Abstract
Understanding the role of horizontal gene transfer (HGT) in adaptation is a key challenge in evolutionary biology. In microbes, an important mechanism of HGT is prophage acquisition (phage genomes integrated into bacterial chromosomes). Prophages can influence bacterial fitness via the transfer of beneficial genes (including antibiotic‐resistance genes, ARGs), protection from superinfecting phages, or switching to a lytic lifecycle that releases free phages infectious to competitors. We expect these effects to depend on environmental conditions because of, for example, environment‐dependent induction of the lytic lifecycle. However, it remains unclear how costs/benefits of prophages vary across environments. Here, studying prophages with/without ARGs in Escherichia coli, we disentangled the effects of prophages alone and adaptive genes they carry. In competition with prophage‐free strains, benefits from prophages and ARGs peaked in different environments. Prophages were most beneficial when induction of the lytic lifecycle was common, whereas ARGs were more beneficial upon antibiotic exposure and with reduced prophage induction. Acquisition of prophage‐encoded ARGs by competing strains was most common when prophage induction, and therefore free phages, were common. Thus, selection on prophages and adaptive genes they carry varies independently across environments, which is important for predicting the spread of mobile/integrating genetic elements and their role in evolution.
Keywords: Antibiotic resistance, fitness, lysogen, mobile genetic elements, prophage, temperate phage
A major challenge in evolutionary biology is to understand the role of horizontal gene transfer (HGT) in adaptation (Boto 2010). This question is relevant for different types of organisms, including prokaryotes and eukaryotes (Keeling and Palmer 2008), and answering it requires that we understand how natural selection acts on horizontally acquired genetic variation across different environmental conditions. In bacteria, infectious viruses (phages) are key agents of horizontal gene transfer (Touchon et al. 2017). Some temperate phages exist exclusively episomally, whereas others can integrate into bacterial chromosomes as prophages and modify the bacterial phenotype (lysogenic conversion). This may be beneficial to bacteria, depending on environmental conditions (Waldor and Mekalanos 1996). However, some environmental conditions lead to prophage induction (switching back to the lytic cycle), killing the host cell, and releasing free phages. Our understanding of when prophages will be costly/beneficial to their bacterial hosts, and how these costs/benefits vary across environmental conditions, remains limited (Touchon et al. 2017).
A second important reason to investigate selection on prophages is they can transfer antibiotic resistance genes (ARGs). This may happen via generalized or specialized transduction (Blahova et al. 1999; Gabashvili et al. 2020), or by lysogenic conversion, when ARGs are directly encoded on the phage genome (Brenciani et al. 2010; Calero‐Caceres et al. 2019). Whether or not lysogenic conversion by means of ARG‐encoding prophages is an important driver of antibiotic resistance in nature is unclear. Although a recent viral metagenome (virome) analysis suggests ARGs can be encoded on prophages (Moon et al. 2020), another study suggested ARGs on prophages are less common than previously thought, and showed experimentally that predicted phage‐encoded ARGs often failed to confer resistance (Enault et al. 2017). Thus, quantifying the net fitness effects of prophages with/without ARGs for bacteria across different environments would improve both our basic understanding of the role of phage‐encoded DNA in evolution, and their potential contribution to the spread of antibiotic resistance.
We hypothesize that the net fitness effects of both prophages and adaptive genes they encode (here, ARGs) vary strongly depending on environmental conditions. The benefit of carrying a prophage‐encoded ARG relative to costs of expressing it will be higher in the presence of the selecting antibiotic, whereas in the absence of the antibiotic, expression of the ARG may have a net negative effect on bacterial fitness (a “cost of resistance”; Andersson and Levin 1999). In some conditions, we expect such costs to be compounded by prophages switching back to the lytic cycle and killing the host (Casjens 2003; Paul 2008). This switch from the lysogenic to the lytic cycle can happen spontaneously or in response to environmental conditions, such as sub‐inhibitory antibiotic concentrations that trigger the bacterial SOS response (Otsuji et al. 1959). Although lysis is detrimental for individual cells, the prophage‐carrying population might in fact benefit from the release of free phages, because they may infect and kill competing cells that are susceptible to the released phages in the same microbial community (Brown et al. 2006; Selva et al. 2009; Li et al. 2017). Whether a condition that induces the lytic cycle is costly or beneficial for the host population is predicted to depend on the balance between the costs of lysis within the prophage‐carrying population and the benefit that comes from lysing susceptible competitors (Touchon et al. 2017). The ultimate impact of prophages on their host's fitness is complicated further by the possibility that any free phages released via lysis can potentially transfer ARGs to competing phage‐susceptible cells, further modulating competition between bacterial populations. Consequently, we expect the fitness of lysogens relative to prophage‐free strains to vary depending on antibiotic concentration and other environmental factors influencing prophage induction and lysogeny.
Fitness costs and benefits associated with prophages and prophages that encode adaptive genes, such as ARGs remain incompletely understood. This might be due, among other reasons, to the difficulty of directly comparing prophage‐free bacteria with equivalent strains carrying prophages, both with and without ARGs. Consequently, the following questions remain unanswered: (1) How does the net fitness effect of prophage carriage vary across different antibiotic concentrations? (2) Are these effects specific to prophages that encode ARGs? (3) Are prophages more beneficial in environmental conditions where lysis is more frequent? (4) In which environmental conditions is the acquisition of ARG‐carrying prophages by susceptible strains most likely? To answer these questions, we determined fitness costs and benefits associated with ARGs encoded on naturally occurring prophages of Escherichia coli. To test whether our results are repeatable across different genotypes and resistance genes, we used three common resistance genes encoded on four different lambdoid phages. We used strains carrying these prophages, with and without ARGs that we inserted into the prophages, in various combinations (12 different lysogens). We competed each lysogen against an antibiotic‐ and phage‐susceptible strain (more than 600 competitions in total) in multiple environmental conditions (presence and absence of selective antibiotics at various concentrations, and of a compound that induces the lytic cycle of the prophage). This allowed us to differentiate between the effects of prophages alone and prophages carrying ARGs on their bacterial host's fitness, and to do so across antibiotic concentrations and conditions associated with increased lysis and free phage production. We found that the fitness benefits from prophages and prophage‐encoded ARGs peaked in different environmental conditions, suggesting that in some conditions prophages may increase in frequency because of beneficial genes they carry, and in others because of the benefits of prophage carriage itself, such as lysis of susceptible competitors.
Material and Methods
ORGANISMS
We used E. coli K‐12 MG1655 (hereafter wild‐type, WT), which has originally been cured of active prophages (Hayashi et al. 2006), and 12 different lysogens constructed from this strain (Table 1). Specifically, we used four different prophages, lambda+ (originally obtained as strain JL573 from J.W. Little, University of Arizona), HK022, Phi80 (both originally obtained from M.E. Gottesman, Columbia University), and mEp234 (originally obtained from I.‐N. Wang, SUNY Albany). The prophage in each lysogen encoded either no antibiotic resistance gene (ARG) or one of three different ARGs conferring resistance to ampicillin (amp), kanamycin (kan), or chloramphenicol (cm).
Table 1.
Bacterial strains. The Wild Type (WT) is E. coli K‐12 MG1655. All other strains are lysogens constructed from this strain. Antibiotic resistance phenotype abbreviations: amp ‐ ampicillin, cm ‐ Chloramphenicol, kan ‐ kanamycin. Resistance gene abbreviations: bla ‐ beta‐lactamase, cat ‐chloramphenicol acetyltransferase, neo ‐ aminoglycoside phosphotransferase from Tn5
| Strain | Prophage | Antibiotic resistance phenotype (resistance gene) | Gene deleted to insert resistance marker |
|---|---|---|---|
| WT | none | none | none |
| P1 | lambda+ | none | none |
| P4 | HK022 | none | none |
| P7 | Phi80 | none | none |
| P19 | mEp234 | none | none |
| 140 | lambda+ | kan (neo) | bor |
| 143 | HK022 | kan (neo) | cor |
| 144 | Phi80 | kan (neo) | cor |
| 146 | mEp234 | kan (neo) | cor |
| 151 | lambda+ | cm (cat) | bor |
| 153 | HK022 | cm (cat) | cor |
| 154 | Phi80 | cm (cat) | cor |
| 160 | lambda+ | amp (bla) | bor |
To be able to compare different ARGs within the same phage type and vice versa, phages carrying an ARG were constructed using the first step (exchange of a gene with a resistance cassette) of a λ Red‐promoted gene replacement (Datsenko and Wanner 2000). Resistance cassettes were amplified from plasmids pKD3 (cat, chloramphenicol resistance), pKD4 (neo, kanamycin resistance), and pGEM®‐T Easy (bla, ampicillin resistance) with primers that carry 5’ homology extensions (Table S1). Primers were designed to omit the FRT‐sites on pKD3 and pKD4. Lysogens were then transformed with PCR products by electroporation. To enable a recombination event with the prophage, lysogens carried the plasmid pKD46, a temperature‐sensitive low‐copy number plasmid that expresses the λ Red genes under the control of an arabinose‐inducible promoter and were induced with 20 mM arabinose prior to electroporation. Transformed cells were subsequently plated on the respective antibiotic and successful transformants were isolated, induced and the recombineered phages were inserted into fresh WT cells.
