Abstract
Background & Aims
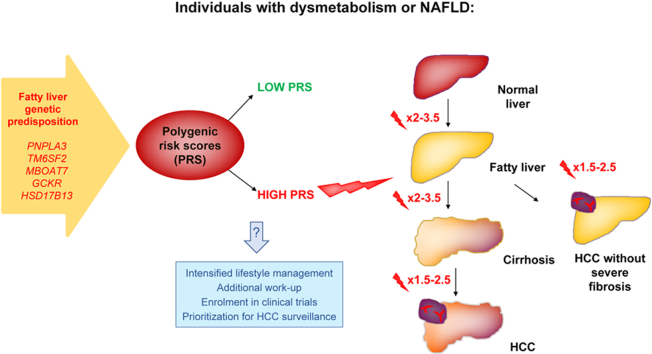
Hepatocellular carcinoma (HCC) risk stratification in individuals with dysmetabolism is a major unmet need. Genetic predisposition contributes to non-alcoholic fatty liver disease (NAFLD). We aimed to exploit robust polygenic risk scores (PRS) that can be evaluated in the clinic to gain insight into the causal relationship between NAFLD and HCC, and to improve HCC risk stratification.
Methods
We examined at-risk individuals (NAFLD cohort, n = 2,566; 226 with HCC; and a replication cohort of 427 German patients with NAFLD) and the general population (UK Biobank [UKBB] cohort, n = 364,048; 202 with HCC). Variants in PNPLA3-TM6SF2-GCKR-MBOAT7 were combined in a hepatic fat PRS (PRS-HFC), and then adjusted for HSD17B13 (PRS-5).
Results
In the NAFLD cohort, the adjusted impact of genetic risk variants on HCC was proportional to the predisposition to fatty liver (p = 0.002) with some heterogeneity in the effect. PRS predicted HCC more robustly than single variants (p <10-13). The association between PRS and HCC was mainly mediated through severe fibrosis, but was independent of fibrosis in clinically relevant subgroups, and was also observed in those without severe fibrosis (p <0.05). In the UKBB cohort, PRS predicted HCC independently of classical risk factors and cirrhosis (p <10-7). In the NAFLD cohort, we identified high PRS cut-offs (≥0.532/0.495 for PRS-HFC/PRS-5) that in the UKBB cohort detected HCC with ~90% specificity but limited sensitivity; PRS predicted HCC both in individuals with (p <10-5) and without cirrhosis (p <0.05).
Conclusions
Our results are consistent with a causal relationship between hepatic fat and HCC. PRS improved the accuracy of HCC detection and may help stratify HCC risk in individuals with dysmetabolism, including those without severe liver fibrosis. Further studies are needed to validate our findings.
Lay summary
By analyzing variations in genes that contribute to fatty liver disease, we developed two risk scores to help predict liver cancer in individuals with obesity-related metabolic complications. These risk scores can be easily tested in the clinic. We showed that the risk scores helped to identify the risk of liver cancer both in high-risk individuals and in the general population.
Keywords: Biomarker, Cirrhosis, Genetics, Hepatic fat, Non-alcoholic fatty liver disease
Graphical abstract
Highlights
-
•
Genetic predisposition to liver fat accumulation predisposes to cirrhosis and HCC.
-
•
Hepatic fat promotes carcinogenesis, partly via fibrosis.
-
•
Polygenic risk scores may improve HCC risk stratification during dysmetabolism.
See Editorial, pages 769–770
Introduction
Hepatocellular carcinoma (HCC) is now the third leading cause of cancer-related mortality worldwide.1,2 The prevalence of non-alcoholic fatty liver disease (NAFLD; also known as metabolic dysfunction-associated fatty liver disease when associated with dysmetabolism)3 has increased in parallel with the growing burden of obesity and type 2 diabetes (T2D) to become a leading cause of HCC.[4], [5], [6], [7] About 21–33% of the general population is affected by NAFLD, and the proportion of individuals progressing to severe liver fibrosis and HCC is projected to increase in the near future.8 Current guidelines recommend HCC surveillance in patients with NAFLD and cirrhosis, and that it should be considered in those with advanced liver fibrosis.9 However, no reliable biomarker is yet available to stratify HCC risk in patients without severe fibrosis, accounting for a large fraction of HCC cases in individuals with dysmetabolism.9,10 The high prevalence of NAFLD and the evidence that HCC frequently arises in individuals unaware of their risk make classical HCC surveillance strategies impractical, resulting in delayed diagnosis and unfavourable prognosis.11 Thus, non-invasive biomarkers to identify patients at risk of NAFLD-related HCC onset are urgently needed.
Hepatic fat content has a strong inherited component.12 Variants in genes involved in the regulation of hepatic lipid metabolism, such as in patatin-like phospholipase domain-containing protein 3 (PNPLA3), transmembrane 6 superfamily member 2 (TM6SF2), membrane bound O-acyltransferase domain containing 7 (MBOAT7) and glucokinase regulator (GCKR), predispose to NAFLD, hepatic fibrosis, and HCC in the presence of environmental triggers.[12], [13], [14], [15], [16] Conversely, a splice variant in 17β-hydroxysteroid dehydrogenase type 13 (HSD17B13) prevents severe fibrosis and HCC development.17 We previously developed a robust polygenic risk score (PRS) of hepatic fat content (termed PRS-HFC) and showed that the impact of genetic risk variants on fibrosis is proportional to that on hepatic fat, consistent with hepatic fat accumulation being a driver of liver disease.18 Recently, Stender et al. confirmed that an unweighted PRS based on PNPLA3-TM6SF2-HSD17B13 predicted cirrhosis and HCC in Europeans.19
Here we hypothesized that liver fat promotes HCC in individuals with NAFLD and dysmetabolism. Mendelian randomization is considered the most appropriate epidemiological tool to assess causality when randomized controlled trials are not feasible. Therefore, we examined the impact of the previously developed PRS-HFC based on well-characterized risk variants that can be evaluated in the clinic on HCC in at-risk individuals and in the general population. We also performed a further adjustment for HSD17B13 (termed PRS-5). Next, we identified PRS thresholds able to identify with good specificity a subset of individuals with NAFLD and dysmetabolism at high risk of HCC. Finally, we showed that PRS predicted HCC irrespective of severe liver fibrosis.
