Abstract
Oncocytomas represent a subset of benign pituitary adenomas that are characterized by significant mitochondrial hyperplasia. Mitochondria are key organelles for energy generation and metabolic intermediate production for biosynthesis in tumour cells, so understanding the mechanism underlying mitochondrial biogenesis and its impact on cellular metabolism in oncocytoma is vital. Here, we studied surgically resected pituitary oncocytomas by using multi-omic analyses. Whole-exome sequencing did not reveal any nuclear mutations, but identified several somatic mutations of mitochondrial DNA, and dysfunctional respiratory complex I. Metabolomic analysis suggested that oxidative phosphorylation was reduced within individual mitochondria, and that there was no reciprocal increase in glycolytic activity. Interestingly, we found a reduction in the cellular lactate level and reduced expression of lactate dehydrogenase A (LDHA), which contributed to mitochondrial biogenesis in an in vitro cell model. It is of note that the hypoxia-response signalling pathway was not upregulated in pituitary oncocytomas, thereby failing to enhance glycolysis. Proteomic analysis showed that 14-3-3η was exclusively overexpressed in oncocytomas, and that 14-3-3η was capable of inhibiting glycolysis, leading to mitochondrial biogenesis in the presence of rotenone. In particular, 14-3-3η inhibited LDHA by direct interaction in the setting of complex I dysfunction, highlighting the role of 14-3-3η overexpression and inefficient oxidative phosphorylation in oncocytoma mitochondrial biogenesis. These findings deepen our understanding of the metabolic changes that occur within oncocytomas, and shine a light on the mechanism of mitochondrial biogenesis, providing a novel perspective on metabolic adaptation in tumour cells.
Keywords: proteomics, metabolomics, whole-exome sequencing, metabolic reprogramming, mtDNA mutation, pituitary adenoma
Introduction
Metabolic reprogramming is a hallmark of cancer [1]. Mitochondria play a central role in the metabolic reprogramming of cancer cells [2]. Unlike mitochondria in normal cells, those in cancer cells are frequently dysfunctional and modulate nearly all aspects of the malignant characteristics of cancer [3]. Mitochondrial biogenesis can drive cancer cell proliferation and metastasis [4,5], and modulating mitochondrial biogenesis may help in cancer therapy [6,7]. However, the mechanisms by which mitochondrial biogenesis are initiated and driven are largely unknown.
Pituitary oncocytomas represent a benign variant of pituitary adenoma characterized by an abundance of mitochondria. The oncocytic phenotype of pituitary tumours was first described in 1973 in null-cell adenomas [8]. Although several hormonally active oncocytomas have been reported in the literature, these tumours tend to occur in older individuals who harbour non-functioning pituitary adenomas [9]. Oncocytomas and null-cell adenomas have some similar characteristics, including low levels of cytoplasmic organelles and small secretory granules [10]. However, oncocytomas show a significant degree of mitochondrial hyperplasia [10], which is quite rare in other subtypes of pituitary adenoma. Oncocytomas have been reported to occur in adenomas of the thyroid, parathyroid, kidney, and salivary gland, all of which have secretory functions [11]. Although mitochondrial DNA (mtDNA) mutations have been reported in thyroid, renal and pituitary oncocytomas [12-14], the genomic landscape and metabolic profile of pituitary oncocytomas have not been described.
Mitochondria play critical roles in tumour initiation and progression via ATP generation, and provide necessary intermediates for various biosynthetic pathways [15]. Furthermore, mitochondrial dysfunction can lead to metabolic reprogramming, thereby affecting the malignancy of a given tumour [16]. Thus, pituitary oncocytomas provide a unique opportunity to study the role of mitochondria in tumour metabolism and mitochondrion-related cellular biology. For this purpose, an understanding of the mechanisms underpinning mitochondrial biogenesis in oncocytomas is vital. Although mtDNA mutations, especially in genes encoding complex I subunits, have been revealed in oncocytomas with different origins [13,17], the mutations themselves were insufficient for mitochondrial proliferation [18]. Similarly to what has been found in skeletal and cardiac myocytes, which have high energy demands, mitochondrial biogenesis in these cells is more probably a compensatory mechanism. The endocrine and exocrine organs where oncocytomas commonly arise also need much energy or intermediates for hormonal biosynthesis and secretion. However, tumour cells are prone to, or can be easily reprogrammed to, use glycolysis under certain conditions, such as hypoxia, to meet their energy demands [19]. Therefore, we hypothesized that, in pituitary oncocytomas, dysfunctional mitochondria cause inefficient oxidative phosphorylation, necessitating the upregulation of glycolysis. Once glycolysis is functioning at a maximal rate, or is restricted for some reason, we theorized that mitochondrial biogenesis functions as the only means of energy compensation.
Herein, we investigated pituitary oncocytomas by using multi-omic approaches, and showed mtDNA mutation-related mitochondrial dysfunction. Unexpectedly, we found that glycolysis was not appropriately upregulated, and that there was a particularly low level of lactate in oncocytoma tissue. We confirmed that aberrant glycolysis was crucial for mitochondrial biogenesis in an in vitro cell model with impaired mitochondria, as observed in oncocytoma. We also found that 14-3-3η, which is a potential candidate responsible for LDHA inhibition and glycolysis interference in oncocytomas, was intimately involved in mitochondrial biogenesis.
Materials and methods
Oncocytoma specimens
All tumour samples were obtained following trans-sphenoidal surgery performed at Beijing Tiantan Hospital. Fresh tumour samples were frozen at −80 °C in isopentane and stored in liquid nitrogen. The clinical and pathological data of all patients are shown in supplementary material, Table S1. This study was approved by the ethics committee of Beijing Tiantan Hospital (ethics approval number: KY2013-015-02). Informed consent was obtained from all of the enrolled subjects, and the study was performed in compliance with the principles governed by the Declaration of Helsinki.
Whole-exome sequencing, nanoscale liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS), and metabolomic analyses
These procedures are detailed in supplementary material, Supplementary materials and methods.
Reverse transcription quantitative polymerase chain reaction
For mRNA quantification, purified total RNA (1 μg) from oncocytoma tissue was reverse-transcribed into cDNA with a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. YWHAH, VDAC1, VDAC2, VDAC3, MFN2, OPA1, PPARGC1A and PPARGC1B were selected for reverse transcription quantitative polymerase chain reaction analysis. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an endogenous control for normalizing the expression level of the target genes. For mtDNA copy number quantification, genomic DNA was extracted from tissue or cells with a Genomic DNA isolation Kit (Qiagen, Germantown, MD, USA). MT-TL1 and PLOG were used as an mtDNA indicator and a nuclear DNA control, respectively. All primers are listed in supplementary material, Table S2. In total, 200 ng of cDNA or genomic DNA was used for quantitative polymerase chain reaction (qPCR), which was performed with a StepOne Real-Time PCR System and KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Wilmington, MA, USA).
Cell culture and reagents
HEK293T, 786-O, HeLa and Hep3B cells were purchased from American Type Culture Collection (Manassas, VA, USA), and were cultured in Dulbecco’s modified E agle’s m edium ( DMEM) ( Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco). Rotenone (100 nM), oxamate (50 mM), 2-deoxy-d-glucose (2-DG) (5 mM), cobalt chloride (CoCl2) (500 μM) and desferrioxamine (DFO) (100 μM) were purchased from Sigma-Aldrich (St Louis, MO, USA).
