Abstract
Purpose of the review:
Immunotherapy strategies alternative to current antiretroviral therapies will need to address viral diversity while increasing the immune system’s ability to efficiently target the latent virus reservoir. Antibody-based molecules can be designed based on broadly neutralizing and non-neutralizing antibodies that target free virions and infected cells. These multispecific molecules, either by IgG-like or non-IgG-like in structure, aim to target several independent HIV-1 epitopes and/or engage effector cells to eliminate the replicating virus and infected cells. This detailed review is intended to stimulate discussion on future requirements for novel immunotherapeutic molecules.
Recent findings:
Bispecific (bs−) and trispecific antibodies (tsAbs) are engineered as a single molecules to target two or more independent epitopes on the HIV-1 Envelope (Env). These Ab-based molecules have increased avidity for Env, leading to improved neutralization potency and breadth compared to single parental Abs. Furthermore, bs- and tsAbs that engage cellular receptors with one arm of the molecule help concentrate inhibitory molecules to the sites of potential infection and facilitate engagement of immune effector cells and Env-expressing target for their elimination.
Summary:
Recently engineered Ab-based molecules of different sizes and structures show promise in vitro or in vivo and are encouraging candidates for HIV treatment.
Keywords: HIV, bispecific, trispecific, Ab-based molecules, cure
INTRODUCTION
The treatment of HIV-1 infection with antiretroviral therapy (ART) has been effective in controlling virus replication, delaying disease progression, and reducing HIV-1 transmission (1). However, life-long daily administration of ART can cause drug-related toxicities (2). Infusion of broadly neutralizing antibodies (bNAbs) as an alternative therapeutic strategy to ART offers several advantages that include lower toxicity, improved pharmacokinetics (3, 4), Fc-mediated effector functions (5-7) to eliminate infected cells, and diversity of treatment options for patients not responding to ART (8, 9). Infusion of single bNAb or combination of two bNAbs which target independent sites on the HIV-1 Envelope (Env) spike, has mediated suppression of viremia (10-13) and delayed virus rebound during analytical treatment interruption (ATI) (8, 10, 11, 13-17). However, it has also been shown that outgrowth of pre-existent resistant viral variants (11-13, 16, 17) or the development of resistance (15, 16, 18) limits the efficacy of bNAb immunotherapy. Therefore, a combination of bNAbs is necessary for broader coverage of the epidemic and to prevent development of escape mutations following treatment pressure (19). Selected individual and combinations bNAbs currently tested in clinical trials are listed in Table 1.
Table 1.
Antibodies and antibody-based molecules in clinical trials and preclinical development.
| Type | bNAbs | Epitope | Clinical trial number | Interaction with cellular receptor |
Effector cells recruitment |
Phase | |||
|---|---|---|---|---|---|---|---|---|---|
| mAbs in clinical trial | |||||||||
| mAb | VRC01 | CD4bs | NCT02591420 | Fc | CD8, NK, monocytes | 1, Recruiting | |||
| mAb | VRC07-523LS | CD4bs | NCT03739996 | Fc | CD8, NK, monocytes | 2, Recruiting | |||
| 2 mAbs | VRC01+10-1074 | CD4bs+V3 glycan | NCT03831945 | Fc | CD8, NK, monocytes | 1, Recruiting | |||
| 2 mAbs | VRC01+10-1074 | CD4bs+V3 glycan | NCT03707977 | Fc | CD8, NK, monocytes | 1, 2, Recruiting | |||
| 2 mAbs | PGT121+VRC07-523LS | V3 glycan+CD4bs | NCT03721510 | Fc | CD8, NK, monocytes | 1/2a Recruiting | |||
| 2 mAbs | 3BNC117+10-1074 | CD44bs+V3 glycan | NCT03571204 | Fc | CD8, NK, monocytes | 1, Recruiting | |||
| 3 mAbs | PGT121+VRC07-523LS+PGDM1400 | V3 glycan+CD4bs+V2 apex | NCT03721510 | Fc | CD8, NK, monocytes | 1/2a Recruiting | |||
| Type | bNAbs | Epitope | # of HIV isolates in panel |
Neutr/ killing IC50, μg/mL |
Breadth, % |
Interaction with cellular receptor |
Effector cells recruitment |
Size, kDa |
Ref. |
| IgG-like | |||||||||
| CrossMAb | 3BNC117/10-1074 | CD4bs/V3 glycan | 219 | 0.11 | 95.8 | Fc | CD8, NK, monocytes | 160-180 | (23) |
| PG16/10-1074 | V2 glycan/V3 glycan | 219 | 0.192 | 87.5 | Fc | ||||
| PG16/PGT121 | V2 glycan/V3 glycan | 219 | 0.207 | 91.4 | Fc | ||||
| PG16/PGT128 | V2 glycan/V3 glycan | 219 | 0.167 | 88 | Fc | ||||
| PGT151/35O22 | (gp120-gp41 interface)x2 | 219 | 0.22 | 72.5 | Fc | ||||
| 3BNC117/PGT135 | CD4bs/V3 glycan | 219 | 0.036 | 93.3 | Fc | ||||
| 8ANC195/PGT128 | gp120-gp41 interface/V3 glycan | 219 | 0.0844 | 90.5 | Fc | ||||
| PGT151/10-1074 | gp120-gp41 interface/V3 glycan | 219 | 0.041 | 85.7 | Fc | ||||
| CrossMAb | Cap256.VRC26/10-1074 (BICM-1A) | V2 glycan/V3 glycan | 15 | n.i. | n.i. | Fc | CD8, NK, monocytes | 150 | (26**) |
| scFv-Fc | Cap256.VRC26/10-1074 (BISC-1A) | V2 glycan/V3 glycan | 15 | 0.0235 | 66.7 | Fc | CD8, NK, monocytes | 150 | (26**) |
