Abstract
ABCC6 mediates release of ATP from hepatocytes into the blood. Extracellularly, ATP is converted into the mineralization inhibitor pyrophosphate. Consequently, inactivating ABCC6 mutations give low plasma pyrophosphate underlying the ectopic mineralization disorder pseudoxanthoma elasticum. How ABCC6 mediates cellular ATP release is unknown. Mechanistic studies are hampered because fluorophores attached to ABCC6’s N- or C-terminus result in intracellular retention and degradation. Here we describe that intramolecular introduction of fluorophores yields fully functional ABCC6 fusion proteins. An ABCC6 variant with the catalytic glutamate of the second nucleotide-binding domain mutated, correctly routed to the plasma membrane, but was inactive. Finally, N-terminal His10 or FLAG tags did not affect the activity of fusion proteins, allowing their purification for biochemical characterization. Hence, these fusion proteins provide excellent tools to study ABCC6 biology.
Keywords: ABC transporter, pseudoxanthoma elasticum, fluorescent fusion protein, purification, cellular ATP efflux
Introduction
Inactivating mutations in ABCC6 underlie the progressive autosomal recessive mineralization disorder pseudoxanthoma elasticum (PXE, OMIM #264800) (1–3), which manifests with lesions in skin, eyes and vascular system. PXE is rare and affects approximately 1 in 25,000–50,000 newborns. There is currently no specific or effective therapy for patients with this condition (4).
ABCC6 encodes ATP-binding cassette subfamily C member 6 (ABCC6) and is primarily expressed in the liver (5), with lower levels found in the kidneys. Importantly, ABCC6 is virtually absent in the soft tissues affected by PXE. In the hepatocytes of the liver, ABCC6 mediates the release of nucleoside triphosphates, predominantly ATP, into the bloodstream (6, 7). Notably, only a very small fraction of the total hepatic ATP pool is released into blood circulation (0.003–0.015 %/min) (6, 8) and ABCC6-mediated ATP release will therefore not substantially affect intracellular ATP concentrations. Outside the hepatocytes, yet within the liver vasculature, released ATP is rapidly converted into adenosine-monophosphate (AMP) and the calcification inhibitor pyrophosphate (PPi) by the ecto-enzyme ectonucleotide pyrophosphatase phosphodiesterase 1 (ENPP1). In the absence of functional ABCC6, plasma concentrations of the mineralization inhibitor PPi are reduced with approximately 60% (6, 9) and this explains the soft tissue mineralization that manifests in PXE. The extracellular AMP that is also formed by ENPP1 activity is converted by NT5E/CD73 into inorganic phosphate (Pi) and adenosine (6). The latter is reabsorbed by hepatocytes (10) and erythrocytes (11) and can be used to resynthesize ATP.
Published data firmly link ABCC6 to the release of ATP from cells. The mechanistic details of ABCC6-mediated ATP efflux are, however, still elusive. Most ABC proteins belonging to the C-branch of the ABC superfamily of membrane proteins function as efflux transporters, and use the energy generated by intracellular ATP hydrolysis at their nucleotide-binding domains (NDBs) to move specific substrates over the plasma membrane, often against significant concentration gradients. There are exceptions, however, including ABCC8 and ABCC9, which regulate the ATP-dependent gating of complex potassium channels (12) and the chloride channel cystic fibrosis transmembrane conductance regulator (CFTR, ABCC7) (13).
The gold standard to determine if a compound is a substrate of an ABC transporter is to determine if it is translocated in vesicular uptake assays. Standard vesicular uptake assays make use of vesicles generated of all cellular membranes, including plasma membrane and intracellular membranes, of cells overproducing the transporter of interest. Several intracellular organelles however, contain carriers for ATP (14, 15) and this is an important source of background in ATP uptake assays. Moreover, the high concentration of ATP that ABC transporters need at their nucleotide-binding domains to drive transport, further adds to the high background in vesicular ATP-uptake assays. Dual-Color Fluorescence-Burst Analysis (DCFBA) is a novel technique that allows monitoring substrate translocation by ABC transporters at the single proteo-liposome level and intrinsically has an excellent signal-to noise-ratio (16). DCFBA relies on the coincidence of fluorescence from spectrally well separated fluorophores in the confocal volume of a microscope. Labeling of ABCC6 with a fluorophore and using, for instance, fluorescent ATP analogs, or an intra-liposomal fluorescent ATP sensor (17), would allow applying DCFBA to study ATP transport by ABCC6. Generation of functional fluorescent fusion proteins can also be used to monitor the synthesis, intracellular trafficking and interaction with other cellular factors of ABCC6 in real time in living cells.
Whereas many ABCC proteins remain fully functional when tagged with N- or C-terminal fluorophores (18–20), addition of bulky amino acid sequences at these positions in ABCC6, results in intracellular retention and loss of functionality. Here we describe that introduction of monomeric fluorophores with excellent spectral characteristics at several positions within the ABCC6 amino acid sequence yields fusion proteins that are fully active. We anticipate that these fluorescently labeled ABCC6-fusion proteins provide excellent tools to study intracellular trafficking and unraveling the molecular details of ABCC6-mediated cellular ATP release by DCFBA.
Materials and Methods
Generation of constructs encoding fluorescent rat Abcc6 fusion proteins
Uracil-Specific Excision Reagent (USER) cloning (21) was used to introduce DNA sequences encoding fluorescent moieties and affinity tags to rat Abcc6 (rAbcc6) in the Gateway entry vector pEntr223 (7). cDNA sequences were amplified using Phusion U PCR master mix (Thermo Scientific). The sequences encoding the His10 and FLAG tags, the HRV C3 protease cleavage site, the fluorescent proteins and the linkers additional to the E1426Q point mutation were introduced into- or flanking the rAbcc6 sequence to generate constructs depicted in Figure 1B. All primers and templates used for USER cloning are listed in Table 1. cDNAs encoding the fluorescent proteins mNeonGreen (L40C-Crispr EFS.mNeon) and mScarlet (pmScarlet-I-C1) were obtained from Addgene. PCR fragments were purified using the Nucleospin gel and PCR cleanup kit (Macherey-Nagel) and assembled using the USER enzyme mix (New England Biolabs), according to the instructions of the manufacturer. Resulting circular constructs were verified by Sanger sequencing and transformed into competent E. coli DH5alpha cells.
Figure 1. Overview of the rAbcc6 fusion protein constructs.
