographic knee OA (undergoing TKR) compared to patients with K/L grade 1 or 2 radiographic knee OA (28). To determine whether systemic levels of miR‐34a‐5p are altered in OA, we performed miRNA qRT‐PCR analysis on plasma samples from TKR patients and healthy controls (Supplementary Table 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). Significantly higher levels of miR‐34a‐5p were detected in plasma from TKR patients compared to healthy controls (Figure 1A). Since these cohorts were not age‐matched, we conducted a Pearson correlation analysis between age and plasma miR‐34a‐5p levels in healthy controls and TKR patients collectively. We identified a weak‐to‐moderate positive correlation between age and plasma miR‐34a‐5p in our cohort (r = 0.3488, r2 = 0.1216, [95% confidence interval 0.1257–0.5383]; P = 0.0029); however, only 12% of the variation in expression can be explained by age (Supplementary Figure 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract).
Figure 1.

Local and systemic increases in microRNA‐34a‐5p (miR‐34a‐5p) in patients with late‐stage radiographic knee osteoarthritis (OA; who underwent total knee replacement [TKR]) and in the knee joints of mice with OA induced by destabilization of the medial meniscus (DMM). A, Expression of miR‐34a‐5p in plasma from healthy controls (n = 34) and patients with late‐stage knee OA (TKR; n = 37), as determined by quantitative reverse transcriptase–polymerase chain reaction. B, Expression of miR‐34a‐5p in normal cadaveric cartilage (n = 8) and knee OA (TKR) articular cartilage (n = 11). C, Expression of miR‐34a‐5p in synovial tissue from patients with early radiographic knee OA (Kellgren/Lawrence [K/L] grade 0 or 1; n = 7) and patients with OA with a K/L grade of 3 or 4 (TKR; n = 7). D, Safranin O histologic staining (top) and in situ hybridization (ISH; bottom) of miR‐34a‐5p in control non‐degenerated and degenerated knee OA cartilage from TKR patients. E, ISH quantification of the percentage of miR‐34a‐5p positively stained cells from control non‐degenerated (n = 4) and degenerated (n = 4) human knee cartilage. F, ISH of miR‐34a‐5p in knee sections obtained from mice 10 weeks after sham or DMM surgery. Bottom rows show higher‐magnification views (original magnification × 40) of the 2 boxed areas in the top row (original magnification x 4) showing regions of the tibial plateau articular cartilage; top right panels show higher‐magnification views (original magnification × 10) of the boxed area showing the medial anterior aspect of the synovial lining. Arrows indicate miR‐34a‐5p positively stained chondrocytes. G, ISH quantification of percentage of miR‐34a‐5p positively stained cells in mouse knee articular cartilage, synovium, and the meniscus 10 weeks after sham surgery (n = 4) or DMM surgery (n = 4). In A–C, E, and G, data were log‐transformed prior to analysis. Each symbol represents an individual subject; horizontal lines and error bars show the mean ± SEM. * = P < 0.05; ** = P < 0.01, by Student’s unpaired 2‐tailed t‐test.
We then explored whether miR‐34a‐5p expression was dysregulated locally in joint tissues. Expression of miR‐34a‐5p was significantly increased in articular cartilage and synovial tissue from TKR patients compared to normal cadaveric cartilage and synovial tissue from patients with early radiographic knee OA (K/L grade 0 or 1), respectively (Figures 1B and C). ISH of human TKR cartilage revealed a significantly higher percentage of miR‐34a‐5p–positive chondrocytes in degenerated, proteoglycan‐depleted articular cartilage compared to control non‐degenerated cartilage (Figures 1D and E).
To investigate whether miR‐34a‐5p expression is also increased in a mouse model of knee OA, OA was surgically induced in mice by DMM. The severity of cartilage pathology in mouse knee joints 10 weeks after DMM surgery was quantified using the OARSI grading system (32) and compared to that in control mice subjected to sham surgery (Supplementary Figure 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). ISH of mouse knee joints subjected to DMM and harvested 4 and 10 weeks after surgery showed significantly more miR‐34a‐5p–positive cells in the synovial lining, menisci, and articular cartilage at 10 weeks and in the synovial lining and menisci at 4 weeks compared to tissues from mice subjected to sham surgery (Figures 1F and G and Supplementary Figure 3, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). Taken together, these results indicate that miR‐34a‐5p expression is increased locally in OA knee joint tissues from both humans and mice, and its expression is also increased systemically in late‐stage human knee OA.
Altered expression of key OA phenotypic markers in human chondrocytes treated with miR‐34a‐5p mimic or miR‐34a‐5p ASO
Since we observed increased miR‐34a‐5p expression in OA articular cartilage from humans and mice, we next used human OA chondrocyte cultures from the cartilage of TKR patients to examine the mechanistic contribution of miR‐34a‐5p to OA pathology. Human OA chondrocytes (obtained from patients undergoing TKR) were treated with 100 nM miR‐34a‐5p mimic or control oligonucleotide for 24 hours (Supplementary Figure 4A, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract) to examine the expression of key markers involved in ECM composition, as well as cartilage catabolic, inflammatory, autophagic, and hypertrophic mechanisms. MiR‐34a‐5p mimic treatment significantly decreased the expression of the key ECM markers COL2A1 and ACAN, and the pro‐autophagy marker ATG5, compared to control oligonucleotide treatment, while the autophagy markers ATG3 and ULK1 did not show significant changes (Figure 2A). Conversely, miR‐34a‐5p mimic treatment significantly increased the expression of key catabolic markers (MMP13 and ADAMTS5), an inflammatory marker (IL1B), and a hypertrophic marker (COL10A1) in OA chondrocytes compared to control oligonucleotide treatment.
Figure 2.

Modulation of the expression of key OA markers in chondrocytes and fibroblast‐like synoviocytes (FLS) in vitro by miR‐34a‐5p. A and B, Expression of mRNA for anabolic markers (COL2A1 and ACAN), autophagy markers (ATG5, ATG3, and ULK1), catabolic markers (MMP13 and ADAMTS5), an inflammatory marker (IL1B), and a hypertrophy marker (COL10A1) in human OA chondrocytes treated with 100 nM miR‐34a‐5p mimic (A) or miR‐34a‐5p antisense oligonucleotide (ASO) (B) for 24 hours compared to human OA chondrocytes treated with control oligonucleotide, as determined by quantitative reverse transcriptase–polymerase chain reaction (qRT‐PCR). C and D, Expression of mRNA for an extracellular matrix component (COL1A1), a myofibroblast marker (ACTA2), autophagy markers (ATG5, ATG3, and ULK1), a profibrotic cytokine (TGFB), and proinflammatory cytokines (TNF and IL6) in human OA FLS treated with 100 nM miR‐34a‐5p mimic (C) or miR‐34a‐5p ASO (D) for 24 hours compared to human OA FLS treated with control oligonucleotides, as determined by qRT‐PCR. Relative expression data were log‐transformed prior to analysis. Each symbol represents an individual patient sample; horizontal lines and error bars show the mean ± SEM (n = 7–9 patients per group). * = P < 0.05; ** = P < 0.01, by Student’s unpaired 2‐tailed t‐test. See Figure 1 for other definitions.