| Cap256.VRC26/PGT121 (BISC-1B) | V2 glycan/V3 glycan | 15 | 0.0246 | 73.3 | Fc | ||||
| Cap256.VRC26/PGT128 (BISC-1C) | V2 glycan/V3 glycan | 15 | 0.0127 | 80 | Fc | ||||
| PGT145/10-1074 (BISC-2A) | V2 apex/V3 glycan | 9 | 0.051 | 88.9 | Fc | ||||
| PGT145/PGT121 (BISC-2B) | V2 apex/V3 glycan | 9 | 0.162 | 22.2 | Fc | ||||
| PGT145/PGT128 (BISC-2C) | V2 apex/V3 glycan | 9 | 0.022 | 88.9 | Fc | ||||
| CrossMAb | VRC01/10E8 | CD4bs/MPER | 206 | 0.187 | 30 | Fc | CD8, NK, monocytes | 150 | (25) |
| VRC01/PGT121 | CD4bs/V3 glycan | 206 | 0.092 | 48 | Fc | ||||
| VRC01/PG9-16 | CD4bs/V2 glycan | 206 | 0.048 | 56 | Fc | ||||
| 10E8/PG9-16 | MPER/V2 glycan | 206 | 0.123 | 43 | Fc | ||||
| scFv-(G4S)n-IgG | VRC01/PGT121 | CD4bs/V3 glycan | 208 | 0.38 | 95.1 | Fc | CD8, NK, monocytes | (28**) | |
| CrossMab | iMab-CAP256 | αCD4/V2 glycan | 20 | 0.00079 | 100 | single Fc chain | reduced, CD8, NK, monocytes | 190 | (30*) |
| 10E08-iMab | MPER/ αCD4 | 14 | 0.0026 | 100 | single Fc chain | ||||
| PG9-iMab | V2 glycan/ αCD4 | 14 | 0.0079 | 92.8 | single Fc chain | ||||
| CrossmAb | 10E8/iMab (NCT03875209) | MPER/αCD4 | 118 | 0.002 | 100 | single Fc chain | reduced, CD8, NK, monocytes | 150 | (33) |
| PGT145/iMab | MPER/αCD4 | n.i. | n.i. | n.i. | single Fc chain | ||||
| 3BNC117/iMab | MPER/αCD4 | n.i. | n.i. | n.i. | single Fc chain | ||||
| PGT128/iMab | MPER/αCD4 | n.i. | n.i. | n.i. | single Fc chain | ||||
| PGT151/iMab | MPER/αCD4 | n.i. | n.i. | n.i. | single Fc chain | ||||
| CrossmAb | 10E8/P140 | MPER/αCCR5 | 118 | 0.001 | 99 | none | None | 150 | (33) |
| PGT145/P140 | MPER/αCCR5 | n.i. | n.i. | n.i. | none | ||||
| 3BNC117/P140 | MPER/αCCR5 | n.i. | n.i. | n.i. | none | ||||
| PGT128/P140 | MPER/αCCR5 | n.i. | n.i. | n.i. | none | ||||
| PGT151/P140 | MPER/αCCR5 | n.i. | n.i. | n.i. | none | ||||
| scFv-(G4S)n-IgG | PG9-iMab | V2 glycan/αCD4 | 118 | 0.004 (<10μg/mL) | 100 | none | none | 204 | (29) |
| PG9-PRO | V2 glycan/αCCR5 | 118 | n.i. | none | |||||
| PG16-iMAbs | V2 glycan/αCD4 | 118 | 0.0027 (<10μg/mL) | 100 | none | ||||
| VHH CrossMAb | J3/2E7 | CD4bs/gp41 | 8 | n.i. | n.i | Fc | CD8, NK, monocytes | 80 | (34*) |
| DART® | A32xCD3 MP3 | CD4i | 22 | n.i. | n.i. | Fc | CD8, NK, monocytes | 107 | (40**) |
| non-IgG-like | |||||||||
| DART® | A32xCD3 | CD4i | 22 | 160–230 pg/ml | n.i. | αCD3 | CD8 | 56 | (40**, 42**) |
| 7B2xCD3 | gp41 | 22 | 160–230 pg/ml | n.i. | αCD3 | ||||
| PGT121xCD3 | V3 glycan | 22 | n.i. | n.i. | αCD3 | ||||
| PGT145xCD3 | V2 apex | 22 | n.i. | n.i. | αCD3 | ||||
| VRC01xCD3 | CD4bs | 22 | n.i. | n.i. | αCD3 | ||||
| 10E8xCD3 | MPER | 22 | n.i. | n.i. | αCD3 | ||||
| BiTE® | CD4-CD3 | CD4 | 1 | 0.048 | n.i. | αCD3 | CD8 | 54 | (38**) |
| VRC01-CD3 | CD4bs | 1 | 2.86 | n.i. | αCD3 | ||||
| B12-CD3 | CD4bs | 1 | 0.0038 | n.i. | αCD3 | ||||
| scFv-Fab | 10E8-N6 | MPER/CD4bs | 109 | 0.0685 | 100 | none | none | 54 | (27*) |
| m36.4-PRO | gp120 C3/αCCR5 | 117 | 0.0131 | 100 | none | 75 | |||
| PGDM1400-PRO 140 | V2 apex/αCCR5 | 117 | 0.0225 | 96 | none | 54 | |||
| Bi-scFvs | PGT121-VRC01 | V3 glycan/CD4bs | 208 | 0.4 | 94.7 | none | none | 54 | (28**) |
| tsAb | |||||||||
| CrossMAb-scFv | VRC01-PGT121-10E8 | CD4bs/V3 glycan/MPER | 208 | 0.071 | 99.5 | Fc | CD8, NK, monocytes | 200 | (28**) |
| scFv tandem-Fc | 10E8Fab-PGT121fv-PGDM1400fv.V8 | MPER/V3 glycan/V2 apex | 27 | 0.1116 | 89 | Fc | CD8, NK, monocytes | 150 | (27*) |
| scFv tandem | 10E8-PGT121-PGDM1400 | MPER/V3 glycan/V2 apex | 117 | 0.0135 | 99 | none | none | 100 | (27*) |
| PRO 140-PGDM1400-PGT121 | αCCR5/V2 apex/V3 glycan | 117 | 0.026 | 97 | none | 100 | |||
| 10E8-PGDM1400-PRO | MPER/V2 apex/αCCR5 | 117 | 0.071 | 98 | none | 100 | |||
| CODV-IgG | VRC01/PGDM1400-10E8 (NCT03705169) | CD4bs/V2 apex/MPER | 208 | 0.039 | 98 | Fc | CD8, NK, monocytes | (49**) | |
| N6/PGDM1400-10E8v4 | CD4bs/V2 apex/MPER | 208 | 0.02 | 99.5 | Fc | ||||
| BiTE® | CD4-17b-CD3 | CD4/αCCR5 | 1 | 0.037 | αCD3 | CD8 | 80 | (38**) | |
IC50, the half maximal inhibitory concentration. BiTE, bispecific T-cell engager; bNAbs, broadly neutralizing antibodies; Fc, fragment crystallizable region; n.i.-not indicated; NK, natural killer; scFv, single-chain fragment variable; tsAb, trispecific antibody; MPER, membrane-proximal external region.
Bispecific (bs−) and trispecific (ts−) bNAbs are single molecules designed to simultaneously bind two or three distinct antigens, respectively. Engineered bsAbs and tsAbs represent a promising alternative to bNAb combination therapy by pursuing multiple targets on the Env protein. This approach may provide increased breadth to overcome HIV’s diversity and to cover natural resistance. Moreover, these Ab-based molecules can overcome complications related to infusion of multiple bNAbs that include costs associated with preclinical testing, manufacturing, and delivery. These molecules and their mechanisms of actions are the scope of the current review.