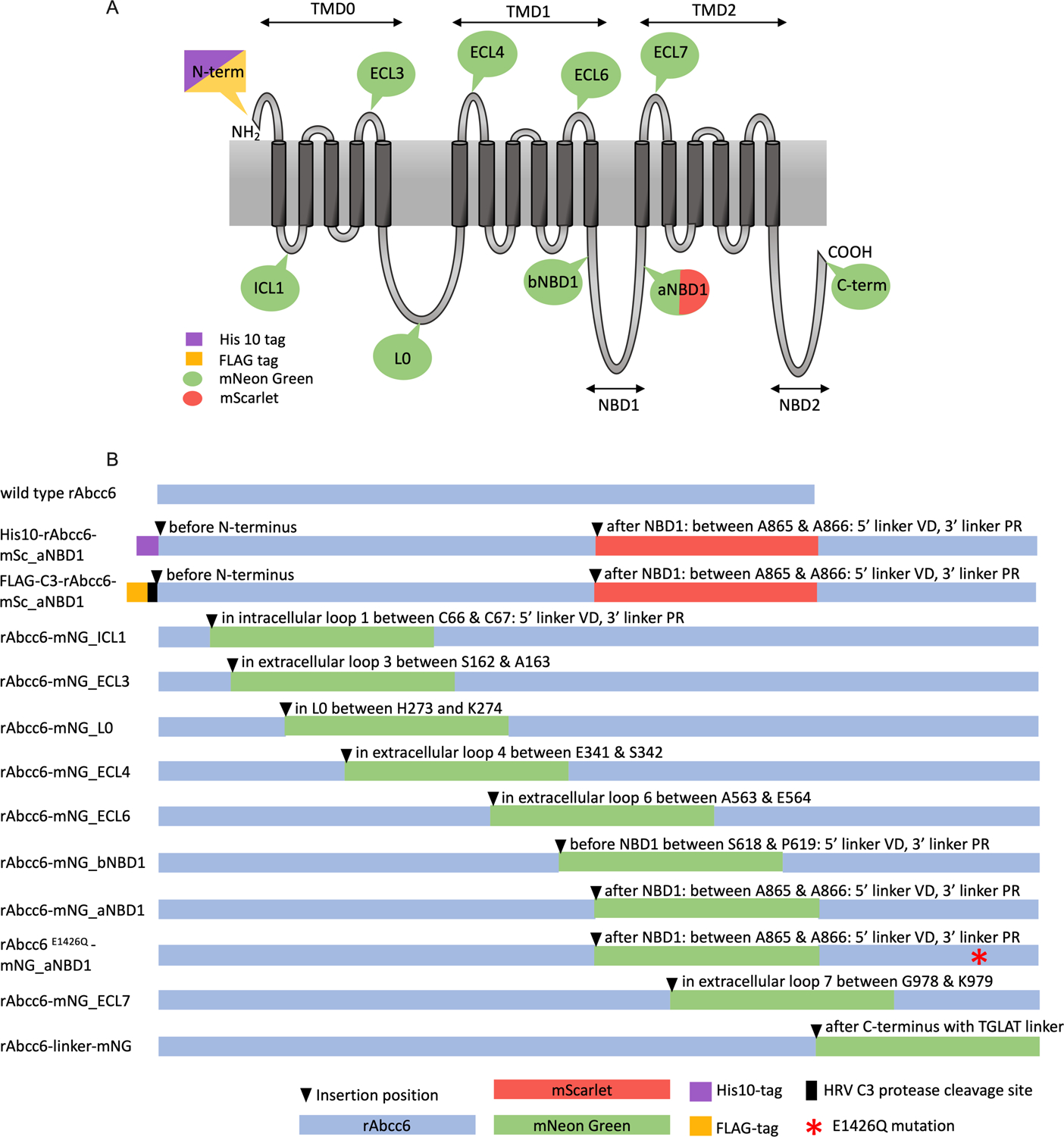
A: Schematic representation of the membrane topology of rAbcc6 with the locations of the fluorophores and tags. B: Schematic overview of the generated fusion proteins indicating the positions where the mNeonGreen or mScarlet fluorophores were introduced. His10, FLAG and HRV C3 protease cleavage sites are also depicted as is the mutation of the Walker B catalytic glutamate residue (E1426Q).
Table 1.
USER PCR primers and templates used in the generation of the rAbcc6 fusion constructs.
| construct | template | forward primer | reverse primer |
|---|---|---|---|
| His10-rAbcc6 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ATGGTGGUGGTGATGATGATGCATGCCAACTTTTTTGTACAA |
| pDonr223_rAbcc6 | ACCACCAUCATCATCATGAGTCCTGCGCAGGCCTG | AGCACACAUTCTCCACCACAGAGGTATTC | |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| His10-rAbcc6-mSc_aNBD1 | pDonr223_His10_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACAGCAUGTGCTTCTCCTTCTCCTT |
| pmSc-I C1 | ATGCTGUGGACATGGTGAGCAAGGGCGAG | ACTGGTGGCUGCTCTGGGCTTGTACAGCTCGTCCATGC | |
| pDonr223_His10_rAbcc6 | AGCCACCAGUGATGACCTTGGAGGCTTT | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| Flag-C3-rAbcc6-mSc-1_aNBD1 | pDonr223_rAbcc6 clone 6.3 | AGGCGCAGCGGUCGGGCTGAACGGGGGGTT | AGCTTGUCGTCATCGTCTTTGTAGTCCATGCCAACTTTTTTGTA CAAAGTTGGCA |
| pDonr223_rAbcc6 clone 6.3 | ACAAGCUGGAGGTGCTGTTCCAGGGTCCCAACGG AGAGCACTCAATGGCCACGC | AACTCCUGCAGGGAAGCTGGCCACATCA | |
| pDonr223_His10_rAbcc6_mSc aNBD1 | AGGAGTUCACACCCCAGTAGGGGAGC | ACCGCTGCGCCUTATCCGGTAACTATCGTC | |
| rAbcc6-mNG_ICL1 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACCATGUCCACGCAGCCATGGCGATGGAT |
| pDonr223_rAbcc6 | AGCCCAGAUGCTACCTCCGGATGTCCC | AGCACACAUTCTCCACCACAGAGGTATTC | |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | ACATGGUGTCCAAGGGCGAAGAGGACAACA | ATCTGGGCUTGTACAGCTCGTCCATGCCCATCA | |
| rAbcc6-mNG_ECL3 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACACCAUGGAGGCCTGCTGGACAGT |
| pDonr223_rAbcc6 | AGGGAGCUTCCGCCAGGAGCCCCTCC | AGCACACAUTCTCCACCACAGAGGTATTC | |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | ATGGTGUCCAAGGGCGAAGAGGACA | AGCTCCCUGCCTTGTACAGCTCGTCCATG | |
| rAbcc6-mNG_L0 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACCATGUGCCCCGGCAGCTCACTGA |
| pDonr223_rAbcc6 | AGGGCACAGUGGTATGGGGACCCCCGAGACAGA | AGCACACAUTCTCCACCACAGAGGTATTC | |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | ACATGGUGTCCAAGGGCGAAGAGGA | ACTGTGCCCUTTCTTGTACAGCTCGTCCATG | |
| rAbcc6-mNG_ECL4 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACCATCUCGAGGTCGCCCATGAACT |
| pDonr223_rAbcc6 | ACAAGTCCUCGGCTTGGACGGGCTGGC | AGCACACAUTCTCCACCACAGAGGTATTC | |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | AGATGGUGTCCAAGGGCGAAGAGGACAAC | AGGACTTGUACAGCTCGTCCATGCCCATCA | |
| rAbcc6-mNG_ECL6 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACCATCGCAUCCATGGCGTTGTCCTCTG |
| pDonr223_rAbcc6 | AGAAGGCGUTTGTGACGCTCACGGTGCTC | AGCACACAUTCTCCACCACAGAGGTATTC | |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | ATGCGATGGUGTCCAAGGGCGAAGAGGA | ACGCCTTCUCCTTGTACAGCTCGTCCATG | |
| rAbcc6-mNG_bNBD1 | pDonr223_rAbcc6 | AGACCCUCCAGATGCTCCTCGAAGGATCGA | ATGTCCACACUCAAGACCATGCCATTGGGGTCTA |
| L40C-Crispr EFS.mNeon | AGTGTGGACAUGGTGTCCAAGGGCGAAGA | AGGGTCUGGGCTTGTACAGCTCGTCCATG | |
| rAbcc6-mNG_aNBD1 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | ACAGCAUGTGCTTCTCCTTCTCCTT |
| pDonr223_rAbcc6 | AGCCACCAGUGATGACCTTGGAGGCTTT | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | ATGCTGUGGACATGGTGTCCAAGGGCGAA | ACTGGTGGCUGCTCTGGGCTTGTACAGCTCGTCCATG | |
| rAbcc6E1426Q-mNG_aNBD1 | pDonr223_rAbcc6_mNG_aNBD1 | AGGCGCAGCGGUCGGGCTGAACGGGGGGTT | AGCACACAUTCTCCACCACAGAGGTATTC |
| pDonr223_rAbcc6_mNG_aNBD1 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | AGTGGCTUGATCCAGGATGAGGATCTGGGTTTTCCGGAGAAG | |
| pDonr223_rAbcc6_mNG_aNBD1 | AAGCCACUGCCTCCGTGGACCCAGGGA | ACCGCTGCGCCUTATCCGGTAACTATCGTC | |
| rAbcc6-mNG_ECL7 | pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | AGCACACAUTCTCCACCACAGAGGTATTC |
| pDonr223_rAbcc6 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | ACCATCCCAUCCACGACCGGGTCGTCGG | |
| pDonr223_rAbcc6 | AGAAGCAGAUGCATTCAGCCCTGCGTGGC | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| L40C-Crispr EFS.mNeon | ATGGGATGGUGTCCAAGGGCGAAGAGGACAACA | ATCTGCTTCUTGTACAGCTCGTCCATGCCCATCA | |
| rAbcc6-TGLAT-mNG | pDonr223_rAbcc6_TGLAT_TEV_GFP_His10 | ATGTGTGCUTCAGGCAGGAGCTGGATCTG | ACCATTGUAGCAAGCCCGGTGGCTAG |
| pDonr223_rAbcc6_TGLAT_TEV_GFP_His10 | ACAAGACUAGTCGTGGTTCTCATCATC | AAAGAACAUGTGAGCAAAAGGCCAGCAA | |
| pDonr223_rAbcc6 | ATGTTCTTUCCTGCGTTATCCCCTG | AGCACACAUTCTCCACCACAGAGGTATTC | |
| L40C-Crispr EFS.mNeon | ACAATGGUGTCCAAGGGCGAAGAGGACAAC | AGTCTTGUACAGCTCGTCCATGCCCATCA |
Cell culture and selection of cell lines overproducing fluorescent rAbcc6 fusion proteins
