Abstract
SERRATE (SE) plays critical roles in RNA metabolism and plant growth regulation. However, its function in stress–response processes remains largely unknown. Here, we examined the regulatory role of SE using the se-1 mutant and its complementation line under saline conditions. The expression of SE was repressed by salt treatment at both mRNA and protein levels. After treatment with different NaCl concentrations, the se-1 mutants showed increased sensitivity to salinity. This heightened sensitivity was evidenced by decreased germination, reduced root growth, more serious chlorosis, and increased conductivity of the mutants compared with the wild type. Further analysis revealed that SE regulates the pre-mRNA splicing of several well-characterized marker genes associated with salt stress tolerance. Our data thus imply that SE may function as a key component in plant response to salt stress by modulating the splicing of salt stress-associated genes.
Keywords: SERRATE, Salt stress, Pre-mRNA alternative splicing
1. Introduction
Plants live in fixed locations and are constantly threatened by various fluctuating abiotic environmental factors, such as salinity, drought, and low temperatures. These abiotic stresses greatly limit crop productivity worldwide, causing more than a 50% reduction of average yields for most major crop plants (Bray et al., 2000). Among these abiotic stresses, salt stress is one of the most serious threats to global agriculture and is becoming more and more widespread across the world. High salinity induces both osmotic and ionic stresses on crop plants, and thereby imposes a negative effect on their growth, development, and ultimately yields (Yang and Guo, 2018).
Plants have successfully evolved intricate self-protective mechanisms to adapt to salt stress conditions (Bohnert et al., 1995; Munns and Tester, 2008; Tuteja, 2007; Wang et al., 2003; Zhu, 2001). Previous studies have demonstrated that plant salt-tolerance mechanisms mainly comprise salt-stress sensing and signaling, pathways involving Na+ transport and detoxification, epigenetic chromatin modification, and accumulation of organic osmolytes (Deinlein et al., 2014). Studies have also indicated that both transcriptomic changes and alternative mRNA splicing play vital roles in mediating plant adaption to environmental stresses (Barbazuk et al., 2008; Hugouvieux et al., 2001; Kong et al., 2014; Kornblihtt et al., 2013; Zhu, 2001). Upon exposure to salt stress, plants must efficiently alter their physiological and biochemical processes to improve ionic and osmotic homeostasis, reduce oxidative damage, and adjust cell division and expansion to control plant growth under this particular condition (Zhu, 2001). Elucidation of the mechanisms underlying salt stress response will contribute to the genetic engineering of salt-tolerant plants.
Arabidopsis SERRATE (SE) encodes a zinc finger protein with 720 amino acids and is predicted to contain four domains, including a single C2H2-type zinc-finger domain near its C-terminus and multiple bipartite nuclear localization motifs at its N-terminus (Machida et al., 2011). SE controls leaf development, meristem activity, inflorescence architecture, and developmental phase transitions, and may also participate in modulation of gene expression via chromatin modification (Prigge and Wagner, 2001; Grigg et al., 2005). The SE mutant se-1 displays a pleiotropic phenotype and is hypersensitive to abscisic acid during germination (Prigge and Wagner, 2001; Bezerra et al., 2004). Studies have demonstrated that SE functions as a critical component of microRNA biogenesis and fine-tune primary-microRNA processing through physical interaction with HYPONASTIC LEAVES 1(HYL1) and CHR2 (Lobbes et al., 2006; Yang et al., 2006; Wang et al., 2018). SE was also shown to physically interact with cap-binding protein 80 (CBP80) and CBP20 (Laubinger et al., 2008; Raczynska et al., 2014), as well as Negative on TATA less2 (AtNot2) (Wang et al., 2013), to cooperatively modulate pre-mRNA alternative splicing. SE is thus thought to function as a critical component in both alternative splicing of pre-mRNA and biogenesis of microRNA. SE can further modulate the transcription of intronless genes (Speth et al., 2018). One recent study has also demonstrated that SE may function as the substrate of SnRK2 kinases to link miRNA biogenesis with ABA signaling (Yan et al., 2017). Although SE has been shown to participate in plant stress responses (Bezerra et al., 2004; Zhang et al., 2008), the exact molecular mechanism underlying its regulatory effect on these responses remains to be elucidated.
