Highlights
-
•
CAD is a pandemic that can be prevented. Conventional risk factors are inadequate to detect who is at risk early in the asymptomatic stage.
-
•
Genetic risk for CAD can be determined at birth, and those at highest genetic risk have been shown to respond to lifestyle changes and statin therapy with a 40% to 50% reduction in cardiac events.
-
•
Genetic risk stratification for CAD should be brought to the bedside in an attempt to prevent this pandemic disease.
Key Words: cardiovascular genetics, coronary artery disease, genetic risk score for CAD, genome-wide association studies, prevention of CAD
Abbreviations and Acronyms: ACC, American College of Cardiology; AHA, American Heart Association; ANRIL, antisense non-coding RNA in the INK4 Locust; bp, base pair; CAD, coronary artery disease; GRS, genetic risk score; GWAS, genome-wide association study; LDL-C, low-density lipoprotein cholesterol; MR, Mendelian randomization; SNP, single nucleotide polymorphism
Summary
Coronary artery disease (CAD) is a pandemic disease that is highly preventable as shown by secondary prevention. Primary prevention is preferred knowing that 50% of the population can expect a cardiac event in their lifetime. Risk stratification for primary prevention using the American Heart Association/American College of Cardiology predicted 10-year risk based on conventional risk factors for CAD is less than optimal. Conventional risk factors such as hypertension, cholesterol, and age are age-dependent and not present until the sixth or seventh decade of life. The genetic risk score (GRS), which is estimated from the recently discovered genetic variants predisposed to CAD, offers a potential solution to this dilemma. The GRS, which is derived from genotyping the population with a microarray containing these genetic risk variants, has indicated that genetic risk stratification based on the GRS is superior to that of conventional risk factors in detecting those at high risk and who would benefit most from statin therapy. Studies performed in >1 million individuals confirmed genetic risk stratification is superior and primarily independent of conventional risk factors. Prospective clinical trials based on risk stratification for CAD using the GRS have shown lifestyle changes, physical activity, and statin therapy are associated with 40% to 50% reduction in cardiac events in the high genetic risk group (20%). Genetic risk stratification has the advantage of being innate to an individual’s DNA, and because DNA does not change in a lifetime, it is independent of age. Genetic risk stratification is inexpensive and can be performed worldwide, providing risk analysis at any age and thus has the potential to revolutionize primary prevention.
Central Illustration
The approach to any common disease is from the perspective of detection, prevention, and specific management once the event occurs. Coronary artery disease (CAD), which occurs commonly throughout the world, has long been recognized to be associated with factors that can be prevented. Risk factors for disease are usually categorized into 3 classes: genetic susceptibility; environmental exposure; and lifestyle factors. The environmental and lifestyle components of this triad for CAD have been well-documented since the 1960s, starting with the Framingham study (1). Guidelines have been standardized for their detection and reduction of their associated risk for CAD (2).
Several randomized placebo-controlled clinical trials have confirmed that a reduction in these risk factors for CAD is associated with a corresponding reduction in cardiac events and mortality (3, 4, 5). This was comprehensively reviewed by The Cholesterol Treatment Trialist, which did a meta-analysis that involved >170,000 individuals and analyzed the safety and efficacy of statin therapy (6,7). In the United States, over the past 30 years, reduction of risk from environmental and lifestyle risk factors has been the predominant reason for a 50% reduction in cardiac events and mortality (8,9).
Epidemiologists have claimed for some time, based on family history (10) and studies on identical twins (11), that genetic risk factors account for 40% to 60% of the predisposition for CAD (12). Discovery and isolation of these genetic variants predisposed to CAD were enabled by the development of new technologies in the early part of this century (13, 14, 15, 16, 17). In this review, we summarize these large studies, their research implications for the pathogenesis of CAD, and recent attempts to incorporate these genetic variants into a risk score to risk stratify for primary and secondary prevention of CAD. The application of a risk score based on an individual’s DNA has several advantages over conventional risk factors because it is independent of age and can be determined at birth or anytime thereafter. The advantages and limitations of a genetic risk score (GRS) for CAD are summarized. The quality and accuracy of GRSs are still in evolution but could be enhanced by clinical application (Central Illustration).
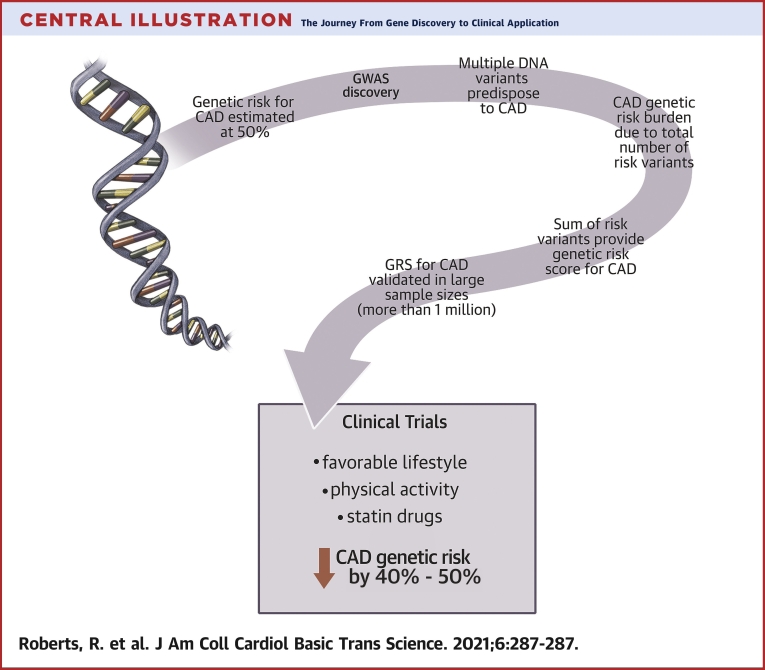
Central Illustration.
The Journey From Gene Discovery to Clinical Application
A brief summary from genetics to clinical applicationis shown. Genome wide association studies made possible an unbiased search for genetic variants predisposing to coronary artery disease (CAD). The total risk burden was summarized in a single number referred to as the Genetic Risk Score (GRS). The GRS was utilized to stratify for risk of CAD. Prospective clinical trials showed lifestyle changes and drug therapy decreased genetic risk by 40% to 50%.
Rare Single Gene Disorders
Variations in the DNA sequence that are disease related occur in a continuum from rare to common. However, because of the earlier clinical detection of the phenotypes and detection through pedigrees, we refer to inherited diseases as those that are rare versus those that are common. Rare disorders by definition occur in <1% of the population and are usually associated with high penetrance. These genes were discovered through a technique referred to as genetic linkage analysis (18). This requires a pedigree of 2 or 3 generations of several individuals affected with the disease. A DNA marker, detected in those affected with the disease (more often than by chance), indicates it is in close physical proximity to the gene responsible for the disease. The DNA region is then cloned and sequenced to identify the responsible mutation, a process referred to as positional cloning. Using this approach, genes responsible for multiple rare cardiovascular inherited disorders were discovered, including familial hypertrophic cardiomyopathy (19,20), and Wolff-Parkinson-White syndrome (21). The last 2 decades of the 20th century were a golden era for these rare diseases and provided tremendous insight into their pathogenesis and management. It is claimed of the 8,315 rare inherited disorders, a responsible gene has been discovered for >5,482 (22). Expression of the responsible mutant gene as a transgene in animal models for rare cardiovascular disorders [e.g., familial hypertrophic cardiomyopathy (23) or Wolff-Parkinson-White syndrome (24)] induces the expected human phenotype in the offspring. Thus, these disorders are often referred to as single-gene disorders because the mutant gene is both necessary and sufficient to induce the phenotype.
Polygenic Disorders: Genome-Wide Association Studies
The predisposition of common disorders, such as CAD and diabetes type 2, are all due to multiple DNA risk variants and are referred to as polygenic disorders. This is in keeping with the hypothesis that the DNA risk variants transmitting predisposition for polygenic disorders are multiple, with each having only minimal risk. Identification of these DNA risk variants requires a different approach than that of collecting pedigrees and analyzing for genetic linkage. The preferred unbiased approach is that of a case−control association study based on an analysis of markers distributed throughout the genome (25). This is, in principle, simple; the frequency of DNA markers is determined by genotyping individuals known to have the disease and genotyping control subjects. Those DNA markers that occur more frequently in cases than controls would be designated in a DNA sequence that contains an increased risk for the disease. The marker itself is unlikely to be the cause of increased risk but is in close physical proximity to the causative sequence. The approach requires DNA markers evenly distributed throughout the human genome at intervals of not more than 3,000 to 6,000 base pairs (bp). This requires 500,000 to 1 million markers to meet this requirement.
Several advancements in technology occurred to make the case−control association study approach practical. In 2001, the human genome (13,17) was sequenced, followed by the HapMap Project (16), which made available millions of informative polymorphic DNA markers spanning the human genome. These markers are single nucleotide polymorphisms (SNPs) that are fairly evenly distributed throughout the genome at approximately 1 SNP/1,000 bp. The number of SNPs per human genome is fairly consistent at approximately 5 million (16,26,27). SNPs were encrypted on computerized microarrays and used as the template for platforms designed to perform high-throughput genotyping (15). The first of these studies was performed for age-related macular degeneration (28). Genome-wide association studies (GWASs) have become a major international focus over the past decade. The National Human Genome Research Instituteve GWAS catalog indicates >5,687 GWASs have been performed, involving >71,000 variant trait associations and published in >3,000 publications (29). These studies have greatly enhanced the architecture of genetic predisposition for CAD and many other polygenic diseases.