To determine whether miR‐34a‐5p inhibition could modulate the expression of OA markers in human chondrocytes, cells were treated with 100 nM miR‐34a‐5p ASO or control oligonucleotide (Supplementary Figure 4A). Quantitative RT‐PCR analysis revealed that treatment with miR‐34a‐5p ASO for 24 hours significantly increased the expression of cartilage ECM genes COL2A1 and ACAN, and autophagy genes ATG5, ATG3, and ULK1, in human OA chondrocytes. No significant differences in COL10A1, MMP13, ADAMTS5, or IL1B expression were observed in miR‐34a‐5p ASO–treated OA chondrocytes compared to control oligonucleotide–treated cells (Figure 2B).
Taken together, these results indicate that miR‐34a‐5p mimic promotes the expression of key cartilage destructive markers, while miR‐34a‐5p ASO treatment promotes the expression of key cartilage ECM/anabolic markers in human OA chondrocytes.
Altered expression of synovitis‐associated markers in OA FLS treated with miR‐34a‐5p mimic or miR‐34a‐5p ASO
We next investigated the in vitro effects of miR‐34a‐5p modulation via mimic or ASO on human OA FLS (Supplementary Figure 4B). Following treatment with mimic or ASO, expression levels of key proinflammatory cytokines (TNF and IL6), a profibrotic cytokine (transforming growth factor β [TGFβ]), a key ECM marker (COL1A1), a myofibroblast marker (α‐smooth muscle actin [α‐SMA], encoded by ACTA2 gene), and autophagy markers (ATG5, ATG3, and ULK1) were assessed (2, 33). In FLS treated with miR‐34a‐5p mimic, expression of COL1A1, ACTA2, TGFB, IL6, TNF, and autophagy genes ATG5 and ULK1, was increased compared to FLS treated with control oligonucleotide (Figure 2C). In contrast, miR‐34a‐5p ASO–treated FLS showed a significant decrease in COL1A1, ACTA2, TGFB, TNF, ATG5, and ATG3 expression (Figure 2D). ULK1 expression levels were moderately, but not significantly, reduced in response to miR‐34a‐5p ASO (P = 0.0509), while IL6 expression levels did not change significantly. Taken together, these data show that miR‐34a‐5p may contribute to synovial pathology during OA by promoting the expression of key inflammatory, ECM, profibrotic, and autophagy markers, while miR‐34a‐5p ASO reduces the expression of some of these synovitis‐associated markers in vitro.
OA‐like phenotype in the knee joints of C57BL/6J mice treated with miR‐34a‐5p mimic
We next investigated the in vivo effects of in vivo–grade miR‐34a‐5p mimic injected once into the knees of 10‐week‐old male C57BL/6J mice. We found that a single intraarticular injection of miR‐34a‐5p mimic (5 μg) resulted in reduced proteoglycan staining, reduced chondrocyte cellularity, cartilage fissuring and fibrillation, increased OARSI score, and increased expression of key apoptotic markers (PARP p85 and caspase 3) compared to control oligonucleotide treatment (Figures 3A–E and O). Furthermore, the synovium of miR‐34a‐5p mimic–treated mice showed increased histologic evidence of synovial thickening and ECM deposition compared to controls, as assessed by Masson’s trichrome histochemical staining (Figure 3F). The synovitis score was found to be higher following in vivo miR‐34a‐5p mimic injection compared to control oligonucleotide injection (Figure 3G). Thus, our in vivo data supported our in vitro findings of the pathologic effects of increased miR‐34a‐5p expression on knee joint tissues and demonstrated that a single intraarticular injection of miR‐34a‐5p mimic has destructive effects on the joint.
Figure 3.
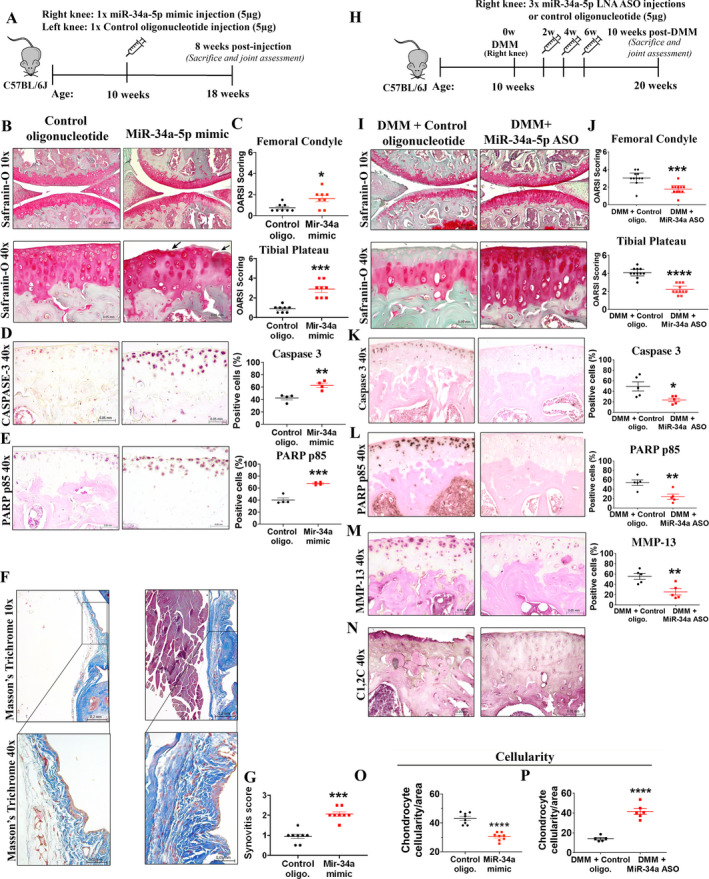
Intraarticular injection of miR‐34a‐5p mimic promotes knee OA development in mice, while intraarticular delivery of miR‐34a‐5p locked nucleic acid (LNA)–antisense oligonucleotide (ASO) protects against DMM‐induced OA. A, Schematic illustration showing the experimental design for injection of in vivo–grade miR‐34a‐5p mimic or control oligonucleotide (control oligo) into the knees of male C57BL/6J mice. B and C, Safranin O staining of (B) and Osteoarthritis Research Society International (OARSI) scores for (C) the femoral condyle and tibial plateau obtained from mice 8 weeks after injection with control oligonucleotide or miR‐34a‐5p mimic. Arrows show cartilage fissuring and fibrillation. D and E, Immunohistochemical (IHC) staining and quantification of caspase 3 (D) and poly(ADP‐ribose) polymerase (PARP) p85 (E) in knee joints from mice injected with control oligonucleotide or miR‐34a‐5p mimic. F and G, Masson’s trichrome histologic staining of the medial anterior aspect of the mouse synovium (F) and synovitis scores for (G) mice injected with control oligonucleotide or miR‐34a‐5p mimic. Bottom row in F shows higher‐magnification views of the boxed areas in the top row. H, Schematic illustration showing the experimental design for injection of in vivo–grade miR‐34a‐5p LNA‐ASO or control oligonucleotide into the knees of mice subjected to DMM. I and J, Safranin O staining of (I) and OARSI scores for (J) the medial femoral condyle and tibial plateau of mice subjected to DMM and injected with control oligonucleotide or miR‐34a‐5p LNA‐ASO. Mice were assessed 10 weeks after DMM. K–M, IHC staining and quantification of caspase 3 (K), PARP p85 (L), and matrix metalloproteinase 13 (MMP‐13) (M) in the knee joints of mice subjected to DMM and injected with control oligonucleotide or miR‐34a‐5p LNA‐ASO. N, IHC staining