MitoTracker Orange staining and flow cytometry
Cells were treated as indicated, and were stained with MitoTracker Orange (Thermo Fisher Scientific) for 15 min. Fluorescence intensity was detected by flow cytometry at a wavelength of 576 nm. Data were analysed with FlowJo (FlowJo, Ashland, OR, USA), and the number of mitochondria was calculated from the area under the curve with the same cell number.
Oxygen consumption and extracellular acidification assessment
The oxygen consumption rate (OCR) and extracellular acidification r ate ( ECAR) w ere a ssessed with the Seahorse XFe96 Analyzer (Agilent Technologies, Santa Clara, CA, USA). Cells were seeded in the manufacturer-provided 96-well plate at a density of 30 000 cells per well (11 or 12 wells). Glycolytic activity and glycolytic capacity were assessed with a Seahorse Glycolysis Stress Test Kit (Agilent Technologies); mitochondrial respiration was assessed with a Cell Mito Stress Test Kit (Agilent Technologies).
Immunoblotting, immunohistochemistry, and coimmunoprecipitation (CoIP)
Immunoblotting was performed as described previously [20]. In brief, proteins were extracted with a total protein extraction kit (Millipore, Billerica, MA, USA), and the concentration was measured with the bicinchoninic acid protein assay kit (Pierce, Rochford, IL, USA). Total proteins (50 μg) were subjected to immunoblotting. Enhanced chemiluminescence was performed according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Piscataway, NJ, USA) for visualization. The final d ata w ere subjected to grey scale scanning and semiquantitative analysis with ImageJ software (NIH, Bethesda, MD, USA). Primary antibodies anti-5′-AMP-activated protein kinase-α (AMPKα) (#2532 s, 1:1000), anti-phosphorylated AMPKα (p-AMPKα) (Thr172; #2531 s, 1:1000), anti-hypoxia-inducible factor-1α (HIF-1α) (#14179 s, 1:1000), anti-lactate dehydrogenase (LDH) A (#3582 s, 1:1000), anti-c-Myc (#9420 s, 1:1000), anti-haemagglutinin (HA) (#3724 s, 1:1000), anti-14-3-3η (#9640 s, 1:1000), anti-β-actin (#4970 s,1:1000) and anti-GAPDH (#2118 s, 1:1000) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-mitochondria (#ab92824, 1:1000), anti-LDHB (#ab85319, 1:1000) and anti-PGC1β (#ab176328, 1:1000) were purchased from Abcam (Cambridge, MA, USA). Anti-FLAG antibodies (#F7425, 1:1000) were from Sigma-Aldrich.
For CoIP, cells were treated as indicated, and protein was extracted. CoIP was performed with anti-LDHA antibody (#MA5-17247, 2ug; Thermo Fisher Scientific) and the Dynabeads Protein G for Immunoprecipitation Kit (Thermo Fisher Scientific). For each sample, 2 μg of mouse anti-LDHA antibody was used. BS3 (Thermo Fisher Scientific) was used as a crosslinker. Immunoblotting was performed as described above. For the LDHA ubiquitination assay, the deubiquitinase inhibitor N-ethylmaleimide (5mM; Sigma-Aldrich) was added to lysis buffer to prevent deubiquitination before immunoprecipitation. Alternatively, anti-FLAG M2 antibodies conjugated with Sepharose beads (Cell Signaling Technology) were used for CoIP according to the manufacturer’s instructions.
Construct preparation and transfection
The coding sequence of human YWHAH was cloned from pEX-2TK-14-3-3η-GST [21] (a gift from M. Yaffe, Addgene plasmid #13277) into pcDNA4 by the use of BamHI and EcoRI endonucleases and quick ligase (New England Biolabs, Ipswich, MA, USA). The LDHA-FLAG plasmid was purchased from Sino Biological (Beijing, China). Point mutations (p.S137A and p.T248A) were generated with the Q5 Site-Directed Mutagenesis Kit (New England Biolabs), and sequences were verified by Quintarabio (Berkeley, CA, USA). Transfection of 1 μg of each plasmid was performed with Lipofectamine 2000 (Thermo Fisher Scientific) 1 night prior to any chemical treatment.
Respiratory chain complex I activity assay
Complex I activity was detected with the Complex I Enzyme Activity Microplate Assay Kit (Abcam), according to the manufacturer’s instruction. In brief, proteins were isolated from 200 mg of fresh tissue, and were diluted to the desired concentration. In total, 200 μl of diluted sample was used. Absorbance was detected at 450 nm with a multimode microplate reader (Tecan, Männedorf, Zürich, Switzerland).
Gene set enrichment analysis
We submitted 408 genes whose products were found to be significantly different between oncocytoma and normal pituitary tissue to gene set enrichment analysis (software.broadinstitute.org/gsea), and 396 Entrez genes were identified. Hallmark gene sets in MSigDB were chosen for overlap computation. The top 10 results with FDR q-values of <0.05 were used in our study.
LDH enzyme activity assay
LDH enzyme activity was detected with the Lactate Dehydrogenase Activity Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Tissue (30 mg) samples were rapidly homogenized on ice in 0.5 ml of buffer, and centrifuged at 10 000 × g for 15 min to remove insoluble materials. Samples (2 μl) were used in each well of a 96-well plate, and were tested in duplicate. Absorbance was detected at 450 nm with a microplate reader.
Lactate measurements
The concentration of lactate released by cells was determined with the Lactate Colorimetric/Fluorometric Assay Kit (BioVision, Milpitas, CA, USA). After cells had been treated with rotenone for 48 h, cells were cultured in FBS-free DMEM overnight. Medium was collected, and l(+)-lactate was detected with a colourimetric assay by use of a standard curve, according to the kit manufacturer’s instructions.
Statistical analysis
In vitro data are presented as mean±standard deviation. Ex vivo data are presented as mean±standard error of the mean. Statistical analyses were performed with GraphPad Prism 6 (GraphPad, San Diego, CA, USA). Variables were analysed with an unpaired Student’s t-test for comparison between two groups unless otherwise indicated. For all statistical analyses, a P value of <0.05 was considered to be statistically significant.