Genetic predisposition, or lack of predisposition for a disease, is determined by the DNA sequences transmitted to offspring at conception. Genes are classically defined as DNA sequences that code for proteins. This definition was developed before we knew much regarding the remainder of the sequences in the total genome. It is now expected that >90% of the genome is functional and influences the expression directly or indirectly of genes upstream and downstream, including across other chromosomes. Sequences that code for proteins account for >1% of the DNA in the human genome. All genes have multiple forms, referred to as alleles, as do DNA sequences in the nonprotein-coding regions. These alleles differ from each other and their parent sequence, usually by only a single nucleotide substitution mutation. The frequency of these alleles in the population varies from rare to common. Despite the minor sequence difference between alleles, its function is often altered, which for disease-related alleles could lead to being neutral or associated with an increased or decreased predisposition. Each individual has a maximum of 2 copies of each allele, 1 from each parent. The particular allele occupying any chromosomal locus of an individual is determined at conception. The use of the term allele in this review refers to alternative forms of any DNA sequence, which may or may not be a part of a protein-coding sequence. Those DNA sequences shown to be associated with increased risk for a disease are referred to as genetic risk variants.
Genetic Risk Variants For CAD
The first genetic risk variant for CAD, 9p21, was discovered in 2007, simultaneously and independently by 2 groups (30,31), followed shortly by confirmation from 2 other groups (32,33). Over the next 2 years, several groups throughout the world confirmed the association between the 9p21 genetic risk variant and CAD (34). The reported features of the 9p21 genetic risk variant were similar throughout the world, except in Africans, who had the 9p21 variant is not a risk factor (35, 36, 37). The 9p21 genetic risk variant was shown to be common, being present in approximately 75% of the population. The risk for CAD is mediated independently of conventional risk factors for CAD. Subsequent efforts by groups such as the Myocardial Infarction Genetics Consortium (38) identified 2 novel genetic risk variants for CAD, followed by a variant at 3q22 found by Erdmann et al. (39), the SLC22A3 variant found by Trégouët et al. (40), and the 12q24 variant found by Gudbjartsson et al. (41). The Coronary Artery Disease Genetics Consortium (42) identified 5 novel genetic risk variants for CAD. The characteristics of the genetic variant 9p21, along with the subsequent 10 genetic risk variants predisposed to CAD, were consistent with the hypothesis that common polygenic disorders are due to multiple commonly occurring alleles, each associated with only minimal risk.
The associated minimal risk of each variant strongly indicated there were many more, and the search would have to be intensified. It would require a greater density of DNA markers and massive sample sizes. This led to the formation of an international consortium, with >80,000 cases and control subjects, the CARDIoGRAM (Coronary Artery Disease Genome-Wide Replication, and Meta-analysis) Consortium (43). The GWAS approach used millions of SNPs as markers with increased sample sizes. This required statistical correction, which with 1 million SNPs and a Bonferroni correction, demanded a p value of 0.00000005 that became known as genome-wide significance (44). In addition, replication in an independent population was required.
Over the next 10 years, international consortium and individual efforts discovered 171 genetic risk variants that are of genome-wide significance and have been replicated in an independent population. A total of 163 of these genetic risk variants for CAD were carefully tabulated in the review by Erdmann et al. (45). The additional 8 variants are summarized in Table 1. It is expected there are many more to be identified. There are many other genetic risk variants for CAD that are not of genome-wide significance but are highly significant in that they have <5% false positives as determined by the false discovery rate (39,45, 46, 47).
Table 1.
Eight Newly Discovered Genetic Risk Variants
| Position | Lead SNP | RAF | OR | Gene(s) at Locus | Ref. # | |
|---|---|---|---|---|---|---|
| 1 | 6p21.3 | rs3869109 | G (0.55) | 1.16 | HLC-C,HLA-B, HCG27 | (140) |
| 2 | chr2: 19809058 | rs2123536 | T (0.39) | 1.25 | TTC32-WDR35 | (141) |
| 3 | chr4: 156730909 | rs1842896 | T (0.76) | 1.23 | GUCY1A3 | (141) |
| 4 | chr6: 32449331 | rs9268402 | G (0.59) | 1.17 | C6orf10 | (141) |
| 5 | chr12: 88605319 | rs7136259 | T (0.39) | 1.21 | ATP2B1 | (141) |
| 6 | 6p24.1 | rs6903956 | A (0.08) | 1.51 | C6orf105 | (142) |
| 7 | chr3: 17099388 | rs4618210 | A (0.45) | 0.91 | PLCL2 | (143) |
| 8 | chr19: 2111529 | rs3803915 | A (0.21) | 0.89 | AP3D1-DOT1L-SF3A2 | (143) |
Since the paper by Erdmann et al. (45), there has been 8 newly discovered genetic risk variants predisposed to coronary artery disease.
OR = odd's ratio; RAF = relative allele frequency; SNP = single nucleotide polymorphism.
Need for Genetic Risk Stratification
CAD is a slow-growing disease that usually takes decades to manifest clinical features. This process is influenced by several known environmental and lifestyle risk factors. Decreasing environmental or lifestyle risk has been shown to be extremely effective, particularly for secondary prevention. Secondary prevention is activated when an individual has a clinical event (e.g., myocardial infarction or angina). In contrast, primary prevention, by definition, does not have such defining signals. Unfortunately, the conventional risk factors for CAD are age-dependent and are only detected late in life, which is less than optimal for primary prevention. The potential to markedly attenuate this disease, if not eliminate it, is robust with the currently existing therapeutic armamentarium. Modification of risk factors clearly would have to be initiated early in life, and, in some cases, even before adolescence. In women with hormonal protection during the pre-menopausal stage, it is perhaps possible to delay primary prevention until the fourth or fifth decade, and still prevent the development of clinical sequelae (46,47).
The predominant culprit in the pathogenesis of coronary atherosclerosis is the plasma low-density lipoprotein cholesterol (LDL-C) and the duration of exposure. Every additional 10 years of exposure to plasma LDL-C is associated with a doubling of morbidity and mortality (48). The Cardiology Practice Guidelines have established a plasma LDL-C of ≤70 mg/dl as the target for primary or secondary prevention of CAD. However, the plasma LDL-C of a woman in her fourth decade averages 121 mg/dl, whereas a man in his fourth decade averages 146 mg/dl, which is nearly twice that of the desired target (49). Should we treat everyone with statin therapy? Primary prevention based on conventional risk factors requires treatment with statin therapy in 100 individuals to prevent a single cardiac event (50). Only one-half are expected to benefit because only 50% will develop a cardiac event.
To avoid the cost of treating everyone and inappropriately treating one-half of the population who will not benefit beckons the issues to risk stratify. The Cardiology Practice Guidelines were set up as an attempt to avoid treating everyone with the hope of selecting those who would most benefit from preventive therapy. Using the conventional risk factors, namely, age, cholesterol, diabetes, obesity, blood pressure, sedentary way of life, and smoking, a simple calculation was devised to be applicable to anyone from age 40 to 75 years, taking into account that everyone at age 40 years will have increased plasma LDL-C. The American College of Cardiology/The American Heart Association (AHA/ACC) adopted the method of the Pooled Cohort Equation, which calculates risk for 10 years based on these risk factors (51). Anyone with a 10-year risk of ≥7.5% are candidates for preventive therapies.
A 40-year-old woman with the risk profile of Caucasian, blood pressure of 120/70 mm Hg, total cholesterol of 240, LDL-C 180 mg/dl, high-density lipoprotein (HDL) 30 mg/dl, non-smoker, and without a history of diabetes has a 10-year risk of 1.3% based on the Pooled Cohort Equation. This is far below the level of 7.5% required to recommend preventive therapy. Nevertheless, a plasma LDL-C of 180 mg/dl is >2-fold higher than the desired level, and despite the lack of other risk factors, it is likely to contribute to CAD. It appears to be a missed opportunity for primary prevention of CAD in women with these clinical findings. It is well-documented that the incidence of heart disease is the same in women as in men but is delayed by approximately 10 years. If we are to take advantage of the opportunity for primary prevention in pre-menopausal women, it would be greatly enhanced with a method of risk detection that is independent of age.
Mendelian randomization (MR) has shown that individuals with a minimal decrease in LDL-C due to genotypes inherited at conception are associated with a 50% reduction in cardiac risk per mmol/l reduction in LDL-C (52). This is approximately 3-fold greater than the benefit observed in clinical trials that enroll individuals with an average age of 60 years. To be effective, primary prevention in men has to occur in their 20s or 30s for risk stratification for early intervention (53). Because CAD is now a pandemic disease, attempts to halt its spread will require earlier primary prevention in both men and women, preferably based on a risk assessment that is independent of age. Risk assessment for CAD, based on genetic risk, is independent of age because DNA does not change in an individuals’ lifetime. This is discussed in further detail in subsequent sections.
A Treasure Trove For New Drugs And Research-Functional Analysis of Genetic Risk Variants
Genetics have been most favorable for CAD (54). Families with familial hypercholesterolemia provided strong evidence for cholesterol being a causative agent (55). The development of the first statin drug and its lowering of cholesterol in patients with familial hypercholesterolemia was associated with a corresponding reduction in cardiac mortality and morbidity. The statin class of drugs is currently our main therapy in both primary and secondary prevention of CAD. Approximately a decade ago, a new armamentarium in the prevention of CAD was introduced; PCSK9 inhibitors were stimulated yet again by a genetic observation. The discovery of a gain-of-function mutation in the PCSK9 gene was observed to be associated with increased risk for myocardial infarction (56). This was followed by the discovery of a loss-of-function in the PCSK9 gene that was associated with decreased risk for myocardial infarction (57). PCSK9 inhibitors are now part of the routine clinical management and prevention of CAD (58).
