Abstract
Cells must build and maintain at least one membrane that surrounds essential cellular components and provides structural integrity. Gram-negative bacteria possess an inner membrane, which separates the aqueous cytoplasmic and periplasmic compartments, and an outer membrane, which surrounds the periplasm. The outer membrane is an asymmetric bilayer with phospholipids in its inner leaflet and lipopolysaccharides in its outer leaflet. This structure provides cellular integrity and prevents the entry of many toxic compounds into the cell. Constructing the outer membrane is challenging, since its lipid constituents must be synthesized within the inner membrane, transported across the periplasm, and ultimately assembled into an asymmetric structure. This review highlights major recent advances in our understanding of the mechanism and structure of the intermembrane, multi-protein machine that transports lipopolysaccharide across the cell envelope. Although our understanding of phospholipid transport is very limited, we also provide a brief update on this topic.
Keywords: LPS, Lpt, diderm, inter-membrane transport, glycerophospholipid
Introduction
The outer membrane (OM) of Gram-negative bacteria is an asymmetrical lipid bilayer [1]. Its inner leaflet is built with phospholipids, while the glycolipid lipopolysaccharide (LPS, Fig. 1A) is the main lipid component of its outer leaflet (Fig. 1B) [2]. The tight packing of LPS molecules at the cell surface and the large hydrophilic moiety of the glycolipid make a strong permeability barrier against small, nonpolar molecules [3]. As a result, Gram-negative bacteria are naturally more resistant than monodermic bacteria to many antimicrobials, and developing antibiotics that can cross the OM has proven very difficult [4].
Figure 1: The Gram-negative cell envelope and LPS transport.
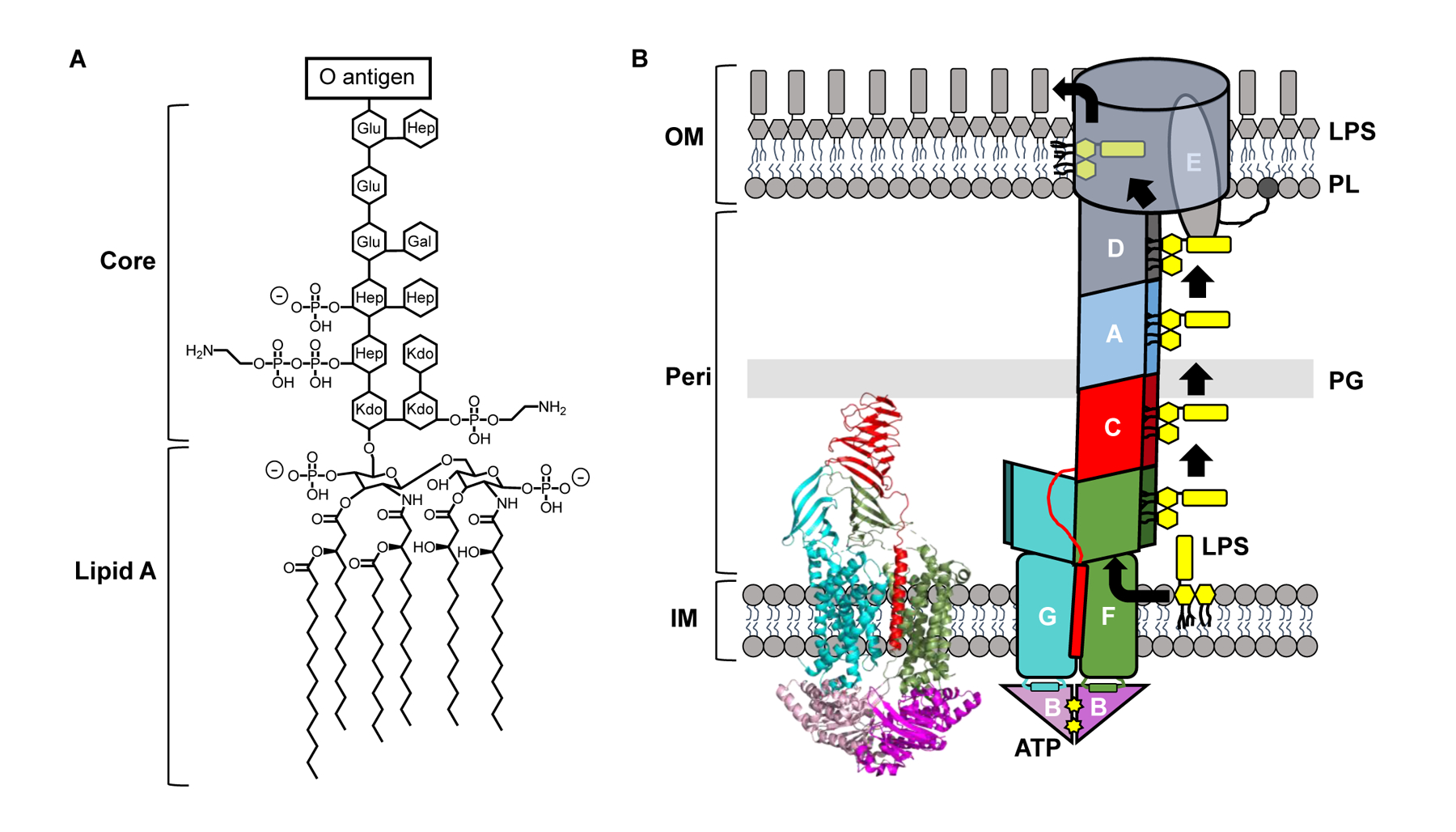
A) Structure of LPS from Escherichia coli K-12. B) ATP utilization by LptB2FGC drives the transport of newly synthesized LPS (yellow) across the periplasmic compartment. LptB2FGC is depicted both as a crystal structure (PDB ID: 6MJP) derived from Vibrio cholerae (left) and as a cartoon (right). The crystal structure carries the catalytically inactive LptB/E163Q variant. LPS, lipopolysaccharide. PL, phospholipid. PG, peptidoglycan. OM, outer membrane. IM, inner membrane. Peri, periplasm. Kdo, keto-deoxyoctulosonate. Hep, heptose. Glu, glucose. Gal, galactose.
