Abstract
Rheumatoid arthritis (RA) is a chronic immune-mediated disease that primarily affects the synovium of diarthrodial joints. During the course of RA, the synovium transforms into a hyperplastic invasive tissue that causes destruction of cartilage and bone. Fibroblast-like synoviocytes (FLS), which form the lining of the joint, are epigenetically imprinted with an aggressive phenotype in RA and have an important role in these pathological processes. In addition to producing the extracellular matrix and joint lubricants, FLS in RA produce pathogenic mediators such as cytokines and proteases that contribute to disease pathogenesis and perpetuation. The development of multi-omics integrative analyses have enabled new ways to dissect the mechanisms that imprint FLS, have helped to identify potential FLS subsets with distinct functions and have identified differences in FLS phenotypes between joints in individual patients. This Review provides an overview of advances in understanding of FLS biology and highlights omics approaches and studies that hold promise for identifying future therapeutic targets.
Rheumatoid arthritis (RA) is an immune-mediated synovial disease caused by a complex interaction between genetic and environmental factors1. Although systemic immune dysregulation and autoimmunity occur in RA, the clinical manifestations are primarily synovial inflammation and joint damage2. Considerable advances in targeted therapy have improved outcomes, but a notable percentage of affected individuals still experience persistent inflammation and progressive disability3. Abnormal adaptive immunity, including mucosal immune responses that begin years before the onset of classifiable disease, is now recognized as a driving force in the evolution of RA from preclinical disease to overt synovitis4. However, the role of stromal elements, most notably fibroblast-like synoviocytes (FLS), in RA pathogenesis, has also gained attention, and targeting FLS is emerging as an attractive therapeutic approach in RA5-7.
FLS are highly specialized mesenchymal cells found in the synovium of diarthrodial joints. The synovium consists of two layers, the intimal lining layer and the sublining layer, with FLS primarily residing in the former compartment. In the healthy joint, the intimal lining forms a thin porous barrier at the interface between the sublining and the synovial fluid space8. FLS control the composition of the extracellular matrix (ECM) and of the synovial fluid, thereby lubricating and nourishing cartilage surfaces. In RA, however, FLS have unique aggressive behaviours that have an active role in disease pathogenesis and progression5,9.
Although cytokines and growth factors are important stromal cell regulators, FLS in RA are not simply ‘passive responders’ that react to the inflammatory milieu9-11. These cells are epigenetically imprinted with an activated and aggressive phenotype that operates independently of the inflammatory stimuli. Cultured FLS from patients with RA have autonomous pathogenic features that are maintained after many months in tissue culture or after implantation into mice9,12.
The mechanisms that imprint FLS are only partially understood, but high-throughput omics technologies are creating new ways to dissect these processes. Data from studies using new genomic methodology show that epigenetic mechanisms have a critical function in orchestrating the aggressive phenotype of RA FLS. Epigenetic patterns in FLS also change as disease evolves from early to established RA and suggest that, like adaptive immunity, FLS abnormalities in RA are not fixed but are influenced by the local environment13.
In this Review, we summarize the functional characteristics of FLS in health and in disease and subsequently focus on new data showing how epigenetic imprinting modifies the phenotype of FLS during RA. Understanding the unique genomic abnormalities and integrating diverse datasets should help to define the pathogenesis of RA and identify novel non-obvious targets for the treatment of this disease.
FLS physiology in the healthy joint
The synovial intimal lining is composed of two main types of synoviocytes: FLS (also known as type B synoviocytes) and macrophage-like synoviocytes (also known as type A synoviocytes). This thin and delicate structure sits on a bed of connective tissue known as the sublining layer, which also contains fibroblasts in addition to fat cells, macrophages and blood vessels14.
The synovial intimal lining has long been considered a loose association of cells serving as an ineffective barrier owing to a lack of classical adhesion structures such as tight junctions, desmosomes and a true basement membrane8,15-17. However, this paradigm has been challenged, with some evidence suggesting a more organized structure18. In 2019, a report identified a layer of macrophages adjacent to the FLS of the intimal lining. These macrophages arise from interstitial macrophages residing in the sublining layer, express proteins associated with tight junctions and are thus proposed to be ‘barrier forming’19, which is not a typical macrophage function. Additional studies are needed to validate these very interesting results.
FLS contribute directly to the synovial fluid composition by producing hyaluronic acid and other joint lubricants such as lubricin (also known as proteoglycan 4)20. The synovial fluid provides nourishment to the underlying articular cartilage and decreases the adherence of cells and proteins21,22. The synovial fluid also contains proteins and constituents of blood plasma, as well as limited numbers of leukocytes. As the cartilage lacks its own blood supply, these leukocytes are thought to pass into the synovial fluid through the synovial intimal lining23. FLS also help to shape and maintain the synovial ECM by producing matrix components (such as fibronectin, collagens, tenascin, proteoglycans and laminin) and ECM-degrading enzymes (such as proteases, matrix metalloproteinases (MMPs) and cathepsins).
FLS physiology in RA
The destruction of cartilage and non-osseous support structures of the joint in RA can be largely ascribed to effects mediated by FLS24-27. In the rheumatoid joint, the number of FLS increases considerably and contributes to the transformation of the synovial lining from a delicate structure into an invasive hyperplastic tissue mass known as a pannus9 (FIG. 1). RA FLS proliferate in culture when exposed to platelet-derived growth factor, transforming growth factor-β, TNF, or IL-1β, each of which is produced by immune cells present in the inflamed joint. RA FLS can proliferate in an anchorage-independent manner and have impaired contact inhibition, which is reminiscent of transformed cells28. However, RA FLS in the joint have a limited capacity to proliferate in situ and their expansion is, in part, a result of a low rate of apoptosis due to increased expression of pro-survival factors (reviewed elsewhere29). Compared with other cell types, RA FLS are also resistant to endoplasmic reticulum stress-induced apoptosis, probably because of increased autophagy and proteasomal activity30,31.
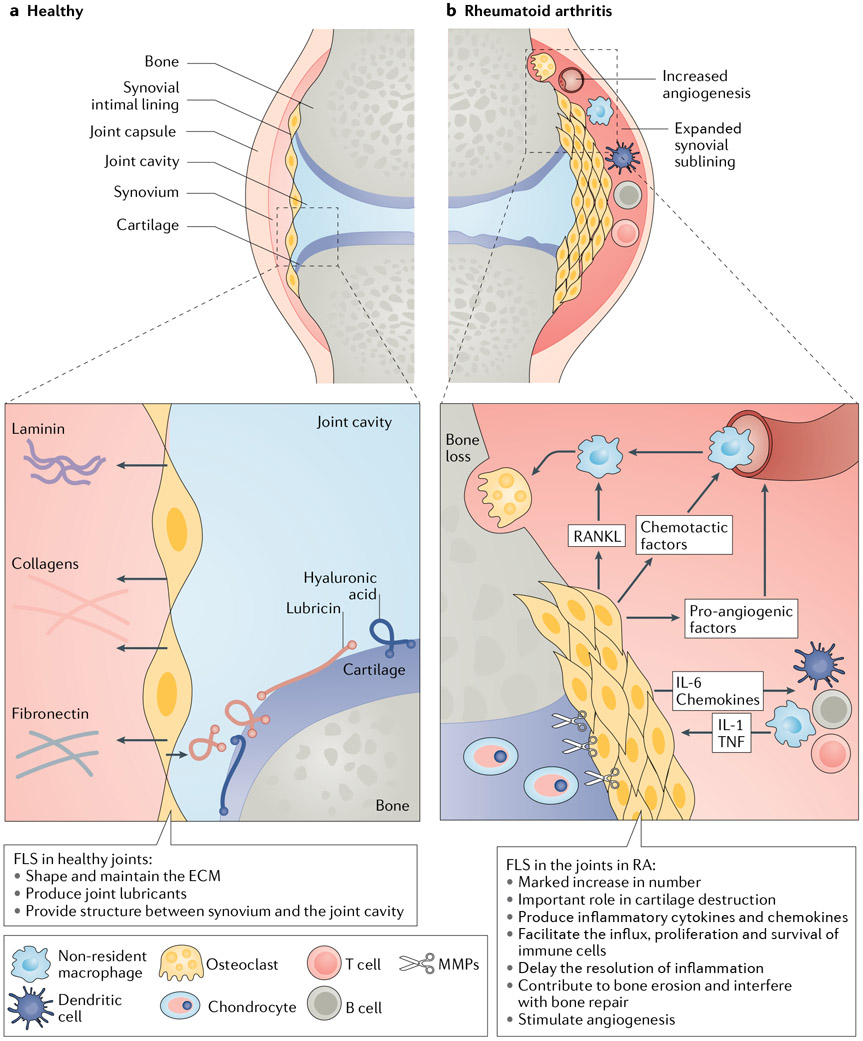
Fig. 1 ∣. The synovial joint in health and in RA.
a ∣ In the healthy joint, the synovial intimal lining is loosely organized and only one or two cell layers deep. Fibroblast-like synoviocytes (FLS) in the synovial intimal lining produce joint lubricants such as hyaluronic acid and lubricin. FLS also help to shape the extracellular matrix (ECM) by producing various matrix components, such as type IV collagen. b ∣ In rheumatoid arthritis (RA), the synovial intimal lining of the joint greatly expands and is transformed into an invasive hyperplastic pannus. The FLS express matrix metalloproteinases (MMPs) and are important contributors to the destruction of cartilage and non-osseous support structures. These cells help to promote and maintain joint inflammation by producing a repertoire of cytokines (such as IL-6), chemokines (such as CXC-chemokine ligand 10) and pro-angiogenic factors (such as VEGF). FLS in RA also contribute to bone erosion by facilitating osteoclastogenesis and by inhibiting bone repair.
Some evidence suggests that FLS in RA can arise from local epithelial to mesenchymal transition, an essential developmental process in the formation of complex tissues that is thought to occur in adult tissues after epithelial stress32. Another source of FLS expansion is pluripotent mesenchymal stem cells that migrate from the bone marrow and into the synovium where they differentiate into FLS. An influx of such blood-borne mesenchymal precursors precedes inflammation in the mouse model of collagen-induced arthritis, suggesting that this influx of cells contributes to the initiation of joint inflammation33. Healthy and mature FLS are often regarded as resident cells that remain in their local environment bound to the ECM34. Interestingly, studies in mice suggest that FLS also have migratory potential and can ‘metastasize’ to distant joints in vivo, potentially spreading disease from joint to joint35. However, migration of FLS from one joint to another has not been shown in humans.
At the pannus–cartilage interface of the rheumatoid joint, FLS-mediated overproduction of MMPs, such as MMP1, MMP3 and MMP13, damage the collagen-rich structures of the joint tissues and enable FLS invasion24-27,36,37. In synovial tissue samples from patients with new-onset RA (that is, <1 week since onset of symptoms), the expression of MMPs is already high in the synovial intimal lining38. The expansion of FLS in RA correlates with the duration of the disease, the amount of macrophage infiltration into the synovium and the severity of the cartilage erosions39,40. Studies in mice also show that activation of FLS is sufficient and in some models, such as human TNF transgenic mice and mice with collagen antibody-induced arthritis, FLS are indispensable for triggering arthritis41,42.
Metabolic regulation
An emerging characteristic of FLS in RA is their ability to reprogram their own cellular metabolism. Metabolic profiling using mass spectrometry detected alterations in glycolysis, the pentose phosphate pathway and amino acid metabolism in FLS during RA compared with FLS during osteoarthritis (OA)43. Increased glycolysis in RA FLS is of particular interest. Although glycolysis is less efficient than oxidative phosphorylation, it is the preferred source of ATP under hypoxic conditions44. Hypoxia-inducible factor 1α (HIF1α) is an inducer of glycolysis and its expression in RA FLS is linked to aggressive features such as migration and invasion45. Furthermore, several genes transcriptionally regulated by HIF1α and genes involved in glycolysis are upregulated in RA FLS, such as HK2 and SLC1A1 (REFS46,47). The contribution of dysregulated FLS metabolism to RA pathogenesis has been reviewed extensively elsewhere48,49.
Immune regulation
Although FLS in the healthy joints have modest immuneregulatory functions, FLS in RA have emerged as important immune modulators in pathogenesis through secreting factors such as IL-6 and through direct cell–cell interactions (reviewed elsewhere50). These cells actively facilitate the influx, proliferation and survival of immune cells as well as joint angiogenesis by producing a repertoire of cytokines, chemokines and pro-angiogenic factors5,25,51. Recruitment of macrophages, mast cells, T cells, B cells and dendritic cells expands the sublining layer of the synovium and helps to maintain and promote joint inflammation6,52. RA FLS also delay the resolution of inflammation by inhibiting apoptosis of pathogenic cells. For example, RA FLS prolong T cell survival by expressing type I interferons and support neutrophil survival by secreting granulocyte–macrophage colony-stimulating factor53,54.
Crosstalk with B cells.