Results
Frequent mtDNA mutations lead to mitochondrial dysfunction
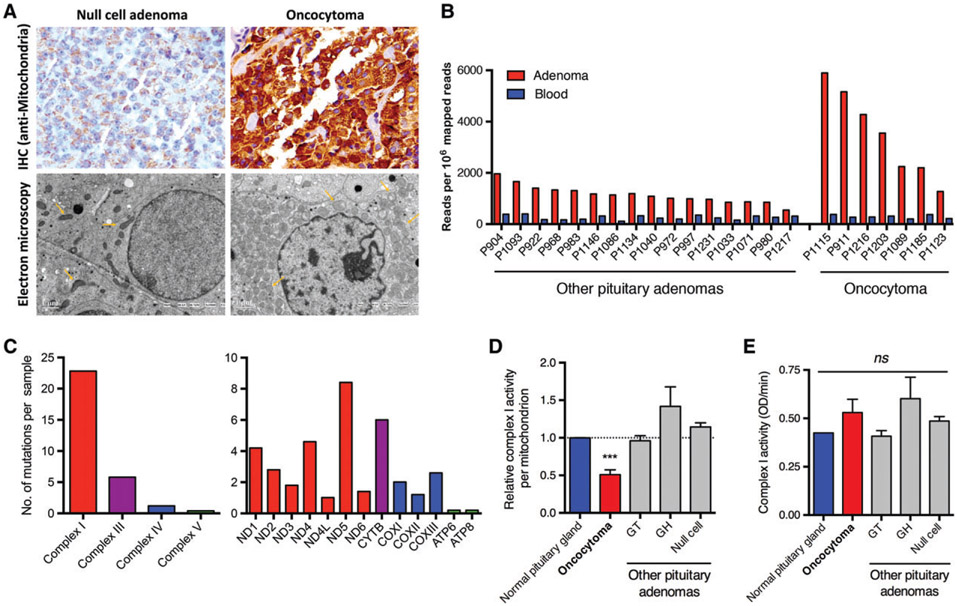
In order to investigate mitochondrial homeostasis, 52 pathologically confirmed pituitary oncocytoma specimens from patients were analysed (supplementary material, Table S1). As compared with normal pituitary specimens, oncocytomas showed intense mitochondrial staining without spindle cell morphology on immunohistochemical staining (Figure 1A, upper panel). Electron microscopy confirmed that pituitary oncocytomas harboured increased numbers of round, abnormally shaped mitochondria (Figure 1A, lower panel). To determine whether any genomic mutations were associated with the unusual mitochondrial phenotype observed in resected oncocytomas, whole-exome sequencing was performed in seven pituitary oncocytomas and 16 pituitary adenomas without oncocytic transformation. As expected, oncocytomas had increased mtDNA copy numbers as compared with other pituitary adenomas (Figure 1B). These results were confirmed by excluding mitochondrial–nuclear DNA fusion [22] (supplementary material, Table S3) and by copy number ratio analysis of MT-TL1 (an mtDNA gene) relative to PLOG (a nuclear DNA gene). The MT-TL1/PLOG ratio was increased in pituitary oncocytomas as compared with normal tissue as measured by qPCR (supplementary material, Figure S1A).
Figure 1.
An increased number of dysfunctional mitochondria in pituitary oncocytomas. (A) Immunohistochemistry (IHC) and electron microscopy showed a dramatic increase in the number of abnormally shaped mitochondria within pituitary oncocytomas as compared with null-cell adenomas. (B) An increased copy number of mtDNA was identified in oncocytomas as compared with other pituitary adenomas and peripheral blood as judged from next-generation sequencing data. (C) The distribution of somatic mtDNA mutations in oncocytomas (n = 5). (D) Respiratory complex I activity was impaired in oncocytomas (n = 10) as compared with normal pituitaries (n = 3), null-cell adenomas (n = 10), gonadotrophin (GT)-secreting adenomas (n = 2), and growth hormone (GH)-secreting adenomas (n = 2). Data were normalized to the mtDNA copy number of each sample. (E) Respiratory complex I activity per cell. ***p < 0.001. ns, not significant.
Unfortunately, we failed to detect any nuclear somatic mutations, which have been reported in renal oncocytoma, pituitary spindle cell oncocytoma, and other pituitary adenomas [14,23,24], in our oncocytoma samples. However, frequent mtDNA mutations (averaging 30 mutations per patient) were identified (supplementary material, Table S4). Consistent with other oncocytic tumours [13,17,25], these somatic mutations were mainly located in genes that encode subunits of respiratory complex I (Figure 1C). As a consequence, the activity of complex I normalized per mitochondrion was significantly decreased in oncocytic tissue (Figure 1D). At the cellular level, however, complex I activity was restored, owing to a compensatory expansion of the number of mitochondria (Figure 1E). In agreement with this, overexpression of genes encoding voltage-dependent anion channels situated in the mitochondrial outer membrane was detected (supplementary material, Figure S1B). In addition, the expression of mitochondrial fusion-related genes such as MFN2 and OPA1 was downregulated (supplementary material, Figure S1C), suggesting inhibition of mitochondrial fusion similar to that in renal oncocytoma [14].
Metabolism is significantly altered because of mitochondrial dysfunction in pituitary oncocytomas
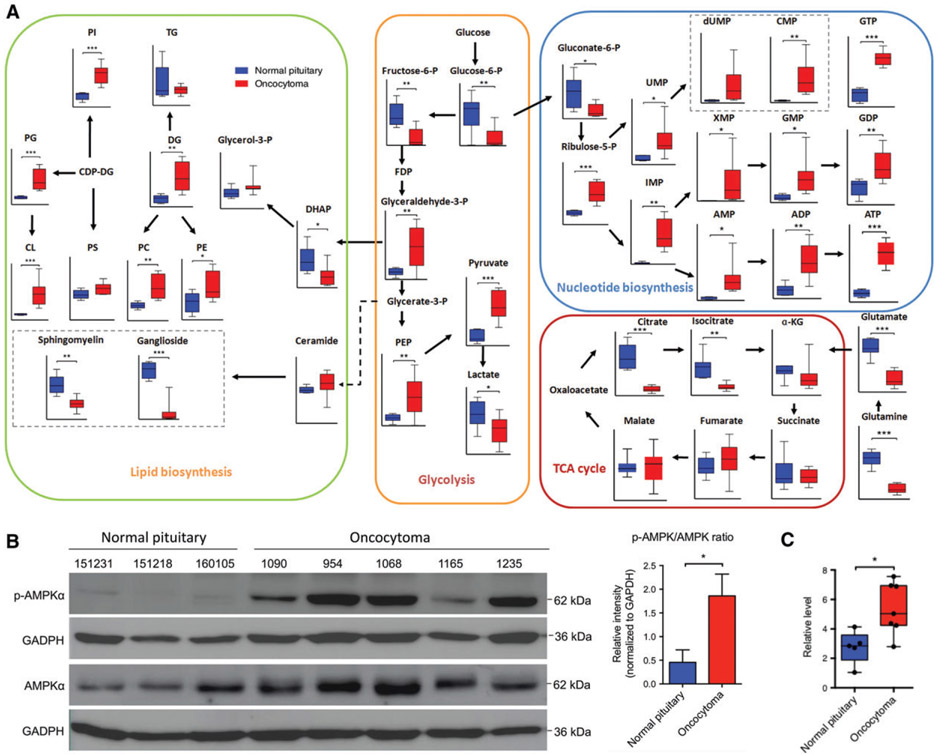
As the compensatory effect hypothesis has previously been suggested to explain mitochondrial hyperplasia in oncocytomas [11], we utilized metabolomics analysis to investigate whether, and to what degree, mitochondrial hyperplasia compensated for mitochondrial dysfunction. As compared with normal tissue, oncocytomas had diminished tricarboxylic acid (TCA) cycle flux and enhancement of pentose phosphate pathway metabolism and phospholipid synthesis (Figure 2A; supplementary material, Figure S2A,B). Decreased amino acid synthesis and increased fatty acid oxidation were also evident in oncocytomas (supplementary material, Figure S2C-E). The impaired oxidative phosphorylation and enhanced synthesis of phospholipids and nucleotides seemed to be consequences of mitochondrial dysfunction and biogenesis (Figure 2A). In addition, upregulation of p-AMPKα (Thr172) suggested that individual oncocytoma cells were still short of energy (Figure 2B). An increased NAD+/NADH ratio and a relatively low α-ketoglutarate (α-KG)/succinate ratio also suggested partial rather than complete loss of complex I activity (Figure 2C; supplementary material, Figure S2F) [11,16], which was again compensated for by an increased number of mitochondria. These results implied that the signal of mitochondrial biogenesis from the existing dysfunctional mitochondria, although not fully clear, was still sustained. Unexpectedly, we noticed disrupted glycolysis in pituitary oncocytomas, which showed a decreased level of lactate (Figure 2A); this was further confirmed by an independent assay performed on another 10 cases of pituitary oncocytoma (supplementary material, Figure S3A).
