Abstract
The glutamate receptor Delta 1 (GluD1) is strongly expressed in the striatum. Knockout of GluD1 expression in striatal neurons elicits cognitive deficits and disrupts the thalamostriatal system in mice. To understand the potential role of GluD1 in the primate striatum, we compared the cellular and subcellular localization of striatal GluD1 immunoreactivity (GluD1-IR) in mice and monkeys. In both species, striatal GluD1-IR displayed a patchy pattern of distribution in register with the striosome/matrix compartmentation, but in an opposite fashion. While GluD1 was more heavily expressed in the striosomes than the matrix in the monkey caudate nucleus, the opposite was found in the mouse striatum. At the electron microscopic level, GluD1-IR was preferentially expressed in dendritic shafts (47.9 ± 1.2%), followed by glia (37.7 ± 2.5%), and dendritic spines (14.3 ± 2.6%) in the matrix of the mouse striatum. This pattern was not statistically different from the labeling in the striosome and matrix compartments of the monkey caudate nucleus, with the exception of a small amount of GluD1-positive unmyelinated axons and axon terminals in the primate striatum. Immunogold staining revealed synaptic and perisynaptic GluD1 labeling at putative axo-dendritic and axo-spinous glutamatergic synapses and intracellular labeling on the surface of mitochondria. Confocal microscopy showed that GluD1 is preferentially co-localized with thalamic over cortical terminals in both the striosome and matrix compartments. These data provide the anatomical substrate for a deeper understanding of GluD1 regulation of striatal glutamatergic synapses, but also suggest possible extrasynaptic, glial and mitochondrial GluD1 functions.
Keywords: striosome, matrix, patch, cerebellin, thalamostriatal, parafascicular, RRIDs: AB_2571757, AB_2314065, AB_2716850, AB_2571759, AB_2301751, AB_2315569
Graphical Abstract
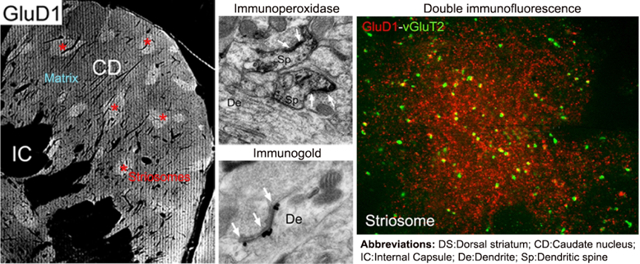
GluD1 is more strongly expressed in the striosome than in the matrix compartment of the monkey caudate nucleus. At the subcellular level, GluD1 is found at asymmetric glutamatergic synapses on dendrites and spines. Double immunofluorescence data suggest an association between GluD1 profiles and vGluT2-containing thalamic terminals in both striatal compartments.
Introduction
Copy number variation (CNV), single nucleotide polymorphism and missense mutation studies have shown a strong association between disruptions of the GRID1 gene, that codes for GluD1, and various neuropsychiatric and cognitive disorders in humans (Glessner et al., 2009; Smith et al., 2009; Treutlein et al., 2009; Greenwood et al., 2011; Nord et al., 2011; Edwards et al., 2012; Griswold et al., 2012; Turner et al., 2017). Altered GluD1 expression was also found in neurons derived from patients with Rett syndrome and in a mouse model of Rett syndrome (Livide et al., 2015; Patriarchi et al., 2016). In line with these human data, recent evidence indicates that deletion of GluD1 leads to deficits in fear conditioning, aberrant emotional and social behaviors and depressive-like behaviors in mice (Yadav et al., 2012; Yadav et al., 2013; Benamer et al., 2018; Nakamoto et al., 2020a). Together, these findings suggest that GluD1 may be a highly vulnerable target for neuropsychiatric disorders.
GluD1, and its family member GluD2, are categorized as ionotropic receptors, but do not exhibit the typical fast ligand-gated ion flow (Kakegawa et al., 2011). In the cerebellar cortex, GluD2 is responsible for the development, maintenance and regeneration of parallel fiber synapses on Purkinje cells (Kashiwabuchi et al., 1995; Hirano, 2012; Yuzaki, 2012; Ichikawa et al., 2016; Berridge et al., 2018; Pernice et al., 2019; Nakamoto et al., 2020b). In line with these observations, recent studies showing that ablation of GluD1 results in changes in dendritic spine density and morphology of pyramidal neurons within the medial prefrontal cortex and CA1 region of the hippocampus, support a similar role of GluD1 in maintaining synaptic integrity in forebrain nuclei (Gupta et al., 2015).
Glutamate delta receptors, like GluD1, exhibit their synaptic effects by collaborating with presynaptic proteins to build a molecular bridge between terminals and postsynaptic elements. Cerebellin-1 (Cbln1) is a presynaptically released glycoprotein first discovered in the cerebellum, where it works in concert with postsynaptic GluD2 and presynaptic protein Neurexin (Nxn), to serve as a crucial synaptic organizer of Purkinje cells (Hirai et al., 2005; Matsuda et al., 2010; Uemura et al., 2010). Nxn is a presynaptic organizer protein that binds to postsynaptic and intermediary elements (Sudhof, 2017; 2018). Together, GluD2, Nxn, and Cbln1 help the development and maintenance of synapses through the formation of tripartite molecular complexes that anchor pre- and post-synaptic elements (Uemura et al., 2010). There is evidence that Cbln1 and Nxn form such synaptic bridges with GluD1 in the mouse striatum (Krishnan et al., 2017; Yuzaki, 2018; Liu et al., 2020).
Our current understanding of GluD1 function in the CNS is hampered by the limited knowledge of its subcellular localization within the mammalian brain. Although some electron microscopic data of GluD1 localization have been reported for the mouse cerebellar cortex and hippocampal formation (Konno et al., 2014; Hepp et al., 2015; Nakamoto et al., 2020b), such information is lacking for the striatum. In a recent study, Liu and colleagues (2020) demonstrated strong GluD1 expression within the mouse dorsal striatum and provided evidence for a GluD1-mediated regulation of the thalamostriatal projection from the parafascicular nucleus (Pf). They further showed that knockout of striatal GluD1 expression elicits cognitive deficits (Liu et al. 2020). To help determine the underlying substrates of these behavioral observations and to extend our understanding of GluD1 function in the primate brain, we undertook a detailed light and electron microscopic analysis of the cellular and sub-cellular localization of GluD1 in the mouse and monkey dorsal striatum.
Materials and Methods
Animals
Five wild-type male mice from Creighton University School of Medicine and three adult male rhesus macaque monkeys from the Yerkes National Primate Research Center breeding colony were used in this study (Table 1). Mice and monkeys were deeply anesthetized with isoflurane or an overdose of pentobarbital (100 mg/kg), respectively, before intracardiac perfusion. Thereafter, animals were perfused with PBS or Ringer solution followed by a mixture of paraformaldehyde (4%) and glutaraldehyde (0.1%). After perfusion, the brains were taken out from the skull, cut in 10–15 mm thick blocks and post-fixed in 4% paraformaldehyde for 24 hours. The tissue blocks were then cut in 60 μm-thick coronal sections with a vibrating microtome. The housing, feeding, and experimental conditions used in these studies followed the guidelines for animal use and welfare set by the National Institutes of Health (National Research Council), and have been approved by Emory and Creighton University’s Institutional Animal Care and Use Committees (IACUC).
Table 1:
Code, sex and age of animals used in the study
| Animal ID | Gender | Age | Analysis type |
|---|---|---|---|
| Mouse | |||
| RM-12 | Male | 45 days | LM/EM (P+G) |
| RM-74 | Male | 54 days | LM/EM (P+G) |
| RM-75 | Male | 51 days | LM/EM (P) |
| RM-76 | Male | 51 days | LM/EM (P+G) |
| RM-119 | Male | 79 days | LM/EM (P) |
| Monkey | |||
| MR-271 | Male | 2 years 9 months | LM/EM (P) |
| MR-272 | Male | 3 years 9 months | LM/EM (P+G) |
| MR-292 | Male | 2 years 6 months | LM/EM (P) |
Abbreviations: LM/EM: Light microscopy/Electron microscopy; P: Immunoperoxidase; G: Pre-embedding Immunogold
Antibodies
All commercially available primary antibodies used in this study have been well characterized and are listed in the Research Resources Identifiers (RRIDs) Portal (Table 2). The specificity of the GluD1 antibody has been further validated in our previous study by the lack of staining in the striatum of GluD1 KO mice (Liu et al. 2020).
Table 2:
List of primary antibodies used in the study
| Antibody-Cat. number | Host | Vendor | Dilution | RRIDs |
|---|---|---|---|---|
| GluD1-Af1390 | Rabbit | Frontiers Inst. Company, Ltd | 1:5000 | AB_2571757 |
| GluD1-GluD1C-GP1 | Guinea pig | Frontiers Inst. Company, Ltd | 1:1000 | AB_2571759 |
| Calbindin D28k-C9848 | Mouse | Sigma Aldrich Company, Ltd | 1:4000 | AB_2314065 |
| Mu-opiate Receptor-Ab1580 | Rabbit | Millipore | 1:10000 | AB_2716850 |
| vGluT1-Ab5905 | Guinea pig | Millipore | 1:1000 | AB_2301751 |
| vGluT2-VGT2-6 | Rabbit | MAb Technologies | 1:1000 | AB_2315569 |
Light Microscopy (LM) Immunocytochemistry
We first sought to examine GluD1 immunoreactivity at the LM level to determine if it followed any regional or compartmental pattern of distribution in the mouse and monkey striatum. Series of brain sections from the pre-commissural striatum of the mouse RM-119 and the monkey MR-272 were immunostained for GluD1. Based on the rhesus monkey and mouse brain atlases (Paxinos et al., 2000; Paxinos and Franklin, 2001), the monkey sections were chosen from anteroposterior (AP) stereotaxic coordinates 20 to 15, while the mice sections ranged from 5.0 to 4.0. To determine if GluD1 immunoreactivity displayed any relationships with the striosome/matrix striatal compartments, adjacent sections were labeled for either calbindin D28k (CaB, in monkey) or mu opioid receptors (MOR, in mice) to delineate the striatal compartments. The tissue was first placed in a sodium borohydride solution (1% in phosphate buffered saline, PBS 0.01 M, pH 7.4) for 20 minutes before being washed five times in PBS. Sections were then submerged in a pre-incubation solution for 60 minutes at room temperature (RT). The pre-incubation solution consisted of 1% normal animal serum (goat for GluD1/MOR and horse for CaB 1% Bovine Serum Albumin (BSA), PBS, and a 0.3% Triton-X-100 solution. After this pre-incubation, sections were incubated in the primary antibody solution of 1% normal animal serum, 1% BSA, primary antibody, and 0.3% Triton at RT for 24 hours. Following the primary antibody incubation, sections were washed in PBS and placed for 90 minutes at RT in a secondary antibody solution consisting of 1% normal animal serum, 1% BSA, secondary biotinylated antibody raised against the primary antibodies (goat anti-rabbit IgG Vector laboratories, RRID: AB_2313606 for GluD1 and MOR immunostaining, and goat anti-mouse IgG, Vector laboratories, RRID: AB_2827937), and 0.3% Triton-X-100. Sections were then washed thoroughly in PBS before being placed in an avidin-biotin-peroxidase complex (ABC, Vector Laboratories, Burlingame, CA USA) solution for 90 minutes. The tissue was rinsed twice with PBS and once with tris(hydroxymethyl)aminomethane (TRIS, 0.05M, pH 7.6) before being transferred into a solution containing 0.01M imidazole, 0.005% hydrogen peroxide, and 0.025% 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma, St. Louis, MO) in Tris for 10 minutes The DAB reaction was stopped by several PBS washes, sections were mounted onto slides and cover slipped.