HEK293 cells were cultured as described previously (22). cDNAs encoding the various rAbcc6 fusion proteins were subcloned into a Gateway-compatible pQCXIP expression vector (7) using LR Clonase-II (Thermo Scientific) and transfected into HEK293 cells using the calcium phosphate precipitation method. Levels of the various fusion proteins in clones resistant to 2 µg puromycin/ml medium were determined by immunoblot analysis, using the antibodies given below. Clones expressing high amounts of the rAbcc6 fusion proteins were also selected based on the fluorescent signal detected in a Flex Station 3 microplate reader (Molecular Devices), using the Exλ/Emλ of 506nm/517nm and 569nm/594nm for mNeonGreen and for mScarlet, respectively.
In preliminary experiments we found that ABCC6-dependent ATP efflux and extracellular PPi formation was highest when HEK293 cells had formed completely confluent monolayers. In addition, background ATP release from HEK293 parental cells was lowest under these conditions. Therefore, PPi accumulation in 24-hour medium samples of HEK293 cells was determined in 96-well plates containing confluent monolayers. Incubation was started by replacing the culture medium with 100 µl fresh medium. Medium was collected 24 hours later and frozen at −20 °C until analysis. Real-time ATP efflux assays (see below), cells were performed using confluent monolayers of HEK293 cells grown on poly-D-lysine-coated black 96-well plates with transparent bottom as described (7, 22).
Immunoblot analysis
Cell lysates were prepared as previously described (22). Five (5) µg of total protein was separated on a 7.5% SDS-polyacrylamide gel (Bio-Rad) and transferred to a PVDF membrane using the Trans Blot Turbo system (Bio-Rad). Wild type and mutant rAbcc6 was detected with the polyclonal K14 rabbit anti-rAbcc6 antibody (kind gift of Dr. Bruno Stieger) diluted 1:3000 and HRP-conjugated donkey anti-rabbit secondary antibody (1:5000, SA1200, Fisher Scientific). His10 tagged rAbcc6 fusion proteins were also detected with the anti-His6 mouse monoclonal antibody (1:250, MA1–21315, Thermo Fisher), followed by incubation with HRP-conjugated anti-mouse secondary antibody (1:5000). Anti-α-tubulin (1:1000, Sc-23948, Santa-Cruz Biothechnology) was used as loading control, with HRP-conjugated polyclonal rabbit anti-mouse IgG employed as secondary antibody (1:5000, P0161, Dako). Antibody binding was visualized by ECL (Pierce Western blotting substrate, Thermo Scientific).
Analysis of the subcellular localization of the fluorescent rAbcc6 fusion proteins in HEK293 cells
HEK293 cells overexpressing the fluorescent rAbcc6 fusion proteins were grown for 2 days on poly-D-lysine coated Ibi-Treat 1.5 µ-Slide 4 well chamber slides (80426, Ibidi). The subcellular localization of the fluorescent rAbcc6 fusion proteins was analyzed by two point-scanning laser confocal microscope Nikon Eclipse Ti equipped with a Nikon A1R+. For live cell imaging a temperature-controlled humidifying chamber at 37°C with 5% CO2 and a plan Apoλ 60X oil objective with a 3X optical zoom was used. For mNeonGreen and mScarlet we used 490.1 and 561.3 nm excitation and 525/50 and 595/50 nm emission filters, respectively. Images were acquired with the pinhole set to 1 airy unit.
Analysis of the subcellular localization of unmodified rAbcc6 in HEK293 cells
HEK293 cells overproducing unmodified rAbcc6 were seeded on ibi-Treat 1.5 µ-Slide 4 well chamber slides (80426, Ibidi) as described above. Cells were fixed in 4% PFA and subsequently in −20 °C cold methanol for 5 min. Slides were blocked with Protein Block solution (BioGenex) for 60 min and incubated with the polyclonal rabbit anti-rAbcc6 antibody K14 diluted 1:100 for 60 min. Next, slides were incubated with A488-conjugated anti-rabbit secondary antibody (A11008, Fisher Scientific) diluted 1:1000 for 60 min. The intracellular localization of the wild type, unmodified, rAbcc6 was visualized essentially as described above, but now using a Plan Fluor 40x Oil DIC H H2 objective with a 3x optical zoom. 490.1 nm excitation and 525/50 nm emission filters were used, with the pinhole set to 1 airy unit.
Quantification of pyrophosphate in culture medium and real-time ATP efflux assays
Quantification of PPi in culture medium and real-time ATP efflux assays were performed as described previously (22). In the real-time ATP efflux assay, firefly luciferase and luciferin are added to the culture medium. Any ATP released by the cells will be converted by the luciferase into a light signal that is detected by luminometry in a Flex Station Pro (Molecular Devices). This approach allows following ATP release in living cells in real time. Any manipulation of cells results in ATP efflux that is independent of ABCC6. To reduce background ATP concentrations, cells were first incubated for 60 minutes at 27 °C, a temperature at which there is low ABCC6-dependent ATP efflux activity, resulting in degradation of extracellular ATP. After these 60 minutes, the temperature was increased to 37 °C, resulting in ABCC6 becoming fully active and start of cellular ATP release.