Patients and methods
Study cohorts
The NAFLD case-control cross-sectional cohort included 1,699 unrelated individuals with NAFLD of European ancestry, who were evaluated from 2008 until 2019 at Italian and UK centres for suspected liver disease (from simple steatosis to severe fibrosis and HCC), or who underwent liver biopsy during bariatric surgery. Liver damage was assessed by histology, except when clear clinical or radiological signs of cirrhosis were detected (n = 342, 20.1%). Part of this cohort has previously been described.14,15,20,21 NAFLD,3 severe fibrosis,22 HCC,23 and selection of 865 controls24,25 were defined as reported in the supplementary methods. We also considered an independent NAFLD validation cohort of 427 German individuals. The demographic and clinical data of these individuals are shown in Table S1.
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, approved by the Fondazione IRCCS Ca’ Granda, and ratified by the participating centres. Informed written consent was obtained from each participant.
The UK Biobank (UKBB) has data (including baseline assessment, physical measures, biological samples, and genetic data) from >500,000 individuals recruited between 2006 and 2010 and aged 40–69 years26 (ukbiobank.ac.uk). During follow-up, information about health-related outcomes have been collected from national datasets. Data used in this study were obtained from the UKBB under Application Number 37142. Selection criteria and HCC and diabetes definitions are reported in the supplementary methods.27 Subsequently, individuals with concomitant viral hepatitis (ICD-10 code B15-B19) were excluded (n = 535, 0.15%) for a sensitivity analysis presented in the supplementary results. This left us with 364,048 individuals. Clinical features are shown in Table S1 and Table S2.
Genotyping
Study participants were genotyped for rs738409 (PNPLA3 I148M variant), rs58542926 (TM6SF2 E167K), rs641738 C>T MBOAT7, rs1260326 (GCKR P446L) and rs72613567 (HSD17B13:TA),12 as specified in Supplementary Methods.
Statistical analysis
For descriptive statistics, categorical variables are shown as number and proportion. Continuous variables are shown as mean (SD) or median (IQR), as appropriate.
Observational associations were performed by fitting data to generalized linear models. Logistic regression models were fit to examine binary traits, and the association between PRS and liver disease was adjusted for age, sex, BMI and T2D, with or without further adjustment for the presence of severe fibrosis stage (F3-F4).
To estimate the causal relationship between genetically determined predisposition to accumulate liver fat and HCC, we used the most established risk variants for hepatic fat content as instruments in a Mendelian randomization analysis, as reported in the supplementary methods.12,18,28 We used the PRS-HFC, a robust genetic instrument calculated by summing the number of steatosis-predisposing alleles in PNPLA3-TM6SF2-MBOAT7-GCKR weighted by their effect size on hepatic fat content, quantified by the reference standard in the general population.18 We next developed a modified NAFLD score adjusted for the rs72613567 HSD17B13 variant17 (PRS-5: available in 2,532, 98.7%, coefficient: -0.361). We reported the association of both instruments with phenotypes throughout the study, as PRS-HFC is a proxy for genetic predisposition to accumulate liver fat, while PRS-5 considers all variants robustly associated with NAFLD at the time of study planning.12
The causal effect of genetic predisposition to NAFLD on HCC was estimated by instrumental variable regression analysis (using the AER package in R), with NAFLD as an explanatory variable, and HCC as the outcome.29 To further account for the possible pleiotropy of the genetic variants considered, we also considered, in sensitivity analyses, robust Mendelian randomization approaches by the MendelianRandomization R package.30
The main goal was to determine the thresholds in the PRS able to identify individuals at higher genetic risk of HCC. Diagnostic accuracy of PRS was evaluated by receiver-operating characteristic curves, and the best cut-off identified as the point with maximum (sensitivity+specificity-1).
Statistical analysis was carried out using the JMP Pro 14.0 Statistical Analysis Software (SAS Institute, Cary, NC), and R statistical analysis software version 3.5.2 (http://www.R-project.org/). p values <0.05 (two tailed) were considered significant.
Results
Causal relationship between hepatic fat and HCC in the NAFLD cohort
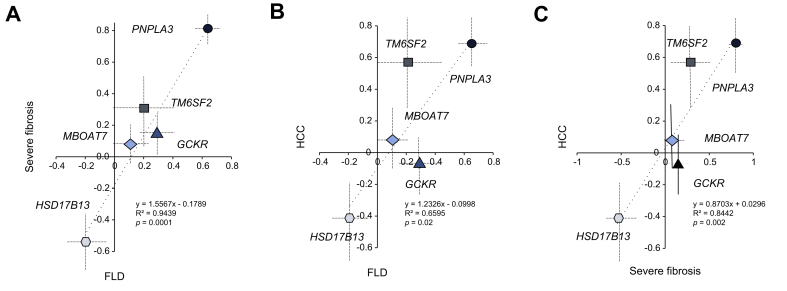
We first examined the relationship between the impact of genetic risk variants on NAFLD, severe liver fibrosis and HCC (Fig. 1). The increase in the risk of HCC conferred by risk variants was proportional to the increase in the risk of NAFLD (p = 0.02). There was a direct relationship between the risk conferred to fatty liver disease (FLD) and severe fibrosis (p = 0.0001), and between severe fibrosis and HCC (p = 0.002).
Fig. 1.
Correlation between the impact of genetic risk variants in PNPLA3, TM6SF2, MBOAT7, GCKR, and HSD17B13 on the risk of FLD, severe fibrosis and HCC in the NAFLD cohort.
Correlations coefficients and 95% CIs at generalized linear regression models are reported.(A) Correlation between the impact on the risk of FLD and severe fibrosis; (B) correlation between the impact on the risk of FLD and HCC; (C) correlation between the impact on the risk of severe fibrosis and HCC. p values were determined at generalized linear regression analysis. FLD, fatty liver disease; GCKR, glucokinase regulator; HCC, hepatocellular carcinoma; HSD17B13, 17β-hydroxysteroid dehydrogenase type 13; MBOAT7, membrane bound O-acyltransferase domain containing 7; NAFLD, non-alcoholic fatty liver disease; PNPLA3, patatin-like phospholipase domain-containing protein 3; TM6SF2, transmembrane 6 superfamily member 2.
