Abstract
The objective of this study was to conduct a meta-analysis to systematically summarize and investigate the association of miRNA-124 rs531564, miRNA-218 rs11134527, miRNA-146a rs2910164, miRNA-196a2 rs11614913, and miRNA-499 rs3746444 polymorphisms with cervical cancer. A systematic review was performed to identify relevant studies using Embase and PubMed databases. A chi-square-based Q-test combined with the inconsistency index (I2) was used to check the heterogeneity between studies. A total of six case-control studies on rs2910164 and rs11614913, 4 studies on rs3746444 and rs11134527, and three studies on rs531564 were included. No evidence of association was found between miR-146a rs2910164, miR-196a2 rs11614913, miRNA-499 rs3746444, and miR-218 rs11134527 polymorphisms and cervical cancer risk in all the genetic models. The miR-124 rs531564 polymorphism was associated with a statistically increased risk of cervical cancer in a homozygote model (CC vs. GG: OR = 2.87, 95% CI: 1.40-5.91, PH = 0.887), dominant model (GC/CC vs. GG: OR = 1.38, 95% CI: 1.07-1.80, PH = 0.409), and recessive model (CC vs. GC/GG: OR = 2.26, 95% CI: 1.58-3.23, PH = 0.979). However, this finding should be interpreted with caution for limited samples and heterogeneity. Large-scale and well-designed studies are needed to validate our result.
1. Introduction
Cervical cancer (CC) is the fourth most common cancer in females, with 570,000 new cases and 311,000 deaths estimated for 2018 worldwide [1]. Cervical cancer is a multistep process involving the transformation of the normal cervical epithelium to cervical intraepithelial neoplasia that is subsequently transformed to cervical cancer [2]. Due to cervical cytology screening, cervical cancer can be detected at an early stage, greatly reducing the incidence and mortality. However, cervical cancer is still one of the deadliest female-specific cancers due to its tendency to metastasize and recur after treatment. It is well established that the persistence of Human Papillomavirus (HPV) infection is the main cause of cervical cancer and is indeed deemed as a necessary cause for the disease. However, evidence suggests that genetic risk factors also play a crucial role in the pathogenesis of cervical cancer [3–5].
MicroRNAs (miRNAs) are short nonprotein-coding small RNAs of approximately 23 nucleotides that mediate posttranscriptional regulation through base pairing to partially complementary sites of the target mRNA and promote translational repression or messenger RNA degradation [6, 7]. Increasing evidences show that single nucleotide polymorphisms (SNPs) or variation in miRNA sequence could potentially alter various biological processes by influencing target selection of miRNAs and is closely associated with various human diseases [8]. The relationship between genetic mutations in microRNA and cancer susceptibility has been getting increasing attention. Several recent studies have indicated that miRNA-related SNPs can remarkably alter the biogenesis and function of the corresponding miRNAs and may function as oncogenic or antioncogenic molecules in human cancers [9, 10].
Recently, though a number of studies have been conducted to investigate the association of miR-124 G>C (rs531564), miR-218 A>G (rs11134527), miR-146a G>C (rs2910164), miR-196a2 T>C (rs11614913), and miR-499 A>G (rs3746444) polymorphisms with cervical cancer [2, 11–20], the results were inconclusive. Therefore, a meta-analysis was performed to evaluate the association between these five SNPs and the risk of cervical cancer.
2. Methods and Materials
2.1. Publication Search
The PubMed and Embase databases were searched by two independent investigators covering all papers published up to December 27, 2020. The following terms were used: “cervical or CIN”, “microRNA or miRNA or microRNAs”, and “polymorphism or polymorphisms”. The references of the retrieved articles and review articles were also searched manually for further relevant studies. All eligible studies met the following criteria: (1) full-text study, (2) case-control design, (3) analysis of the relationship between miRNA polymorphisms and the risk of cervical cancer, (4) studies focusing on human beings, and (5) sufficient data to calculate risk estimates: the odds ratio (OR) with 95% confidence interval (CI) and a P value. The selection of the study was completed independently by two investigators (Liu and Dong) according to the inclusion criteria.
2.2. Data Extraction
Each publication was sought for the following information: the first author's surname, the publication year, country of origin, ethnicity, genotyping method, the number of cases and controls, and Hardy-Weinberg equilibrium (HWE). According to the amount of the sample, we defined sample size as the large sample size (≥200) and small sample size (<200).
Two reviewers (Liu and Dong) extracted the eligible research data repeatedly. Disagreement between two reviewers was discussed with another reviewer (Wang) until a consensus is reached.
When Hardy-Weinberg equilibrium (HWE) in the control was not reported, the χ2 test was used to evaluate the HWE of polymorphism in the control group. If P < 0.05, it is considered deviated from HWE.
2.3. Methodological Quality Assessment
The quality of the selected studies was assessed according to the methodological quality assessment scale which was mentioned in prior meta-analysis [22] (see Table S1).
In this scale, five items were evaluated, including (1) the representativeness of the subjects, (2) the ascertainment of cervical cancer, (3) the control of the genotyping methods, (4) HWE in controls, and (5) assessment of association. The quality score ranged from 0 to 9, and a high score indicated good quality of the study. Studies with a score less than 5 were removed from the subsequent analyses.
2.4. Statistics
We conducted our meta-analysis based on the PRISMA checklists and followed the guideline [23]. The strength of association was assessed between miRNA polymorphisms and the risk of cervical cancer by crude OR corresponding to 95% CI [24]. Pooled ORs were performed for allelic comparison (miR-146a: G versus C, miR-196a: T versus C, miR-499: A versus G, miR-218: A versus G, and miR-124: G versus C), heterozygote model (miR-146a: GC versus GG, miR-196a: TC versus TT, miR-499: AG versus AA, miR-218: AG versus AA, and miR-124: GC versus GG), homozygote model (miR-146a: CC versus GG, miR-196a: CC versus TT, miR-499: GG versus AA, miR-218: GG versus AA, and miR-124: CC versus GG), dominant model (miR-146a: CC+GC versus GG, miR-196a: TC+CC versus TT, miR-499: AG+GG versus AA, miR-218: AG+GG versus AA, and miR-124: GC+CC versus GG), and recessive model (miR-146a: CC versus CG+GG, miR-196a: CC versus TC+TT, miR-499: GG versus AG+AA, miR-218: GG versus AG+AA, and miR-124: CC versus GC+GG), respectively.
A chi-square-based Q-test combined with the inconsistency index (I2) was used to check the heterogeneity between studies [25]. The random-effects model was used when heterogeneity tests yielded significant results (P < 0.1 or I2 ≥ 50%) [26, 27]. Besides, subgroup analyses were stratified by sample size and HWE. Publication bias was assessed by visual inspection of funnel plots and using Egger's test. Sensitivity analyses were performed to identify individual studies' effect on pooled results and test the reliability of results. P value of Egger's test <0.05 was considered representative of statistically significant publication bias. All statistical analyses were carried out using the STATA 12.0 software (Stata Corp., College Station, TX, USA).