Results and discussion
Although ABCC6 has been firmly linked to the efflux of ATP and other nucleoside triphosphates (6, 7), the molecular details of ABCC6-mediated ATP efflux are elusive. We set out to develop fluorescently labeled, functionally active, ABCC6 fusion-proteins for future biochemical and intracellular trafficking studies. Rat Abcc6 (rAbcc6) was used for our studies as in our hands it exhibited higher activity than its human orthologue. The higher activity of rAbcc6 will especially benefit anticipated future biochemical characterization of the protein.
ABCC6’s closest relative, ABCC1, tolerates the addition of GFP to its C-terminus, but not to its N-terminus (18–20). Most likely, addition of bulky groups to the extracellular N-terminus results in intracellular routing problems and subsequent degradation. We nevertheless started by generating constructs encoding fluorescent groups at the C- and N-terminus of rAbcc6, but none of these expressed at reasonable levels in HEK293 cells. Nor did these fusion proteins route to the plasma membrane and support cellular ATP release (data not shown). From these data we concluded that, different from ABCC1, the C-terminus is crucial for ABCC6 function. This notion is supported by the fact that several single amino acid mutations in the C-terminal 10 amino acids of ABCC6 are pathogenic (23). An explanation for these results is that ABCC6 function requires complexing with other proteins, including chaperones, for which its C-terminus is essential.
We next systematically introduced the mNeonGreen (mNG) fluorophore at several positions within the rAbcc6 protein sequence: in 4 extracellular and 3 intracellular loops, at altogether 8 different positions (Fig. 1A). Fig. 1B shows a schematic representation and the used nomenclature for the generated rAbcc6 fusion proteins. In addition to constructs encoding the green fluorescent mNeonGreen fusion protein, we also generated 2 constructs encoding rAbcc6 fusion proteins containing the red mScarlet (mSc) fluorophore. The monomeric ~27 kDa mNeonGreen (24) and ~26 kDa mScarlet (25) were chosen for our studies because of their excellent fluorescent characteristics i.e.: high brightness, good photostability and large Stokes shifts. Properties that, for instance, allow super-resolution optical microscopy (26). For anticipated future purification studies, sequences encoding an N-terminal His10 or FLAG-tag were added to some constructs (Fig 2, S2). Finally, to provide a negative control for functional studies, an inactive variant of the rAbcc6 fusion protein with mNeonGreen positioned after the first nucleotide binding domain was generated by changing the catalytic glutamate in NBD2 into a glutamine (E1426Q, position in non-modified rAbcc6).
Figure 2: Expression of the rAbcc6 fusion proteins in HEK293 cells.
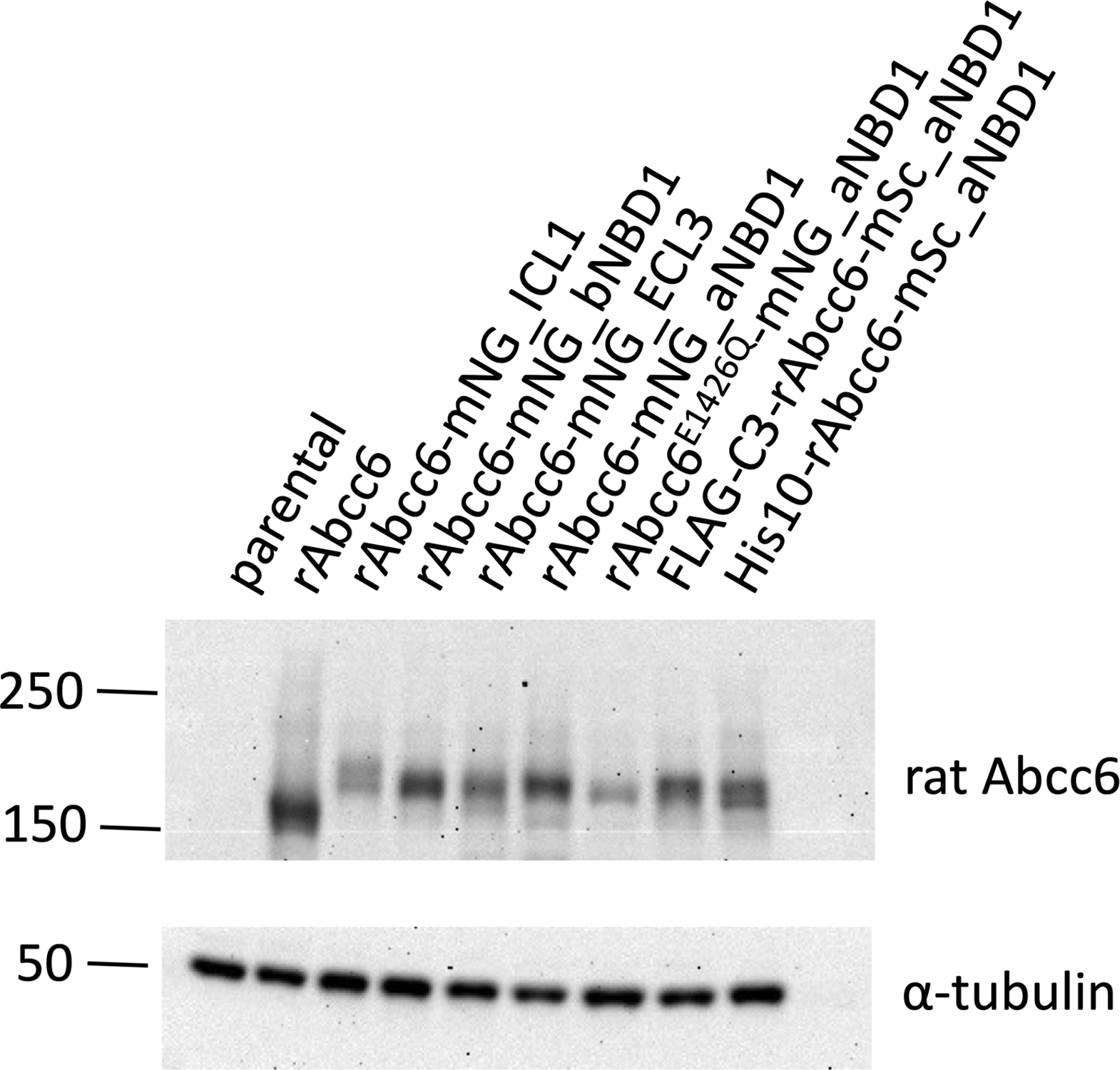
Immunoblot analysis showing relative protein levels of unmodified rAbcc6 and the rAbcc6 fluorescent fusion proteins in HEK293 cells. The polyclonal K14 rabbit anti-rAbcc6 antibody (1:3000x) and HRP-conjugated anti-rabbit secondary antibody (1:5000x) were used to detect rAbcc6.
Expression of constructs encoding mNeonGreen in the extracellular loops 4 (ECL4), 6 (ECL6) and 7 (ECL7) of rAbcc6 resulted in low abundance of the fusion proteins in the HEK293 cells. The extracellular loops of rAbcc6 are short. Possibly, insertion of long amino acid sequences at these positions leads to folding- or intracellular routing problems and, consequently, intracellular degradation. The L0 loop of ABCC6 is predicted to be highly unstructured and it came as a surprise that insertion of mNeonGreen in this loop also yielded low levels of protein in HEK293 cells. Several intramolecular positions in rAbcc6 did tolerate insertion of fluorophores, however. These positions included the first intracellular loop (ICL1) and sequences preceding or following the first nucleotide binding domain (NBD1) of rAbcc6 (Fig. 2, S2). A construct encoding a fusion protein in which mNeonGreen was positioned in the third extracellular loop (ECL3) of rAbcc6 also expressed at reasonable levels. Of note, we tested several independent HEK293 clones for each construct and show the results of clones producing highest fusion protein levels.