Instrumental variable regression adjusted for age, sex, BMI and T2D showed that NAFLD was causally associated with HCC (beta +0.30 ± 0.06, odds ratio (OR) 1.35, 1.18–1.58, p = 1∗10-5 for PRS-HFC; beta +0.29 ± 0.07, OR 1.27, 1.10–1.45, p = 1∗10-5 for PRS-5). The association coefficient was attenuated by 37–41%, but remained statistically significant, after further correction for severe liver fibrosis (p <0.05). Formal mediation analysis is reported in the supplementary results.
Estimation of causality by a range of modern Mendelian randomization approaches, taking into account the possible pleiotropic effects of the genetic instruments (e.g. a direct impact on HCC not mediated by NAFLD), and other sensitivity analyses were generally consistent with a causal effect of NAFLD on HCC, and are reported in the supplementary results, Fig. S1 and Table S3.
PRS-HFC and PRS-5 predict the full spectrum of NAFLD
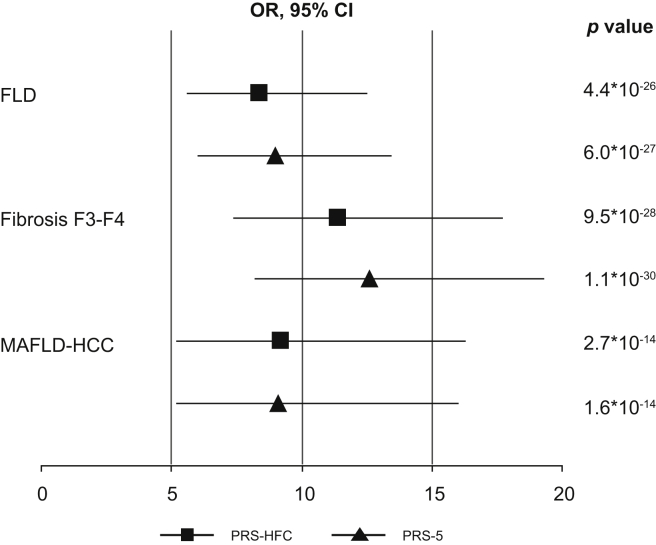
The impact of PRS on the full spectrum of liver disease in the NAFLD cohort is reported in Fig. 2 and Table 1 (upper panel). PRS were associated with a ~12-fold increased OR of severe fibrosis (p <10-27 for both) and a ~9-fold increased OR of HCC (OR 9.2, 5.2–16.3, p = 2.7∗10-14 for PRS-HFC; and OR 9.1, 5.2–16.0, p = 1.6∗10-14 for PRS-5). The association was independent of age, sex, BMI and T2D (p <0.01 for both), but not of severe fibrosis (p >0.1). In the NAFLD cohort, there was no significant interaction between PRS and BMI, T2D or HOMA-IR (homeostasis model assessment of insulin resistance) in determining HCC risk (p >0.1). Similar results were obtained in the German NAFLD cohort (see supplementary results).
Fig. 2.
Impact of the PRS-HFC and the PRS-5 on the full spectrum of FLD in the NAFLD cohort, as evaluated by logistic regression analysis.
FLD, fatty liver disease; HCC, hepatocellular carcinoma; HFC, hepatic fat content; MAFLD, metabolic associated fatty liver disease; NAFLD, non-alcoholic fatty liver disease; OR, odds ratio (at logistic regression analysis); PRS, polygenic risk score.
Table 1.
PRS-HFC and PRS-5 are independent predictors of liver disease in the NAFLD and in the UKBB cohorts.
| Univariate analysis |
Model 1∗ |
Model 2∗∗ |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| p value | OR | 95% CI | p value | OR | 95% CI | p value | OR | 95% CI | |
| NAFLD cohort | |||||||||
| PRS- HFC | |||||||||
| FLD | 4.4∗10–26 | 8.4 | 5.7–12.5 | 9.2∗10–21 | 10.1 | 6.2–16.5 | – | – | – |
| Fibrosis F3-F4 | 9.5∗10–28 | 11.4 | 7.4–17.7 | 7.0∗10–13 | 7.5 | 4.3–13.0 | – | – | – |
| HCC | 2.7∗10–14 | 9.2 | 5.2–16.3 | 3.6∗10–3 | 3.0 | 1.4–6.4 | 1.5∗10–1 | 1.8 | 0.8–3.9 |
| PRS-5 | |||||||||
| FLD | 6.0∗10–27 | 9.0 | 6.0–13.4 | 1.5∗10–21 | 10.7 | 6.6–17.3 | – | – | – |
| Fibrosis F3-F4 | 1.1∗10–30 | 12.6 | 8.2–19.3 | 1.0∗10–15 | 9.4 | 5.4–16.2 | – | – | – |
| HCC | 1.6∗10–14 | 9.1 | 5.2–16.0 | 1.7∗10–3 | 3.3 | 1.6–6.9 | 1.3∗10–1 | 1.8 | 0.8–4.1 |
| UKBB cohort | |||||||||
| PRS-HFC | |||||||||
| Cirrhosis | 7.3∗10–32 | 4.1 | 3.2–5.1 | 6.4∗10–33 | 4.2 | 3.3–5.3 | – | – | – |
| HCC | 4.8∗10–15 | 11.1 | 6.1–20.4 | 5.0∗10–15 | 11.1 | 6.1–20.2 | 1.5∗10–6 | 4.6 | 2.5–8.6 |
| PRS-5 | |||||||||
| Cirrhosis | 1.4∗10–36 | 4.4 | 3.5–5.6 | 3.3∗10–37 | 4.5 | 3.6–5.7 | – | – | |
| HCC | 8.6∗10–17 | 11.9 | 6.6–21.3 | 1.2∗10–16 | 11.7 | 6.54–21 | 5.9∗10–7 | 4.8 | 2.6–8.9 |
At logistic regression adjusted for age, sex, BMI, T2D, and further adjusted for ethnicity (PC1:10), array batch, assessment centre in the UKBB cohort.