DETERMINATION OF OPTIMAL ANTIBIOTIC CONCENTRATIONS TO USE IN COMPETITION EXPERIMENTS
We measured bacterial growth of each strain across an antibiotic gradient to identify suitable antibiotic concentrations for the competition assays and to determine whether prophage carriage alone (without ARGs) had an effect on antibiotic resistance. Growth assays were performed for each strain at 37°C in triplicates in 96‐well plates containing 10 different antibiotic concentrations, no‐drug controls, and contamination controls (medium only). At the beginning (t = 0) and after 20 h (t = 20), we measured optical density at 600 nm (OD600). We fitted a Hill‐function to the data (Regoes et al. 2004) of the change in optical density (ODt = 20 – ODt = 0) as a function of antibiotic concentration and extracted different inhibitory concentrations (ICs), at which growth relative to that observed for the same strain in the absence of the antibiotic was reduced by 20% (IC20), 30% (IC30), and 50% (IC50). We confirmed that the ARGs encoded on prophages conferred antibiotic resistance to lysogens by observing much higher ICs for ARG‐carrying lysogens compared to the WT (Fig. S1). In preliminary competition assays, we observed the extinction of the WT at concentrations higher than IC50. We therefore performed the main competition experiment at the following antibiotic concentrations: high (IC50 for the WT; Kan: 2.17 μg/ml, Cm: 1.31 μg/ml, Amp: 4.41 μg/ml), intermediate (IC30 for the WT; Kan: 1.55 μg/ml, Cm: 1.08 μg/ml, Amp: 3.52 μg/ml), and low (IC20 for the WT; Kan: 0.565 μg/m, Cm: 0.623 μg/ml, Amp: 1.8 μg/ml).
COMPETITION ASSAYS
To determine the costs and benefits for bacteria of carrying prophages we performed competition assays between each lysogen (Table 1) and a phenotypically marked version of the WT. This competing strain is susceptible to each phage and antibiotic, and is labeled with a fluorescent yellow‐super‐fluorescent protein (SYFP) marker (Gullberg et al. 2014). For the eight lysogens where the prophage encoded an ARG (Table 1), we assayed each lysogen with and without the respective antibiotic at each concentration. For each ARG‐carrying lysogen, we also assayed a corresponding ARG‐free lysogen (Table 1; carrying the same prophage but without an ARG) in the same conditions. We tested each of these combinations of lysogen (n = 8), ARGs (with/without) and antibiotic concentration (zero, low, intermediate, high) in the presence and absence of a compound that induces the lytic cycle (mitomycin C, at a final concentration of 0.5 μg/ml). With five independent biological replicates of each strain, each originating from a single colony, this gave 640 competitions in total. We also accounted for the effect of the SYFP marker carried by the WT, by measuring the fitness of the SYFP‐labeled WT relative to the unlabelled WT in each of the tested environmental conditions. We then subtracted the difference in relative fitness between both WT strains from the fitness of each lysogen relative to the YFP‐labeled WT for each environmental condition.
For each treatment group, five independent biological replicates of each competing strain derived from five different colonies were inoculated in 5 ml of LB and grown overnight at 37°C in a shaking incubator (180 rpm). The following day we determined the optical density of each overnight culture at 600 nm and adjusted the OD of each culture to the lowest measured OD, to obtain similar concentrations of bacteria per culture. Then, cultures of each lysogen were mixed 1:1 (v:v) with the SYFP‐labelled WT, diluted 1:1000 into the respective environment (combination of antibiotic concentration and mitomycin C) in a 96‐well microplate to initiate each competition culture, and incubated overnight at 37°C. At the start of each competition and after 24 h, absolute cell densities of the two competing populations were determined by flow cytometry (Novosampler Pro; ACEA Biosciences Inc., San Diego CA), where fluorescent (WT) and non‐fluorescent cells (lysogens) were counted, similarly to past work on costs and benefits of antibiotic resistance (Qi et al. 2016). Competitive fitness was estimated as selection rate constant, r, calculated as the difference of the realized Malthusian parameters between competing strains (Lenski et al. 1991):
DETECTION OF PROPHAGE ACQUISITION BY THE WILD TYPE
We plated the competing‐cultures after 24 h onto non‐selective and selective agar‐plates (supplemented with the respective antibiotic) to determine the presence and frequency of newly antibiotic‐resistant WT clones (which could grow on antibiotic plates and were distinguished from the competing strain by yellow fluorescence). We did this only for competition cultures involving ARG‐carrying lysogens. To determine whether newly antibiotic‐resistant colonies had acquired antibiotic resistance by lysogenization (acquiring the prophage), all antibiotic‐resistant WT clones were subsequently checked for the presence of the respective prophage using prophage‐induction by mitomycin C. This works by incubating 100 μl of bacterial culture with 0.5 μg/ml mitomycin C for 4 hours in a spectrophotometer, measuring optical density periodically. If prophages have been acquired, we expect the population to crash and eventually clear completely due to prophage induction and lysis, which in these conditions typically occurs after ∼1‐2 h (Refardt 2011). The supernatant of these mitomycin C‐induced cultures was further spotted on a lawn of susceptible E. coli K12 MG1655, to confirm the presence of free phages in the induced cultures (see also below in section “Production of free phages”). We also used a second test for evidence of prophage acquisition, by assaying resistance of each putative new lysogen to a virulent mutant of the respective prophage (prophage acquisition is expected to confer immunity against superinfection by the same phage genotype). We did this by cross‐streaking each putative new lysogen over a line of the respective lytic phage (Refardt 2011), and inferred resistance when new lysogens showed no visible sign of inhibition by phage after 24 h incubation.
We estimated the relative fitness of the new lysogens (WT clones that acquired antibiotic resistance via lysogenization during the competition assay) by competition assays as above. We did this for each new lysogen against the ancestral WT (telling us about the net effect of acquiring the prophage including the ARG), the ancestral lysogen without the respective ARG (telling us about the effect of the ARG), and the ancestral lysogen with the ARG (which we would expect to have similar fitness to the new lysogens). Competitive fitness assays were performed in triplicate in the absence of antibiotics and at the IC20 of the respective antibiotic as above.
PRODUCTION OF FREE PHAGES
Lysogens can release free phages due to spontaneous prophage induction or in response to environmental stress, such as exposure to mitomycin C (Pricer and Weissbach 1964). In addition to the competition assays, we determined the production of free phages for each tested lysogen in the same eight environments used during competition assays (four antibiotic concentrations each tested in the presence and absence of mitomycin C) by means of standard spot‐assays (Carlson 2005) in a separate experiment. Overnight cultures of each lysogen (with and without ARGs), grown from single colonies, were diluted 1/1000 in 5 ml LB and incubated for 2 h to bring cultures into exponential growth. Each of these 2 h‐growth cultures was adjusted to a similar optical density (OD 600 nm) to achieve approximately equal cell densities, before 180 μl was transferred to a 96‐well microplate. After 4 h (when induction had completed, which was visible by complete clearance of mitomycin C‐treated cultures), we diluted each culture 1/10 in SM‐Buffer, added a drop of chloroform, and centrifuged for 10 min at 5000 × g to remove bacteria and isolate any free phages. To estimate the abundance of free phages in each culture, we then spotted a dilution series of these lysates on double‐agar plates containing the WT in the overlay agar. Plates were incubated overnight at 37°C and plaque‐forming units (PFU) were counted the following day, with three replicates per treatment.