Here, we examined SE expression levels during salt stress and showed them to be repressed by high salinity at both mRNA and protein levels. Phenotype analysis showed that the se-1 mutant exhibited hypersensitivity to salt stress during almost all developmental stages. Further investigation indicated that SE directly affected the pre-mRNA splicing of several salt stress-associated genes, such as RD29A, RD29B, RD20, and ADH1. Our findings demonstrate that SE functions positively in salt stress response, possibly by modulating pre-mRNA splicing of salt stress-associated genes.
2. Materials and methods
2.1. Materials and plant growth conditions
Chemicals and reagents used in this study were purchased from TaKaRa Biotechnology Co. Ltd (Tokyo) or Shanghai Sangon Biotechnology Co. Ltd (Shanghai, China). The Arabidopsis thaliana ecotype Columbia was used throughout this study. Seeds of se-1 were kindly gifted by Wagner D.R. (Prigge and Wagner, 2001). The Arabidopsis plants were grown in a tissue culture room at 22 °C with a 16 h light/8 h dark photoperiod.
2.2. Abiotic stress treatments
Seedlings grown on 1/2 MS plates or soil were used for salt treatment.
Seeds were sowed on 1/2 MS plates containing different concentrations of NaCl and then the plates were kept for 3 days in the dark at 4 °C to break dormancy (stratification) and transferred thereafter to a tissue culture room at 22 °C under 180 μE m2·sec−1 light with a 16-h light/8-h dark photo-period. Four-week-old seedlings grown in soil were watered with 250 mM NaCl. Whole seedlings were photographed at given times after salt treatment.
2.3. qRT-PCR
qRT-PCR was performed as described by Chen et al. (2013). The gene-specific primers for quantitative RT-PCR were SE-F (5′ -GGAGAGGTGGACCTGCCCCTT -3′) and SE-R (5′- ACGGTCACTTCTTCCTCTGGAGC -3′), ACTIN2-F (5′-TGTGCCAATCTACGAGGGTTT-3′) and ACTIN2-R (5′-TTTCCCGCTCTGCTGTTGT-3′).
2.4. Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed as described by Chen et al. (2010). The gene-specific primers for semi-quantitative RT-PCR were as follows: RD29A-F(5′ -TCAAACAGAGGAACCACCACTCAACA -3′) and RD29A-R(5′ -GTAATCGGAAGACACGACAGGAAAC -3′), RD29B–F(5′ -CACAGTTGACACGTCCTTATGGTCATGA -3′) and RD29B-R(5′ -TAGGTTTACCCGTTACACCACCTCTCA -3′), RD20-F(5′-GGAGAGGCAGAGGCTTTGGC-3′) and RD20-R(5′ -CGGAAGTGTAACGTAGCTGAACG-3′) and ADH1-F(5′- GTCTACCACCGGACAGATTATTCGA-3′) and ADH1-R(5′ -GGCAACACATGATCTCCTGGCTG -3′).
2.5. Northern blot analysis
Northern blot analysis was performed as described by Chen et al. (2010).
2.6. GUS staining
Histochemical detection of GUS activity was performed as described by Chen et al. (2013).
2.7. GFP fluorescence observation
Seven-day-old 35SPro:SE-GFP transgenic plants were treated with 150 mM NaCl and the GFP fluorescence was observed after treatment for 8 h and 24 h, respectively. Leica fluorescence microscopy was utilized to see the GFP fluorescence.
2.8. Western blotting
Fourteen-day-old 35SPro:SE-GFP transgenic seedings were harvested after treatment with NaCl and total proteins were extracted using the protein extraction buffer containing 100 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, 1% SDS, 1×complete protease inhibitor cocktail (Roche) and 50 μM MG132. The anti-GFP (Sigma–Aldrich) and anti-Actin (Affinity) antibody were used to detect SE-GFP and Actin, respectively.
2.9. Relative electrolyte leakage and chlorophyll content measurement
The leaves of 4-week-old plants that were treated with 250 mM NaCl were harvested for relative electrolyte leakage measurement as described by Jiang et al. (2007). The leaves of salt-treated plants were used for chlorophyll extraction and determination according to the method described by Lichtenthaler (1987).
2.10. Construction of SEPro:SE fusion and 35SPro:SE-GFP fusion
For complementation analysis, the SE promoter was followed the SE complete 3780bp genomic DNA sequence and the 35S PolyA terminer and the resulting SEPro:SE fusion fragments were finally cloned into the Arabidopsis transformation vector pOCA28. Similarly, the SE cDNA sequence was first fused with GFP and then the SE-GFP fusion was finally cloned into the Arabidopsis transformation vector pOCA30.