3. Results
3.1. Study Characteristics
A total of 247 studies were acquired from Embase and PubMed databases (Embase: 98 and PubMed: 149). The detailed screening process is shown in Figure 1. After reading the titles and the abstracts, 235 articles were excluded, of which 61 were duplicate ones, 149 had no relation to this topic, 22 were reviews or meta-analyses, and 3 were conference abstracts. Finally, 12 eligible case-control studies were included in our meta-analysis (Table 1) [10–21].
Figure 1.
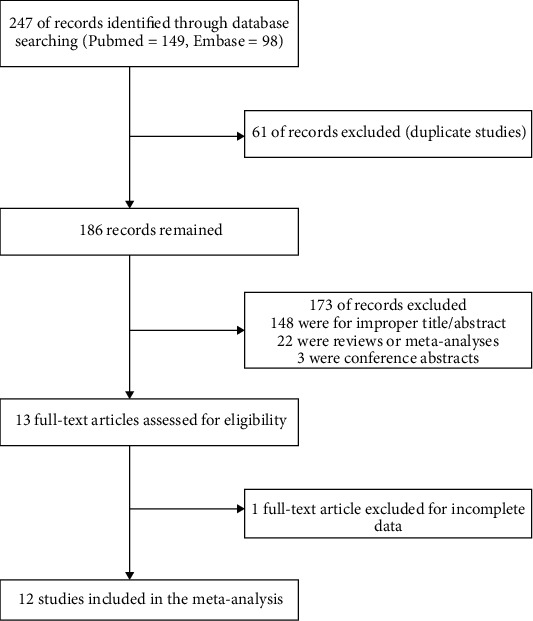
Flow chart of study selection.
Table 1.
Characteristics of studies included in the meta-analysis.
| Study ID | Year | Country | Ethnicity | Genotyping methods | Sample size (case/control) | Case | Control | P for HWE | Quality | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-146a rs2910164 G>C | CC | CG | GG | CC | CG | GG | |||||||
| Bin Zhou | 2011 | China | Asian | PCR-RFLP | 266/309 | 70 | 113 | 43 | 116 | 159 | 34 | 0.060 | 9 |
| Shizhi Wang | 2019 | China | Asian | TaqMan | 954/1339 | 318 | 475 | 141 | 471 | 631 | 212 | 0.978 | 9 |
| Nisha Thakur | 2019 | India | Asian | PCR-RFLP | 150/150 | 21 | 49 | 80 | 28 | 49 | 73 | 0.001 | 8 |
| Cong Yue | 2011 | China | Asian | PCR-RFLP | 447/443 | 105 | 224 | 118 | 150 | 206 | 87 | 0.285 | 9 |
| Shruti SrivaStava | 2017 | India | Asian | PCR-RFLP | 184/164 | 18 | 85 | 81 | 8 | 72 | 84 | 0.130 | 9 |
| Guange Chen | 2020 | China | Asian | TaqMan | 290/445 | 118 | 123 | 49 | 152 | 209 | 80 | 0.580 | 9 |
| miR-196a2 rs11614913 T>C | TT | CT | CC | TT | CT | CC | |||||||
| Bin Zhou | 2011 | China | Asian | PCR-RFLP | 266/309 | 57 | 123 | 46 | 82 | 169 | 58 | 0.077 | 9 |
| Shizhi Wang | 2019 | China | Asian | TaqMan | 954/1339 | 271 | 464 | 194 | 424 | 629 | 269 | 0.201 | 9 |
| Zhiling Yan | 2019 | China | Asian | TaqMan | 547/567 | 117 | 277 | 153 | 153 | 282 | 132 | 0.926 | 9 |
| Nisha Thakur | 2019 | India | Asian | PCR-RFLP | 150/150 | 17 | 58 | 75 | 57 | 51 | 42 | 0.0001 | 8 |
| Shruti SrivaStava | 2017 | India | Asian | PCR-RFLP | 184/164 | 71 | 93 | 20 | 62 | 81 | 21 | 0.492 | 9 |
| Guange Chen | 2020 | China | Asian | TaqMan | 290/445 | 105 | 125 | 58 | 140 | 220 | 80 | 0.691 | 9 |
| miR-499 rs3746444 A>G | AA | AG | GG | AA | AG | GG | |||||||
| Bin Zhou | 2011 | China | Asian | PCR-RFLP | 266/309 | 134 | 84 | 8 | 223 | 71 | 15 | 0.005 | 8 |
| Shizhi Wang | 2019 | China | Asian | TaqMan | 954/1339 | 675 | 228 | 27 | 946 | 339 | 35 | 0.485 | 9 |
| Shruti SrivaStava | 2017 | India | Asian | PCR-RFLP | 184/164 | 26 | 78 | 80 | 54 | 76 | 34 | 0.449 | 9 |
| Nisha Thakur | 2019 | India | Asian | PCR-RFLP | 150/150 | 25 | 47 | 78 | 21 | 49 | 80 | 0.005 | 8 |
| miR-218 rs11134527 A>G | AA | AG | GG | AA | AG | GG | |||||||
| Ting-Yan Shi | 2013 | China | Asian | TaqMan | 1565/1391 | 588 | 752 | 225 | 512 | 638 | 241 | 0.083 | 9 |
| Xiaoyi Zhou | 2010 | China | Asian | PCR-RFLP | 703/713 | 268 | 316 | 101 | 247 | 339 | 127 | 0.568 | 9 |
| Li Chuanyin | 2017 | China | Asian | TaqMan | 609/583 | 233 | 294 | 92 | 242 | 273 | 68 | 0.497 | 9 |
| Guange Chen | 2020 | China | Asian | TaqMan | 290/445 | 93 | 123 | 61 | 185 | 160 | 85 | <0.001 | 8 |
| miR-124 rs531564 G>C | CC | CG | GG | CC | CG | GG | |||||||
| Xingdong Xiong | 2014 | China | Asian | PCR-LDR | 208/107 | 91 | 15 | 1 | 151 | 51 | 6 | 0.507 | 9 |
| Henghui Wu | 2014 | China | Asian | PCR-LDR | 158/260 | 134 | 22 | 2 | 184 | 66 | 10 | 0.194 | 9 |
| Li Chuanyin | 2017 | China | Asian | TaqMan | 609/583 | 17 | 144 | 448 | 7 | 118 | 458 | 0.846 | 9 |
In six studies, genotype frequencies of rs531564, rs11134527, rs2910164, rs11614913, and rs3746444 were presented separately, so each of them was considered as a separate study in this meta-analysis. Therefore, all the six included studies containing 2291 cases and 2850 controls for rs2910164, six studies containing 2291 cases and 2850 controls for rs11614913, four studies involving 1554 cases and 1962 controls for rs3746444, four studies containing 3167 cases and 3132 controls for rs11134527, and three studies containing 975 cases and 950 controls for rs531564 were finally analyzed in our meta-analysis. For these five miRNA in this meta-analysis, the subjects in all included studies were of the Asian population.