ABCC6 needs to be localized in the plasma membrane (27) to exert its function in cellular ATP release. We, therefore, next studied the subcellular localization of the generated fusion proteins in HEK293 cells. Localization was compared to that of unmodified rAbcc6 detected with the K14 antibody (Fig. 3A). As expected, much of the unmodified rAbcc6 was detected in the plasma membrane of the HEK293 cells, although there was also some intracellular staining, similar to what has previously been published for human ABCC6 (27, 28). Possibly, retention of unmodified rAbcc6 in intracellular compartments is related to the high level of overexpression achieved in the HEK293 cell system (See also S1).
Figure 3: Subcellular localization of the rAbcc6 fusion proteins in HEK293 cells.
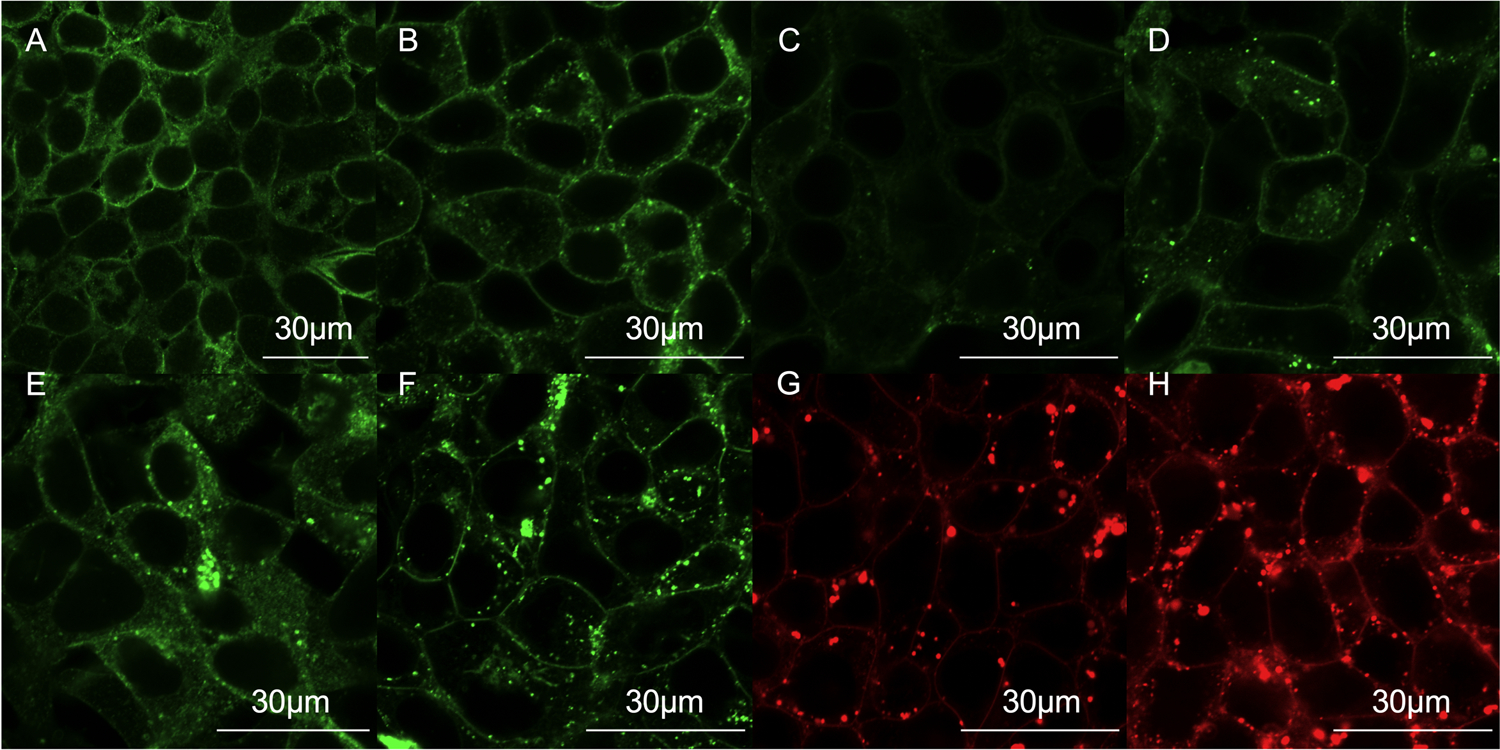
A: representative image of the subcellular localization of the wild type, unmodified rAbcc6 protein overexpressed in HEK293 cells by immunofluorescence with an 40X objective and 3X optical zoom. B-H: representative images of the subcellular localization of the fluorescent fusion proteins by live cell confocal microscopy Panels B-E show the localization of the green fluorescent rAbcc6 fusion proteins with an 60X objective and 3X optical zoom (B: rAbcc6-mNG_ICL1; C: rAbcc6-mNG_ECL3; D: rAbcc6-mNG_bNBD1; E: rAbcc6-mNG_aNBD1. Panel F: rAbcc6E1426Q-mNG_aNBD1). Panels G and H show representative images of the red fluorescent rAbcc6 fusion proteins (G: His10-rAbcc6_mSc_aNBD1; H: FLAG-C3-rAbcc6-mSc_aNBD1). All scale bars represent 30 µm.
Live-cell confocal microscopy was used to determine the subcellular localization of the fusion proteins (Fig. 3B–H). All rAbcc6 fusion proteins that showed reasonable expression on immunoblot analysis, showed some plasma membrane localization. Especially fusion proteins with mNeonGreen inserted in the first intracellular loop (rAbcc6-mNG_ICL1) or before or after the first NBD (rAbcc6-mNG_aNBD1) showed good plasma membrane localization. Of the two mScarlet fusion proteins, the FLAG-tagged version of rAbcc6 (FLAG-C3-rAbcc6-mSc_aNBD1) showed better plasma membrane localization than the His10-tagged version. Plasma membrane localization was less convincing for the rAbcc6 fusion protein with mNeonGreen inserted in the third extracellular loop (rAbcc6-mNG_ECL3). Intriguingly, although the catalytically-dead E1426Q mutant variant of the rAbcc6 fusion protein with mNeonGreen inserted after NBD1 (rAbcc6E1426Q-mNG_aNBD1) showed lower expression on immunoblot analysis, the relative fraction that ended up in the plasma membrane seemed to be increased, with relatively less cytoplasmic localization.
In conclusion, several of the rAbcc6 fusion proteins correctly routed to the plasma membrane of HEK293 cells very similar to unmodified rAbcc6. The intracellular punctuated signal that was detected for example for FLAG-C3-rAbcc6-mSc_aNBD1 (Fig. 3 and Video. S1) indicates incorporation of the fusion proteins into vesicular-like structures in the cytoplasm of as yet unknown origin.