3. Results
3.1. Basal expression of SE
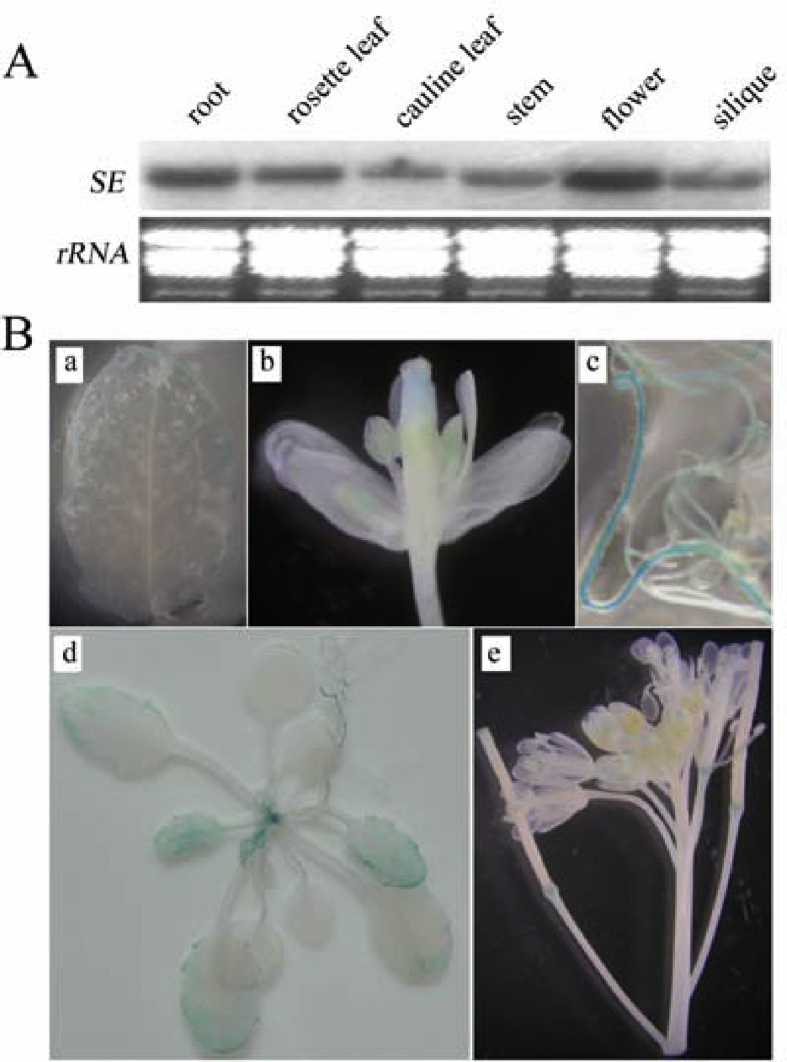
Previous studies have demonstrated that alternative splicing of mRNA plays vital roles in the modulation of plant adaption to various environmental stresses. We investigated SE to better understand this process. We first analyzed SE basal expression. Although SE expression has been previously analyzed in different plant tissues by reverse transcription-PCR (RT-PCR) (Yang et al., 2006), we re-examined its expression using northern blotting and the GUS reporter gene to uncover a more detailed pattern. Similar to the previous report, SE was expressed in all examined plant tissues, with the highest levels of SE mRNA detected in flowers (Fig. 1A). GUS staining further revealed the tissue-specific expression of the SE. As shown in Fig. 1B, GUS activity was detected in leaves, shoot apices, roots, and flowers. In junior leaves, GUS activity was detected throughout the whole leaf. In senior leaves, however, GUS activity was detected only on the leaf margins, especially at the tip, and no GUS activity was detected in mature leaves. In inflorescences, GUS staining was detected in developing flowers, especially in young siliques and pollen. In mature siliques, however, GUS staining was detected only at the junction of siliques and pedicels. These results suggest that SE is constitutively expressed in various organs and may participate in both plant development processes and stress responses.
Fig. 1.
Basal expression of SERRATE (SE). (A) Expression pattern of SE in different organs using northern blotting analysis. (B) β-Glucuronidase (GUS) staining of SE. a. Leaf; b. Flower; c. Root; d. 20-day-old seedling; e. Inflorescence.