for C1,2C in the knee joints of mice subjected to DMM and injected with control oligonucleotide (n = 5) or miR‐34a‐5p LNA‐ASO (n = 5). O and P, Chondrocyte cellularity per unit area in mice injected with control oligonucleotide or miR‐34a‐5p mimic (O) and in mice subjected to DMM and injected with control oligonucleotide or LNA‐ASO (P). In C–E, G, J–M, O, and P, each symbol represents an individual mouse; horizontal lines and error bars show the mean ± SEM (n = 8 mice per group in C, G, and O; 4 mice per group in D and E; 11 mice per group in J; 5 mice per group in K–M; and 6 mice per group in P). * = P < 0.05; ** = P < 0.01; *** = P < 0.001; **** = P < 0.0001, by Mann‐Whitney U test in C, G, and J and by Student’s unpaired 2‐tailed t‐test in D, E, K, L, M, O, and P. See Figure 1 for other definitions.
Cartilage‐protective effects of miR‐34a‐5p LNA‐ASO in the mouse DMM model of knee OA
Taken together, our in vitro and in vivo data showed that miR‐34a‐5p mimic promoted the development of an OA‐like phenotype. Therefore, we next hypothesized that targeting miR‐34a‐5p using an in vivo–grade LNA‐ASO would protect against OA development. Five micrograms of in vivo–grade miR‐34a‐5p LNA‐ASO or control oligonucleotide was injected into the mouse knee joints subjected to DMM at 2, 4, and 6 weeks after DMM surgery (Figure 3H). Mouse knee joints were collected 10 weeks after DMM surgery for histologic and immunohistochemical analyses. Injections of miR‐34a‐5p LNA‐ASO protected articular cartilage against degeneration, evidenced by reduced proteoglycan loss, chondrocyte loss, cartilage fibrillation, and OARSI score, as well as reduced expression of caspase 3, PARP p85, MMP‐13, and collagen breakdown product (C1,2C) compared to control oligonucleotide injection (Figures 3I–N and P). Histopathologic changes in the degree of synovitis of miR‐34a‐5p LNA‐ASO–injected mice compared to control‐injected mice were not significantly different (Supplementary Figure 5A, Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). Thus, our preclinical data support the therapeutic potential of intraarticular delivery of miR‐34a‐5p LNA‐ASO to protect against DMM‐induced cartilage damage.
Increased circulating levels of miR‐34a‐5p in obese compared to nonobese patients with knee OA
Increased circulating levels of miR‐34a‐5p have been associated with obesity, the most modifiable risk factor for OA (34); thus, we next tested whether obese OA (TKR) patients exhibited a differential expression profile of miR‐34a‐5p compared to nonobese OA (TKR) patients (Supplementary Table 1). Obese TKR patients (body mass index [BMI] ≥30) had significantly higher miR‐34a‐5p plasma levels than nonobese TKR patients (BMI 18.9–29.9) (Figure 4A). Despite the obese TKR group expressing significantly higher miR‐34a‐5p than the nonobese TKR group, we did observe heterogeneity in miR‐34a‐5p expression levels in both of these groups.
Figure 4.
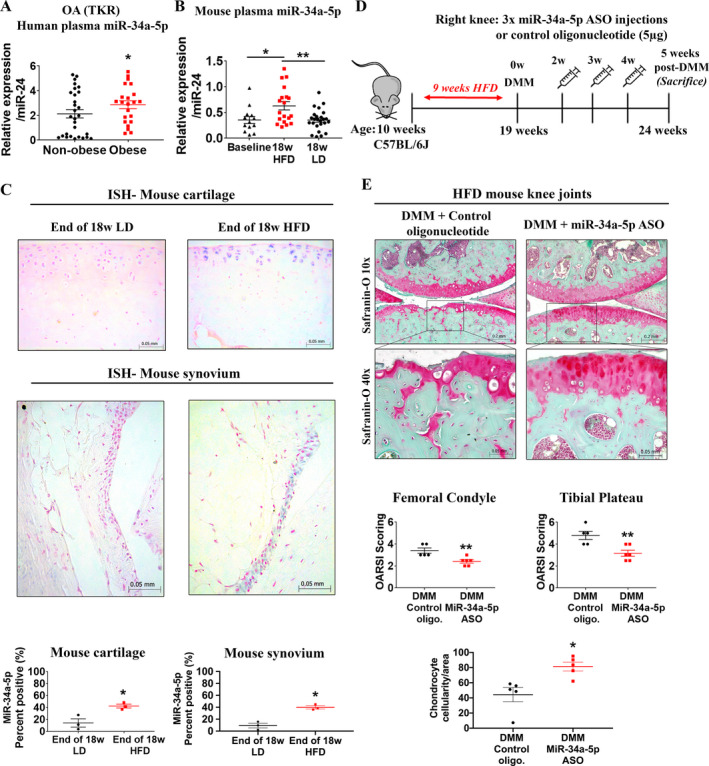
Expression of miR‐34a‐5p is increased during obesity in humans and mice, and miR‐34a‐5p locked nucleic acid (LNA)–antisense oligonucleotide (ASO) treatment imparts cartilage‐protective effects in a high‐fat diet (HFD)–induced accelerated knee OA model in mice. A, Expression of miR‐34a‐5p in the plasma of nonobese patients (n = 29) and obese patients (n = 22) with knee OA (TKR), as determined by quantitative reverse transcriptase–polymerase chain reaction. B, Expression of miR‐34a‐5p in the plasma of 10‐week‐old C57BL/6J mice (baseline; n = 13), mice fed a high‐fat diet for 18 weeks (n = 19), and mice fed a lean diet (LD) for 18 weeks (n = 25). In A and B, relative expression data were log‐transformed prior to analysis. Each symbol represents an individual mouse; horizontal lines and error bars show the mean ± SEM. * = P < 0.05; ** = P < 0.01, by two‐way analysis of variance and Tukey’s multiple comparisons test. C, ISH and percentage of miR‐34a‐5p positively stained cells in mouse articular cartilage and synovium (medial compartment of the knee) at the end of 18 weeks of a lean diet or a high‐fat diet. Data were log‐transformed prior to analysis. Each symbol represents an individual mouse; horizontal lines and error bars show the mean ± SEM (n = 3 mice per group). * = P < 0.05, by Student’s unpaired 2‐tailed t‐test. D, Schematic illustration showing the experimental design for intraarticular injection of miR‐34a‐5p LNA‐ASO or control oligonucleotide into mice fed a high‐fat diet and subjected to DMM surgery. E, Safranin O staining of and Osteoarthritis Research Society International (OARSI) scores for the medial femoral condyle and tibial plateau from mice fed a high‐fat diet, subjected to DMM, and injected with control oligonucleotide or miR‐34a‐5p LNA‐ASO. The chondrocyte cellularity per unit area of mice fed a high‐fat diet, subjected to DMM, and injected with control oligonucleotide or miR‐34a‐5p LNA‐ASO is also shown. Mice were assessed 5 weeks after DMM surgery. Each symbol represents an individual mouse; horizontal lines and error bars show the mean ± SEM (n = 5 mice injected with control oligonucleotide and 6 mice injected with miR‐34a‐5p LNA‐ASO). * = P < 0.05; ** = P < 0.01, by Mann‐Whitney U test for OARSI score; by Student’s unpaired 2‐tailed t‐test for chondrocyte cellularity. See Figure 1 for other definitions.