Further adjusted for the presence of severe fibrosis (stage F3-F4) in the NAFLD cohort or diagnosis of cirrhosis in the UKBB cohort. HCC, hepatocellular carcinoma; NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; PRS-HFC, polygenic risk score of hepatic fat content considering variants in PNPLA3-TM6SF2-MBOAT7-GCKR; PRS-5, polygenic risk score considering 5 risk variants (further adjusted for HSD17B13 variation); T2D, type 2 diabetes.
Similarly, PRS were associated with cirrhosis and HCC in the UKBB cohort, both in the overall cohort and in individuals without chronic viral hepatitis (Table 1, bottom panel). In this latter group, PRS were associated with >15-fold increased OR of HCC (OR 15.3, 8.1–28.7, p = 2.6∗10-17 for PRS-HFC; and OR 15.9, 8.6–29.1, p = 4.4∗10-19 for PRS-5). The association between PRS and HCC remained significant after adjustment for cirrhosis and was more robust in individuals without chronic viral hepatitis (OR 6.6, 3.4–12.7, p = 1.7∗10-8 for PRS-HFC; and OR 6.9, 3.6–13.1, p = 4.7∗10-9 for PRS-5). These data are consistent with a causative effect of genetic predisposition to hepatic fat accumulation on carcinogenesis, which is partially mediated by severe fibrosis.
Diagnostic accuracy of PRS for HCC and identification of thresholds associated with high genetic risk
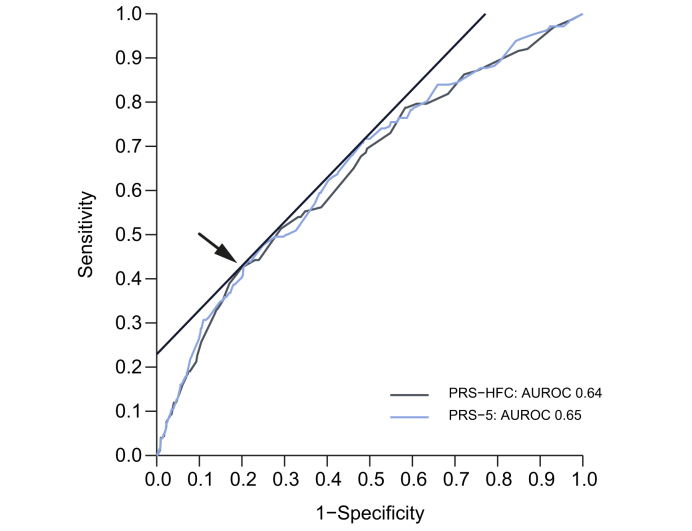
In the NAFLD cohort, the area under the receiver-operating characteristic curve (AUROC) for HCC was 0.64 for PRS-HFC and 0.65 for PRS-5 (Table 2). A value of ≥0.532 was identified as the best single cut-off for PRS-HFC, with 43% sensitivity and 80% specificity. For PRS-5, the corresponding cut-off (43% sensitivity and 79% specificity) was ≥0.495 (Table 2 and Fig. 3). Hereinafter, we define PRS-HFC ≥0.532 and PRS-5 ≥0.495 as ‘positive’ tests. In the UKBB cohort, both PRS had the same AUROC (0.63), and high scores had a lower sensitivity but higher specificity than in the NAFLD cohort (27% and 90%, respectively) (Table 3).
Table 2.
Diagnostic accuracy of PRS-HFC (n = 2,564) and PRS-5 (n = 2,245) for HCC in the NAFLD cohort.
| PRS-HFC | PRS-5 | |
|---|---|---|
| AUROC | 0.64 | 0.65 |
| Diagnostic threshold | 0.532 | 0.495 |
| Prevalence (%) | 569 (22.2) | 580 (22.9) |
| OR (95% CI) | 3.0 (2.2–3.9) | 2.9 (2.1–3.8) |
| p value∗ | 3.7∗10–14 | 8.1∗10–13 |
| Sensitivity (95% CI) | 0.43 (0.37–0.49) | 0.43 (0.37–0.50) |
| Specificity (95% CI) | 0.80 (0.78–0.81) | 0.79 (0.77–0.81) |
| PPV (95% CI) | 0.17 (0.14–0.20) | 0.16 (0.13–0.19) |
| NPV (95% CI) | 0.93 (0.92–0.94) | 0.94 (0.93–0.95) |
| LR+ (95% CI) | 2.13 (1.79–2.52) | 2.06 (1.74–2.54) |
| LR- (95% CI) | 0.71 (0.64–0.80) | 0.72 (0.64–0.81) |
At logistic regression analysis. HCC, hepatocellular carcinoma; LR+, positive likelihood ratio; LR-, negative likelihood ratio; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value; PRS-HFC, polygenic risk score of hepatic fat content considering variants in PNPLA3-TM6SF2-MBOAT7-GCKR; PRS-5, polygenic risk score considering 5 risk variants (further adjusted for HSD17B13 variation).
Fig. 3.
Comparison of the diagnostic accuracy of the PRS-HFC and the PRS-5 for HCC in the NAFLD cohort.
The AUROC of the 2 PRS to predict HCC and the optimal diagnostic thresholds are shown. HCC, hepatocellular carcinoma; HFC, hepatic fat content; NAFLD, non-alcoholic fatty liver disease; PRS, polygenic risk score.
Table 3.