For future biochemical studies functionality of the fusion proteins is crucial and we, therefore, determined if the rAbcc6 fusion proteins supported cellular ATP release, just like unmodified rAbcc6 (Fig. 4A). Four of the fusion proteins, rAbcc6-mNG_ICL1, rAbcc6-mNG_bNBD1, rAbcc6-mNG_aNBD1 and FLAG-C3-rAbcc6-mSc_aNBD1, supported robust ATP release and behaved very similar to unmodified rAbcc6 when produced in HEK293 cells. Somewhat lower ATP release was seen in cells containing rAbcc6 with mNeonGreen inserted in ECL3 or if the fusion protein contained an N-terminal His10 tag, in addition to an mScarlet fluorophore positioned after NBD1 (His10-rAbcc6_mSc_aNBD1). Low routing to the plasma membrane of these fusion proteins (Fig. 3) most likely underlies the reduced ATP efflux. ATP release was completely absent from HEK293 cells producing the catalytically dead walker B mutant rAbcc6E1426Q-mNG_aNBD1, despite high amounts of this fusion protein were detected in the plasma membrane (Fig. 3F).
Figure 4. Functional analysis of the fluorescent rAbcc6 fusion proteins.
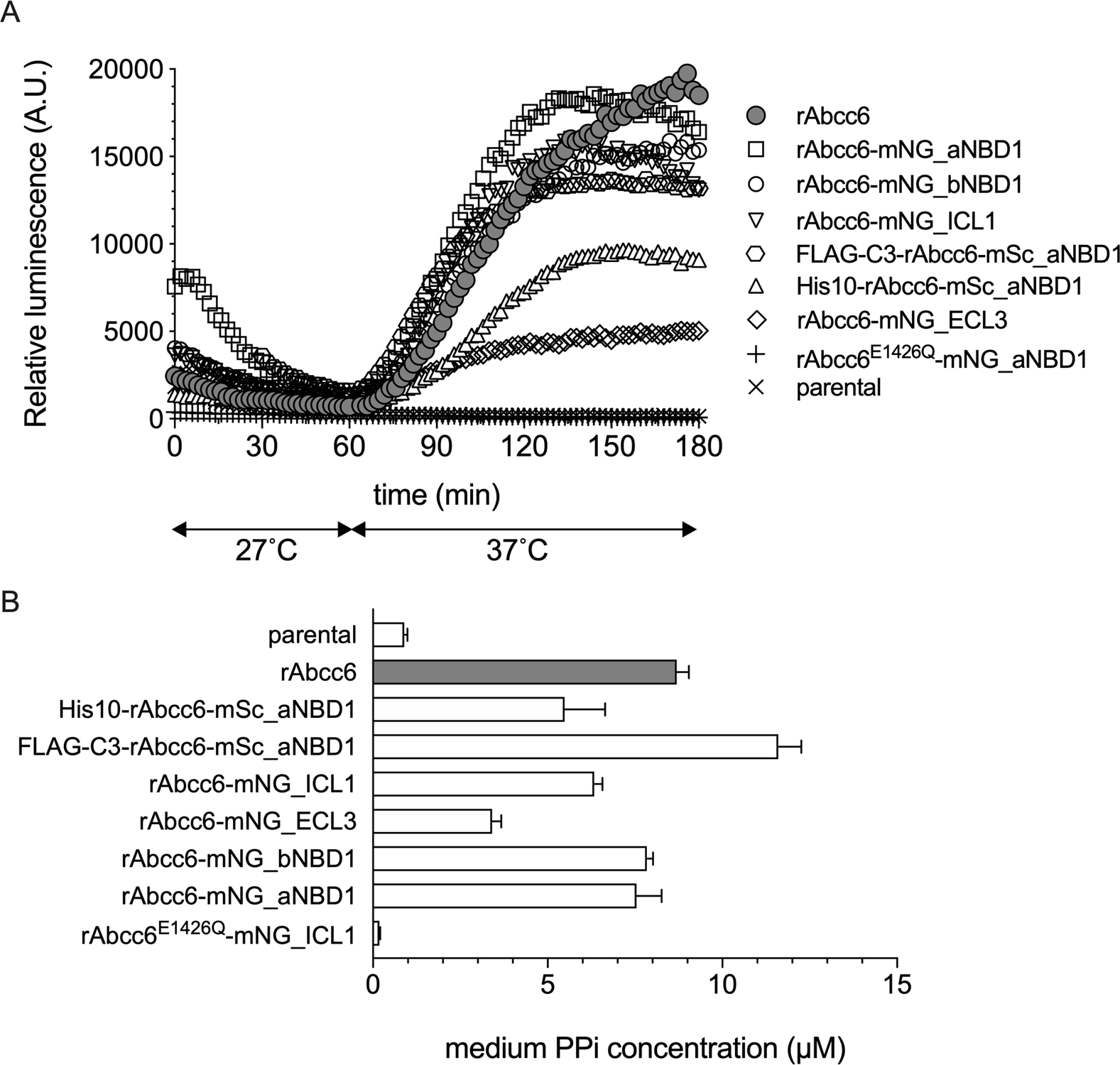
A: ATP efflux was followed in real time for 3 hours using a luciferase/luciferin-based detection system. Data represent the mean of an experiment performed in quadruple. B: PPi accumulation in 24-hour cell culture medium samples are given. Data are presented as mean +/− SE of an experiment performed in quadruple. Representative examples of at least 2 independent experiments are shown in A and B.
HEK293 cells rapidly convert any extracellular ATP into AMP and PPi (6, 7, 22) and we used PPi as an additional marker for ABCC6 activity. PPi concentrations were quantified in 24-hour medium samples collected from the various cell lines. The results of these experiments mimicked our findings in the real-time ATP efflux assay, with HEK293 cells producing rAbcc6-mNG_ICL1, rAbcc6-mNG_bNBD1, rAbcc6-mNG_aNBD1 and FLAG-C3-rAbcc6-mSc_aNBD1 having similar levels of PPi in their culture medium as cells containing unmodified rAbcc6. The rAbcc6-mNG_ECL3 fusion protein also exhibited decreased activity in these experiments. The HEK293 cells overproducing the rAbcc6 fusion protein containing the E1426Q mutation (rAbcc6E1426Q-mNG_aNBD1) had PPi levels in their medium that were comparable to those found in medium of HEK293 parental cells, demonstrating a complete absence of activity.
In conclusion, our data indicate fluorophores, and potentially other functional groups, can be introduced within the rAbcc6 sequence in the first intracellular loop and before and after the first NBD (NBD1), without compromising functionality. Insertion at these sites results in proteins showing similar expression and activity as unmodified rAbcc6. Introduction of mNeonGreen in the third extracellular loop of rAbcc6 resulted in reduced plasma membrane localization and, consequently, functionality when expressed in HEK293 cells. The fusion protein with mNeonGreen inserted in the third extracellular loop might nevertheless be useful, for instance when a functional group is needed in an extracellular domain of ABCC6. Notably, immunoblot analysis of rat liver and HEK293-rAbcc6 cells indicate several-fold higher expression of rAbcc6 in our HEK293-rAbcc6 cells than in rat hepatocytes (S1). The somewhat lower protein levels found for the rAbcc6-mNG_ECL3 construct might therefore still allow studying ABCC6 function. There are currently no antibodies available recognizing extracellular domains of ABCC6. Our data indicate that extracellular loop 3 (ECL3) should allow introduction of affinity tags to, for instance, reliably determine ABCC6 plasma membrane localization.
