Abstract
Plants are primary producers of food and oxygen on Earth and will likewise be indispensable to the establishment of large-scale sustainable ecosystems and human survival in space. To contribute to the understanding of how plants respond to spaceflight stress, we examined the significance of the unfolded protein response (UPR), a conserved signaling cascade that responds to a number of unfavorable environmental stresses, in the model plant Arabidopsis thaliana. To do so, we performed a large-scale comparative transcriptome profiling in wild type and various UPR-defective mutants during the SpaceX-CRS12 mission to the International Space Station. We established that orbital culture substantially alters the expression of hundreds of stress-related genes compared with ground control conditions. Although expression of those genes varied in the UPR mutants on the ground, it was largely similar across the genotypes in the spaceflight condition. Our results have yielded new information on how plants respond to growth in orbit and support the hypothesis that spaceflight induces the activation of signaling pathways that compensate for the loss of UPR regulators in the control of downstream transcriptional regulatory networks.
Key Words: Spaceflight, Unfolded protein response, Arabidopsis, Microgravity.
1. Introduction
Extraterrestrial habitation and prolonged space travel require successful plant growth to recreate livable environments for humans (Ferl et al., 2002; Massa et al., 2016; Zhou et al., 2019). Studies over the past 70 years have sought to develop a better understanding of how plants are affected by and adapt to the significant stresses imposed by spaceflight (e.g., microgravity, radiation, vibration, limited exchange of gases), which can affect plant development and yield (Paul et al., 2013). Recent iterations of the sophisticated chamber hardware for plant growth housed on the International Space Station (ISS) have allowed for multigenerational plant growth in space and analyses of plant responses to this environment (Massa et al., 2016). However, these facilities are insufficient for large-scale plant growth on extraterrestrial environments due to their size and resource cost. Therefore, generation of germplasm adapted to stresses experienced during growth in extraterrestrial environments is a critical contribution to the realization of sustainable plant cultivation in space.
The unfolded protein response (UPR) is a signaling cascade that responds to a number of unfavorable environmental and cellular stresses. The UPR is generally activated by a buildup of unfolded proteins in the endoplasmic reticulum (ER), a condition known as ER stress (Ron and Walter, 2007). The ER stress sensors conserved across metazoans and plants include the ER-associated protein kinase and ribonuclease inositol-requiring enzyme 1 (IRE1) and ER membrane-tethered transcription factors (TFs) (metazoan activating transcription factor 6 [ATF6] and plant basic leucine zipper 17 [bZIP17] and bZIP28). Activation of IRE1 leads to unconventional splicing of an intron from the mRNA of an IRE1-downstream bZIP-TF (metazoan X-box binding protein 1 and plant bZIP60). The UPR TFs are translocated to the nucleus to control expression of UPR target genes and restore ER homeostasis (Koizumi et al., 2001; Chen and Brandizzi, 2012; Halbleib et al., 2017; Kim et al., 2018; Ruberti et al., 2018; Tam et al., 2018; Mishiba et al., 2019; Pastor-Cantizano et al., 2019; Pu et al., 2019). Insufficient UPR leads to the actuation of cell death (Ron and Walter, 2007; Walter and Ron, 2011).
In terrestrially grown plants, the UPR is a key mediator of responses to a variety of stresses, including heat, pathogen, and high light/singlet oxygen (Deng et al., 2011; Moreno et al., 2012; Guillemette et al., 2014; Zhang et al., 2015, 2017; Pastor-Cantizano et al., 2019; Beaugelin et al., 2020). Additionally, analyses of higher order UPR mutants have demonstrated that the UPR regulators are necessary for post-embryonic growth and reproductive development in Arabidopsis under unstressed conditions as well (Chen and Brandizzi, 2012; Kim et al., 2018; Mishiba et al., 2019; Pu et al., 2019). Therefore, a better understanding of the UPR can enable efforts to potentiate plant stress responses and improve plant yield.
Given the broad responsiveness of the UPR to environmental stresses, we hypothesized that the UPR effectors could coordinate gene expression reprogramming in spaceflight stress conditions. To test this hypothesis, we analyzed global gene expression changes in wild type (WT) Arabidopsis as well as loss-of-function mutants of IRE1 (ire1a ire1b, herein dubbed ire1), bZIP28, and bZIP60 (single and double mutants: bzip28, bzip60, bzip28 bzip60), cultivated in orbit during the SpaceX CRS12 mission to the ISS. We used these genetic backgrounds to identify genes controlled jointly or specifically by the UPR sensors and UPR TFs and define the extent to which the known signaling pathways of the UPR functionally interact in a whole organism under microgravity-associated conditions. We established that, in space and on ground, gene expression undergoes a substantial reprogramming on a genome-scale. Growth in orbit substantially altered the expression of thousands of genes associated with significant biological traits compared with ground controls. However, while many of these spaceflight-responsive genes were regulated uniquely in certain UPR mutants compared with WT in the ground control, such a genotype-specific regulation was not observed in the spaceflight condition. These observations not only provide new insight into how plants respond to spaceflight, but also establish that spaceflight-induced transcriptional responses mitigate the need for the gene-regulatory networks controlled by the UPR sensors.
2. Materials and Methods
2.1. Launch hardware and experimental timeline
This flight experiment utilized 4 Biological Research In a Canister (BRIC) containing a total of 22 Petri Dish Fixation Unit (PDFU) hardware (Wells et al., 2001) to cultivate sterile dark-grown seedlings germinated aboard the ISS for a 14 day period. PDFU actuation chambers were loaded with a tissue fixative (RNAlater; Invitrogen) to preserve samples at the conclusion of the flight experiment. An identical set of samples was prepared and grown on Earth with a 2-day offset at Kennedy Space Center (KSC) ISS Environmental Simulator to allow for data transmission and reproduction of incubation conditions experienced by flight samples in orbit. HOBO data loggers equipped with temperature sensors were integrated into two of the four BRICs to record temperatures experienced by samples during the experiment for post hoc analysis. Launch samples (i.e., Arabidopsis seeds) were integrated into BRIC flight hardware in a sterile hood ∼48 h before the August 14, 2017, launch of SpaceX CRS12 spacecraft. Integrated science/hardware was kept at 4°C to maintain seed dormancy before packing in cold storage bags while being loaded onto Dragon capsule and during launch. After docking, samples were removed from cold storage bags by ISS astronauts, warmed to ambient ISS temperature, allowing seed germination and experiment initiation in the BRIC. After 14 days, ISS astronauts actuated PDFUs, which were then incubated at room temperature for a further 3 h before being transferred to the ISS −80 MELFI freezer. Samples were kept at approximately −80°C until BRICs were conditioned to −32°C in double cold bag storage (Hutchison and Campana, 2009) and stowed in the Dragon capsule before undocking and atmospheric reentry on September 16. After returning to KSC, samples were stored at −80°C until de-integration of the flight and ground samples. De-integration occurred on November 1, 2017.