Double immunofluorescence and confocal images protocol
Two striatal monkey sections (vibratome, 60 μm-thick) from anteroposterior (AP) stereotaxic coordinates 20 to 15, were double immunostained using specific antibodies to label GluD1 and vGluT1 (rabbit GluD1, Af1390; dilution 1:500, and guinea pig vGluT1, RRID:AB_2301751; dilution 1:1000) or GluD1 and vGluT2 (guinea pig GluD1; dilution 1:1000; RRID:AB_2571759, and rabbit vGluT2; dilution 1:1000; MAb technologies, Atlanta, GA; RRID:AB_2315569). Briefly, sections were incubated in sodium borohydride (1%) in PBS for 20 minutes and repeatedly rinsed in PBS. The sections were placed in a pre-incubation solution containing normal donkey serum (NDS; 5%), BSA (1%) and Triton-X (0.3%) for 1 hour and then into an incubation solution containing the two specific primary antibodies (GluD1 and vGluT1 or GluD1 and vGluT2), NDS (1%) and BSA (1%) overnight and then washed in PBS. The sections were then incubated in a solution containing the corresponding secondary antibodies (diluted 1:100) conjugated with rhodamine (GluD1) or FITC (vGluT1 or vGluT2), NDS (1%) and BSA (1%) for 1 hour, rinsed in PBS, mounted with Vectashield (Vector Laboratories) and stored at 4°C. Sections were scanned with a Leica confocal (DM5500B; Leica Microsystems, Bannockburn) and a Hamamatsu camera using Simple PCI software was used to acquire the images. The quantitative analysis for the immunolabeled structures was done using the NIH FIJI software.
Electron Microscopy Immunocytochemistry
Pre-embedding Immunoperoxidase
Brain sections from the pre-commissural striatum of 3 mice and 3 monkeys were processed for GluD1 localization at the electron microscopic level (Tables 1,2). The tissue was first placed into a sodium borohydride solution (1% in PBS) for 20 minutes and washed several times in PBS. Sections were then submerged in a cryoprotectant solution (25% sucrose and 10% glycerol in PB 0.1M, pH 7.4) at RT for 20 minutes before being taken out and placed into a −80°C freezer for another 20 minutes. Following this, sections were reintroduced to a 100% cryoprotectant solution for 10 minutes, which was then substituted by diluted cryoprotectant solutions. Sections were immersed in a pre-incubation solution for 60 minutes at RT. From this point, our staining methods were the same as those described above for the LM immunocytochemistry, with the exception that Triton-X-100 was omitted from all incubation solutions, and the primary incubation period lasted for 48 hours instead of 24 hours. Following the DAB reaction, sections were transferred to a phosphate buffer solution (PB, 0.1M, pH 7.4). The tissue was then post-fixed in a 1% osmium tetroxide solution for 20 minutes. Following washes in PB, the samples were dehydrated in a stepwise manner in 50–100% alcohol solutions before being placed in propylene oxide. Uranyl acetate (1%) was added to the 70% alcohol to increase contrast in the electron microscope. The dehydrated sections were embedded in epoxy resin (Durcupan, ACM; Fluka, Buchs, Switzerland) for 12 hours, mounted onto oil-coated slides and cover-slipped before being baked at 60°C for 48 hours. Blocks of striatal tissue were then taken out from the slide with a razor blade, glued on resin blocks and cut in ultrathin sections (70 nm-thickness) using an ultramicrotome (Ultra-cut T2; Leica, Germany Leica) and mounted onto single slot Pioloform-coated copper grids.
Pre-embedding Immunogold
To help further assess the subcellular and subsynaptic localization of GluD1, tissue sections from RM-12, RM-74, RM-76 and MR-272 were processed for pre-embedding immunogold staining. Sections first underwent the same sodium borohydride, cryoprotectant and freeze-thawing treatments as described in the pre-embedding immunoperoxidase EM section. They were then placed in a pre-incubation solution for 60 minutes at RT and washed multiple times in TBS-gelatin (0.24g TRIS, 0.88g NaCl, 100μL fish gelatin, 100mL of distilled water, pH 7.6). This was followed by a pre-incubation in a PBS solution that contained 5% non-fat dry milk for 30 minutes at RT. Sections were then immersed in the GluD1 primary antibody solution containing TBS-gelatin and 1% non-fat dry milk for 24 hours at RT, which was followed by thorough washes in TBS-gelatin and a 2-hour incubation at RT in a gold-conjugated goat anti-rabbit secondary antibody (1:100-Frontiers Institute Company Ltd, Ishikari, Japan) solution containing 1% non-fat dry milk and 98% TBS-gelatin. After rinses in TBS-gelatin, sections were transferred to a 2% aqueous acetate buffer solution (pH 7.0) before being processed with a HQ silver developing kit (Nanoprobes Inc., Yaphank, NY, USA) inside a dark room for 7–10 minutes. The reaction was stopped by repeated washes in acetate buffer. Sections were then gradually transferred to a PB solution (0.05M, pH 7.4) by stepwise substitution of acetate buffer with PB. They were then post-fixed with osmium, dehydrated and embedded in resin, as described for the immunoperoxidase staining, except that 0.5% osmium solution was used for 10 minutes and the 70% alcohol/uranyl acetate treatment was reduced to 10 minutes.
Data Analysis
Light Microscopy Image Analysis
Six pairs of serially cut sections from the striatum (AP 20–15 according to Paxinos et al. 2000) of monkey MR-272 immunostained for either GluD1 or CaB were digitally scanned by an Aperio ScanScope CS system (Aperio Technologies, Vista, CA) and analyzed using ImageScope software (Aperio Technologies). The pattern of GluD1 and CaB labeling in the pre-commissural caudate nucleus from these pairs of sections was assessed and qualitatively compared to determine the relationships between GluD1-enriched striatal regions with striosomes defined by their lack of CaB immunostaining (Gerfen et al., 1985, Cote et al. 1991). Using the same approach, three pairs of sections through the pre-commissural striatum (AP 5.0–4.0 according to Paxions and Franklin, 2001) of the mouse RM-119 were immunostained for either GluD1 or MOR. From this material, the distribution pattern of striatal regions with low GluD1 expression was qualitatively compared with the localization of striosomes identified by strong MOR staining (Herkenham and Pert, 1981).
Electron Microscopy Image Analysis
Immunoperoxidase-stained Tissue:
All tissue sections were examined under a JEOL JEM 1011 transmission electron microscope and images were acquired using an Erlagshen ES1000W Gatan Camera. Based on our light microscopic data indicating a differential level of GluD1 expression in the striosome and matrix striatal compartments (Fig. 1A,B) in mice and monkeys, blocks of tissue for EM analysis were chosen from either the striosome or the matrix compartment of the monkey pre-commissural caudate nucleus or only from the matrix compartment in mice. The delineation of striosomes and matrix in the GluD1-immunostained sections was based on adjacent sections immunostained for calbindin (for monkey) or MOR (for mice).
Figure 1: Striosome/Matrix Compartmentation of GluD1 in the monkey and mouse striatum.
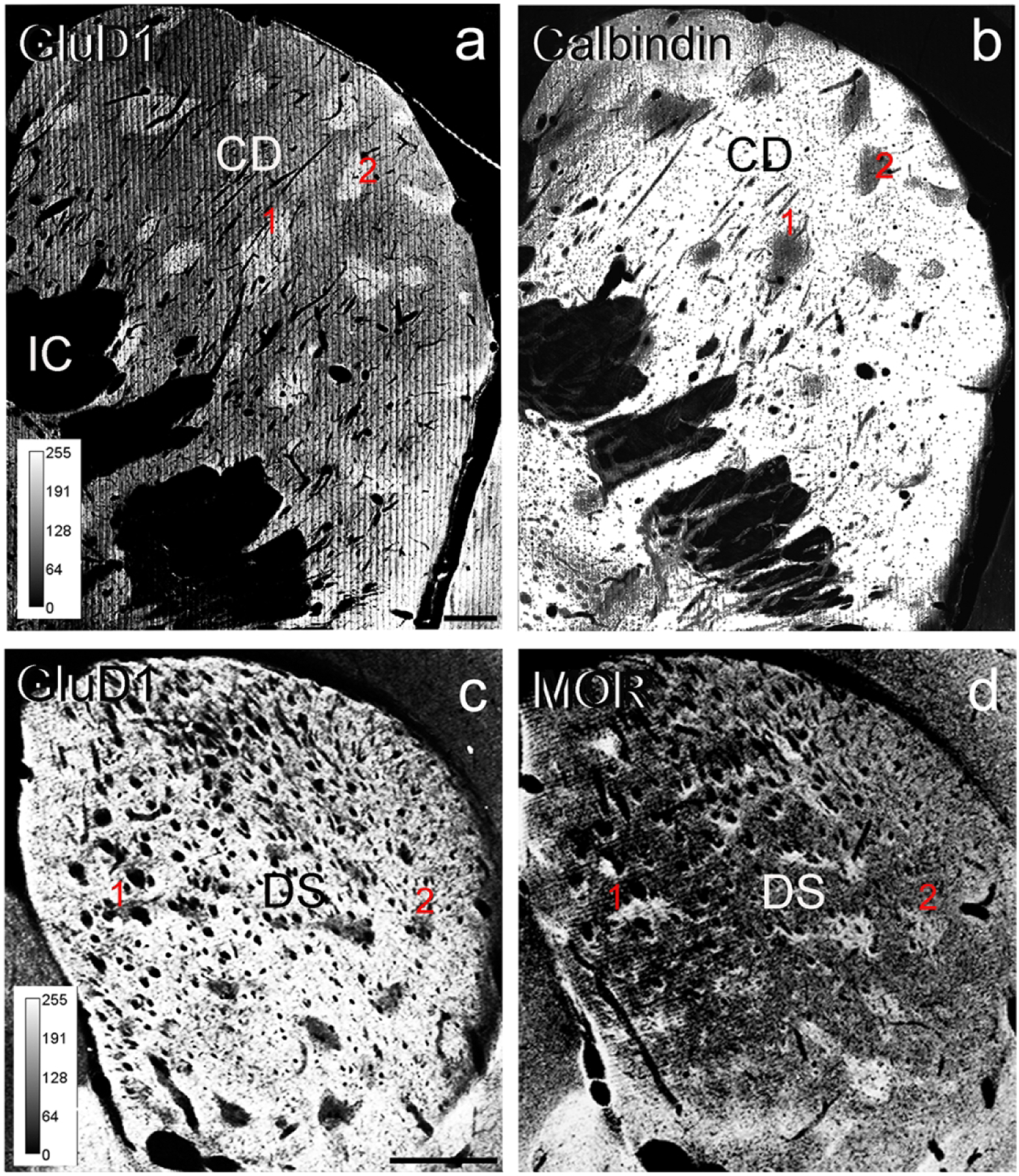
Light micrographs of GluD1 immunoperoxidase staining in the pre-commissural caudate nucleus of a monkey (a) and the pre-commissural dorsal striatum of a mouse (c). (b) and (d) illustrate adjacent sections immunostained for calbindin D28k (b) or MOR (d) to differentiate the striosomes from the matrix compartments of the monkey and mouse striatum, respectively. Note the close correspondence between dense patches of GluD1 immunostaining in a with calbindin-immunonegative striosomes in b. In mouse, areas of low GluD1 labeling in c correspond to MOR-enriched striosomes in d. In both sets of sections, red numbers 1 and 2 indicate corresponding striosomes. The colors in all panels have been inverted, and the levels of immunoreactivity correspond to the intensity scales shown in panel (a). Abbreviations: CD: caudate nucleus, DS: dorsal striatum, IC: internal capsule. Scale bar in a (applies to b) = 1 mm. Scale bar in c (applies to d) = 1 mm.
In the EM, 50–60 micrographs of randomly distributed GluD1-immunostained elements were taken at 25,000X from both the striosome and the matrix compartments of the pre-commissural caudate nucleus of monkeys (N=3 animals; n=324 images) or from the matrix compartment only in the pre-commissural dorsal striatum of mice (N=3 animals; n=157 images). As such, a total of 6 blocks of striatal tissue were analyzed from three monkeys and three blocks of tissue were used from three mice. For each block, data were collected from a single grid. From these images analyzed with the Gatan Digital Micrograph software, GluD1-labeled elements were counted and categorized as spine, dendrite, glia, axon or terminal based on their ultrastructural features (Peters et al. 1991). The mean percentages of each category of GluD1-labeled profiles was then calculated by dividing the number of specific labeled neuronal or glial structures by the total number of GluD1-labeled elements in each striatal compartment.