ENGINEERING MULTI-SPECIFIC AB-BASED MOLECULES FOR HIV CURE
Bispecific-Abs are designed to either recognize two distinct HIV-1 Env epitopes via the single chain variable fragments (scFv) of two independent bNAbs or to engage cellular receptors with one scFv and a single HIV-1 Env epitope with another scFv. The scFvs of these molecules are either joined by a single fragment constant region (Fc) to form a traditional Y-shaped Ab structure or are connected via a linker. Thus, bsAb molecules can be separated into a class of IgG-like molecules and a class of non-IgG-like molecules. When designing Ab-based molecules, the optimal molecules for HIV treatment and cure should combine bNAbs that display adequate coverage of the viral swarms representing the latent HIV-1 reservoir (20) and can be present in sufficient concentrations at the site of viral reactivation in order to be effective. Ab-based molecules that have neutralizing function can bind free virions during acute infection, or post LRA-treatment. This will neutralize the virus and prevent re-infection of other target cells. The summary of recently developed Ab-based molecules is shown in Table 1. These molecules were tested against different panels of HIV- isolates with different assay platforms. Therefore, the neutralization breadth and potency or cell-mediated killing of each Ab-based molecule is reflective of the utilized panel. It should be noted that not every molecule was tested for Fc-mediated functions, which could be relevant for eradication of the reservoir (21). The potency of the molecules discussed here is based on their neutralizing functions unless otherwise stated.
IgG-like bsAbs
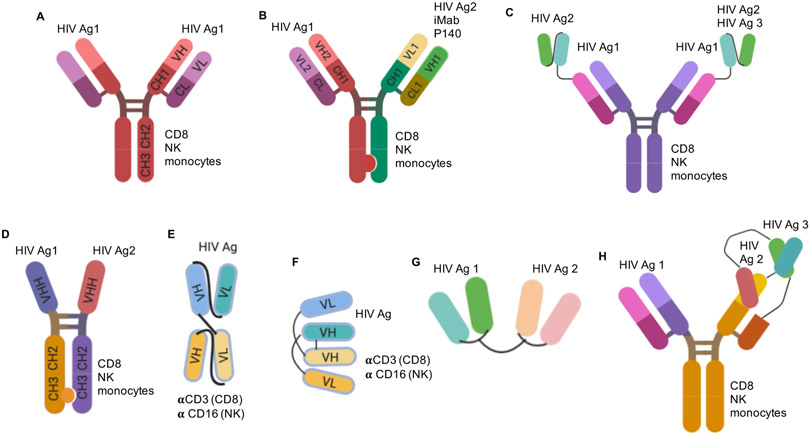
CrossMAb technology has allowed combining scFvs with a single Fc chain from two distinct Abs to form a traditional Y-shaped Ab structure. “Knob-in-hole” modification of Fc regions favors the formation of heavy chain heterodimers of desired bsAb (22). Meanwhile, the “crossover” of CL and CH1 sequences in one arm of the Ab favors correct heavy (H) and light (L) chain pairings in both arms (22). This allows for the generation of a typical monoclonal Ab (mAb) in terms of mass and architecture with association of the desired H and L chains (23, 24*, 25, 26**) (Figure 1 A-B). Another approach is to exchange the scFv of one bNAb with the scFv of another bNAb (26**), or fuse it to a full length bNAb via a flexible (G4S)n linker in a scFv tandem format (27*, 28**) (Figure 1C). Both of these types of bsAbs demonstrated an increase in neutralization breadth and potency compared to the single parental Abs (Table 1). In addition, they have a functional Fc region that can engage Fcγ-receptors-bearing effectors to mediate lysis of HIV-infected target cells or phagocytosis of virions.
Figure 1. Structures and functions of multi-specific Ab-based molecules.
These constructs represent bsAbs and tsAbs that simultaneously bind with antigen-binding domain to multiple independent HIV-1 antigens (HIV Ags) on virions or HIV-1-infected target cells and/or cellular receptors. Most of the presented constructs (with an exception of G) are able to bind to effector cells and induce cell-mediated effector functions. The binding to effector cells is facilitated via Ab Fc region (CD8, NK and/or monocytes) or αCD3 and αCD16 arms to recruit CD8 and NK cells, respectively. (A) classical IgG, (B) CrossMAb with “knob-in-hole” mutations, (C) scFv-(G4S)n-IgG, (D) VHH CrossMAb, (E) BiTE® (Bi-specific T cell Engager), (F) DART® (Dual Affinity Re-Targeting molecule), (G) scFv-scFv, (H) CODV-Ig. Created with BioRender.com.
Enhancement in neutralization potency appeared to be more pronounced when combining scFvs targeting HIV-1 Env epitopes with those targeting host-cell receptors CD4 or CCR5 using a CrossMAb approach (27*, 29, 30*) (Figure 1B). iMab is a mAb that binds to domain 2 of human cellular receptor CD4, on the opposite side of gp120 and MHC class II binding, and potently inhibits HIV-1 entry via a noncompetitive mechanism (29, 31). PRO 140 is a mAb targeting cellular receptor CCR5 and prevents it from binding to gp120 (32). The proposed mechanism of enhanced potency relies on anchoring of the bsAbs by binding the cellular receptors, to both effectively concentrate inhibitory molecules at the cell surface and to better engage the Env epitopes during virus–cell interaction (29, 33). One bsAb of this class, 10E8.4/iMab, is in a phase 1 clinical trial (NCT03875209, Table 1).
Another format of bsAbs uses the heavy chain of llama-derived heavy-chain-only antibodies (VHH), which are therefore smaller than typical IgG molecules (34*). The small size of VHH (13-15 kDa) allows them to bind the cavities that are difficult to reach for traditional Abs (150 kDa), and for the CDR3 loop to protrude further within the cavity to reach neutralizing epitopes on the Env. These epitopes are hidden on native HIV-1 Env trimers by conformation or glycosylation, and Abs of large size or with short CDR3 length fail to reach these neutralizing epitopes. Using covalently linked VHH with “knob-in-hole” technology also increases potency due to an increase in avidity (35, 36) (Figure 1D). VHH Abs can also be pared with those binding the CD4bs or the co-receptor binding site. The arms of multispecific VHH Abs can be joined by flexible (G4S)7 linkers to allow plasticity of the molecule and for the arm to reach their target epitopes. Unlike other bsAbs paired with iMab or PRO 140, VHH bsAbs combined with other llama-isolated Abs targeting these receptors, demonstrated increased breadth but not potency (37). FcR-mediated functions of these bsAbs depend on their structure.