REDUCTION IN BACTERIAL GROWTH CAUSED BY FREE PHAGES
We measured the ability of free phages to inhibit population growth of the WT by determining the reduction in bacterial growth (RBG) in liquid culture, adapted from (Poullain et al. 2008). To do this we first isolated free phages from each lysogen (Table 1), by induction as described above using 0.5 μg/ml mitomycin C in the absence of antibiotics. We then incubated cultures of the WT in 96‐well plates, with each culture containing either no antibiotics or one of the three antibiotics (ampicillin, chloramphenicol, or kanamycin) at IC20, IC30, or IC50. Each culture was inoculated at a concentration of ×∼1.3 × 108 CFU/ml of bacteria, to which we added ∼6.7×108 PFU/ml of one of the phages or no phage (control). We then estimated bacterial abundance by measuring optical density at 600 nm at t = 0 h and again at t = 24 h of static incubation at 37°C, with three replicates per treatment. The reduction in bacterial growth was calculated as:
STATISTICAL ANALYSES
All analyses were performed in R version 3.5.3.
Relative fitness
Our experimental design was not fully factorial (our strains represent a subset of all possible phage×ARG combinations; Table 1). We, therefore, split the dataset by antibiotic, which allowed us to fit a fully factorial linear model within each antibiotic treatment. In a first model, we took lysogen fitness relative to the WT in the absence of mitomycin C as the response variable, biological replicate as a random effect, and as fixed effects, we included antibiotic concentration (zero, IC20, IC30, IC50), ARG (with/without), and phage (Lambda, HK022, Phi80 or mEp234 for kanamycin; Lambda, HK022 or Phi for chloramphenicol; phage not included for ampicillin because only Lambda was represented with this antibiotic; Table 1). In a second model, we included the treatment groups with mitomycin C added, and mitomycin C (presence/ absence) as a fixed factor. In both models, we initially included all interaction terms, before reducing each model by sequentially removing non‐significant interactions.
To separate the fitness effects of prophage carriage from the fitness effects of ARGs, we estimated the average fitness effect of each prophage in each treatment group (combination of antibiotic, antibiotic concentration, mitomycin C, and prophage type) as the mean selection rate constant for the ARG‐free lysogen relative to the WT. We then estimated the average fitness effect of each prophage‐encoded ARG in each treatment group as the difference in mean selection rate constant between each ARG‐carrying lysogen and the corresponding ARG‐free lysogen in the same treatment group. That is, the fitness effect of the ARG here is the additional cost/benefit of the ARG in competition with a prophage‐free strain, after accounting for any fitness effect of the prophage the ARG is carried on (measured separately with the equivalent ARG‐free lysogen). Next, to determine which treatment groups were associated with similar fitness benefits of prophages and ARGs, we performed a pairwise‐distance cluster analysis robust to outliers. We clustered treatment groups according to the variables (i) mean prophage fitness effect and (ii) mean ARG fitness effect. We did this separately for each antibiotic. Preliminary analysis of within‐cluster sums of squares for different levels of k (the number of clusters) suggested that two to five clusters gave good levels of explanatory power for each antibiotic, so we proceeded with k‐means clustering using km = 3 in the R package ComplexHeatmap.
Fitness costs of ARGs without antibiotics/mitomycin C
We tested for an average effect of ARGs on bacterial fitness in the absence of antibiotics and mitomycin C using a pairwise t‐test comparing lysogens with versus without ARGs across all phage and ARG combinations. To determine the smallest fitness effect detectable with this test, we used power analysis with the function power.t.test, with power = 0.8, p = 0.05, SD = 0.74 and n = 8. This revealed that this test would detect significant fitness effects greater than r = 0.85.
Production of free phages in each environmental condition and absolute cell densities
Absolute cell densities during the pairwise competitions and free phage production of each lysogen in the same conditions were analyzed following the same approach as for relative fitness data, using a log‐transformation for free phage abundances (measured in PFU/ml). We further performed a correlation analysis to determine whether the average relative fitness of lysogens in different treatment groups was correlated with the amount of free phages produced by lysogens in the same conditions (but measured in a separate experiment).
Phage predation rate
We determined differences in RBG for the different phages for each antibiotic using a linear model with antibiotic concentration, presence/absence of the respective ARG, phage type, and all their interactions as fixed effects, reducing the model as above.
Lysogenization
Using data on the fraction of competition assays where we observed new lysogens, we used logistic regression to estimate the probability for the WT population to acquire an ARG‐carrying prophage (lysogenization) in each environmental condition (treatment group). We took presence/absence of new lysogens in each competition culture as the dependent variable and presence/absence of mitomycin C, antibiotic concentration (zero, IC20, IC30, or IC50), and phage type as independent variables. We pooled the data from the different antibiotic treatments here, because we found no effect of antibiotic (and therefore the type of resistance gene) on the probability to acquire an ARG‐carrying prophage (glm: χ2(df = 2) = 0.7, p = 0.71). The full model was analyzed by a generalized linear model and Analysis of Deviance, for which we assumed deviance change to be approximately χ2 distributed.
Relative fitness of WT‐lysogens
To analyse the fitness of new lysogens (WT colony isolates that acquired an ARG‐carrying prophage during the competition experiment), we used a linear mixed effects model (package: nlme, function lme) for each antibiotic (Amp, Cm, and Kan) with prophage type, presence/absence of antibiotics (IC20), and competitor (Phage‐and‐ARG‐free WT, ARG‐free lysogen, and ARG‐carrying lysogen) and their interactions as fixed effects, and lysogen as a random effect.
Growth rate in monoculture
We used the SummarizeGrowth function implemented in the R package growthcurver (Sprouffske and Wagner 2016) to estimate the intrinsic growth rate for each strain (Table 1) in the absence of antibiotics or mitomycin C. We used Welch's pairwise t‐tests with sequential Bonferroni correction to determine whether individual lysogens had different growth rates compared to the WT.
Results
NET BENEFITS OF ARG‐ENCODING PROPHAGES INCREASE AT HIGHER ANTIBIOTIC CONCENTRATIONS
In the absence of antibiotics and mitomycin C, prophage‐carrying strains (lysogens) generally had a competitive fitness greater than or similar to the WT (selection rate constant ≥0; Fig. 1A). In these conditions, we did not detect an average difference between lysogens where the prophage encoded an ARG and those where it did not (paired t‐test: t7 = 0.45, p = 0.67; Fig. 1A). Note we do not exclude costs of ARGs too small for our method to detect (see Methods) or for individual lysogens, although in only three out of eight cases estimated fitness was lower with the ARG than without. Together, this suggests that in general, neither ARGs nor prophages were costly in the absence of antibiotics. This was supported by experiments in monoculture, where we observed similar growth rates for lysogens compared to the WT (mean difference of lysogen growth rate from WT = 3.8% ±16.6% (s.d.); Figure S2).
Figure 1.