Figure 2.
An aberrant metabolic phenotype in oncocytomas with inhibited oxidative phosphorylation and inappropriately downregulated glycolysis. (A) Metabolomic analysis revealed reduced TCA cycle flux, increased nucleotide and phospholipid synthesis, and inappropriately downregulated glycolysis with low lactate levels in oncocytomas (n = 7 and n = 10 for normal pituitary and oncocytoma, respectively). (B) Oncocytomas had an elevated p-AMPKα/AMPKα ratio (n = 3 and n = 5 for normal pituitaries and oncocytomas, respectively). (C) The NAD+/NADH ratio was significantly higher in oncocytomas (n = 7) than in the normal pituitary (n = 5). *p < 0.05; **p < 0.01; ***p < 0.001. CDP-DG, Cytidine Diphosphate Diacylglycerol; CL, Cardiolipin; DG, Diacylglycerol; DHAP, Dihydroxyacetone phosphate ; FDP, Fructose Diphosphate ; PC, Phosphatidylcholines; PE, Phosphatidylethanolamine; PEP, Phosphoenolpyruvate; PG, Phosphatidylglycerol; PI, Phosphatidylinositol; PS, Phosphatidylserine ; TG, Triglycerid.
Prevention of compensatory glycolysis is critical for mitochondrial biogenesis in cells harbouring dysfunctional mitochondria
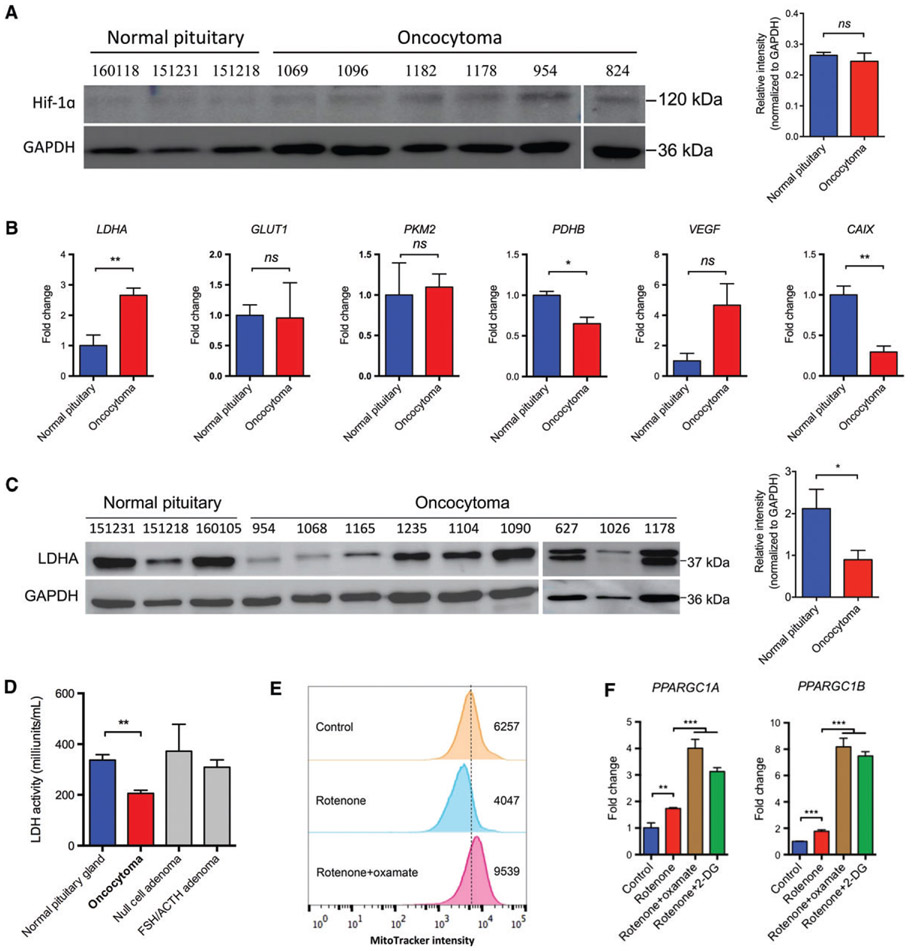
Glycolysis can be aerobic, as observed with the Warburg effect, or be anaerobic under hypoxic conditions [26]. Stabilization of HIF-1α is common in tumours [27], and can be affected by mitochondrial dysfunction [11,28]. However, we found that HIF-1α was not accumulated in oncocytomas as compared with normal pituitary tissue (Figure 3A), which is consistent with a previous study [29]. Moreover, there was no broad upregulation of HIF-1α-associated downstream genes, suggesting that HIF-1α signalling was not activated in oncocytomas (Figure 3B). Notably, the protein level of LDHA tended to be decreased in oncocytomas, with a concurrent increase in its mRNA level, probably because of a negative feedback effect (Figure 3B,C). However, there was no significant change in LDHB expression, despite a slight decrease in the mRNA level (supplementary material, Figure S3B,C). Consistent with these results, we further observed that LDH activity was reduced in oncocytomas but not in other pituitary adenomas (Figure 3D).
Figure 3.
Glycolysis inhibition in oncocytomas contributed to mitochondrial biogenesis in an in vitro model. (A) Oncocytomas did not have elevated HIF-1α expression as compared with normal pituitaries (n = 3 and n = 5 for normal pituitaries and oncocytomas, respectively). (B) With the exception of LDHA, HIF-1α downstream targets were not upregulated in oncocytomas (n = 3 and n = 5 for the normal pituitaries and oncocytomas, respectively). (C) LDHA expression was significantly inhibited in oncocytomas as compared with normal glands (n = 3 and n = 9 for normal pituitaries and oncocytomas, respectively). (D) As compared with normal tissue (n = 3), oncocytomas (n = 4) showed decreased LDH activity, unlike null-cell adenomas (n = 4) and follicle-stimulating hormone (FSH)/adrenocorticotropic hormone (ACTH)-secreting adenomas (n = 4). (E) HEK293T cells were treated with rotenone (100 nM) and oxamate (50 mM) for >48 h, and stained with MitoTracker Orange probes. Flow cytometry showed higher intensity in the same number of cells. Numbers show the area under the curve of each sample. (F) HEK293T cells treated with rotenone and oxamate or 2-DG (5 mM) showed enhanced transcription of PPARGC1A and PPARGC1B. *p < 0.05; **p < 0.01; ***p < 0.001. CAIX, carbonic anhydrase IX; GLUT1, glucose transporter 1; ns, not significant; PDHB, pyruvate dehydrogenase-β; PKM2, pyruvate kinase muscle isozyme M2; VEGF, vascular endothelial growth factor.