FLS in RA can extend the lifespan of B cells through the production of IL-6, vascular cell adhesion molecule 1 (VCAM1), CXC-chemokine ligand 12 (CXCL12), B cell activating factor (BAFF, also known as TNFSF13B) and a proliferation-inducing ligand (APRIL, also known as TNFSF13)55-57. RA FLS also contribute to the differentiation and activation of B cells, which can then produce a variety of autoantibodies58,59. For example, BAFF and APRIL production by FLS is induced by Toll-like receptor 3 ligands, and Toll-like receptor 3 stimulation enhances the capacity of RA FLS to promote B cell differentiation57. Signalling between FLS and B cells in the RA joint is bidirectional. FLS express a modified 75-kD isoform of osteopontin (OPN) that supports FLS–B cell interactions. In FLS–B cell co-cultures, the 75-kD OPN-positive FLS produce a higher amount of IL-6 than 75-kD OPN-negative FLS or 75-kD OPN-positive FLS cultured alone60. FLS therefore participate in complex networks that support B cell differentiation and activation, which can, in turn, enhance adaptive immune responses in the joint.
Crosstalk with T cells.
FLS in RA also serve as antigenpresenting cells to T cells and can internalize neutrophil extracellular traps containing citrullinated peptides. Those peptides can then be presented to adaptive immune cells to amplify the local inflammatory response61,62. Similar to B cells, T cells can influence the function of RA FLS. Co-culture of FLS with resting T cells can induce IL-6, IL-8 and prostaglandin expression by RA FLS63. This effector function is further enhanced in the presence of IL-17 (REF.64). Furthermore, cytokine-activated T cells, such as T cells that have been stimulated with TNF, IL-6 or IL-2, can activate FLS via membrane-bound TNF65.
Crosstalk with monocytes and macrophages.
FLS in RA can attract monocytes from the vasculature by secreting chemotactic factors such as CC-chemokine ligand 2 (CCL2; also known as MCP1), CCL5, CCL8, CXCL5 and CXCL10 (REF.5). The recruited cells, particularly following differentiation into macrophages, are the most prominent source of TNF and IL-1β in the rheumatoid synovium, which in turn activate FLS to produce pro-inflammatory cytokines, chemokines and tissue-destructive factors such as IL-6, IL-8 and MMPs66-70. A genome-wide RNA analysis even detected a biphasic gene expression programme induced by TNF in RA FLS71. This programme consisted of an initial primarily unstable transcriptome that progressively switched to a very stable transcriptome comprising a number of genes, including IL6, CXCL8 and PTGS2 (encoding prostaglandin G/H synthase 2).
Soluble factors from RA FLS induced by TNF suppress TNF-induced expression of type I interferon regulated genes in macrophages by suppressing activation of Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signalling72. Prostaglandins produced by RA FLS work in concert with pro-inflammatory factors to shift macrophages towards a state characterized by high expression of pro-heparin-binding EGF-like growth factor (HBEGF) and pro-inflammatory genes such as IL1B and CXCL2. These HBEGF+ macrophages induce FLS invasiveness73. RANKL expressed by RA FLS can also stimulate osteoclastogenesis by macrophages and osteoclast activation74. Furthermore, RA FLS interfere with the repair of bone erosions by suppressing the activation of osteoblasts through secretion of Dicckopf-1, a regulatory molecule in the Wnt pathway that inhibits osteoblast function75.
Crosstalk with endothelial cells.
FLS in RA also regulate the influx of the inflammatory infiltrate by engaging in crosstalk with neighbouring vascular endothelial cells. Following activation (for example, in response to inflammation in the rheumatoid joint), the expression of cell adhesion molecules on endothelial cells is increased, which facilitates the capture, rolling and arrest of immune cells from the vasculature and the transmigration of immune cells into tissue76. When co-cultured with endothelial cells, FLS from the inflamed joints of patients with advanced RA increase the expression of adhesion molecules on endothelial cells, promoting adhesion of lymphocytes to the endothelial cells77. By contrast, the effect of FLS from the inflamed joints of patients with resolving RA or very early RA on the ability of co-cultured endothelial cells to interact with lymphocytes was similar to the effect of FLS from non-inflamed tissue. Importantly, when adding inflammatory cytokines such as IL-6 to this model, FLS from non-inflamed or resolving tissue inhibit lymphocyte adhesion, whereas FLS from very early RA or advanced RA support lymphocyte adhesion77.
Thus, as RA evolves, the FLS lose their immunoprotective capability and acquire a stimulatory phenotype in the later stages of the disease. These findings suggest that FLS in RA have transitional and immunomodulatory properties, but that these properties are outcome specific and stage specific and that FLS from different disease stages are functionally distinct77. Interestingly, as discussed in the “FLS phenotypes” section below, emerging data suggest that FLS isolated from the same synovial tissue also have functional heterogeneity.
Transcriptome patterns.
An analysis that combined RNA sequencing (RNA-seq) data with histopathology data identified at least three possible synovial patterns in patients with treatment-naive RA: a fibroblastic pattern that included a lack of an immune cell infiltrate; a macrophage-rich myeloid pattern characterized by enrichment of macrophages or monocytes; and a lympho-myeloid pattern characterized by aggregates of B and T cells78,79. Individual joints can also have varying patterns, and whether the pattern is consistent from one joint to another in a particular patient, or whether the pattern changes at different stages of the disease, is unclear. Ongoing studies are investigating whether the histological pattern correlates with clinical features or response to therapy.
FLS phenotypes
The development of therapeutic interventions directed at immune cells has been greatly facilitated by the discovery of cell-specific and lineage-specific markers6. Considerable effort has been expended to identify such markers for RA FLS, which have an extensive repertoire of surface receptors and markers. Some of these markers are general fibroblast markers such as type IV and type V collagens, and some are differentially expressed between different anatomical compartments in the synovium5,80.
The FLS surface marker that has garnered the most attention is cadherin 11 (CDH11). CDH11 is critical for homotypic aggregation of FLS from the synovial lining in vitro and in vivo. In laminin-containing micromass cultures, FLS migrate to the surface of the micromass and form a lining-like cellular organization81. These cells also recruit macrophages from the interior of the micromass to the ‘lining’8,81. CDH11-deficient mice have a hypoplastic synovium, and FLS isolated from these mice fail to form a lining layer at the micromass surface82,83. CDH11 extracellular domains can also be shed from the surface of FLS and can modulate the signalling and activation of neighbouring cells84. The importance of CDH11+ FLS in the destructive processes of RA was shown in the K/BxN serum transfer mouse model of arthritis, in which the deletion of CDH11 reduced cartilage erosion and joint inflammation82. CDH11 is a relatively specific marker for FLS of the synovial lining but is also expressed by osteoblasts and by some fibroblasts in the synovial sublining9. Furthermore, CDH11 has also been detected in other tissues such as in fibroblasts in the lung during idiopathic lung disease85.
Other markers found on FLS are less specific to FLS, are expressed by other cell lineages and are increased in RA. For example, podoplanin (PDPN) is a transmem-brane glycoprotein that is expressed in several human cancers86,87. PDPN is also highly expressed in FLS of the invading synovial tissue in RA, whereas FLS in the OA synovium are predominantly PDPN-negative80. This observation has proved useful when using novel transcriptomic and cellular profiling technologies to investigate FLS from RA synovium.
Heterogeneity in FLS surface marker expression as well as in FLS morphology, transcriptome and functions have been reported88-92. On the basis of these traits, different FLS or fibroblast subsets have been proposed by several groups (see TABLE 1). For example, a single-cell RNA-seq analysis of the RA synovium identified two main fibroblast phenotypes: a CD55+ population in the lining and a CD90+ population in the sublining90. The CD55+ fibroblasts were enriched for HAS1 (encoding a hyaluronan synthase), as well as genes associated with endothelial cell proliferation and regulation of reactive oxygen species responses. The CD90+ fibroblasts were enriched for genes related to MMP expression and organization of the ECM90. Another analysis that incorporated immunohistochemistry data on the expression of CD248 (also known as endosialin), PDPN, CD90 (also known as THY1) and VCAM1 in the RA synovium identified potential RA FLS subsets with functional differences, namely a PDPN+ subset in the lining layer and a CD248+ subset in the sublining92. Interestingly, in the severe combined immunodeficiency mouse model of cartilage destruction, the PDPN+ fibroblasts were the subset of fibroblasts that attached to, invaded and degraded cartilage92.
Table 1 ∣.
Examples of potential fibroblast-like synoviocyte and fibroblast phenotypes
| Subset | Location | Characteristics | Publication |
|---|---|---|---|
| CD34−CD90− | Lining | Low expression of TNFRSF11B (a gene that inhibits osteoclastogenesis) | Mizoguchi et al. (2018)89 |
| Increased osteoclastogenesis in vitro | |||
| High expression of MMP1, MMP3, PRG4, HAS1 and CD55 | |||
| This subset is less abundant in RA than in OA | |||
| Most of the cells are positive for CDH11 | |||
| CD34+ | Lining and sublining | Increased expression of genes involved in fibroblast migratory responses (CTHRC1, TWIST1, POSTN, LOXL2, PDGFRB and MMP14) | |
| Increased invasive and migratory properties in vitro | |||
| High secretion of IL-6, CXCL12 and CCL2 in vitro after stimulation with TNF | |||
| Increased ability to recruit peripheral blood monocytes in vitro | |||
| Most of the cells are positive for CDH11 | |||
| CD34−CD90+ | Sublining, surrounding blood vessels (in OA) or capillary structures (in RA) | Enrichment in the expression of mitotic and proliferative genes | |
| Increased expression of genes involved in fibroblast migratory response (CTHRC1, TWIST1, POSTN, LOXL2, PDGFRB and MMP14) | |||
| Increased expression of RANKL (a gene involved in osteoclastogenesis) | |||
| Low expression of TNFRSF11B (a gene that inhibits osteoclastogenesis) | |||
| Increased osteoclastogenesis in vitro | |||
| Increased invasive and migratory properties in vitro | |||
| Expanded 3-fold in the synovium in RA compared with in OA | |||
| In RA, most of the cells are positive for CDH11 | |||
| The proportion of this subset correlates positively with the proportion of leukocytes, synovitis and synovial hypertrophy | |||
| CD55+ | Lining | Enrichment in expression of HAS1 | Stephenson et al. (2018)90 |
| Enrichment in the expression of genes involved in pathways associated with endothelial cell proliferation and regulation of reactive oxygen species responses | |||
| CD90+ | Sublining, surrounding large vessels | Enrichment in the expression of genes involved in pathways associated with MMP activity and organization of the extracellular matrix | |
| PDPN+ | RA lining | Attach to, invade and degrade cartilage in the severe combined immunodeficiency mouse model of cartilage degradation | Croft et al. (2016)92 |
| Increased expression of PDPN after TNF and IL-1 stimulation in vitro | |||
| CD248+ | RA sublining | Increased expression of CD248 after TGFβ1 stimulation in vitro | |
| CD34+ | Sublining | Express genes related to the extracellular matrix | Zhang et al. (2019)91 |
| HLA-DRhi | Sublining | Expanded by >15-fold in RA in synovial tissue containing high levels of leukocyte infiltration compared with synovial tissue in OA | |
| Express genes related to the extracellular matrix | |||
| Express genes related to MHC class II presentation and the IFNγ-mediated signalling pathway (such as IFI30) | |||
| High expression of CXCL12 and CXCL9 | |||
| IL6 expression is restricted to this subset | |||
| DKK3+ | Sublining | Express genes related to the extracellular matrix | |
| High expression of DKK3, CADM1 and COL8A2 | |||
| CD55+ | Lining | Less abundant in RA in synovial tissue containing high levels of leukocyte infiltration than in synovial tissue in OA | |
| High expression of lubricin (encoded by PRG4) | |||
| High expression of DNASE1L3, a gene whose loss of function is associated with RA | |||
| FAPα+THY+ | Sublining | Immune-effector profile | Croft et al. (2019)93 |
| High expression of genes encoding cytokines and chemokines (including IL6, LIF, IL33 and IL34) | |||
| Transfer of these cells into mice with serum transfer-induced arthritis exacerbates disease | |||
| Transfer of these cells into mice with collagen-induced arthritis increases CD4+T cell, neutrophil and macrophage infiltration and reduces FOXP3+ regulatory T cells | |||
| FAPα+THY− | Lining | Bone-effector profile | |
| High expression of CCL9 and TNFSF11, potent inducers of osteoclast activity | |||
| High expression of MMP3, MMP9 and MMP13 | |||
| Surface expression of RANKL | |||
| Secrete high levels of RANKL | |||
| Have an increased RANKL:osteoprotegerin ratio | |||
| Stimulate osteoclast differentiation and activation in vitro | |||
| Mediate bone and cartilage damage and promote osteoclast activity when transferred into the joints of mice with serum transfer-induced arthritis |
CCL, CC-chemokine ligand; CXCL, CXC-chemokine ligand; MMP, matrix metalloproteinase; OA, osteoarthritis; RA, rheumatoid arthritis; TGF, transforming growth factor.