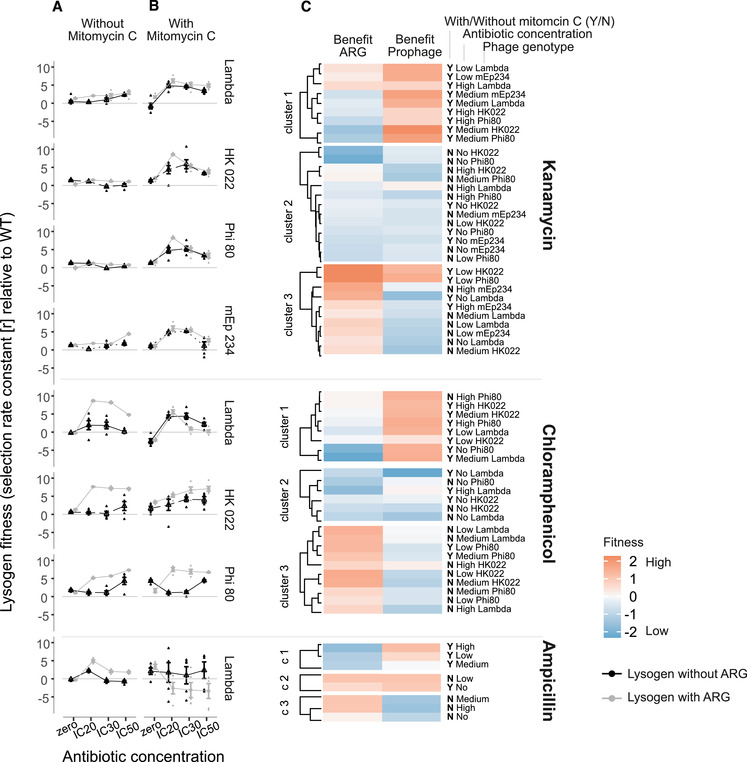
Competitive fitness of lysogens relative to the antibiotic‐susceptible and phage‐susceptible Wild Type in the (A) absence and (B) presence of mitomycin C. Each row of panels shows data from a different combination of antibiotic (given at right of panel C) and prophage type (given at right of panel B). Shown are single data points as well as means ± s.e. from six replicate populations. Black/grey points indicate lysogens without/with ARGs, assayed in competition with the WT at different antibiotic concentrations (x‐axis). Lysogens have higher fitness than the WT when the selection rate constant r > 0. (c) Benefits of ARGs (left column) and prophages (right column) in different treatment groups (labeled at the right of the heatmap), clustered by k‐means clustering (see Methods). Data have been scaled using the scale function implemented in R prior to the cluster analysis.
Upon addition of antibiotics (but without mitomycin C), average lysogen fitness increased (antibiotic concentration effect in linear model for mitomycin C‐free treatment groups ‐ ampicillin treatment group: F3,28 = 104.255, p < 0.001; chloramphenicol treatment group: F3,108 = 38.81, p < 0.001; kanamycin treatment group: F3,148 = 6.36, p < 0.001; Fig. 1A). This effect was driven by lysogens with ARG‐carrying prophages, indicating resistance genes became advantageous at higher antibiotic concentrations (antibiotic concentration × ARG interaction in linear model for mitomycin C‐free treatment groups ‐ ampicillin: F3,28 = 18.03, p < 0.001, chloramphenicol: F3,108 = 25.58, p < 0.001, kanamycin: F1,148 = 5.14, p = 0.002; Fig. 1A). By contrast, only a minority of ARG‐free lysogens showed notable fitness increases, and only at certain antibiotic concentrations (Fig. 1A). While the increasing advantage of ARGs was qualitatively consistent across all ARG‐encoding prophages, the average fitness advantage of the ARG (relative to ARG‐free versions of the same prophages) was higher for chloramphenicol‐ and ampicillin‐resistance than for kanamycin‐resistance (Fig. 1A).
BENEFITS OF ARGS AND PROPHAGES PEAK IN DIFFERENT ENVIRONMENTAL CONDITIONS
For most lysogens addition of mitomycin C had a relatively strong, positive effect on average lysogen fitness relative to the WT at non‐zero antibiotic concentrations (Fig. 1B). This was the case for all lysogens in the kanamycin treatment group (mitomycin C × antibiotic concentration interaction for kanamycin treatment group: F3, 250 = 114.56, p < 0.001; Fig. 1B) and for some lysogens in the chloramphenicol treatment group (F3, 186 = 0.62, p = 0.6; significant only at antibiotic concentration = IC30: Estimate = 2.88, t = 2.65, p = 0.009). In the ampicillin treatment group, addition of mitomycin C did not result in higher average lysogen fitness at non‐zero concentrations (Fig. 1B), as it did in the kanamycin‐treatment lysogens.
The increasing benefits of ARGs at non‐zero antibiotic concentrations observed in the absence of mitomycin C (Fig. 1A) were modified by addition of mitomycin C (mitomycin C × ARG × antibiotic concentration interaction for chloramphenicol: F3,186 = 8.71, p < 0.001; for kanamycin: F3,250 = 9.55, p < 0.001; Fig. 1B). This was because in the absence of mitomycin C and at non‐zero antibiotic concentrations, carriage of an ARG had a larger effect on lysogen fitness than carrying a prophage did (in Cluster 3, there was a higher average benefit of ARGs than prophages, and these treatment groups were mostly mitomycin C‐free; Fig. 1C). In contrast, in the presence of mitomycin C, the fitness benefits of ARGs relative to prophages were less pronounced or even absent, and lysogen fitness was primarily determined by prophage carriage (in Cluster 1 there was a higher benefit of prophages than ARGs, and mitomycin C was present in most of these treatment groups; Fig. 1C). Crucially, this pattern of relatively large benefits of prophage carriage in the presence of mitomycin C, and of ARG carriage in the absence of mitomycin C and presence of antibiotics, was consistent across all three antibiotics (Fig. 1C). The reduced benefit of ARG carriage was particularly pronounced in the presence of ampicillin and mitomycin C, where AmpR‐lysogens were at a disadvantage relative to the WT (Fig, 1B and C).
We observed no significant variation of mean lysogen fitness among the four different prophage types (lambda, HK022, Phi80, mEp234) in the presence of kanamycin (prophage type effect for kanamycin: F3, 250 = 1.59, p = 0.19). However, in the presence of chloramphenicol, average lysogen fitness varied among prophage types (prophage type effect for chloramphenicol: F2,186 = 40.88, p < 0.001; Fig. 1). This indicates the variation of lysogen fitness across different types of ARG‐carrying prophages depends on which type of antibiotic resistance we look at.
INCREASED LYSOGEN FITNESS IN ENVIRONMENTAL CONDITIONS THAT SUPPORT HIGHER FREE‐PHAGE PRODUCTION
When grown in monoculture, almost all lysogens produced free phages in every experimental environment (except for prophage lambda carrying the ampicillin resistance gene; Figure S3). Addition of mitomycin C significantly increased the average amount of released phages (effect of mitomycin C: ampicillin treatment group: F1,38 = 61.44, p < 0.001; chloramphenicol treatment group: F1,107 = 25.84, p < 0.001; kanamycin treatment group: F1,110 = 81.39, p < 0.001; Fig. 2). This indicates that, in the competition experiments above, addition of mitomycin C was probably also associated with increased free phage production. Consistent with these treatments being associated with relatively high lysogen fitness in the competition experiment, we observed a positive association across all treatment groups between average lysogen fitness in the competition experiment and the concentration of free phages measured in monoculture (r = 0.2, p = 0.02; Fig. S4). Note while free phage abundance was strongly influenced by mitomycin C, it also varied depending on which phages were present, being on average higher in cultures including ARG‐carrying lysogens (Fig. S3). Bacterial densities in those cultures were relatively high compared to cultures with ARG‐free lysogens (linear model: F1,958 = 34.503, p < 0.001), which potentially explains the increased total free phage production we observed.
Figure 2.
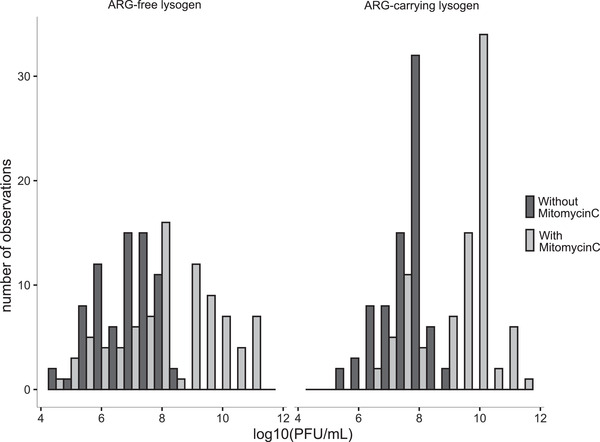
Production of free phages in the presence (grey) and absence (black) of mitomycin C. The number of cultures (y‐axis) with each amount of produced phages (x‐axis) is shown for competition cultures involving ARG‐free lysogens (left panel) and ARG‐carrying lysogens (right panel). Further details, including variation depending on all of the factors we tested and among replicates within each treatment group, are shown in Figure S3.