The molecular details of ABCC6-mediated cellular ATP release are unknown. Although most of the available data indicate ABCC6 is a genuine efflux transporter (29), it cannot be excluded that a mechanism different from direct transport underlies ABCC6-dependent cellular ATP release. Several pathways have been described that allow cells to release ATP into the extracellular environment, of which exocytosis is probably the best-known mechanism (30). Exocytosis does not underlie ABCC6-mediated ATP release, as we found that potent inhibitors of the exocytotic pathway, bafilomycin, nocodazol and Brefeldin A, do not inhibit ATP release (data not shown).
Due to the huge concentration gradient over the plasma membrane, theoretically a channel would do to get ATP into the extracellular environment. ABCC6 is a specific conduit for NTPs, but there are no channels known that are selective for NTPs (31). Our data on the rAbcc6E1426Q mutant also indicate ABCC6 does not function as an ATP channel. In all known ABC transporters mutation of the equivalent catalytic glutamate residue leads to a complete loss of ATP hydrolytic activity and completely wipes out transport function (32, 33). In contrast, the only ABC protein that functions as a channel, ABCC7/CFTR, adapts an open conformation when this catalytic glutamate residue is mutated, resulting in a constant flow of chloride ions out of the cell (13). Our finding that rAbcc6E1426Q, despite routing to the plasma membrane, does not support cellular ATP release, therefore provides indirect evidence that ABCC6 is not an ATP channel.
An attractive hypothesis is that ABCC6 functions as an ATP-dependent ATP efflux pump. The following observations support this hypothesis: First, most members of the C-branch of the ABC superfamily, including ABCC6’s closest homologue ABCC1, are bona fide organic anion efflux transporters and also ABCC6 has been shown to transport a few organic anions (5, 34), albeit sluggishly. Second, the ATP efflux rates found in HEK293-ABCC6 cells (6) are compatible with direct transport as these are very similar to the rates by which ABCC1 pumps morphine-3-glucuronide out of HEK293 cells (35). Third, ATP efflux from ABCC6-containing cells can be blocked by the general ABCC inhibitors benzbromarone, indomethacin and MK571 (data not shown). Fourth, there are intriguing differences in the nucleotide-binding properties between ABCC6 and ABCC1 as shown by the work of Gros and coworkers (36, 37): Under conditions that support ATP hydrolysis (i.e. the presence of substrate, 37 °C), the nucleotide binding domains (NBDs) of ABC transporters like ABCC1 can be labeled with the photoactive ATP analogue 8-azido-ATP. Labeling depends on the presence of phosphate analogs, such as orthovanadate or beryllium fluoride, to trap 8-azido-ADP at the NBDs. ABCC6 is, however, also labeled in the absence of phosphate analogues. Moreover, although phosphate analogs increase 8-azido-ATP labeling of ABCC6, unlike ABCC1 this does not require the addition of a specific substrate (36, 37). An attractive explanation for these results is that 8-azido-ATP binds not only to the NBDs of ABCC6, but also to its substrate binding/translocation site.
Despite of the fact that most data indicate ABCC6 transports ATP, vesicular transport experiments, the gold standard to identify substrates of ABC transporters (38), so far failed to directly demonstrate ABCC6-dependent transport of radiolabeled ATP into inside-out vesicles. We anticipate the fusion proteins described here can be used to study rAbcc6-dependent ATP transport using Dual-Color Fluorescence-Burst Analysis (DCFBA), which allows monitoring transport at the single proteo-liposome level (16). DCFBA requires fluorescent labelling of the protein of interest to study the transport of molecules labeled with a second, spectrally non-overlapping fluorophore. We now aim to optimize the purification and subsequent reconstitution in liposomes of the fusion proteins, and test in DCFBA experiments if ATP is a transported substrate of ABCC6.
We foresee that fluorescently labeled ABCC6 not only has applicability in basic research but can also be used in (pre)clinical studies to follow the behavior of ABCC6 containing pathogenic missense mutations. Several of these mutations are known to negatively affect plasma membrane targeting, without compromising ABCC6’s intrinsic ATP translocating activity. Chemical chaperones, such as 4-phenylbutyrate have been suggested as allele-specific therapies for PXE caused by these mis-localized mutants by correcting routing of ABCC6 to the plasma membrane (28). Fluorescently tagged ABCC6 mutants would facilitate high-throughput screening for candidate molecules that act as pharmacological chaperones.
Conclusion
We have generated and characterized a handful of fluorescent rAbcc6 fusion proteins for cell biological and biochemical applications including protein purification. We have shown, that the first intracellular loop and the loop harboring the first nucleotide binding domain are the best intracellular sites to insert functional moieties, as they allow generation of fully functional active fusion proteins. The generated proteins were not only expressed at comparable levels to non-modified rAbcc6 but also routed to the plasma membrane similarly and were fully functional, independent of the nature of the green or red fluorophore used. Collectively, information on suitable sites to insert various functional moieties, might provide additional tools to further studies on the intriguing, but complex, ABCC6 function.
Finally, our work also provides mechanistic insights into the operation of ABCC6, as we show that catalytic activity at NBD2 is needed for ABCC6 function, different at variance of the only known ABC protein functioning as a channel, ABCC7. Of note, this suggests that ABCC6 is not a channel for ATP.
Supplementary Material
Acknowledgments
Funding sources and
FSz received financial support from the Fulbright Visiting Scholar Program sponsored by the U.S. Department of State and a mobility grant from the Hungarian Academy of Sciences. Further funding for this work was provided by PXE International and the National Institutes of Health Grant R01AR072695 (KvdW).
Abbreviations
- PXE
pseudoxanthoma elasticum
- NBD
nucleotide binding domain
- ABCC6
ATP-binding cassette subfamily C member 6
- PPi
pyrophosphate
- ENPP1
ectonucleotide pyrophosphatase phosphodiesterase 1
- CFTR
cystic fibrosis transmembrane conductance regulator
- DCFBA
Dual-Color Fluorescence-Burst Analysis
- USER cloning
Uracil-Specific Excision Reagent cloning
- HEK293
Human Embryonic Kidney cell line 293
Footnotes
disclosure of conflicts of interest
The authors declare no conflict of interest.