Immunogold-stained Tissue:
In the immunogold-stained tissue, neuronal structures had to contain a minimum of three immunogold particles to be categorized as GluD1-immunoreactive, while a single gold particle was sufficient to categorize glial processes as GluD1-immunoreactive because of their small size. A total of 16–18 images of randomly distributed GluD1-labeled structures were taken at 25,000X-40,000X from the striatum of 3 mice and one monkey to corroborate the GluD1 localization seen in the immunoperoxidase-stained material and to further assess its subcellular localization. Due to the small sample size, only a qualitative description of the immunogold labeling is presented in this study.
Statistical Analysis
Inter-individual differences in the relative percentages of GluD1-positive elements between animals of the same group (mouse vs monkey and striosome vs matrix) were tested using one-way ANOVA in Sigmaplot 14.0 software. All data are presented as an average percentage value ± Standard Error of the Mean (SEM). Error bars account for variance between animals for each region of interest. Mice and monkey data were compared using two-sample t-tests to verify if there were any statistically significant species difference in the distribution of GluD1.
Results
GluD1 is Differentially Expressed in the Striosome and Matrix Compartments of Mice vs Monkeys
Overall, a heterogeneous pattern of striatal GluD1 immunostaining was found in the three monkeys and three mice used in this study. To further determine if this heterogeneity was related to the striosome/matrix compartmentation of the striatum, the overall distribution of striatal GluD1 immunoreactivity was analyzed from immunoperoxidase-stained tissue in mice (N=1 animal; n=6 sections) and monkeys (N=1 animal; n=12 sections). As depicted in Figure 1, the pattern of striatal GluD1 immunostaining was heterogeneous in the dorsal striatum of both species. In the monkey pre-commissural caudate nucleus, small areas of strong GluD1 immunolabeling lay within a diffuse, less intensely stained, neuropil (Fig. 1a), a pattern reminiscent of the striosome/matrix striatal compartmentation (Graybiel, 1990). Thus, to determine if the GluD1-enriched areas corresponded to the striosome striatal compartment, we compared the distribution of GluD1 with that of CaB immunostaining in adjacent sections. As depicted in Figure 1a,b, there was a complete registration between dense GluD1-immunostained regions and CaB-negative striosomes (Gerfen et al., 1985, Graybiel, 1990, Cote et al. 1991) in the pre-commissural striatum, thereby indicating that GluD1 immunoreactivity is differentially expressed between the striosomes and extrastriosomal matrix of the primate striatum. This pattern of staining was similar across the three monkeys used in this study
Although the pattern of GluD1 immunostaining was also heterogeneous in the mouse striatum, the relationship with the striosome and matrix compartments was opposite to that shown in monkeys (Fig. 1c,d). As illustrated in Figure 1c, the mouse dorsal striatum neuropil was enriched in GluD1 immunostaining, except for small pockets of low immunoreactivity. When compared with adjacent sections immunostained for MOR, a commonly used marker of striosomes in rodents (Herkenham and Pert, 1981; Graybiel, 1990), the areas of low GluD1 immunoreactivity were in register with the striatal areas enriched in MOR, thereby suggesting that the striosome compartment displays a lower level of GluD1 immunoreactivity than the matrix compartment in the pre-commissural mouse striatum (Fig. 1c,d). These findings demonstrate that GluD1 expression follows the striosome/matrix compartmentation in both the monkey and mouse dorsal striatum, but according to a species-specific inverse relationship.
Electron Microscopic Localization of GluD1 in the Mouse and Monkey Striatum-Immunoperoxidase Staining
Our next set of experiments aimed at assessing the subcellular localization of GluD1 in the mouse and monkey dorsal striatum using quantitative EM analysis of immunoperoxidase-stained tissue. Based on our LM data, we sought to compare the ultrastructural localization of GluD1 in the striosome and matrix compartments of the monkey striatum to determine if the higher level of striosomal GluD1 immunoreactivity depicted in Fig 1a was related to any differences in the ultrastructural localization of GluD1 between the two striatal compartments. In monkeys, this analysis was performed on tissue from the pre-commissural caudate nucleus, where the striosomes are best delineated from the matrix. In mice, our EM analysis was focused on matrix tissue from the pre-commissural dorsal striatum, because the striosomes expressed very low levels of GluD1 immunoreactivity.
The pattern of GluD1 immunoreactivity seen within specific neuronal elements was largely the same between striosome and matrix regions and across animal species (Figs 2,3). Within dendritic shafts, dense peroxidase staining, that occasionally diffused into synaptic clefts, was often aggregated at the post-synaptic densities (PSDs) of asymmetric synapses (Fig. 3c,d). We also encountered dendritic profiles with dense GuD1 staining along the plasma membrane at sites devoid of clear synaptic contacts (Fig. 3a). In other instances, the peroxidase deposit was in the intracellular compartment, adjacent to the external membrane of mitochondria (Figs 2a–c; 3a,e). In labeled spines, the extent of GluD1 immunoreactivity was variable, ranging from nearly completely filled spine heads and necks (Fig. 2d) to restricted staining associated with the PSD of asymmetric axo-spinous synapses (Fig. 3a). GluD1-immunoreactive unmyelinated axonal profiles, which were only found in the monkey striatum, were identified as darkly stained circular structures with microtubules surrounded by other unlabeled axons (Fig. 3e). Although rare, GluD1-positive terminals with light plasma membrane staining were occasionally seen within the monkey caudate. Glial GluD1-immunostained processes were sometimes located near GluD1-labeled synapses (Figs 2c; 3a,c) or in the striatal neuropil without any obvious associations with specific neuronal elements (Figs 2a,b; 3b).
Figure 2: GluD1-immunoreactive elements in the mouse striatum.
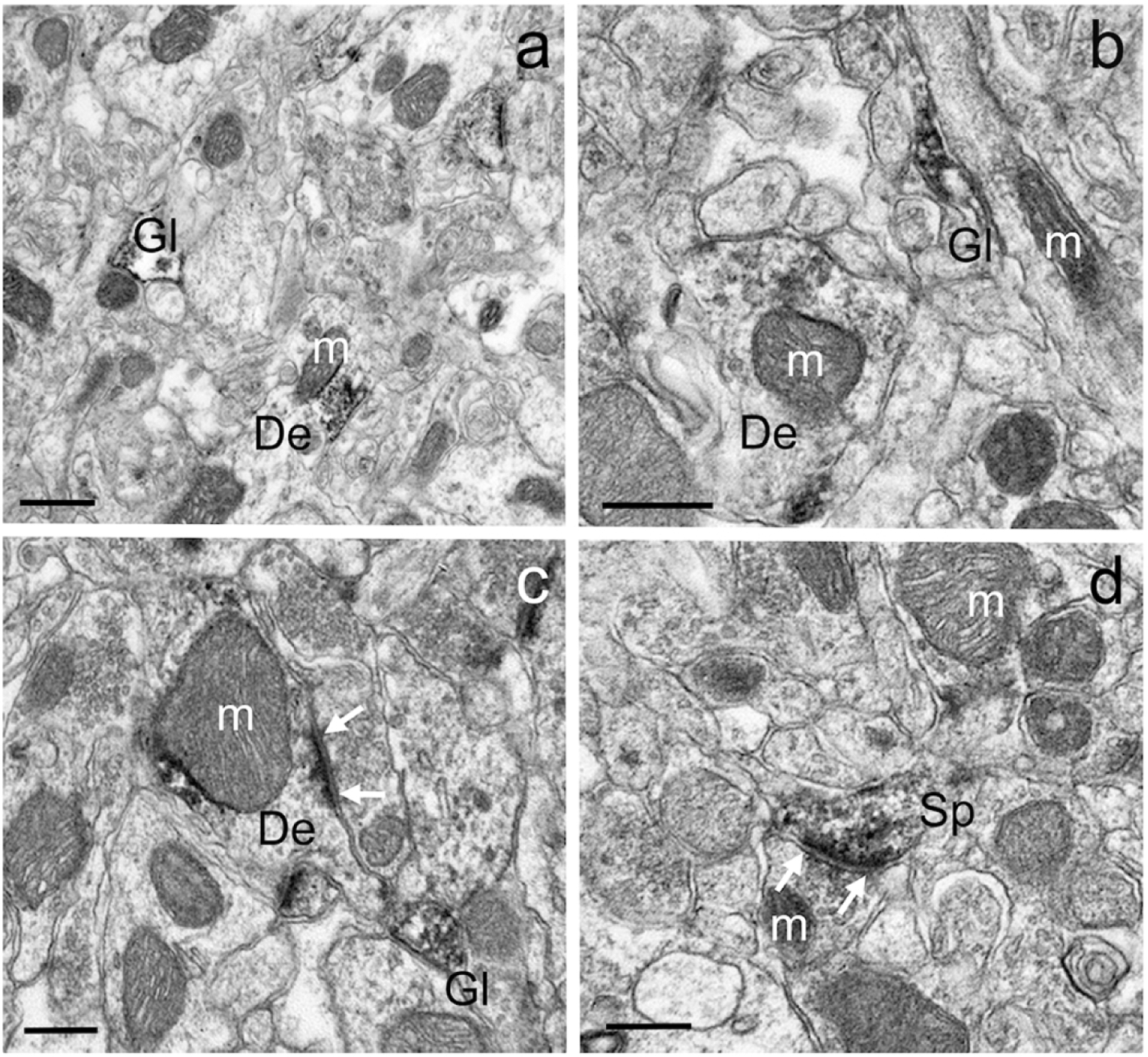
Examples of different GluD1-immunoreactive dendrites (a-c), spines (d) and glia (a,b) in the mouse dorsal striatum. In c and d, white arrows indicate asymmetric axo-dendritic (c) and axo-spinous (d) synapses. Note some GluD1 peroxidase staining associated with the surface of mitochondria in a-c. Abbreviations: Sp: spine, De: dendrite, Gl: glia and m: mitochondria. Scale bar in a = 0.50μm. Scale bar in b = 0.35μm. Scale bar in c = 0.25μm. Scale bar in d = 0.25μm.
Figure 3: GluD1-immunoreactive elements in the monkey striatum.
Examples of GluD1-positive neuronal and glial structures in the monkey striatum. White arrows indicate asymmetric synapses with dense aggregates of GluD1 imunolabeling in their close vicinity. Abbreviations: Sp: spine, De: dendrite, Gl: glia, m: mitochondria and Ax: unmyelinated axon. Scale bar in a (applies to b-e) = 0.30μm.
Inter-individual differences in the relative distribution of GluD1 between animals of the same group (striosome vs matrix) were tested using one-way ANOVA. Because this analysis did not reveal any significant within-group differences, quantitative data reported in Figure 4 are average percentages ± SEM of pooled data from 3 mice and 3 monkeys per group. Overall, the present findings from the mouse striatum confirm data from our recent study (Liu et al. 2020). In brief, GluD1 immunoreactivity in the mouse dorsal striatum was preferentially associated with dendritic shafts (47.9 ± 1.2% of total labeled structures) and glia (37.7 ± 2.5%), but significantly less abundant in dendritic spines (14.3 ± 2.6%) (Fig. 4a). A comparable trend was found in both the striosome and matrix compartments within the pre-commissural caudate nucleus in monkeys. In the striosome compartments, GluD1 was also preferentially found in dendritic shafts (44.4 ± 2.8% of total labeled elements) and glia (36.4 ± 0.4%) compared to spines (12.5 ± 2.7%), axon terminals (1.8 ± 0.3%) and unmyelinated axons (4.8 ± 7.2%) (Fig. 4b). Overall, the pattern was the same in the matrix, i.e dendritic shafts accounted for the largest proportion of GluD1-labeled structures (49.0 ± 1.5%) followed by glia (33.8 ± 3.9%), spines (12.7 ± 1.5%), axons (7.2 ± 1.5%) and axon terminals (1.2 ± 0.3%) (Fig. 4b). A two-sample t-test analysis of the relative proportion of GluD1-labeled elements between the striosome and matrix compartments of the pre-commissural caudate from 3 monkeys did not reveal any significant difference between the two compartments (dendrites p=0.870; glia p=0.548; spines p=0.953; axons p=0.243; terminals p=0.268). A two-sample t-test comparing the relative proportion of GluD-1-labeled elements between the matrix compartments of mice (N=3) and monkeys (N=3) also yielded no statistically significant differences (dendrites p=0.140; glia p=0.451; spines p=0.633).