In the competition experiments, mitomycin C also caused a significant reduction in total bacterial population densities, by more than one order of magnitude compared to mitomycin C‐free treatments (mean ± SD = 8.48 × 107 ± 8.45 × 106 cells/ml with versus 1.25 × 109 ± 7.23 × 107 cells/ml without; linear model ‐ ampicillin treatment group: t = −6.3, p < 0.001; chloramphenicol treatment group: t = −3.95, p < 0.001; kanamycin treatment group: t = −2.03, p = 0.04; Fig. S5). This reduction in population density could have resulted from direct killing or growth inhibition by mitomycin C (for both the lysogen and the WT in each competition). Alternatively, it could have resulted from the effects of prophage induction and lysis (of prophage carrying cells upon switching to the lytic cycle, and of WT cells by released free phages). We found growth of the prophage‐free WT was not strongly reduced by mitomycin C at this concentration in pure culture (Fig. S6). Prophage‐carrying strains showed temporal growth dynamics upon exposure to mitomycin C in pure culture (Fig. S6) consistent with prophage induction and abrupt lysis of a large fraction of the population, rather than constant growth inhibition by mitomycin C independent of the prophage. This suggests the reduced population densities in the presence of mitomycin C in competition cultures were probably caused by prophage induction, resulting in cell death of the induced lysogens and subsequent killing of the competing WT population by released phages. In support, using reduction in bacterial growth (RBG) assays, we confirmed the wild type was highly susceptible to growth inhibition by free phages (Fig. S7). Thus, experimental conditions supporting increased free phage production by lysogens were associated with increased competitive fitness of lysogens relative to the WT, even though both strains reached lower absolute abundances compared to when free phages were less abundant (Fig. S5).
LIKELIHOOD OF ARG‐TRANSFER BY PROPHAGES VARIES WITH ENVIRONMENTAL CONDITIONS
In 16 out of 320 pairwise competitions involving ARG‐carrying lysogens, we detected putative new lysogens (descended from the prophage‐free WT strain) that appeared to have acquired ARG‐carrying prophages from the competing lysogen populations (Table 2). Note, we carried out these additional experiments only for competition cultures involving ARG‐carrying lysogens. The acquisition of ARG‐carrying prophages was evidenced by the WT strain becoming antibiotic resistant, becoming inducible by mitomycin C (including the production of free phages that could subsequently infect a susceptible WT clone), and becoming resistant to a virulent mutant of the respective prophage (see Methods). 15 of these 16 WT lysogens emerged in the presence of antibiotics (antibiotic concentration effect in logistic regression: χ23 = 8.52, p = 0.04). Thirteen of the WT lysogens were detected in the presence of mitomycin C (mitomycin C effect in logistic regression: χ21 = 8.36, p = 0.004). The combination of intermediate antibiotic concentrations (IC20 and IC30) and the presence of mitomycin C were the same environmental conditions where lysogen fitness peaked in the competition experiments. These mostly fall into cluster one, representing environmental conditions where prophage carriage was more beneficial than ARG carriage (Fig. 1C). Finally, most new (WT) lysogens emerged in cultures with lambda prophages (12 out of 16), suggesting the probability of prophage‐mediated ARG‐transfer to a susceptible strain differs across prophage types (effect of prophage type in logistic regression: χ23 = 12.73, p = 0.005, Table 2).
Table 2.
Treatment groups of all 16 populations where putative WT‐lysogens (that appeared to have acquired a prophage during pairwise competition) were detected. We only looked at competition cultures involving ARG‐carrying lysogens. Frequency: colony count for new WT lysogens in each culture as a percentage of the total colony count for the WT population
| Phage genotype | Mitomycin C | Antibiotic | Concentration | Cluster | Frequency (%) |
|---|---|---|---|---|---|
| Lambda | Y | amp | IC50 | 1 | 0.06 |
| Lambda | Y | amp | IC30 | 1 | 0.06 |
| Lambda | Y | amp | IC30 | 1 | 0.08 |
| Lambda | Y | cm | IC30 | 1 | 0.06 |
| Lambda | Y | cm | IC30 | 1 | 0.16 |
| Lambda | Y | cm | IC30 | 1 | 0.05 |
| Lambda | Y | cm | IC20 | 1 | 0.09 |
| Lambda | Y | cm | IC20 | 1 | 0.08 |
| Lambda | Y | cm | IC20 | 1 | 0.04 |
| Lambda | Y | cm | IC50 | 2 | 0.37 |
| Lambda | Y | kan | IC50 | 1 | 0.04 |
| Lambda | Y | kan | IC30 | 1 | 0.04 |
| Lambda | Y | kan | IC30 | 1 | 6.67 |
| HK022 | N | kan | IC20 | 2 | 25 |
| Phi80 | Y | kan | IC30 | 1 | 15.38 |
| mEp234 | N | kan | 0 | 2 | 0.24 |
| HK022 | N | cm | IC30 | 3 | 0 |
ACQUISITION OF ARG‐CARRYING PROPHAGES INCREASES BACTERIAL FITNESS IN THE PRESENCE OF ANTIBIOTICS
To determine how acquisition of ARG‐carrying prophages in our main experiment affected bacterial fitness in these conditions, we carried out pairwise competitions between the new WT‐lysogens (isolated from the competition experiment) and each of three different ancestral strains: (1) phage‐free‐ARG‐free WT, (2) ARG‐free lysogens with the same type of prophage, (3) ARG‐carrying lysogens with the same type of prophage and ARG. We found that the fitness of new WT‐lysogens relative to competing bacteria was affected by increasing antibiotic concentration, but this effect depended on the identity of the competing bacteria (competitor × environment interaction: ampicillin treatment group: F2,50 = 8.53, p < 0.001; chloramphenicol treatment group: F2,157 = 134.99, p<0.001; kanamycin treatment group: F2,184 = 50.17, p<0.001; Fig. 3, and Figure S8). When they competed with either WT bacteria or ARG‐free lysogens (Fig. 3 left and middle columns), new WT lysogens had a fitness advantage in the presence but not the absence of antibiotics. By contrast, when they competed with ARG‐carrying lysogens (Fig. 3 right column), new WT lysogens had similar competitive fitness across the two environmental conditions. Thus, new WT lysogens behaved similarly to the bacteria they acquired prophages from, with a fitness advantage contingent on antibiotics and the presence of antibiotic‐susceptible competitors.
Figure 3.
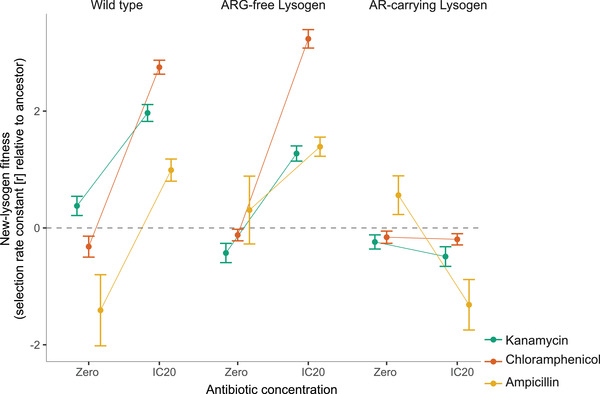
Competitive fitness of new WT‐lysogens relative to each of three competing ancestral strains (left: in competition with the phage‐free, ARG‐free Wild Type; middle: in competition with an ARG‐free lysogen with the same prophage type; right: in competition with an ARG‐carrying lysogen with the same prophage type). Fitness was measured in the absence and presence of antibiotics (x‐axis). Different colors correspond to different ARGs (see legend). Each line gives the mean ± s.e. across all of the new‐WT lysogens we isolated for each ARG; individual reaction norms for every new WT‐lysogen (each isolated from one of the 16 populations where we detected new lysogens, Table 2) are given in Figure S8. New WT‐lysogens have a higher fitness than the competing strain when r > 0.