Other groups have reported great diversity of FLS, especially in the sublining compartment, with variable results depending on whether bulk transcriptomics or single-cell RNA-seq was used. For example, CD34−CD90− cells were identified primarily in the synovial lining, CD34−CD90+ cells were found exclusively in the sublining and CD34+ cells were localized to both the lining and sublining in one study89. In patients with OA, the CD34−CD90+ cells were located surrounding larger blood vessels in the synovium, whereas these cells were markedly expanded in the synovium in patients with RA and were located in the perivascular zone surrounding capillaries. The proportion of the CD34−CD90+ cells correlated positively with the proportion of leukocytes and the extent of synovitis in the RA synovium. Furthermore, this cell phenotype was associated with increased osteoclastogenesis, invasion and migration in vitro89. Another study characterized four different fibroblast phenotypes, including one CD55+ lining population- and three sublining populations91. One of the sublining populations had high expression of the MHC class II HLA-DR and is of particular interest, as these cells are expanded over 15-fold in RA and are a major source of the pro-inflammatory cytokine IL-6 (REF.91).
Different fibroblast phenotypes can be identified from the same datasets depending on the questions asked. This idea is exemplified by a study from 2019 (REF.93). In this study, the investigators found that the expression of fibroblast activation protein-α (FAPα), a cell-membrane dipeptidyl peptidase, is higher in both synovial tissue and FLS from patients with active RA than in patients with RA in whom the joint inflammation has resolved93. This finding led the researchers to suspect that FAPα expression could be associated with a pathogenic FLS phenotype, and further investigation identified two different pathogenic FAPα+ subsets in RA synovium. Interestingly, these pathogenic cells could be identified in part by reanalysing previous data91. The FAPα+CD90+ subset is located in the sublining and has an immune-effector profile characterized by high expression of a number of cytokines and chemokines, including IL-6, IL-33 and IL-34. The FAPα+CD90− subset is located in the lining and has a bone effector profile that includes high expression of inducers of osteoclast activity (CCL9 and TNFS11) and MMPs involved in cartilage degradation (MMP3, MMP9 and MMP13)93, indicating that the cells mediate bone and cartilage damage.
It is unclear whether these different populations are true subsets that have a fixed phenotype or whether the phenotype of FLS can be plastic and influenced by the microenvironment, resulting in variation in the relative abundance of the different putative phenotypes. Similar plasticity is well-known for macrophages, which align their function with signals in their tissue microenvironment, assuming a wide spectrum of phenotypes19. Single-cell transcriptomics and mass cytometry data do suggest that the FLS ‘subsets’ are a continuum and that some groups of cells, such as PDPN+ cells and CD34−CD90+ cells, might actually be extreme phenotypes with transcriptomes that simply reflect the local environment88. That possibility, which is favoured by some, would also help to explain why many groups identify different types of ‘subsets’ depending on the methodology employed.
Genetic modification of FLS in RA
Somatic mutations in a variety of genes could contribute to the altered phenotype of FLS in RA. Transition mutations, in particular, could be caused by reactive oxygen species and reactive nitrogen species in the highly inflamed joint94 (FIG. 2a). For example, several groups have identified mutations in TP53, which encodes cellular tumour antigen p53, in RA95-97. p53, also known as ‘the guardian of the genome’, maintains genome integrity and prevents proliferation of cells with damaged DNA95. In normal cells, the expression and activity of p53 are carefully restrained, but in response to DNA damage or other toxic stimuli, p53 is quickly stabilized and activated98. The expression of p53 is increased in the synovium in RA94, as well as in cancer99, owing to a prolonged half-life of mutated p53. Dominant-negative mutations in p53 could protect FLS from apoptosis and contribute to FLS invasiveness96,100,101. Gain-of-function mutations, such as V600R, in the proto-oncogene RAF1 also occur in FLS in RA102. This oncogene encodes the serine/threonine-protein kinase BRAF and the V600R mutation causes constitutive activation of the mitogen-activated protein kinase (MAPK) pathway. Antibodies directed against a mutated isoform of citrullinated vimentin have also been reported in individuals with RA, which could be explained by missense mutations in RA FLS increasing vimentin antigenicity103. Mitochondrial DNA mutations are also well documented in RA FLS104,105. For example, FLS in RA have approximately twice the number of mutations in MT-ND1, encoding mitochondrial NADH dehydrogenase 1, than FLS in OA104. Some of these mutations result in changes that could potentially be recognized by the cell as non-self when presented by MHC molecules104.
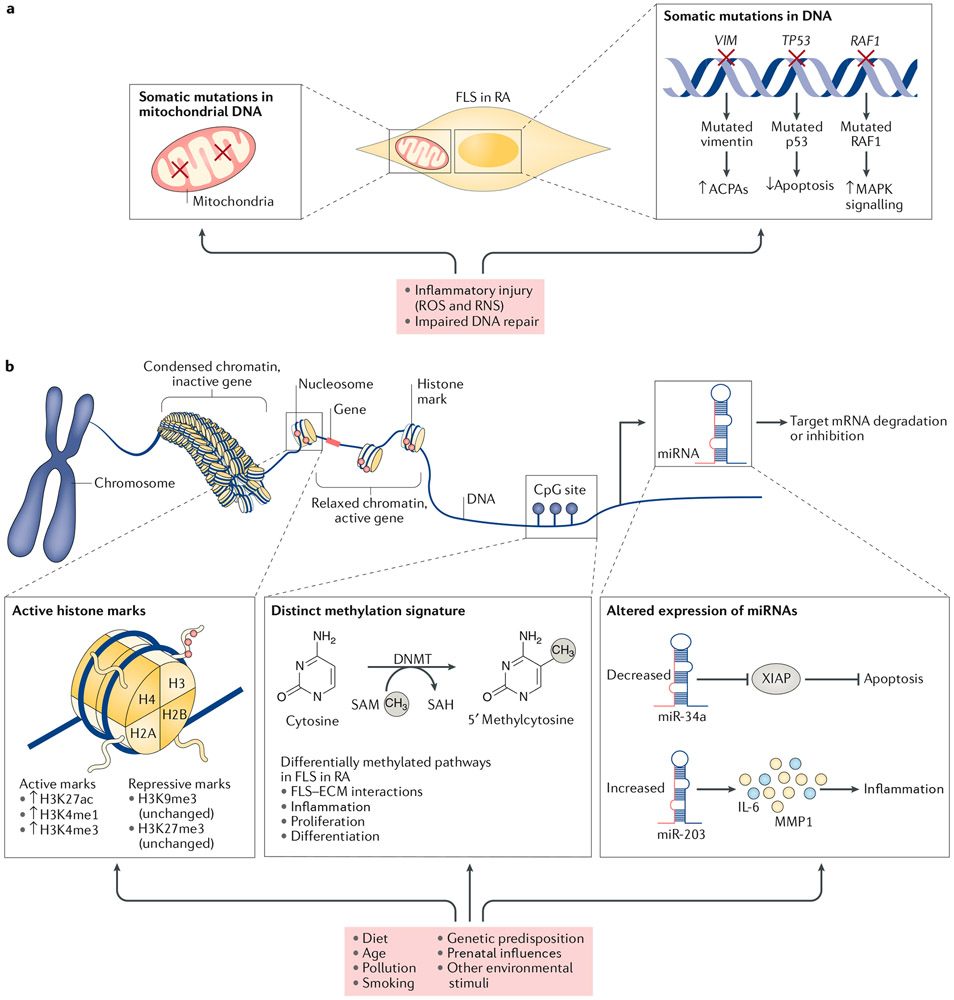
Fig. 2 ∣. Genetic and epigenetic mechanisms involved in FLS imprinting in RA.
Genetic and epigenetic modifications in fibroblast-like synoviocytes (FLS) in rheumatoid arthritis (RA) can be inherited or influenced by environmental factors, such as diet, inflammation, pollution and smoking. a ∣ The inflammatory milieu in the rheumatic joint can lead to increased production of reactive oxygen species (ROS) and reactive nitrogen species (RNS), which can alter the expression of proteins involved in DNA repair and might result in genetic imprinting of the FLS through somatic mutations in DNA and mutations in the mitochondrial DNA (as indicated by red crosses). b ∣ Histone modifications regulate the accessibility of the transcriptional machinery to gene promoters. Certain histone marks are associated with active transcription, whereas others are associated with transcriptional repression. In RA, changes in histone modifications (as indicated by the arrows or parentheses) strongly bias FLS towards increased transcription of pathogenic genes. DNA methylation occurs by the addition of a methyl group to a cytosine base where cytosine is followed by guanine (CpG sites). Methylation of the DNA in promoter regions is associated with a closed chromatin structure and transcriptional repression, whereas low levels of methylation in the promoter region favour an open chromatin structure and gene transcription. RA FLS have a distinct methylation pattern that is linked to their aggressive phenotype. RA FLS also have alterations in microRNA (miRNA) expression. miRNAs can cleave or inhibit target mRNA, and abnormal miRNA expression in RA FLS has been linked to increased resistance to apoptosis and increased production of IL-6 and matrix metalloproteinases (MMPs). ACPAs, anti-citrullinated protein/peptide antibodies; DNMT, DNA-methyltransferase; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; RAF1, RAF proto-oncogene serine/threonine-protein kinase; XIAP, X-linked inhibitor of apoptosis protein.
Epigenetic imprinting of FLS in RA
Genomics studies have defined a strong genetic component to RA that accounts for ~50–60% of the disease risk variance106,107. In addition to the most prominent genetic risk loci, the HLA-DR region, over 100 single nucleotide polymorphisms (SNPs) are associated with RA106,108. A number of genes associated with these SNPs have biological relevance in RA FLS, such as PTPN11 (encoding tyrosine-protein phosphatase non-receptor type 11 (PTPN11)) and LBH (encoding protein LBH (LBH))109,110. Despite the strong genetic component of RA, the disease concordance rate for RA for identical twins is only 12–15%111,112, suggesting that the environment and other stochastic influences have an important role in disease risk and severity113.
Epigenetic modifications can function as a bridge between the environment and the genome through chemical modification of chromatin or other mechanisms that can influence gene activity and expression without changing the DNA sequence114,115 (FIG. 2b). Epigenomic profiling in autoimmunity is still nascent, but intriguing data from candidate gene and unbiased approaches show that an altered epigenomic landscape contributes to the biology of FLS in RA116,117.
DNA methylation
One of the most frequently studied epigenetic mechanisms in RA FLS is DNA methylation, which occurs through the addition of a methyl group from S-adenosyl methionine to a cytosine base in DNA sites where the cytosine is followed by a guanine base (known as CpG sites). This process is mediated by DNA methyltransferases. Methylation patterns can be vertically transmitted from the mother cell to the daughter cell, or DNA methylation can occur de novo in response to cellular stress118. DNA methylation occurs throughout the genome, but its functional effects are best understood in promoter regions where high amounts of DNA methylation are associated with a closed chromatin structure and transcriptional repression. Conversely, low levels of methylation in a promoter region favour an open chromatin structure, enabling the formation of transcriptional complexes and gene transcription119. Methylation of CpG sites in the protein-coding region of the gene, in enhancers or in introns commonly occurs, but its effects on gene transcription are variable120. DNA methylation has an important part in silencing transposable elements (or ‘jumping genes’), which account for approximately 40% of the human genome. Loss of methylation at these elements is common in cancer and contributes to genomic instability and disturbance of DNA repair121.
Initial studies of the FLS methylome in RA quantified the overall methylation state of the cells and the data suggested that FLS are globally hypomethylated in RA. Indeed, suppression of methylation using the DNA methylation inhibitor 5-azacytidine in vitro directly affects the phenotype of normal FLS, resulting in an aggressive phenotype that mimics FLS in RA122. Furthermore, candidate gene approaches identified several genes that are differentially methylated in RA, such as DR3, which has a hypermethylated promoter region in RA123.
Subsequent larger unbiased genome-wide studies have revealed the presence of a distinct methylation pattern in RA FLS. A genome-wide study found no overall difference in levels of methylation in FLS in RA compared with FLS in OA, although 1,859 CpG sites were differentially methylated (732 hypomethylated and 1,127 hypermethylated)124. These differentially methylated regions corresponded to pathways related to FLS–ECM interactions, inflammation, proliferation and differentiation125 (FIG. 2b). Additional unbiased independent datasets confirmed and extended the DNA methylation signature126,127. Interestingly, the pattern of differential methylation in RA FLS overlapped with the differential methylation pattern in peripheral blood CD4+ naïve T cells in RA. These data suggest that disease-associated methylome signatures occur in cells that are more accessible than FLS128. In 2018, whole genome bisulfite sequencing was used to compare patterns of methylated loci in RA FLS and non-RA FLS and further confirmed the presence of a DNA methylation signature in RA FLS (as discussed in the section on “Integrating epigenomic datasets”)129.