Elevated miR‐34a‐5p expression levels in the circulation and knee joint tissues of mice fed a high‐fat diet
Since we observed increased miR‐34a‐5p expression in obese OA (TKR) patients compared to nonobese OA (TKR) patients, we measured plasma levels of miR‐34a‐5p in a mouse model of obesity induced by a high‐fat diet. Ten‐week‐old (baseline) male C57BL/6J mice were fed a high‐fat diet for 18 weeks and were significantly heavier than mice fed a lean diet for the same duration (Supplementary Figures 6A and B, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). Quantitative RT‐PCR analysis revealed that plasma miR‐34a‐5p levels were significantly increased after 18 weeks of a high‐fat diet compared to both baseline and 18 weeks of a lean diet (Figure 4B).
To understand molecular changes in the knee joints of mice with diet‐induced obesity, we determined whether a high‐fat diet altered local knee joint miR‐34a‐5p expression. Following 18 weeks of a high‐fat diet or a lean diet, mouse knee joints were collected and miR‐34a‐5p expression was evaluated using ISH. Consistent with increased plasma levels, miR‐34a‐5p expression was significantly increased in the cartilage and synovium of mice fed a high‐fat diet compared to mice fed a lean diet (Figure 4C). Thus, our findings indicate that elevated miR‐34a‐5p expression was induced by high‐fat diet–induced obesity both systemically in circulation and locally in mouse knee joint tissues.
Cartilage‐protective effects of miR‐34a‐5p LNA‐ASO in mice fed a high‐fat diet and subjected to DMM surgery
Since we observed increased miR‐34a‐5p expression in the knee joints of mice fed a high‐fat diet, we next tested the therapeutic effects of miR‐34a‐5p LNA‐ASO in a preclinical model of severe OA by subjecting mice with high‐fat diet–induced obesity to DMM surgery (35). Mice fed a high‐fat diet for 18 weeks, subjected to DMM surgery, and intraarticularly injected with 5 μg of miR‐34a‐5p LNA‐ASO or control oligonucleotide at 2, 4, and 6 weeks after DMM surgery showed a severe OA phenotype with marked depletion of articular cartilage exposing the subchondral bone (Supplementary Figure 6). Therefore, the extent of joint damage rendered it impossible to evaluate the therapeutic effects of miR‐34a‐5p LNA‐ASO. To circumvent this, 10‐week‐old mice were fed a high‐fat diet for 9 weeks (as opposed to 18 weeks), subjected to DMM surgery at 19 weeks of age, and then given weekly (as opposed to biweekly) injections of 5 μg of miR‐34a‐5p LNA‐ASO or control oligonucleotide at 2, 3, and 4 weeks after DMM surgery.
Mice fed a high‐fat diet for 9 weeks were significantly heavier and had significantly higher fasting blood glucose levels than mice at baseline (Supplementary Figures 6D and E). Joints were collected at 5 weeks (rather than 10 weeks) after DMM surgery to capture early changes with or without LNA‐ASO treatment (Figure 4D). Histopathologic assessment of control oligonucleotide–injected knees from mice fed a high‐fat diet revealed extensive proteoglycan loss, enlarged empty chondrocyte lacunae, deep cartilage fissures, and erosion at 5 weeks after DMM surgery (Figure 4E). Conversely, we observed significant protection against both articular cartilage damage and chondrocyte loss in miR‐34a‐5p LNA‐ASO–injected mice fed a high‐fat diet compared to controls (Figure 4E). We did not, however, observe differences in the severity of synovitis between the 2 groups (Supplementary Figure 5B). Overall, intraarticular miR‐34a‐5p LNA‐ASO injections imparted cartilage‐protective effects in both a moderately severe OA mouse model (DMM surgery) and a severe, accelerated OA mouse model (high‐fat diet plus DMM surgery).
Characterization of miR‐34a–KO mice
To identify potential downstream signaling mechanisms through which miR‐34a‐5p operates within articular cartilage, homozygous miR‐34a (which includes both miR‐34a‐5p and miR‐34a‐3p) global KO and WT mice were bred from heterozygous KO mice, as described in Supplementary Methods (Figures 5A and B and Supplementary Figure 7A, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). KO of miR‐34a was confirmed by genotyping (Figure 5C) and qRT‐PCR analysis of mouse chondrocytes (Figure 5D). Quantitative RT‐PCR analysis showed that chondrocytes from miR‐34a–KO mice exhibited significantly increased Col2a1 and Acan expression compared to WT mice (Figure 5E), suggesting that genetic ablation of miR‐34a enhances expression levels of mRNA for major ECM components of the articular cartilage.
Figure 5.