2.2. Germplasm and culture conditions
Arabidopsis thaliana seeds of the following genotypes were used for flight and ground controls: WT (Col-0 ecotype), atire1 (Nagashima et al., 2011; Chen and Brandizzi, 2012), bzip60 (Moreno et al., 2012), bzip28 (Gao et al., 2008), and bzip28 bzip60 (Deng et al., 2013). Petri dishes (60 mm) were prepared with 6.7 mL of sterile ½ Murashige and Skoog media supplemented with Gamborg's B5 Vitamins (PhytoTechnology Laboratories), 0.5% sucrose (Sigma-Aldrich), 0.4% Phytagel (Sigma-Aldrich), pH adjusted to 5.7. In a sterile hood, seeds of WT and UPR-mutant genotypes were surface sterilized with one wash of 70% ethanol, one wash of 50% bleach containing 0.5% Tween 20, and then nine additional washes with sterile H2O distilled twice. After the final wash was removed, seeds were resuspended in 1.5 mL sterile water for wet plating by using a 1 mL pipette equipped with sterile filter tip. For each experimental unit (flight and ground control), five plate replicates of WT and bzip28/bzip60 genotypes and four plate replicates of atire1, bzip28, and bzip60 genotypes were prepared. Each plate replicate contained 70–80 seeds evenly spaced in a grid pattern on the plate surface. The Petri dishes were sealed with Parafilm (Heathrow Scientific), and individual plates were then wrapped twice with sterile aluminum foil. Individually wrapped plates were grouped by BRIC configuration and wrapped together with two more layers of sterile aluminum foil before sample removal from the sterile hood. Plates were placed at 4°C until the integration of samples into science hardware the following morning.
2.3. Sample processing and experimental material assessment
Samples of flight and ground control experiments were preserved in RNAlater in situ and kept at −80°C (see the Results section for experimental timeline). The Petri dishes were removed from packaging and thawed in groups of three to prevent excess exposure to room temperature during sample collection. After removing most of the RNAlater from the plates, sterile forceps were used to transfer seedlings from the plates to microcentrifuge tubes containing two glass beads. Seedlings were then frozen in liquid nitrogen. This procedure was carried out quickly to maximize RNA recovery. Accordingly, only some pictures were taken of plates before extraction for example purposes. Most pictures were taken after the bulk of the sample was removed, with the remaining seedlings also imaged for post hoc inspection. All plates were free from any visible evidence of bacterial or fungal contamination. Frozen samples were ground to a powder with a Retsch Mixer Mill (Retsch, Haan, Germany). RNA was extracted from tissues by using a NucleoSpin RNA Plant Kit (Macherey-Nagel) according to the manufacturer's instructions including DNase digestion. The overall quality and RNA integrity number (RIN) of RNA samples were assessed with Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA).
2.4. Library preparation, sequencing, and bioinformatics analysis
RNA-sequencing (RNA-seq) libraries were constructed by using the Illumina TruSeq Stranded mRNA Library (Illumina, San Diego, CA) and sequenced in single-end mode on the Illumina HiSeq 4000 platform (50-nt) at Research Technology Support Facility Genomics Core at Michigan State University. For each library, read quality was assessed with the FastQC (version 0.11.3) software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were cleaned for quality and adapter sequences with Cutadapt (version1.8.1) using a minimum base quality 20 retaining reads with a minimum length of 30 nucleotides after trimming (Martin, 2011). Quality-filtered reads were aligned to the Col-0 reference genome (TAIR10) with Bowtie (version 2.3.1) and TopHat (version 2.1.1) with a 10 bp minimum intron length and 15,000 bp maximum intron length (Langmead and Salzberg, 2012; Kim et al., 2013). Fragments per kilobase exon model per million mapped reads (FPKM) were measured by using TAIR10 gene model annotation with Cufflinks (version 1.3.0) (Trapnell et al., 2010). The log2 transformed and normalized gene expression levels (FPKM +1) were used for correlation analysis (Spearman's rank correlation coefficient) between biological replicates and principal component analysis (PCA). Per-gene read counts were identified with HTSeq (version 0.6.1p1) in the union mode with a minimum mapping quality of 20 with stranded reverse counting (Anders et al., 2015). Differential gene expression analysis was performed in four biological replicates (for WT and bzip28 bzip60, selected based on the correlation with other biological replicates) with DESeq2 (version 1.16.1) within R (version 3.4.0) based on a comparison of spaceflight to ground with adjusted p-value <0.01 and absolute log2-transformed fold change >1.5 (Love et al., 2014). Genes of which the total count across all samples is <100 were not included in the analysis. Gene Ontology (GO) overrepresentation was performed with PANTHER (Fisher's exact type with false discovery rate correction) (www.pantherdb.org) (Mi et al., 2019a, 2019b). K-means clustering analysis on average FPKM values from biological replicates was performed using the Morpheus tool (Morpheus, https://software.broadinstitute.org/morpheus). The optimal number of K-means clusters was determined with the factoextra package in R.
3. Results
3.1. Spaceflight alters the growth of seedlings independent of an intact UPR signaling
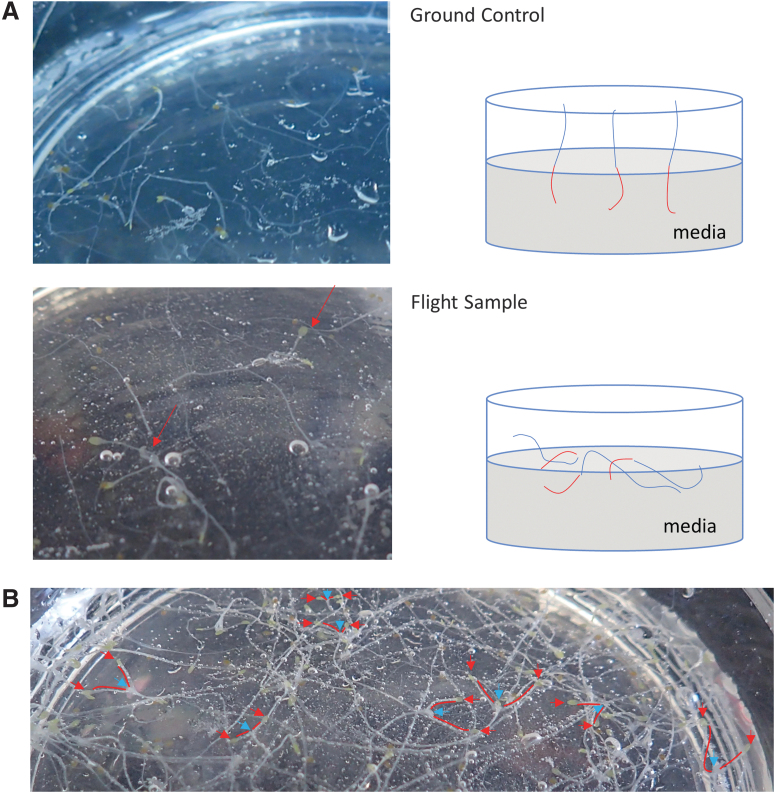
When we inspected the WT and UPR-mutant seedlings (atire1, bzip60, bzip28, and bzip28 bzip60) of the ground control and flight samples at the completion of the mission, we found that in the ground control samples, etiolated hypocotyls (i.e., pale and elongated due to the lack of light) were above the surface of the solidified media while roots had penetrated the growth medium perpendicular to the surface (Fig. 1A). However, in flight samples, we found that etiolated hypocotyls as well as roots had generally penetrated the growth medium regardless of genotype tested (Fig. 1A). Interestingly, we also observed that cotyledon petioles of flight sample seedlings were elongated (Fig. 1B) compared with ground control seedlings (Fig. 1A). Overall, these observations are consistent with plant growth in the darkness and space, conditions leading to elongated hypocotyls and petioles, and a lack of directional growth, respectively (Paul et al., 2017). These observations also suggest that the UPR unlikely exerts a noticeable role in growth direction in response to altered gravity levels.