The TaqMan assay and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) were used for genotyping to determine the SNPs in the included studies. HWE of genotype distribution in the controls was tested in all studies, and two articles were not consistent with HWE. And according to the methodology quality assessment, all the studies were above a score of 6.0 and recruited into the following analyses.
3.2. Association between miRNA-146a rs2910164 Polymorphism and Cervical Cancer Susceptibility
The association strength between miRNA-146a rs2910164 polymorphism and cervical cancer risk is shown in Table 2. As shown in Table 2, significant association was not identified in any genetic model (C vs. G: OR = 1.08, 95% CI 0.90-1.30, PH = 0.001, Figure 2; GC vs. GG: OR = 0.94, 95% CI 0.76-1.15, PH = 0.140; CC vs. GG: OR =0.85, 95% CI 0.57-1.26, PH = 0.001; GC/GG vs. CC: OR = 0.90, 95% CI 0.70-1.15, PH = 0.014; and GG vs. GC/CC: OR = 0.90, 95% CI 0.68-1.19, PH = 0.002).
Table 2.
Summary of pooled ORs in the meta-analysis.
| N | OR | P H | OR | P H | OR | P H | OR | P H | OR | P H | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA-146a | C/G | GC/GG | CC/GG | GC+CC/GG | CC/GC+GG | ||||||
| Overall | 6 | 1.08 (0.90-1.30) | 0.001 | 0.94 (0.76-1.15) | 0.140 | 0.85 (0.57-1.26) | 0.001 | 0.90 (0.70-1.15) | 0.014 | 0.90 (0.68-1.19) | 0.002 |
| Sample size | |||||||||||
| Large | 4 | 1.13 (0.91-1.40) | 0.001 | 0.88 (0.67-1.16) | 0.072 | 0.77 (0.49-1.20) | 0.001 | 0.83 (0.60-1.15) | 0.012 | 0.86 (0.64-1.16) | 0.003 |
| Small | 2 | 0.96 (0.59-1.57) | 0.038 | 1.08 (0.78-1.50) | 0.390 | 1.22 (0.37-4.04) | 0.029 | 1.06 (0.67-1.69) | 0.132 | 1.17 (0.40-3.41) | 0.043 |
| HWE | |||||||||||
| P > 0.05 | 5 | 1.06 (0.86-1.31) | 0.000 | 0.93 (0.73-1.19) | 0.083 | 0.88 (0.56-1.39) | 0.000 | 0.91 (0.67-1.22) | 0.007 | 0.93 (0.69-1.27) | 0.001 |
| P < 0.05 | 1 | 1.24 (0.88-1.74) | — | 0.91 (0.55-1.52) | — | 0.68 (0.36-1.31) | — | 0.83 (0.53-1.31) | — | 0.71 (0.38-1.32) | — |
| miRNA-196a2 | C/T | TC/TT | CC/TT | TC+CC/TT | CC/TC+TT | ||||||
| Overall | 6 | 0.83 (0.67-1.04) | 0.000 | 1.19 (0.90-1.58) | 0.001 | 1.41 (0.94-2.10) | 0.000 | 1.28 (0.93-1.75) | 0.000 | 1.23 (0.97-1.57) | 0.021 |
| Sample size | |||||||||||
| Large | 4 | 0.92 (0.84-1.02) | 0.284 | 1.06 (0.86-1.31) | 0.106 | 1.19 (1.00-1.41) | 0.369 | 1.10 (0.90-1.33) | 0.112 | 1.12 (0.97-1.29) | 0.676 |
| Small | 2 | 0.62 (0.22-1.78) | 0 | 1.91 (0.52-7.08) | 0.001 | 2.24 (1.32-15.50) | 0.000 | 2.12 (0.44-10.21) | 0.000 | 1.49 (0.49-4.52) | 0.006 |
| HWE | |||||||||||
| P > 0.05 | 5 | 0.93 (0.85-1.02) | 0.336 | 1.06 (0.89-1.26) | 0.182 | 1.16 (0.99-1.37) | 0.394 | 1.08 (0.92-1.28) | 0.173 | 1.10 (0.96-1.27) | 0.681 |
| P < 0.05 | 1 | 0.36 (0.26-0.51) | — | 3.81 (1.97-7.37) | — | 5.99 (3.09-11.59) | — | 4.80 (2.62-8.76) | — | 2.57 (1.59-4.15) | — |
| miRNA-499 | G/A | AG/AA | GG/AA | AG+GG/AA | GG/AG+AA | ||||||
| Overall | 4 | 0.76 (0.50-1.15) | 0 | 1.34 (0.82-2.20) | 0.001 | 1.42 (0.61-3.30) | 0 | 1.42 (0.83-2.42) | 0 | 1.27 (0.68-2.35) | 0.002 |
| Sample size | |||||||||||
| Large | 2 | 0.86 (0.58-1.27) | 0.025 | 1.34 (0.65-2.75) | 0.001 | 1.03 (0.66-1.60) | 0.705 | 1.28 (0.70-2.35) | 0.003 | 0.99 (0.64-1.53) | 0.413 |