Angiopoietin-like 3 is known to inhibit lipoprotein and endothelial lipase, which leads to increased levels of triglyceride and other lipids (59, 60, 61). Individuals inheriting a loss of function in the gene encoding angiopoietin-like 3 experience a 41% lower risk of CAD due to low plasma levels of LDL-C and triglycerides (62,63). Inhibition of angiopoietin-like 3 by pharmacological means is also associated with lower plasma levels of LDL-C independent of the LDL-C receptor. This led to development of an antibody, evinacumab, which inhibits angiopoietin-like 3. It has undergone clinical trials, and phase III trials performed recently in individuals with homozygous familial hypercholesterolemia showed a 41% reduction in LDL-C (64). Thus, a new mechanism has been exploited to add to the armamentarium of drug therapy to decrease LDL-C independent of cholesterol (statins) or removal by the LDL receptor (PCSK9 inhibitors). Genetics has again contributed significantly to the future prevention and management of CAD.
More than one-half of the genetic risk variants do not mediate their risk through known conventional risk factors (34). This would indicate that several factors, yet to be discovered, are involved with the pathogenesis of CAD. This is inspirational fodder for the investigator. Elucidation of the mechanisms whereby they mediate their risk enable the development of drugs based on novel targets. The thrust of research in this field is still primitive, in part, because of the lack of tools to pursue such a task. First, the lead SNP signifies a risk region that contains thousands of bp of which only ≥1 bp are the causative risk mutation. The efforts to map and sequence these regions are expensive, at a ratio of approximately 30 to 1 over the cost of genotyping. Even after massive sequencing, it will be a prohibitive task. This is made more difficult because no 1 risk variant is necessary or sufficient to induce the phenotype of CAD; therefore, researchers cannot depend on a conventional knockout or transgenic animal models to confirm or disprove causality. Furthermore, >80% of the risk variants for CAD, as for other polygenic diseases, are located in nonprotein-coding sequences (34) that are inherently unfriendly to functional analysis. Their location in nonprotein-coding regions relegates risk variants to a regulatory role, namely, influencing the expression of protein-coding sequences that can be hundreds of thousands of bp upstream or downstream (cis-acting) or even on other chromosomes (trans-acting). This task is further complicated by the recent observation that suggested that only approximately one-third of causal genes are the nearest genes to the GWAS hit (65,66). One approach is to express these sequences in various in vitro experiments to seek a clue to their function. Investigators are pursuing means to place the risk variant sequences and their proximal genes into biological pathways in the hopes of determining their function (45).
Despite the difficulties, research is extremely active in pursuing the function of genetic risk variants, as can be exemplified by discussion of a few of these variants located at loci 9p21, 1p13, 15q24, and 6p24. The 9p21 risk variant for CAD was the first genetic risk variant to be discovered for CAD and has undergone extensive research. Multiple studies confirmed 9p21 mediates its risk for CAD, independent of known risk factors. Fine mapping of the 9p21 risk variant region showed that it is confined to 58,000 bp (67). This 58,000 bp does not contain any protein-coating sequences but does overlap with a long noncoding RNA of 126,000 bp referred to as an antisense noncoding RNA in the INK4 Locus (ANRIL) (68,69). ANRIL has no known function, but its expression induces the expression of 3 nearby well-known genes, CDKN2a, CDKN2b, and ARF, which encode for cyclin-dependent kinase inhibitors. An attractive hypothesis is that the 9p21 risk variant reduces the expression of ANRIL and the kinase inhibitors that enable the growth of vascular atheroma. Several studies provided support for this hypothesis (70,71). Unfortunately, other studies showed lack of correlation between the 9p21 risk variant and expression of cyclin-dependent kinase inhibitors (72, 73, 74).
Visel et al. (75) pursued studies of the 9p21 variant in a mouse model. This corresponds to a 70,000 bp region in the mouse. This region was knocked out, and the resulting phenotype showed excessive proliferation of smooth muscle cells and several neoplasms. However, none of the vessels showed fatty streaks or any evidence of coronary atherosclerosis. The absence of atherosclerosis might be, in part, because the mouse was resistant to atherosclerosis or the lack of the 9p21 risk variant, because there was only a 50% sequence homology between the human 9p21 58,000 bp and the 70,000 bp region of the mouse. Studies suggested that the 9p21 risk variant did not appear in the genome until that of higher primates (67), and thus, would not be expected to give an atherosclerotic phenotype in the mouse.
Harismendy et al. (76) performed a series of studies that concluded that the 9p21 risk variant mediated its risk for CAD through interferon-γ. However, we subsequently showed that the effects of interferon-γ on CDKN2a/b were independent of the 9p21 risk variant (77), which was confirmed recently by Erridge et al. (78). Recent studies involved the deletion of the 58-kb locus-induced pluripotent stem cells derived from individuals homozygous for the nonrisk haplotype. Expression of this region in the smooth muscle cells suggested the 9p21 risk variants were related to adhesion, contraction, and proliferation (79).
Although the mechanism of the 9p21 risk variant remains unknown, clinical observation suggests that it acts through the vessel wall and predisposes to the development of atheroma. This was confirmed by the deCode group, which found that the 9p21 risk variant was also associated with an increased risk for abdominal aortic aneurysms and intracranial aneurysms (80). There was no association between the 9p21 risk variant and myocardial infarction (81,82). This strongly indicated 9p21 was not associated with plaque rupture or thrombosis. Several studies indicated the 9p21 risk variant was associated with progression of coronary atherosclerosis (81,83,84). The 9p21 risk variant was predisposed to atheroma in the coronaries and ballooning in the aortic aneurysm, which suggested a defect in the vessel wall and led to these 2 different abnormalities.
The SORT1 gene located near the 1p13 locus is known to increase the risk for CAD approximately 15% to 20% per copy and causes a 10% to 15% increase in plasma LDL-C. SORT1 is highly expressed in the liver and primary hepatocytes (85,86). Studies have been controversial, showing increased expression of very LDL in the mouse, and other studies showing decreased very LDL secretion (87, 88, 89). Because of these confusing results, SORT1, as a risk variant for CAD, has received less attention.
A third risk variant for CAD located on chromosome 6p24 has at least 2 candidate causal genes, PHACTR1 (phosphatase and actin regulator 1) and EDN1 (endothelin 1). Fine mapping of this region strongly suggest endothelin 1 is more likely to be the candidate because it is known to be involved with vascular biology. Nevertheless, studies show that PHACTR1 also has vascular properties, but conflicting results have been observed in comparing PHACTR1 versus EDN1 on expression of vascular tissue. It remains to be determined which of these 2 genes are likely to be the causal candidate for CAD predisposition (90,91).
The risk variant for CAD on chromosome 15q25 harbors the gene ADAMTS7 (A disintegrin and metalloproteinase with thrombospondin motifs-7). This variant is associated with reduced protein maturation, proteinase activity, and also migration of vascular smooth muscle cells in vitro (92). Knockout experiments in the mouse show reduced atherosclerosis and decreased cell proliferation in response to vascular injury (93,94). These results need to be confirmed and are still in process.
In a recent review, Erdmann et al. (45) compiled a listing of the known most proximal genes of the SNP that show peak association. This is shown in Figure 1. Several variants suggest inflammation is a risk for CAD, including the major histocompatibility complex variant and others shown in the diagram. A recent gene-tissue expression study showed expression of HLA-C (Major Histocompatibility Complex, Class I, C) in the coronary artery (95), which lent further support to a role for chronic inflammation in the development of atherosclerosis. This is in keeping with other experimental and clinical evidence collected over the years that signifies a role for inflammation (96). The recent clinical trial CANTOS (Canakinumab Anti-Inflammatory Thrombosis Outcomes Study) (96) confirmed that interleukin-1β is involved as 1 of the causative factors of coronary atherosclerosis.
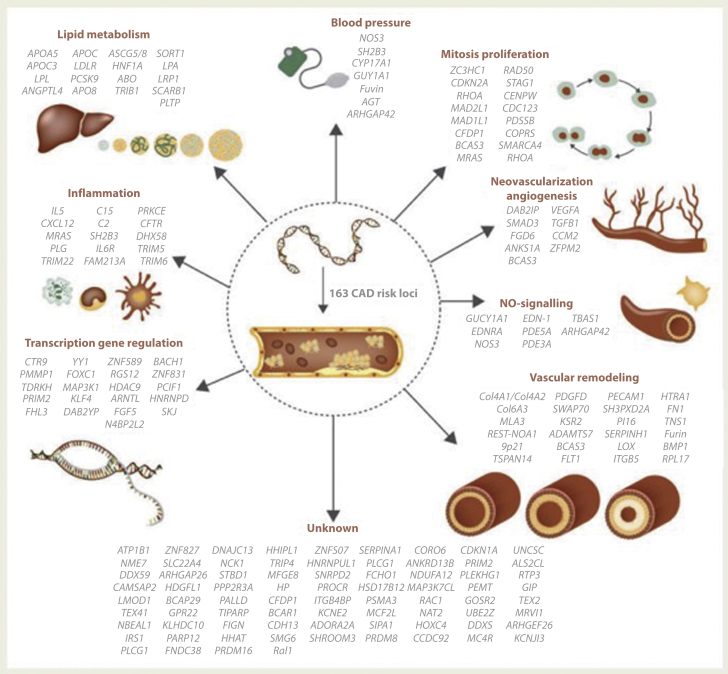
Figure 1.