FIG. 1.
(A) Representative ground and flight sample plates were imaged after the bulk of etiolated hypocotyls, and RNAlater was removed to ensure maximal RNA integrity. Remaining seedlings were used for post hoc analysis of morphological/growth differences. (B) Left: Example plate of a WT flight sample, which was imaged after thawing, but before seedling removal. Individual seedlings that could be distinguished from the bulk were marked with a blue arrow to mark the shoot meristem. The cotyledons of elongated petioles were marked with a red arrow and a red line used to connect the cotyledon to the meristem of the same seedling. Petioles were elongated compared with ground sample petioles [representative morphology of ground control presented in (A)]. WT, wild type.
3.2. Spaceflight results in an increase of total RNA
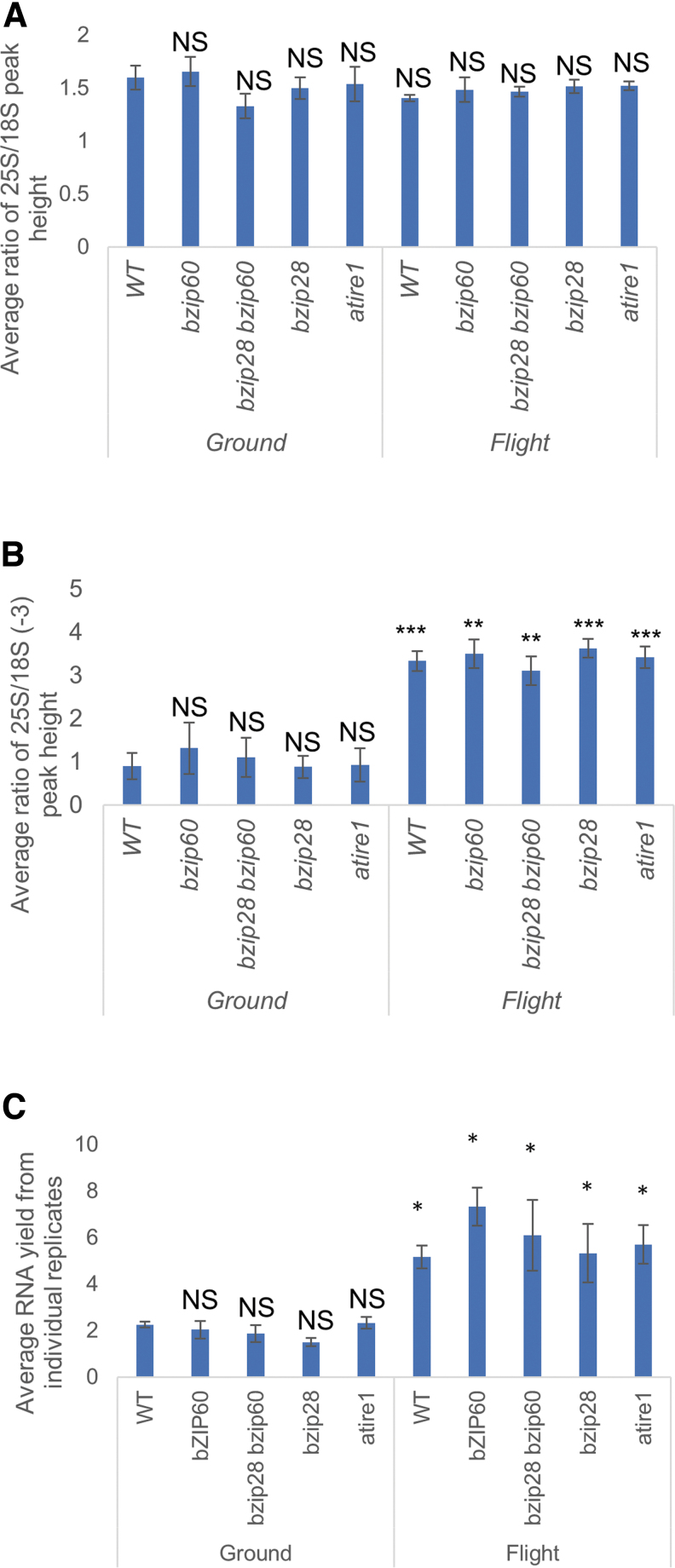
RNA degradation has previously been observed in independent Arabidopsis spaceflight experiments performed in BRIC-PDFUs (Paul et al., 2012; Johnson et al., 2017). To test the RNA quality of our samples, we measured RIN as an indicator of overall RNA quality (RIN, 1 = low quality; 10 = high quality) of each sample and compared size peaks of 25S and 18S ribosomal RNA (rRNA) among samples (Mueller et al., 2004). Note that these measurements include smaller plastid rRNA peaks, which lower the maximum RIN value to around 8, independent of RNA quality (Babu and Gassmann, 2016). We established that all flight samples had RIN values between 7.5 and 8.0, indicating that RNA was of high quality. For ground samples, RIN values were found to be between 4.0 and 7.7, which would ordinarily indicate mild degradation of some samples. However, closer analyses revealed that, in nearly all ground samples (77%), the RIN algorithm failed to identify the correct peaks. In these samples, the algorithm identified the 18S rRNA peak as the 25S rRNA peak, and a putative organelle rRNA peak (Babu and Gassmann, 2016) as the 18S rRNA (Supplementary Fig. S1). Therefore, to compare RNA quality between treatments and genotypes from the Bioanalyzer data outputs, we used the ratio of 25S/18S peak heights as a substitute measure. Because the 25S peak height is reduced more quickly than the 18S peak in RNA-degrading conditions (e.g., elevated temperature, exogenous RNases, endogenous apoptotic RNase activity) (Mueller et al., 2004; Babu and Gassmann, 2016), a decreased 25S/18S ratio would indicate RNA degradation. We observed no significant differences of the 25S/18S ratio across all genotypes (Fig. 2A); therefore, both flight and ground samples had no significant RNA degradation.
FIG. 2.
(A) Average ratio of 25S/18S peak heights as determined from Bioanalyzer traces from each sample was used as a secondary measure of RNA quality due to the RIN algorithms incorrect identification of the appropriate peaks. (B) The relative content of rRNA found in each genotype in both conditions was determined by comparing the height of the 25S peak with an organellular rRNA peak [18S (−3)], which was found in each sample. (C) Average RNA yields from each genotype from flight and ground samples. Statistical significance determined by Welch's t-test, p-value represented by not significant (NS) = >0.05; * = <0.05; ** = <0.005; *** = <0.0005. RIN, RNA integrity number.