| Small | 2 | 0.68 (0.27-1.72) | 0 | 1.34 (0.52-3.48) | 0.035 | 2.01 (0.35-11.56) | 0 | 1.58 (0.44-5.63) | 0.002 | 1.67 (0.55-5.05) | 0.001 |
| HWE | |||||||||||
| P > 0.05 | 2 | 0.67 (0.28-1.58) | 0 | 1.36 (0.61-3.00) | 0.007 | 2.28 (0.52-9.98) | 0 | 1.64 (0.54-5.01) | 0 | 1.80 (0.69-4.74) | 0.006 |
| P < 0.05 | 2 | 0.86 (0.55-1.36) | 0.048 | 1.33 (0.56-3.16) | 0.029 | 0.84 (0.50-1.43) | 0.886 | 1.26 (0.59-2.70) | 0.035 | 0.89 (0.60-1.34) | 0.583 |
| miRNA-218 | G/A | AG/AA | GG/AA | AG+GG/AA | GG/AG+AA | ||||||
| Overall | 4 | 0.98 (0.83-1.15) | 0.003 | 1.07 (0.89-1.29) | 0.053 | 1.01 (0.73-1.40) | 0.006 | 1.06 (0.86-1.31) | 0.010 | 0.97 (0.76-1.23) | 0.035 |
| Sample size | |||||||||||
| Large | 4 | 0.98 (0.83-1.15) | 0.003 | 1.07 (0.89-1.29) | 0.053 | 1.01 (0.73-1.40) | 0.006 | 1.06 (0.86-1.31) | 0.010 | 0.97 (0.76-1.23) | 0.035 |
| Small | 0 | — | — | — | — | — | — | — | — | — | — |
| HWE | |||||||||||
| P > 0.05 | 3 | 1.03 (0.88-1.21) | 0.019 | 0.99 (0.88-1.13) | 0.318 | 0.92 (0.66-1.28) | 0.021 | 0.97 (0.82-1.16) | 0.091 | 0.92 (0.69-1.24) | 0.031 |
| P < 0.05 | 1 | 0.79 (0.63-0.98) | — | 1.53 (1.09-2.15) | — | 1.43 (0.94-2.16) | — | 1.49 (1.09-2.05) | — | 1.15 (0.79-1.66) | — |
| miRNA-124 | C/G | GC/GG | CC/GG | GC+CC/GG | CC/GC+GG | ||||||
| Overall | 3 | 0.58 (0.40-0.83) | 0.097 | 1.27 (0.97-1.66) | 0.899 | 2.87 (1.40-5.91) | 0.887 | 1.38 (1.07-1.80) | 0.409 | 2.26 (1.58-3.23) | 0.979 |
| Sample size | |||||||||||
| Large | 2 | 0.46 (0.32-0.66) | 0.858 | 1.70 (0.47-6.17) | 0.967 | 3.63 (1.04-12.63) | 0.996 | 3.13 (0.90-10.85) | 0.995 | 2.24 (1.51-3.31) | 0.860 |
| Small | 1 | 0.75 (0.59-0.95) | — | 1.25 (0.95-1.64) | — | 2.48 (1.02-6.04) | — | 1.32 (1.01-1.72) | — | 2.36 (0.97-5.74) | — |
| HWE | |||||||||||
| P > 0.05 | 3 | 0.58 (0.40-0.83) | 0.097 | 1.27 (0.97-1.66) | 0.899 | 2.87 (1.40-5.91) | 0.887 | 1.38 (1.07-1.80) | 0.035 | 2.26 (1.58-3.23) | 0.979 |
| P < 0.05 | 0 | — | — | — | — | — | — | — | — | — | — |
Figure 2.
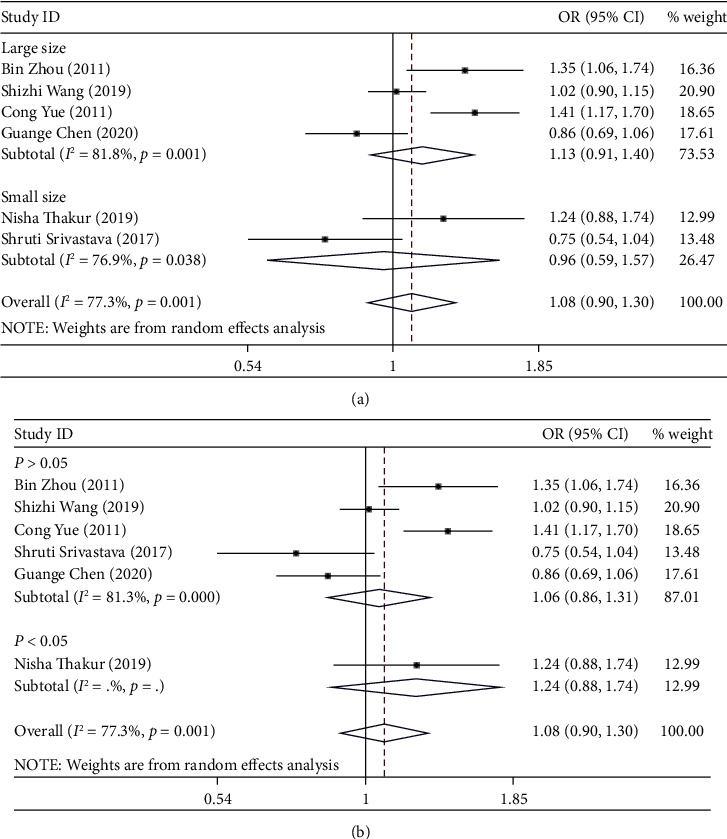
(a) Forest plot of allele comparison of miRNA-146a rs2910164 for overall comparison (C versus G). (b) Forest plot of allele comparison of miRNA-146a rs2910164 for overall comparison (C versus G).
Next, subgroup analysis was conducted according to sample size and HWE. In the large sample size and HWE group, a significant association was also not identified in any genetic model. And the heterogeneity is still high. When we delete three articles (Shizhi Wan, Shruti SrivaStava, and Guange Chen), heterogeneity is significantly reduced (Table 3). A significantly increased risk of cervical cancer susceptibility was identified in allelic comparison (C vs. G: OR = 1.36, 95% CI: 1.19-1.56, PH = 0.800), while there is a significantly decreased risk of cervical cancer susceptibility in heterozygote comparison (GC vs. GG: OR = 0.76, 95% CI: 0.60-0.97, PH = 0.379), homozygote model (CC vs. GG: OR = 0.53, 95% CI: 0.40-0.70, PH = 0.683), dominant model (GC/CC vs. GG: OR = 0.68, 95% CI: 0.54-0.85, PH = 0.406), and recessive model (CC vs. GC/GG: OR = 0.66, 95% CI: 0.53-0.82, PH = 0.638).
Table 3.
Summary of pooled ORs after deleting some articles.