Oxidative phosphorylation and glycolysis are two major sources of ATP production. We assumed that, when mitochondrial dysfunction was present, glycolysis would be upregulated to compensate for diminished ATP production resulting from ineffective oxidative phosphorylation. However, if glycolysis was concurrently inhibited because of decreased LDH activity, we conjectured that cells could be forced to initiate mitochondrial biogenesis to compensate for functionally deficient mitochondria. To test this hypothesis, we established an in vitro cell model of oncocytoma by using HEK293T cells, as no pituitary oncocytoma cell lines are currently available, and it was extremely difficult to acquire sufficient primary pituitary oncocytoma cells from patients. We cultured cells with a low dose of rotenone, a complex I inhibitor, to generate cells with dysfunctional mitochondria similar to those observed in oncocytoma. When these cells were further cultured with oxamate, which is a structural analogue of pyruvate and an LDH inhibitor, mitochondrial biogenesis was induced, as confirmed by MitoTracker Orange staining (Figure 3E). Similarly, when LDHA expression was knocked down with small interfering RNA (siRNA) in cells, we detected a 50% increase in the number of mitochondria in the presence of rotenone as compared with those cells transfected with negative control siRNA. Consistently, expression of both PPARCG1A and PPARCG1B, which are two critical genes for mitochondrial biogenesis, were upregulated in the presence of rotenone and oxamate (Figure 3F). Similar results were obtained when glycolysis was disrupted by the glucose analogue 2-DG (Figure 3E). These results suggest that impairment of the glycolytic pathway is necessary for mitochondrial biogenesis in the context of mitochondrial dysfunction.
14-3-3η is overexpressed in pituitary oncocytomas with dysfunctional mitochondria, and inhibits compensatory glycolysis
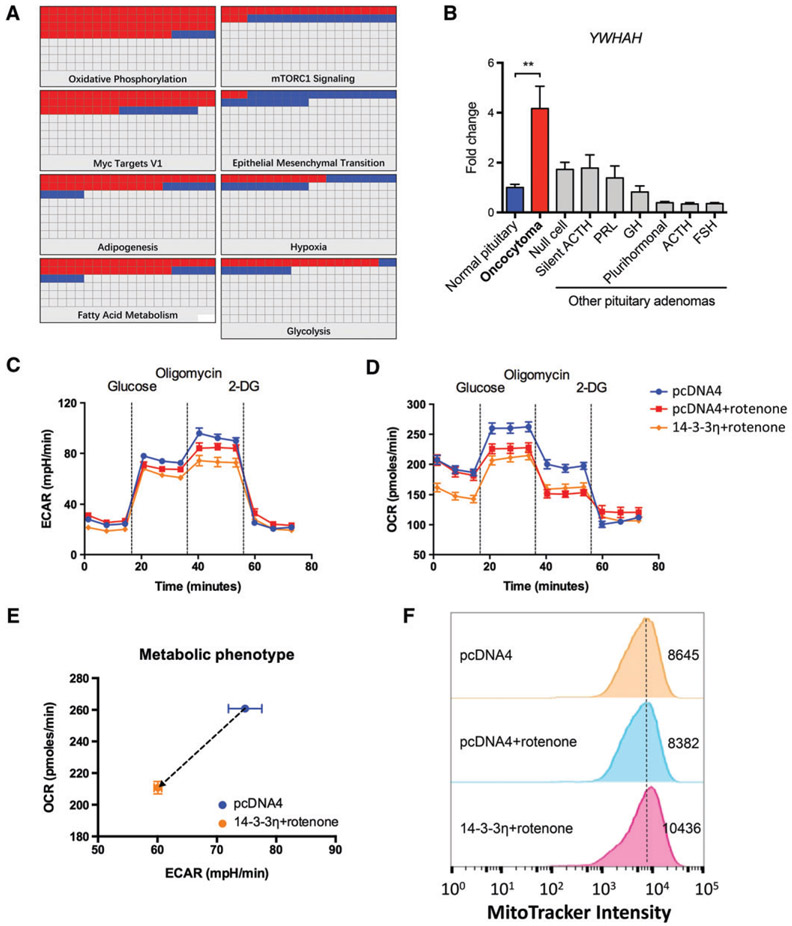
Given a compensated activity of complex I in an individual cell, an increased NAD+ level, and a relatively low α-KG level, a partial loss of function in complex I was inferred. According to Gasparre’s hypothesis [11,16], we should observe accumulation of HIF-1α and enhanced glycolysis; this was not the case in our pituitary oncocytomas. We thus inferred that other causes, such as alterations in the levels of certain regulatory proteins, may be responsible for failure of glycolytic induction. We sought to identify the inhibitor of glycolysis in pituitary oncocytomas by using proteomic approaches. The expression patterns of several genes involved in oxidative phosphorylation, fatty acid metabolism, hypoxia and glycolysis were significantly altered in oncocytomas when analysed by nanoLC-MS/MS (Figure 4A; supplementary material, Tables S5 and S6). This was consistent with the results of our prior metabolic analysis.
Figure 4.
14-3-3η is a putative inhibitor of glycolysis and is critical for mitochondrial biogenesis. (A) Proteomic study showed significant changes in several signalling pathways, including oxidative phosphorylation, hypoxia, and glycolysis. (B) YWHAH was exclusively overexpressed in oncocytoma; n = 5, n = 10, n = 7, n = 5, n = 7, n = 8, n = 10, n = 5 and n = 10 for normal tissue, oncocytoma, null-cell adenoma, silent adrenocorticotropic hormone (ACTH) adenoma, prolactin (PRL)-secreting adenoma, growth hormone (GH)-secreting adenoma, plurihormonal adenoma, ACTH-secreting adenoma, and follicle-stimulating hormone (FSH)-secreting adenoma, respectively. (C-E) In the HEK293T model, 14-3-3η transfection inhibited glycolysis and mitochondrial respiration when complex I was inhibited by rotenone (100 nM). (F) Flow cytometry showed that 14-3-3η led to an increased number of mitochondria in the HEK293T cells with pharmacologically induced oxidative phosphorylation dysfunction. **p < 0.01. mTORC1, mammalian target of rapamycin complex 1.
Previous studies have shown that a member of the 14-3-3 chaperone family can inhibit glycolysis in tumour cells [30]. We observed, through our proteomic analysis, that two 14-3-3 family members, 14-3-3η and 14-3-3θ, were overexpressed in pituitary oncocytomas (data not shown), but only 14-3-3η was exclusively upregulated in oncocytomas among various subtypes of pituitary adenoma, as confirmed by mRNA level quantification (Figure 4B). Using the mitochondria-impaired HEK293T model, we found that 14-3-3η was able to inhibit both glycolysis and mitochondrial respiration (Figure 4C-E; supplementary material, Figure S4). We also detected mitochondrial biogenesis in cells harbouring rotenone-induced inefficient mitochondria and transfected with 14-3-3η (Figure 4F). These findings suggested that concurrent 14-3-3η overexpression and complex I impairment was responsible for mitochondrial biogenesis in pituitary oncocytoma.