Interestingly, by comparing the 25S peak height with one of the two other peaks near the 18S peak [i.e., a putative organellular rRNA peak, designated 18S (−3); Supplementary Fig. S1] in each sample, it was clear that the relative ratio of 25S to 18S (−3) was significantly lower in the ground samples compared with flight samples (Fig. 2B and Supplementary Fig. S1). This low ratio indicates that the ground samples were depleted of nuclear-encoded rRNAs (i.e., 25S and 18S rRNAs) compared with other RNA species. This observation is also consistent with our findings that our ground samples contained significantly less total RNA [of which rRNA is a significant fraction (Lodish, 2000)] than flight samples (Fig. 2C). Because we extracted RNA from a similar number of seedlings from flight samples and ground samples, our results indicate that flight samples contained larger amount of total RNA compared with ground samples, which is likely to be caused by elevated levels of nuclear encoded rRNA in flight samples, and unlikely to be due to RNA degradation of the ground samples.
3.3. Global transcriptomic analyses indicate that gene expression reprogramming in response to spaceflight depends partially on intact UPR signaling
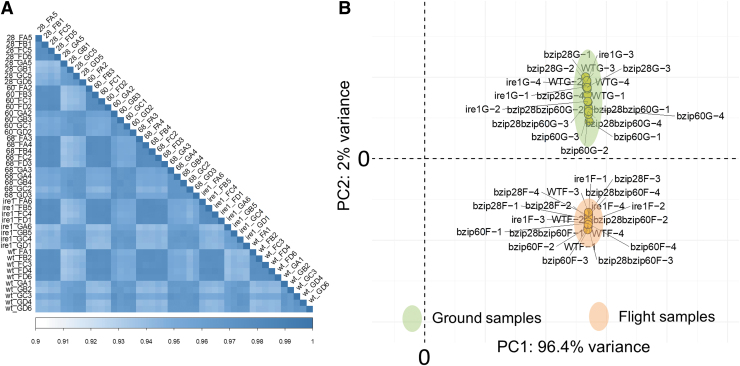
Having established that the total RNA from ground and flight samples was of acceptable quality, we next proceeded to RNA-seq to investigate the impact of spaceflight on global gene expression changes in the UPR mutants. In RNA-seq library preparation, mRNA was enriched by purification to efficiently remove rRNA (Zhao et al., 2018) and mitigate potential sequencing bias due to higher rRNA levels in flight samples. We obtained an average of ∼32 million reads per sample, of which 95–99% were successfully mapped to the Arabidopsis reference genome (Supplementary Fig. S2). Spearman's rank correlation coefficients calculated between biological replicates showed a high reproducibility of our RNA-seq data set (Fig. 3A). Furthermore, PCA exhibited a strong separation of ground samples from flight samples (Fig. 3B) and indicated that ground samples located more closely to each other than the flight samples. Overall, these analyses further supported a statistical robustness of the RNA-seq and justified further investigation.
FIG. 3.
(A) Spearman's correlation coefficients demonstrate a close relationship between biological replicates. (B) Principal component analysis demonstrates a clear separation between flight and ground samples.
To investigate gene expression changes in response to spaceflight, we identified differentially expressed genes (DEGs) in each genotype by comparing gene expression values in ground and flight samples (Supplementary Table S1). A total of 3465 genes were classified as DEGs in at least one of the tested genotypes. WT had the largest number of DEGs (upregulated DEGs in flight compared with ground, n = 1675; downregulated DEGs, n = 831) among the genotypes tested. The bzip28 bzip60 mutant had the smallest number of DEGs (upregulated DEGs, n = 1293; downregulated DEGs, n = 562) (Fig. 4A, B). In all genotypes, the number of upregulated DEGs was higher than that of downregulated DEGs (WT, 2.02-fold; atire1, 1.98-fold; bzip28, 2.28-fold; bzip60, 1.80-fold; bzip28 bzip60, 2.30-fold), indicating a higher impact of spaceflight on inducing gene expression rather than suppressing it. While the identity of 34.8% (783/2249) of upregulated DEGs and 27.5% (335/1217) of downregulated DEGs overlapped across all genotypes, relatively smaller numbers of DEGs were found to be genotype-specific, ranging from 30 (downregulated exclusively in bzip28 bzip60) to 207 (upregulated in WT).
FIG. 4.
Differential expression analysis using the four biological replicates with the highest correlation was performed via HTseq v0.6.1pl and DESeq v2. For each genotype and for genes, DEGs in flight samples relative to ground samples were determined using a strict criterion: adjusted p-value <0.01; |log2FC| >1.5. Total number of upregulated (A) and downregulated (B) DEGs in each background was analyzed to determine what proportion was shared between the different genotypes. DEG, differentially expressed gene.
In summary, based on the verified number of upregulated and downregulated DEGs in flight samples compared with ground samples across genotypes, the UPR mutants had consistently fewer overall DEGs than WT, indicating that the UPR could play at least a partial a role in regulating the transcriptional reprogramming in space compared with ground control.
3.4. Biological pathways connected to the DEGs between flight and ground
To gain insights in the biological pathways altered in spaceflight in our experimental setup, we performed separate GO analyses on upregulated and downregulated DEGs in WT (Supplementary Table S2), generating a list of parental-GO terms (more general, represented by a larger number of genes in the reference gene set) and cognate child-GO terms (more specific, smaller numbers of genes in the reference gene set) (Fig. 5). Intriguingly, we found that stress-responsive genes (e.g., “response to abscisic acid,” “response to hypoxia,” “response to water deprivation,” and “response to oxidative stress”) as well as genes involved in physiological responses often associated with stress response adaptation were enriched in downregulated DEGs in WT. This result is consistent with previous studies that reported downregulation of water stress-related genes when using Arabidopsis BRIC-PDFU microarray transcriptomes (Johnson et al., 2017) and that found abscisic acid response and water stress response overrepresentation in misregulated DEGs in Col-0 WT with RNA-seq (Choi et al., 2019). In addition, we observed that metabolic processes associated with stress adaptation (Hildebrandt Tatjana et al., 2015; Batista-Silva et al., 2019), including amino acid catabolism and sucrose starvation response, were overrepresented in flight-downregulated DEGs. Ribosome biogenesis, translation, and gene expression processes were highly underrepresented in this category, that is, they were more likely to be upregulated by flight or remain unchanged. By further analyzing the normalized gene expression values (FPKM), we also found that ribosome biogenesis and rRNA processing GO terms appeared significantly overrepresented in genes upregulated by >2-fold changes (flight/ground). These, however, were not considered as DEGs based on the strict statistical criteria applied in our analyses (see the Materials and Methods section). The lower FPKMs of ribosome biogenesis-related genes in ground control samples are consistent with our observations that the ground control samples were partially depleted of 25S and 18S rRNA compared with flight samples (Fig. 2C).
FIG. 5.
Representative biological processes gene ontologies (GO terms) over- or underrepresented by upregulated or downregulated DEGs in the WT background. GO, Gene Ontology.