| N | OR | P H | OR | P H | OR | P H | OR | P H | OR | P H | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA-146a | C/G | GC/GG | CC/GG | GC+CC/GG | CC/GC+GG | ||||||
| Overall | 6 | 1.08 (0.90-1.30) | 0.001 | 0.94 (0.76-1.15) | 0.140 | 0.85 (0.57-1.26) | 0.001 | 0.90 (0.70-1.15) | 0.014 | 0.90 (0.68-1.19) | 0.002 |
| After deleting three articles | 3 | 1.36 (1.19-1.56) | 0.800 | 0.76 (0.60-0.97) | 0.379 | 0.53 (0.40-0.70) | 0.683 | 0.68 (0.54-0.85) | 0.406 | 0.66 (0.53-0.82) | 0.638 |
| miRNA-499 | G/A | AG/AA | GG/AA | AG+GG/AA | GG/AG+AA | ||||||
| Overall | 4 | 0.76 (0.50-1.15) | 0.000 | 1.34 (0.82-2.20) | 0.001 | 1.42 (0.61-3.30) | 0.000 | 1.42 (0.83-2.42) | 0.000 | 1.27 (0.68-2.35) | 0.002 |
| After deleting two articles | 2 | 1.04 (0.90-1.21) | 0.730 | 0.93 (0.77-1.12) | 0.674 | 0.97 (0.65-1.46) | 0.514 | 0.94 (0.79-1.13) | 0.633 | 1.01 (0.72-1.42) | 0.673 |
| miRNA-218 | G/A | AG/AA | GG/AA | AG+GG/AA | GG/AG+AA | ||||||
| Overall | 4 | 0.98 (0.83-1.15) | 0.003 | 1.07 (0.89-1.29) | 0.053 | 1.01 (0.73-1.40) | 0.006 | 1.06 (0.86-1.31) | 0.010 | 0.97 (0.76-1.23) | 0.035 |
| After deleting two articles | 2 | 1.11 (1.02-1.21) | 0.395 | 0.97 (0.85-1.10) | 0.214 | 0.79 (0.66-0.94) | 0.594 | 0.92 (0.81-1.04) | 0.235 | 0.80 (0.68-0.94) | 0.982 |
3.3. Association between miR-196a2 rs11614913 Polymorphism and Cervical Cancer Susceptibility
Significant association was not identified in any genetic model (T vs. C: OR = 0.83, 95% CI: 0.67-1.04, PH = 0.000, Figure 3; TC vs. TT: OR = 1.19, 95% CI: 0.90-1.58, PH = 0.001; CC vs. TT: OR = 141, 95% CI: 0.94-2.10, PH = 0.000; TC/CC vs. TT: OR = 1.28, 95% CI: 0.93-1.75, PH = 0.000; and CC vs. TC/TT: OR = 1.23, 95% CI: 0.97-1.29, PH = 0.021) when all eligible studies were pooled.
Figure 3.
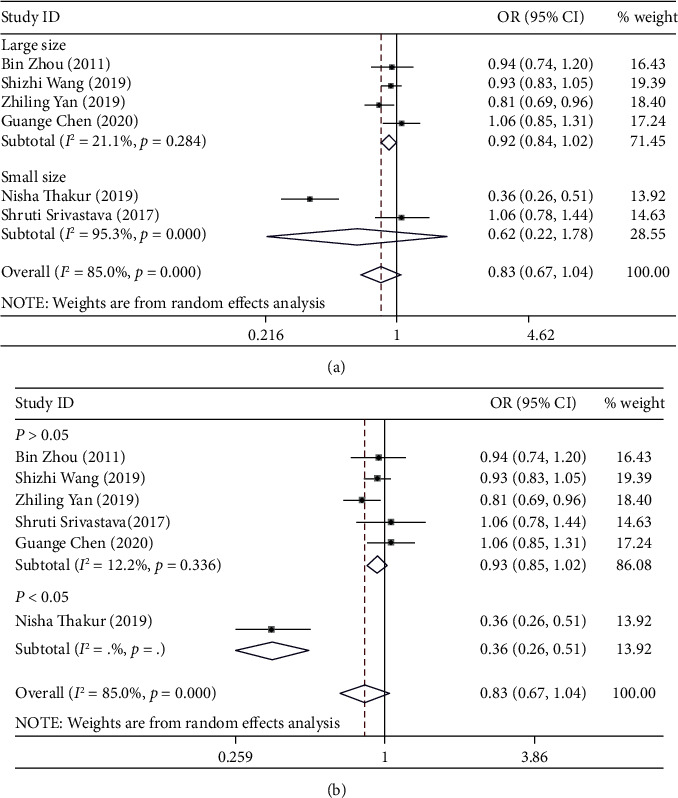
(a) Forest plot of allele comparison of miRNA-196a2 rs11614913 for overall comparison (T versus C). (b) Forest plot of allele comparison of miRNA-196a2 rs11614913 for overall comparison (T versus C).
Next, subgroup analysis was conducted according to sample size and HWE. In large sample size, heterogeneity is significantly reduced. A significantly increased risk of cervical cancer susceptibility was identified in homozygote comparison (CC vs. TT: OR = 1.19, 95% CI: 1.00-1.41, PH = 0.369). In the HWE group, heterogeneity is also significantly reduced. But significant association was not identified in any genetic model.
3.4. Association between miR-499 rs3746444 Polymorphism and Cervical Cancer Susceptibility
Significant association was not identified in any genetic model (G vs. A: OR = 0.76, 95% CI: 0.50-1.15, PH = 0.000, Figure 4; AG vs. AA: OR = 1.34, 95% CI: 0.82-2.20, PH = 0.001; GG versus AA: OR = 1.42, 95% CI: 0.61-3.30, PH = 0.000; AG/GG vs. AA: OR = 1.42, 95% CI: 0.83-2.42, PH = 0.000; and GG vs. AG/AA: OR = 1.27, 95% CI: 0.68-2.35; PH = 0.002).
Figure 4.
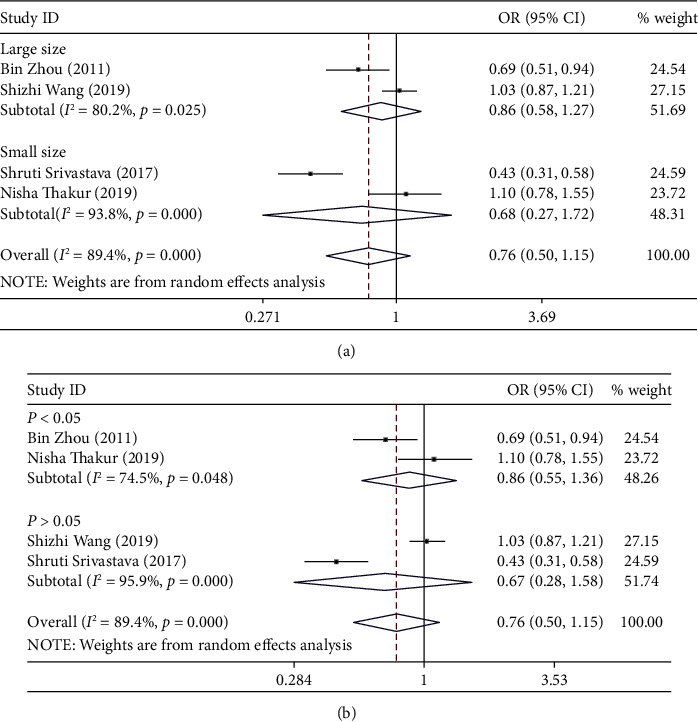
(a) Forest plot of allele comparison of miRNA-499 rs3746444 for overall comparison (G versus A). (b) Forest plot of allele comparison of miRNA-499 rs3746444 for overall comparison (G versus A).