14-3-3η overexpression and complex I inactivation together lead to decreased LDHA expression
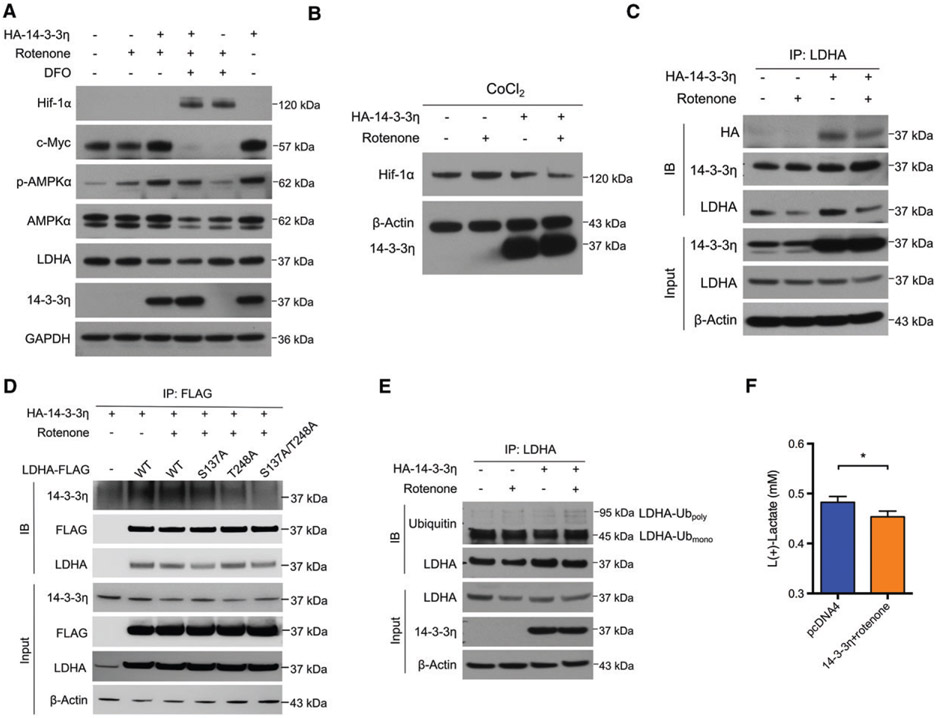
To investigate the potential role of 14-3-3η in mitochondrial biogenesis, we investigated whether 14-3-3η could inhibit LDHA. In the in vitro cell model, we found that 14-3-3η dramatically inhibited LDHA expression when rotenone was present (Figure 5A). The increased expression of p-AMPKα and c-Myc seen in samples treated with 14-3-3η and rotenone was also in accordance with the changes observed in oncocytoma specimens (Figure 2C; supplementary material, Figure S5A). In addition, 14-3-3η restored the expression of PGC-1β that was inhibited by rotenone and inhibited HIF-1α to prevent LDHA induction (Figure S5B). The inhibitory effect of 14-3-3η and rotenone on LDHA expression persisted under hypoxic conditions (Figure 5A). Furthermore, 14-3-3η was able to decrease the expression of HIF-1α by itself and in conjunction with rotenone (Figure 5B). Thus, 14-3-3η may lead to impairment of hypoxia-induced glycolysis secondary to decreased HIF-1α expression even when oncocytomas are exposed to hypoxia resulting from relatively poor vascularization or pseudo-hypoxia resulting from dysfunctional oxidative phosphorylation. Moreover, transfection of 14-3-3η selectively promoted transcription of PPARGC1B but not of PPARGC1A (supplementary material, Figure S5C,D), which was similar to observations made in human oncocytoma samples (supplementary material, Figure S5E). Similarly, transcription of LDHA in our in vitro cell model was also increased, as observed in human oncocytoma samples (Figure 3B; supplementary material, Figure S5F). The driver of upregulated LDHA transcription is probably mediated by c-Myc rather than HIF-1α in oncocytomas, or related to a negative feedback loop induced by 14-3-3η-mediated degradation of LDHA. Both the inhibited LDHA expression and decreased lactate production induced by 14-3-3η and rotenone were also observed in other cell lines (supplementary material, Figure S6A,B). Similarly to HEK293T cells, 786-O and HeLa cells showed compromised mitochondrial respiration and glycolytic capacity when 14-3-3η and rotenone were present (supplementary material, Figure S6C).
Figure 5.
14-3-3η repressed LDHA expression in the presence of rotenone. (A) HEK293T cells were transfected with vector or HA-14-3-3η plasmids, and treated with rotenone (100 nM) for 48 h. Concurrent 14-3-3η overexpression and rotenone treatment induced overexpression of c-Myc and p-AMPKα, and reduced expression of LDHA. LDHA expression remained inhibited in the presence of DFO (100 μM)-induced HIF-1α upregulation. (B) HEK293T cells were transfected with vector or HA-14-3-3η plasmids, and treated with rotenone (100 nM) with or without CoCl2 (500 μM) for 48 h. 14-3-3η decreased CoCl2-induced HIF-1α upregulation. (C) HEK293T cell lysates were immunoprecipitated by anti-LDHA antibodies, and an interaction between HA-14-3-3η and LDHA was detected. (D) HEK293T cells were treated as indicated for 48 h. CoIP was performed with anti-FLAG antibodies. (E) HEK293T cells were treated as indicated for 48 h, and MG-132 (10 μM) was added for another 6 h. Cell lysates were immunoprecipitated by anti-LDHA antibodies, and ubiquitinated LDHA was detected. (F) HEK293T cells were treated as indicated for 48 h, and the concentration of lactate in the medium was determined. n = 3. *p < 0.05. IB, immunoblotting; IP, immunoprecipitation; WT, wild type.
As members of the 14-3-3 family commonly function by interacting with their target proteins, and could interfere with the ubiquitination of proteins such as c-Myc [30], we hypothesized that 14-3-3η could also bind to LDHA and affect LDHA degradation through a similar mechanism. Two phosphorylated sites (CKLLIV[S]NPVD and IKLKGY[T]SWAI) in LDHA were predicted to be recognized by 14-3-3η according to 14-3-3-Pred (www.compbio.dundee.ac.uk/1433pred) [31]. Indeed, we detected an interaction between 14-3-3η and LDHA (Figure 5C). When one or two of the phosphorylation sites were mutated, the 14-3-3η–LDHA interaction was impaired to varying degrees (Figure 5D). Furthermore, this interaction upregulated the level of polyubiquitinated LDHA in our in vitro model (Figure 5E). Consistent with a previous report, monoubiquitinated LDHA was predominant in cells [32], and was not significantly changed. Accordingly, 14-3-3η overexpression reduced lactate production in HEK293T cells (Figure 5F). Consistent with this, the NADH level in these cells was found to be increased (probably because of decreased lactate production via LDH) when 14-3-3η was overexpressed, and was reduced by rotenone (supplementary material, Figure S5G). These results suggest that 14-3-3η could expedite LDHA degradation by promoting its polyubiquitination.
Discussion
Oncocytoma represents an unusual type of tumour that lies approximately between normal tissue and malignant cancers in the spectrum of neoplasms. It can also be a special stage in tumourigenesis, although most cancers do not show such a stage. Although oncocytomas with different tissue origins share the same characteristics of mtDNA mutations and markedly active mitochondrial biogenesis, the underlying mechanisms can be distinct. For instance, Golgi disassembly and defective elimination of mitochondria lead to renal oncocytoma [14], whereas mitochondrial biogenesis activator-related copy number gains cause thyroid oncocytoma [33]. In pituitary oncocytomas, however, we found glycolysis inhibition, particularly 14-3-3η-induced LDH inactivation leading to mitochondrial biogenesis. Currently, the reasons for 14-3-3η overexpression in pituitary oncocytomas are unknown. As we did not find an increased copy number of YWHAH by whole-exome sequencing, we inferred that epigenetic alteration of YWHAH or any of the 14-3-3η inducers may be at play. Nevertheless, further investigations are warranted.