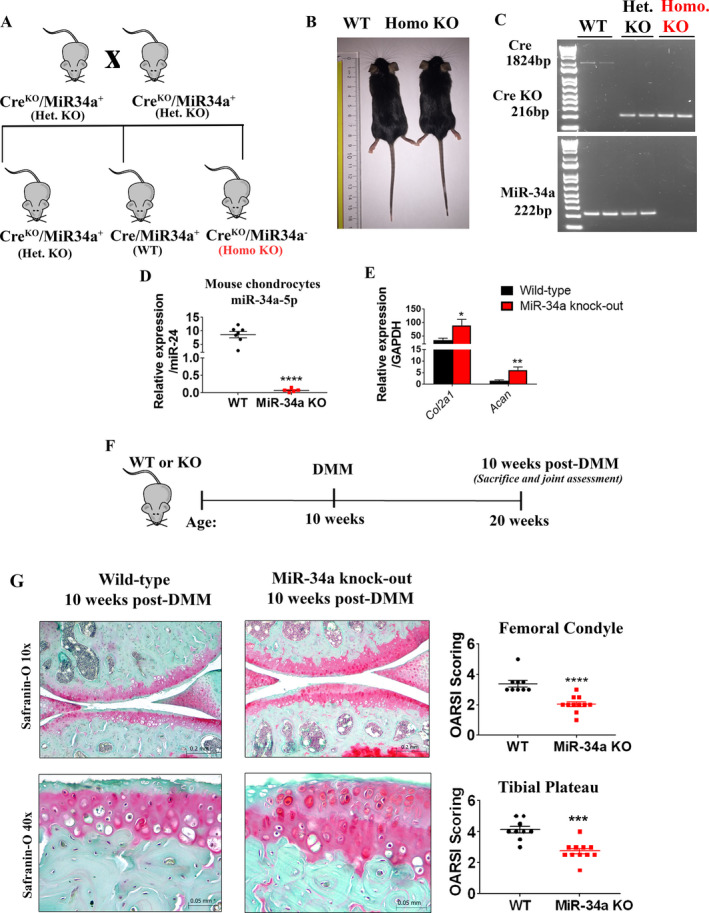
Genetic ablation of miR‐34a protects mice against DMM‐induced cartilage damage. A, Schematic illustration of the breeding strategy used to generate miR‐34a homozygous (homo)–knockout (KO) and wild‐type (WT) mice. het = heterozygous. B, Lengths of 10‐week‐old WT and miR‐34a homozygous–KO mice. C, Cre and miR‐34a genotyping of mouse genomic DNA by polymerase chain reaction. D, Relative expression of miR‐34a‐5p in mouse chondrocytes isolated from articular cartilage of 5‐week‐old WT and miR‐34a homozygous–KO mice, determined by quantitative reverse transcriptase–polymerase chain reaction (qRT‐PCR). Relative expression data were log‐transformed. Each symbol represents an individual mouse; horizontal lines and error bars show the mean ± SEM (n = 7 WT mice and 5 KO mice). **** = P < 0.0001, by Student’s unpaired 2‐tailed t‐test. E, Relative expression of Col2a1 and Acan in chondrocytes from 5‐week‐old WT mice and miR‐34a homozygous–KO mice, determined by qRT‐PCR. Relative expression data were log‐transformed. Bars show the mean ± SEM (n = 7 WT mice and 9 KO mice). * = P < 0.05; ** = P < 0.01, by Student’s unpaired 2‐tailed t‐test. F, Schematic illustration of the experimental design for DMM surgery of male WT or miR‐34a–KO mice. G, Safranin O staining of and Osteoarthritis Research Society International (OARSI) scores for the medial femoral condyle and tibial plateau from WT mice and KO mice 10 weeks after DMM surgery. Each symbol represents an individual mouse; horizontal lines and error bars show the mean ± SEM (n = 9 WT mice and 11 KO mice). *** = P < 0.001; **** = P < 0.0001, by Mann‐Whitney test. See Figure 1 for other definitions.
Reduced severity of cartilage degeneration in miR‐34a–KO mice in vivo
We next determined the effect of miR‐34a genetic ablation on the severity of cartilage degeneration by subjecting WT and miR‐34a–KO mice to DMM surgery (Figure 5F), followed by histopathologic assessments and OARSI scoring. Safranin O staining of 10‐week‐old WT and miR‐34a–KO mouse knee joints before DMM surgery showed no joint histomorphometric differences (Supplementary Figure 7B). Similar to miR‐34a‐5p LNA‐ASO intraarticular intervention, articular cartilage was significantly protected against DMM‐induced damage at the medial tibial plateau and femoral condyle in miR‐34a–KO mice compared to WT mice 10 weeks after DMM surgery, as demonstrated by OARSI scoring (Figure 5G). As observed with miR‐34a‐5p LNA‐ASO treatment, the degree of synovitis in miR‐34a–KO and WT mice was comparable 10 weeks after DMM surgery (Supplementary Figure 5C).
Downstream signaling network of miR‐34a‐5p in articular chondrocytes, determined by RNA sequencing
In order to profile the miR‐34a‐5p downstream signaling network, articular cartilage was collected and pooled from the hips and knees of 5‐week‐old miR‐34a–KO and WT mice, and chondrocytes were subjected to RNA sequencing (Figure 6A). RNA sequencing data from primary mouse KO and WT chondrocytes identified 175 differentially expressed genes (84 up‐regulated and 91 down‐regulated) (Figure 6B; Supplementary Table 3, Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). Since we used a KO of miR‐34a, which involves genetic deletion of both ‐5p and ‐3p strands, the next steps were used to specifically identify the target genes of miR‐34a‐5p with both mouse and human relevance. Subsequent analysis of the top 1% of putative targets of human miR‐34a‐5p and miR‐34a‐3p (obtained using mirDIP [36]) identified 1,330 and 9 putative targets for each miRNA, respectively. Of the 1,339 total human putative targets of miR‐34a, only 6 overlapped with the list of up‐regulated genes and 4 overlapped with the down‐regulated genes identified from sequenced RNA transcripts in miR‐34a–KO mouse compared to WT mouse articular chondrocytes. Focusing on up‐regulated genes, the 6 putative targets were exclusively found in the list of miR‐34a‐5p human target genes (Figure 6C), which included Pparg, Cadm1, Abcc5, Reck, Maoa, and Adgrg2. The remaining 169 differentially expressed genes in KO mouse compared to WT mouse chondrocytes could be modulated by miR‐34a‐5p or miR‐34a‐3p in vivo; however, given that only the top 1% of putative targets were considered, it is unknown whether other genes are also direct targets of miR‐34a in humans.
Figure 6.