Diagnostic accuracy of PRS-HFC ≥0.532 and PRS-5 ≥0.495 for HCC in the UKBB cohort (overall, in non-cirrhotic individuals and in participants stratified by the presence of obesity or T2D).
| Overall | No cirrhosis | BMI ≥30 | T2D | |
|---|---|---|---|---|
| PRS-HFC ≥0.532 | ||||
| Cases, n | 198 | 95 | 88 | 81 |
| PRS-HFC median (IQR) cases | 0.337 (0.128–0.595) | 0.266 (0.128–0.394) | 0.4 (0.192–0.604) | 0.394 (0.240–0.603) |
| Controls, n | 358,126 | 356,725 | 86,116 | 25,051 |
| PRS-HFC median (IQR) controls | 0.193 (0.126–0.394) | 0.193 (0.126–0.394) | 0.193 (0.126–0.394) | 0.193 (0.126–0.394) |
| AUROC (PRS-HFC) | 0.63 | 0.55 | 0.69 | 0.70 |
| Positive PRS prevalence (%) | 35,734 (11.1) | 35,458 (11.0) | 8,497 (10.9) | 2,741 (12.2) |
| OR (95% CI) | 3.3 (2.4–4.5) | 1.8 (1.1–3.1) | 5.2 (3.4–8.1) | 4.4 (2.7–6.9) |
| p value∗ | 1.0∗10–13 | 2.7∗10–2 | 8.3∗10–14 | 3.6∗10–10 |
| Sensitivity, % | 27% | 17% | 36% | 35% |
| Specificity, % | 90% | 90% | 90% | 89% |
| PPV | 0.01 | 0.01 | 0.01 | 0.01 |
| NPV | 1.00 | 1.00 | 1.00 | 1.00 |
| LR+ | 2.69 | 1.70 | 3.70 | 3.19 |
| LR- | 0.81 | 0.92 | 0.71 | 0.73 |
| PRS-5 ≥0.495 | ||||
| Cases, n | 197 | 95 | 87 | 80 |
| PRS-5 median (IQR) cases | 0.292 (0.126–0.524) | 0.193 (0.063–0.394) | 0.394 (0.161–0.597) | 0.394 (0.193–0.587) |
| Controls N | 356,746 | 355,355 | 85,803 | 24,959 |
| PRS-5 median (IQR) controls | 0.174 (0.063–0.337) | 0.174 (0.063–0.337) | 0.167 (0.063–0.337) | 0.191 (0.063–0.337) |
| AUROC (PRS-5) | 0.63 | 0.54 | 0.69 | 0.71 |
| Positive PRS prevalence (%) | 34,673 (10.8) | 34,405 (10.7) | 8,217 (10.6) | 2,644 (11.8) |
| OR (95% CI) | 3.4 (2.5–4.7) | 1.9 (1.1–3.2) | 5.5 (3.6–8.5) | 4.6 (2.9–7.3) |
| p value∗ | 1.9∗10–14 | 2.0∗10–2 | 1.7∗10–14 | 8.9∗10–11 |
| Sensitivity, % | 27% | 17% | 37% | 35% |
| Specificity, % | 90% | 90% | 90% | 90% |
| PPV | 0.01 | 0.01 | 0.01 | 0.01 |
| NPV | 1.00 | 1.00 | 1.00 | 1.00 |
| LR+ | 2.77 | 1.74 | 3.86 | 3.34 |
| LR- | 0.81 | 0.92 | 0.70 | 0.73 |
At logistic regression analysis. Polygenic risk scores values are reported as median (IQR). HCC, hepatocellular carcinoma; LR+, positive likelihood ratio; LR-, negative likelihood ratio; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value; PRS-HFC, polygenic risk score of hepatic fat content considering variants in PNPLA3-TM6SF2-MBOAT7-GCKR; PRS-5, polygenic risk score considering 5 risk variants (further adjusted for HSD17B13 variation); T2D, type 2 diabetes.
Both PRS were able to predict HCC more robustly than single variants, with PRS-5 conferring a slight improvement over PRS-HFC (Table S4). Positive PRS tests were associated with an ~3-fold increased HCC risk both in the NAFLD (p <10-12) and the UKBB (p ≤10-13) cohorts, reaching almost a 4-fold increased risk (p <10-14) for non-viral HCC (Table 2, Table 3); in the UKBB cohort, the association was independent of cirrhosis (p <10-5 in the overall cohort and p <10-7 in non-viral cohort; Table S4).
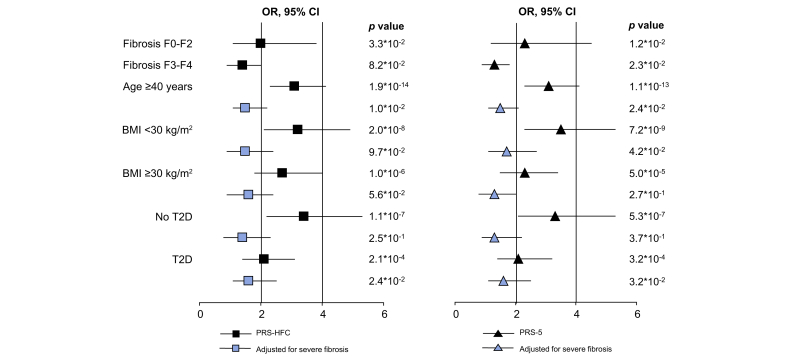
In the NAFLD cohort, the prevalence of positive tests was 22.2% for PRS-HFC and 22.9% for PRS-5. Positive PRS were associated with a 3-fold higher risk of HCC (Table 2; p <10-12 for both). We next tested whether the association of HCC-positive tests was independent of severe fibrosis. PRS tests also predicted HCC in patients without severe fibrosis (OR >2.0, 1.1–3.8, p = 3.3∗10-2 for PRS-HFC; and OR 2.3, 1.2–4.5, p = 1.2∗10-2 for PRS-5; Table S5), and improved HCC detection in individuals aged over 40 years independently of severe fibrosis (OR 1.5, 1.1–2.2, p = 1.0∗10-2 for PRS-HFC; and OR 1.5, 1.1–2.1, p = 2.4∗10-2 for PRS-5). Additional sensitivity analyses and validation in the German cohort are presented in Table S5 and the supplementary results (Fig. 4).
Fig. 4.
Association of PRS-HFC ≥0.532 and PRS-5 ≥0.495 with HCC in individuals included in the NAFLD cohort at logistic regression analysis.
Stratified by the presence of the main risk factors (fibrosis severity, age, BMI, T2D). HCC, hepatocellular carcinoma; HFC, hepatic fat content; NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; PRS, polygenic risk score; T2D, type 2 diabetes.