As glycolysis is favoured by most cancers [34], the observation of reduced glycolysis in pituitary oncocytoma was surprising, particularly in the setting of the mitochondrial dysfunction seen in pituitary oncocytoma. In this case, pituitary oncocytomas not only harbour impaired mitochondria, but are also unable to rely on glycolysis as a source of ATP production. Therefore, mitochondrial biogenesis appeared to be the only means to satisfy the cellular energy requirements. In our in vitro model constructed from HEK293T cells, we saw very similar changes, including mitochondrial biogenesis, when both respiratory complex I and LDH were inhibited, suggesting that this mechanism of mitochondrial biogenesis is relatively conserved even in cells with distinct origins. Although some investigators demonstrated that mitochondrial dysfunction alone was capable of repressing HIF-1α synthesis or inducing HIF-1α accumulation in certain types of cell [28,35], at least in pituitary oncocytomas, induction of HIF-1α was not the underlying mechanism. The opposite effects on HIF-1α expression may be due to the distinctly different mtDNA mutations, various levels of mitochondrial and cellular metabolites, and disparate involvement of other signalling pathways that can regulate HIF-1α.
Overexpression of 14-3-3η in oncocytomas, exclusively among all types of pituitary adenoma, compels us to hypothesize that it plays a special role in oncocytomas. The 14-3-3 family has seven members, and can impact on cell fate and cancer development [36]. Distinct roles of 14-3-3 members in glycolysis have been reported. Generally, 14-3-3 family members promotes glycolysis in various ways [37], but some of them, such as 14-3-3σ, can repress glycolysis [30]. The roles of 14-3-3η in glycolysis are less well understood. Our study establishes the first link between 14-3-3η and glycolysis mediated through LDHA.
Although 14-3-3 family members are able to interact with a diverse array of proteins, no 14-3-3η–LDHA binding has been previously reported. We showed physical interaction between the two proteins, and identified two domains that are probably responsible for this interaction. As the two predicted binding sites of 14-3-3η in LDHA reside in the NAD+/substrate-binding domain, the interaction between LDHA and 14-3-3η may compromise the enzymatic activity of LDHA, resulting in decreased LDH production and increased LDHA degradation. In particular, the phosphorylation of Thr248, which is located in one of the predicted interaction domains (IKLKGY[T]SWAI), has been previously reported in LDHA [37]. Furthermore, a recent report showed that LDHA phosphorylation at Tyr10 resulted in LDHA activation and cancer progression [38]. However, the effects of phosphorylation at other sites (www.phosphosite.org; Cell Signaling Technology) in LDHA remain poorly understood and require in-depth study.
In conclusion, our study reveals a metabolic profile of pituitary oncocytomas that adopt mitochondrial biogenesis as a compensatory mechanism for impaired glycolysis in the setting of dysfunctional oxidative phosphorylation (Figure 6). We suggest that the absence of HIF-1α overexpression and the concurrent inhibition of LDHA are responsible for impaired glycolysis. We further showed that impaired glycolysis and oxidative phosphorylation were sufficient to induce mitochondrial biogenesis in vitro. We also identified 14-3-3η as a potential glycolytic inhibitor, resulting in metabolic reprogramming and inducing mitochondrial biogenesis in pituitary oncocytomas by inhibiting LDHA expression. Our findings provide a novel understanding of the mechanism by which mitochondrial hyperplasia occurs in pituitary oncocytomas, and offer potential treatment strategy for other dysfunctional mitochondria-related diseases by targeting compensatory mitochondrial biogenesis.
Figure 6.
A scheme showing the rationale of mitochondrial biogenesis in pituitary oncocytomas. Oxidative respiration of mitochondria in oncocytoma cells is partially impaired because of mtDNA mutations and consequent complex I disassembly Overexpression of 14-3-3η inhibits glycolysis by inactivating LDH. The tumour cells survive by turning to mitochondrial biogenesis because of failed induction of glycolysis.
Supplementary Material
Supplementary materials and methods
Figure S1. Increased mitochondrial number in pituitary oncocytoma, related to Figure 1
Figure S2. Metabolic characterization of oncocytoma, related to Figure 2
Figure S3. Lactate levels and LDHB expression, related to Figure 3
Figure S4. 14-3-3η inhibited glycolysis and oxidative phosphorylation, related to Figure 4
Figure S5. The mRNA levels of PPGCAR1 and LDHA, and others, related to Figure 5
Figure S6. Reduced LDHA expression and lactate production in other cancer cell lines, related to Figures 4 and 5
Table S1. Clinical characteristics of patients with pituitary oncocytomas
Table S2. Primers used in the qPCR study
Table S3. All fusion genes based on DNA whole-exome sequencing data
Table S4. mtDNA mutations detected in each patient with pituitary oncocytomas
Table S5. Gene set enrichment analysis
Table S6. The differentially expressed proteins and the top 10 Hallmark gene sets
Acknowledgements
We thank Mioara Larion from the Neuro-Oncology Branch, Center for Cancer Research, National Cancer Institute for help with metabolomics analysis and interpretation. YZ is supported by the Research Special Fund for Public Welfare Industry of Health (201402008) and the National High Technology Research and Development Programme of China (2014AA020610). JF is supported by Beijing Municipal Administration of Hospitals ‘Youth’ Programme (QML20160506) and the National Natural Science Foundation of China (81702455). GX is supported by the National Natural Science Foundation of China (81472374 and 21435006). QZ, QS, CM, HW, MJS, GD, MRG, DMP and ZZ are supported by the Intramural Research Program of the National Institute of Neurological Disorder and Stroke and National Cancer Institute at the National Institutes of Health.
Footnotes
No conflicts of interest were declared.
References
*Cited only in supplementary material.