Figure 4: Relative Distribution of GluD1-immunostained elements.
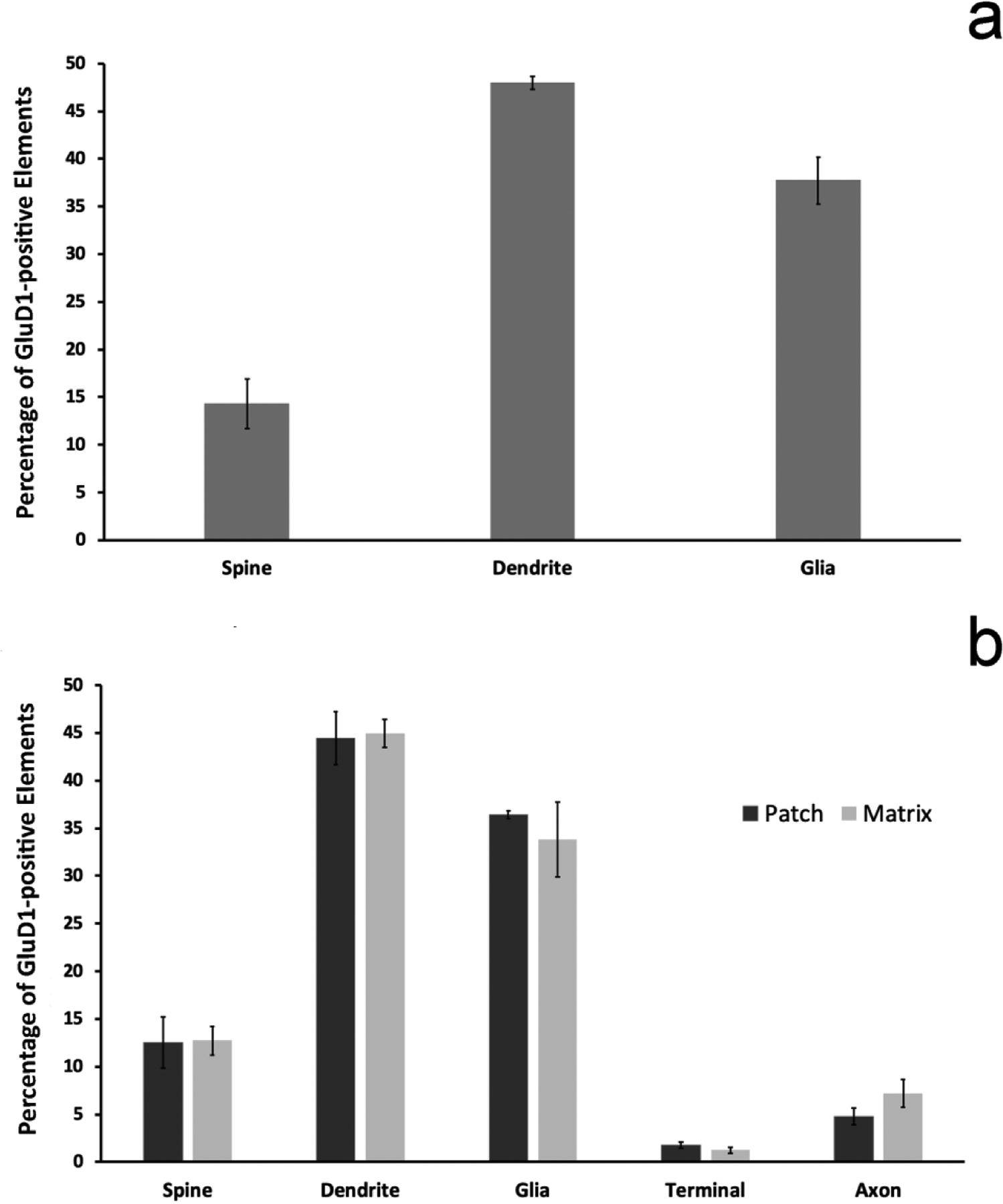
(a) shows the mean relative percentages (+/− SEM) of GluD1-positive neuronal and glial structures in the matrix compartment of the mouse dorsal striatum (N=3 animals; n=157 images). (b) illustrates the relative percentage of GluD1-immunostained elements in the striosome vs matrix compartments of the monkey caudate nucleus (N=3 animals; n=324 images). There is no statistically significant difference in the relative percentages of the different neuronal and glial elements between the striosome and matrix compartments (two sample t-test; dendrites p=0.870; glia p=0.548; spines p=0.953; axons p=0.243; terminals p=0.268). There is also no statistically significant difference between the percentages of GluD1-immunoreactive elements of the mouse vs the monkey matrix compartment (two-sample t-test; dendrites p=0.140; glia p=0.451; spines p=0.633).
Electron Microscopic Localization of GluD1 in the Mouse and Monkey Striatum-Immunogold Staining
In a third set of experiments, we sought to further enhance our knowledge of the subcellular (and subsynaptic) localization of GluD1 in the mouse and monkey striatum using the pre-embedding immunogold approach. However, due to technical challenges in using the antibodies to reliably assess the localization of GluD1 with gold particles in many of our animals, results reported in this part of study were collected from the striatal tissue of one monkey (MR-272) and three mice (RM-12, RM-74, RM-76). A lower cut-off of three or more immunogold particles per neuronal elements was arbitrarily set to differentiate immunoreactive from non-immunoreactive neuronal elements, while in glia, a single gold particle was considered as an evidence of GluD1 immunoreactivity because of their small size. Due to the low number of animals that could be used in these experiments, statistical analyzes were not performed.
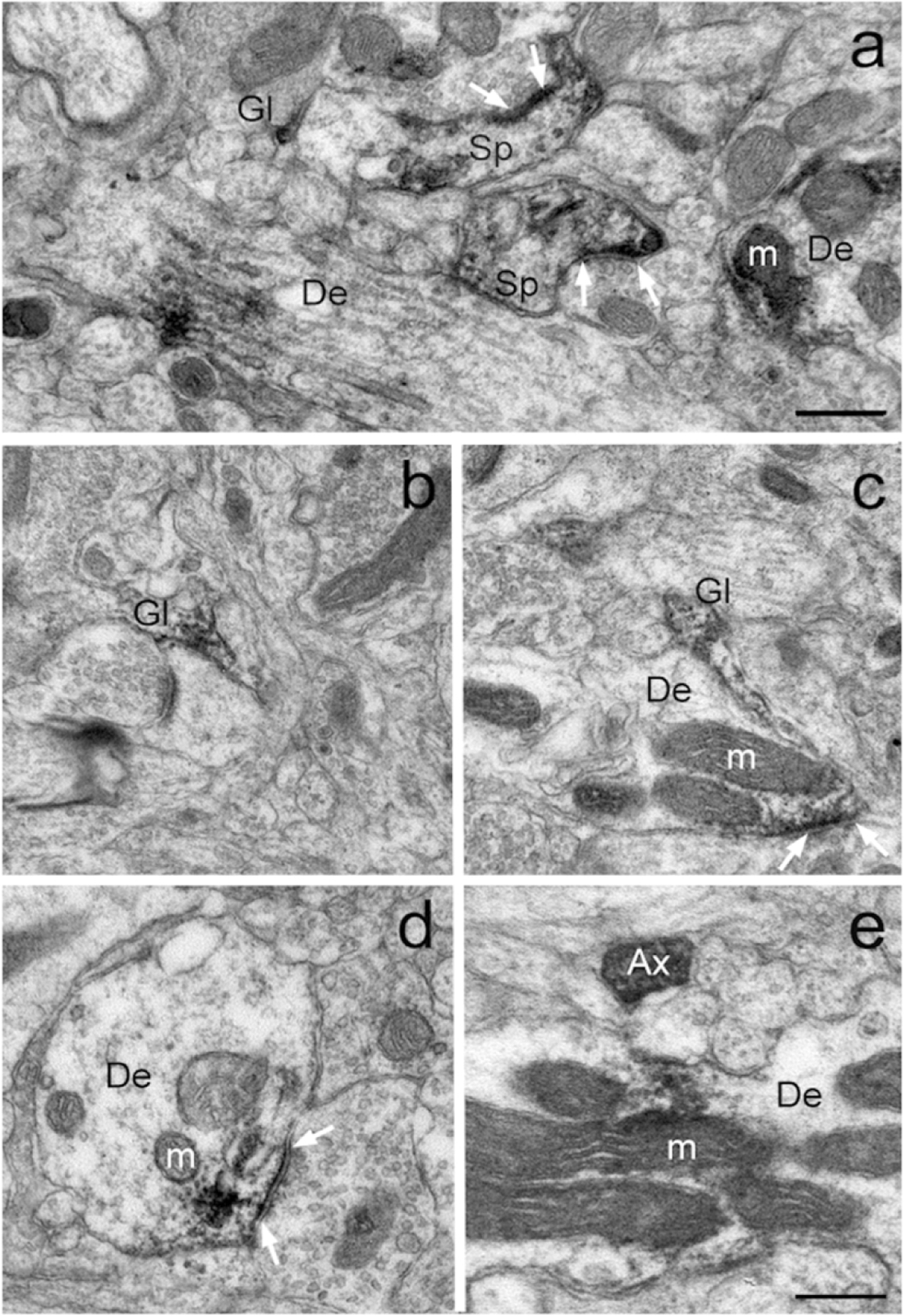
In line with the immunoperoxidase data, the immunogold labeling was found in dendritic shafts, spines and glial processes in the mouse and monkey striatum. As depicted in Figure 5, both plasma membrane-bound and intracellular gold particles were found in GluD1-positive dendrites and spines. In some instances, the plasma membrane-bound gold labeling was aggregated at the edges and in the main body of asymmetric synapses (Fig. 5a–g) or located at non-synaptic sites along the plasma membrane (Fig. 5a,b), while the intracellular labeling was commonly seen in close apposition with the membrane of mitochondria (Fig. 5a,d) or attached to microtubules (Fig. 5f). Because of their small diameter relative to the size of the silver-intensified gold particles, glial processes contained GluD1 gold labeling that filled the whole processes without any clear distinction between the plasma membrane-bound vs intracellular compartment (Fig. 5b).
Figure 5: Immunogold localization of GluD1 in the mouse and monkey striatum.