Next, subgroup analysis was conducted according to sample size and HWE. In the large sample size and HWE group, a significant association was also not identified in any genetic model. The heterogeneity in the HWE group is still high, and in a large sample size, the heterogeneity is high except the homozygote model and recessive model. When we delete the two articles (Bin Zhou and Shruti SrivaStava), heterogeneity is significantly reduced in all models (Table 3). However, significant association was also not identified in any genetic model.
3.5. Association between miR-218 rs11134527 Polymorphism and Cervical Cancer Susceptibility
Significant association was not identified in any genetic model (G vs. A: OR = 0.98, 95% CI: 0.83-1.15, PH = 0.003; AG vs. AA: OR = 1.07, 95% CI: 0.89-1.29, PH = 0.053, Figure 5; GG vs. AA: OR = 1.01, 95% CI: 0.73-1.40, PH = 0.006; AG/GG vs. AA: OR = 1.06, 95% CI: 0.86-1.31, PH = 0.091; and GG vs. AG/AA: OR = 0.97, 95% CI: 0.76-1.23, PH = 0.035).
Figure 5.
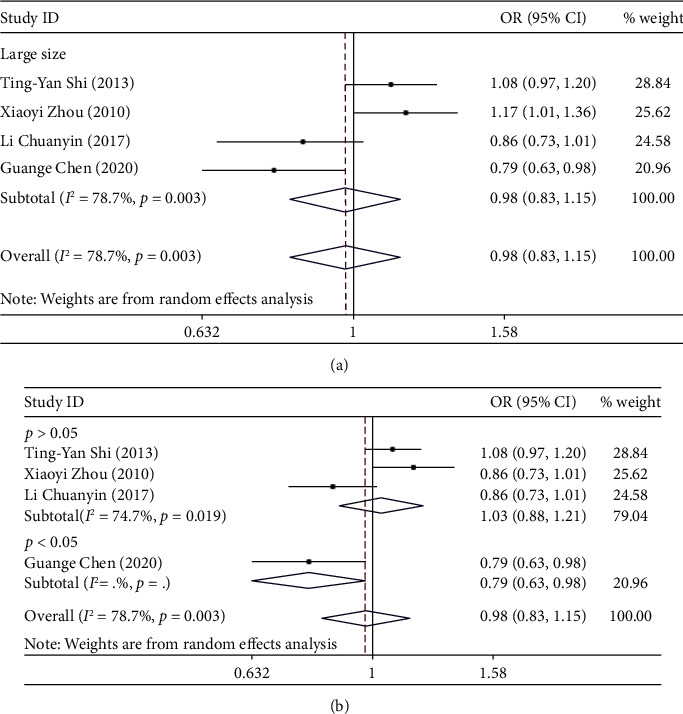
Forest plot of allele comparison of miR-218 rs11134527 for overall comparison (G versus A). (b) Forest plot of allele comparison of miR-218 rs11134527 for overall comparison (G versus A).
The sample size of the 4 included articles is all large size and subgroup analysis was conducted according to HWE. In the HWE group, heterogeneity is also significantly reduced. But significant association was not identified in any genetic model. When we delete two articles (Li Chuanyin and Guange Chen), heterogeneity is significantly reduced in all models (Table 3). We observed a significantly increased risk of cervical cancer susceptibility in allelic comparison (G versus A: OR = 1.11, 95% CI: 1.02-1.21, PH = 0.395), while there is a significantly decreased risk of cervical cancer susceptibility in the homozygote model (GG vs. AA: OR = 0.79, 95% CI: 0.66-0.94, PH = 0.594) and recessive model (GG vs. AG/AA: OR = 0.80, 95% CI: 0.68-0.94, PH = 0.982). Significant association was not identified in heterozygote comparison (AG vs. AA: OR = 0.97, 95% CI: 0.85-1.10; PH = 0.214) and dominant model (GG/AG vs. AA: OR = 0.92, 95% CI: 0.81-1.04, PH = 0.235).
3.6. Association between miR-124 rs531564 Polymorphism and Cervical Cancer Susceptibility
We observed a significantly increased risk of cervical cancer susceptibility in the homozygote model (CC vs. GG: OR = 2.87, 95% CI: 1.40-5.91, PH = 0.887, Figure 6), dominant model (GC/CC vs. GG: OR = 1.38, 95% CI: 1.07-1.80, PH = 0.409), and recessive model (CC vs. GC/GG: OR = 2.26, 95% CI: 1.58-3.23, PH = 0.979), while there is a significantly decreased risk of cervical cancer susceptibility in allelic comparison (C vs. G: OR = 0.58, 95% CI: 0.40-0.83, PH = 0.097). Significant association was not identified in heterozygote comparison (GC vs.GG: OR = 1.27, 95% CI: 0.97-1.66, PH = 0.899).
Figure 6.
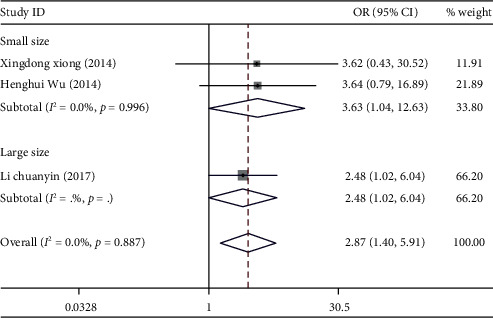
Forest plot of homozygote comparison of miR-124 rs531564 for overall comparison (CC versus GG).
Next, subgroup analysis was conducted according to sample size. In a large sample size, a significantly increased risk of cervical cancer susceptibility could be identified in the homozygote model (CC vs. GG: OR = 3.63, 95% CI: 1.04-12.63, PH = 0.998) and recessive model (CC vs. GC/GG: OR = 2.24, 95% CI: 1.51-3.31, PH = 0.86), while there is a significantly decreased risk of cervical cancer susceptibility in allelic comparison (C vs. G: OR = 0.46, 95% CI: 0.32-0.66; PH = 0.858). Significant association was not identified in heterozygote comparison (GC vs.GG: OR = 1.27, 95% CI: 0.97-1.66, PH = 0.899) and dominant model (GC/CC vs. GG: OR = 3.13, 95% CI: 0.90-10.85, PH = 0.995).
3.7. Sources of Heterogeneity
Since significant heterogeneity was found for miR-146 (rs2910164), miR-196a2 (rs11614913), miR-499 (rs3746444), and miR-218(rs11134527), we conducted a subgroup analysis by sample size, and the results showed that the sample size is the source of heterogeneity of miR-196a2 (rs11614913) in each genetic model. However, for other SNPs, heterogeneity was still high in large sample size. However, when we delete some articles, we found that the heterogeneity is significantly reduced.
3.8. Sensitivity Analysis
The influence of each study on the pooled ORs in this meta-analysis was examined by excluding one study at a time using STATA 12.0 software. The results showed that no significant alteration in the pooled ORs was found in any of the genetic models for the five SNPs.