Discussion
We found fitness benefits resulting from prophage carriage peaked in different experimental environments compared to fitness advantages of prophage‐encoded adaptive genes (here antibiotic resistance genes, ARGs). Prophage‐encoded ARGs, which were not associated with fitness costs in the absence of antibiotics on average (Andersson and Levin 1999), were more beneficial in environmental conditions where prophage induction was less frequent but antibiotics were present (cluster 3 in Fig. 1C). In contrast, prophages were more beneficial than ARGs in environmental conditions where induction of the lytic cycle was more frequent (cluster 1 in Fig. 1C). This beneficial effect of prophage carriage is likely driven by the release of free phages which, as determined in a separate experiment, was greatest in treatment groups where lysogen fitness was highest. Upon prophage induction, we expect the released phage particles infect and lyse competing, phage‐susceptible WT cells (as observed in pure cultures ‐ Figure S7; for a detailed review see (Harrison and Brockhurst 2017)). By selectively killing phage susceptible competitors, prophages can therefore increase the competitive fitness of their hosts (Bossi et al. 2003; Brown et al. 2006; De Paepe et al. 2016; Haaber et al. 2016). However, this benefit was (1) only visible at the population level, as individual cells are presumably killed upon induction of the lytic cycle (Fig. S5) and (2) depended on being surrounded by competitors susceptible to the released phages, as lysogens did not grow better than prophage‐free strains in monoculture without antibiotics and mitomycin C (Figure S2). We observed formation of new WT lysogens in a small fraction of populations, indicating released phages can also drive genetic exchange.
The competitive advantage of lysogens over prophage‐free competitors we observed supports the notion of a mutualistic, rather than parasitic, relationship between prophages and bacteria (Bondy‐Denomy and Davidson 2014; Nanda et al. 2015; Fillol‐Salom et al. 2019). In our experiments, the benefits of prophages appeared to result not only from their carriage of ARGs, but the release of free phages that then lysed competing bacteria. Such killing of phage‐susceptible competitors has been observed in other systems, where the release of free phages indirectly benefits their carriers during the bacterial competition (Selva et al. 2009; Burns et al. 2015; Li et al. 2017) and facilitates microbial invasion (Brown et al. 2006). Released phages can also integrate into the chromosomes of competing bacteria, as observed in our new WT lysogens. In this case, the competitor becomes resistant to the phage and the beneficial effect of the phage to its original host population disappears. One way to overcome this resistance evolution is polylysogeny (carriage of more than one prophage) in some bacteria (Burns et al. 2015). In this case, when the competitor becomes resistant to one of the released phages, other released phages might still be able to kill the competitor. Thus, the prevalence of prophages in bacterial genomes may reflect a combination of direct competitive benefits in some environments and their role in the exchange of genetic material (Jain et al. 1999; Ochman et al. 2000; Koonin et al. 2001). Indeed, prophages can constitute up to 20% of bacterial genomes and contribute significantly to sequence diversity among isolates (Casjens 2003). For instance, pathogenic E. coli serogroup O157 contains between 15 and 18 different prophages, which account for most of the increased genome size (5.5 Mb) compared to K‐12 MG1655 (4.6 Mb, (Perna et al. 2001)).
An important implication of our results is that we found only a few experimental conditions where both ARGs and prophages were beneficial (left and right panels both red in Fig. 1C). In other words, in most environmental conditions it was advantageous for bacteria to have a prophage, an ARG, or neither, but not both a prophage and an ARG. This is consistent with, and helps to explain, metagenomic evidence that prophages rarely encode ARGs (Enault et al. 2017). An interesting avenue for future research would be to look for comparative evidence of whether bacterial strains with ARGs and prophages at separate loci gain similar fitness benefits compared to those with ARG‐encoding prophages.
One limitation of our study is that we used engineered lysogens instead of naturally isolated lysogens. However, this approach enabled us to disentangle the effects of prophages and ARGs, and to show that this applied across multiple phage types and antibiotics. Despite this, we observed variation of lysogen fitness across phage types in our chloramphenicol treatment, and a cost of prophage carriage for lambda lysogens in the presence of ampicillin and mitomycin C. This suggests lysogen fitness is also influenced by an interaction between the type of prophage and the type of antibiotic. There are multiple possible mechanisms by which such an interaction could arise, and an interesting avenue for future work would be to investigate this for a much larger range of prophage types and antibiotics, including multiple antibiotics from each class. We nevertheless speculate that the relatively high fitness of prophage‐free bacteria in the presence of ampicillin may reflect the mechanism of ampicillin resistance. For β‐lactams such as ampicillin, resistance is often mediated via β‐lactamases that deactivate the antibiotic, reducing the effective concentration and potentially diminishing inhibitory effects on susceptible cells in mixed cultures (Medaney et al. 2016). We also note that we performed our experiments in plain LB, without additional magnesium and calcium ions that would promote phage adsorption. Infection and lysis/lysogeny of susceptible cells might therefore be even more effective in conditions supporting higher adsorption rates per bacterium‐virus encounter.
Some key properties of our experimental conditions are nevertheless likely to also apply in nature. Many pathogenic E. coli harbor lambdoid prophages (Casjens 2003), and co‐occurrence of closely related enterobacteria that differ in their prophage repertoire are common in gastrointestinal tracts. Moreover, in animal farms, sub‐MIC concentrations of antibiotics are likely common as a result of antibiotic application as growth promoters and prophylactics (Van Boeckel et al. 2015). Thus, we can expect conditions that would favor bacterial acquisition of such ARG‐carrying prophages to be widespread. Understanding the drivers of lysogen fitness, such as variable free phage production depending on abiotic conditions as we observed, is therefore important for predicting the risk that prophage‐encoded ARGs could spread from agricultural or natural environments to human microbiota or pathogens.
Another key implication of our results is that lysogenization of phage‐susceptible cells by ARG‐carrying prophages appears to be most likely in the presence of antibiotics and in conditions where free phage abundance is high. There are at least two possible explanations. First, the relatively high benefits of acquiring prophages and ARGs in these conditions presumably increased their final abundance and the probability of us detecting them. For ARGs, we observed stronger fitness benefits in the presence of antibiotics, both in our main competition assay and in experiments with new WT lysogens. Independently of ARGs, the benefits of prophage acquisition were higher in the presence of mitomycin C and antibiotics. This may result from the relatively high abundances of free phages in these treatment groups, indicated by our phage‐production experiment. In such conditions, we can expect the benefits of prophage acquisition stemming from immunity to super‐infection (Lwoff 1953; van Houte et al. 2016) to be relatively high. Consistent with this, previous work showed that prophages can enhance lysogen competitiveness by lysing phage‐susceptible competitors (Gama et al. 2013; Davies et al. 2016). Note that, as well as mitomycin C, other antibiotics or drugs can induce the lytic cycle and trigger the release of free phages. Even though we did not observe prophage induction upon exposure to any of the three antibiotics used in the present study, this has been observed for instance for prophages of E. coli O157:H7 (Zhang et al. 2000), Staphylococcus aureus (Ubeda et al. 2005; Maiques et al. 2006) and the virus‐like gene transfer agent VSH‐1 in the swine pathogen Brachyspira hyodysenteriae (Matson et al. 2005; Stanton et al. 2008). We therefore speculate that in other systems, exposure to antibacterial compounds can promote new lysogens, either via direct selection for prophage‐encoded resistance genes or indirectly by enhancing free phage production, which in turn selects for prophage‐derived resistance to superinfection.
A second, non‐exclusive explanation for the bias of new lysogen formation toward antibiotic‐ and mitomycin C treatments is that the frequency of lysogenization events of competing cells per phage‐bacteria contact may be higher in these treatment groups. Lysogeny is controlled by both, the phage and the host (Herskowitz and Hagen 1980). For instance, if bacteria are growing in sub‐optimal/stressful conditions, such as low temperatures (Obuchowski et al. 1997) or under starvation, lambda is biased toward the lysogenic over the lytic cycle (Herskowitz and Hagen 1980). Similarly, when the multiplicity of infection (MOI) is high, lysogeny is favored (Kourilsky 1973; Kobiler et al. 2005). We observed trends consistent with these results. Lysogenization of the phage‐susceptible WT population was favored under those environmental conditions that resulted in a high number of free infecting phage particles and at sub‐inhibitory antibiotic concentrations.