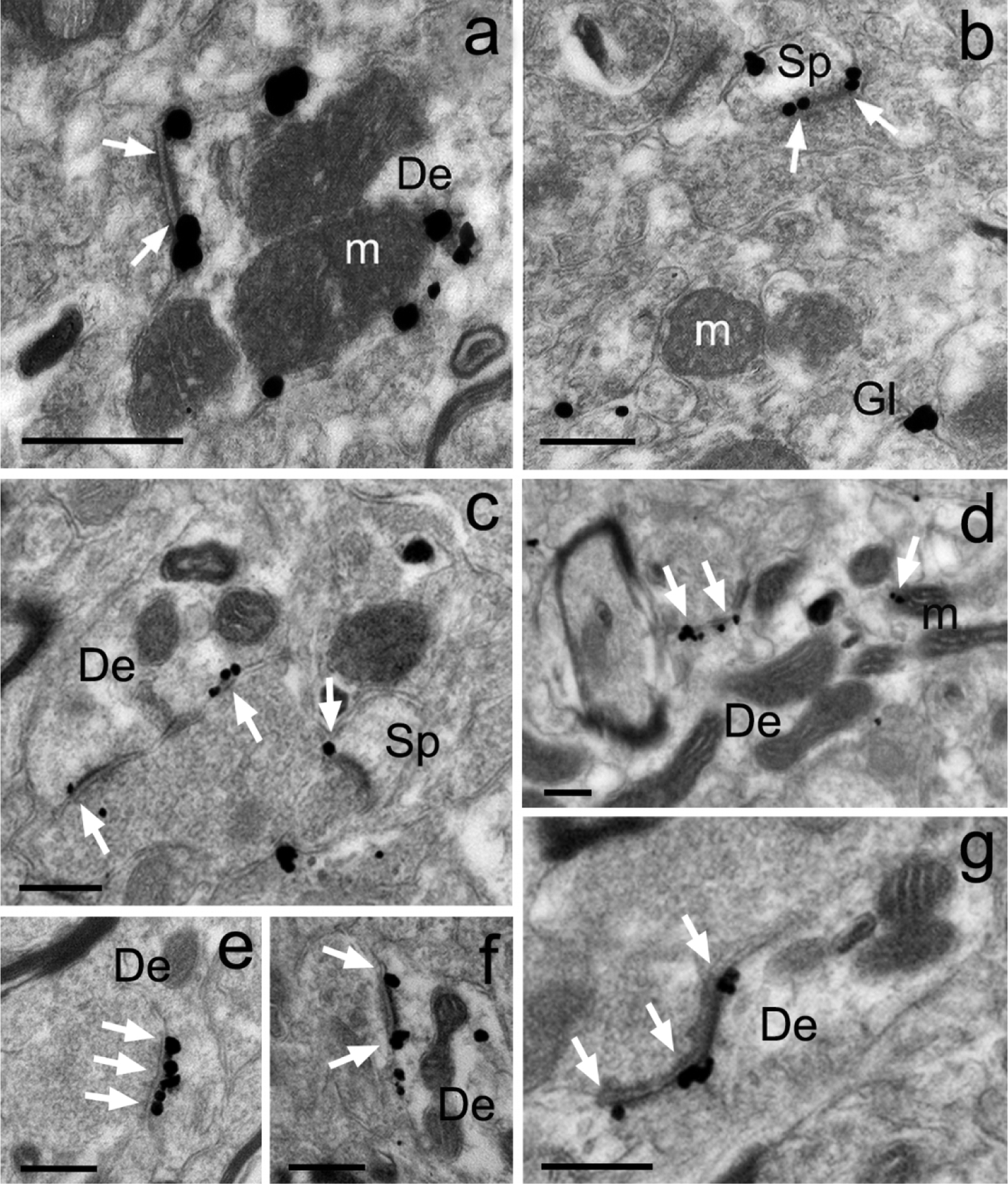
Examples of GluD1-immunoreactive elements in the mouse (a,b) and monkey (c-g) pre-commissural striatum as revealed with the pre-embedding immunogold technique. (a-b) Peri-synaptic gold labeling at an asymmetric axo-dendritic (a) and axo-spinous (b) synapse. Intracellular gold particles closely apposed to the external membrane of a mitochondria are also depicted in panel “a”. (c-g) Examples of peri-synaptic (c,f,g) and synaptic (e,g) GluD1 immunolabeling at asymmetric synapses in the monkey caudate nucleus. White arrows indicate GluD1-labeled asymmetric synapses. Abbreviations: Sp: spine, De: dendrite, Gl: glia and m: mitochondria. Scale bars = 0.25μm in all panels.
Confocal analysis of relationships between vGluT1- and vGluT2-immunoreactive terminals and GluD1-immunoreactive profiles
To determine if the relationships between vGluT1 or vGluT2 terminals and GluD1-immunoreactive profiles reported in our recent mouse study (Liu et al., 2020) was also seen in monkeys, we processed striatal tissue from one monkey for double immunofluorescence, and assessed the extent of co-localization between vGluT1- or vGluT2- and GluD1-immunoreactive profiles in the monkey caudate nucleus. Although we did not use a specific marker of striosomes in this analysis, we could identify striatal areas significantly more enriched than others in GluD1 immunoreactivity, which we considered as putative striosomes. Based on this assumption, our data confirmed that there is a higher level of co-localization of vGluT2/GluD1, than vGluT1/GluD1 in both the putative striosomal and extrastriosomal regions of the monkey striatum (Figs. 6–8 and Table 3).
Figure 6: Confocal images of GluD1/vGluT1 in the monkey striatum.
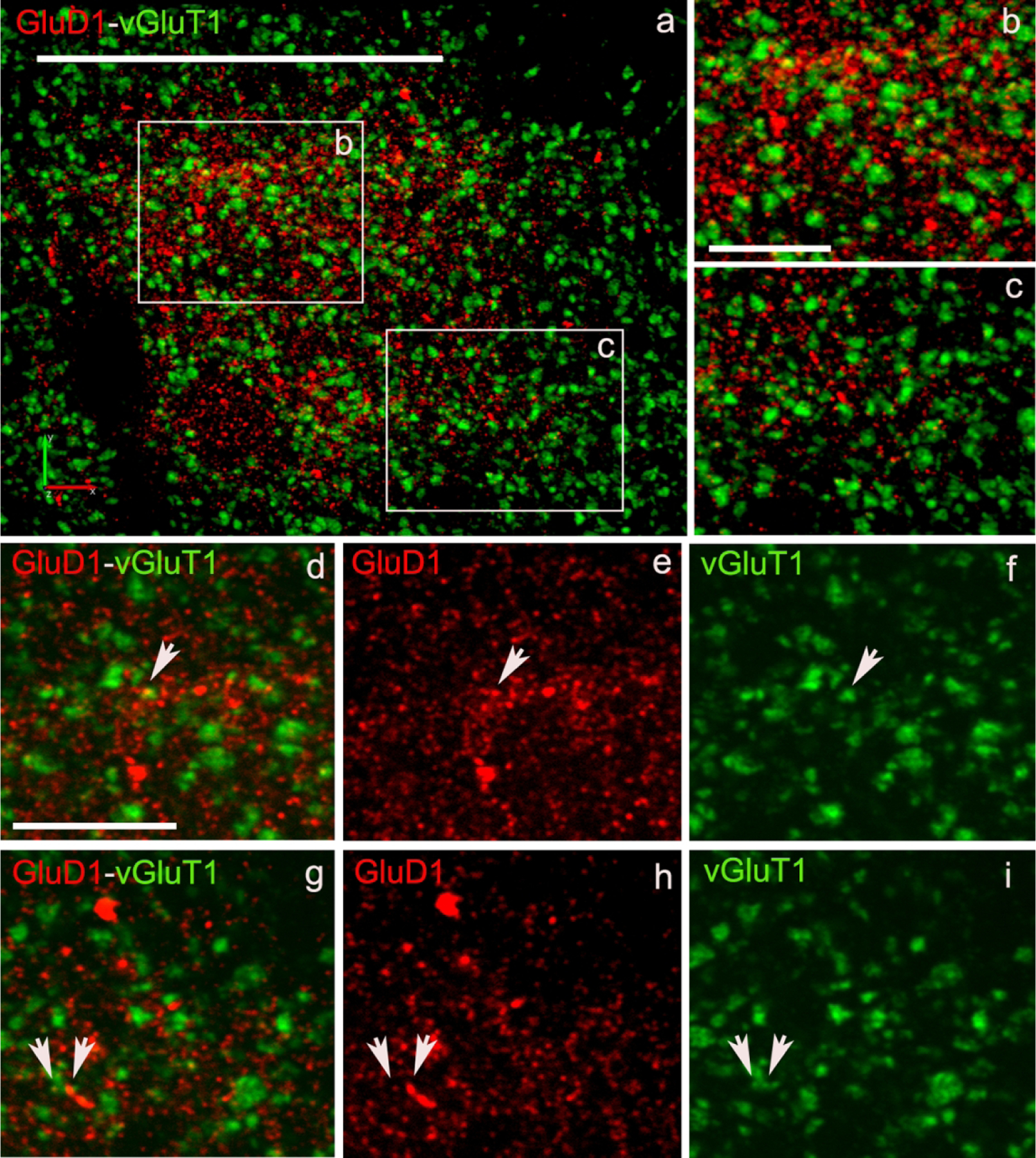
(a) Low magnification image of a putative striosome (enriched in GluD1 immunoreactivity) in the caudate nucleus of monkey striatum double immunostained for GluD1 (red) and vGluT1 (Figure 6) or vGluT2 (Figure 7) (green). (b,c) High magnification images of boxed areas marked in panel a. (d,g) Colocalization of GluD1 and vGluT1/vGluT2 in double immunostained “terminal-like” structures (puncta). The same puncta structures (arrows in d-i) are identified in single immunofluorescence images for GluD1 (e,h) and vGluT1 or vGluT2 (f,i). Scale bar in a = 50μm in b (applies to c) and d (applies to e-i) = 10μm.
Figure 8: Quantitative analysis of double immunostained confocal images.
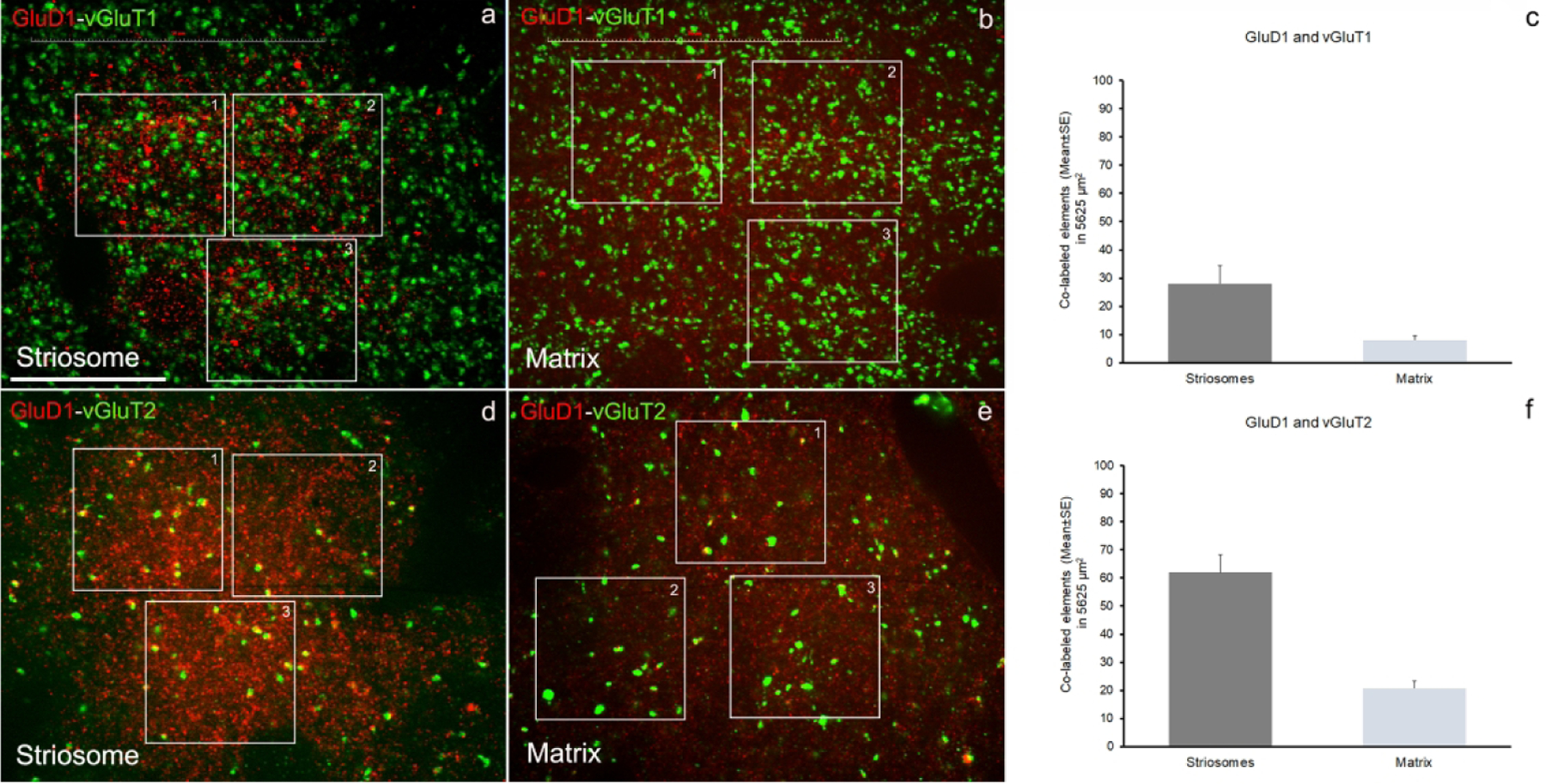
Quantification of co-labeled puncta structures in double immunostained sections have been done in striatal areas (caudate) with high GluD1-immunofluorescence (striosome-like) (a, d) and areas with low GluD1-immunofluorescence (matrix-like) (b,e). The quantitative results for co-labeled elements for GluD1/vGluT1 (c) and GluD1/vGluT2 (d) in striosome- and matrix-like areas have been obtained from three areas/image (boxed areas) and three images/striosome- and /matrix-like areas for each double immunostaining, GluD1/vGluT1 or GluD1/vGluT2. The data are shown as the mean value (±SE) of co-labeled elements in the total area analyzed (5625 μm2). The detailed quantitative data for co-labeled puncta elements and single labeled structures are shown in the Table 3. Scale bar in a = 25μm.