3.2. Expression of SE under salinity stress
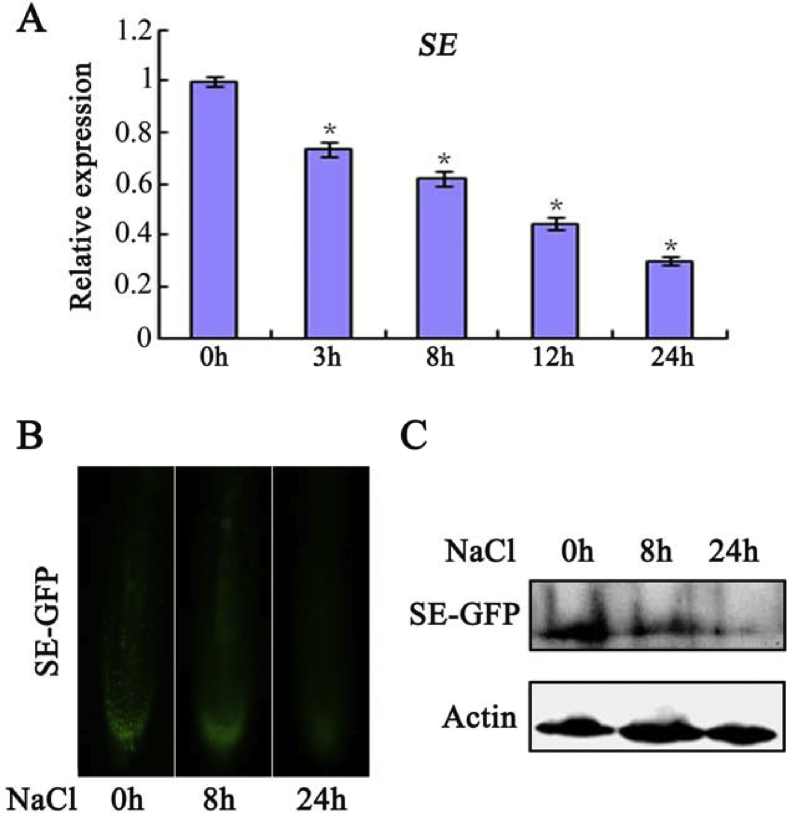
To better determine the role of SE, we further analyzed its expression pattern under saline conditions. Samples taken at various time points from wild-type (WT) NaCl-treated plants and controls were analyzed by quantitative real-time PCR (qRT-PCR). SE expression gradually decreased during the course of the salt treatment (Fig. 2A). To further reveal SE expression patterns, the stability of the SE protein during treatment was determined by GFP observation using root tips of NaCl-treated 35SPro:SE-GFP transgenic plants (Fig. 2B). The GFP signal was clearly detected before treatment, but gradually destabilized during the process of salt stress. Similarly, western blotting analysis showed that salt stress promotes SE degradation (Fig. 2C). SE was thus repressed at both mRNA and protein levels by salt stress, implying its involvement in stress response.
Fig. 2.
Expression of SERRATE (SE) after NaCl treatment. (A) Real-time qPCR analysis of transcript levels of SE after 150 mM NaCl treatment. Values are means ± SD of three independent biological replicates. ∗P < 0.05, Student's t-test compared with controls. (B) GFP fluorescence in transgenic 35SPro:SE-GFP plants after 150 mM NaCl treatment for 8 and 24 h respectively. (C) Western-blot analysis of SE-GFP protein accumulation in transgenic 35SPro:SE-GFP plants after 150 mM NaCl treatment for 8 and 24 h respectively. An anti-GFP antibody was used to detect SE-GFP. Actin was used as an internal control.