The methylation profile of FLS in RA is quite stable in tissue culture and is consistent with functional observations showing that the abnormal RA FLS phenotype persists in vitro and in vivo when implanted into mice for many months126. Cytokines such as IL-1 and TNF could contribute to the RA pattern of differential methylation, as this pattern is reproduced in non-RA FLS following treatment with these cytokines in vitro125. However, the effects of these cytokines are transient and the pattern reverts back to ‘normal’ when the cytokines are removed, suggesting that other factors are required to imprint cells permanently.
Given that FLS from patients in the early stages of RA are not readily available, most studies of DNA methylation in RA FLS have been performed on cells derived from patients with longstanding RA. Thus, much of the kinetics of epigenetic remodelling during the evolution of the disease remains uncertain. However, the few methylation studies published13, which include some studies of FLS from patients with early RA, strongly suggest that differential methylation of RA FLS occurs early in the disease course and evolves as the disease progresses. One study compared differentially methylated loci in FLS from patients with different forms of arthritis13. Interestingly, the principal component analysis showed that DNA methylation patterns in FLS from patients with early or late RA formed clusters that were near, but distinct, from each other and from that of other forms of inflammatory arthritis, and were clearly segregated from the DNA methylation patterns of FLS from patients with OA. The total genomic methylation level in FLS from patients with longstanding RA was slightly lower than that in FLS from patients with early RA. A total of 5,469 genes were differentially methylated, and several pathways were enriched for hypomethylated genes in FLS from patients with longstanding RA. These pathways included integrin signalling, retinoic acid receptor activation and Wnt/β-catenin signalling13.
A more recent study has focused on DNA promoter methylation in FLS from healthy individuals and from patients with very early, resolving or established RA130. Differentially methylated promoter sites were present in FLS from patients with RA, even in the very early stages of disease, compared with FLS from healthy individuals, and occurred in promoters of genes involved in pathways related to the actin cytoskeleton, CDH, integrin and Wnt signalling, as well as antigen presentation130. Taken together with the observations that the DNA methylation pattern in FLS in early RA can be distinguished from the DNA methylation pattern in late RA, these results could help to explain the more aggressive phenotype observed in FLS from patients with established RA and suggest that epigenomic modifications can be plastic under some circumstances rather than fixed13,131. Altered DNA methylation in RA FLS is probably not only a consequence of the inflammatory milieu in the joint but might also function to promote initiation and progression of the disease130.
MicroRNAs
MicroRNAs (miRNAs) are non-coding RNAs that exert epigenetic control of gene expression through cleaving or inhibiting the target mRNA132. miRNAs could have an important role in the genomic risk of RA, particularly as genome-wide association data indicate that the majority of RA risk loci are located in non-coding regions of DNA133,134. Several miRNAs are associated with RA and can modify FLS function. For example, miR-34a targets the anti-apoptotic protein X-linked inhibitor of apoptosis protein (XIAP) and is downregulated in RA owing to DNA methylation of the miRNA promoter135. XIAP expression, in turn, contributes to RA by promoting resistance of FLS to apoptosis135. Other miRNAs, such as miR-203, are overexpressed in RA FLS compared with in OA FLS and have a pro-inflammatory function. miR-203 expression is linked with the aggressive phenotype of RA FLS, including the increased production of MMP1 and IL-6 by these cells, and treatment of FLS with the methylation inhibitor 5-azacytidine increases the expression of miR-203, providing a link between DNA methylation, miRNA expression and RA pathogenesis136.
The expression of some miRNAs is directly influenced by cytokines produced in the rheumatoid synovium. For example, miR-155 is overexpressed in RA FLS and the expression of this miRNA is induced in these cells after exposure to TNF, IL-1, lipopolysaccharides, polyinosinic:polycytidylic acid or bacterial lipoprotein137. miR-155-deficient mice are resistant to collagen-induced arthritis and these mice have no signs of inflammatory cell infiltrates in the synovium138. In K/BxN serum transfer-induced arthritis, the generation of osteoclasts is reduced in miR-155-deficient mice, which results in decreased local bone destruction; however, the severity of joint inflammation in miR-155-deficient mice and wild type mice is similar139. More complete descriptions of individual miRNAs in RA FLS and their functional effects are discussed elsewhere114,127,140,141.
Histone modifications
Various post-translational modifications of the N-terminal tail residues of histone proteins serve as epigenetic marks that regulate the accessibility of the transcriptional machinery to gene promoters. For example, trimethylation of histone 3 (H3) at lysine 4 (H3K4me3) is associated with an open chromatin and active transcription, whereas trimethylation of lysine 27 (H3K27me3) is associated with a closed chromatin conformation and transcriptional repression. A study investigating histone marks at the promoters of MMP-encoding genes in FLS from patients with RA or OA found that levels of H3Kme3 in the promoters of MMP1, MMP3, MMP9 and MMP13 were increased in RA FLS compared with in OA FLS, whereas levels of H3K27me3 in the promoters of MMP1 and MMP9 were decreased142. Furthermore, in a genome-wide analysis of the histone landscape in FLS in RA, histone marks associated with an open chromatin structure and gene expression, such as acetylation of H3 at lysine 27 (H3K27ac), H3K4me3 and monomethylation of H3 at lysine 4 (H3K4me1), were increased in RA FLS compared with in OA FLS129. Notably, the genome-wide expression of histone marks associated with gene repression, such as H3K27me3 and trimethylation of H3 at lysine 9 (H3K9me3), was similar in OA FLS and in RA FLS. These latter results suggest that the highly active transcriptional apparatus in RA in FLS is primarily caused by changes in histone modifications that promote transcription rather than changes in histone modifications that repress transcription (FIG. 2b).
Of the various forms of histone modification implicated in RA, the involvement and function of histone acetylation and its related enzymatic machinery are most frequently studied. Acetylation of lysine residues on histones neutralizes the positive charge of the amino acid residue, which weakens the binding between the histone and the negatively charged DNA, favouring an open chromatin structure and promoting transcription143. Histone acetylation is a reversible process: the addition of an acetyl group (acetylation) is mediated by histone acetyl transferases, whereas the removal of this group (deacetylation) is mediated by a family of histone deacetylates (HDACs). BET proteins can recognize acetylated lysine residues and couple these markers to the transcriptional machinery144.
There are four different classes of HDACs: class I (HDAC1-3 and HDAC8), class II (HDAC4-7, HDAC9 and HDAC10), class III sirtuins (SIRT1–7) and class IV (HDAC11)145. An analysis of the various class I, class II and class IV HDACs found that the expression of HDAC1 was increased in RA FLS compared with in OA FLS, whereas the expression of the other HDACs was unchanged. The increased expression of HDAC1 was associated with increased proliferation and survival of RA FLS146 but decreased MMP1 production146. By contrast, the expression of the class III HDAC SIRT1 is lower in RA FLS than in OA FLS147. The expression of SIRT1 is also downregulated in the joints of mice with collagen-induced arthritis compared with healthy mice148. Given that SIRT1 can regulate metalloproteinase production, this change could result in increased MMP1 and MMP3 expression.
Histone acetylation is modulated by pro-inflammatory cytokines present in the joint in RA. For example, TNF stimulation increases histone 4 acetylation in FLS, which is accompanied by increased chromatin accessibility and a prolonged inflammatory response67. Histone acetylation caused by chronic exposure of TNF primes FLS for activation and leads to enhanced inflammatory response to a second hit, such as exposure to IFNγ149. In RA synovial tissue, the expression of HDAC1, HDAC2 and HDAC3 correlates with the expression of TNF, whereas the expression of HDAC5 correlates negatively with both disease activity and IL-6 expression150,151. Interestingly, IL-1 and TNF selectively suppress the expression of HDAC5 in FLS from patients with RA, promoting nuclear localization of the transcription factor interferon regulatory factor 1 and transcription of a number of type 1 interferon response genes151. Thus, HDAC regulation is a dynamic process and is linked to pathogenic mechanisms in RA; blocking cytokines with therapeutics could have complex effects on transcription owing to modification of the epigenome.
Integrating epigenomic datasets
The studies described in the previous sections focused largely on candidate gene approaches or assessment of individual epigenomic marks. However, these marks do not function independently and can function in concert as part of the broader epigenomic landscape. Initial integrative analyses of combined epigenomic datasets of RA FLS were relatively simple and relied on identifying overlap between these datasets, such as overlap in DNA methylation patterns, gene expression and RA risk alleles. For example, datasets containing differentially expressed genes, differentially methylated loci and RA genome-wide association study risk alleles were compared to define a group of ‘triple evidence’ genes that were abnormal in all three datasets152.
The triple evidence group included not only genes already implicated in RA, such as CSF2 and HLA-DQA1, but also genes that had an unknown function in RA, such as ELMO1 and LBH. Subsequent analysis found that ELMO1 was highly expressed in RA FLS compared with in OA FLS, and was involved in FLS migration and invasion152. The methylation study was expanded with methylation data from enhancer regions, which again implicated LBH as well as PTPN11 as ‘multi-evidence’ genes109,153. Follow-up studies found that knockdown of LBH in FLS in vitro blocks cell cycle progression and promotes accumulation of DNA damage, whereas deletion of LBH exacerbates disease in mice with K/BxN serum transfer-induced arthritis154,155. PTPN11 is highly expressed in FLS from patients with RA and promotes FLS invasiveness156. Hypermethylation of two CpGs in an intronic enhancer region in PTPN11 promotes PTPN11 transcription by increasing enhancer activation by endogenous glucocorticoids109. Analyses such as these can, therefore, identify possible pathogenic genes in RA that can be prioritized and studied.
More recently, the global epigenomic landscape of FLS in RA was mapped by integrating diverse multi-plexed epigenomic data into a single analysis using a novel algorithm129. This method was the first to integrate whole-genome bisulfite sequencing, assay for transposase-accessible chromatin using sequencing (ATAC-seq), RNA-seq and chromatin immunoprecipitation sequencing data. Regions with similar epigenomic profiles could be grouped, revealing 125 distinct clusters (out of >350,000 possible combinations) in FLS. 13 of these clusters were enriched for epigenomic regions that were differentially modified in FLS from patients with RA compared with FLS from individuals with OA. The differentially marked regions predominantly corresponded to enhancers and promoters.
Further analysis revealed differentially modified pathways in 8 of the 13 enriched clusters, largely involving inflammation, the immune response, ECM regulation and cell migration. However, a number of unexpected pathways also emerged, such as the ‘Huntington’s Disease Signalling’ pathway, which includes a variety of genes implicated in the regulation and processing of the protein Huntingtin. siRNA-mediated knockdown of HIP1 (encoding HIP1, a particularly prominent protein in this pathway) in RA FLS decreased their invasion in an in vitro invasion model (a model that correlates with in vivo cartilage and joint damage157,158), providing biological validation of this differentially modified pathway129. Interestingly, a SNP in LBH was one of 10 unique RA-associated SNPs that overlapped with the differentially modified epigenomic regions, providing further evidence that this unbiased method has biological relevance129. In addition, HIP1 was independently validated as a potential therapeutic target in RA by a classical unbiased genetics approach159.
Joint-specific FLS features
Traditional views of RA suggest that the pathogenesis, histology and gene expression patterns are similar in affected joints at different locations160. This view was challenged when studies investigating methylation patterns in FLS found differences in the methylome signatures and transcriptome on the basis of the joint of origin161,162. The researchers noted two general types of joint-specific differences. The first difference involved genes related to cell differentiation and development, such as HOX family genes and genes involved in Wnt signalling (FIG. 3). These differences were not disease specific and occurred in both patients with OA and in patients with RA. Epigenetic imprinting probably occurs in cells in the joint to support the unique biomechanical features of each joint location. Whether this imprinting occurs in the bone marrow as mesenchymal stem cells migrate to the correct joint or whether imprinting occurs after these cells arrive at a joint is unclear. The latter scenario seems more likely because mechanical stimuli can downregulate the expression of genes, such as MMP1 and PTG2S, and the production of prostaglandin E2 in FLS163.