We also found that GO terms enriched in upregulated DEGs included biological processes that have been noted in previous Arabidopsis spaceflight transcriptome analyses, such as secondary metabolite biosynthesis associated with defense responses (Johnson et al., 2017; Choi et al., 2019). Furthermore, we verified the dichotomous occurrence of the “absence of light” and “response to red or far red (R/FR) light response” GO terms in the downregulated and upregulated gene sets, respectively. The overrepresentation of photosynthetic components in upregulated DEGs is partially consistent with the results of a previous BRIC-PDFU experiment showing light/high light response and some photosynthesis-related genes to be differentially regulated in a subset of the tested genotypes (Choi et al., 2019). In addition to the findings consistent with previous spaceflight reports, we also observed a significant enrichment of DNA repair, DNA replication, and cell cycle pathways in the upregulated DEGs, which could be possibly associated with exposure of flight samples, but not ground samples, to ionizing radiation during spaceflight.
Together, these results indicate that in our experimental conditions, spaceflight globally affects gene expression changes associated with a broad array of significant biological processes, largely identified also in previous spaceflight transcriptome studies (Johnson et al., 2017; Choi et al., 2019).
3.5. The UPR regulators exert a minor but significant role on gene expression in spaceflight
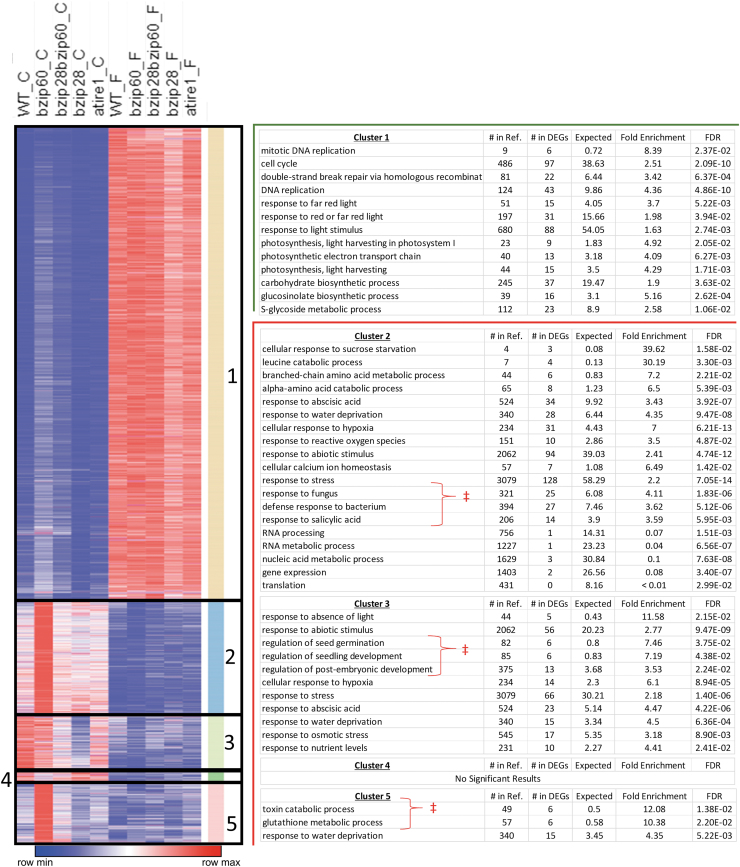
Next, we aimed to gain insights into the transcriptome changes caused by the absence of an intact UPR signaling in both ground and spaceflight conditions. To address it, we performed K-means clustering analysis on FPKM values for all DEGs (n = 3465) obtained in at least one genotype (Fig. 6 and Supplementary Tables S3 and S5) and then performed GO analysis to correlate the expression signature of each cluster with biological functions (Supplementary Table S4). Our clustering analysis suggested that variations in the identified DEGs between different genotypes (Fig. 4) were primarily the result of variable FPKM values in the ground control samples (Fig. 6). In flight, the FPKM values were roughly equalized to insignificantly different levels of expression across all genotypes. Of the 3465 DEGs, in the ground samples, 517 DEGs had FPKM values that were significantly different (p ≤ 0.01) in at least one UPR mutant compared with WT; however, in flight samples, only 144 genes had FPKM values that were significantly different in at least one UPR-mutant genotype compared with WT (Fig. 6 and Supplementary Fig. S3).
FIG. 6.
K-means clustering analysis of all 3465 DEGs in at least 1 background. Each row represents fragments per kilobase exon model per million mapped reads values of an individual gene averaged between biological replicates. For each row, blue represents the minimum relative expression value, red the maximum expression value, and white is the middle value. For each cluster, GO biological process analysis of DEGs demonstrates a role for UPR regulation of ground control stress responses, which are largely repressed by flight. UPR, unfolded protein response.
The largest cluster (Cluster 1 DEGs; n = 2299) consisted of DEGs upregulated to varying degrees in most of the genotypes tested. As such, a nearly identical set of GO terms that were enriched in upregulated DEGs in WT (Fig. 5) were also enriched in Cluster 1. We then compared the FPKM values of DEGs in ground samples across genotypes and found that 39.1% and 19.8% of DEGs in Cluster 1 were significantly different in the bzip60 and bzip28 bzip60, respectively, compared with WT, whereas only 4.6% and 2.9% of DEGs were significantly different in bzip28 and atire1, respectively, compared with WT (Supplementary Fig. S4). Overall, these results indicate that bZIP60 has functions that are independent of IRE1 and bZIP28, which in turn are required to downregulate Cluster 1 DEGs in ground control conditions. These observations are in accordance with the findings in ground conditions that bZIP28 and bZIP60 control some UPR target genes in an independent manner (Ruberti et al., 2018).
In Cluster 2 (DEGs; n = 546), expression of DEGs was induced exclusively in ground bzip60 compared with other ground genotypes while highly suppressed to similar levels of expression in flight samples of all genotypes. As such, the proportion of DEGs in bzip60, whose expression was significantly different from WT, was much higher (17%) than other genotypes (bzip28 bzip60, 0.9%; bzip28, 1.0%; atire1, 1.2%) (Supplementary Fig. S4). We reasoned that bZIP28 and bZIP60 could have a negative feedback relationship in which the absence of bZIP28 suppressed the effect of bzip60 mutation exclusively in the ground condition. The overrepresented GO terms in Cluster 2 were largely similar to the GO terms enriched in the downregulated DEGs genes in the WT genotype: “response to abscisic acid,” “response to water deprivation,” and “response to hypoxia” (Fig. 5). In addition, Cluster 2 DEGs showed significant enrichment of GO terms associated with biotic stress responses that were not found in analyses of the WT genotype (Fig. 5) or found to be strongly enriched in the any of the other clusters (Fig. 6 and Supplementary Table S4). These results indicate that bZIP60 may have repressive roles in regulating genes involved in both abiotic and biotic stress responses.
The gene expression pattern of Cluster 3 (DEGs; n = 274) was also characterized largely by genes with lower expression values in spaceflight compared with ground control across genotypes, except for bzip28. However, contrasting with Cluster 2, the FPKM values in the bzip60 genotype was not different from WT FPKM values; only 3.2% of DEGs showed significant different FPKM compared with WT (Supplementary Fig. S4). Instead, the absence of bZIP28 (i.e., in the bzip28 mutant) had a higher impact on gene expression in the ground condition compared with other mutants (Supplementary Fig. S4). Interestingly, these Cluster 3 DEGs had intermediate FPKM values in the bzip28 bzip60 genotype in ground control compared with the extremes of the bzip60 and bzip28 single mutants, indicating an antagonistic regulation of bZIP28 and bZIP60 on these genes in the ground condition, which was largely compensated for in the bzip28 bzip60 genotype.