Conclusions
We provide empirical evidence that the benefits to lysogens of carrying prophages and adaptive genes encoded thereon vary independently across environmental conditions. Prophages have fitness benefits independent of any ARGs they carry, which are probably driven by the release of free phages that lyse competing phage‐susceptible cells, and consequently, vary with environmental factors affecting induction of the lytic cycle. Disentangling the benefits of prophages and ARGs improves our understanding of selection on prophages in the presence of phage‐susceptible competitors (Gama et al. 2013; Davies et al. 2016). More generally, our results provide new information about the evolutionary biology of mobile genetic elements, by showing that selection on MGEs themselves and adaptive genes they carry can vary independently across environmental conditions. This helps to explain the mixed evidence of whether prophages frequently encode ARGs. Finally, we found the horizontal transfer of phage‐encoded resistance genes to phage‐ and antibiotic susceptible strains were relatively common for certain prophage types and at non‐zero (but sub‐MIC) concentrations of antibiotics. This improves our understanding of the conditions in which ARG‐encoding phages are likely to spread, both vertically and horizontally, in bacterial communities.
AUTHOR CONTRIBUTIONS
C.C.W. obtained the funding, C.C.W. and A.R.H. designed the study, D.R. constructed lysogens, C.C.W. performed experimental work, C.C.W. analyzed the data, and C.C.W. and A.R.H. interpreted the data and wrote the manuscript. All authors commented on the final version of the manuscript.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
DATA ARCHIVING
Data have been deposited at dryad: https://doi.org/10.5061/dryad.dfn2z350c
Associate Editor: E. Top
Handling Editor: T. Chapman
Supporting information
Table S1. Primer sequences that were used to amplify resistance cassettes and exchange them with non‐essential genes in prophages.
Figure S1. Bacterial growth measured at multiple concentrations of three antibiotics (given in panel labels at top).
Figure S2. Growth rate [r] of each lysogen in the absence of antibiotics and mitomycin C, arranged by prophage type (x‐axis) and antibiotic resistance gene (see legend).
Figure S3. Free phage abundance in pure cultures of lysogens in the absence (left column) and presence (right column) of mitomycin C at different antibiotic concentrations (x‐axis).
Figure S4. Correlation between log‐transformed free phage abundance (measured after four hours in pure cultures of lysogens; y‐axis) and lysogen relative fitness (measured in competition with the prophage‐free WT; x‐axis).
Figure S5. Bacterial population densities in competitions between (a) ARG‐free lysogens and Wild Type bacteria and (b) ARG‐carrying lysogens and Wild Type bacteria.
Figure S6. Population density of each strain over time (20 h) in the presence of mitomycin C (0.5 μg/ml).
Figure S7. Reduction in bacterial growth (1‐RBG) imposed by free phages for each phage genotype (columns, labelled at top) for each antibiotic (rows, labelled at right), without an ARG = black, with ARG = grey at four antibiotic concentrations (x‐axis).
Figure S8. Competitive fitness of new WT‐lysogens relative to each of three competing ancestral strains (left: phage‐free, ARG‐free Wild Type; middle: ARG‐free lysogen with the same prophage type; right: ARG‐carrying lysogen with the same prophage type), in the absence and presence of antibiotics (x‐axis).
ACKNOWLEDGMENTS
We thank Iris Wurmitzer for help in the laboratory and Tobias Bergmiller for support on recombineering. This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska‐Curie grant agreement No 794447‐ProphARG given to CCW and from the Swiss National Science Foundation (grant number PZ00P3_179743) given to CCW. Construction of phages and lysogens was carried out under the project ‘Genetic Diversity of Hosts and Parasites’ (grant no. 0‐21248‐08), funded by the Competence Center Environment and Sustainability of the ETH, given to Sebastian Bonhoeffer.
LITERATURE CITED
- Andersson, D. I. , and Levin B. R.. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489‒493. [DOI] [PubMed] [Google Scholar]
- Blahova, J. , Kralikova K., Krcmery V., and Jezek P.. 1999. Transduction of antibiotic resistance in Pseudomonas aeruginosa: Relationship between lytic and transducing activity of phage isolate AP‐423. Acta. Virol. 43:395‒398. [PubMed] [Google Scholar]
- Bondy‐Denomy, J. , and Davidson A. R.. 2014. When a Virus is not a Parasite: The Beneficial Effects of Prophages on Bacterial Fitness. J. Microbiol. 52:235‒242. [DOI] [PubMed] [Google Scholar]
- Bossi, L. , Fuentes J. A., Mora G., and Figueroa‐Bossi N.. 2003. Prophage contribution to bacterial population dynamics. J. Bacteriol. 185:6467‒6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto, L. 2010. Horizontal gene transfer in evolution: facts and challenges. P. Roy. Soc. B‐Biol. Sci. 277:819‒827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenciani, A. , Bacciaglia A., Vignaroli C., Pugnaloni A., Varaldo P. E., and Giovanetti E.. 2010. Phi m46.1, the Main Streptococcus pyogenes Element Carrying mef(A) and tet(O) Genes. Antimicrob. Agents. Ch. 54:221‒229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. P. , Chat L. L.e., De Paepe M., and Taddei F.. 2006. Ecology of microbial invasions: Amplification allows virus carriers to invade more rapidly when rare. Curr. Biol. 16:2048‒2052. [DOI] [PubMed] [Google Scholar]
- Burns, N. , James C. E., and Harrison E.. 2015. Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol. Appl. 8:346‒351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero‐Caceres, W. , Ye M., and Balcazar J. L.. 2019. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 27:570‒577. [DOI] [PubMed] [Google Scholar]
- Carlson, K. 2005. Working with bacteriophages: common techniques and methodological approaches. Pp. 429–484 in Kutter E., and Sulakvelidze A., eds. Bacteriophages: Biology and applications. GRC Press, Boca Raton, Florida. [Google Scholar]
- Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277‒300. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. , and Wanner B. L.. 2000. One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. P. Natl. Acad. Sci. USA. 97:6640‒6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, E. V. , James C. E., Kukavica‐Ibrulj I., Levesque R. C., Brockhurst M. A., and Winstanley C.. 2016. Temperate phages enhance pathogen fitness in chronic lung infection. Isme. J. 10:2553‒2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe, M. , Tournier L., Moncaut E., Son O., Langella P., and Petit M. A.. 2016. Carriage of. Latent Virus Is Costly for Its Bacterial Host due to Frequent Reactivation in Monoxenic Mouse Intestine. Plos. Genetics. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enault, F. , Briet A., Bouteille L., Roux S., Sullivan M. B., and Petit M. A.. 2017. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. Isme J 11:237‒247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillol‐Salom, A. , Alsaadi A., de Sousa J. A. M., Zhong L., Foster K. R., Rocha E. P. C., Penades J. R., Ingmer H., and Haaber J.. 2019. Bacteriophages benefit from generalized transduction. Plos. Pathogens. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili, E. , Osepashvili M., Koulouris S., Ujmajuridze L., Tskhitishvili Z., and Kotetishvili M.. 2020. Phage Transduction is Involved in the Intergeneric Spread of Antibiotic Resistance‐Associated bla(CTX‐M), mel, and tetM Loci in Natural Populations of Some Human and Animal Bacterial Pathogens. Curr. Microbiol. 77:185‒193. [DOI] [PubMed] [Google Scholar]
- Gama, J. A. , Reis A. M., Domingues I., Mendes‐Soares H., Matos A. M., and Dionisio F.. 2013. Temperate Bacterial Viruses as Double‐Edged Swords in Bacterial Warfare. PLoS One. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg, E. , Albrecht L. M., Karlsson C., Sandegren L., and Andersson D. I.. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. Mbio 5:e01918‒01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaber, J. , Leisner J. J., Cohn M. T., Catalan‐Moreno A., Nielsen J. B., Westh H., Penades J. R., and Ingmer H.. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, E. , and Brockhurst M. A.. 2017. Ecological and Evolutionary Benefits of Temperate Phage: What Does or Doesn't Kill You Makes You Stronger. BioEssays : news and reviews in molecular, cellular and developmental biology 39:1700112. [DOI] [PubMed] [Google Scholar]