In the UKBB cohort (see Table 3, Tables S6 and S7), the prevalence of positive tests was 11.1% for PRS-HFC and 10.8% for PRS-5. Positive PRS tests had a 27% sensitivity and 90% specificity and were associated with a ~2-fold higher OR of HCC in individuals without cirrhosis (p <0.05) and with a ~3-fold higher OR in those with cirrhosis (p <10-5). Importantly, they showed higher performance in individuals with obesity/T2D (OR 5.2/4.4, respectively; Table 3). The diagnostic accuracy of positive PRS tests in other clinically relevant subgroups is reported in the supplementary results, but it was higher in individuals with obesity and/or T2D (~40% sensitivity and 90% specificity). This observation is consistent with the presence of a significant interaction between PRS and BMI, probably mediated by FLD in determining HCC in individuals without chronic viral hepatitis (p = 0.02 and p = 0.012 for PRS-HFC and PRS-5, respectively).
Discussion
In this study, we used genetic instruments to a) examine whether weighted PRS, reflecting the genetic predisposition to accumulate liver fat and develop NAFLD, predict HCC development in at-risk individuals and in the general population, and b) evaluate the causal relationship between NAFLD and HCC. Next, we determined the diagnostic accuracy of PRS thresholds to indicate increased genetic risk of HCC. Despite limitations related to the potential heterogeneity of the metabolic and carcinogenic effects of the genetic variants, results were generally consistent with the presence of a causal link between hepatic fat accumulation and HCC. Upon confirmation by further studies, this relationship would suggest that approaches/drugs aiming to reduce liver fat may contribute to prevent HCC.[4], [5], [6], [7],31 As we exploited simple yet very robust PRS based on a limited number of validated risk variants that can be used irrespective of the presence of liver disease in individuals with dysmetabolism, these data may provide a new instrument that, upon refinement and further testing, may guide a cost-effective surveillance for HCC.
We focused our analyses on individuals with dysmetabolism and specifically those with NAFLD or metabolic risk factors but no severe liver fibrosis. This is because a) they represent a large fraction of the general population and b) there is no accurate biomarker to predict HCC in these individuals. We did not exclude those with moderate alcohol intake in the general population because of the difficulties in assessing average alcohol intake and the synergic effect of alcohol with dysmetabolism in the pathogenesis of FLD. In individuals with dysmetabolism, who are mostly unaware that they have liver disease, HCC surveillance is not currently performed because of the lack of cost-effective approaches. Therefore, we identified PRS thresholds, which showed a high specificity for HCC, allowing the identification of a subset of individuals who may benefit from further refinement of HCC risk stratification by the combined used of other biomarkers and imaging approaches. Furthermore, these individuals at high genetic risk may benefit from lifestyle and pharmacological approaches to halt liver disease progression.
The first main study finding was that genetic predisposition to liver fat accumulation results in an increased HCC risk, consistently with causation, both in at-risk individuals with NAFLD and in the general population, where the effect of PRS on HCC risk was larger in those with dysmetabolism. An association consistent with a causal relationship between genetic predisposition to hepatic fat and HCC was also confirmed in the NAFLD cohort by modern robust Mendelian randomization approaches that take into account the heterogeneity of the effects of the genetic instruments, e.g. the possibility that they promote liver cancer independently of the impact on liver fat.30 The only exception to the direct correlation between the risk of NAFLD and that of HCC was related to GCKR variation, which decreases T2D risk, and may therefore have led to an underestimation of the causal relationship between hepatic fat and HCC.12 Secondly, we showed that the association between genetic predisposition to hepatic fat accumulation and HCC was partly, but not completely, mediated by the promotion of severe fibrosis. This result is consistent with the clinical observation that HCC occurs in individuals without severe liver fibrosis and suggests that liver fat accumulation directly favours hepatic carcinogenesis. The significant interaction between PRS and adiposity on HCC risk at the population level is also consistent with a causal association of NAFLD with HCC.30 However, the present analysis was not able to discriminate whether quantitative or qualitative changes in liver fat content or lipotoxicity predispose to HCC.
Despite the robust statistical association, and the fact that PRS predicted HCC more accurately than single variants reported in clinical guidelines,32 as expected PRS alone had a moderate accuracy to predict HCC (AUROC 0.65 in the NAFLD cohort, 0.70 in individuals with T2D in the UKBB cohort). However, PRS are easily determined by a simple, once-in-a-lifetime blood test, are independent of fluctuations of environmental triggers, and predict the future development of cirrhosis, which frequently precedes HCC. Therefore, we reasoned that if we could find a threshold to identify HCC with a high predictive value, this may be the first step towards a cost-effective HCC surveillance to a subset of individuals with inborn and acquired risk for this disease. We therefore selected the best PRS thresholds (i.e. ≥0.532/0.495 for PRS-HFC/PRS-5) to predict HCC in the NAFLD cohort. Positive tests were associated with a >3-fold higher adjusted OR of HCC, as confirmed in an independent German NAFLD cohort and in the UKBB cohort. In particular, positive PRS tests conferred a >5-fold higher risk in obese individuals from the general population. In the UKBB cohort, ~11% of individuals had high PRS, which despite limited sensitivity had a good specificity for detection of non-viral HCC (~90%). In individuals with dysmetabolism from the general population, a clinically relevant setting where the prevalence of HCC was relatively low, PRS reached a higher accuracy to detect HCC (OR >4.4). The improved sensitivity (~40%, positive likelihood ratio ~3.7-3.9) is consistent with the synergic effect between obesity and genetic predisposition in determining NAFLD.33
PRS predicted HCC independently of the presence of severe fibrosis in the NAFLD cohort and of cirrhosis in the UKBB cohort, especially in younger patients and in those with T2D. Therefore, more accurate evaluation of liver damage and possibly HCC surveillance may be recommended for individuals with dysmetabolism older than 40 years. Although selecting a subset at risk based on a high PRS will not detect a large fraction of HCC cases in individuals without clinical evidence of cirrhosis, it would still represent a step forward compared to the current absence of any surveillance in this group. Alternatively, PRS could be integrated with classical risk factors and other biomarkers for repeated evaluations of HCC risk in individuals with dysmetabolism, but the relative cost-effectiveness of these strategies remains to be determined.