3.3. Hypersensitivity of se-1 to NaCl during seed germination and post-germination early growth
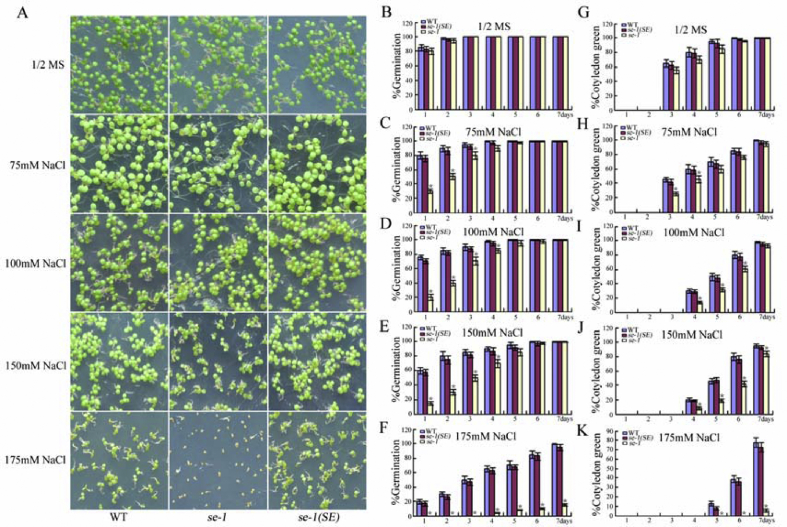
To determine the role of SE in seed germination, seeds of the WT, the mutant se-1, and one complementation line, se-1(SE), were first germinated on 1/2 MS medium containing either 75, 100, 150, or 175 mM NaCl and then their germination rates were compared. No significant difference was observed in germination among se-1, WT, and se-1(SE) (Fig. 3A and B) on the control 1/2 MS medium. But in the presence of NaCl, se-1 germinated later than WT and se-1(SE) (Fig. 3A and C–F). After 1 day, 80, 77, and 60% of WT seeds germinated on the 1/2 MS medium containing 75, 100, or 150 mM NaCl, respectively. However, the germination rates of the se-1 mutant were only about one-third that of the WT under these conditions. More notably, 20% of the WT seeds still germinated, but no se-1 seeds germinated on 1/2 MS medium containing 175 mM NaCl (Fig. 3A and F). Similarly, the early seedling growth of se-1 was also slower than that of the WT (Fig. 3G–K). After 7 days on 1/2 MS medium containing 175 mM NaCl, 80% of WT but only 5% of se-1 seedlings had green cotyledons (Fig. 3A and K). These results indicate that se-1 is hypersensitive to salt stress during seed germination.
Fig. 3.
NaCl dose-response analysis of germination and post-germination early growth in wild-type (WT), se-1, and se-1(SE) seedlings. (A) Germination rates of WT, se-1, and se-1(SE) after NaCl treatment. (B–F) Statistics of WT, se-1, and se-1(SE) germination rates after NaCl treatment. (G–K) Calculations for WT, se-1, and se-1(SE) green cotyledons after NaCl treatment. Values are means ± SD of three independent biological replicates. ∗P < 0.05, Student's t-test compared with controls.
In addition to the germination test, we investigated primary root elongation of se-1 under salinity stress conditions. Roots of se-1 seedlings showed enhanced sensitivity to salt compared with WT seedlings over the same period (Fig. 4A and B). As shown in Fig. 3, Fig. 4, transformation of the SE restored the mutant's hypersensitivity to salt stress to levels similar to those found in WT, indicating that the SE genomic construct can fully complement the se-1 mutant for salt sensitivity. These results imply that se-1 is also hypersensitive to salt stress during post-germination growth.
Fig. 4.
Hypersensitivity of se-1 to NaCl during post-germination growth. (A) The phenotype of representative seedlings after treatment with different NaCl concentrations for 9 days. (B) The root length of representative seedlings after treatment with different NaCl concentrations. Values are means ± SD of three independent biological replicates. ∗P < 0.05, Student's t-test compared with controls.
3.4. Enhanced sensitivity of se-1 to NaCl stress during the vegetative growth stage
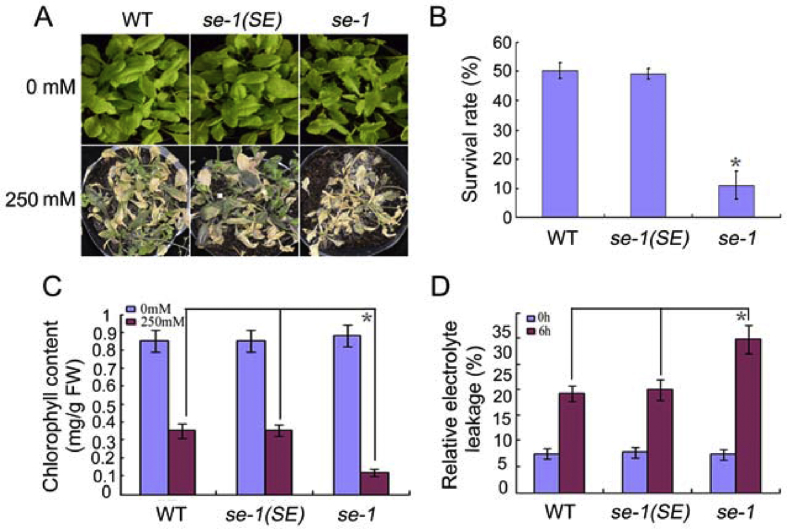
To further determine the function of SE in salt stress response, we evaluated WT, se-1, and se-1(SE) plants for their responses to NaCl stress in the vegetative growth stage. We subjected soil-grown, 4-week-old seedlings of these three genotypes to 250 mM NaCl treatment. Following the salt treatment, as shown in Fig. 5A, the se-1 plants displayed more serious and accelerated chlorosis and wilting phenotypes compared with WT and se-1(SE) plants. The survival rate of se-1 plants was also dramatically lower than both WT and se-1(SE) plants (Fig. 5B). Thus, mutation of SE caused enhanced sensitivity to NaCl stress.