- Hayashi, K. , Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S., Ohtsubo E., Baba T., Wanner B. L., Mori H., et al. 2006. Highly accurate genome sequences of Escherichia coli K‐12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006 0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I. , and Hagen D.. 1980. The lysis‐lysogeny decision of phage lambda: explicit programming and responsiveness. Annual review of genetics 14:399‒445. [DOI] [PubMed] [Google Scholar]
- Jain, R. , Rivera M. C., and Lake J. A.. 1999. Horizontal gene transfer among genomes: The complexity hypothesis. P. Natl. Acad. Sci. USA. 96:3801‒3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, P. J. , and Palmer J. D.. 2008. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9:605‒618. [DOI] [PubMed] [Google Scholar]
- Kobiler, O. , Rokney A., Friedman N., Court D. L., Stavans J., and Oppenheim A. B.. 2005. Quantitative kinetic analysis of the bacteriophage lambda genetic network. Proc. Natl. Acad. Sci. U. S. A. 102:4470‒4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E. V. , Makarova K. S., and Aravind L.. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709‒742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourilsky, P. 1973. Lysogenization by bacteriophage lambda. I. Multiple infection and the lysogenic response. Mol. Gen. Genet. 122:183‒195. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E. , Rose M. R., Simpson S. C., and Tadler S. C.. 1991. Long‐Term Experimental Evolution in Escherichia‐Coli .1. Adaptation and Divergence during 2,000 Generations. Am. Nat. 138:1315‒1341. [Google Scholar]
- Li, X. Y. , Lachnit T., Fraune S., Bosch T. C. G., Traulsen A., and Sieber M.. 2017. Temperate phages as self‐replicating weapons in bacterial competition. J. R. Soc. Interface. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwoff, A. 1953. Lysogeny. Bacteriol. Rev. 17:269‒337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiques, E. , Ubeda C., Campoy S., Salvador N., Lasa I., Novick R. P., Barbe J., and Penades J. R.. 2006. beta‐lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 188:2726‒2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson, E. G. , Thompson M. G., Humphrey S. B., Zuerner R. L., and Stanton T. B.. 2005. Identification of genes of VSH‐1, a prophage‐like gene transfer agent of Brachyspira hyodysentetiae. J. Bacteriol. 187:5885‒5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaney, F. , Dimitriu T., Ellis R. J., and Raymond B.. 2016. Live to cheat another day: bacterial dormancy facilitates the social exploitation of beta‐lactamases. Isme J 10:778‒787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, K. , Jeon J. H., Kang I., Park K. S., Lee K., Cha C. J., Lee S. H., and Cho J. C.. 2020. Freshwater viral metagenome reveals novel and functional phage‐borne antibiotic resistance genes. Microbiome. 8:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, A. M. , Thormann K., and Frunzke J.. 2015. Impact of Spontaneous Prophage Induction on the Fitness of Bacterial Populations and Host‐Microbe Interactions. J. Bacteriol. 197:410‒419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuchowski, M. , Shotland Y., Koby S., Giladi H., Gabig M., Wegrzyn G., and Oppenheim A. B.. 1997. Stability of CII is a key element in the cold stress response of bacteriophage lambda infection. J. Bacteriol. 179:5987‒5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H. , Lawrence J. G., and Groisman E. A.. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299‒304. [DOI] [PubMed] [Google Scholar]
- Otsuji, N. , Sekiguchi M., Iijima T., and Takagi Y.. 1959. Induction of Phage Formation in the Lysogenic Escherichia‐Coli K‐12 by Mitomycin‐C. Nature 184:1079‒1080. [DOI] [PubMed] [Google Scholar]
- Paul, J. H. 2008. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? Isme. J. 2:579‒589. [DOI] [PubMed] [Google Scholar]
- Perna, N. T. , Plunkett G., Burland V., Mau B., Glasner J. D., Rose D. J., Mayhew G. F., Evans P. S., Gregor J., Kirkpatrick H. A., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157 : H7. Nature 409:529‒533. [DOI] [PubMed] [Google Scholar]
- Poullain, V. , Gandon S., Brockhurst M. A., Buckling A., and Hochberg M. E.. 2008. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution. 62:1‒11. [DOI] [PubMed] [Google Scholar]
- Pricer, W. E., Jr. , and Weissbach A.. 1964. The Effect of Lysogenic Induction with Mitomycin C on the Deoxyribonucleic Acid Polymerase of Escherichia Coli K12‐Lambda. J. Biol. Chem. 239:2607‒2612. [PubMed] [Google Scholar]
- Qi, Q. , Toll‐Riera M., Heilbron K., Preston G. M., and MacLean R. C.. 2016. The genomic basis of adaptation to the fitness cost of rifampicin resistance in Pseudomonas aeruginosa. Proceedings. Biological. sciences. 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refardt, D. 2011. Within‐host competition determines reproductive success of temperate bacteriophages. Isme J. 5:1451‒1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regoes, R. R. , Wiuff C., Zappala R. M., Garner K. N., Baquero F., and Levin B. R.. 2004. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob. Agents Chemother. 48:3670‒3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva, L. , Viana D., Regev‐Yochay G., Trzcinski K., Corpa J. M., Lasa I., Novick R. P., and Penades J. R.. 2009. Killing niche competitors by remote‐control bacteriophage induction. P. Natl. Acad. Sci. USA. 106:1234‒1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske, K. , and Wagner A.. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton, T. B. , Humphrey S. B., Sharma V. K., and Zuerner R. L.. 2008. Collateral effects of antibiotics: Carbadox and metronidazole induce VSH‐1 and facilitate gene transfer among Brachyspira hyodysenterae strains. Appl. Environ. Microb. 74:2950‒2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon, M. , de Sousa J. A. M., and Rocha E. P. C.. 2017. Embracing the enemy: the diversification of microbial gene repertoires by phage‐mediated horizontal gene transfer. Curr. Opin. Microbiol. 38:66‒73. [DOI] [PubMed] [Google Scholar]
- Ubeda, C. , Maiques E., Knecht E., Lasa I., Novick R. P., and Penades J. R.. 2005. Antibiotic‐induced SOS response promotes horizontal dissemination of pathogenicity island‐encoded virulence factors in staphylococci. Mol. Microbiol. 56:836‒844. [DOI] [PubMed] [Google Scholar]
- Van Boeckel, T. P. , Brower C., Gilbert M., Grenfell B. T., Levin S. A., Robinson T. P., Teillant A., and Laxminarayan R.. 2015. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A. 112:5649‒5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte, S. , Buckling A., and Westra E. R.. 2016. Evolutionary Ecology of Prokaryotic Immune Mechanisms. Microbiol. Mol. Biol. R. 80:745‒763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor, M. K. , and Mekalanos J. J.. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910‒1914. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , McDaniel A. D., Wolf L. E., Keusch G. T., Waldor M. K., and Acheson D. W.. 2000. Quinolone antibiotics induce Shiga toxin‐encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664‒670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences that were used to amplify resistance cassettes and exchange them with non‐essential genes in prophages.
Figure S1. Bacterial growth measured at multiple concentrations of three antibiotics (given in panel labels at top).
Figure S2. Growth rate [r] of each lysogen in the absence of antibiotics and mitomycin C, arranged by prophage type (x‐axis) and antibiotic resistance gene (see legend).
Figure S3. Free phage abundance in pure cultures of lysogens in the absence (left column) and presence (right column) of mitomycin C at different antibiotic concentrations (x‐axis).
Figure S4. Correlation between log‐transformed free phage abundance (measured after four hours in pure cultures of lysogens; y‐axis) and lysogen relative fitness (measured in competition with the prophage‐free WT; x‐axis).
Figure S5. Bacterial population densities in competitions between (a) ARG‐free lysogens and Wild Type bacteria and (b) ARG‐carrying lysogens and Wild Type bacteria.
Figure S6. Population density of each strain over time (20 h) in the presence of mitomycin C (0.5 μg/ml).
Figure S7. Reduction in bacterial growth (1‐RBG) imposed by free phages for each phage genotype (columns, labelled at top) for each antibiotic (rows, labelled at right), without an ARG = black, with ARG = grey at four antibiotic concentrations (x‐axis).
Figure S8. Competitive fitness of new WT‐lysogens relative to each of three competing ancestral strains (left: phage‐free, ARG‐free Wild Type; middle: ARG‐free lysogen with the same prophage type; right: ARG‐carrying lysogen with the same prophage type), in the absence and presence of antibiotics (x‐axis).
Data Availability Statement
Data have been deposited at dryad: https://doi.org/10.5061/dryad.dfn2z350c