Suggested Pathways for Involvement of Genetic Risk Variants
Various organs or pathways associated with 163 mapped genetic risk loci for CAD are shown in the figure. Reprinted with permission by Erdmann et al. (45). CAD = coronary artery disease.
The availability of multiple genetic risk variants has enabled MR to be used to determine whether associations are causative (97). Using genetic variants that increase plasma HDL cholesterol as the variable in MR studies, we cast considerable doubt on the dogma that HDL is protective of CAD (98). This was confirmed in several other studies (99, 100, 101, 102, 103, 104). In contrast, MR studies that used genetic variants that influenced plasma triglyceride levels or plasma LDL-C strongly indicated that they were causative for CAD (98). Several favorable and potential targets for the development of drugs to treat CAD and myocardial infarction were excluded by MR studies and included fibrinogen, folic acid, uric acid, and C-reactive protein (97). In selecting a target for the development of novel drugs, it is strongly recommended, when possible, to perform MR to confirm that the target is causative of the disease. The association of biomarkers with a disease does not necessarily reflect causality. A good example of this is plasma C-reactive protein, a marker for inflammation, that increases in concentration in the presence of inflammation. Inflammation is 1 of the causative factors for CAD, thus, increased C-reactive protein reflects the tendency for increased CAD. Nevertheless, this immune response has nothing to do with causing coronary atherosclerosis. Confirmation by MR of a biomarker or drug target as non-causative can save millions of dollars and years spent on a large clinical trial. A confounding factor for MR may be pleiotropy, in which the genetic variant affects multiple traits (97); nevertheless, some techniques can minimize limitations (e.g., the latent causal variable model) (105).
Development of a GRS
Perhaps the most well-known approach to modeling the contribution of common genetic variations predisposed to CAD is risk stratification based on the GRS. In brief, the CAD GRS is derived from a list of common genetic variants that have risk-weighted associations with CAD. The weight of each risk variant is multiplied by the number of variants at that site (0, 1, or 2), and the final score is simply the sum of the weighted dosage for each risk variant included in the GRS. This sum is typically represented as a natural log for the purposes of distributing individuals into percentiles for risk stratification.
The potential use of CAD GRS for clinical risk stratification has fueled interest in the methods behind GRS development (50,106,107). Although there is no standardized procedure for developing a CAD GRS for clinical use, establishing a CAD GRS for clinical use can be broken down into 3 basic steps: 1) selection of the CAD risk variants to be included in the microarray; 2) genotyping the individual’s DNA using the microarray; and 3) formation of an informatics pipeline to generate the GRS.
The CAD GRS can be estimated from variants using weights for CAD previously established by GWASs. Recently discovered genetic risk variants can be added by imputation. As discussed earlier, the most recent CAD GWAS brought the number of CAD risk variants with genome-wide significance to 171 (45). Previous studies described the application of CAD GRS derived from these variants (106,108). Recently, methods were used to develop the GRS based on millions of variants by relaxing the thresholds for genome-wide significance and by using software that accounted for linkage disequilibrium and estimated the proportion of SNPs with non-zero weights (109). One such genome-wide CAD GRS included 6.6 million variants and demonstrated superior performance to the CAD GRS that used less CAD risk variants, although it was genome-wide significant (5 × 10−8) (109). In both cases, the risk variants, along with their respective weights derived from GWASs, are readily available (108, 109, 110).
A detailed description of the procedure for generating a novel CAD GRS from a CAD GWAS has been previously described (111). However, as CAD GRS moves closer toward the reality of clinical implementation, it is reasonable to expect that a standardized list of variants, shown to demonstrate the best performance for the intended use, will be established and made readily available. However, before the clinical implementation of CAD GRS, a number of issues must be addressed; namely, that the current CAD GRS showed variable performance when applied to populations of non-European ancestry. This issue of disparities in accuracy of the CAD GRS is largely due to most of the risk variants being derived from GWASs performed in populations of predominantly European descent and the lack of well-powered studies conducted in globally diverse populations (112). Further work is needed to ensure that progress made in CAD GRS development does not increase disparities in the treatment of CAD.
Once the method of score generation has been determined, a procedure for genotyping patients and then processing that information into a GRS must be developed. If the CAD GRS being calculated has a limited number of risk variants, such as a CAD GRS that contains 50 SNPs, then calculating the GRS is simple. However, using the 6.6 million SNP genome-wide CAD GRS will require the use of larger microarrays and the use of software that can perform genotype imputation. Briefly, genotype imputation is a computational procedure that uses statistical inference to approximate the identity of variants not covered by the array being used. A number of software programs exist for genotype imputation. A few of the commonly used ones include IMPUTE2, BEAGLE, and Minimac 4 (113). The choice of software will depend upon the needs of the study, because each program will differ slightly in the time it takes to process each sample, the central processing unit it requires, and its average imputation quality.
For the purposes of setting up a CAD GRS, it will be necessary to ensure sufficient imputation accuracy of all variants to be included in the GRS, which will depend on the variants included in the array, the imputation software selected, and the reference panel used by the imputation software. The reference panel enables imputation software to infer the identity of genetic variants untyped by the array used to genotype the patient (16,27). Older reference panels, such as those from the HapMap project or 1000 Genome Project, have struggled to generate acceptable imputation accuracy in ethnically diverse samples, particularly those of African ancestry (114,115). However, the newest reference panel developed by the TOPMed (Trans-Omics for Precision Medicine) group was derived from 60,039 individuals of diverse ancestry who underwent deep whole-genome sequencing and demonstrated marked progress in this area (115). Notably, the TOPMed reference panel demonstrated an improvement of 0.26 to 0.92 in the average imputation quality in African ancestry genomes (115). These improvements mark an important step toward closing the gap in the variable performance of CAD GRS in multiethnic populations.
Once a patient’s sample has been received and genotyped, the genetic data will need to move through an informatics pipeline that can process the data into a CAD GRS. In short, from start to finish, a pipeline for a genome-wide CAD GRS consists of: 1) genotyping of the sample; 2) imputation; and 3) score calculation. During the set up of the pipeline, a series of quality control checks need to be performed to ensure that results are accurate and reliable. Because the anticipated use of a CAD GRS for risk stratification, further work will also be necessary to evaluate the validity of the CAD GRS before it can be brought to the clinic (116). Standardization of CAD GRS development and delivery will need to occur before clinical implementation.
Validation of a GRS
Development of a GRS was 1 of the goals derived from the GWAS search for risk variants predisposed to CAD. Risk stratification based on genetic risk was not age-dependent and could be obtained as early as at birth. This would be a major advantage for primary prevention of CAD as discussed previously (see the section on Need for Genetic Risk Stratification). The initial efforts using only the 9p21 risk variant were discouraging (117). Predictability of cardiac events became more promising as more risk variants became available, and the total burden of genetic risk for CAD was shown to be proportional to the number of risk variants (34). A GRS based on 12 risk variants predisposed to CAD (118) was encouraging, although the benefit compared with conventional risks factors was clinically insignificant. The sustained discovery effort provided many more DNA risk variants, which enabled the development of a much improved GRS.
This was tested by Mega et al. (106) in 2015 using a large study with a sample size of 48,421 individuals. This clinical trial consisted of 2 primary prevention trials (JUPITER [Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin] and ASCOT [Anglo-Scandinavian Cardiac Outcomes Trial]) and 2 secondary prevention trials (CARE [Cholesterol and Recurrent Events] and PROVE-IT-TIMI [the Pravastatin or Atorvastatin Evaluation and Infection Therapy–Thrombolysis in Myocardial Infarction]). These trials were completed and showed that statin therapy was effective for primary and secondary prevention (106). Baseline blood samples were available and were genotyped for 27 genetic risk variants predisposed to CAD. The GRS predicted cardiac events independent of traditional risk factors, including family history. Furthermore, it predicted who would benefit most from statin therapy. The number needed to treat to prevent 1 cardiac event was decreased 3-fold in the primary prevention trials. The risk gradient across the GRS was significant: to prevent 1 cardiac event in 10 years, 66 subjects with low genetic risk needed to be treated, 42 subjects in those with intermediate risk needed to be treated, and 25 subjects with high genetic risk needed to be treated in JUPITER, and 57, 47, and 20 subjects needed to be treated, respectively, in ASCOT (106). Thus, the GRS not only provided risk stratification regarding prognosis but also detected those who benefitted most from the proven preventive drug for CAD, namely, statins (119). In a similar randomized clinical trial termed WOSCOPS (West of Scotland Coronary Prevention Study) (120), 57 genetic risk variants were genotyped in a sample size of 10,456 individuals. The results showed that the high genetic risk group had a reduction of cardiac events of 44% versus 24% in the remaining groups. The number needed to treat to prevent 1 cardiac event in the high-risk group was 13 subjects versus 38 subjects in all other groups. These results confirmed the greater power of genetic risk stratification other than that of traditional risk factors. It is unusual for a single biomarker to have both predictive and prognostic properties (121).