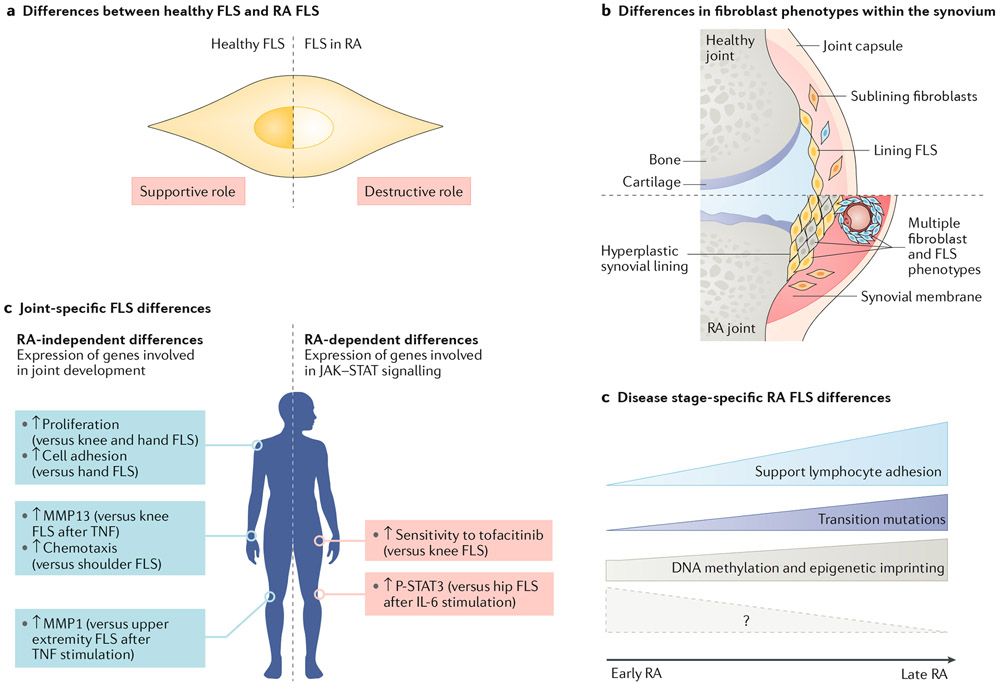
Fig. 3 ∣. Diversity of FLS in RA.
a ∣ Fibroblast-like synoviocytes (FLS) in healthy joints and FLS in joints affected by rheumatoid arthritis (RA) are very different. For example, healthy FLS mainly have a supportive role in the synovium and support structures. RA FLS, on the other hand, have an aggressive, imprinted phenotype and have a destructive role. b ∣ Fibroblasts, including FLS, within the same joint have a variety of phenotypes, especially in RA. These phenotypes might represent true subsets and/or these phenotypes might reflect the local environment in which they reside. c ∣ FLS show spatial heterogeneity with biological differences between various joints. These differences can include RA-independent differences in the expression of genes involved in cell differentiation such as HOX and WNT genes (blue boxes; examples from Frank-Bertoncelj et al.162) or RA-dependent differences in the expression of genes, including genes involved in cytokine signalling through Janus kinase (JAK)–signal transducer and activator of transcription (STAT) (red boxes; examples from Hammaker et al.164). Arrows indicate changes in levels of transcripts or functions for the indicated joint compared with other joints. d ∣ The function of RA FLS varies depending on the stage of disease. FLS in the late stage of RA promote lymphocyte adhesion to endothelial cells and can have genetic alterations. Differential methylation of DNA in RA FLS occurs early in disease but evolves as the disease progresses. Additional studies on FLS in very early RA are needed to dissect some initial pathogenic pathways (shown by the question mark); future omics analyses will hopefully shed light on these processes.
The second group of joint-specific marks were specific to RA and persisted even when the cell differentiation and proliferation pathways were filtered out of the analysis (FIG. 3). These disease-specific differences involved cytokine signalling pathways such as the JAK–STAT pathway131,161. STAT3 phosphorylation after stimulation with IL-6 in vitro is higher in FLS of the knee than FLS of the hip and correlates with levels of JAK1 in the FLS from the two locations164. Knee FLS are also less sensitive to the JAK inhibitor tofacitinib compared with hip FLS164. Hence, differences in JAK–STAT signalling and sensitivity to JAK inhibitors could contribute to the variable responses at different joint locations in patients with RA being treated with this class of drugs. Other groups have also shown that FLS from different locations are functionally distinct, including having differential responses to TNF and having distinct adhesive, proliferative and matrix-degrading characteristics162.
Most studies of joint-specific differences in FLS have focused mainly on FLS derived from the knee and hip joints, but other joints also have location-specific biology. An unsupervised hierarchical cluster analysis of RNA-seq data on FLS from the knees, hands and shoulders found that the cells segregate according to anatomical joint location. In addition to confirming distinct patterns of HOX and WNT gene expression, the data suggested that the non-coding transcriptome differed the most between the various joints162. The lncRNAs HOTAIR and HOTTIP were the most differentially expressed transcripts between the upper versus lower extremity FLS and between the distal versus proximal FLS, respectively. The HOX gene signature shared similar features to the embryonic positional HOX gene expression pattern along the proximal–distal and anterior–posterior developmental axes165-167.
Targeting FLS to treat RA
Despite great advances in RA therapy, many patients with RA have persistent disease. The current treatment approaches primarily focus on altering adaptive or innate immune responses by targeting pro-inflammatory cytokines, B cells or T cells3. Some of these drugs can affect the aggressive RA FLS phenotype, most notably cytokine or signal transduction inhibitors that can decrease the activation state of RA FLS. RA FLS have some imprinted or autonomous phenotypes that can persist even when removed from the cytokine-rich environment of the joint. This aggressive phenotype, together with the effects that RA FLS have on their microenvironment, can be summarized into hallmarks that distinguish RA FLS from healthy FLS (FIG. 4). As seen for cancer, we propose that these features can provide organizing principles for understanding how to target these cells in RA168,169. In this section, we discuss some potential therapeutic strategies, including approaches that target FLS metabolism, FLS surface markers, signalling pathways that contribute to the invasive and migratory potential of FLS or FLS apoptosis and the epigenetic signature of FLS.
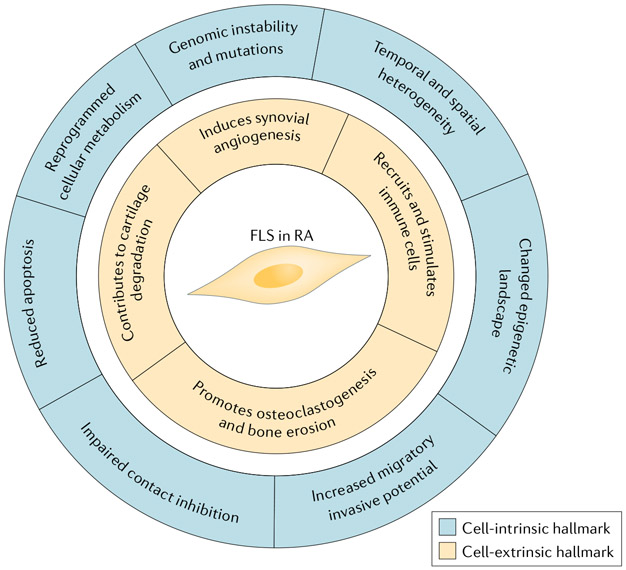
Fig. 4 ∣. The hallmark features of FLS in RA.
Fibroblast-Like synoviocytes (FLS) in rheumatoid arthritis (RA) have many features that distinguish them from FLS in healthy joints. The figure shows some major cell-intrinsic hallmarks of FLS in RA (intracellular features; outer circle) and cell-extrinsic hallmarks of FLS in RA (effects on the local tissues; inner circle). These features can either be imprinted, leading to permanent changes in FLS function, or can be part of a reversible response to the inflammatory environment.
Strategies that directly target FLS
An advantage of FLS-targeted therapies is that they could potentially be used in combination with immune suppression with limited added effect on host defence170. A variety of FLS-directed strategies have been evaluated, although none has been validated in the clinic to date. A discussion of all potential RA FLS targets is beyond the scope of this Review; instead, we describe some representative examples in this section.
Metabolism.
Resetting the dysregulated metabolic profile of RA FLS offers opportunities for disease modulation and the restoration of homeostasis171. Compared with FLS in OA, the balance between oxidative phosphorylation and glycolysis is altered in FLS in RA and is shifted towards glycolysis47. Interestingly, some current RA therapies, such as methotrexate and leflunomide, already target metabolic pathways such as purine or pyrimidine metabolism171. Furthermore, the JAK inhibitor tofacitinib modulates RA FLS metabolism by inducing oxidative phosphorylation while reducing the expression of the glycolysis inducer HIF1α and the glycolytic enzyme hexokinase 2 (HK2)172. HK2 is over-expressed in FLS from patients with RA compared with in FLS from patients with OA and silencing of HK2 in these cells inhibits FLS invasion and migration in vitro; furthermore, deletion of HK2 reduces bone and cartilage damage in mice with K/BxN serum transfer-induced arthritis46. FLS from patients with RA also express higher amounts of the glucose transporter, GLUT1, compared with FLS from patients with OA47. In vitro, glucose deprivation or incubation with glycolytic inhibitors reduces cytokine production, proliferation and migration of RA FLS; furthermore, in vivo, the glycolysis inhibitor 3-bromopyruvate reduces arthritis severity in the K/BxN serum-induced mouse model of arthritis47,48.
Surface markers and FLS phenotypes.
The potential existence of pathogenic FLS phenotypes could contribute to disease stratification and enable a more selective approach to targeting synovial mesenchymal cells in RA. Strategies for targeting FLS-specific surface receptors, such as CDH11, are already under evaluation. Indeed, CDH11-deficient mice are resistant to inflammatory arthritis82. A phase I trial of a monoclonal antibody directed at CDH11 (RG6125) was completed, but a phase II trial and subsequent development in RA were discontinued in 2018 owing to a lack of efficacy173. In addition, the data discussed above suggesting the existence of certain fibroblast or FLS phenotypes that contribute to synovitis creates opportunities for targeting cells on the basis of their cell surface phenotype (see TABLE 1). One concern with this approach is that the markers that distinguish these subsets, such as CD90, CD34 and PDPN, can be expressed on many cell lineages. Future studies will potentially identify surface proteins that are more specific to the pathogenic cells and could be therapeutically targeted.
Intracellular proteins and signal transduction.
Several members of the protein tyrosine phosphatase family (PTPs) are expressed in FLS and some, including PTPσ, PTPκ and PTPα, contribute to the aggressive phenotype of RA FLS, such as their migratory and invasive properties174-176. Interestingly, the expression of PTPs is induced in FLS in preclinical models of arthritis and blocking PTPs, such as PTPσ, can ameliorate arthritis in the K/BxN serum transfer model174. Hence, PTPs in FLS are a promising therapeutic target in RA. Historically, targeting the PTP family has been challenging because of the difficulty in designing selective inhibitors. Initial efforts mainly focused on developing inhibitors that target the PTP active site, a largely unsuccessful strategy hindered by the high charge and high level of active-site conservation among PTP family members. However, recent developments have enabled the design of compounds that achieve a high degree of selectivity for individual PTPs (reviewed elsewhere177).
MAPKs are highly activated in RA FLS, but targeting the downstream effector MAPKs, such as p38, has had limited success in human clinical trials. The lack of efficacy of this approach could be owing to the involvement of these kinases in feedback regulatory mechanisms that decrease pro-inflammatory signalling178-180. Members that are upstream in the kinase cascade of the MAPK family, such as MKK3, MKK6 and MAP3K5, are alternative targets that have been explored in preclinical models181. MAP3K5 is hypomethylated in RA FLS compared with in OA FLS and pro-inflammatory cytokines induce the expression of MAP3K5 in these cells through the activation of RelA124,182. Treatment with a small-molecule inhibitor of MAP3K5 (GS-627) reduces invasion, migration and proliferation of RA FLS in vitro182. Furthermore, Map3k5−/− mice are protected from K/BxN serum-induced arthritis183, and treatment with GS-627 also protects against joint damage and inflammation in rats with collagen-induced arthritis182.
Gene therapy approaches can also be used to specifically target or induce intracellular signalling in RA FLS. For example, genes can be delivered that induce apoptosis in RA FLS, such as with the intra-articular delivery of vectors containing PUMA, a down-stream effector of p53 and an effective inducer of apoptosis184,185. Because FLS do not express the adenovirus receptor, gene transfer approaches have been challenging. However, the development of a novel adenovirus–baculovirus construct enabled more efficient transfection than previous attempts, and the use of this vector to deliver PUMA directly into the joints led to improved arthritis in rats with adjuvant-induced arthritis185. Gene therapy approaches that involve the injection of FLS transduced in culture to express cytokine inhibitors such as IL-1RA have also been explored186. However, developing drugs that are locally delivered to target FLS (such as by intra-articular injection) is complicated in RA because treating an individual joint might not improve the large number of joints affected in this disease.
Targeting the imprinted FLS signature
The abnormal epigenetic landscape of RA FLS suggests that targeting differentially regulated genes or pathways could have therapeutic potential. Individual epigenetic marks and integrative analyses can identify and prioritize previously under-appreciated genes and pathways that might be amenable to drug development. None of these potential targets has progressed to clinical trials yet, although several targets, such as HIP1, ELMO1 and LBH, have shown preclinical efficacy. The DNA methylation profile of patients with early RA also suggests that disease mechanisms vary according to the stage of disease owing to an evolving epigenetic pattern. Thus, distinct therapeutic targets could be identified based on the stage of disease. miRNAs that are dysregulated and implicated in RA can also be targeted. Findings from animal models suggest that promoting the expression of protective miRNAs that are downregulated in RA FLS could decrease disease severity187.