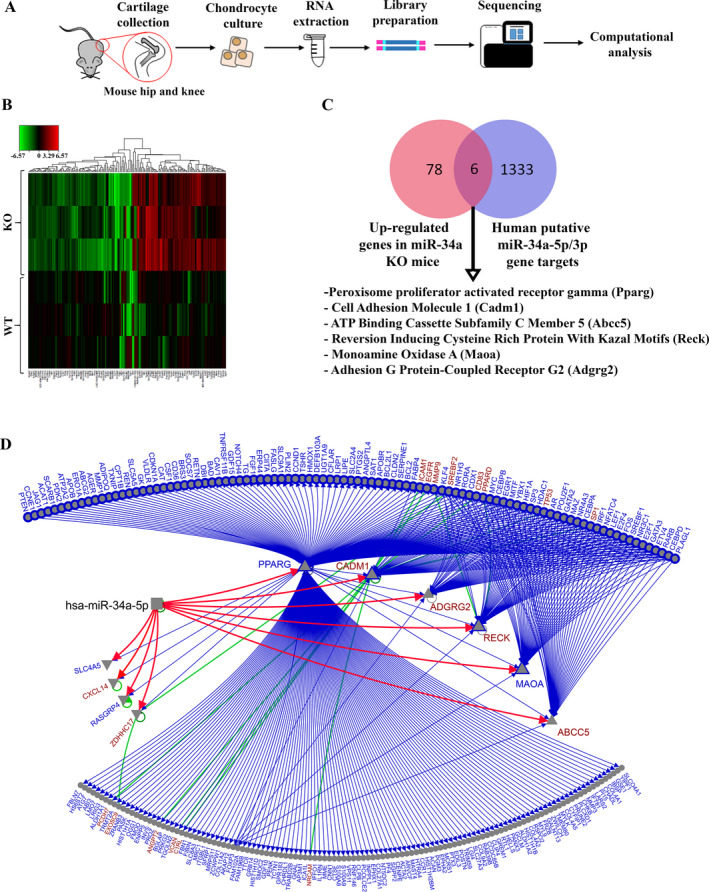
RNA sequencing of wild‐type (WT) and microRNA‐34a–knockout (miR‐34a–KO) mouse chondrocytes identifies a miR‐34a‐5p signaling network. A, Schematic illustration of the experimental design for chondrocyte collection from mouse hip and knee articular cartilage, sample preparation, and RNA sequencing. B, Heatmap representing differentially expressed genes in miR‐34a–KO mouse chondrocytes compared to WT mouse chondrocytes. Each row represents an individual sample (n = 3 WT mice and 3 KO mice), and each column represents a gene transcript. The scale represents log fold‐change of KO transcript expression relative to the corresponding mean of the WT group in each gene. C, Venn diagram illustrating the overlap of 6 genes between 84 up‐regulated miR‐34a–KO mouse chondrocyte genes identified from RNA sequencing and 1,339 mirDIP‐derived human putative miR‐34a‐5p and miR‐34a‐3p direct gene targets. The 6 overlapping genes identified are all predicted targets of miR‐34a‐5p in humans. D, Combined transcription factor (TF) and protein–protein interaction (PPI) and miR‐34a‐5p target network for the differentially expressed genes in miR‐34a–KO mouse chondrocytes. Red directed lines are from mirDIP, blue directed lines are from the TF network, and green (undirected) lines are PPIs. Blue nodes are genes from the TF network only, while red nodes are connected by both TF and PPIs. Outlined nodes are linked by validated edges. Triangles pointing up represent up‐regulated genes, while triangles pointing down represent down‐regulated genes.
Integrative network analysis of the 6 up‐regulated and 4 down‐regulated differentially expressed genes (identified in mouse and common to human putative miR‐34a‐5p target genes) delineated the possible direct and indirect links on miR‐34a‐5p (red lines in Figure 6D), protein–protein interactions (green undirected lines in Figure 6D), and transcription factor connections (blue directed lines in Figure 6D) among the differentially expressed genes identified using RNA sequencing (bioinformatic resources used for network integration and visualization listed in Supplementary Table 4, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41552/abstract). It should be noted that while Pparg was identified as one of the 6 putative targets of miR‐34a‐5p, it was undetected in the WT mouse chondrocytes and poorly expressed in 2 of 3 KO mouse chondrocytes subjected to RNA sequencing. Overall, we identified 6 human putative gene targets of miR‐34a‐5p; however, further validation is needed to determine which of these genes are direct targets of miR‐34a‐5p in articular cartilage.
DISCUSSION
This study elucidated the role and therapeutic potential of miR‐34a‐5p, which is up‐regulated in OA and disrupts articular cartilage homeostasis. Our in vitro data show that miR‐34a‐5p promotes the expression of key catabolic markers and reduces the expression of key ECM markers in human OA chondrocytes. Interestingly, the expression of ECM and autophagy markers was increased in miR‐34a‐5p ASO–treated OA chondrocytes; however, the expression levels of MMP13, ADAMTS5, IL1B, and COL10A1 were not significantly altered, suggesting that targeting miR‐34a‐5p using an ASO is insufficient to reduce the expression of some of these cartilage‐destructive markers in vitro. Our in vitro data also suggest that miR‐34a‐5p contributes to synovial pathology by promoting the expression of key inflammatory, ECM, profibrotic, and autophagy markers in OA FLS. Interestingly, miR‐34a‐5p mimic treatment reduced the expression of autophagy markers in OA chondrocytes; however, the same treatment in OA FLS resulted in an increase in autophagy markers. In rheumatoid arthritis, synovial hyperproliferation and thickening is facilitated in part by increased autophagy and correlates with disease activity (37). Thus, in OA, miR‐34a‐5p could play a role in synovitis partly by increasing autophagy gene expression. These differences in how OA chondrocytes and FLS respond to miR‐34a‐5p mimic or miR‐34a‐5p ASO suggest differential regulation of cellular metabolism processes by miR‐34a‐5p in different cell types.
Intraarticular injection of in vivo–grade miR‐34a‐5p mimic induced an OA‐like phenotype associated with cartilage damage, proteoglycan loss, increased expression of apoptotic markers, and synovial ECM deposition. However, the diffusion of the mimic within the joint space and its uptake by intraarticular tissues in contact with the synovial fluid still remains to be investigated. Future studies could use a fluorescently labeled miR‐34a‐5p mimic to better understand its uptake mechanism.
Our in vivo mouse studies using ISH demonstrate an increased percentage of cells positive for miR‐34a‐5p in the synovium, meniscus, and articular cartilage of mouse knee joints subjected to DMM. In knee OA (TKR) patients, not only is miR‐34a‐5p increased in synovial fluid (28), but we determined miR‐34a‐5p is also increased locally in joint tissues and systemically in circulation. Due to the challenges of obtaining samples from healthy, aged individuals with no history of musculoskeletal disease, our healthy controls and TKR patients were not age‐matched, a limitation of our study. Although beyond the scope of our study, future investigations should be conducted to comprehensively identify associations between levels of miR‐34a‐5p and age, sex, comorbidities, and patient‐reported pain, which could provide greater insight into its clinical utility as a biomarker.