Non-IgG-like bsAbs
Besides targeting CD4 and CCR5 receptors necessary for HIV-1 entry, bispecific molecules that engage other cellular receptors have been designed. Bispecific T cell engagers (BiTEs®) bind to CD3 or CD16 with one arm and to HIV-1 Env with another (38) (Figure 1E). A similar concept, Dual Affinity Re-Targeting (DART®) molecules demonstrated higher KD with CD3, improved stability and half-life of the molecule due to the disulfide linking of two arms and, therefore, improved ability to engage target CD4 T cells and effector (CD8) cells (39) (Figure 1F, Table 1). In vitro studies showed DART molecules retained the neutralization breadth and potency of the Ab component (40, 41). Importantly, in absence of the Fc-region these molecules mediate lysis of HIV-1-infected cells, measured in vitro and ex vivo. Using αCD3 and αCD16 arms these molecules recruiting cytotoxic CD8+ T cells or Natural Killer (NK) cells, respectively, to the infected cells expressing the HIV-1 Env, leading to elimination of HIV-infected CD4 T cells (40-43). In addition, these molecules are smaller in size compared to traditional Abs, have better potential to penetrate tissues and a reduced production cost. Currently, enrolment into a phase 1 clinical trial with MGD014 DART, which targets the C1C2 HIV-1 Env epitope with one arm and CD3 with the other, is ongoing (NCT03570918). Nevertheless, the original BiTEs® and DART® molecules had limited in vivo pharmacokinetics (bioavailability, solubility, stability, and half-life) compared to traditional Abs (44, 45). To improve half-life, MGD011 DART® intended for B-cell malignancies, was engineered with Fc region (46). Similar approach can potentially be utilized in the design of anti-HIV DART molecules. The BiTE® Blinatumomab, intended for treatment of acute lymphoid leukemia, has been reported to induce immune activation by cytokines (47). This is one of the most serious side effects, although new technologies allow the production of BiTE® molecules with improved pharmacokinetic properties and decreased toxicity (48).
Another format of bsAbs is a tandem single chain variable fragment (scFv1- scFv2). These bsAbs are similar in structure to BiTE® or DART® molecules but target two distinct HIV-1 antigens with each arm (Figure 1G). These bsAbs demonstrated increased neutralization breadth and potency compared to the parental Abs (27*, 28**). However, have poor pharmacokinetics, and do not have an Fc region and thus they lack FcR-mediated function necessary to eliminate infected cells.
Trispecific Abs
In the past three years, several novel tsAbs have been designed (27*, 28**, 38**, 49). A tsAb targeting MPER, V3 glycan and V2 apex 10E8Fab- PGT121fv-PGDM1400fv.V8.4DS, known as SAR441236, protected non-human primates (NHP) against a mucosal challenge with multiple SHIVs, demonstrating superior breadth compared to the parental bNAbs (49**). This tsAb was designed as a PGDM1400 Ab with one Fab was switched to the VRC01 Fab, and the scFv of the other PGDM1400 Fab was linked to the scFv of 10E8.4 in a reverse-order tandem-forming Cross-Over Dual Variable (CODV) Ig (Figure 1H). SAR441236 is currently being tested in a phase 1 clinical trial (NCT03705169). The enhancement in neutralization potency of tsAbs was attributed to improved avidity that allows for simultaneous epitope engagement on the same Env (25, 28**). TsAbs that have an Fc region or αCD3 arm are also able to recruit effector cells and mediate killing of HIV-1-infected cells.
It is important to note that non-IgG-like bsAbs and tsAbs that lack Fc region are cleared from the body by renal cells (50) or undergo FcRn-mediated recycling (51). The unnatural architecture of many scFv-format Ab-based molecules may also lead to anti-drug antibody responses in vivo.
Design challenges: spatial orientation and linker length
Not all HIV-1 Envs expressed on the surface of infected cells are trimers. Therefore, the scFvs that target epitopes based on trimeric assembly of HIV-1 Env will have limited efficacy based on trimeric Env expression (26**). In addition, Env detected on the virion or infected cell surface is sparse and not evenly distributed. Thus, it is possible that two Envs will never be in close enough proximity to be engaged by one bs- or tsAbs molecule at the same time and rather there will be engagement of different epitopes on the same Env (52, 53). Proximity of epitopes is important when engineering bsAbs or tsAbs: the linker between the arms ensures the proper binding of epitopes and the orientations of the VH and VL regions of the scFvs in the bs- or tsAbs can influence the potency of such molecules. The linker length must be sufficiently flexible to allow each arm to bind its epitope but not impair folding and assembly of the rest of the molecule, nor to be cleaved during manufacturing or by the host proteases. Several studies have reported on modifications in the linker between scFvs (49**), as well as with the rest of the molecule (23, 26**, 29, 34*, 54, 55).
Fc-mediated function of Ab-based molecules
In addition to preventing primary infections, bsAbs can prevent cell-to-cell transmission, which likely mediates a significant fraction of viral spread (56). Thus, targeting the cell receptor with one arm could potentially block cell-mediated spreading of infection. Abs can also eliminate infected cells via Fc-mediated functions that include antibody dependent cellular cytotoxicity (ADCC), antibody dependent cellular phagocytosis (ADCP), complement-dependent cytotoxicity (CDC) and, perhaps, trogocytosis. ADCC activities have been correlated with slow disease progression in HIV-1-infected individuals (57-60). In addition, the Fc region has been shown to enhance protective efficacy in vivo (5, 61). Therefore, the Fc region was added to several non-IgG-like molecules to improve overall therapeutic potential of the molecule, such as DART® A32xCD3 MP3 and scFv tandem-Fc 10E8Fab-PGT121fv-PGDM1400fv.V8 (Table 1).
An important factor to consider in the design of Ab-based molecules is modifications in the Fc region to improve Fc-mediated functions of Abs (62). Among those modifications are the triple S298A/E333A/K334A (AAA) (63) and S239D/I332E/A330L (64) amino acid mutations previously reported to augment antibody ability to bind to FcγRIIIa and to enhance ADCC activity. Ramadoss et al. demonstrated fusion of the Fc region of anti-HIV Abs to the scFv of the anti-CD16 Ab, NM3E2, which increased binding to FcγRIIIA on NK cells with subsequent enhancement in killing of infected cells (65). In addition, utilization of an IgG3 Fc backbone instead of IgG1, 2 or 4 allows greater flexibility due to a longer hinge region of IgG3 (23, 66, 67): artificial modification of hinge length has been shown to increase ADCP function of Abs (68). Lastly, glycosylation of the Fc region has also been shown to increase Ab effector functions (69-72). Besides effector function, M428L/N434S (referred to as LS) mutations in Fc improves in vivo stability and the half-life of Abs (3).