Table 3:
Confocal quantitative analysis
| GluD1-vGluT1 | GluD1-vGluT2 | ||||||
|---|---|---|---|---|---|---|---|
| STRIOSOMES | STRIOSOMES | ||||||
| Total Area Analyzed per Image = A1+A2+A3 = 1875 μm2; Total Area (3 images) = 5625 μm2 | Total Area Analyzed per Image = A1+A2+A3 = 1875 μm2; Total Area (3 images) = 5625 μm2 | ||||||
| Counts from GluD1-vGluH (ImageJ) | GluD1 | vGluT1 | GluD1+vGluT1 | Counts from GluD1-vGluT2 (ImageJ) | GluD1 | vGluT2 | GluD1+vGluT2 |
| confo_14_v1 | 832 | 575 | 23 | confo_7_v2 | 715 | 128 | 50 |
| confo_9_v1 | 991 | 668 | 41 | confo_8_v2 | 712 | 104 | 64 |
| confo_2_v1 | 790 | 549 | 21 | confo_15_v2 | 632 | 120 | 72 |
| Totals (number in 5625 μm2) | 2613 | 1792 | 85 | Totals (number in 5625 μm2) | 2059 | 352 | 186 |
| Mean | 871 | 597 | 28 | Mean | 686 | 117 | 62 |
| SE | 27.2 | 36.1 | 6.4 | SE | 27.2 | 7.1 | 6.4 |
| MATRIX | MATRIX | ||||||
| Total Area Analyzed per Image = A1+A2+A3 = 1875 μm2; Total Area (3 images) = 5625 μm2 | Total Area Analyzed per Image = A1+A2+A3 = 1875 μm2: Total Area (3 images) = 5625 μm2 | ||||||
| Counts from GluD1-vGluT2 (ImageJ) | GluD1 | vGluT1 | GluD1+vGluT1 | Counts from GluD1-vGluT2 (ImageJ) | GluD1 | vGluT2 | GluD1+vGluT2 |
| confo_10_v1 | 289 | 458 | 5 | confo_4_v2 | 398 | 38 | 16 |
| confo_13_v1 | 617 | 474 | 10 | confo_10_v2 | 477 | 43 | 21 |
| confo_15_v1 | 710 | 633 | 9 | confo_14_v2 | 367 | 57 | 25 |
| Totals (number in 5625 μm2) | 1616 | 1565 | 24 | Totals (number in 5625 μm2) | 1242 | 138 | 62 |
| Mean | 539 | 522 | 8 | Mean | 414 | 46 | 20.7 |
| SE | 127.7 | 55.8 | 1.5 | SE | 32.8 | 5.7 | 2.6 |
Discussion
Our findings reveal that GluD1 is differentially expressed in the striosome and matrix compartments of the mouse and monkey striatum, a pattern reminiscent of many other striatal neurochemicals and neurotransmitter receptors. However, GluD1 distribution differs from other striatal chemicals because it displays an opposite species-specific pattern of expression between the two compartments. This differential compartmental distribution is not reflected by significant differences in the overall pattern of GluD1 localization between striosomes and matrix in the monkey striatum, ie in both compartments, dendritic shafts and glia are the most common GluD1-containing structures followed by spines, unmyelinated axons and scarce terminals. This pattern of distribution is also similar to that of GluD1-immunoreactive profiles in the matrix of the mouse dorsal striatum. As expected, the subcellular localization of GluD1immunoreactivity suggests that the receptor plays a role at glutamatergic axo-spinous and axo-dendritic synapses. However, the localization of GluD1 immunoreactivity at non-synaptic sites along neuronal plasma membranes, in glia or on the external membrane of mitochondria, suggests that striatal GluD1 may contribute to a wide range of neuronal and glial functions in the rodent and primate striatum.
Differential Striosome/Matrix Distribution of GluD1 Immunoreactivity
Our findings indicate that the striosomes are more enriched in GluD1 immunoreactivity than the matrix in the monkey striatum, while the opposite is true in mice, ie the bulk of GluD1 immunoreactivity is localized in the matrix compartment. To our knowledge, a species difference in the striosome/matrix compartmentation of neurochemicals between normal mouse and monkeys using similar immunohistochemical approaches and antibodies has not been described before (Graybiel, 1990; Crittenden and Graybiel, 2011). However, previous data reported different effects of striatal dopamine depletion on the changes in GluR1 AMPA receptor subunit expression in the striosomes and matrix between primate and rodent models of Parkinson’s disease (Porter et al., 1994; Betarbet et al., 2000). Although the functional significance of this potential species difference in striosome/matrix GluD1 expression remains unclear, it is noteworthy because of the known hodological differences between the two compartments and their differential responses to drug treatments and brain disorders (Crittenden and Graybiel, 2011). Our EM data demonstrate that the differences in GluD1 immunoreactivity between the striosome and the matrix compartments in the monkey striatum cannot be explained by a differential pattern of distribution of GluD1 between neuronal and glial elements. However, we cannot rule out that the relative prevalence of the various GluD1-immunoreactive profiles is higher in the striosomes than in the matrix compartment, because our material did not allow us to assess the relative density of labeled vs unlabeled elements in each compartment, due to potential false negative results.
Another important consequence of the striosome/matrix compartmentation of GluD1 relates to the differential sources of pre-synaptic afferents that could functionally interact with post-synaptic GluD1. As described in previous studies, GluD1 exhibits its synaptic effects by collaborating with other presynaptic proteins to build a molecular bridge between terminals and postsynaptic elements. In the cerebellar cortex, cerebellin-1 (Cbln1) and Neurexin (Nxn), in concert with GluD2, are crucial synaptic organizers of Purkinje cells (Hirai et al., 2005; Hirano, 2012; Otsuka et al. 2016). Recent evidence indicates that Cbln1 and Nxn form such synaptic bridges with GluD1 in the mouse striatum (Yuzaki. 2018; Liu et al. 2020). Although Nxn is ubiquitously distributed in the mouse CNS, Cbln1 displays a much more restricted pattern of localization. Among the Cbln1-enriched brain regions that contribute significant afferent projections to the striatum is the parafascicular (Pf) nucleus of the thalamus (Herkenham and Pert. 1981; Miura et al., 2006; Wei et al., 2007; Kusnoor et al. 2009; Kusnoor et al. 2010; Smith et al. 2014). Besides the cerebellar cortex, thalamic Pf neurons are among the most strongly enriched subcortical neurons in Cbln1 mRNA and protein (Miura et al., 2006; Wei et al., 2007; Kusnoor et al. 2009; Kusnoor et al. 2010). Knowing that the Pf projects exclusively to the matrix compartment of the rodent and primate striatum (Herkenham and Pert, 1981, Smith et al., 2014), it is likely to be the main source of Cbln1-containing terminals in the striatal matrix. Tract tracing data have indeed demonstrated that most Pf-thalamostriatal neurons express Cbln1 immumunoreactivity in mice (Kusnoor et al, 2010). In line, with these findings, preliminary EM data have suggested that Cbln1-positive terminals in mouse striatum display the ultrastructural features and patterns of synaptic connections that resemble those of Pf-striatal terminals (Kusnoor et al., 2010). The anatomical and functional interactions between striatal GluD1 and Pf-striatal terminals were further confirmed by recent data showing that vGluT2-containing thalamic terminals, but not vGuT1-positive cortical terminals, are in close contact with GluD1-immunoreactive striatal profiles (Liu et al., 2020). In addition, selective knock out of striatal GluD1 led to a profound loss of vGluT2-positive thalamostriatal terminals in mice (Liu et al., 2020). Altogether, these data provide a strong anatomical foundation for functional interactions between post-synaptic GluD1 and pre-synaptic Pf-striatal Cbln1 in the striatal matrix compartment, but they do not fully address the issue about the potential sources of pre-synaptic Cbln1 in the monkey striosomes.
Because Pf projections target almost exclusively the matrix compartment in monkeys (Sadikot et al., 1992; Smith et al, 2004, 2014), this thalamic nucleus cannot be considered as the source of Cbln1 in the striosomes. Given Cbln1 mRNA and protein localization in the rodent brain (Pang et al., 2000; Miura et al., 2006; Wei et al., 2007) and the known origins of inputs to striosomes (Gerfen, 1984; Gerfen, 1989; Graybiel, 1990; Smith et al., 2016), some potential sources of Cbln1 to this striatal compartment could be the retrosplenial cortex, non-Pf anterior and midline thalamic nuclei and the bed nucleus of stria terminalis. However, this remains highly speculative without a detailed analysis of the striatal projections of Cbln1-containing cortical and subcortical neurons.
Our confocal microscopy data extend our recent mouse findings (Liu et al., 2020) to nonhuman primates, such that in both species thalamostriatal vGluT2-containing terminals are more frequently co-localized with GluD1-immunoreactive profiles than vGluT1-positive corticostriatal terminals. Our data suggest that this difference is found in both striatal compartments in monkeys. Because the CM/Pf complex projects exclusively to the matrix in mice and monkeys, our observations raise the possibility that thalamic afferents, other than those from the CM/Pf, may be associated with GluD1 in the monkey striosome compartment. Future double immuno-electron microscopic studies using immunoperoxidase and immunogold as markers of vGluTs and GluD1, combined with specific markers of striosomes, are needed to fully address this issue.
The lack of a map of the neuronal localization Cbln1 in the primate brain is another major limitation that hampers advancing knowledge in this field. One may also consider the possibility that striosomal GluD1 interacts with Cerebellin-2 (Cbln2), which displays a far more widespread distribution than Cbln1 at the cortical level (Miura et al., 2006), including limbic cortices known as major sources of inputs to striosomes (Gerfen 1984, 1989; Graybiel 1990; Crittenden and Graybiel, 2011). However, because GluD1 shows a weak affinity for Cbln2 (Wei et al. 2012), the likelihood that Cbln2 may be the key partner of GluD1 in striosomes is unlikely. Finally, the likelihood that GluD1 in striosomes interacts with other, yet unknown, presynaptic proteins to mediate different functions than that of synaptic organizers cannot be ruled out.
GluD1 Expression in Glia and Mitochondria
Our immunoperoxidase and immunogold data additionally showed that GluD1 is expressed in glial processes throughout the monkey caudate and mouse dorsal striatum. Although the role of GluD1 in glia remains unknown, recent findings have shown that GluD1 mRNA is strongly expressed in certain types of glia – specifically in oligodendrocyte precursor cells (OPC), also called NG2+ cells (Larson et al. 2016; Saunders et al. 2018). Although NG2+ cells often differentiate into myelinating oligodendrocytes throughout the brain (Antel et al. 2019; Berry et al. 2020), this is not their only fate. These cells are also found in regions devoid of myelin, suggesting that they potentially have other functions (Larson et al., 2016). There is evidence that NG2+ cells are a unique population of glial cells that exhibit membrane and synaptic properties as well as gene expression patterns commonly seen in neurons (Larson et al., 2016). In addition, this cell type receives direct synaptic inputs from glutamatergic cells (Bergles et al. 2000; Ziskin et al 2007; Bergles et al. 2010). Although much remains to be known about the role of these neuro-glia connections, authors have speculated that they may be part of the NG2+ cells differentiation process (Kukley et al. 2010; Hill and Nishiyama. 2014). Knowing the importance of GluD1 in synaptic development and maintenance, the presence of GluD1 within NG2+ cells could have to do with their differentiation process (Gupta et al. 2015; Hepp et al. 2015). Further co-localization, molecular and functional studies are needed to further address this issue.