We also identified that miR‐34a‐5p expression is elevated in obese TKR patients compared to nonobese TKR patients, as well as in mice with obesity induced by a high‐fat diet. Our preclinical evidence further support cartilage‐protective effects of miR‐34a‐5p LNA‐ASO treatment in both a moderately severe OA mouse model (DMM surgery), and a severe and accelerated OA mouse model (high‐fat diet plus DMM surgery). Whether the cartilage‐protective effects are a direct result of articular cartilage uptake of the LNA‐ASO or a secondary effect due to synovial tissue uptake warrants further investigation. However, our findings are consistent with previous studies showing cartilage‐protective effects of miR‐34a antagomir or lentiviral vector encoding miR‐34a in attenuating surgically induced OA in rats (29, 31).
The role of miR‐34a in OA pathophysiology was further supported by our in vivo data showing that miR‐34a–KO mice subjected to DMM surgery exhibit protection from cartilage degeneration. Notably, the degree of cartilage protection observed in the miR‐34a–KO mice, which has a deletion of both miR‐34a‐5p and miR‐34a‐3p strands, that were subjected to DMM surgery was similar to that observed with intraarticular miR‐34a‐5p LNA‐ASO injections after DMM surgery in C57BL/6J mice. This suggests that miR‐34a‐5p, as opposed to miR‐34a‐3p, is specifically involved in cartilage degeneration; however, further experiments distinguishing roles and functions of miR‐34a‐5p and miR‐34a‐3p in OA pathogenesis should be conducted to determine potential differential contributions.
While ASO treatment exhibited cartilage‐protective effects, we observed no beneficial effect on the degree of synovitis in our preclinical mouse models. Despite our attempts to comprehensively evaluate articular cartilage and synovial changes, future studies should include evaluation of subchondral bone in order to assess the effects of miR‐34a‐5p LNA‐ASO on other joint tissues.
In this study, we subjected mice to DMM surgery at 10 weeks of age (38, 39, 40), except for in the high‐fat diet model (in which mice were subjected to DMM at 19 weeks of age). It is now recommended that DMM be performed on skeletally mature mice at ~12 weeks of age or older in order to better translate results to adult humans (41). While our findings remain plausible, recent recommendations should be considered for future investigations.
MiR‐34a–KO mouse chondrocytes expressed higher levels of articular cartilage ECM components (Col2a1 and Acan) compared to WT chondrocytes. Unbiased screening using RNA sequencing of WT and miR‐34a–KO mouse chondrocytes identified 6 differentially expressed mouse genes (Cadm1, Abcc5, Reck, Maoa, Adgrg2, and Pparg) that overlapped with mirDIP‐predicted miR‐34a‐5p putative direct targets in humans. Although we have identified a putative miR‐34a‐5p signaling network in this study, further validation is required to determine whether these genes are direct targets of miR‐34a‐5p in articular cartilage.
To conclude, we show that increased levels of miR‐34a‐5p have joint destructive effects and can be targeted using ASO technology to attenuate cartilage degeneration during OA.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Kapoor had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Endisha, Kapoor.
Acquisition of data
Endisha, Datta, Sharma, Nakamura, Rossomacha, Younan, Lively, Potla, Shestopaloff, Krawetz, Mahomed, Jurisica, Gandhi, Kapoor.
Analysis and interpretation of data
Endisha, Datta, Rossomacha, Younan, Ali, Tavallaee, Lively, Potla, Shestopaloff, Rockel, Krawetz, Mahomed, Jurisica, Gandhi, Kapoor.
Supporting information
Supplementary Material
Acknowledgments
The authors would like to acknowledge Kim Perry, Amanda Weston, and the clinical research team of the Division of Orthopedics at Toronto Western Hospital for the collection of human samples.
Supported in part by grants to Dr. Kapoor from the Canadian Institute of Health Research (project grant 173389), the Canadian Epigenetics, Environment, and Health Research Consortium of the Canadian Institutes of Health Research (grant 431893), the Canada Research Chairs Program (grant 950‐232237), the Natural Sciences and Engineering Research Council of Canada (grant 06360), the Canadian Foundation for Innovation (grant 35171), the Krembil Foundation, the Toronto General and Western Hospital Foundation, and the University Health Network Arthritis Program. Dr. Endisha is recipient of a Krembil Research Institute postdoctoral fellowship award. Dr. Jurisica’s work was supported in part by funding from the Ontario Research Fund (RDI grant 34876), Natural Sciences Research Council (NSERC grant 203475), Canada Foundation for Innovation (grants 225404 and 30865), and IBM.
No potential conflicts of interest relevant to this article were reported.
References
- 1. Grotle M, Hagen KB, Natvig B, Dahl FA, Kvien TK. Obesity and osteoarthritis in knee, hip and/or hand: an epidemiological study in the general population with 10 years follow‐up. BMC Musculoskelet Disord 2008;9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kapoor M, Martel‐Pelletier J, Lajeunesse D, Pelletier JP, Hassan F. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol 2011;7:33–42. [DOI] [PubMed] [Google Scholar]
- 3. Caramés B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging‐related loss is linked with cell death and osteoarthritis. Arthritis Rheum 2010;62:791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas CM, Fuller CJ, Whittles CE, Sharif M. Chondrocyte death by apoptosis is associated with cartilage matrix degradation. Osteoarthritis Cartilage 2007;15:27–34. [DOI] [PubMed] [Google Scholar]
- 5. Kim HA, Lee YJ, Soong SC, Choe KW, Song YW. Apoptotic chondrocyte death in human osteoarthritis. J Rheumatol 2000;27:455–62. [PubMed] [Google Scholar]
- 6. O'Brien MA, Moravec RA, Riss TL. Poly (ADP‐ribose) polymerase cleavage monitored in situ in apoptotic cells. Biotechniques 2001;30:886–91. [DOI] [PubMed] [Google Scholar]
- 7. Song RH, Tortorella MD, Malfait AM, Alston JT, Yang Z, Arner EC, et al. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS‐4 and ADAMTS‐5. Arthritis Rheum 2007;56:575–85. [DOI] [PubMed] [Google Scholar]
- 8. Neuhold LA, Killar L, Zhao W, Sung ML, Warner L, Kulik J, et al. Postnatal expression in hyaline cartilage of constitutively active human collagenase‐3 (MMP‐13) induces osteoarthritis in mice. J Clin Invest 2001;107:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitchell PG, Magna HA, Reeves LM, Lopresti‐Morrow LL, Yocum SA, Rosner PJ, et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase‐13 from human osteoarthritic cartilage. J Clin Invest 1996;97:761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loeuille D, Chary‐Valckenaere I, Champigneulle J, Rat AC, Toussaint F, Pinzano‐Watrin A, et al. Macroscopic and microscopic features of synovial membrane inflammation in the osteoarthritic knee: correlating magnetic resonance imaging findings with disease severity. Arthritis Rheum 2005;52:3492–501. [DOI] [PubMed] [Google Scholar]