Targeting tissue reservoirs of HIV
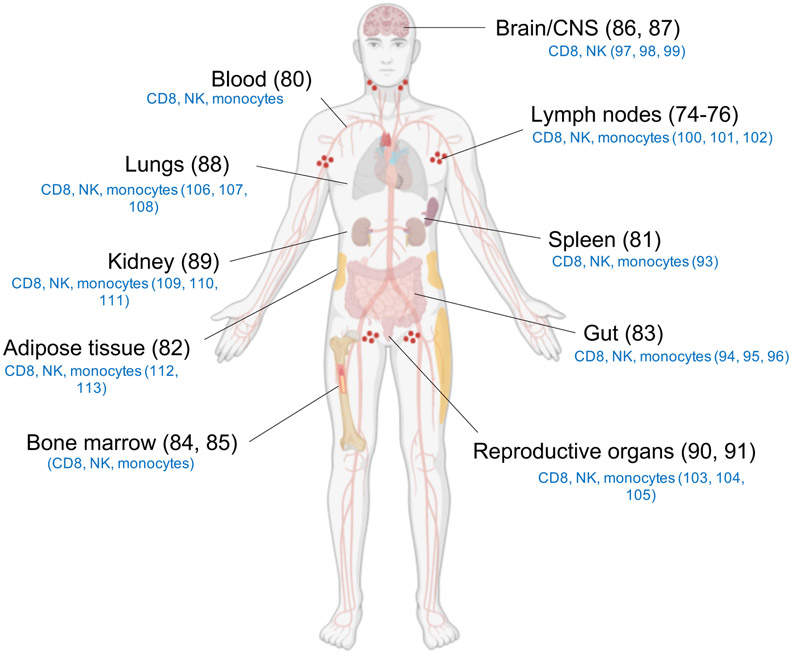
Several studies have shown that B cell follicles, and germinal centers (GCs) in particular, are major sites for HIV-1 reservoir establishment (73). Low levels of HIV-1 replication in lymphatic tissues may also contribute to the persistence of the HIV-1 reservoir (74-76). To overcome this low level expression in the tissues, Latency Reversing Agents (LRAs) have been identified and used to induce proviral transcription in latently infected cells (41, 77) with consequent expression of viral antigens on the cell surface that can be targeted by cytotoxic effector cells. Originally termed the ‘shock and kill’ strategy, this combination of LRAs, ART and virus-induced immune responses has proven to be limited by the ability of the LRAs to induce sufficient virus replication and/or of the cytotoxic effector cells to reach the sites of replications (78, 79). Therefore, the efficacy of Ab-based immunotherapies depends on the recruitment of effector cells in immunologically privileged areas. Besides blood (80) and lymphoid tissues, the HIV-1 reservoir may be found in spleen (81), adipose tissue (82), gut (83), bone marrow (84, 85), CNS (86, 87), lungs (88), kidney (89) and in reproductive organs (90, 91) (Figure 2). ART can penetrate these tissues to various degrees and prevent viral replication but will not eliminate the HIV-1-infected cells (92). Ab-based molecules with effector functions could have a potential to engage tissue resident effector cells (Figure 2) to eliminated infected cells (93-113). In addition to whole virions, shed HIV Env gp120 monomers have been reported to accumulate in lymphoid tissues and other organs during chronic HIV-infection (114). The impact of Abs and Ab-based molecules binding to shed gp120 has not been reported and could be addressed upon completion of ongoing clinical trials with individual or combinations of bNAbs for HIV prevention and treatment. When designing novel Ab-based molecules, the binding to shed gp120 and other “off target” effects should be considered and evaluated in in vitro and in vivo models.
Figure 2. Tissue reservoirs of HIV.
Accumulation of HIV tissue reservoir. Resident effector cells (indicated in blue) that may be utilized in cell-mediated lysis of infected target cells. Created with BioRender.com.
CONCLUSION
Engineered Ab-based molecules for treatment of HIV have advantages over single Ab or combination of Abs that are mainly related to the ability to target different epitopes using a single molecule which increases breadth and prevents viral escape. These molecules will also provide advantages related to cost effective production and clinical administration, including infusion platforms and dose intervals. Small Ab-based molecules may have improved delivery to the tissues, where reservoir-bearing cells are concentrated. Ab-based molecules will ultimately be required to engage effector cellular subsets at sites of virus replication for improved viral clearance. A critical aspect of the efficacy of engineered antibody-based immunothereapy for cure of HIV-1 will depend on new and improved strategies aimed at reactivating the integrated virus in order to detect the infected cells (115).
It is possible, that one molecule may not have all the necessary features to improve beyond traditional mAbs. Therefore, it will be important to further analyze combinations of IgG-like and non-IgG like Ab-based molecules with complementary functions and different structures to achieve a functional cure.
Key points.
Abs-based molecules are promising therapeutic candidates for a long-term control and treatment of HIV-1 infection
bsAbs or tsAbs demonstrate increased antiviral potency compared to single parental Abs
Engagement of cell-mediated immunity is crucial for elimination of infected cells
Combination of Ab-based molecules with complementary functions should be considered for HIV therapy and cure
Acknowledgments
The authors thank Justin Pollara, Dieter Mielke, Sherry Stanfield-Oakley and Junsuke Nohara for providing insightful comments in preparation of the review.
Financial support and Sponsorship
This review here was supported, in part, by CARE, a Martin Delaney Collaboratory program, and the National Institute of Allergy and Infectious Diseases, National Institute of Neurological Disorders and Stroke (NINDS), National Institute on Drug Abuse (NIDA), and National Institute of Mental Health (NIMH) of the National Institutes of Health (grant number 1UM1AI126619); partially supported by NIAID (P01-AI120756-01A1), the Immunology Core of the Duke University Center for AIDS Research (CFAR; 5P30 AI064518), and the External Quality Assurance Program Oversight Laboratory (EQAPOL) (HHSN272201000045C and HHSN272201700061C). Dr. Tuyishime was supported by the NIH Ruth L. Kirschstein National Research Service Award (NRSA) 5T32AI007392.
Footnotes
Conflicts of interest
G. F. has patents submitted on HIV antibodies and DART® molecules described in this article. M.T. reports no conflicts of interest.
References
Papers of particular interest, published within the 48 months of the review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Cohen MS. Successful treatment of HIV eliminates sexual transmission. Lancet (London, England). 2019. [DOI] [PubMed] [Google Scholar]
- 2.Reust CE. Common adverse effects of antiretroviral therapy for HIV disease. Am Fam Physician. 2011;83(12):1443–51. [PubMed] [Google Scholar]
- 3.Ko SY, Pegu A, Rudicell RS, Yang ZY, Joyce MG, Chen X, et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514(7524):642–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rath T, Baker K, Dumont JA, Peters RT, Jiang H, Qiao SW, et al. Fc-fusion proteins and FcRn: structural insights for longer-lasting and more effective therapeutics. Crit Rev Biotechnol. 2015;35(2):235–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158(6):1243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu CL, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper-Stromberg A, et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science (New York, NY). 2016;352(6288):1001–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsons MS, Chung AW, Kent SJ. Importance of Fc-mediated functions of anti-HIV-1 broadly neutralizing antibodies. Retrovirology. 2018;15(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-On Y, Gruell H, Schoofs T, Pai JA, Nogueira L, Butler AL, et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nature medicine. 2018;24(11):1701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagh K, Bhattacharya T, Williamson C, Robles A, Bayne M, Garrity J, et al. Optimal Combinations of Broadly Neutralizing Antibodies for Prevention and Treatment of HIV-1 Clade C Infection. PLoS pathogens. 2016;12(3):e1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP Jr, Buckley N, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522(7557):487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nature medicine. 2017;23(2):185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Science translational medicine. 2015;7(319):319ra206. [DOI] [PubMed] [Google Scholar]
- 13.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature. 2018;561(7724):479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, et al. Strain-Specific V3 and CD4 Binding Site Autologous HIV-1 Neutralizing Antibodies Select Neutralization-Resistant Viruses. Cell host & microbe. 2015;18(3):354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu CL, Lorenzi JC, et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature. 2016;535(7613):556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, et al. Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption. The New England journal of medicine. 2016;375(21):2037–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11(6):615–22. [DOI] [PubMed] [Google Scholar]
- 18.Mehandru S, Vcelar B, Wrin T, Stiegler G, Joos B, Mohri H, et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. Journal of virology. 2007;81(20):11016–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Mendoza C Risk of HIV Escape using Sub-Optimal Antiretroviral Dual or Monotherapy. AIDS Rev. 2016;18(4):223. [PubMed] [Google Scholar]
- 20.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1(1):e13–21. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari G, Pollara J, Tomaras GD, Haynes BF. Humoral and Innate Antiviral Immunity as Tools to Clear Persistent HIV Infection. The Journal of infectious diseases. 2017;215(suppl_3):S152–s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(27):11187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bournazos S, Gazumyan A, Seaman MS, Nussenzweig MC, Ravetch JV. Bispecific Anti-HIV-1 Antibodies with Enhanced Breadth and Potency. Cell. 2016;165(7):1609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.*.Wagh K, Seaman MS, Zingg M, Fitzsimons T, Barouch DH, Burton DR, et al. Potential of conventional & bispecific broadly neutralizing antibodies for prevention of HIV-1 subtype A, C & D infections. PLoS pathogens. 2018;14(3):e1006860.This work demonstrated that one bsAb would not be optimal against all HIV-1 subtypes and specific bsAb are higly efficacious against different subtypes.