A more intriguing approach is to remodel the RA epigenome to return it to a ‘normal’ state. Although epigenetic changes are long-lived, targeting the epigenetic machinery, such as histone-modifying enzymes, could alter the epigenome landscape. One challenge is the ubiquitous expression of histone-modifying enzymes and the lack of specificity of these enzymes for RA-related genes. In addition, some of these enzymes also modify non-histone proteins and multiple enzymes can modify the same histone residue143. Even so, some small molecule inhibitors of HDACs have proven effective in animal models of arthritis188; furthermore, an oral inhibitor of class I and class II HDACs (ITF2357, or Givinostat) has been tested in a phase II trial for the treatment of systemic-onset juvenile idiopathic arthritis, showing some clinical efficacy189-194. HDAC inhibitors seem to work by modulating the acetylation status of histones, although their precise mechanism of action remains largely unknown195. For example, ITF2357 has anti-inflammatory effects on RA FLS, which are mediated by suppressing transcription of cytokines195. This inhibitor also accelerates the decay of mRNA transcripts encoding pro-inflammatory mediators, such as IL-6, IL-8 and PTGS2 (REFS195,196).
A report in 2019 noted that RA FLS have a set of 280 TNF-inducible genes expressed with prolonged kinetics that escape repression owing to persistent H3K27 acetylation and increased chromatin accessibility of their regulatory elements197. The regulatory elements for these ‘fibroblast sustained genes’ are enriched for binding motifs for nuclear factor-κB, interferon regulatory factors and AP1, transcription factors that have a known role in RA synovitis. Hence, targeting TNF or targeting histone acetylation ‘reader’ proteins such as the BET proteins could potentially modulate persistent FLS activation by modifying the effects of these histone marks.
BET inhibitors have promising effects in animal models of arthritis and in RA FLS. For example, the BET inhibitor I-BET151 suppresses the production of cytokines and MMPs by RA FLS following in vitro stimulation with TNF or IL-1β and also reduces the proliferation and chemo-attractant potential of these cells198. Another BET inhibitor, JQ1, decreases RA FLS proliferation and production of pro-inflammatory cytokines (such as TNF, IL-1β, IL-6 and IL-8) and MMPs (such as MMP1, MMP3 and MMP13) in vitro199. JQ1 also downregulates TNF-induced nuclear factor-κB-dependent transcription in RA FLS and protects mice from collagen-induced arthritis199,200.
In contrast to histone modifiers, the number of enzymes controlling DNA methylation is quite limited and offers fewer opportunities to target FLS in RA selectively (reviewed elsewhere201). However, DNA methyltransferase inhibitors are being studied in oncology202. These compounds could be evaluated in RA if the safety profile is acceptable for non-oncological indications.
Future perspectives
New unbiased efforts that integrate large datasets and clinical phenotypes will be important for disease stratification, for the identification of cell subsets and for high-lighting potential therapeutic targets203. Understanding how and when the epigenetic imprinting of FLS in RA occurs could also define how and when the epigenomic landscape is remodelled and how to individualize therapy204.
The alteration of causative risk alleles that are important for pathogenic FLS biology through genetic manipulation is theoretically possible using gene editing. Newly developed CRISPR–Cas9 genome editing technologies that enable the conversion of C–G base pairs to T–A base pairs are promising tools for in vivo base correction205-207. For example, CRISPR–Cas-based genome editing was used in combination with highly specific guide RNAs to rescue the disease phenotype in a mice model of the autosomal recessive liver disease phenylketonuria by changing a single base208.
Another exciting development in the past few years is the newly launched Human Biomolecular Atlas Program, a NIH-sponsored consortium that intends to use multi-omic information to develop a widely accessible framework for comprehensively mapping the human body at single-cell resolution209. Ultimately, the goal is to provide 3D tissue maps that reveal the organization of tissues. If successful, this effort should have an invaluable effect on the advancement of human biology and precision medicine, and could help to fully elucidate the multiple functions of FLS in healthy and RA joints.
Conclusion
FLS are important contributors to the pathogenesis of RA and have a disease-specific imprinted phenotype that evolves as the disease progress. FLS in RA also have temporal and spatial heterogeneity; this heterogeneity leads to biological differences between different joints and between rheumatoid and non-rheumatoid joints, between early-stage and late-stage disease, and potentially between different FLS subsets. Prior research on FLS in RA focused mainly on candidate gene approaches, but broader unbiased datasets are now being probed for new and unanticipated promoters of disease. These large-scale approaches for studying FLS from different joints and from patients at various disease stages, including single cell transcriptomics analyses, should identify new therapeutic targets and help to facilitate individualized approaches to treating RA.
Key points.
Rheumatoid arthritis (RA) is a complex immune-mediated disease with clinical manifestations primarily involving synovial inflammation and joint damage.
Fibroblast-like synoviocytes (FLS) contribute to the pathogenesis of RA and are epigenetically imprinted with an aggressive phenotype in RA.
Synovial fibroblasts, including FLS, can have distinct phenotypes with different functional characteristics.
Epigenetic mechanisms associated with FLS imprinting in RA include alterations in DNA methylation, histone modifications and microRNA expression.
Integration of data from multi-omics analyses is needed to improve the characterization of FLS in RA.
Therapies that target FLS are emerging as promising therapeutic tools, raising hope for future applications in RA.
RNA sequencing.
(RNA-seq). A technique that measures the quantity and sequences of RNA in a biological sample, using next-generation sequencing.
Transposable elements.
DNA sequences that can move (transpose) to a new position in the genome, which can affect the activity of nearby genes.
Enhancer.
Short sequences of regulatory DNA elements that, when bound by transcription factors, can promote transcription of a particular gene by enhancing the activity of the gene promoter through physical interactions in cis.
Whole-genome bisulfite sequencing.
A technique for assessing genome-wide DNA methylation, using sodium bisulfite treatment and DNA sequencing.
Assay for transposase-accessible chromatin using sequencing.
A technique that identifies areas of open chromatin in the genome that are accessible to transcription factors, using a transposase and DNA sequencing.
Chromatin immunoprecipitation sequencing.
A technique that identifies the DNA binding sites in the genome for a particular protein of interest, using antibodies and DNA sequencing.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Rheumatology thanks G. Kalliolias, K. Reedquist, C. Ospelt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Klareskog L, Catrina AI & Paget S Rheumatoid arthritis. Lancet 373, 659–672 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Arnett FC et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 31, 315–324 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Choy EH, Kavanaugh AF & Jones SA The problem of choice: current biologic agents and future prospects in RA. Nat. Rev. Rheumatol 9, 154–163 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Holers VM et al. Rheumatoid arthritis and the mucosal origins hypothesis: protection turns to destruction. Nat. Rev. Rheumatol 14, 542–557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartok B & Firestein GS Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol. Rev 233, 233–255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filer A The fibroblast as a therapeutic target in rheumatoid arthritis. Curr. Opin. Pharmacol 13, 413–419 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Firestein GS Biomedicine. Every joint has a silver lining. Science 315, 952–953 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Valencia X et al. Cadherin-11 provides specific cellular adhesion between fibroblast-like synoviocytes. J. Exp. Med 200, 1673–1679 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottini N & Firestein GS Duality of fibroblast-like synoviocytes in RA: passive responders and imprinted aggressors. Nat. Rev. Rheumatol 9, 24–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blewis ME et al. Interactive cytokine regulation of synoviocyte lubricant secretion. Tissue Eng. Part A 16, 1329–1337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McInnes IB & Schett G Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol 7, 429–442 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Firestein GS Invasive fibroblast-like synoviocytes in rheumatoid arthritis. Passive responders or transformed aggressors? Arthritis Rheum. 39, 1781–1790 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Ai R et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. 67, 1978–1980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veale D & Firestein GS in Kelley and Firestein’s textbook of rheumatology 10th edn, (eds Firestein GS et al. ) 20–33 (Elsevier, 2016). [Google Scholar]
- 15.Lever JD & Ford EH Histological, histochemical and electron microscopic observations on synovial membrane. Anat. Rec 132, 525–539 (1958). [DOI] [PubMed] [Google Scholar]
- 16.Barland P, Novikoff AB & Hamerman D Electron microscopy of the human synovial membrane. J. Cell Biol 14, 207–220 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castor CW The microscopic structure of normal human synovial tissue. Arthritis Rheum. 3, 140–151 (1960). [DOI] [PubMed] [Google Scholar]
- 18.Culemann S et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 572, 670–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckley CD Macrophages form a protective cellular barrier in joints. Nature 572, 590–592 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Jay GD, Britt DE & Cha CJ Lubricin is a product of megakaryocyte stimulating factor gene expression by human synovial fibroblasts. J. Rheumatol 27, 594–600 (2000). [PubMed] [Google Scholar]
- 21.Jay GD & Waller KA The biology of lubricin: near frictionless joint motion. Matrix Biol. 39, 17–24 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Swann DA, Silver FH, Slayter HS, Stafford W & Shore E The molecular structure and lubricating activity of lubricin isolated from bovine and human synovial fluids. Biochem. J 225, 195–201 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firestein GS Etiology and pathogenesis of rheumatoid arthritis, In Kelley and Firestein’s Textbook of Rheumatology 10th edn, (eds Firestein GS et al. ) 1115–1166 (Elsevier, Philadelphia, 2017) [Google Scholar]
- 24.Sabeh F, Fox D & Weiss SJ Membrane-type I matrix metalloproteinase-dependent regulation of rheumatoid arthritis synoviocyte function. J. Immunol 184, 6396–6406 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Noss EH & Brenner MB The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunol. Rev 223, 252–270 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Muller-Ladner U, Pap T, Gay RE, Neidhart M & Gay S Mechanisms of disease: the molecular and cellular basis of joint destruction in rheumatoid arthritis. Nat. Clin. Pract. Rheumatol 1, 102–110 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Burrage PS, Mix KS & Brinckerhoff CE Matrix metalloproteinases: role in arthritis. Front. Biosci 11, 529–543 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Lafyatis R et al. Anchorage-independent growth of synoviocytes from arthritic and normal joints. Stimulation by exogenous platelet-derived growth factor and inhibition by transforming growth factor-β and retinoids. J. Clin. Invest 83, 1267–1276 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korb A, Pavenstadt H & Pap T Cell death in rheumatoid arthritis. Apoptosis 14, 447–454 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Ospelt C, Gay RE, Gay S & Klein K Dual role of autophagy in stress-induced cell death in rheumatoid arthritis synovial fibroblasts. Arthritis Rheumatol. 66, 40–48 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Shin YJ et al. Autophagy induction and CHOP under-expression promotes survival of fibroblasts from rheumatoid arthritis patients under endoplasmic reticulum stress. Arthritis Res. Ther 12, R19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steenvoorden MM et al. Transition of healthy to diseased synovial tissue in rheumatoid arthritis is associated with gain of mesenchymal/fibrotic characteristics. Arthritis Res. Ther 8, R165 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marinova-Mutafchieva L, Williams RO, Funa K, Maini RN & Zvaifler NJ Inflammation is preceded by tumor necrosis factor-dependent infiltration of mesenchymal cells in experimental arthritis. Arthritis Rheum. 46, 507–513 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Corr M & Zvaifler NJ Mesenchymal precursor cells. Ann. Rheum. Dis 61, 3–5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefevre S et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat. Med 15, 1414–1420 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolboom TC et al. Invasive properties of fibroblast-like synoviocytes: correlation with growth characteristics and expression of MMP-1, MMP-3, and MMP-10. Ann. Rheum. Dis 61, 975–980 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller MC et al. Membrane type 1 matrix metalloproteinase is a crucial promoter of synovial invasion in human rheumatoid arthritis. Arthritis Rheum. 60, 686–697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smolen JS et al. Rheumatoid arthritis. Nat. Rev. Dis. Primers 4, 18001 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Izquierdo E et al. Synovial fibroblast hyperplasia in rheumatoid arthritis: clinicopathologic correlations and partial reversal by anti-tumor necrosis factor therapy. Arthritis Rheum. 