- 1.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 2012; 21: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa MI, Rodrigues AS, Pereira S, et al. Mitochondrial mechanisms of metabolic reprogramming in proliferating cells. Curr Med Chem 2015; 22: 2493–2504. [DOI] [PubMed] [Google Scholar]
- 3.Boland ML, Chourasia AH, Macleod KF. Mitochondrial dysfunction in cancer. Front Oncol 2013; 292: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Outschoorn UE, Pavlides S, Sotgia F, et al. Mitochondrial biogenesis drives tumor cell proliferation. Am J Pathol 2011; 178: 1949–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeBleu VS, O’Connell JT, Gonzalez-Herrera KN, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 2014; 16:992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G, Frederick DT, Wu L, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest 2016; 126: 1834–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pal HC, Prasad R, Katiyar SK. Cryptolepine inhibits melanoma cell growth through coordinated changes in mitochondrial biogenesis, dynamics and metabolic tumor suppressor AMPKalpha1/2-LKB1. Sci Rep 2017; 7: 1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacs K, Horvath E. Pituitary ‘chromophobe’ adenoma composed of oncocytes. A light and electron microscopic study. Arch Pathol 1973; 95: 235–239. [PubMed] [Google Scholar]
- 9.Niveiro M, Aranda FI, Paya A, et al. Oncocytic transformation in pituitary adenomas: immunohistochemical analyses of 65 cases. Arch Pathol Lab Med 2004; 128: 776–780. [DOI] [PubMed] [Google Scholar]
- 10.Greenman Y, Melmed S. Diagnosis and management of nonfunctioning pituitary tumors. Ann Rev Med 1996; 47: 95–106. [DOI] [PubMed] [Google Scholar]
- 11.Gasparre G, Romeo G, Rugolo M, et al. Learning from oncocytic tumors: why choose inefficient mitochondria? Biochim Biophys Acta 2011; 1807: 633–642. [DOI] [PubMed] [Google Scholar]
- 12.Kurelac I, MacKay A, Lambros MB, et al. Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumorigenesis that correlate with low genomic instability in pituitary adenomas. Hum Mol Genet 2013; 22: 226–238. [DOI] [PubMed] [Google Scholar]
- 13.Gasparre G, Porcelli AM, Bonora E, et al. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci U S A 2007; 104: 9001–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi S, Tolkunov D, Aviv H, et al. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep 2015; 13: 1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012; 12: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasparre G, Porcelli AM, Lenaz G, et al. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol 2013; 5: pii: a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayr JA, Meierhofer D, Zimmermann F, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res 2008; 14: 2270–2275. [DOI] [PubMed] [Google Scholar]
- 18.Carelli V, Maresca A, Caporali L, et al. Mitochondria: biogenesis and mitophagy balance in segregation and clonal expansion of mitochondrial DNA mutations. Int J Biochem Cell Biol 2015; 63: 21–24. [DOI] [PubMed] [Google Scholar]
- 19.Eales KL, Hollinshead KE, Tennant DA. Hypoxia and metabolic adaptation of cancer cells. Oncogenesis 2016; 5: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Bai X, Chen W, et al. Wnt/β-catenin signaling enhances hypoxia-induced epithelial–mesenchymal transition in hepatocellular carcinoma via crosstalk with Hif-1α signaling. Carcinogenesis 2013; 34: 962–973. [DOI] [PubMed] [Google Scholar]
- 21.Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 1997; 91: 961–971. [DOI] [PubMed] [Google Scholar]
- 22.Ju Y, Tubio J, Mifsud W, et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res 2015; 25: 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller MB, Bi WL, Ramkissoon LA, et al. MAPK activation and HRAS mutation identified in pituitary spindle cell oncocytoma. Oncotarget 2016; 7: 37054–37063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song ZJ, Reitman ZJ, Ma ZY, et al. The genome-wide mutational landscape of pituitary adenomas. Cell Res 2016; 26: 1255–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasparre G, Hervouet E, de Laplanche E, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet 2008; 17: 986–995. [DOI] [PubMed] [Google Scholar]
- 26.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004; 4: 891–899. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003; 3: 721–732. [DOI] [PubMed] [Google Scholar]
- 28.Hsu CC, Wang CH, Wu LC, et al. Mitochondrial dysfunction represses HIF-1alpha protein synthesis through AMPK activation in human hepatoma HepG2 cells. Biochim Biophys Acta 2013; 1830: 4743–4751. [DOI] [PubMed] [Google Scholar]
- 29.Porcelli AM, Ghelli A, Ceccarelli C, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet 2010; 19: 1019–1032. [DOI] [PubMed] [Google Scholar]
- 30.Phan L, Chou PC, Velazquez-Torres G, et al. The cell cycle regulator 14-3-3sigma opposes and reverses cancer metabolic reprogramming. Nat Commun 2015; 6: 7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madeira F, Tinti M, Murugesan G, et al. 14-3-3-Pred: improved methods to predict 14-3-3-binding phosphopeptides. Bioinformatics 2015; 31: 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onishi Y, Hirasaka K, Ishihara I, et al. Identification of monoubiquitinated LDH-A in skeletal muscle cells exposed to oxidative stress. Biochem Biophys Res Commun 2005; 336: 799–806. [DOI] [PubMed] [Google Scholar]
- 33.Kurelac I, de Biase D, Calabrese C, et al. High-resolution genomic profiling of thyroid lesions uncovers preferential copy number gains affecting mitochondrial biogenesis loci in the oncocytic variants. Am J Cancer Res 2015; 5: 1954–1971. [PMC free article] [PubMed] [Google Scholar]
- 34.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 2016; 41: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun W, Zhou S, Chang SS, et al. Mitochondrial mutations contribute to HIF1alpha accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenase kinase 2 in head and neck squamous cell carcinoma. Clin Cancer Res 2009; 15: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 2009; 19: 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleppe R, Martinez A, Doskeland SO, et al. The 14-3-3 proteins in regulation of cellular metabolism. Semin Cell Dev Biol 2011; 22: 713–719. [DOI] [PubMed] [Google Scholar]
- 38.Jin L, Chun J, Pan C, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene 2017; 36: 3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Zhu X, He F, Zeng H, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 2014; 46: 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis 2009; 30: 2967–2975. [DOI] [PubMed] [Google Scholar]
- *41.Karp NA, Lilley KS. Design and analysis issues in quantitative proteomics studies. Proteomics 2007; 7: 42–50. [DOI] [PubMed] [Google Scholar]
- *42.Geng X, Wang G, Qin Y, et al. iTRAQ-based quantitative proteomic analysis of the initiation of head regeneration in planarians. PLoS One 2015; 10: e0132045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Hsieh HC, Chen YT, Li JM, et al. Protein profilings in mouse liver regeneration after partial hepatectomy using iTRAQ technology. J Proteome Res 2009; 8: 1004–1013. [DOI] [PubMed] [Google Scholar]
- *44.Sandberg A, Lindell G, Kallstrom BN, et al. Tumor proteomics by multivariate analysis on individual pathway data for characterization of vulvar cancer phenotypes. Mol Cell Proteomics 2012; 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Li Y, Ruan Q, Ye G, et al. A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring. J Chromatogr A 2012; 1255: 228–236. [DOI] [PubMed] [Google Scholar]
- *46.Ye G, Liu Y, Yin P, et al. Study of induction chemotherapy efficacy in oral squamous cell carcinoma using pseudotargeted metabolomics. J Proteome Res 2014; 13: 1994–2004. [DOI] [PubMed] [Google Scholar]
- *47.Zhou Y, Song RX, Zhang ZS, et al. The development of plasma pseudotargeted GC-MS metabolic profiling and its application in bladder cancer. Anal Bioanal Chem 2016; 408: 6741–6749. [DOI] [PubMed] [Google Scholar]
- *48.Zeng J, Kuang H, Hu C, et al. Effect of bisphenol A on rat metabolic profiling studied by using capillary electrophoresis time-of-flight mass spectrometry. Environ Sci Technol 2013; 47: 7457–7465. [DOI] [PubMed] [Google Scholar]
- *49.Hoene M, Li J, Häring HU. The lipid profile of brown adipose tissue is sex-specific in mice. Biochim Biophys Acta 2014; 1842: 1563–1570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Figure S1. Increased mitochondrial number in pituitary oncocytoma, related to Figure 1
Figure S2. Metabolic characterization of oncocytoma, related to Figure 2
Figure S3. Lactate levels and LDHB expression, related to Figure 3
Figure S4. 14-3-3η inhibited glycolysis and oxidative phosphorylation, related to Figure 4
Figure S5. The mRNA levels of PPGCAR1 and LDHA, and others, related to Figure 5
Figure S6. Reduced LDHA expression and lactate production in other cancer cell lines, related to Figures 4 and 5
Table S1. Clinical characteristics of patients with pituitary oncocytomas
Table S2. Primers used in the qPCR study
Table S3. All fusion genes based on DNA whole-exome sequencing data
Table S4. mtDNA mutations detected in each patient with pituitary oncocytomas
Table S5. Gene set enrichment analysis
Table S6. The differentially expressed proteins and the top 10 Hallmark gene sets