Similar to Clusters 2 and 3, Cluster 4 (DEGs; n = 55) contained genes, whose expression exhibited overall lower FPKMs in spaceflight compared with ground across genotypes. However, Cluster 4 showed a unique pattern: the expression of the DEGs in this cluster was significantly lower in bzip60 compared with WT and the other genotypes in the ground condition (Supplementary Fig. S4) and showed no prominent differences across genotypes in the spaceflight condition. Although Cluster 4 was not significantly represented by any biological process GO terms, a closer analysis revealed that 22% of all DEGs in this cluster were encoded on the mitochondrial genome; mitochondria-encoded genes comprise only 0.5% of all protein-coding genes in the Arabidopsis genome annotation (https://www.arabidopsis.org) and 0.3% of DEGs called in this study.
Cluster 5 (DEGs; n = 291) showed a gene expression pattern similar to Cluster 2 with significantly higher levels in the bzip60 genotype compared with the other genotypes in the ground control samples (Supplementary Fig. S4). In this Cluster, the DEGs were more affected by the absence of bZIP28 (bzip28 bzip60 and bzip28) in the ground condition compared with Cluster 2. The GO term “response to water deprivation,” which was found to be enriched in Cluster 2, was significantly enriched in Cluster 5. In addition, relatively narrow child GO terms “toxin catabolic process” and “glutathione metabolic process,” which were not enriched in other clusters, were enriched in Cluster 5.
Overall, by comparing the bzip60 and bzip28 single mutants with the bzip28 bzip60 double mutant, our transcriptomic profiling provides evidence for a highly complex, unconventional, regulatory relationship between bZIP60 and bZIP28 under the conditions experienced by ground control seedlings. Furthermore, a small number of significant differences between WT and the UPR mutants in spaceflight were found, supporting a small but significant role of the UPR in gene expression in spaceflight.
4. Discussion
In this study, we examined the transcriptional responses of WT and seedlings with a compromised UPR to spaceflight to set the foundations to manipulate a critical growth and stress signaling pathway for improving plant adaptation to extraterrestrial environments. We utilized the BRIC-PDFU sterile plant culture hardware during the SpaceX CRS12 mission to compare the transcriptional responses with the spaceflight between A. thaliana WT and mutants defective in one or more components of the UPR, namely the TFs bZIP60, bZIP28, and the ER-resident kinase/ribonuclease IRE1.
The BRIC-PDFU hardware has been employed in a number of dark-grown Arabidopsis transcriptome experiments toward different aims (Paul et al., 2012; Kwon et al., 2015; Johnson et al., 2017; Choi et al., 2019). Variability in technical experimental details and limited overlap between spaceflight/ground DEGs has been verified even between the same WT control genotype in simultaneous experiments (Johnson et al., 2017). However, some broad biological pathways have been found to be induced or repressed in response to spaceflight, including cell wall modification, response to light/high light, oxidative stress, osmotic stress response, heat shock, and biotic defense/secondary metabolite synthesis (Paul et al., 2012; Johnson et al., 2017; Choi et al., 2019). Many of these responses were also noted in our study, including the downregulation of water stress response in space, which has been identified in four separate experiments (Johnson et al., 2017; Choi et al., 2019). However, we also observed correlative differences in seedling growth and an overall gene expression landscape not noted in previous studies. Our ground control seedlings had grown in a predictable manner, consistent with the expected morphology of terrestrially grown Arabidopsis etiolated hypocotyls (Fig. 1). Flight sample growth was also largely in line with expectations for seedlings grown in microgravity, including the observed petiole elongation, which was shared in all flight samples. Indeed, a petiole elongation of flight samples was also present in the images published of dark-grown Arabidopsis experiments in the Col-0 background in previous experiments (Johnson et al., 2015; Paul et al., 2017).
This growth phenotype is consistent with low R/FR ratio and shade avoidance syndrome mediated by phytochrome signaling (Franklin, 2008). The correlative responses observed in the transcriptome analysis (Fig. 4) support that differentially regulated growth phenotypes and the large transcriptional rearrangements in flight during our experiment might have been mediated by phytochrome-related signaling, which is also known to constitutively repress abscisic acid signaling (Yang et al., 2016), and were found to be repressed in our flight samples. However, these differences in the response to light and the increased expression of photosynthetic components between ground and flight samples are anomalous when considering the spaceflight culture methods used in this study. BRIC-PDFUs are autoclavable, black polymer containers, which are sealed with metal lids during science integration, and allow injection of the chemical preservative without opening the unit. After being sealed on August 12, the seeds and seedlings germinated from these seeds in the BRICs were not exposed to light during the launch and the 14-day growth period on the ISS. Therefore, the dichotomous occurrence of the “absence of light” overrepresentation in downregulated and of “response to R/FR light response” in upregulated DEGs is unlikely to be the result of actual differential exposure to light.
One likely explanation is related to the hypocotyl-tissue media contact that occurred in our flight samples, which lacked a clear growth vector in microgravity, but had not occurred in our ground samples where roots grew perpendicular into the media. In previous experiments by Johnson et al. (2015) and Paul et al. (2017), the dark-grown ground control plates were oriented vertically and both sample sets displayed petiole elongation, although the precise differences in length or extent of petiole elongation between flight and ground were not quantified. How media contact could induce R/FR light signaling in the dark is not immediately obvious; however, earlier studies showed that media containing sucrose modulated the R/FR signaling mediated by phytochrome A, promoting a red light response (Dijkwel et al., 1997). Coincidentally, the higher rRNA and RNA content observed in flight samples compared with ground control samples in our study (Fig. 2) is also consistent with an increased exposure of cells to sucrose or glucose, which is known to induce RNA accumulation, rRNA transcription, and ribosome biogenesis in plants (Kojima et al., 2007; Ishida et al., 2016), yeast (Kunkel et al., 2019), and mammals (Hannan et al., 2003). Nonetheless, we cannot rule out the remote possibility that our observations may be influenced by possible interactions between ionizing radiation and phytochrome R/FR signaling. For example, low-dose gamma (γ) irradiation of lettuce seeds was found to mimic the effects of FR deactivation of red light-activated phytochromes (Hsiao and Vidaver, 1974). Additionally, a structural study of the bacterial phytochrome from the radiation-resistant bacteria Deinococcus radiodurans established that X-ray radiation induced deprotonation of chromophore in the inactive phytochrome, a biochemical step thought to be involved in light-induced activation of this protein (Li et al., 2015). Although the dose required to deprotonate 50% of the phytochrome (Li et al., 2015) was orders of magnitude larger than that expected to be experienced during our experimental period on the ISS, differences between prokaryotic and eukaryotic phytochromes could affect relevant properties of a hypothetical phytochrome–radiation interaction.