- 11. Sutton S, Clutterbuck A, Harris P, Gent T, Freeman S, Foster N, et al. The contribution of the synovium, synovial derived inflammatory cytokines and neuropeptides to the pathogenesis of osteoarthritis [review]. Vet J 2009;179:10–24. [DOI] [PubMed] [Google Scholar]
- 12. Peffers MJ, Balaskas P, Smagul A. Osteoarthritis year in review 2017: genetics and epigenetics. Osteoarthritis Cartilage 2018;26:304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cong L, Zhu Y, Tu G. A bioinformatic analysis of microRNAs role in osteoarthritis. Osteoarthritis Cartilage 2017;25:1362–71. [DOI] [PubMed] [Google Scholar]
- 14. Lagos‐Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853–8. [DOI] [PubMed] [Google Scholar]
- 15. Beyer C, Zampetaki A, Lin NY, Kleyer A, Perricone C, Iagnocco A, et al. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis 2015;74:e18. [DOI] [PubMed] [Google Scholar]
- 16. Diaz‐Prado S, Cicione C, Muiños‐López E, Hermida‐Gómez T, Oreiro N, Fernández‐López C, et al. Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC Musculoskelet Disord 2012;13:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Almeida RC, Ramos YF, Mahfouz A, den Hollander W, Lakenberg N, Houtman E, et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann Rheum Dis 2019;78:270–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther 2010;12:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Endisha H, Rockel J, Jurisica I, Kapoor M. The complex landscape of microRNAs in articular cartilage: biology, pathology, and therapeutic targets [review]. JCI Insight 2018;3:e121630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swingler TE, Wheeler G, Carmont V, Elliott HR, Barter MJ, Abu‐Elmagd M, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum 2012;64:1909–19. [DOI] [PubMed] [Google Scholar]
- 21. Grillari J, Grillari‐Voglauer R. Novel modulators of senescence, aging, and longevity: small non‐coding RNAs enter the stage. Exp Gerontol 2010;45:302–11. [DOI] [PubMed] [Google Scholar]
- 22. Yudoh K, Nguyen VT, Nakamura H, Hongo‐Masuko K, Kato T, Nishioka K. Potential involvement of oxidative stress in cartilage senescence and development of osteoarthritis: oxidative stress induces chondrocyte telomere instability and downregulation of chondrocyte function. Arthritis Res Ther 2005;7:R380–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nugent M. MicroRNAs: exploring new horizons in osteoarthritis [review]. Osteoarthritis Cartilage 2016;24:573–80. [DOI] [PubMed] [Google Scholar]
- 24. Zhao Z, Dai XS, Wang ZY, Bao ZQ, Guan JZ. MicroRNA‐26a reduces synovial inflammation and cartilage injury in osteoarthritis of knee joints through impairing the NF‐κB signaling pathway. Biosci Rep 2019;39:BSR20182025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25. He X, He L, Hannon GJ. The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res 2007;67:11099–101. [DOI] [PubMed] [Google Scholar]
- 26. Yamakuchi M, Ferlito M, Lowenstein CJ. MiR‐34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 2008;105:13421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamakuchi M, Lowenstein CJ. MiR‐34, SIRT1 and p53: the feedback loop. Cell Cycle 2009;8:712–5. [DOI] [PubMed] [Google Scholar]
- 28. Li YH, Tavallaee G, Tokar T, Nakamura A, Sundararajan K, Weston A, et al. Identification of synovial fluid microRNA signature in knee osteoarthritis: differentiating early‐ and late‐stage knee osteoarthritis. Osteoarthritis Cartilage 2016;24:1577–86. [DOI] [PubMed] [Google Scholar]
- 29. Yan S, Wang M, Zhao J, Zhang H, Zhou C, Jin L, et al. MicroRNA‐34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int J Mol Med 2016;38:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abouheif MM, Nakasa T, Shibuya H, Niimoto T, Kongcharoensombat W, Ochi M. Silencing microRNA‐34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 2010;49:2054–60. [DOI] [PubMed] [Google Scholar]
- 31. Zhang W, Hsu P, Zhong B, Guo S, Zhang C, Wang Y, et al. MiR‐34a enhances chondrocyte apoptosis, senescence and facilitates development of osteoarthritis by targeting DLL1 and regulating PI3K/AKT pathway. Cell Physiol Biochem 2018;48:1304–16. [DOI] [PubMed] [Google Scholar]
- 32. Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage 2006;14:13–29. [DOI] [PubMed] [Google Scholar]
- 33. Remst DF, Blom AB, Vitters EL, Bank RA, van den Berg WB, Blaney Davidson EN, et al. Gene expression analysis of murine and human osteoarthritis synovium reveals elevation of transforming growth factor β–responsive genes in osteoarthritis‐related fibrosis. Arthritis Rheumatol 2014;66:647–56. [DOI] [PubMed] [Google Scholar]
- 34. Hijmans JG, Diehl KJ, Bammert TD, Kavlich PJ, Lincenberg GM, Greiner JJ, et al. Influence of overweight and obesity on circulating inflammation‐related microRNA. Microrna 2018;7:148–54. [DOI] [PubMed] [Google Scholar]
- 35. Datta P, Zhang Y, Parousis A, Sharma A, Rossomacha E, Endisha H, et al. High‐fat diet‐induced acceleration of osteoarthritis is associated with a distinct and sustained plasma metabolite signature. Sci Rep 2017;7:8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tokar T, Pastrello C, Rossos AE, Abovsky M, Hauschild AC, Tsay M, et al. MirDIP 4.1‐integrative database of human microRNA target predictions. Nucleic Acids Res 2018;46:D360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu L, Wang H, Wu Y, He Z, Qin Y, Shen Q. The autophagy level is increased in the synovial tissues of patients with active rheumatoid arthritis and is correlated with disease severity. Mediators Inflamm 2017;2017:7623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poole R, Blake S, Buschmann M, Goldring S, Laverty S, Lockwood S, et al. Recommendations for the use of preclinical models in the study and treatment of osteoarthritis. Osteoarthritis Cartilage 2010;18 Suppl 3:S10–6. [DOI] [PubMed] [Google Scholar]
- 39. Vasheghani F, Zhang Y, Li YH, Blati M, Fahmi H, Lussier B, et al. PPARγ deficiency results in severe, accelerated osteoarthritis associated with aberrant mTOR signalling in the articular cartilage. Ann Rheum Dis 2015;74:569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lietman C, Wu B, Lechner S, Shinar A, Sehgal M, Rossomacha E, et al. Inhibition of Wnt/β‐catenin signaling ameliorates osteoarthritis in a murine model of experimental osteoarthritis. JCI Insight 2018;3:e96308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ratneswaran A, Beier F. An approach towards accountability: suggestions for increased reproducibility in surgical destabilization of medial meniscus (DMM) models [editorial]. Osteoarthritis Cartilage 2017;25:1747–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