63, 2575–2583 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Redlich K & Smolen JS Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat. Rev. Drug Discov 11, 234–250 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Armaka M, Ospelt C, Pasparakis M & Kollias G The p55TNFR-IKK2-Ripk3 axis orchestrates arthritis by regulating death and inflammatory pathways in synovial fibroblasts. Nat. Commun 9, 618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armaka M et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med 205, 331–337 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn JK et al. GC/TOF-MS-based metabolomic profiling in cultured fibroblast-like synoviocytes from rheumatoid arthritis. Joint Bone Spine 83, 707–713 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Epstein T, Gatenby RA & Brown JS The Warburg effect as an adaptation of cancer cells to rapid fluctuations in energy demand. PLoS One 12, e0185085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hua S & Dias TH Hypoxia-inducible factor (HIF) as a target for novel therapies in rheumatoid arthritis. Front. Pharmacol 7, 184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustamante MF et al. Hexokinase 2 as a novel selective metabolic target for rheumatoid arthritis. Ann. Rheum. Dis 77, 1636–1643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Carbonell R et al. Critical role of glucose metabolism in rheumatoid arthritis fibroblast-like synoviocytes. Arthritis Rheumatol. 68, 1614–1626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Oliveira PG, Farinon M, Sanchez-Lopez E, Miyamoto S & Guma M Fibroblast-like synoviocytes glucose metabolism as a therapeutic target in rheumatoid arthritis. Front. Immunol 10, 1743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bustamante MF, Garcia-Carbonell R, Whisenant KD & Guma M Fibroblast-like synoviocyte metabolism in the pathogenesis of rheumatoid arthritis. Arthritis Res. Ther 19, 110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox DA, Gizinski A, Morgan R & Lundy SK Cell-cell interactions in rheumatoid arthritis synovium. Rheum. Dis. Clin. North Am 36, 311–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szekanecz Z, Besenyei T, Paragh G & Koch AE New insights in synovial angiogenesis. Joint Bone Spine 77, 13–19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buckley CD Why does chronic inflammation persist: an unexpected role for fibroblasts. Immunol. Lett 138, 12–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckley CD et al. Fibroblasts regulate the switch from acute resolving to chronic persistent inflammation. Trends Immunol. 22, 199–204 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Filer A et al. Differential survival of leukocyte subsets mediated by synovial, bone marrow, and skin fibroblasts: site-specific versus activation-dependent survival of T cells and neutrophils. Arthritis Rheum. 54, 2096–2108 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reparon-Schuijt CC et al. Regulation of synovial B cell survival in rheumatoid arthritis by vascular cell adhesion molecule 1 (CD106) expressed on fibroblast-like synoviocytes. Arthritis Rheum. 43, 1115–1121 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Burger JA, Zvaifler NJ, Tsukada N, Firestein GS & Kipps TJ Fibroblast-like synoviocytes support B-cell pseudoemperipolesis via a stromal cell-derived factor-1- and CD106 (VCAM-1)-dependent mechanism. J. Clin. Invest 107, 305–315 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bombardieri M et al. A BAFF/APRIL-dependent TLR3-stimulated pathway enhances the capacity of rheumatoid synovial fibroblasts to induce AID expression and Ig class-switching in B cells. Ann. Rheum. Dis 70, 1857–1865 (2011). [DOI] [PubMed] [Google Scholar]
- 58.de Brito Rocha S, Baldo DC & Andrade LEC Clinical and pathophysiologic relevance of autoantibodies in rheumatoid arthritis. Adv. Rheumatol 59, 2 (2019). [DOI] [PubMed] [Google Scholar]
- 59.van Venrooij WJ, van Beers JJ & Pruijn GJ Anti-CCP antibody, a marker for the early detection of rheumatoid arthritis. Ann. N. Y. Acad. Sci 1143, 268–285 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Take Y et al. Specifically modified osteopontin in rheumatoid arthritis fibroblast-like synoviocytes supports interaction with B cells and enhances production of interleukin-6. Arthritis Rheum. 60, 3591–3601 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Carmona-Rivera C et al. Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis. Sci. Immunol 2, eaag3358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tran CN et al. Presentation of arthritogenic peptide to antigen-specific T cells by fibroblast-like synoviocytes. Arthritis Rheum. 56, 1497–1506 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Yamamura Y et al. Effector function of resting T cells: activation of synovial fibroblasts. J. Immunol 166, 2270–2275 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Chabaud M et al. Human interleukin-17: a T cell-derived proinflammatory cytokine produced by the rheumatoid synovium. Arthritis Rheum. 42, 963–970 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Tran CN et al. Molecular interactions between T cells and fibroblast-like synoviocytes: role of membrane tumor necrosis factor-α on cytokine-activated T cells. Am. J. Pathol 171, 1588–1598 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naylor AJ, Filer A & Buckley CD The role of stromal cells in the persistence of chronic inflammation. Clin. Exp. Immunol 171, 30–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee A et al. Tumor necrosis factor α induces sustained signaling and a prolonged and unremitting inflammatory response in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 65, 928–938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Migita K et al. TNF-α-induced miR-155 regulates IL-6 signaling in rheumatoid synovial fibroblasts. BMC Res. Notes 10, 403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiao Z et al. Notch signaling mediates TNF-α-induced IL-6 production in cultured fibroblast-like synoviocytes from rheumatoid arthritis. Clin. Dev. Immunol 2012, 350209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perlman H et al. IL-6 and matrix metalloproteinase-1 are regulated by the cyclin-dependent kinase inhibitor p21 in synovial fibroblasts. J. Immunol 170, 838–845 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Loupasakis K et al. Tumor Necrosis Factor dynamically regulates the mRNA stabilome in rheumatoid arthritis fibroblast-like synoviocytes. PLoS One 12, e0179762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donlin LT, Jayatilleke A, Giannopoulou EG, Kalliolias GD & Ivashkiv LB Modulation of TNF-induced macrophage polarization by synovial fibroblasts. J. Immunol 193, 2373–2383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuo D et al. HBEGF+ macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci. Transl. Med 11, eaau8587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones DH, Kong YY & Penninger JM Role of RANKL and RANK in bone loss and arthritis. Ann. Rheum. Dis 61 (Suppl. 2), ii32–ii39 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diarra D et al. Dickkopf-1 is a master regulator of joint remodeling. Nat. Med 13, 156–163 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Klein D The tumor vascular endothelium as decision maker in cancer therapy. Front. Oncol 2, eaag3358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Filer A et al. Identification of a transitional fibroblast function in very early rheumatoid arthritis. Ann. Rheum. Dis 76, 2105–2112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis MJ et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 28, 2455–2470.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Humby F et al. Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients. Ann. Rheum. Dis 78, 761–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ekwall AK et al. The tumour-associated glycoprotein podoplanin is expressed in fibroblast-like synoviocytes of the hyperplastic synovial lining layer in rheumatoid arthritis. Arthritis Res. Ther 13, R40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kiener HP et al. Synovial fibroblasts self-direct multicellular lining architecture and synthetic function in three-dimensional organ culture. Arthritis Rheum. 62, 742–752 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Lee DM et al. Cadherin-11 in synovial lining formation and pathology in arthritis. Science 315, 1006–1010 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Chang SK, Gu Z & Brenner MB Fibroblast-like synoviocytes in inflammatory arthritis pathology: the emerging role of cadherin-11. Immunol. Rev 233, 256–266 (2010). [DOI] [PubMed] [Google Scholar]
- 84.Noss EH et al. Evidence for cadherin-11 cleavage in the synovium and partial characterization of its mechanism. Arthritis Res. Ther 17, 126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vandooren B et al. Tumor necrosis factor α drives cadherin 11 expression in rheumatoid inflammation. Arthritis Rheum. 58, 3051–3062 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Wicki A et al. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell 9, 261–272 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Martin-Villar E et al. Characterization of human PA2.26 antigen (T1α-2, podoplanin), a small membrane mucin induced in oral squamous cell carcinomas. Int. J. Cancer 113, 899–910 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Donlin LT et al. Methods for high-dimensonal analysis of cells dissociated from cyropreserved synovial tissue. Arthritis Res. Ther 20, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mizoguchi F et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun 9, 789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stephenson W et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun 9, 791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang F et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Croft AP et al. Rheumatoid synovial fibroblasts differentiate into distinct subsets in the presence of cytokines and cartilage. Arthritis Res. Ther 18, 270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Croft AP et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tak PP, Zvaifler NJ, Green DR & Firestein GS Rheumatoid arthritis and p53: how oxidative stress might alter the course of inflammatory diseases. Immunol. Today 21, 78–82 (2000). [DOI] [PubMed] [Google Scholar]
- 95.Lane DP Cancer. p53, guardian of the genome. Nature 358, 15–16 (1992). [DOI] [PubMed] [Google Scholar]
- 96.Firestein GS et al. Apoptosis in rheumatoid arthritis: p53 overexpression in rheumatoid arthritis synovium. Am. J. Pathol 149, 2143–2151 (1996). [PMC free article] [PubMed] [Google Scholar]
- 97.Yamanishi Y et al. p53 tumor suppressor gene mutations in fibroblast-like synoviocytes from erosion synovium and non-erosion synovium in rheumatoid arthritis. Arthritis Res. Ther 7, R12–R18 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Horn HF & Vousden KH Coping with stress: multiple ways to activate p53. Oncogene 26, 1306–1316 (2007). [DOI] [PubMed] [Google Scholar]
- 99.Theoret MR et al. Relationship of p53 overexpression on cancers and recognition by anti-p53 T cell receptor-transduced T cells. Hum. Gene Ther 19, 1219–1232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han Z, Boyle DL, Shi Y, Green DR & Firestein GS Dominant-negative p53 mutations in rheumatoid arthritis. Arthritis Rheum. 42, 1088–1092 (1999). [DOI] [PubMed] [Google Scholar]
- 101.Pap T, Aupperle KR, Gay S, Firestein GS & Gay RE Invasiveness of synovial fibroblasts is regulated by p53 in the SCID mouse in vivo model of cartilage invasion. Arthritis Rheum. 44, 676–681 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Weisbart RH et al. BRAF drives synovial fibroblast transformation in rheumatoid arthritis. J. Biol. Chem 285, 34299–34303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bang H et al. Mutation and citrullination modifies vimentin to a novel autoantigen for rheumatoid arthritis. Arthritis Rheum. 56, 2503–2511 (2007). [DOI] [PubMed] [Google Scholar]
- 104.Da Sylva TR, Connor A, Mburu Y, Keystone E & Wu GE Somatic mutations in the mitochondria of rheumatoid arthritis synoviocytes. Arthritis Res. Ther 7, R844–R851 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maximo V, Soares P, Lima J, Cameselle-Teijeiro J & Sobrinho-Simoes M Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: a study with emphasis on Hurthle cell tumors. Am. J. Pathol 160, 1857–1865 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Okada Y et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Viatte S, Plant D & Raychaudhuri S Genetics and epigenetics of rheumatoid arthritis. Nat. Rev. Rheumatol 9, 141–153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim K, Bang SY, Lee HS & Bae SC Update on the genetic architecture of rheumatoid arthritis. Nat. Rev. Rheumatol 13, 13–24 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Maeshima K et al. Abnormal PTPN11 enhancer methylation promotes rheumatoid arthritis fibroblast-like synoviocyte aggressiveness and joint inflammation. JCI Insight 10.1172/jci.insight.86580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsuda S et al. Regulation of the cell cycle and inflammatory arthritis by the transcription cofactor. J. Immunol 199, 2316–2322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aho K, Koskenvuo M, Tuominen J & Kaprio J Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol 13, 899–902 (1986). [PubMed] [Google Scholar]
- 112.MacGregor AJ et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 43, 30–37 (2000). [DOI] [PubMed] [Google Scholar]
- 113.Svendsen AJ et al. On the origin of rheumatoid arthritis: the impact of environment and genes-a population based twin study. PLoS One 8, e57304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Doody KM, Bottini N & Firestein GS Epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes. Epigenomics 9, 479–492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jablonka E & Raz G Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol 84, 131–176 (2009). [DOI] [PubMed] [Google Scholar]
- 116.Allis CD & Jenuwein T The molecular hallmarks of epigenetic control. Nat. Rev. Genet 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Karouzakis E, Gay RE, Gay S & Neidhart M Epigenetic deregulation in rheumatoid arthritis. Adv. Exp. Med. Biol 711, 137–149 (2011). [DOI] [PubMed] [Google Scholar]
- 118.Schubeler D Function and information content of DNA methylation. Nature 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 119.Robertson KD DNA methylation and human disease. Nat. Rev. Genet 6, 597–610 (2005). [DOI] [PubMed] [Google Scholar]
- 120.Jin B, Li Y & Robertson KD DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2, 607–617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kulis M & Esteller M DNA methylation and cancer. Adv. Genet 70, 27–56 (2010). [DOI] [PubMed] [Google Scholar]
- 122.Karouzakis E, Gay RE, Michel BA, Gay S & Neidhart M DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 60, 3613–3622 (2009). [DOI] [PubMed] [Google Scholar]