Both our immunoperoxidase and immunogold labeling demonstrated extrasynaptic and intracellular GluD1expression near mitochondrial membranes in dendritic shafts. To our knowledge, there is no significant literature about GluD1 functions in mitochondria. However, in line with our EM-observations, preliminary pull-down assays and mass spectroscopy data suggest interaction of GluD1 with mitochondrial proteins (Dravid et al., unpublished data).
Concluding Remarks
Our level of knowledge of the role of GluD1 in the CNS is still in its infancy. Data presented in this study set the foundation for further studies of striatal GluD1 function. Combined with recent findings (Liu et al., 2020), the present data further indicate that GluD1 is located to subserve regulatory functions of thalamostriatal glutamatergic synapses in mouse. However, they also suggest that GluD1 functions in the striatum may go beyond the regulation of thalamic afferents, particularly in the striosome compartment of the monkey striatum. Furthermore, EM evidence for expression of GluD1 immunoreactivity in glia and mitochondria provides additional targets through which GluD1 may regulate normal striatal functions. Future studies that examine possible changes in GluD1 expression in animal models of basal ganglia disorders and postmortem human material are warranted to determine the possible contribution of GluD1 dysregulation in brain diseases.
Figure 7: Confocal images of GluD1/vGluT2 in the monkey striatum.
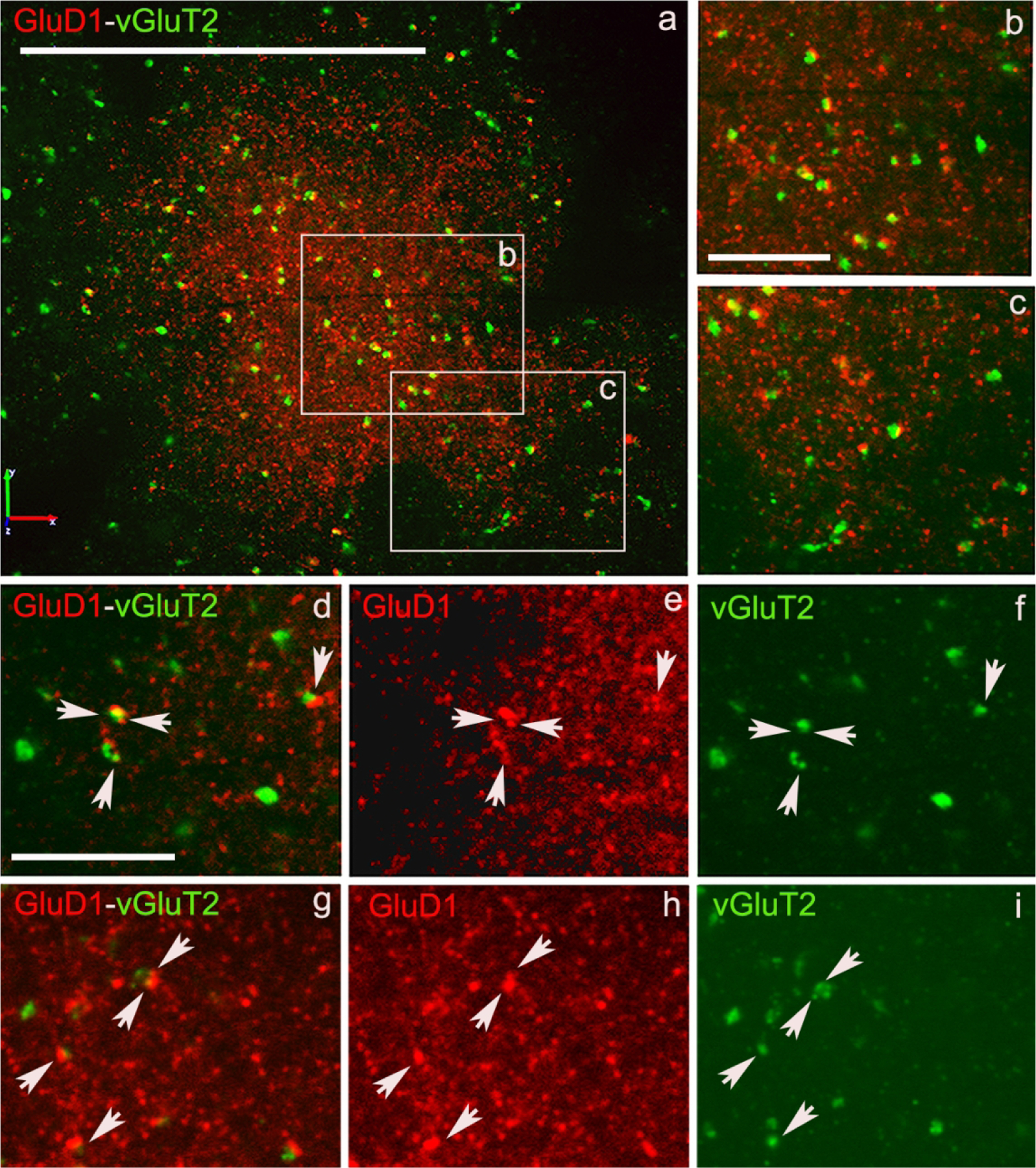
(a) Low magnification image of a putative striosome (enriched in GluD1 immunoreactivity) in the caudate nucleus of monkey striatum double immunostained for GluD1 (red) and vGluT1 (Figure 6) or vGluT2 (Figure 7) (green). (b,c) High magnification images of boxed areas marked in panel a. (d,g) Colocalization of GluD1 and vGluT1/vGluT2 in double immunostained “terminal-like” structures (puncta). The same puncta structures (arrows in d-i) are identified in single immunofluorescence images for GluD1 (e,h) and vGluT1 or vGluT2 (f,i). Scale bar in a = 50μm in b (applies to c) and d (applies to e-i) = 10μm.
Acknowledgements:
The authors thank Susan Jenkins and Jean-Francois Pare for technical assistance. This work was supported by NIH grants R01 MH116003 and P50 NS098685, the Yerkes National Primate Center NIH/ORIP base grant P51 OD011132, and by the NSF1456818 and Stem Cell 2019-05 grants.
Footnotes
Data Availability Statement: The data that support the findings of this study are available from the corresponding author upon reasonable request
Conflict of Interest: The authors declare nonfinancial competing interests or other conflicts of interests for this article.
References
- Antel JP, Lin YH, Cui Q-L, Pernin F, Kennedy TE, Ludwin SK, & Healy LM (2019). Immunology of oligodendrocyte precursor cells in vivo and in vitro. Journal of Neuroimmunology, 331, 28–35. 10.1016/j.jneuroim.2018.03.006 [DOI] [PubMed] [Google Scholar]
- Benamer N, Marti F, Lujan R, Hepp R, Aubier TG, Dupin A. a. M., Frébourg G, Pons S, Maskos U, Faure P, Hay YA, Lambolez B, & Tricoire L (2018). GluD1, linked to schizophrenia, controls the burst firing of dopamine neurons. Molecular Psychiatry, 23(3), 691–700. 10.1038/mp.2017.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Jabs R, & Steinhäuser C (2010). Neuron-glia synapses in the brain. Brain Research Reviews, 63(1–2), 130–137. 10.1016/j.brainresrev.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Roberts JDB, Somogyi P, & Jahr CE (2000). Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature, 405(6783), 187–191. 10.1038/35012083 [DOI] [PubMed] [Google Scholar]
- Berridge G, Menassa DA, Moloney T, Waters PJ, Welding I, Thomsen S, Zuberi S, Fischer R, Aricescu AR, Pike M, Dale RC, Kessler B, Vincent A, Lim M, Irani SR, & Lang B (2018). Glutamate receptor δ2 serum antibodies in pediatric opsoclonus myoclonus ataxia syndrome. Neurology, 91(8), e714–e723. 10.1212/WNL.0000000000006035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry K, Wang J, & Lu QR (2020). Epigenetic regulation of oligodendrocyte myelination in developmental disorders and neurodegenerative diseases. F1000Research, 9. 10.12688/f1000research.20904.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Porter RHP & Greenamyre TJ (2000) GluR1 glutamate receptor subunit is regulated differentially in the primate basal ganglia following nigrostriatal dopamine denervation. Journal of Neurochemistry, 74, 1166–1174. [DOI] [PubMed] [Google Scholar]
- Côté P-Y, Sadikot AF, & Parent A (1991). Complementary Distribution of Calbindin D-28k and Parvalbumin in the Basal Forebrain and Midbrain of the Squirrel Monkey. European Journal of Neuroscience, 3(12), 1316–1329. 10.1111/j.1460-9568.1991.tb00064.x [DOI] [PubMed] [Google Scholar]
- Crittenden JR, & Graybiel AM (2011). Basal Ganglia Disorders Associated with Imbalances in the Striatal Striosome and Matrix Compartments. Frontiers in Neuroanatomy, 5. https://doi.org/10.3389/fnana.2011.00059 Electrophysiological properties of NG2+ cells: Matching physiological studies with gene expression profiles . (n.d.). Retrieved April 5, 2020, from https://doi.org/10.3389/fnana.2011.00059https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4792778/Electrophysiological properties of NG2+ cells: Matching physiological studies with gene expression profiles . (n.d.). Retrieved April 5, 2020, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4792778/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI Jr., Schuckit MA, Porjesz B & Dick DM (2012) Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet, 22, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, (1984) The neostriatal mosaic: Compartmentalization of corticostriatal input and striatonigral output systems. Nature, 311, 461–464. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Baimbridge KG, & Miller JJ (1985). The neostriatal mosaic: Compartmental distribution of calcium-binding protein and parvalbumin in the basal ganglia of the rat and monkey. Proceedings of the National Academy of Sciences of the United States of America, 82(24), 8780–8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR (1989). The neostriatal mosaic: Striatal patch-matrix organization is related to cortical lamination. Science, 246(4928), 385–388. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD & Hakonarson H (2009). Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature, 459, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel Ann M. (1990). Neurotransmitters and neuromodulators in the basal ganglia. Trends in Neurosciences, 13(7), 244–254. 10.1016/0166-2236(90)90104-I [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Green MF, Gur RE, Gur RC, Hardiman G, Kelsoe JR, Leonard S, Light GA, Nuechterlein KH, Olincy A, Radant AD, Schork NJ, Seidman LJ, Siever LJ, Silverman JM, Stone WS, Swerdlow NR, Tsuang DW, Tsuang MT, Turetsky BI, Freedman R & Braff DL (2011) Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry, 168, 930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, Jaworski JM, Salyakina D, Konidari I, Whitehead PL, Wright HH, Abramson RK, Williams SM, Menon R, Martin ER, Haines JL, Gilbert JR, Cuccaro ML & Pericak-Vance MA (2012) Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet, 21, 3513–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Yadav R, Pavuluri R, Morley BJ, Stairs DJ, & Dravid SM (2015). Essential role of GluD1 in dendritic spine development and GluN2B to GluN2A NMDAR subunit switch in the cortex and hippocampus reveals ability of GluN2B inhibition in correcting hyperconnectivity. Neuropharmacology, 93, 274–284. 10.1016/j.neuropharm.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepp R, Hay YA, Aguado C, Lujan R, Dauphinot L, Potier MC, Nomura S, Poirel O, El Mestikawy S, Lambolez B, & Tricoire L (2015). Glutamate receptors of the delta family are widely expressed in the adult brain. Brain Structure & Function, 220(5), 2797–2815. 10.1007/s00429-014-0827-4 [DOI] [PubMed] [Google Scholar]
- Herkenham M, & Pert CB (1981). Mosaic distribution of opiate receptors, parafascicular projections and acetylcholinesterase in rat striatum. Nature, 291(5814), 415–418. 10.1038/291415a0 [DOI] [PubMed] [Google Scholar]
- Hill RA, & Nishiyama A (2014). NG2 Cells (Polydendrocytes): Listeners to the Neural Network with Diverse Properties. Glia, 62(8), 1195–1210. 10.1002/glia.22664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M & Morgan JI (2005) Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci, 8, 1534–1541. [DOI] [PubMed] [Google Scholar]
- Hirano T (2012) Glutamate-receptor-like molecule GluRdelta2 involved in synapse formation at parallel fiber-Purkinje neuron synapses. Cerebellum, 11, 71–77. [DOI] [PubMed] [Google Scholar]
- Ichikawa R, Sakimura K, & Watanabe M (2016). GluD2 Endows Parallel Fiber–Purkinje Cell Synapses with a High Regenerative Capacity. Journal of Neuroscience, 36(17), 4846–4858. 10.1523/JNEUROSCI.0161-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Miyoshi Y, Hamase K, Matsuda S, Matsuda K, Kohda K, Emi K, Motohashi J, Konno R, Zaitsu K & Yuzaki M (2011) D-serine regulates cerebellar LTD and motor coordination through the delta2 glutamate receptor. Nat Neurosci, 14, 603–611. [DOI] [PubMed] [Google Scholar]
- Kashiwabuchi N, Ikeda K, Araki K, Hirano T, Shibuki K, Takayama C, Inoue Y, Kutsuwada T, Yagi T, Kang Y & et al. (1995) Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell, 81, 245–252. [DOI] [PubMed] [Google Scholar]