- 25.Asokan M, Rudicell RS, Louder M, McKee K, O'Dell S, Stewart-Jones G, et al. Bispecific Antibodies Targeting Different Epitopes on the HIV-1 Envelope Exhibit Broad and Potent Neutralization. Journal of virology. 2015;89(24):12501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.**.Davis-Gardner ME, Alfant B, Weber JA, Gardner MR, Farzan M. A Bispecific Antibody That Simultaneously Recognizes the V2- and V3-Glycan Epitopes of the HIV-1 Envelope Glycoprotein Is Broader and More Potent than Its Parental Antibodies. mBio. 2020;11(1).This group engineered novel bsAbs with improved neutralization potency by varying the linker between the arms within a bsAb. This work demonstrated key details in engineering of novel class of anti-HIV Ab-based molecules.
- 27.*.Khan SN, Sok D, Tran K, Movsesyan A, Dubrovskaya V, Burton DR, et al. Targeting the HIV-1 Spike and Coreceptor with Bi- and Trispecific Antibodies for Single-Component Broad Inhibition of Entry. Journal of virology. 2018;92(18).This work demonstrated a huge variety of bsAbs and tsAbs anti-HIV-1 constructs with different orientation of variable regions in respect to other Fabs within a construct and with their respective binding sites.
- 28.**.Steinhardt JJ, Guenaga J, Turner HL, McKee K, Louder MK, O'Dell S, et al. Rational design of a trispecific antibody targeting the HIV-1 Env with elevated anti-viral activity. Nat Commun. 2018;9(1):877.This work demonstrated engineering of novel anti-HIV tsAb with exceptional in vitro neutralization potency representing a promising candidate for treatment of HIV infection.
- 29.Pace CS, Song R, Ochsenbauer C, Andrews CD, Franco D, Yu J, et al. Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(33):13540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.*.Moshoette T, Ali SA, Papathanasopoulos MA, Killick MA. Engineering and characterising a novel, highly potent bispecific antibody iMab-CAP256 that targets HIV-1. Retrovirology. 2019;16(1):31.This work demonstrated the evaluation of novel bsAb, where enhanced neutralization potency was reflective of a specific quaternary V1V2 epitope.
- 31.Burkly LC, Olson D, Shapiro R, Winkler G, Rosa JJ, Thomas DW, et al. Inhibition of HIV infection by a novel CD4 domain 2-specific monoclonal antibody. Dissecting the basis for its inhibitory effect on HIV-induced cell fusion. Journal of immunology (Baltimore, Md : 1950). 1992;149(5):1779–87. [PubMed] [Google Scholar]
- 32.Trkola A, Ketas TJ, Nagashima KA, Zhao L, Cilliers T, Morris L, et al. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. Journal of virology. 2001;75(2):579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Yu J, Lanzi A, Yao X, Andrews CD, Tsai L, et al. Engineered Bispecific Antibodies with Exquisite HIV-1-Neutralizing Activity. Cell. 2016;165(7):1621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.*.Strokappe NM, Hock M, Rutten L, McCoy LE, Back JW, Caillat C, et al. Super Potent Bispecific Llama VHH Antibodies Neutralize HIV via a Combination of gp41 and gp120 Epitopes. Antibodies (Basel). 2019;8(2).This work combined engineering of bsAbs with improved knob-in-hole technology utilizing higly potent small llama-based VHH bNAbs.
- 35.Hultberg A, Temperton NJ, Rosseels V, Koenders M, Gonzalez-Pajuelo M, Schepens B, et al. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PloS one. 2011;6(4):e17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jähnichen S, Blanchetot C, Maussang D, Gonzalez-Pajuelo M, Chow KY, Bosch L, et al. CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(47):20565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matz J, Kessler P, Bouchet J, Combes O, Ramos OH, Barin F, et al. Straightforward selection of broadly neutralizing single-domain antibodies targeting the conserved CD4 and coreceptor binding sites of HIV-1 gp120. Journal of virology. 2013;87(2):1137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.**.Brozy J, Schlaepfer E, Mueller CKS, Rochat MA, Rampini SK, Myburgh R, et al. Antiviral Activity of HIV gp120-Targeting Bispecific T Cell Engager Antibody Constructs. Journal of virology. 2018;92(14).This work demonstrated engineering of novel BiTEs with potent cell-mediated antiviral activity.