Fig. 5.
Salt sensitivity of soil-grown se-1. (A) The phenotype of 4-week-old plants after treatment with 250 mM NaCl for 18 days. (B) Relative survival rate of 4-week-old plants treated with 250 mM NaCl for 2 weeks. (C) Chlorophyll contents of 4-week-old plants treated with 250 mM NaCl for 2 weeks. (D) Relative electrolyte leakage of 4-week-old plants treated with 250 mM NaCl for 6 h. (B–C) Values are means ± SD of three independent biological replicates. ∗P < 0.05, Student's t-test compared with controls.
Because relative electrolyte leakage (REL) is an important index reflecting cell membrane damage under disadvantageous conditions, we compared REL differences of seedlings after NaCl treatment. Following treatment of 4-week-old seedlings with 250 mM NaCl for 6 h, their leaves were measured for REL. Under salt stress conditions, leakage levels of the se-1 mutants were higher than those of WT and se-1(SE) (Fig. 5C).
We also quantified differences in chlorophyll content among WT, se-1, and se-1(SE) plants. Four-week-old seedlings were first treated with 250 mM NaCl for 2 weeks, and then the total chlorophyll contents were determined. As shown in Fig. 5D, se-1 plants accumulated the least amount of chlorophyll of the three genotypes. These results also indicate that mutation of SE could confer to plants higher sensitivity to NaCl stress.
3.5. SE modulation of pre-mRNA alternative splicing of salt stress-associated genes
SE was previously reported to play critical roles in pre-mRNA alternative splicing. We therefore speculated that SE may participate in salt stress response by modulating pre-mRNA alternative splicing of salt stress-associated genes, thereby causing se-1 to exhibit more pronounced intolerance to salt. To verify this possibility, we compared alternative splicing of the pre-mRNA of several well-characterized marker genes in WT, se-1, and se-1(SE) plants by RT-PCR. As shown in Fig. 6, a minor band and a slower-moving major band were detected in se-1 mutant plants. The minor band was also present in WT and se-1(SE) plants, whereas the major band was barely detected. These results demonstrate that splicing of RD29A, RD29B, RD20, and ADH1 pre-mRNA is altered in the se-1 mutant, possibly explaining why se-1 exhibits a more pronounced intolerance to salt than do WT and se-1(SE) under salt stress conditions.
Fig. 6.