Using a microarray that contained 49,310 risk variants derived from CARDIoGRAMplusC4D consortium, Abraham et al. (122) risk stratified 5 prospective cohorts, 3 from the FINRISK (a large Finnish population survey on risk factors on chronic, noncommunicable diseases) group and 2 from the Framingham Heart Study for a combined sample size of 16,082 subjects. The GRS was strongly associated with incident cardiac events and essentially independent of conventional risk factors, including family history. Integration of the GRS with the Framingham Risk Score or the ACC/AHA Pooled Cohort Equations improved the 10-year risk prediction for CAD events.
Abraham et al. (122) showed 49,310 risk variants had greater power to risk stratify CAD then the previous efforts that used 27 or 57 genetic risk variants predisposed to CAD. We have been consistently reminded that the genetic risk variants of known genome-wide significance only account for a small fraction of the expected inherited predisposition for CAD. If a less statistically stringent correction was used, namely, the false discovery rate, hundreds of other genetic risk variants could be associated with predisposition for CAD. Abraham et al. (122) used 49,310 genetic risk variants and accounted for 27% of inherited predisposition, a much larger proportion than using the genome-wide significant risk variants predisposed to CAD. There was a concern from the beginning that requiring 5 • 10−8, as proposed by the Bonferroni correction, might be too stringent. This led to the use of other less stringent statistical correction methods to select genetic risk variants predisposed to CAD.
Inouye et al. (123) developed a microarray with 1.7 million SNPs, which represented risk variants of genome-wide significance and others with a false discovery rate of <5%. They genotyped a population of 500,000 from the UK Biobank and observed that the top 20% with the highest GRS had a 4-fold increased risk of CAD. The GRS was relatively independent of conventional risk factors. The microarray of 1.7 million SNPs was superior to the previous microarray, which contained 49,310 variants.
Khera et al. (124) used a computerized algorithm, LDpred (109) to select 6.6 million risk variants predisposed to CAD. A pruning analysis was performed to assure all risk variants were in linkage disequilibrium to avoid redundancy (125). The total number of genetic risk variants contributing to predisposition of CAD might well be only a few thousand. However, the statistical programs were such that weakly associated genetic risk variants contributed to the GRS, whereas those that contributed zero risk did not dilute the predictive power (125). Khera et al. (124) genotyped a population of 288,978 from the UK Biobank and observed the top 8% with the highest GRS had a 3-fold increased risk of CAD. The top 0.5% had a 5-fold increased risk for CAD. Phenotypic analysis showed conventional risk factors would not detect most individuals with a high genetic risk for CAD. There were only 20% with hypercholesterolemia, 35% with a family history, and 28% with hypertension. They concluded, as did previous studies, that the GRS was superior to and relatively independent of conventional risk factors.
Although multiple studies, as discussed, have shown the GRS to be superior to conventional risk factors for risk stratification of CAD, there are 2 studies with contrasting results. Both studies were performed with a microarray using >6 million genetic risk variants predisposed to CAD. Mosley et al. (126) genotyped 2 American populations of European descent in the ARIC (Atherosclerosis Risk In Community) (n = 4,847) and the MESA (Multi-Ethnic Study of Atherosclerosis) (n = 2,390) cohorts. They found a close association in the ARIC cohort between the GRS and coronary heart disease, with an odds ratio of 1.89. Those in the top 20% had a 2.89 increased risk for coronary heart disease versus the bottom 80% percentile. Analysis of incident cardiac events in the follow-up of ARIC and MESA showed good correlation, with genetic risk in the top 20%, but the odds ratios were less (1.54 and 1.63, respectively). Results of the risk for CAD estimated from the Pooled Cohort Equations added to the GRS did not significantly improve prediction of cardiac events or change the C-statistic in either the ARIC or MESA population. The second study by Elliott et al. (127) genotyped a cohort of 352,660 individuals from the UK Biobank. The GRS showed a strong association with coronary heart disease, but when added to the result of the Pooled Cohort Equations, showed only a modest improvement in the C statistic, from 0.76 to 0.78. Although this was statistically significant, it was probably not of great clinical significance. The conclusion from both studies was that the GRS was equivalent to the risk score obtained from conventional risk factors but added no additional benefit in this group of individuals with a mean age of 60 years. It was difficult to reconcile these results with the multiple other studies that showed superiority of GRS over conventional risk factors. Nevertheless, Elliott et al. (127) recognized the GRS might offer an advantage in predicting CAD in younger age groups who had conventional risk factors have not yet been developed. This is the dilemma for early primary prevention of CAD. The authors agreed a study should be performed to assess the predictive value of GRS in this younger age group.
Two recent trials that assessed the effect of PCSK9 inhibitors further confirmed the predictability of the GRS for CAD events. The FOURIER (Further Cardiovascular Outcomes Research With PCSK9 Inhibition in Subjects With Elevated Risk) trial (128) assessed the benefit of a PCSK9 inhibitor, evolocumab, on cardiac events in a randomized, placebo-controlled trial of 11,953 patients who agreed to have their DNA analyzed. The microarray used in the study contained either 27 genetic risk variants predisposed to CAD or 6 million genetic risk variants. There was a strong correlation between individuals with intermediate and high genetic risk for CAD and cardiac events with 1.23 and 1.65 hazard ratios, respectively. Those who received evolocumab therapy had a 13% relative reduction in the group with conventional risk factors without high genetic risk and a 31% reduction in the high genetic risk group with or without conventional risk factors. Thus, the group with the highest genetic risk had the most benefit from lowering plasma LDL-C due to evolocumab and was independent of conventional risk factors. The microarray with 27 genetic risk variants predisposed to CAD was just as accurate for predicting cardiac events as the microarray with 1 million risk variants. The ODYSSEY (Evaluation of Cardiovascular Outcomes After an Acute Coronary Syndrome During Treatment With Alirocumab) trial (129), a randomized, placebo-controlled study, assessed the effect of alirocumab on cardiac events in 11,953 individuals. The group with the highest GRS had the highest risk for CAD and a 37% reduction in cardiac events by alirocumab versus a 13% reduction in the group with the lowest GRS. These results also indicated that the GRS was superior to conventional risk factor stratification in identifying those at greater risk who would benefit most from cholesterol-lowering medications.
Lifestyle Changes And Drug Therapy Reduce Genetic Risk For CAD
Tikkanen et al. (130) risk-stratified for CAD using a GRS to assess the effect of physical activity on the risk of cardiac events in a large sample size of 468,095 individuals from the UK Biobank. The physical activity performed was that of the handgrip for 3 s and cardiorespiratory fitness determined by oxygen consumption on a stationary bike. The highest genetic risk group, as determined by the GRS, had the most benefit from cardiorespiratory fitness, exhibiting a 49% lower risk for CAD.
A prospective study was performed by Khera et al. (108) involving a sample size of 55,685 individuals. Individuals with a high GRS for CAD (20%) had a 90% higher risk of cardiac events than those with a lower GRS. The group was risk-stratified to compare a healthy lifestyle (no current smoking, no obesity, regular physical activity, and a healthy diet) to that of an unhealthy lifestyle (at least 2 unfavorable features). The group with the highest GRS and a favorable lifestyle experienced 46% fewer cardiac events than those with an unfavorable lifestyle.
The GRS was evaluated in >1 million individuals. It was shown to be more predictive and prognostic than using traditional risk factors for CAD. Using the GRS showed that lifestyle changes, such as exercise, diet, and drugs such as statins, could markedly reduce genetic risk.
Limitations of GRS
The influence of ethnicity and regionalization
The global application of a GRS assumes the alleles-carrying risk variants have a similar frequency across different ethnic populations. This assumption is a concern because the genetic risk variants were discovered primarily in populations of European descent. A brief overview of human evolution supports a common ancestry for many disease-related variants, but not necessarily for all, particularly for ethnic groups such as Africans. It also provides the framework for adopting a cautious approach in other ethnic groups and the necessity for ongoing research.
In support of global applications is our common African ancestry, which has slowly evolved from the chimpanzee over approximately 6 million years. It is reasonable to assume the dynamics of genetic drift would provide an equilibrium of alleles across the populations. However, this was interrupted by the large migration out of Africa approximately 100,000 years ago. This provided a bottleneck to the ebb and flow of genetic drift as a proportion of the population migrated into East and South Asia, whereas others migrated to Southern and Northern Europe. On a global basis, separate alleles may be divided into ancestral (transmitted from other primates) and derived (due to new mutations) alleles. Ancestral alleles will be older, having evolved over >100,000 years ago, whereas derived alleles would be ≤100,000 years old. It is well known that certain bottlenecks can change disease-associated alleles, such as the increased risk for cystic fibrosis in Québécois (131) or increased cardiovascular disease in descendants of the HMS Bounty (132). Polygenic diseases such as CAD are due to multiple alleles with minimal effect sizes and the different frequencies at different loci would be expected to distribute toward the mean. Globally, a similar distribution of allele frequencies (133), with the possible expectation of a reduced genetic load in the African population (134), would be anticipated. Genetic drift is expected to be less in those who stayed in Africa because there is less pressure from natural selection compared with those who moved onto the challenge of adapting to a new environment (135).