- 39.Moore PA, Zhang W, Rainey GJ, Burke S, Li H, Huang L, et al. Application of dual affinity retargeting molecules to achieve optimal redirected T-cell killing of B-cell lymphoma. Blood. 2011;117(17):4542–51. [DOI] [PubMed] [Google Scholar]
- 40.Sloan DD, Lam CY, Irrinki A, Liu L, Tsai A, Pace CS, et al. Targeting HIV Reservoir in Infected CD4 T Cells by Dual-Affinity Re-targeting Molecules (DARTs) that Bind HIV Envelope and Recruit Cytotoxic T Cells. PLoS pathogens. 2015;11(11):e1005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pegu A, Asokan M, Wu L, Wang K, Hataye J, Casazza JP, et al. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun. 2015;6:8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung JA, Pickeral J, Liu L, Stanfield-Oakley SA, Lam CY, Garrido C, et al. Dual-Affinity Re-Targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. The Journal of clinical investigation. 2015;125(11):4077–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollara J, Edwards RW, Jha S, Lam CK, Liu L, Diedrich G, et al. Redirection of Cord Blood T Cells and Natural Killer Cells for Elimination of Autologous HIV-1-Infected Target Cells Using Bispecific DART® Molecules. Front Immunol. 2020;11:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lameris R, de Bruin RC, Schneiders FL, van Bergen en Henegouwen PM, Verheul HM, de Gruijl TD, et al. Bispecific antibody platforms for cancer immunotherapy. Crit Rev Oncol Hematol. 2014;92(3):153–65. [DOI] [PubMed] [Google Scholar]
- 45.Kontermann RE. Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs. 2009;23(2):93–109. [DOI] [PubMed] [Google Scholar]
- 46.Liu L, Lam CK, Long V, Widjaja L, Yang Y, Li H, et al. MGD011, A CD19 x CD3 Dual-Affinity Retargeting Bi-specific Molecule Incorporating Extended Circulating Half-life for the Treatment of B-Cell Malignancies. Clin Cancer Res. 2017;23(6):1506–18. [DOI] [PubMed] [Google Scholar]
- 47.Zhu M, Wu B, Brandl C, Johnson J, Wolf A, Chow A, et al. Blinatumomab, a Bispecific T-cell Engager (BiTE(®)) for CD-19 Targeted Cancer Immunotherapy: Clinical Pharmacology and Its Implications. Clin Pharmacokinet. 2016;55(10):1271–88. [DOI] [PubMed] [Google Scholar]
- 48.Strohl WR, Naso M. Bispecific T-Cell Redirection versus Chimeric Antigen Receptor (CAR)-T Cells as Approaches to Kill Cancer Cells. Antibodies (Basel). 2019;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.**.Xu L, Pegu A, Rao E, Doria-Rose N, Beninga J, McKee K, et al. Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques. Science (New York, NY). 2017;358(6359):85–90.This study demonstarted the design, in vitro and in vivo evaluation of tsAb that is currently tested in Phase I clinical trial.
- 50.Miners JO, Yang X, Knights KM, Zhang L. The Role of the Kidney in Drug Elimination: Transport, Metabolism, and the Impact of Kidney Disease on Drug Clearance. Clin Pharmacol Ther. 2017;102(3):436–49. [DOI] [PubMed] [Google Scholar]
- 51.Roopenian DC, Christianson GJ, Sproule TJ, Brown AC, Akilesh S, Jung N, et al. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. Journal of immunology (Baltimore, Md : 1950). 2003;170(7):3528–33. [DOI] [PubMed] [Google Scholar]
- 52.Stano A, Leaman DP, Kim AS, Zhang L, Autin L, Ingale J, et al. Dense Array of Spikes on HIV-1 Virion Particles. Journal of virology. 2017;91(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiller J, Chackerian B. Why HIV virions have low numbers of envelope spikes: implications for vaccine development. PLoS pathogens. 2014;10(8):e1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galimidi RP, Klein JS, Politzer MS, Bai S, Seaman MS, Nussenzweig MC, et al. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell. 2015;160(3):433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Padte NN, Yu J, Huang Y, Ho DD. Engineering multi-specific antibodies against HIV-1. Retrovirology. 2018;15(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. The Journal of experimental medicine. 2013;210(13):2813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baum LL, Cassutt KJ, Knigge K, Khattri R, Margolick J, Rinaldo C, et al. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. Journal of immunology (Baltimore, Md : 1950). 1996;157(5):2168–73. [PubMed] [Google Scholar]
- 58.Lambotte O, Ferrari G, Moog C, Yates NL, Liao HX, Parks RJ, et al. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS (London, England). 2009;23(8):897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smalls-Mantey A, Doria-Rose N, Klein R, Patamawenu A, Migueles SA, Ko SY, et al. Antibody-dependent cellular cytotoxicity against primary HIV-infected CD4+ T cells is directly associated with the magnitude of surface IgG binding. Journal of virology. 2012;86(16):8672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wren LH, Chung AW, Isitman G, Kelleher AD, Parsons MS, Amin J, et al. Specific antibody-dependent cellular cytotoxicity responses associated with slow progression of HIV infection. Immunology. 2013;138(2):116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis-Gardner ME, Gardner MR, Alfant B, Farzan M. eCD4-Ig promotes ADCC activity of sera from HIV-1-infected patients. PLoS pathogens. 2017;13(12):e1006786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saunders KO. Conceptual Approaches to Modulating Antibody Effector Functions and Circulation Half-Life. Front Immunol. 2019;10:1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem. 2001;276(9):6591–604. [DOI] [PubMed] [Google Scholar]
- 64.Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, et al. Engineered antibody Fc variants with enhanced effector function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(11):4005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramadoss NS, Zhao NQ, Richardson BA, Grant PM, Kim PS, Blish CA. Enhancing natural killer cell function with gp41-targeting bispecific antibodies to combat HIV infection. AIDS (London, England). 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roux KH, Strelets L, Brekke OH, Sandlie I, Michaelsen TE. Comparisons of the ability of human IgG3 hinge mutants, IgM, IgE, and IgA2, to form small immune complexes: a role for flexibility and geometry. Journal of immunology (Baltimore, Md : 1950). 1998;161(8):4083–90. [PubMed] [Google Scholar]
- 67.Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. Journal of immunology (Baltimore, Md : 1950). 1997;159(7):3372–82. [PubMed] [Google Scholar]
- 68.Chu TH, Crowley AR, Backes I, Chang C, Tay M, Broge T, et al. Hinge length contributes to the phagocytic activity of HIV-specific IgG1 and IgG3 antibodies. PLoS pathogens. 2020;16(2):e1008083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lund J, Tanaka T, Takahashi N, Sarmay G, Arata Y, Jefferis R. A protein structural change in aglycosylated IgG3 correlates with loss of huFc gamma R1 and huFc gamma R111 binding and/or activation. Mol Immunol. 1990;27(11):1145–53. [DOI] [PubMed] [Google Scholar]
- 70.Chung AW, Crispin M, Pritchard L, Robinson H, Gorny MK, Yu X, et al. Identification of antibody glycosylation structures that predict monoclonal antibody Fc-effector function. AIDS (London, England). 2014;28(17):2523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomann M, Schlothauer T, Dashivets T, Malik S, Avenal C, Bulau P, et al. In vitro glycoengineering of IgG1 and its effect on Fc receptor binding and ADCC activity. PloS one. 2015;10(8):e0134949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Umaña P, Jean-Mairet J, Moudry R, Amstutz H, Bailey JE. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 1999;17(2):176–80. [DOI] [PubMed] [Google Scholar]
- 73.Chun TW, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nat Immunol. 2015;16(6):584–9. [DOI] [PubMed] [Google Scholar]
- 74.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science (New York, NY). 2009;323(5919):1304–7. [DOI] [PubMed] [Google Scholar]