Interpreting a role for the UPR in the transcriptional response to spaceflight is complicated. We observed clear differences in the number of spaceflight DEGs in the UPR-mutant backgrounds compared with WT (Fig. 4); and only 1118 of the 3465 DEGs were common to all genotypes. This would normally suggest that the transcriptional readjustments that occurred in response to spaceflight were at least partially the result of UPR-dependent processes. However, upon closer analysis of the underlying FPKM values, it became clear that the differences in fold change values (flight/ground) across genotypes were more heavily influenced by the differential regulation of expression in the ground samples by the UPR regulators (Fig. 6). The number of DEGs with FPKM values significantly different from WT FPKM values in at least one UPR-mutant genotype was four times greater in the ground samples compared with flight samples (Supplementary Fig. S3). The heat map visualization of these values (Fig. 6) further suggests that the variations in ground samples expression levels were largely muted by spaceflight, as the endpoint transcript levels in flight samples were nearly uniform in the different genotypes. Overall, this would suggest that the UPR does not have a broad involvement in the response to spaceflight. One explanation for this observation may be related to the concerted downregulation of many stress-responsive processes in the flight samples compared with the ground samples (Fig. 6). In spaceflight conditions, it seems likely that alternative signaling pathways are actuated, which repress the observed stress responses regulated by the UPR. Given the prevalence of starvation responses in ground samples, it is possible that microgravity-induced changes in growth habit provide better nutrient availability (Fig. 7). As such, plants in flight may be able to better handle the stresses imposed by culture conditions, without requiring UPR regulator involvement.
FIG. 7.
(A) Hypothesized regulatory framework for stress-responsive DEGs found to be regulated by the UPR on the ground but not in flight conditions. Possible interactions between bZIP60 and bZIP28 in the regulation of DEGs were found in (B) Clusters 2 and 5, (C) Cluster 3, and (D) Cluster 4.
Nonetheless, the observations related to an interaction between the UPR regulators bZIP60 and bZIP28 and the stresses imposed on ground control seedlings have yielded important information, which should be explored in the future. In the canonical ER stress response induced chemically or via environmental stress, bZIP60 and bZIP28 TFs interact in the nucleus and direct the actions of the COMPASS DNA methylation complex to increase transcription of target genes (Song et al., 2015). Furthermore, these TFs can also bind independently to gene promoters to activate downstream UPR genes, as evidenced by the weaker activation of ER chaperones in the bzip28 bzip60 double mutant compared with either of the bzip28 or bzip60 single mutants (Ruberti et al., 2018; this work). Although it has been suggested that bZIP60 and bZIP28 may also have unique target genes (Pastor-Cantizano et al., 2019), in our ground control samples, the transcriptomic data suggest that bZIP60 and bZIP28 may have a more complex antagonistic relationship in the control of genes related to the response to abscisic acid, hypoxia, water deprivation, and to oxidative stress (Fig. 6). In Clusters 2 and 5, and Cluster 4, we observed that the expression levels of the DEGs were higher or lower than WT in the bzip60 genotype, respectively (Fig. 6 and Supplementary Fig. S3). However, these differences were not observed in the bzip28 bzip60 double mutant as would be expected. Conversely, in Cluster 3, we observed in ground samples that the expression levels of the DEGs were sharply lower in the bzip28 genotype compared with WT. In the bzip28 bzip60 genotype, the genes in Cluster 3 had expression levels that were higher than the bzip28 genotype but were also lower than those of these genes in the WT or bzip60 genotypes. In response to chemically induced UPR conditions, bZIP60 and bZIP28 cooperatively upregulate several UPR genes (Song et al., 2015; Ruberti et al., 2018). In our ground controls, the stress-responsive genes, largely represented by “response to abscisic acid,” “response to water deprivation,” and “response to hypoxia” abiotic stress responses, were regulated by bZIP28 and bZIP60 in a way that suggests that these TFs have antagonistic regulatory effects on these processes (Fig. 7B–D).
The exact nature of the stress experienced by the ground control seedlings would need to be elucidated to better understand the impact of this information on agronomic and/or spaceflight applications. Studies on the effect of plant growth in BRIC-PDFUs have already established that significant stress may be imposed on the seedlings grown in these conditions (Johnson et al., 2015; Basu et al., 2017). Consistent with our results, other BRIC-PDFU transcriptomes have found a downregulation of genes related to water stress responses (Johnson et al., 2017). However, as evaporative water loss from the BRIC-PDFUs is unlikely because they are sealed containers, the strongly represented GO terms related to “response to water deprivation” and “response to abscisic acid” are unlikely to be a direct response to actual water loss from the plates. Instead, it may be possible that the ground seedlings with etiolated hypocotyls that are not in contact with the media or have less overall contact with the media are water stressed compared with flight seedlings that are in direct contact or have penetrated the media. Independent of the underlying stimulus for the observed differences, the possibility that bZIP60 and bZIP28 may have antagonistic interactions related to control of abscisic acid signaling or water deprivation responses should be investigated at the molecular level in the future to improve plant growth and stress responses in space and on the ground.
Data Accessibility
Raw sequencing data have been deposited at the NCBI Gene Expression Omnibus under the accession number GSE148914. Raw sequencing data have also been deposited at the NASA GeneLab data repository (Ray et al., 2018) for spaceflight experiments under the accession number GLDS-321.
Supplementary Material
Abbreviations Used
- ATF6
activating transcription factor 6
- BRIC
Biological Research In a Canister
- bZIP17
basic leucine zipper 17
- bZIP28
basic leucine zipper 28
- bZIP60
basic leucine zipper 60
- DEG
differentially expressed gene
- ER
endoplasmic reticulum
- FPKM
fragments per kilobase exon model per million mapped reads
- GO
Gene Ontology
- IRE1
inositol-requiring enzyme 1
- ISS
International Space Station
- KSC
Kennedy Space Center
- PCA
principal component analysis
- PDFU
Petri Dish Fixation Unit
- R/FR
red/far red
- RIN
RNA integrity number
- RNA-seq
RNA-sequencing
- rRNA
ribosomal RNA
- TF
transcription factor
- UPR
unfolded protein response
- WT
wild type
Authors' Contributions
F.B. designed the experiments. S.Z.-D. and E.A. performed preliminary launch preparations. E.A. executed the experiment. D.K.K. performed bioinformatics analysis. E.A., D.K.K., S.Z.-D, and F.B. analyzed data and wrote the article.
Acknowledgments
We would like to thank the NASA, KSC, and NASA contractor support staff whose diligent work made these experiments possible.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported primarily by NASA NNX12AN71G with contributing support from Training Program in Plant Biotechnology for Health and Sustainability (T32-GM110523), the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494 and DE-SC0018409), the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (award number DE-FG02-91ER20021), National Institutes of Health (GM101038), and AgBioResearch (MICL02598) to F.B.