- Konno K, Matsuda K, Nakamoto C, Uchigashima M, Miyazaki T, Yamasaki M, Sakimura K, Yuzaki M, & Watanabe M (2014). Enriched Expression of GluD1 in Higher Brain Regions and Its Involvement in Parallel Fiber–Interneuron Synapse Formation in the Cerebellum. The Journal of Neuroscience, 34(22), 7412–7424. 10.1523/JNEUROSCI.0628-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Stoppel DC, Nong Y, Johnson MA, Nadler MJ, Ozkaynak E, Teng BL, Nagakura I, Mohammad F, Silva MA, Peterson S, Cruz TJ, Kasper EM, Arnaout R & Anderson MP (2017) Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature, 543, 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukley M, Nishiyama A, & Dietrich D (2010). The Fate of Synaptic Input to NG2 Glial Cells: Neurons Specifically Downregulate Transmitter Release onto Differentiating Oligodendroglial Cells. The Journal of Neuroscience, 30(24), 8320–8331. 10.1523/JNEUROSCI.0854-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnoor SV, Parris J, Muly EC, Morgan JI, & Deutch AY (2010). Extracerebellar role for Cerebellin1: Modulation of dendritic spine density and synapses in striatal medium spiny neurons. The Journal of Comparative Neurology, 518(13), 2525–2537. 10.1002/cne.22350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnoor Sheila V., Muly EC, Morgan JI, & Deutch AY (2009). Is the loss of thalamostriatal neurons protective in parkinsonism? Parkinsonism & Related Disorders, 15(Suppl 3), S162–S166. 10.1016/S1353-8020(09)70806-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson VA, Zhang Y, & Bergles DE (2016). Electrophysiological properties of NG2+ cells: Matching physiological studies with gene expression profiles. Brain Research, 1638(Pt B), 138–160. 10.1016/j.brainres.2015.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shelkar GP, Gandhi PJ, Gawande DY, Hoover A, Villalba RM, Pavuluri R, Smith Y, & Dravid SM (2020). Striatal glutamate delta-1 receptor regulates behavioral flexibility and thalamostriatal connectivity. Neurobiology of Disease, 137, 104746. 10.1016/j.nbd.2020.104746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livide G, Patriarchi T, Amenduni M, Amabile S, Yasui D, Calcagno E, Lo Rizzo C, De Falco G, Ulivieri C, Ariani F, Mari F, Mencarelli MA, Hell JW, Renieri A & Meloni I (2015) GluD1 is a common altered player in neuronal differentiation from both MECP2-mutated and CDKL5-mutated iPS cells. Eur J Hum Genet, 23, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, Watanabe M & Yuzaki M (2010) Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science, 328, 363–368. [DOI] [PubMed] [Google Scholar]
- Miura E, Lijima T, Yuzaki M, Watanabe M (2006) Distinct expression of Cbln family mRNAs in developing and adult mouse brains. European Journal of Neurosscience, 24, 750–760. [DOI] [PubMed] [Google Scholar]
- Nakamoto C, Kawamura M, Nakatsukasa E, Natsume R, Takao K, Watanabe M, Abe M, Takeuchi T, & Sakimura K (2020a). GluD1 knockout mice with a pure C57BL/6N background show impaired fear memory, social interaction, and enhanced depressive-like behavior. PloS One, 15(2), e0229288. 10.1371/journal.pone.0229288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto C, Konno K, Miyazaki T, Nakatsukasa E, Natsume R, Abe M, Kawamura M, Fukazawa Y, Shigemoto R, Yamasaki M, Sakimura K, & Watanabe M (2020b). Expression mapping, quantification, and complex formation of GluD1 and GluD2 glutamate receptors in adult mouse brain. The Journal of Comparative Neurology, 528(6), 1003–1027. 10.1002/cne.24792 [DOI] [PubMed] [Google Scholar]
- Nord AS, Roeb W, Dickel DE, Walsh T, Kusenda M, O’Connor KL, Malhotra D, McCarthy SE, Stray SM, Taylor SM, Sebat J, Network SP, King B, King MC & McClellan JM (2011) Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur J Hum Genet, 19, 727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Konno K, Abe M, Motohashi J, Kohda K, Sakimura K, Watanabe M, & Yuzaki M (2016). Roles of Cbln1 in Non-Motor Functions of Mice. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 36(46), 11801–11816. 10.1523/JNEUROSCI.0322-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Zuo J &Morgan JL (2000) Cbln3, a novel member of the precerebellin family that binds specifically to Cbln1. J. Neurosci 20, 6333–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Amabile S, Frullanti E, Landucci E, Lo Rizzo C, Ariani F, Costa M, Olimpico F, J WH, F MV, Renieri A & Meloni I (2016) Imbalance of excitatory/inhibitory synaptic protein expression in iPSC-derived neurons from FOXG1(+/−) patients and in foxg1(+/−) mice. Eur J Hum Genet, 24, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Huang AW and Toga AW (2000) The Rhesus Monkey Brain in Stereotaxic Coordinates. San Diego, Academic Press. [Google Scholar]
- Paxinos G, Franklin KBJ (2001) The Mouse Brain in Stereotaxic Coordinates. San Diego, Academic Press. [Google Scholar]
- Pernice HF, Schieweck R, Jafari M, Straub T, Bilban M, Kiebler MA, & Popper B (2019). Altered Glutamate Receptor Ionotropic Delta Subunit 2 Expression in Stau2-Deficient Cerebellar Purkinje Cells in the Adult Brain. International Journal of Molecular Sciences, 20(7). 10.3390/ijms20071797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Palay S, & Webster H (1991). The Fine Structure of the Nervous System (3rd ed.). Oxford University Press. [Google Scholar]
- Porter RHP, Greene JG, Higgins DS & Greenamyre JT (1994) Polysynaptic regulation of glutamate receptors and mitochondrial enzyme activities in the basal ganglia of rats with unilateral dopamine depletion. J. Neurosci, 14, 7192–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko E, Wysoker A, Goldman M, Krienen F, de Rivera H, Bien E, Baum M, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, & McCarroll SA (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell, 174(4), 1015–1030.e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot AF, Parent A, Smith Y & Bolam JP (1992) Efferent connections of the centromedian and parafascicular nuclei in primates. A light and electron microscopic study of the thalamostriatal projection in relation to striatal heterogeneity. J. Comp. Neurol, 320, 228–242. [DOI] [PubMed] [Google Scholar]
- Smith JB, Klug JR, Ross DL, Howard CD, Hollon NG, Ko VI, Hoffman H, Callaway EM, Gerfen CR, & Jin X (2016). Genetic-Based Dissection Unveils the Inputs and Outputs of Striatal Patch and Matrix Compartments. Neuron, 91(5), 1069–1084. 10.1016/j.neuron.2016.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Spence MA & Flodman P (2009) Nuclear and mitochondrial genome defects in autisms. Ann N Y Acad Sci, 1151, 102–132. [DOI] [PubMed] [Google Scholar]
- Smith Y, Raju D, Pare J-F & Sidibe M (2004) The thalamostriatal system: A highly specific network of the basal ganglia circuitry. Trends in Neurosci, 27, 520–527. [DOI] [PubMed] [Google Scholar]
- Smith Y, Galvan A, Ellender TJ, Doig N, Villalba RM, Huerta-Ocampo I, Wichmann T, & Bolam JP (2014). The thalamostriatal system in normal and diseased states. Frontiers in Systems Neuroscience, 8, 5. 10.3389/fnsys.2014.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC (2017) Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell, 171, 745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC (2018) Towards an Understanding of Synapse Formation. Neuron, 100, 276–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Mühleisen TW, Frank J, Mattheisen M, Herms S, Ludwig KU, Treutlein T, Schmael C, Strohmaier J, Bösshenz KV, Breuer R, Paul T, Witt SH, Schulze TG, Schlösser RGM, Nenadic I, Sauer H, Becker T, Maier W, … Rietschel M (2009). Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter. Schizophrenia Research, 111(1–3), 123–130. 10.1016/j.schres.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Turner TN, Yi Q, Krumm N, Huddleston J, Hoekzema K, HA FS, Doebley AL, Bernier RA, Nickerson DA & Eichler EE (2017) denovo-db: a compendium of human de novo variants. Nucleic Acids Res, 45, D804–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Lee S-J, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, & Mishina M (2010). Trans-Synaptic Interaction of GluRδ2 and Neurexin through Cbln1 Mediates Synapse Formation in the Cerebellum. Cell, 141(6), 1068–1079. 10.1016/j.cell.2010.04.035 [DOI] [PubMed] [Google Scholar]
- Wei P, Smeyne RJ, Bao D, Parris J & Morgan JI (2007) Mapping of Cbln1-like immunoreactivity in adult and developing mouse brain and its localization to the endolysosomal compartment of neurons. European Journal of Neuroscience, 26, 2962–2978. [DOI] [PubMed] [Google Scholar]
- Wei P, Pattarini R, Rong Y, Guo H, Bansal PK, Kusnoor SV, Deutch AY, Parris J, & Morgan JI (2012). The Cbln family of proteins interact with multiple signaling pathways. Journal of Neurochemistry, 121(5), 717–729. 10.1111/j.1471-4159.2012.07648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ & Dravid SM (2012) Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One, 7, e32969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Hillman BG, Gupta SC, Suryavanshi P, Bhatt JM, Pavuluri R, Stairs DJ & Dravid SM (2013) Deletion of glutamate delta-1 receptor in mouse leads to enhanced working memory and deficit in fear conditioning. PLoS One, 8, e60785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki M (2011). Cbln1 and its family proteins in synapse formation and maintenance. Current Opinion in Neurobiology, 21(2), 215–220. 10.1016/j.conb.2011.01.010 [DOI] [PubMed] [Google Scholar]
- Yuzaki M (2018). Two Classes of Secreted Synaptic Organizers in the Central Nervous System. Annual Review of Physiology, 80(1), 243–262. 10.1146/annurev-physiol-021317-121322 [DOI] [PubMed] [Google Scholar]
- Ziskin JL, Nishiyama A, Rubio M, Fukaya M, & Bergles DE (2007). Vesicular release of glutamate from unmyelinated axons in white matter. Nature Neuroscience, 10(3), 321–330. 10.1038/nn1854 [DOI] [PMC free article] [PubMed] [Google Scholar]