3.9. Publication Bias
Due to the limited number of studies included (<10), no funnel plot or Egger's test was performed.
4. Discussion
In this meta-analysis, the relationship between these five SNPs in microRNAs (rs2910164, rs11614913, rs3746444, rs11134527, and rs531564) and the risk of cervical cancer was evaluated from 11 published studies. We demonstrated that no evidence was found for the association between miRNA-146a rs2910164, miR-196a2 rs11614913, miRNA-499 rs3746444, and miR-218 rs11134527 and cervical cancer risk in any genetic models. The miR-124 rs531564 polymorphism was associated with a statistically increased risk of cervical cancer in the homozygote model, dominant model, and recessive model. However, due to limited samples and heterogeneity, this finding should be interpreted with caution.
Recent studies have demonstrated that miRNAs are abnormally expressed in cervical cancer and play a role in tumor generation [28]. miR-146a (rs2910164) involves a G>C nucleotide substitution, which changes the G:U pair to C:U [29]. Different locations may cause the function of miR-146a to be different, and the binding affinity to the 3′-UTR region of the target gene may also be different [30, 31]. Furthermore, it is reported that miR-146a can restrict the NF-κB signaling pathway mediated by IRAK1 and TRAF6, which is an important signaling factor involved in the occurrence of cancers [32, 33]. Previous studies have shown that miR-124 is closely related to the occurrence and development of cervical cancer [34, 35]. Evidence has been shown that miR-124 rs531564 polymorphism affects miRNA processing, and the G allele affects the circular secondary structure of pre-miR-124 and changes the expression of other miRNAs [36].
In 2013, Yamamoto et al. found that the expression of miR-218 in cervical cancer tissues was significantly lower than that of adjacent noncancerous tissues. The restoration of miR-218 significantly inhibited the proliferation, migration, and invasion of cancer cells [37]. Laminin-5 β3 (LAMB3) has been verified as a transcriptional target of miR-218 [38], and the expression of LAMB3 is increased in the presence of the HPV-16 E6 oncoprotein, and this effect is mediated by miR-218 [19]. The miR-196a2 is located on the human chromosome 12q13.13. In 2016, Torruella-Loran et al. reported that rs11614913 in miR-196a2 has the function of regulating the expression of several genes involved in cancer [39]. Results of a functional study showed that the mutation reduced the efficiency of miRNA precursor processing into its mature form and reduced the ability to regulate target genes [40]. miR-499 is one of the microRNAs that play an important role in posttranscriptional regulation by regulating multiple genes and signal transduction pathways. The miR-499 (rs3746444) polymorphism leads to mismatches from A:U to G:U in the stem structure of the miR-499 precursor. The presence of this mismatch will affect Sox6 and Rod1 genes, which are important for the occurrence of cancers [41, 42].
Significant heterogeneity was found in this meta-analysis for association of miR-218 rs11134527, miR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, and cervical cancer. Therefore, we conducted a subgroup analysis based on sample size and HWE. When we restricted the sample size to the large sample size group and the HWE group, heterogeneity is significantly reduced for miR-196a2 rs11614913. However, the small sample size group of miR-196a2 includes only two studies, and the large sample size group includes three studies. Therefore, further research is needed to confirm these results. Significant heterogeneity was also found for the association of miRNA-146a rs2910164, miR-499 rs3746444, miR-218 rs11134527, and cervical cancer in the large sample size group and the HWE group. However, when we delete some researches in each analysis, heterogeneity is significantly reduced, suggesting that these researches contributed to the source of heterogeneity. However, owing to the limited articles included, further evaluation of the results is still needed.
In the sensitivity analysis, no significant changes were found after omitting one study at a time, indicating that our meta-analysis results are relatively stable and credible.
Our meta-analysis has several advantages. First, this is the first meta-analysis of the association between five miRNA polymorphisms and the susceptibility to cervical cancer. In addition, according to the methodological quality assessment, all included studies are of high quality. Moreover, there are no restrictions in the literature search, so the selection bias is well controlled. Our meta-analysis has some limitations. Firstly, the number of included studies is too few. Secondly, heterogeneity was detected in miR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, and miR-218 rs11134527. After deleting some studies in each analysis, heterogeneity is significantly reduced. The pooled ORs became significant without evidence of heterogeneity. Thirdly, the included studies were all Asian ethnicity; therefore, it is uncertain whether these results can be generalized to other populations. Fourth, publication bias might also exist, because only published studies were included in this meta-analysis, and results that are not statistically significant usually have less chance of publication.
5. Conclusion
In summary, our results suggested that miR-124 rs531564 polymorphism is significantly associated with increased risk of cervical cancer, while miR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, and miR-218 rs2292832 polymorphisms may not be associated with the susceptibility of cervical cancer. However, since few studies have been included, there is not enough data to fully confirm the association between cervical cancer and the five miRNA polymorphisms, so the results should be interpreted with caution.
Acknowledgments
We would like to acknowledge the Department of Obstetrics and Gynecology, Xuanwu Hospital, Capital Medical University, for helping me at work.
Data Availability
The data supporting this meta-analysis are from previously reported studies which have been cited.
Conflicts of Interest
The authors declare that there are no conflicts of interest associated with the manuscript.
Supplementary Materials
Table S1: scale for quality assessment.