It remains to be determined whether disease-associated alleles exhibit the same pattern of distribution and frequency as non-disease−related alleles. Disease-associated alleles may influence survival and natural selection. An analysis (136) showed 69.2% of ancestral risk alleles had a higher frequency in Africans, and 64.5% of derived alleles had a higher frequency in non-Africans. Notably, 44% of all risk alleles discovered are ancestral, meaning they originated >100,000 years ago. The mean frequency of ancestral risk alleles is 9.5% higher in African populations, whereas derived risk allele frequencies are 5.4% less in African populations. Analysis of the 1000 Genome Project showed the frequency of derived alleles are similar across the continents, with Africans more likely to be heterozygous for derived alleles and non-Africans more likely to be homozygous for derived alleles (136). In assessing a GRS for a disease, it may have some validity to separate the world’s population into African and non-African. Kim et al. (136) analyzed 306 GWAS loci for each continental population in the 1000 Genome Project. The highest risk alleles for disease were observed in Africans for metabolic, morphological, neoplastic, and neurological diseases. The African population had lower frequencies for risk alleles associated with cardiovascular disease. The first genetic risk variant discovered for CAD, 9p21 (30,31), transmitted an increased relative risk for CAD of approximately 25% per copy in Europeans. In contrast, 9p21 was not associated with any risk for CAD in the African population (35, 36, 37). The lower frequency of risk alleles for cardiovascular disease is at odds with the clinical observation of a high incidence and prevalence of CAD in Africans. Thus, as we go forward using genetic risk stratification for CAD and other cardiac disorders, it must be born in mind that the risk for CAD may be misestimated in individuals of African descent. Precise genetic risk stratification for diseases may require knowing what proportion of risk alleles is ancestral versus derived.
More than 3,000 GWAS have been performed in the past decade, and the ancestry of the participants were >80% Europeans, and 14% Asians (137). There are limited data on other ethnic groups. The 9p21 risk variant discovered in Europeans is confirmed to be a risk variant in other ethnic groups, including East and South Asians, but not in Africans. Genetic risk stratifying in Africans for CAD may need an enrichment of risk variants for CAD from the African population. The GRS will have to be assessed in the various ethnic groups to determine its overall accuracy.
Data evaluation and replication performed in the same population
Most of the GRSs have been developed with the test set and the validated set selected from the same population (64,65). Any confounding factors, such as ethnicity, have not been detected. It is more desirable for a score to be determined from a test set and validation in an independent population.
Comprehensive Risk Stratification Coupled To Comprehensive Therapy
The recent discovery of genetic risk variants has the potential to predict those at higher risk for ischemic heart disease. The genetic prediction has the same accuracy at birth as it does anytime throughout life. Genetic risk is innate to individuals’ DNA and does not change in a lifetime. It is possible in the future, we will discovery there are exceptions due to epigenetic changes, whereby expression of certain variants may be altered after birth. This makes it possible to risk stratify for CAD in young males, even as teenagers, to determine who would benefit most from primary preventative measures. Similarly, among the pre-menopausal female population, genetic risk stratification could be performed and therapy initiated before the onset of CAD. The schema indicated in Figure 2 would be 1 approach to detect those at greatest risk and who would be more likely to benefit from primary prevention. Use of the GRS and conventional risk stratification could revolutionize primary prevention in both men and women. It is worthy to note that the mechanisms mediating genetic risk are primarily unknown. Elucidating the mechanisms will lead to new drug targets and ultimately novel therapies specific for genetic risk. This will couple comprehensive risk stratification and comprehensive therapy for primary and secondary prevention of CAD.
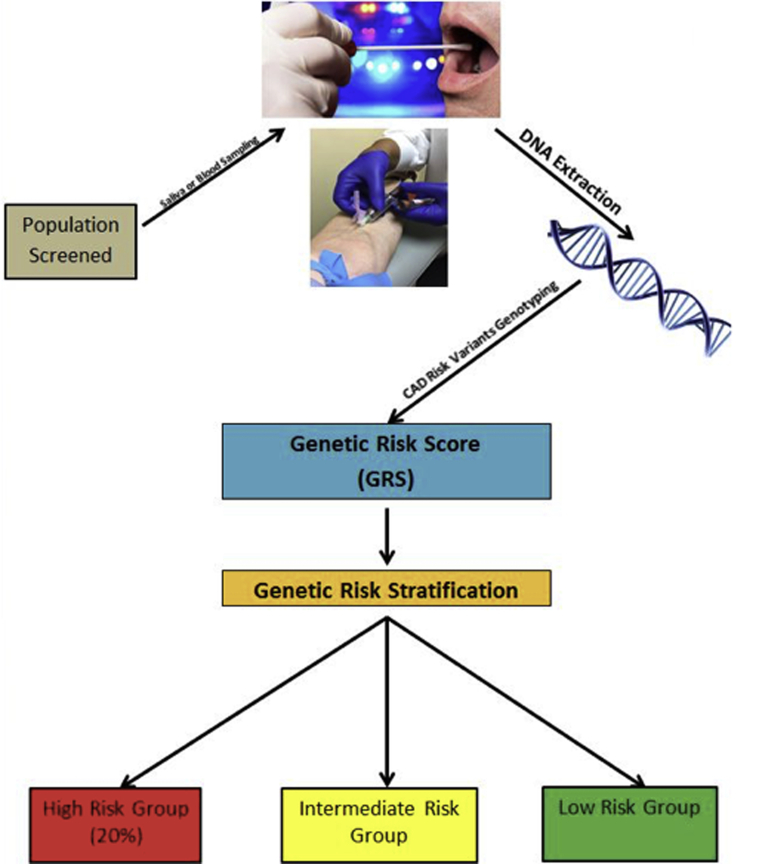
Figure 2.
Genetic Risk Stratification for CAD
A saliva or blood sample can be used to extract the DNA. The DNA is genotyped for risk variants known to predispose to CAD. The genetic risk score is calculated as a single number which is the basis to divide the population into low, intermediate, and high risk. The percentage is an estimate based on previous data. Abbreviation as in Figure 1.
A Future Prospective
CAD is the number 1 killer in the world and primary and secondary prevention have been shown to be effective. It is also known that approximately 50% of individuals in the United States who live out a normal lifespan will experience a cardiac event. It has been proven that lifestyle changes and cholesterol-lowering therapy are safe and effective as primary preventive measures. CAD originates early in life and slowly accumulates over the decades, with clinical manifestations such as myocardial infarction and sudden cardiac death peaking in the sixth decade in men and the seventh decade in women. To prevent these events, we must initiate prevention early in life. Conventional risk factors for CAD are often not present until the sixth or seventh decade and do not correlate with preclinical coronary atherosclerosis (138). In contrast, the GRS, which can be determined any time after birth, shows good correlation with preclinical CAD. The GRS is a very promising approach. There is much to be done to improve the GRS. We do not know the casual sequences, and the SNPs simply serve as proxies for the risk variant sequence. There are many more risk variants yet to be discovered. There are risk variants specific for ethnic groups, which can be imputed onto existing microarrays, as recently shown by Wang et al. (139) for South Asians.
The GRS must be pursued because it offers tremendous hope for a disease that exists throughout the world and can be prevented. Genetic testing and therapy are available everywhere and is inexpensive. Although there are arguments for and against the GRS being used as a clinical tool, we believe the time is now. Plasma cholesterol as a risk factor was recognized in the 1960s and received clinical application as total cholesterol. Clinical application refined it as a risk factor leading to the recognition of LDL-C, HDL-C, and triglycerides. We believe that current use of the GRS in a clinical setting will enhance its development, enable early preventive measures for the 10% to 20% at highest genetic risk, and initiate appropriate primary prevention in the pre-menopausal woman. Acquisition of more knowledge and experience will gradually include more of the population at intermediate risk for CAD.
FUNDING SUPPORT AND AUTHOR DISCLOSURES
The authors have received research funding from the Canadian Institutes of Health Research, CIHR no. MOP P82810, the Canada Foundation for Innovation, the CFI, no. 11966, and the Dignity Health Foundation. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
References
- 1.Kannel W.B., Dawber T.R., Kagan A., Revotskie N., Stokes J., III Factors of risk in the development of coronary heart disease—six-year follow-up experience: the Framingham study. Ann Internal Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 2.Gluckman T.J., Kovacs R.J., Stone N.J., Damalas D., Mullen J.B., Oetgen W.J. The ASCVD risk estimator app: from concept to the current state. J Am Coll Cardiol. 2016;67:350–352. doi: 10.1016/j.jacc.2015.10.068. [DOI] [PubMed] [Google Scholar]
- 3.The Multiple Risk Factor Intervention Trial Group Statistical design considerations in the NHLI multiple risk factor intervention trial (MRFIT) J Chronic Dis. 1977;30:261–275. doi: 10.1016/0021-9681(77)90013-3. [DOI] [PubMed] [Google Scholar]
- 4.Scandinavian Simvastatin Survival Study Group Randomised trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S) Lancet. 1994;344:1383–1389. [PubMed] [Google Scholar]
- 5.Shepherd J., Cobbe S.M., Ford I. Prevention of coronary heart disease with pravastatin in men with hypercholesterolemia. N Engl J Med. 1995;333:1301–1308. doi: 10.1056/NEJM199511163332001. [DOI] [PubMed] [Google Scholar]
- 6.Cholesterol Treatment Trialists’ (CTT) Collaboration. Baigent C., Blackwell L. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376:1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins R., Reith C., Emberson J. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet. 2016;388:2532–2561. doi: 10.1016/S0140-6736(16)31357-5. [DOI] [PubMed] [Google Scholar]
- 8.Lloyd-Jones D.M., Larson M.G., Beiser A., Levy D. Lifetime risk of developing coronary heart disease. Lancet. 1999;353:89–92. doi: 10.1016/S0140-6736(98)10279-9. [DOI] [PubMed] [Google Scholar]
- 9.American Heart Association . American Heart Association; Dallas, TX: 2000. Heart and Stroke Statistical Update. [Google Scholar]
- 10.Myers R.H., Kiely D.K., Cupples L.A., Kannel W.B. Parental history is an independent risk factor for coronary artery disease: the Framingham study. Am Heart J. 1990;120:963–969. doi: 10.1016/0002-8703(90)90216-k. [DOI] [PubMed] [Google Scholar]
- 11.Marenberg M.E., Risch N., Berkman L.F. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1141–1146. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 12.Chan L., Boerwinkle E. Gene-environment interactions and gene therapy in atherosclerosis. Cardiol Rev. 1994;2:130–137. [Google Scholar]
- 13.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 14.International HapMap Consortium. Frazer K.A., Ballinger D.G. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts R. New gains in understanding coronary artery disease, interview with Dr Robert Roberts. Affymetrix Microarray Bull. 2007;3:1–4. [Google Scholar]
- 16.Gibbs R., Belmont J., Hardenbol P. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Lander E., Linton L., Birren B. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Marian A.J., Brugada R., Roberts R. Chapter 55: Mendelian basis of congenital and other cardiovascular disease. In: Fuster V., Harrington R.A., Eapen Z.J., editors. Hurst’s The Heart. 14th Ed. McGraw Hill; New York: 2017. [Google Scholar]
- 19.Hejtmancik J.F., Brink P.A., Towbin J. Localization of gene for familial hypertrophic cardiomyopathy to chromosome 14q1 in a diverse US population. Circulation. 1991;83:1592–1597. doi: 10.1161/01.cir.83.5.1592. [DOI] [PubMed] [Google Scholar]
- 20.Jarcho J.A., McKenna W., Pare J.A. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 21.Gollob M.H., Green M.S., Tang A.S.-L. Identification of a gene responsible for familial Wolff–Parkinson–White syndrome. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 22.Centers for Mendelian Genomics Mendelian traits by the numbers. http://mendelian.org/mendelian-traits-numbers Available at: Accessed October 3, 2020.