Supplementary Material
References
- Anders S, Pyl PT, and Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu CVS and Gassmann M (2016) Assessing integrity of plant RNA with the Agilent 2100 bioanalyzer system. Agil Appl Note Waldbronn 5990–8850EN [Google Scholar]
- Basu P, Kruse CPS, Luesse DR, et al. (2017) Growth in spaceflight hardware results in alterations to the transcriptome and proteome. Life Sci Space Res 15:88–96 [DOI] [PubMed] [Google Scholar]
- Batista-Silva W, Heinemann B, Rugen N, et al. (2019) The role of amino acid metabolism during abiotic stress release. Plant Cell Environ 42:1630–1644 [DOI] [PubMed] [Google Scholar]
- Beaugelin I, Chevalier A, d'Alessandro S, et al. (2020) Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. Plant J 102:1266–1280 [DOI] [PubMed] [Google Scholar]
- Chen Y and Brandizzi F (2012) AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J 69:266–277 [DOI] [PubMed] [Google Scholar]
- Choi WG, Barker RJ, Kim SH, et al. (2019) Variation in the transcriptome of different ecotypes of Arabidopsis thaliana reveals signatures of oxidative stress in plant responses to spaceflight. Am J Bot 106:123–136 [DOI] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu J-X, et al. (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci U S A 108:7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, and Howell SH (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci U S A 110:19633–19638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek PJ, et al. (1997) Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9:583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl R, Wheeler R, Levine HG, et al. (2002) Plants in space. Curr Opin Plant Biol 5:258–263 [DOI] [PubMed] [Google Scholar]
- Franklin KA (2008) Shade avoidance. New Phytologist 179:930–944 [DOI] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, et al. (2008) A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105:16398–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette T, Calmes B, and Simoneau P (2014) Impact of the UPR on the virulence of the plant fungal pathogen A. brassicicola. Virulence 5:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib K, Pesek K, Covino R, et al. (2017) Activation of the unfolded protein response by lipid bilayer stress. Mol Cell 67:673–684 [DOI] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, et al. (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23:8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt Tatjana M, Nunes Nesi A, Araújo Wagner L, et al. (2015) Amino acid catabolism in plants. Mol Plant 8:1563–1579 [DOI] [PubMed] [Google Scholar]
- Hsiao AI and Vidaver W (1974) Phytochrome-mediated germination responses in γ-irradiated lettuce seeds. Plant Physiol 54:72–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison S and Campana S (2009) ISS Payloads Office Cold Stowage Overview. NASA. https://www.nasa.gov/pdf/360392main_P03_0915_JSC.pdf
- Ishida T, Maekawa S, and Yanagisawa S (2016) The pre-rRNA processing complex in Arabidopsis includes two WD40-domain-containing proteins encoded by glucose-inducible genes and plant-specific proteins. Mol Plant 9:312–315 [DOI] [PubMed] [Google Scholar]
- Johnson CM, Subramanian A, Edelmann RE, et al. (2015) Morphometric analyses of petioles of seedlings grown in a spaceflight experiment. J Plant Res 128:1007–1016 [DOI] [PubMed] [Google Scholar]
- Johnson CM, Subramanian A, Pattathil S, et al. (2017) Comparative transcriptomics indicate changes in cell wall organization and stress response in seedlings during spaceflight. Am J Bot 104:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Yamaguchi-Shinozaki K, and Shinozaki K (2018) ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol 176:2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, et al. (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127:949–962 [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, et al. (2007) Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J 49:1053–1063 [DOI] [PubMed] [Google Scholar]
- Kunkel J, Luo X, and Capaldi AP (2019) Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat Commun 10:3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Sparks JA, Nakashima J, et al. (2015) Transcriptional response of Arabidopsis seedlings during spaceflight reveals peroxidase and cell wall remodeling genes associated with root hair development. Am J Bot 102:21–35 [DOI] [PubMed] [Google Scholar]
- Langmead B and Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Burgie ES, Yu T, et al. (2015) X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome. J Am Chem Soc 137:2792–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF (2000) Molecular Cell Biology. W.H. Freeman, New York [Google Scholar]
- Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12 [Google Scholar]
- Massa GD, Wheeler RM, Morrow RC, et al. (2016) Growth chambers on the International Space Station for large plants. Acta Hortic 1134:21–222 [Google Scholar]
- Mi H, Muruganujan A, Ebert D, et al. (2019a) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, et al. (2019b) Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v. 14.0). Nat Protoc 14:703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K-I, Iwata Y, Mochizuki T, et al. (2019) Unfolded protein-independent IRE1 activation contributes to multifaceted developmental processes in Arabidopsis. Life Sci Alliance 2:e201900459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7:e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller O, Lightfoot S, and Schroeder A (2004) RNA integrity number (RIN)–standardization of RNA quality control. Agil Appl Note Publ 1:1–8 [Google Scholar]
- Nagashima Y, Mishiba K-I, Suzuki E, et al. (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Ko DK, Angelos E, et al. (2019) Functional diversification of ER stress responses in Arabidopsis. Trends Biochem Sci 45:123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A-L, Zupanska AK, Ostrow DT, et al. (2012) Spaceflight transcriptomes: unique responses to a novel environment. Astrobiology 12:40–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A-L, Wheeler RM, Levine HG, et al. (2013) Fundamental plant biology enabled by the space shuttle. Am J Bot 100:226–234 [DOI] [PubMed] [Google Scholar]
- Paul A-L, Sng NJ, Zupanska AK, et al. (2017) Genetic dissection of the Arabidopsis spaceflight transcriptome: are some responses dispensable for the physiological adaptation of plants to spaceflight? PLoS One 12:e0180186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Ruberti C, Angelos ER, et al. (2019) AtIRE1C, an unconventional isoform of the UPR master regulator AtIRE1, is functionally associated with AtIRE1B in Arabidopsis gametogenesis. Plant Direct 3:e00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Gebre S, Fogle H, et al. 2018. GeneLab: omics database for spaceflight experiments. Bioinformatics 35:1753–1759 [DOI] [PubMed] [Google Scholar]
- Ron D and Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8:519–529 [DOI] [PubMed] [Google Scholar]
- Ruberti C, Lai Y, and Brandizzi F (2018) Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP 28 and bZIP 60, as well as an antagonizing function of BAX-inhibitor 1 upon the pro-adaptive signaling mediated by bZIP 28. Plant J 93:155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z-T, Sun L, Lu S-J, et al. (2015) Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci U S A 112:2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AB, Roberts LS, Chandra V, et al. (2018) The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev Cell 46:327–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P and Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081. [DOI] [PubMed] [Google Scholar]
- Wells B, McCray RH, Best MD, et al. (2001) A flight-rated petri dish apparatus providing two stage fluid injection for aseptic biological investigations in space. SAE Technical Paper, SAE International [Google Scholar]
- Yang D, Seaton DD, Krahmer J, et al. (2016) Photoreceptor effects on plant biomass, resource allocation, and metabolic state. Proc Natl Acad Sci U S A 113:7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen H, Brandizzi F, et al. (2015) The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet 11:e1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-S, Yang H, Ding L, et al. (2017) Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 29:1007–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhang Y, Gamini R, et al. (2018) Evaluation of two main RNA-seq approaches for gene quantification in clinical RNA sequencing: polyA+ selection versus rRNA depletion. Sci Rep 8:4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Sng NJ, LeFrois CE, et al. (2019) Epigenomics in an extraterrestrial environment: organ-specific alteration of DNA methylation and gene expression elicited by spaceflight in Arabidopsis thaliana. BMC Genomics 20:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data have been deposited at the NCBI Gene Expression Omnibus under the accession number GSE148914. Raw sequencing data have also been deposited at the NASA GeneLab data repository (Ray et al., 2018) for spaceflight experiments under the accession number GLDS-321.