References
- 1.Le Saux O, Urban Z, Tschuch C, Csiszar K, Bacchelli B, Quaglino D, et al. Mutations in a Gene Encoding an ABC Transporter Cause Pseudoxanthoma Elasticum. Nature genetics. 2000;25(2). [DOI] [PubMed] [Google Scholar]
- 2.Ringpfeil F, Lebwohl MG, Christiano AM, Uitto J. Pseudoxanthoma Elasticum: Mutations in the MRP6 Gene Encoding a Transmembrane ATP-binding Cassette (ABC) Transporter. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergen AA, Plomp AS, Schuurman EJ, Terry S, Breuning M, Dauwerse H, et al. Mutations in ABCC6 Cause Pseudoxanthoma Elasticum. Nature genetics. 2000;25(2). [DOI] [PubMed] [Google Scholar]
- 4.Li Q, van de Wetering K, Uitto J. Pseudoxanthoma Elasticum as a Paradigm of Heritable Ectopic Mineralization Disorders: Pathomechanisms and Treatment Development. The American journal of pathology. 2019;189(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belinsky MG, Kruh GD. MOAT-E (ARA) Is a Full-Length MRP/cMOAT Subfamily Transporter Expressed in Kidney and Liver. British journal of cancer. 1999;80(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen RS, Duijst S, Mahakena S, Sommer D, Szeri F, Varadi A, et al. ABCC6-mediated ATP secretion by the liver is the main source of the mineralization inhibitor inorganic pyrophosphate in the systemic circulation-brief report. Arterioscler Thromb Vasc Biol. 2014;34(9):1985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen RS, Kucukosmanoglu A, de Haas M, Sapthu S, Otero JA, Hegman IE, et al. ABCC6 prevents ectopic mineralization seen in pseudoxanthoma elasticum by inducing cellular nucleotide release. Proc Natl Acad Sci U S A. 2013;110(50):20206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallis JL, Gin H, Roumes H, Beauvieux MC. A metabolic link between mitochondrial ATP synthesis and liver glycogen metabolism: NMR study in rats re-fed with butyrate and/or glucose. Nutrition & metabolism. 2011;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Kingman J, van de Wetering K, Tannouri S, Sundberg JP, Uitto J. Abcc6 Knockout Rat Model Highlights the Role of Liver in PPi Homeostasis in Pseudoxanthoma Elasticum. J Invest Dermatol. 2017;137(5):1025–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastor-Anglada M, Pérez-Torras S. Emerging Roles of Nucleoside Transporters. Frontiers in pharmacology. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plagemann PG, Wohlhueter RM, Kraupp M. Adenosine uptake, transport, and metabolism in human erythrocytes. Journal of cellular physiology. 1985;125(2). [DOI] [PubMed] [Google Scholar]
- 12.Bryan J, Muñoz A, Zhang X, Düfer M, Drews G, Krippeit-Drews P, et al. ABCC8 and ABCC9: ABC transporters that regulate K + channels. Pflügers Archiv - European Journal of Physiology. 2006;453(5):703–18. [DOI] [PubMed] [Google Scholar]
- 13.Gadsby DC, Vergani P, Csanády L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440(7083):477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kochendörfer KU, Then AR, Kearns BG, Bankaitis VA, Mayinger P. Sac1p plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. The EMBO journal. 1999;18(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong X, Malhotra R, Guidotti G. ATP uptake in the Golgi and extracellular release require Mcd4 protein and the vacuolar H+-ATPase. The Journal of biological chemistry. 2003;278(35). [DOI] [PubMed] [Google Scholar]
- 16.van den Bogaart G, Kusters I, Velásquez J, Mika JT, Krasnikov V, Driessen AJ, et al. Dual-color Fluorescence-Burst Analysis to Study Pore Formation and Protein-Protein Interactions. Methods (San Diego, Calif). 2008;46(2). [DOI] [PubMed] [Google Scholar]
- 17.Rajendran M, Dane E, Conley J, Tantama M. Imaging Adenosine Triphosphate (ATP). The Biological bulletin. 2016;231(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopal A, Pant AC, Simon SM, Chen Y. In Vivo Analysis of Human Multidrug Resistance Protein 1 (MRP1) Activity Using Transient Expression of Fluorescently Tagged MRP1. 2002. [PubMed]
- 19.Lee SH, Altenberg GA. Expression of functional multidrug-resistance protein 1 in Saccharomyces cerevisiae: effects of N- and C-terminal affinity tags. Biochemical and biophysical research communications. 2003;306(3). [DOI] [PubMed] [Google Scholar]
- 20.Harris MJ, Kuwano M, Webb M, Board PG. Identification of the apical membrane-targeting signal of the multidrug resistance-associated protein 2 (MRP2/MOAT). The Journal of biological chemistry. 2001;276(24). [DOI] [PubMed] [Google Scholar]
- 21.Salomonsen B, Mortensen UH, Halkier BA. USER-derived cloning methods and their primer design. Methods in molecular biology (Clifton, NJ). 2014;1116. [DOI] [PubMed] [Google Scholar]
- 22.Szeri F, Lundkvist S, Donnelly S, Engelke UFH, Rhee K, Williams CJ, et al. The Membrane Protein ANKH Is Crucial for Bone Mechanical Performance by Mediating Cellular Export of Citrate and ATP. PLoS genetics. 2020;16(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favre G, Laurain A, Aranyi T, Szeri F, Fulop K, Le Saux O, et al. The ABCC6 Transporter: A New Player in Biomineralization. Int J Mol Sci. 2017;18(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature Methods. 2013;10(5):407–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindels DS, Haarbosch L, Weeren Lv, Postma M, Wiese KE, Mastop M, et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nature Methods. 2016;14(1):53–6. [DOI] [PubMed] [Google Scholar]
- 26.Tosheva KL, Yuan Y, Pereira PM, Culley S, Henriques R. Between life and death: strategies to reduce phototoxicity in super-resolution microscopy - IOPscience. 2020. [DOI] [PMC free article] [PubMed]
- 27.Pomozi V, Le Saux O, Brampton C, Apana A, Ilias A, Szeri F, et al. ABCC6 is a basolateral plasma membrane protein. Circ Res. 2013;112(11):e148–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomozi V, Brampton C, Szeri F, Dedinszki D, Kozak E, van de Wetering K, et al. Functional Rescue of ABCC6 Deficiency by 4-Phenylbutyrate Therapy Reduces Dystrophic Calcification in Abcc6(−/−) Mice. J Invest Dermatol. 2017;137(3):595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borst P, Váradi A, van de Wetering K. PXE, a Mysterious Inborn Error Clarified. Trends in biochemical sciences. 2019;44(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawada K, Echigo N, Juge N, Miyaji T, Otsuka M, Omote H, et al. Identification of a Vesicular Nucleotide Transporter. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarowski ER. Vesicular and conductive mechanisms of nucleotide release. Purinergic Signal. 82012. p. 359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauna Zuben E., Marianna Müller Xiang-Hong Peng, Ambudkar Suresh V. Importance of the Conserved Walker B Glutamate Residues, 556 and 1201, for the Completion of the Catalytic Cycle of ATP Hydrolysis by Human P-glycoprotein (ABCB1). 2002. [DOI] [PubMed]
- 33.Urbatsch Ina L., Julien Michel, Carrier Isabelle, Rousseau Marc-Etienne, Cayrol Romain a, Gros P. Mutational Analysis of Conserved Carboxylate Residues in the Nucleotide Binding Sites of P-Glycoprotein. 2000. [DOI] [PubMed]
- 34.Ilias A, Urban Z, Seidl TL, Le Saux O, Sinko E, Boyd CD, et al. Loss of ATP-dependent transport activity in pseudoxanthoma elasticum-associated mutants of human ABCC6 (MRP6). J Biol Chem. 2002;277(19):16860–7. [DOI] [PubMed] [Google Scholar]
- 35.van de Wetering K, Zelcer N, Kuil A, Feddema W, Hillebrand M, Vlaming ML, et al. Multidrug resistance proteins 2 and 3 provide alternative routes for hepatic excretion of morphine-glucuronides. Molecular pharmacology. 2007;72(2). [DOI] [PubMed] [Google Scholar]
- 36.Cai J, Daoud R, Alqawi O, Georges E, Pelletier J, Gros P. Nucleotide binding and nucleotide hydrolysis properties of the ABC transporter MRP6 (ABCC6). Biochemistry. 2002;41(25). [DOI] [PubMed] [Google Scholar]
- 37.Cai J, Daoud R, Georges E, Gros P. Functional expression of multidrug resistance protein 1 in Pichia pastoris. Biochemistry. 2001;40(28). [DOI] [PubMed] [Google Scholar]
- 38.Borst P, Evers R, Kool M, Wijnholds J. The multidrug resistance protein family. Biochimica et biophysica acta. 1999;1461(2). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