- 23.Geisterfer-Lowrance A.A.T., Christe M., Conner D.A. A mouse model for familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 24.Sidhu J.S., Rajawat Y.S., Rami T.G. Transgenic mouse model of ventricular preexcitation and atrioventricular reentrant tachycardia induced by an AMP–activated protein kinase loss–of–function mutation responsible for Wolff–Parkinson–White syndrome. Circulation. 2005;111:21–29. doi: 10.1161/01.CIR.0000151291.32974.D5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 26.Altshuler D., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auton A., Abecasis G., Altshuler D. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein R.J., Zeiss C., Chew E.Y. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buniello A., MacArthur J.A.L., Cerezo M. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2018;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPherson R., Pertsemlidis A., Kavaslar N. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helgadottir A., Thorleifsson G., Manolescu A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 32.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samani N.J., Erdmann J., Hall A.S. Genome wide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Assimes T.L., Roberts R. Genetics: implications for prevention and management of coronary artery disease. J Am Coll Cardiol. 2016;68:2797–2818. doi: 10.1016/j.jacc.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 35.Barbalic M., Reiner A.P., Wu C. Genome-wide association analysis of incident coronary heart disease (CHD) in African Americans: a short report. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini N., Hu Y., Reiner A.P. Prospective associations of coronary heart disease loci in African Americans using the MetaboChip: the PAGE study. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lettre G., Palmer C.D., Young T. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe project. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myocardial Infarction Genetics Consortium. Kathiresan S., Voight B.F. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erdmann J., Grosshennig A., Braund P.S. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trégouët D.-A., König I.R., Erdmann J. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 41.Gudbjartsson D.F., Bjornsdottir U.S., Halapi E. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 42.The Coronary Artery Disease (C4D) Genetics Consortium. a genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 43.Preuss M., König I.R., Thompson J.R., CARDIoGRAM Consortium Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: a genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3:475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 45.Erdmann J., Kessler T., Munoz Venegas L., Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. 2018;114:1241–1257. doi: 10.1093/cvr/cvy084. [DOI] [PubMed] [Google Scholar]
- 46.CARDIoGRAMplusC4D Consortium. Deloukas P., Kanoni S. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson C., Goel A., Butterworth A. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 48.Navar-Boggan A.M., Peterson E.D., D’Agostino R.B.S., Neely B., Sniderman A.D., Pencina M.J. Hyperlipidemia in early adulthood increases long-term risk of coronary heart disease. Circulation. 2015;131:451–458. doi: 10.1161/CIRCULATIONAHA.114.012477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swiger K.J., Martin S.S., Blaha M.J. Narrowing sex differences in lipoprotein cholesterol subclasses following mid-life: the very large database of lipids (VLDL-10B) J Am Heart Assoc. 2014;3 doi: 10.1161/JAHA.114.000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 51.American College of Cardiology, American Heart Association ASCVD Risk Estimator. https://tools.acc.org/ldl/ascvd_risk_estimator/index.html#!/calulate/estimator Available at:
- 52.Ference B.A., Majeed F., Penumetcha R., Flack J.M., Brook R.D. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65:1552–1561. doi: 10.1016/j.jacc.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ference B.A., Graham I., Tokgozoglu L., Catapano A.L. Impact of lipids on cardiovascular health. J Am Coll Cardiol. 2018;72:1141. doi: 10.1016/j.jacc.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 54.Roberts R. PCSK9 inhibition–a new thrust in the prevention of heart disease: genetics does it again. Can J Cardiol. 2013;29:899–901. doi: 10.1016/j.cjca.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 55.Brown M.S., Goldstein J.L. Familial hypercholesterolemia: a genetic defect in the low-density lipoprotein receptor. N Engl J Med. 1976;294:1386–1390. doi: 10.1056/NEJM197606172942509. [DOI] [PubMed] [Google Scholar]
- 56.Abifadel M., Varret M., Rabès J.P. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 57.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 58.Stein E.A., Mellis S., Yancopoulos G.D. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 59.Köster A., Chao Y.B., Mosior M. Transgenic angiopoietin-like (ANGPTL)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;146:4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- 60.Mitsuru S., Matsuda M., Hiroaki Y. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 61.Fujimoto K., Koishi R., Shimizugawa T., Ando Y. Angptl3-null mice show low plasma lipid concentrations by enhanced lipoprotein lipase activity. Exp Anim. 2006;55:27–34. doi: 10.1538/expanim.55.27. [DOI] [PubMed] [Google Scholar]
- 62.Dewey F.E., Gusarova V., Dunbar R.L. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Athyros V.G., Katsiki N., Dimakopoulou A., Patoulias D., Alataki S., Doumas M. Drugs that mimic the effect of gene mutations for the prevention or the treatment of atherosclerotic disease: from PCSK9 inhibition to ANGPTL3 inactivation. Curr Pharm Des. 2018;24:3638–3646. doi: 10.2174/1381612824666181009100517. [DOI] [PubMed] [Google Scholar]
- 64.Raal F.J., Rosenson R.S., Reeskamp L.F. Evinacumab for homozygous familial hypercholesterolemia. N Engl J Med. 2020;383:711–720. doi: 10.1056/NEJMoa2004215. [DOI] [PubMed] [Google Scholar]
- 65.Gusev A., Ko A., Shi H. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Z., Zhang F., Hu H. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 67.Jarinova O., Stewart A.F.R., Roberts R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 68.Holdt L.M., Beutner F., Scholz M. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 69.Pasmant E., Laurendeau I., Héron D., Vidaud M., Vidaud D., Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 70.Leeper N.J., Raiesdana A., Kojima Y. Loss of CDKN2B promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. Arterioscler Thromb Vasc Biol. 2013;33:e1–e10. doi: 10.1161/ATVBAHA.112.300399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pilbrow A.P., Folkersen L., Pearson J.F. The chromosome 9p21.3 coronary heart disease risk allele is associated with altered gene expression in normal heart and vascular tissues. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J.B., Deluna A., Mungrue I.N. Effect of 9p21.3 coronary artery disease locus neighboring genes on atherosclerosis in mice. Circulation. 2012;126:1896–1906. doi: 10.1161/CIRCULATIONAHA.111.064881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Folkersen L., Kyriakou T., Goel A. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamsten A., Eriksson P. Quest for genes and mechanisms linking the human chromosome 9p21.3 locus to cardiovascular disease. Circulation. 2012;126:1815–1817. doi: 10.1161/CIRCULATIONAHA.112.136556. [DOI] [PubMed] [Google Scholar]
- 75.Visel A., Zhu Y., May D. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harismendy O., Notani D., Song X. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Almontashiri N.A.M., Fan M., Cheng B.L.M., Chen H.-H., Roberts R., Stewart A.F.R. Interferon-γ activates expression of p15 and p16 regardless of 9p21.3 coronary artery disease risk genotype. J Am Coll Cardiol. 2013;61:143–147. doi: 10.1016/j.jacc.2012.08.1020. [DOI] [PubMed] [Google Scholar]
- 78.Erridge C., Gracey J., Braund P.S., Samani N.J. The 9p21 locus does not affect risk of coronary artery disease through induction of type 1 interferons. J Am Coll Cardiol. 2013;62:1376–1381. doi: 10.1016/j.jacc.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 79.Lo Sardo V., Chubukov P., Ferguson W. Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell. 2018;175:1796–1810.e20. doi: 10.1016/j.cell.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Helgadottir A., Thorleifsson G., Magnusson K.P. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 81.Dandona S., Stewart A.F.R., Chen L. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010;56:479–486. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 82.Reilly M.P., Li M., He J. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ardissino D., Berzuini C., Merlini P.A. Influence of 9p21.3 genetic variants on clinical and angiographic outcomes in early-onset myocardial infarction. J Am Coll Cardiol. 2011;58:426–434. doi: 10.1016/j.jacc.2010.11.075. [DOI] [PubMed] [Google Scholar]