- 123.Takami N et al. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. 54, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 124.Nakano K, Whitaker JW, Boyle DL, Wang W & Firestein GS DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis 72, 110–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakano K, Boyle DL & Firestein GS Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J. Immunol 190, 1297–1303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Whitaker JW et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med. 5, 40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.de la Rica L et al. Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J. Autoimmun 41, 6–16 (2013). [DOI] [PubMed] [Google Scholar]
- 128.Rhead B et al. Rheumatoid arthritis naive T cells share hypermethylation sites with synoviocytes. Arthritis Rheumatol. 69, 550–559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ai R et al. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nat. Commun 9, 1921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Karouzakis E et al. Analysis of early changes in DNA methylation in synovial fibroblasts of RA patients before diagnosis. Sci. Rep 8, 7370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Firestein GS Pathogenesis of rheumatoid arthritis: the intersection of genetics and epigenetics. Trans. Am. Clin. Climatol. Assoc 129, 171–182 (2018). [PMC free article] [PubMed] [Google Scholar]
- 132.Hammond SM An overview of microRNAs. Adv. Drug Deliv. Rev 87, 3–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Farh KK et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Klein K & Gay S Epigenetics in rheumatoid arthritis. Curr. Opin. Rheumatol 27, 76–82 (2015). [DOI] [PubMed] [Google Scholar]
- 135.Niederer F et al. Down-regulation of microRNA-34a* in rheumatoid arthritis synovial fibroblasts promotes apoptosis resistance. Arthritis Rheum. 64, 1771–1779 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Stanczyk J et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 63, 373–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stanczyk J et al. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 58, 1001–1009 (2008). [DOI] [PubMed] [Google Scholar]
- 138.Kurowska-Stolarska M et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl Acad. Sci. USA 108, 11193–11198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bluml S et al. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 63, 1281–1288 (2011). [DOI] [PubMed] [Google Scholar]
- 140.Nakamachi Y et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 60, 1294–1304 (2009). [DOI] [PubMed] [Google Scholar]
- 141.Pandis I et al. Identification of microRNA-221/222 and microRNA-323-3p association with rheumatoid arthritis via predictions using the human tumour necrosis factor transgenic mouse model. Ann. Rheum. Dis 71, 1716–1723 (2012). [DOI] [PubMed] [Google Scholar]
- 142.Araki Y et al. Histone methylation and STAT-3 differentially regulate interleukin-6-induced matrix metalloproteinase gene activation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheumatol. 68, 1111–1123 (2016). [DOI] [PubMed] [Google Scholar]
- 143.Dong X & Weng Z The correlation between histone modifications and gene expression. Epigenomics 5, 113–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nicodeme E et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Angiolilli C, Baeten DL, Radstake TR & Reedquist KA The acetyl code in rheumatoid arthritis and other rheumatic diseases. Epigenomics 9, 447–461 (2017). [DOI] [PubMed] [Google Scholar]
- 146.Horiuchi M et al. Expression and function of histone deacetylases in rheumatoid arthritis synovial fibroblasts. J. Rheumatol 36, 1580–1589 (2009). [DOI] [PubMed] [Google Scholar]
- 147.Niederer F et al. SIRT1 overexpression in the rheumatoid arthritis synovium contributes to proinflammatory cytokine production and apoptosis resistance. Ann. Rheum. Dis 70, 1866–1873 (2011). [DOI] [PubMed] [Google Scholar]
- 148.Woo SJ et al. Myeloid deletion of SIRT1 suppresses collagen-induced arthritis in mice by modulating dendritic cell maturation. Exp. Mol. Med 48, e221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sohn C et al. Prolonged tumor necrosis factor α primes fibroblast-like synoviocytes in a gene-specific manner by altering chromatin. Arthritis Rheumatol. 67, 86–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kawabata T et al. Increased activity and expression of histone deacetylase 1 in relation to tumor necrosis factor-α in synovial tissue of rheumatoid arthritis. Arthritis Res. Ther 12, R133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Angiolilli C et al. Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression. Ann. Rheum. Dis 75, 430–438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Whitaker JW et al. Integrative omics analysis of rheumatoid arthritis identifies non-obvious therapeutic targets. PLoS One 10, e0124254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hammaker D et al. LBH gene transcription regulation by the interplay of an enhancer risk allele and DNA methylation in rheumatoid arthritis. Arthritis Rheumatol. 68, 2637–2645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ekwall AK et al. The rheumatoid arthritis risk gene LBH regulates growth in fibroblast-like synoviocytes. Arthritis Rheumatol. 67, 1193–1202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Matsuda S et al. Regulation of the cell cycle and inflammatory arthritis by the transcription cofactor LBH gene. J. Immunol 199, 2316–2322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stanford SM et al. Protein tyrosine phosphatase expression profile of rheumatoid arthritis fibroblast-like synoviocytes: a novel role of SH2 domain-containing phosphatase 2 as a modulator of invasion and survival. Arthritis Rheum. 65, 1171–1180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tolboom TC et al. Invasiveness of fibroblast-like synoviocytes is an individual patient characteristic associated with the rate of joint destruction in patients with rheumatoid arthritis. Arthritis Rheum. 52, 1999–2002 (2005). [DOI] [PubMed] [Google Scholar]
- 158.Laragione T, Brenner M, Mello A, Symons M & Gulko PS The arthritis severity locus Cia5d is a novel genetic regulator of the invasive properties of synovial fibroblasts. Arthritis Rheum. 58, 2296–2306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Laragione T et al. Huntingtin-interacting protein 1 (HIP1) regulates arthritis severity and synovial fibroblast invasiveness by altering PDGFR and Rac1 signalling. Ann. Rheum. Dis 11, 1627–1635 (2018). [DOI] [PubMed] [Google Scholar]
- 160.Kraan MC et al. Comparison of synovial tissues from the knee joints and the small joints of rheumatoid arthritis patients: implications for pathogenesis and evaluation of treatment. Arthritis Rheum. 46, 2034–2038 (2002). [DOI] [PubMed] [Google Scholar]
- 161.Ai R et al. Joint-specific DNA methylation and transcriptome signatures in rheumatoid arthritis identify distinct pathogenic processes. Nat. Commun 7, 11849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Frank-Bertoncelj M et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun 8, 14852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang P et al. Cyclic mechanical stretch downregulates IL-1β-induced COX-2 expression and PGE(2) production in rheumatoid arthritis fibroblast-like synoviocytes. Connect. Tissue Res 52, 190–197 (2011). [DOI] [PubMed] [Google Scholar]
- 164.Hammaker D et al. Joint location-specific JAK-STAT signaling in rheumatoid arthritis fibroblast-like synoviocytes. ACR Open Rheumatol. 1, 640–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krumlauf R Hox genes in vertebrate development. Cell 78, 191–201 (1994). [DOI] [PubMed] [Google Scholar]
- 166.Zakany J & Duboule D The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev 17, 359–366 (2007). [DOI] [PubMed] [Google Scholar]
- 167.Wang KC et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hanahan D & Weinberg RA The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 169.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 170.Neumann E, Lefevre S, Zimmermann B, Gay S & Muller-Ladner U Rheumatoid arthritis progression mediated by activated synovial fibroblasts. Trends Mol. Med 16, 458–468 (2010). [DOI] [PubMed] [Google Scholar]
- 171.Sanchez-Lopez E, Cheng A & Guma M Can metabolic pathways be therapeutic targets in rheumatoid arthritis? J. Clin. Med 8, E753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.McGarry T et al. JAK/STAT blockade alters synovial bioenergetics, mitochondrial function, and proinflammatory mediators in rheumatoid arthritis. Arthritis Rheumatol. 70, 1959–1970 (2018). [DOI] [PubMed] [Google Scholar]
- 173.Finch R et al. OP0224 results of a phase 2 study of RG6125, an anti-cadherin-11 monoclonal antibody, in rheumatoid arthritis patients with an inadequate response to anti-TNFα therapy. Ann. Rheum. Dis 78, 189 (2019). [Google Scholar]
- 174.Doody KM et al. Targeting phosphatase-dependent proteoglycan switch for rheumatoid arthritis therapy. Sci. Transl. Med 7, 288ra276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stanford SM et al. TGFβ responsive tyrosine phosphatase promotes rheumatoid synovial fibroblast invasiveness. Ann. Rheum. Dis 75, 295–302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stanford SM et al. Receptor protein tyrosine phosphatase α-mediated enhancement of rheumatoid synovial fibroblast signaling and promotion of arthritis in mice. Arthritis Rheumatol. 68, 359–369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stanford SM & Bottini N Targeting tyrosine phosphatases: time to end the stigma. Trends Pharmacol. Sci 38, 524–540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kim C et al. The kinase p38α serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol 9, 1019–1027 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ananieva O et al. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat. Immunol 9, 1028–1036 (2008). [DOI] [PubMed] [Google Scholar]
- 180.Guma M et al. Antiinflammatory functions of p38 in mouse models of rheumatoid arthritis: advantages of targeting upstream kinases MKK-3 or MKK-6. Arthritis Rheum. 64, 2887–2895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yoshizawa T et al. Role of MAPK kinase 6 in arthritis: distinct mechanism of action in inflammation and cytokine expression. J. Immunol 183 1360–1367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nygaard G et al. Regulation and function of apoptosis signal-regulating kinase 1 in rheumatoid arthritis. Biochem. Pharmacol 151, 282–290 (2018). [DOI] [PubMed] [Google Scholar]
- 183.Mnich SJ et al. Critical role for apoptosis signal-regulating kinase 1 in the development of inflammatory K/BxN serum-induced arthritis. Int. Immunopharmacol 10, 1170–1176 (2010). [DOI] [PubMed] [Google Scholar]
- 184.Cha HS, Rosengren S, Boyle DL & Firestein GS PUMA regulation and proapoptotic effects in fibroblast-like synoviocytes. Arthritis Rheum. 54, 587–592 (2006). [DOI] [PubMed] [Google Scholar]
- 185.Hong SS et al. PUMA gene delivery to synoviocytes reduces inflammation and degeneration of arthritic joints. Nat. Commun 8, 146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Evans CH et al. Gene transfer to human joints: progress toward a gene therapy of arthritis. Proc. Natl Acad. Sci. USA 102, 8698–8703 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhu S et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat. Med 18, 1077–1086 (2012). [DOI] [PubMed] [Google Scholar]
- 188.Hammaker D & Firestein GS Epigenetics of inflammatory arthritis. Curr. Opin. Rheumatol 30, 188–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chung YL, Lee MY, Wang AJ & Yao LF A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol. Ther 8, 707–717 (2003). [DOI] [PubMed] [Google Scholar]
- 190.Joosten LA, Leoni F, Meghji S & Mascagni P Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Mol. Med 17, 391–396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nishida K et al. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21WAF1/Cip1 expression. Arthritis Rheum. 50, 3365–3376 (2004). [DOI] [PubMed] [Google Scholar]
- 192.Li M et al. Therapeutic effects of NK-HDAC-1, a novel histone deacetylase inhibitor, on collagen-induced arthritis through the induction of apoptosis of fibroblast-like synoviocytes. Inflammation 36, 888–896 (2013). [DOI] [PubMed] [Google Scholar]
- 193.Lee J et al. A novel histone deacetylase 6-selective inhibitor suppresses synovial inflammation and joint destruction in a collagen antibody-induced arthritis mouse model. Int. J. Rheum. Dis 18, 514–523 (2015). [DOI] [PubMed] [Google Scholar]
- 194.Vojinovic J et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 63, 1452–1458 (2011). [DOI] [PubMed] [Google Scholar]
- 195.Angiolilli C et al. Control of cytokine mRNA degradation by the histone deacetylase inhibitor ITF2357 in rheumatoid arthritis fibroblast-like synoviocytes: beyond transcriptional regulation. Arthritis Res. Ther 20, 148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Grabiec AM, Korchynskyi O, Tak PP & Reedquist KA Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis 71, 424–431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Loh C et al. TNF-induced inflammatory genes escape repression in fibroblast-like synoviocytes: transcriptomic and epigenomic analysis. Ann. Rheum. Dis 78, 1205–1214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Klein K et al. The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann. Rheum. Dis 75, 422–429 (2016). [DOI] [PubMed] [Google Scholar]
- 199.Xiao Y et al. Bromodomain and extra-terminal domain bromodomain inhibition prevents synovial inflammation via blocking IκB kinase-dependent NF-κB activation in rheumatoid fibroblast-like synoviocytes. Rheumatology 55, 173–184 (2016). [DOI] [PubMed] [Google Scholar]
- 200.Mele DA et al. BET bromodomain inhibition suppresses TH17-mediated pathology. J. Exp. Med 210, 2181–2190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tough DF, Tak PP, Tarakhovsky A & Prinjha RK Epigenetic drug discovery: breaking through the immune barrier. Nat. Rev. Drug Discov 15, 835–853 (2016). [DOI] [PubMed] [Google Scholar]
- 202.Subramaniam D, Thombre R, Dhar A & Anant S DNA methyltransferases: a novel target for prevention and therapy. Front. Oncol 4, 80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Najm A et al. Standardisation of synovial biopsy analyses in rheumatic diseases: a consensus of the EUlAr Synovitis and OMERACT Synovial Tissue Biopsy Groups. Arthritis Res. Ther 20, 265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Menche J et al. Integrating personalized gene expression profiles into predictive disease-associated gene pools. NPJ Syst. Biol. Appl 3, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol 35, 371–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Villiger L et al. Treatment of metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med 24, 1519–1525 (2018). [DOI] [PubMed] [Google Scholar]
- 209.HuBMAP Consortium. The human body at cellular resolution: the NIH human biomolecular atlas program. Nature 574, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]