- 75.Siliciano JD, Siliciano RF. HIV-1 eradication strategies: design and assessment. Curr Opin HIV AIDS. 2013;8(4):318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(10):E1126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deeks SG. HIV: Shock and kill. Nature. 2012;487(7408):439–40. [DOI] [PubMed] [Google Scholar]
- 78.Margolis DM, Archin NM, Cohen MS, Eron JJ, Ferrari G, Garcia JV, et al. Curing HIV: Seeking to Target and Clear Persistent Infection. Cell. 2020;181(1):189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L Jr., Ingerman MJ, et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. Jama. 1999;282(17):1627–32. [DOI] [PubMed] [Google Scholar]
- 81.Nolan DJ, Rose R, Rodriguez PH, Salemi M, Singer EJ, Lamers SL, et al. The Spleen Is an HIV-1 Sanctuary During Combined Antiretroviral Therapy. AIDS research and human retroviruses. 2018;34(1):123–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Couturier J, Lewis DE. HIV Persistence in Adipose Tissue Reservoirs. Curr HIV/AIDS Rep. 2018;15(1):60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(51):E4987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McNamara LA, Onafuwa-Nuga A, Sebastian NT, Riddell Jt, Bixby D, Collins KL. CD133+ hematopoietic progenitor cells harbor HIV genomes in a subset of optimally treated people with long-term viral suppression. The Journal of infectious diseases. 2013;207(12):1807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sundstrom JB, Ellis JE, Hair GA, Kirshenbaum AS, Metcalfe DD, Yi H, et al. Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood. 2007;109(12):5293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marban C, Forouzanfar F, Ait-Ammar A, Fahmi F, El Mekdad H, Daouad F, et al. Targeting the Brain Reservoirs: Toward an HIV Cure. Front Immunol. 2016;7:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. Journal of virology. 1991;65(8):3973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Costiniuk CT, Jenabian MA. The lungs as anatomical reservoirs of HIV infection. Rev Med Virol. 2014;24(1):35–54. [DOI] [PubMed] [Google Scholar]
- 89.Canaud G, Dejucq-Rainsford N, Avettand-Fenoël V, Viard JP, Anglicheau D, Bienaimé F, et al. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol. 2014;25(2):407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bagasra O, Farzadegan H, Seshamma T, Oakes JW, Saah A, Pomerantz RJ. Detection of HIV-1 proviral DNA in sperm from HIV-1-infected men. AIDS (London, England). 1994;8(12):1669–74. [DOI] [PubMed] [Google Scholar]
- 91.Launay O, Tod M, Tschöpe I, Si-Mohamed A, Bélarbi L, Charpentier C, et al. Residual HIV-1 RNA and HIV-1 DNA production in the genital tract reservoir of women treated with HAART: the prospective ANRS EP24 GYNODYN study. Antivir Ther. 2011;16(6):843–52. [DOI] [PubMed] [Google Scholar]
- 92.Letendre S, Marquie-Beck J, Capparelli E, Best B, Clifford D, Collier AC, et al. Validation of the CNS Penetration-Effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Arch Neurol. 2008;65(1):65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Langeveld M, Gamadia LE, ten Berge IJ. T-lymphocyte subset distribution in human spleen. Eur J Clin Invest. 2006;36(4):250–6. [DOI] [PubMed] [Google Scholar]
- 94.Poggi A, Benelli R, Venè R, Costa D, Ferrari N, Tosetti F, et al. Human Gut-Associated Natural Killer Cells in Health and Disease. Frontiers in Immunology. 2019;10(961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, et al. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Scientific reports. 2018;8(1):14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bain CC, Schridde A. Origin, Differentiation, and Function of Intestinal Macrophages. Front Immunol. 2018;9:2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smolders J, Heutinck KM, Fransen NL, Remmerswaal EBM, Hombrink P, ten Berge IJM, et al. Tissue-resident memory T cells populate the human brain. Nature Communications. 2018;9(1):4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hao J, Liu R, Piao W, Zhou Q, Vollmer TL, Campagnolo DI, et al. Central nervous system (CNS)-resident natural killer cells suppress Th17 responses and CNS autoimmune pathology. The Journal of experimental medicine. 2010;207(9):1907–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Herz J, Filiano AJ, Smith A, Yogev N, Kipnis J. Myeloid Cells in the Central Nervous System. Immunity. 2017;46(6):943–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu RH, Fang M, Klein-Szanto A, Sigal LJ. Memory CD8+ T cells are gatekeepers of the lymph node draining the site of viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(26):10992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bajénoff M, Breart B, Huang AY, Qi H, Cazareth J, Braud VM, et al. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. The Journal of experimental medicine. 2006;203(3):619–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forkner CE. THE ORIGIN OF MONOCYTES IN CERTAIN LYMPH NODES AND THEIR GENETIC RELATION TO OTHER CONNECTIVE TISSUE CELLS. The Journal of experimental medicine. 1930;52(3):385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davé V, Cardozo-Ojeda EF, Mair F, Erickson J, Woodward-Davis AS, Koehne A, et al. Cervicovaginal tissue residence imprints a distinct differentiation program upon memory CD8 T cells. bioRxiv. 2020:769711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sojka DK, Yang L, Yokoyama WM. Uterine Natural Killer Cells. Front Immunol. 2019;10:960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iijima N, Thompson JM, Iwasaki A. Dendritic cells and macrophages in the genitourinary tract. Mucosal Immunol. 2008;1(6):451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Woodland DL, Scott I. T cell memory in the lung airways. Proc Am Thorac Soc. 2005;2(2):126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cong J, Wei H. Natural Killer Cells in the Lungs. Frontiers in Immunology. 2019;10(1416). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rodero MP, Poupel L, Loyher P-L, Hamon P, Licata F, Pessel C, et al. Immune surveillance of the lung by migrating tissue monocytes. eLife. 2015;4:e07847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Turner J-E, Becker M, Mittrücker H-W, Panzer U. Tissue-Resident Lymphocytes in the Kidney. Journal of the American Society of Nephrology. 2018;29(2):389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Victorino F, Sojka DK, Brodsky KS, McNamee EN, Masterson JC, Homann D, et al. Tissue-Resident NK Cells Mediate Ischemic Kidney Injury and Are Not Depleted by Anti–Asialo-GM1 Antibody. The Journal of Immunology. 2015;195(10):4973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Munro DAD, Hughes J. The Origins and Functions of Tissue-Resident Macrophages in Kidney Development. Front Physiol. 2017;8:837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Q, Wu H. T Cells in Adipose Tissue: Critical Players in Immunometabolism. Frontiers in Immunology. 2018;9(2509). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Navarro A, Marín S, Riol N, Carbonell-Uberos F, Miñana MD. Human adipose tissue-resident monocytes exhibit an endothelial-like phenotype and display angiogenic properties. Stem Cell Res Ther. 2014;5(2):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Santosuosso M, Righi E, Lindstrom V, Leblanc PR, Poznansky MC. HIV-1 envelope protein gp120 is present at high concentrations in secondary lymphoid organs of individuals with chronic HIV-1 infection. The Journal of infectious diseases. 2009;200(7):1050–3. [DOI] [PubMed] [Google Scholar]
- 115.Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, et al. Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature. 2020;578(7793):160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]