References
- 1.Meng S., Fan X., Zhang J., An R., Li S. GJA1 expression and its prognostic value in cervical cancer. BioMed Research International . 2020;2020:10. doi: 10.1155/2020/8827920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava S., Singh S., Fatima N., Mittal B., Srivastava A. N. Pre-microRNA gene polymorphisms and risk of cervical squamous cell carcinoma. Journal of Clinical and Diagnostic Research . 2017;11(9):p. 4. doi: 10.7860/jcdr/2017/25361.10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Freitas A. C., Gurgel A. P., Chagas B. S., Coimbra E. C., do Amaral C. M. Susceptibility to cervical cancer: an overview. Gynecologic Oncology . 2012;126(2):304–311. doi: 10.1016/j.ygyno.2012.03.047. [DOI] [PubMed] [Google Scholar]
- 4.Fang J., Li Y., Zhang J., et al. Correlation between polymorphisms in microRNA-regulated genes and cervical cancer susceptibility in a Xinjiang Uygur population. Oncotarget . 2017;8(19):31758–31764. doi: 10.18632/oncotarget.15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng X., Zhang Y., Yue T., et al. Association between XRCC1 polymorphisms and the risk of cervical cancer: a meta-analysis based on 4895 subjects. Oncotarget . 2017;8(2):2249–2260. doi: 10.18632/oncotarget.13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin L.-C., Xiao G., Zhou R., et al. MicroRNA-361-5p inhibits tumorigenesis and the EMT of HCC by targeting Twist1. BioMed Research International . 2020;2020:14. doi: 10.1155/2020/8891876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K., Tie H., Hu N., et al. Association of two polymorphisms rs2910164 in miRNA-146a and rs3746444 in miRNA-499 with rheumatoid arthritis: a meta-analysis. Human Immunology . 2014;75(7):602–608. doi: 10.1016/j.humimm.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Saunders M. A., Liang H., Li W. H. Human polymorphism at microRNAs and microRNA target sites. Proceedings of the National Academy of Sciences . 2007;104(9):3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin G. A., Croce C. M. MicroRNA-cancer connection: the beginning of a new tale. Cancer Research . 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Huang J., Yang N., et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proceedings of the National Academy of Sciences . 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi T.-Y., Chen X.-J., Zhu M.-L., et al. A pri-miR-218variant and risk of cervical carcinoma in Chinese women. BMC Cancer . 2013;13(1):p. 8. doi: 10.1186/1471-2407-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou B., Wang K., Wang Y., et al. Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Molecular Carcinogenesis . 2011;50(7):499–505. doi: 10.1002/mc.20740. [DOI] [PubMed] [Google Scholar]
- 13.Xiong X., Cheng J., Liu X., Tang S., Luo X. Correlation analysis between miR- 124 rs531564 polymorphisms and susceptibility to cervical cancer. Journal of Southern Medical University . 2014;34(2):210–213. [PubMed] [Google Scholar]
- 14.Wang S., Zhu H., Ding B., et al. Genetic variants in microRNAs are associated with cervical cancer risk. Mutagenesis . 2019;34(2):127–133. doi: 10.1093/mutage/gez005. [DOI] [PubMed] [Google Scholar]
- 15.Thakur N., Singhal P., Mehrotra R., Bharadwaj M. Impacts of single nucleotide polymorphisms in three microRNAs (miR-146a, miR-196a2 and miR-499) on the susceptibility to cervical cancer among Indian women. Bioscience Reports . 2019;39(4):127–133. doi: 10.1042/bsr20180723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H., Zhang J. miR-124 rs531564 polymorphism influences genetic susceptibility to cervical cancer. International Journal of Clinical and Experimental Medicine . 2014;7(12):5847–5851. [PMC free article] [PubMed] [Google Scholar]
- 17.Chuanyin L., Xiaona W., Zhiling Y., et al. The association between polymorphisms in microRNA genes and cervical cancer in a Chinese Han population. Oncotarget . 2017;8(50):87914–87927. doi: 10.18632/oncotarget.21235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue C., Wang M., Ding B., et al. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecologic Oncology . 2011;122(1):33–37. doi: 10.1016/j.ygyno.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 19.Yan Z., Zhou Z., Li C., et al. Polymorphisms in miRNA genes play roles in the initiation and development of cervical cancer. Journal of Cancer . 2019;10(20):4747–4753. doi: 10.7150/jca.33486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X., Chen X., Hu L., et al. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case–control study in Chinese women. Gynecologic Oncology . 2010;117(2):287–290. doi: 10.1016/j.ygyno.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Chen G., Zhang M., Zhu J., et al. Common genetic variants in pre-microRNAs are associated with cervical cancer susceptibility in southern Chinese women. Journal of Cancer . 2020;11(8):2133–2138. doi: 10.7150/jca.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J., Jin M., Zhang M., Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS ONE . 2012;7(1):p. 7. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moher D., Liberati A., Tetzlaff J., Altman D. G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Annals of Internal Medicine . 2009;151(4):264–9, W64. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 24.Vanoverschelde J. L., Robert A. R., Gerbaux A., Michel X., Hanet C., Wijns W. Noninvasive estimation of pulmonary arterial wedge pressure with Doppler transmitral flow velocity pattern in patients with known heart disease. The American Journal of Cardiology . 1995;75(5):383–389. doi: 10.1016/S0002-9149(99)80559-1. [DOI] [PubMed] [Google Scholar]
- 25.Lau J., Ioannidis J. P., Schmid C. H. Quantitative synthesis in systematic reviews. Annals of Internal Medicine . 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 26.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute . 1959;22(4):719–748. [PubMed] [Google Scholar]
- 27.DerSimonian R., Laird N. Meta-analysis in clinical trials revisited. Contemporary Clinical Trials . 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Z. M., Wang X. Regulation of cellular miRNA expression by human papillomaviruses. Biochimica et Biophysica Acta . 2011;1809(11-12):668–677. doi: 10.1016/j.bbagrm.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jazdzewski K., Murray E. L., Franssila K., Jarzab B., Schoenberg D. R., de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proceedings of the National Academy of Sciences . 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J., Ambrosone C. B., DiCioccio R. A., Odunsi K., Lele S. B., Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis . 2008;29(10):1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Y., Cullen B. R. Sequence requirements for micro RNA processing and function in human cells. RNA . 2003;9(1):112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito-Kureha T., Koshikawa N., Yamamoto M., et al. Tropomodulin 1 expression driven by NF-κB enhances breast cancer growth. Cancer Research . 2015;75(1):62–72. doi: 10.1158/0008-5472.CAN-13-3455. [DOI] [PubMed] [Google Scholar]
- 33.Kastrati I., Canestrari E., Frasor J. PHLDA1 expression is controlled by an estrogen receptor-NFκB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene . 2015;34(18):2309–2316. doi: 10.1038/onc.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilting S. M., van Boerdonk R. A., Henken F. E., et al. methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Molecular Cancer . 2010;9(1):p. 167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garzon R., Calin G. A., Croce C. M. MicroRNAs in cancer. Annual Review of Medicine . 2009;2009:199–227. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 36.Wang X., Tang S., le S. Y., et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One . 2008;3(7):p. e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto N., Kinoshita T., Nohata N., et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion by targeting focal adhesion pathways in cervical squamous cell carcinoma. International Journal of Oncology . 2013;42(5):1523–1532. doi: 10.3892/ijo.2013.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: a Cancer Journal for Clinicians . 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 39.Torruella-Loran I., Laayouni H., Dobon B., et al. MicroRNA genetic variation: from population analysis to functional implications of three allele variants associated with cancer. Human Mutation . 2016;37(10):1060–1073. doi: 10.1002/humu.23045. [DOI] [PubMed] [Google Scholar]
- 40.Hoffman A. E., Zheng T., Yi C., et al. MicroRNA miR-196a2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Research . 2009;69(14):5970–5977. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi P., Dou T. H., Geng L., et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Human Immunology . 2010;71(6):621–626. doi: 10.1016/j.humimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 42.Tano K., Mizuno R., Okada T., et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Letters . 2010;584(22):4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: scale for quality assessment.
Data Availability Statement
The data supporting this meta-analysis are from previously reported studies which have been cited.