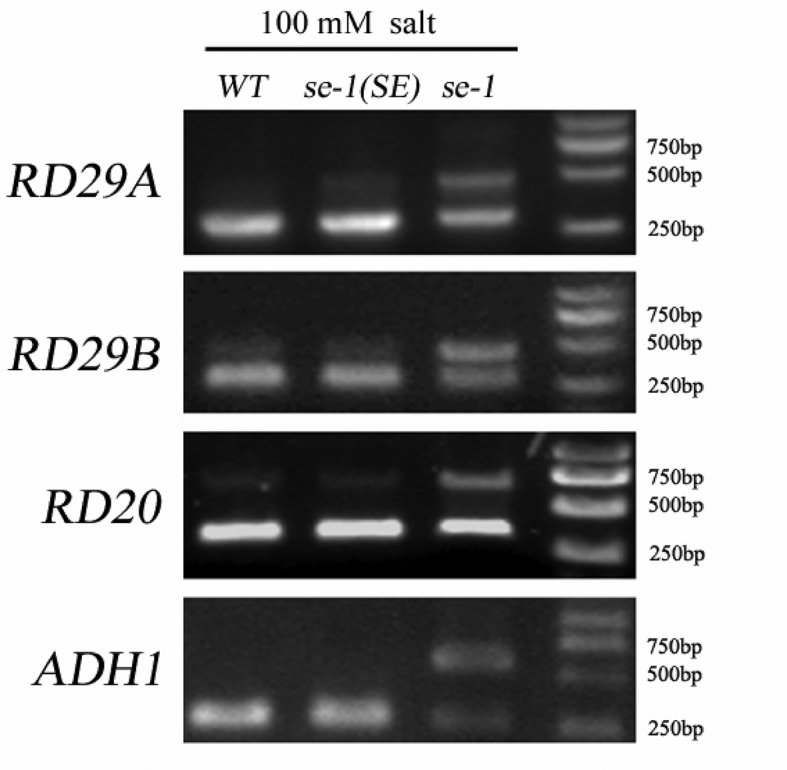
Pre-mRNA alternative splicing of RD29A, RD29B, RD20, and ADH1 upon salinity stress. Seven-day-old wild-type (WT), se-1(SE), and se-1 seedlings were treated with NaCl for 24 h, and their total RNA was then extracted. Agarose gel electrophoresis was used to isolate the alternatively spliced pre-mRNA products. RT-PCR results are shown from left to right for WT, se-1(SE), and se-1.
4. Discussion
As sessile organisms, plants are constantly exposed to various fluctuating abiotic stresses, which may inhibit plant growth and development, or even determine plant species distribution across different ecosystems. Salinity is one of the major abiotic stress factors that dramatically hamper agriculture production worldwide. Although previous studies have demonstrated that Arabidopsis SE functions as a key component in plant growth regulation, little is known about its possible involvement in salt stress response. Here, we provide evidence to show that SE plays an important role in plant response to salt stress. Expression analysis showed that SE was repressed by salt treatment at both mRNA and protein levels, with an enhanced inhibition at the protein level (Fig. 2), indicating the involvement of SE in salt stress response. Phenotypic analysis demonstrated that se-1 mutants are more sensitive to high salinity during seed germination, post-germination early growth, and vegetative growth. Furthermore, complementation experiments showed that transformation of the SE can restore the mutant's hypersensitivity to salt stress to levels similar to those found in WT plants (Fig. 3, Fig. 4, Fig. 5A–C). These results imply that NaCl-repressed SE may function as a positive modulator in plant response to salt stress.
SE has been reported to act as a critical component that modulates gene expression through directing pre-mRNA alternative splicing. SE may thus serve as an important functional protein that participates in pre-mRNA processing to control plant development and responses to various stresses. SE may participate in salt stress response by controlling alternative pre-mRNA splicing of salt stress-associated genes. To gain insight into the molecular basis of SE function in salt stress response, we analyzed the alternative splicing of pre-mRNA of several well-characterized marker genes in WT, se-1(SE), and se-1. Splicing of RD29A, RD29B, RD20, and ADH1 pre-mRNA was altered in the se-1 mutant under salt stress conditions compared with that of WT and se-1(SE) plants (Fig. 6); this change in mRNA splicing compromised the successful accumulation of these stress-associated proteins, ultimately leading the se-1 mutants to exhibit a more pronounced intolerance to high salinity. Consistent with our results, RD29A and several other salt stress-associated genes, such as SRL1, SNRK2.8, and RAP2.7, were also demonstrated to exist alternative splicing in se-1 (Raczynska et al., 2014). Together, these results demonstrate that SE may participate in salt stress response by controlling the pre-mRNA splicing of salt stress-associated genes.
Previous studies have shown that SE and the cap-binding proteins (CBP20 and CBP80) have overlapping functions in pre-mRNA splicing, suggesting the involvement of the two cap-binding proteins in salt stress response. Indeed, similar to our results, CBP20 and CBP80 have also been recently shown to be involved in salt stress response by modulating the pre-mRNA splicing of numerous stress-associated genes (Kong et al., 2014). Furthermore, SE has been shown to physically interact with both CBP20 and CBP80 (Raczynska et al., 2014). On the basis of these results, SE and the cap-binding proteins may have synergistic roles during plant salt stress response. The possibility therefore exists that pre-mRNA alternative splicing may play critical roles in the modulation of plant response to various stresses. Recently, SE was also found to interact with various proteins, including SnRKs and CHR2 (Yan et al., 2017; Wang et al., 2018), indicating that SE may form protein complexes with diverse proteins to fine-tune both development and stress responses.