- 84.Chan K., Patel R.S., Newcombe P. Association between the chromosome 9p21 locus and angiographic coronary artery disease burden: a collaborative meta-analysis. J Am Coll Cardiol. 2013;61:957–970. doi: 10.1016/j.jacc.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Musunuru K., Strong A., Frank-Kamenetsky M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X., Raghavan A., Peters D.T., Pashos E.E., Rader D.J., Musunuru K. Interrogation of the atherosclerosis-associated SORT1 (Sortilin 1) locus with primary human hepatocytes, induced pluripotent stem cell-hepatocytes, and locus-humanized mice. Arterioscler Thromb Vasc Biol. 2018;38:76–82. doi: 10.1161/ATVBAHA.117.310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kjolby M., Andersen O.M., Breiderhoff T. Sort1, Encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 2010;12:213–223. doi: 10.1016/j.cmet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 88.Strong A., Ding Q., Edmondson A.C. Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J Clin Invest. 2012;122:2807–2816. doi: 10.1172/JCI63563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gustafsen C., Kjolby M., Nyegaard M. The hypercholesterolemia-risk gene SORT1 facilitates PCSK9 secretion. Cell Metab. 2014;19:310–318. doi: 10.1016/j.cmet.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Beaudoin M., Gupta R.M., Won H.-H. Myocardial infarction-associated SNP at 6p24 interferes with MEF2 binding and associates with PHACTR1 expression levels in human coronary arteries. Arterioscler Thromb Vasc Biol. 2015;35:1472–1479. doi: 10.1161/ATVBAHA.115.305534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta R.M., Hadaya J., Trehan A. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell. 2017;170:522–533.e15. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pu X., Xiao Q., Kiechl S. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am J Human Genet. 2013;92:366–374. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kessler T., Zhang L., Liu Z. ADAMTS-7 Inhibits re-endothelialization of injured arteries and promotes vascular remodeling through cleavage of thrombospondin-1. Circulation. 2015;131:1191–1201. doi: 10.1161/CIRCULATIONAHA.114.014072. [DOI] [PubMed] [Google Scholar]
- 94.Bauer R.C., Tohyama J., Cui J. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation. 2015;131:1202–1213. doi: 10.1161/CIRCULATIONAHA.114.012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gamazon E.R., Segrè A.V., van de Bunt M. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J Am Coll Cardiol. 2017;70:2278–2289. doi: 10.1016/j.jacc.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roberts R. Mendelian randomization studies promise to shorten the journey to FDA approval. J Am Coll Cardiol Basic Trans Science. 2018;3:690–703. doi: 10.1016/j.jacbts.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Voight B.F., Peloso G.M., Orho-Melander M. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lincoff A.M., Nicholls S.J., Riesmeyer J.S. Evacetrapib and cardiovascular outcomes in high risk vascular disease. N Engl J Med. 2017;376:1933–1942. doi: 10.1056/NEJMoa1609581. [DOI] [PubMed] [Google Scholar]
- 100.Kawashiri M.A., Tada H., Nomura A., Yamagishi M. Mendelian randomization: its impact on cardiovascular disease. J Cardiol. 2018;72:307–313. doi: 10.1016/j.jjcc.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 101.Barter P.J., Caulfield M., Eriksson M. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 102.Schwartz G.G., Olsson A.G., Abt M. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 103.HPS3/TIMI55-REVEAL Collaborative Group. Bowman L., Hopewell J.C., Chen F. Effects of anacetrapib in patients with atherosclerotic vascular disease. N Engl J Med. 2017;377:1217–1227. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 104.Nomura A., Won H.H., Khera A.V. Protein truncating variants at the cholesteryl ester transfer protein gene and risk for coronary heart disease. Circ Res. 2017;121:81–88. doi: 10.1161/CIRCRESAHA.117.311145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Connor L.J., Price A.L. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet. 2018;50:1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mega J.L., Stitziel N.O., Smith J.G. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roberts R., Campillo A., Schmitt M. Prediction and management of CAD risk based on genetic stratification. Trends Cardiovasc Med. 2020;30:328–334. doi: 10.1016/j.tcm.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 108.Khera A.V., Emdin C.A., Drake I. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Khera A.V., Chaffin M., Aragam K.G. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Khera A.V., Chaffin M., Zekavat S.M. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation. 2019;139:1593–1602. doi: 10.1161/CIRCULATIONAHA.118.035658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chatterjee N., Shi J., García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schurz H., Müller S.J., van Helden P.D. Evaluating the accuracy of imputation methods in a five-way admixed population. Front Genet. 2019;10:34. doi: 10.3389/fgene.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hancock D.B., Levy J.L., Gaddis N.C. Assessment of genotype imputation performance using 1000 Genomes in African American studies. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Taliun D., Harris D.N., Kessler M.D. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. bioRxiv. 2019:563866. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Janssens A.C.J.W. Validity of polygenic risk scores: are we measuring what we think we are? Human Mol Genet. 2019;28:R143−50. doi: 10.1093/hmg/ddz205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paynter N.P., Chasman D.I., Buring J.E., Shiffman D., Cook N.R., Ridker P.M. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Davies R.W., Dandona S., Stewart A.F. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ Cardiovasc Genet. 2010;3:468–474. doi: 10.1161/CIRCGENETICS.110.946269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Težak Ž., Kondratovich M.V., Mansfield E. US FDA and personalized medicine: in vitro diagnostic regulatory perspective. Per Med. 2010;7:517–530. doi: 10.2217/pme.10.53. [DOI] [PubMed] [Google Scholar]
- 120.Natarajan P., Young R., Stitziel N.O. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Davies C., Godwin J. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abraham G., Havulinna A.S., Bhalala O.G. Genomic prediction of coronary heart disease. Eur Heart J. 2016;37:3267–3278. doi: 10.1093/eurheartj/ehw450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Inouye M., Abraham G., Nelson C.P. Genomic risk prediction of coronary artery disease in 480,000 adults. J Am Coll Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vilhjálmsson B.J., Yang J., Finucane H.K. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y., Qi G., Park J.-H., Chatterjee N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat Genet. 2018;50:1318–1326. doi: 10.1038/s41588-018-0193-x. [DOI] [PubMed] [Google Scholar]
- 126.Mosley J.D., Gupta D.K., Tan J. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323:627–635. doi: 10.1001/jama.2019.21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Elliott J., Bodinier B., Bond T.A. Predictive accuracy of a polygenic risk score–enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323:636–645. doi: 10.1001/jama.2019.22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marston N.A., Kamanu F.K., Nordio F. Predicting benefit from evolocumab therapy in patients with atherosclerotic disease using a genetic risk score. Circulation. 2020;141:616–623. doi: 10.1161/CIRCULATIONAHA.119.043805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Damask A., Gabriel S.P., Schwartz G.G. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES trial. Circulation. 2020;141:624–636. doi: 10.1161/CIRCULATIONAHA.119.044434. [DOI] [PubMed] [Google Scholar]
- 130.Tikkanen E., Gustafsson S., Ingelsson E. Associations of fitness, physical activity, strength, and genetic risk with cardiovascular disease: longitudinal analyses in the UK Biobank study. Circulation. 2018;137:2583–2591. doi: 10.1161/CIRCULATIONAHA.117.032432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Laberge A.-M., Michaud J., Richter A. Population history and its impact on medical genetics in Quebec. Clin Genet. 2005;68:287–301. doi: 10.1111/j.1399-0004.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 132.Benton M.C., Stuart S., Bellis C. “Mutiny on the Bounty”: the genetic history of Norfolk Island reveals extreme gender-biased admixture. Investig Genet. 2015;6:11. doi: 10.1186/s13323-015-0028-9. [published correction appears in Investig Genet 2015;6:12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lohmueller K.E. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Devel. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 134.Henn B.M., Botigue L.R., Peischl S. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci U S A. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 2005;102:15942. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim M.S., Patel K.P., Teng A.K., Berens A.J., Lachance J. Genetic disease risks can be misestimated across global populations. Genome Biol. 2018;19:179. doi: 10.1186/s13059-018-1561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baber U., Mehran R., Sartori S., Schoos M.M., Sillesen H., Muntendam P. Prevalence, impact, and predictive value of detecting subclinical coronary and carotid atherosclerosis in asymptomatic adults: the BioImage study. J Am Coll Card. 2015;65:1065–1074. doi: 10.1016/j.jacc.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 139.Wang M., Menon R., Mishra S. Validation of a genome-wide polygenic score for coronary artery disease in South Asians. J Am Coll Cardiol. 2020;76:703–714. doi: 10.1016/j.jacc.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davies R.W., Wells G.A., Stewart A.F. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ Cardiovasc Genet. 2012;5:217–225. doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lu X., Wang L., Chen S. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang F., Xu C.-Q., He Q. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 143.Hirokawa M., Morita H., Tajima T. A genome-wide association study identifies PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for myocardial infarction in Japanese. Eur J Hum Genet. 2015;23:374–380. doi: 10.1038/ejhg.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]