Previous studies from our group evaluated the role of PRS in the prediction of HCC.15,16,18 Recently, Gellert-Kristensen et al. tested an unweighted PRS that confirmed a robust association with the risk of HCC in both the Danish general population and the UKBB, based on a smaller number of cases than in the present study.19 Our study is novel in several ways. First, we focused on HCC risk and weighted the impact of a larger panel of variants on FLD, likely providing a more accurate estimate and being able to make inferences on the causal relationship with HCC. Second, we tested the impact of PRS in individuals at high risk of liver disease, stratifying the analyses by liver fibrosis severity. Third, we tested the diagnostic accuracy of PRS in a clinically relevant subgroup of individuals from the general population, in particular those without cirrhosis and with obesity.
Limitations of the present study include the cross-sectional design of the NAFLD cohort, although it included a larger number of HCC cases than previous studies. Individuals without severe fibrosis and controls were younger than those with more advanced disease or HCC, which limited the power to detect and underestimated the impact of genetic factors, as it cannot be ruled out that some participants with positive PRS will progress to advanced liver disease and HCC. Prospective studies will therefore be necessary to confirm the magnitude of the increase in HCC risk conferred by PRS. Importantly, we recently showed that high PRS-HFC predicted HCC incidence in a prospective cohort of patients with cirrhosis in whom HCV had been eradicated by antiviral drugs; the association was independent of classical risk factors including fibrosis severity, improving risk stratification.34 Notably, the best PRS-HFC threshold to discriminate higher HCC risk was superimposable to that identified in the present study in the NAFLD cohort. Lastly, other inherited genetic determinants of the risk of liver damage in the general population may modify HCC risk (e.g. HFE and SERPINA1 mutations for Europeans),35 so that their inclusion in updated PRS may further improve their accuracy, and results may not apply entirely to non-European populations.
In conclusion, and with the limitations highlighted in the discussion, the results of the present study are consistent with a causal role of hepatic fat accumulation in hepatic carcinogenesis. PRS may be useful to non-invasively predict the risk of HCC in individuals with NAFLD and dysmetabolism, independently of severe liver fibrosis, and positive PRS identify a subset of individuals with dysmetabolism at high genetic risk of HCC. Large studies integrating genetic and other biomarkers are necessary to further improve the risk stratification and facilitate the clinical implementation of these findings.
Abbreviations
AUROC, area under the receiver-operating characteristic curve; FLD, fatty liver disease; GCKR, glucokinase regulator; HCC, hepatocellular carcinoma; HFC, hepatic fat content; HSD17B13, 17β-hydroxysteroid dehydrogenase type 13; MBOAT7, membrane bound O-acyltransferase domain containing 7; NAFLD, non-alcoholic fatty liver disease; PC, principal component; PNPLA3, patatin-like phospholipase domain-containing protein 3; PRS, polygenic risk score; TM6SF2, transmembrane 6 superfamily member 2; T2D, type 2 diabetes; UKBB, UK Biobank.
Financial support
This work was supported by project grants from Amgen and sanofi-aventis, the Swedish Research Council [Vetenskapsrådet (VR), 2016-01527], the Swedish state under the agreement between the Swedish government and the county councils (the ALF-agreement) [SU 2018-04276], the Novo Nordisk Foundation Grant for Excellence in Endocrinology [Excellence Project, 9321-430], the Swedish Diabetes Foundation [DIA 2017-205], the Swedish Heart Lung Foundation [20120533], the Wallenberg Academy Fellows from the Knut and Alice Wallenberg Foundation [KAW 2017.0203] (SR); MyFirst Grant AIRC n.16888, Ricerca Finalizzata Ministero della Salute RF-2016-02364358, Ricerca corrente Fondazione IRCCS Ca’ Granda Ospedikale Maggiore Policlinico, the European Union (EU) Programme Horizon 2020 (under grant agreement No. 777377) for the project LITMUS- “Liver Investigation: Testing Marker Utility in Steatohepatitis”, Fondazione IRCCS Ca’ Granda “Liver BIBLE” PR-0391, Fondazione IRCCS Ca’ Granda core COVID-19 Biobank (RC100017A) to LV; European Community’s Seventh Framework Programme (FP7/2001-2013) under grant agreement HEALTH-F2-2009-241762 for the project FLIP. Cancer Research UK (CR UK) centre grant C9380/A18084; programme grant C18342/A23390 and Accelerator award C9380/A26813 to HR. The Swiss National Funds (SNF no. 310030_169196) and the Swiss Foundation for Alcohol Research (SSA) to FS.
Authors’ contributions
LV, SR, SP: study supervision, funding, concept and design, data interpretation; CB, OJ, LV: data analysis; CB, SP, SM, AL, FS, GH, JF, TB, HR, DP, LM, UVG, ALF, QMA: data collection; CB, LV: manuscript drafting; PD, GB, IZ, LS: samples processing and genotyping; VB, RDA, AA, AF, EB, SP.
Data availability statement
The data that support the study findings are available upon reasonable request from the corresponding authors [LV, SR]. The full data are not publicly available due to limitations posed by the informed consent and ethical regulations at some of the participating centres.
Conflict of interest
The authors declare that they have no conflict of interest relevant to the present study. SR has served as a consultant for AstraZeneca, Celgene, Sanofi, Amgen, Akcea Therapeutics, Camp4, AMbys, Medacorp and Pfizer in the past 5 years, and received research grants from AstraZeneca, Sanofi and Amgen. LV has received speaking fees from MSD, Gilead, AlfaSigma and AbbVie, served as a consultant for Gilead, Pfizer, Astra Zeneca, Novo Nordisk, Intercept, Diatech Pharmacogenetics and Ionis Pharmaceuticals, and received research grants from Gilead.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We thank Rosie Perkins, University of Goteborg, for editing the final version of the manuscript.
Footnotes
Author names in bold designate shared co-first authorship.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2020.11.024.
Contributor Information
Stefano Romeo, Email: stefano.romeo@wlab.gu.se.
Luca Valenti, Email: luca.valenti@unimi.it.
Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Canc. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Anstee Q.M., Reeves H.L., Kotsiliti E., Govaere O., Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 3.Eslam M., Newsome P.N., Anstee Q.M., Targher G., Gomez M.R., Zelber-Sagi S. A new definition for metabolic associated fatty liver disease: an international expert consensus statement. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.03.039. [DOI] [PubMed] [Google Scholar]
- 4.Younossi Z., Henry L. Contribution of alcoholic and nonalcoholic fatty liver disease to the burden of liver-related morbidity and mortality. Gastroenterology. 2016;150:1778–1785. doi: 10.1053/j.gastro.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Dyson J., Jaques B., Chattopadyhay D., Lochan R., Graham J., Das D. Hepatocellular cancer: the impact of obesity, type 2 diabetes and a multidisciplinary team. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Baffy G., Brunt E.M., Caldwell S.H. Hepatocellular carcinoma in non-alcoholic fatty liver disease: an emerging menace. J Hepatol. 2012;56:1384–1391. doi: 10.1016/j.jhep.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 7.Park J.W., Chen M., Colombo M., Roberts L.R., Schwartz M., Chen P.J. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int. 2015;35:2155–2166. doi: 10.1111/liv.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes C., Anstee Q.M., Arias-Loste M.T., Bantel H., Bellentani S., Caballeria J. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. J Hepatol. 2018;69:896–904. doi: 10.1016/j.jhep.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 9.Loomba R., Lim J.K., Patton H., El-Serag H.B. AGA clinical practice update on screening and surveillance for hepatocellular carcinoma in patients with nonalcoholic fatty liver disease: expert review. Gastroenterology. 2020 doi: 10.1053/j.gastro.2019.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piscaglia F., Svegliati-Baroni G., Barchetti A., Pecorelli A., Marinelli S., Tiribelli C. Clinical patterns of hepatocellular carcinoma in nonalcoholic fatty liver disease: a multicenter prospective study. Hepatology. 2016;63:827–838. doi: 10.1002/hep.28368. [DOI] [PubMed] [Google Scholar]
- 11.Younossi Z.M., Otgonsuren M., Henry L., Venkatesan C., Mishra A., Erario M. Association of nonalcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004 to 2009. Hepatology. 2015;62:1723–1730. doi: 10.1002/hep.28123. [DOI] [PubMed] [Google Scholar]
- 12.Trepo E., Valenti L. Update on NAFLD genetics: from new variants to the clinic. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Pelusi S., Valenti L. Hepatic fat as clinical outcome and therapeutic target for nonalcoholic fatty liver disease. Liver Int. 2019;39:250–256. doi: 10.1111/liv.13972. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y.L., Patman G.L., Leathart J.B., Piguet A.C., Burt A.D., Dufour J.F. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2013;61:75–81. doi: 10.1016/j.jhep.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 15.Pelusi S., Baselli G., Pietrelli A., Dongiovanni P., Donati B., McCain M.V. Rare pathogenic variants predispose to hepatocellular carcinoma in nonalcoholic fatty liver disease. Sci Rep. 2019;9:3682. doi: 10.1038/s41598-019-39998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donati B., Dongiovanni P., Romeo S., Meroni M., McCain M., Miele L. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7:4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abul-Husn N.S., Cheng X., Li A.H., Xin Y., Schurmann C., Stevis P. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dongiovanni P., Stender S., Pietrelli A., Mancina R.M., Cespiati A., Petta S. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med. 2018;283:356–370. doi: 10.1111/joim.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gellert-Kristensen H., Richardson T.G., Davey Smith G., Nordestgaard B.G., Tybjaerg-Hansen A., Stender S. Combined effect of PNPLA3, TM6SF2, and HSD17B13 variants on risk of cirrhosis and hepatocellular carcinoma in the general population. Hepatology. 2020 doi: 10.1002/hep.31238. [DOI] [PubMed] [Google Scholar]
- 20.Dongiovanni P., Petta S., Maglio C., Fracanzani A., Pipitone R.M., Mozzi E. TM6SF2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 21.Dongiovanni P., Meroni M., Baselli G.A., Mancina R.M., Ruscica M., Longo M. PCSK7 gene variation bridges atherogenic dyslipidemia with hepatic inflammation in NAFLD patients. J Lipid Res. 2019 doi: 10.1194/jlr.P090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddowes P.J., Sasso M., Allison M., Tsochatzis E., Anstee Q.M., Sheridan D. Accuracy of FibroScan controlled attenuation parameter and liver stiffness measurement in assessing steatosis and fibrosis in patients with nonalcoholic fatty liver disease. Gastroenterology. 2019;156:1717–1730. doi: 10.1053/j.gastro.2019.01.042. [DOI] [PubMed] [Google Scholar]
- 23.EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Bedogni G., Bellentani S., Miglioli L., Masutti F., Passalacqua M., Castiglione A. The Fatty Liver Index: a simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006;6:33. doi: 10.1186/1471-230X-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomes Project C., Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. Plos Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150 doi: 10.1053/j.gastro.2016.01.032. 1219-1230 e1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor D.A., Harbord R.M., Sterne J.A., Timpson N., Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 30.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romeo S., Sanyal A., Valenti L. Leveraging human genetics to identify potential new treatments for fatty liver disease. Cell Metab. 2020;31:35–45. doi: 10.1016/j.cmet.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 32.European Association for the Study of the Liver Electronic address eee, European association for the study of D, European association for the study of O. EASL-EASD-EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Stender S., Kozlitina J., Nordestgaard B.G., Tybjaerg-Hansen A., Hobbs H.H., Cohen J.C. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet. 2017;49:842–847. doi: 10.1038/ng.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degasperi E., Galmozzi E., Pelusi S., D'Ambrosio R., Soffredini R., Borghi M. Hepatic fat - genetic risk score predicts hepatocellular carcinoma in HCV cirrhotic patients treated with DAAs. Hepatology. 2020 doi: 10.1002/hep.31500. [DOI] [PubMed] [Google Scholar]
- 35.Valenti L. Uncovering the genetics of cirrhosis: new plots for the usual suspects. Liver Int. 2020;40:281–282. doi: 10.1111/liv.14333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the study findings are available upon reasonable request from the corresponding authors [LV, SR]. The full data are not publicly available due to limitations posed by the informed consent and ethical regulations at some of the participating centres.