5. Conclusions
Salt stress is a major harmful stress factor that dramatically inhibits crop growth and productivity. A comprehensive understanding of the mechanisms mediating salt resistance will contribute to the design of breeding and engineering strategies, ultimately increasing crop production under ever-changing environments. In this study, we demonstrated that SE functions as a positive mediator in plant response to salt stress by regulating pre-mRNA alternative splicing of salt stress-associated genes. Our study has generated additional information on the molecular mechanism of plant response to salt stress.
Author contributions
LGC and DQY conceived and designed the experiments. MHM, QJW and YLC performed the experiments. MHM, QJW and LGC interpreted the data. LGC and MHM wrote the manuscript. All authors reviewed and approved the manuscript.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We thank Wagner D.R. for kindly providing the se-1 mutant seeds.
This work was supported by the National key R & D plan (2016YFD0101006), Natural Science Foundation of China (31671275), Candidates of the Young and Middle-Aged Academic Leaders of Yunnan Province (2015HB094), Yunnan Fundamental Research Projects (grant NO. 2017FB047 and 2019FA010).
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Contributor Information
Diqiu Yu, Email: ydq@xtbg.ac.cn.
Ligang Chen, Email: chenligang@xtbg.ac.cn.
References
- Barbazuk W.B., Fu Y., McGinnis K.M. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- Bezerra I.C., Michaels S.D., Schomburg F.M. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 2004;40:112–119. doi: 10.1111/j.1365-313X.2004.02194.x. [DOI] [PubMed] [Google Scholar]
- Bohnert H.J., Nelson D.E., Jensen R.G. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray E.A., Bailey-Serres J., Weretilnyk E. Responses to abiotic stresses. In: Buchanan B.B., Gruissem W., Jones R.L., editors. Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists; Rockville, MD: 2000. pp. 1158–1249. [Google Scholar]
- Chen L.G., Zhang L.P., Yu D.Q. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant Microbe Interact. 2010;23:558–565. doi: 10.1094/MPMI-23-5-0558. [DOI] [PubMed] [Google Scholar]
- Chen L.G., Zhang L.P., Li D.B. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both a-bscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. 2013;110:E1963–E1971. doi: 10.1073/pnas.1221347110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U., Stephan A.B., Horie T. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg S.P., Canales C., Hay A. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J.M., Schroeder J.I. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Yang B., Harris N.S. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 2007;58:3591–3607. doi: 10.1093/jxb/erm207. [DOI] [PubMed] [Google Scholar]
- Kong X., Ma L., Yang L. Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins Arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 2014;13:2495–2510. doi: 10.1021/pr4012624. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A.R., Schor I.E., Alló M. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. Chlorophylls and carotenoids pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Lobbes D., Rallapalli D., Schmidt D.D. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S., Chen H.Y., Adam Y.Y. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 2011;39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Wagner D.R. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1280. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Stepien A., Kierzkowski D. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2014;42:1224–1244. doi: 10.1093/nar/gkt894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C., Szabo E.X., Martinho C. Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes. Elife. 2018;7 doi: 10.7554/eLife.37078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007;428:419–438. doi: 10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- Wang H., Adhityan A., John S. Modeling of phosphorus dynamics in aquatic sediments:I-model development. Water Res. 2003;37:3928–3938. doi: 10.1016/S0043-1354(03)00304-X. [DOI] [PubMed] [Google Scholar]
- Wang L., Song X., Gu L. NOT2 proteins promote polymerase II-dependent transcription and interact with mu-ltiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25:715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ma Z., Castillo-González Claudia. SWI2/SNF2 atpase CHR2 remodels pri-miRNAs via serrate to impede mirna production. Nature. 2018;557:516–521. doi: 10.1038/s41586-018-0135-x. [DOI] [PubMed] [Google Scholar]
- Yan J., Wang P., Wang B. The SnRK2 kinases modulate mirna accumulation in Arabidopsis. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Guo Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018;60:796–804. doi: 10.1111/jipb.12689. [DOI] [PubMed] [Google Scholar]
- Yang L., Liu Z., Lu F. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Zhang J.F., Yuan L.J., Shao Y. The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 2008;31:562–574. doi: 10.1111/j.1365-3040.2008.01786.x. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]