Both phospholipids and LPS are synthesized at the inner membrane (IM) and must be transported to the OM [5,6]. Synthesis of mature LPS is completed at the periplasmic leaflet of the IM. From there, the essential seven-protein lipopolysaccharide transport (Lpt) complex is responsible for extracting LPS from the IM, transporting it across the periplasm, and inserting it into the outer leaflet of the OM (Fig. 1B) [7–12]. Studies published just in the last three years have uncovered many crucial details about the mechanism of LPS transport to the OM by this trans-envelope machine, particularly at the step of extraction from the IM. Here, we discuss the structural, biochemical, and genetic evidence that have revealed a much clearer understanding of how the Lpt system extracts LPS from the IM through a process energized by ATP. In contrast, the mechanism of phospholipid transport to the OM remains elusive, but we briefly highlight some recent studies proposing some candidate transporters.
Lipopolysaccharide Transport to the Outer Membrane
Lpt: Necessary and sufficient to transport LPS across the cell envelope
Lpt proteins assemble into a complex that spans from the cytoplasm to the OM (Fig. 1B) [13]. At the IM, the LptB2FGC ATP-binding-cassette (ABC) transporter utilizes ATP binding and hydrolysis to extract LPS from the IM [7]. Following extraction, LPS is positioned onto a trans-envelope bridge formed by the seamless association of the C-shaped β-jellyroll folds present in LptF, LptC, LptA, and LptD, providing a structural hydrophobic groove that shields the acyl chains of LPS from the aqueous periplasm [14,15]. Subsequent rounds of ATP binding and hydrolysis from LptB2FGC are thought to push multiple LPS molecules across the bridge until they reach the LptDE OM translocon [13] (Fig. 1B) (see also review about the biogenesis of the LptDE complex by Tomasek and Kahne in this issue [16]). The LptD β-barrel can open at the seams, a feature that is proposed to allow LPS traverse the OM by having its sugars go through the lumen while its acyl chains stay in the hydrophobic core of the membrane [8,17] (Fig. 1B).
This model evolved through the years from data derived from many studies, but received key support in 2018, when the Lpt system was reconstituted in vitro [18]. This technically challenging feat demonstrated that LPS transport from a proteoliposome containing LptB2FGC to another proteoliposome containing LptDE could occur as long as the periplasmic protein LptA bridged the proteoliposomes and ATP was provided. This major accomplishment clearly supported the bridge model and demonstrated that the Lpt system is necessary and sufficient to transport LPS across the envelope in the presence of ATP. Reconstitutions assays also constitute an important technical advance that allows for mechanistically probing the Lpt system and studying inhibitors that could be antibiotics [18,19].
The Atypical Structure of the LptB2FGC Transporter
A key feature of the model for LPS transport is that the LptB2FGC ABC transporter is Lpt’s power engine. The structure of LptB2FGC was much awaited since most ABC transporters function differently, mainly by moving their substrates across a membrane. The first crystal structures of LptB2FG (lacking LptC) were obtained from Klebsiella pneumoniae and Pseudomonas aeruginosa in 2017 [20,21]. These structures revealed a hydrophobic cavity formed by the six transmembrane segments of each LptF and LptG. This cavity was hypothesized to accommodate LPS, and then collapse in order to extract LPS from the IM and place it onto the periplasmic Lpt bridge [22]. We discuss this model in the next section. These structures also confirmed the main sites of interaction between the LptB2 ATPase and its partners LptFG, which had been previously proposed based on genetic and biochemical data [23]. However, these structures did not provide clues about the path that LPS takes through the periplasm, especially since they showed that both LptF and LptG possess periplasmic β-jellyroll domains. This issue was resolved in 2019. Crystal structures of LptB2FGC from Vibrio cholerae and Enterobacter cloacae revealed only the β-jellyroll domain of LptF connected to that of LptC [15]. Cross-linking experiments demonstrated that these two domains indeed interact in cells. So far, an interaction between the β-jellyrolls of LptG and LptC has not been detected. These data suggest that, after extraction, LPS travels to the β-jellyroll of LptF and then to that of LptC [15]. In agreement, site-specific cross-linking showed that LPS interacts with the hydrophobic interior of the β-jellyrolls of LptF and LptC (but not LptG). Furthermore, this study revealed a surprising structural feature of this transporter also seen in an accompanying study showing cryogenic electron microscopy (cryo-EM) structures of the Escherichia coli LptB2FGC complex [15,24]. Both studies reported that the N-terminal transmembrane α-helix (TM) of LptC (henceforth, TMC) is inserted in the wall of the cavity formed between the TMs of LptFG (Fig. 1B), an unprecedented deviation from the typical structure of ABC transporters [15,24]. This finding is also paradoxical, since the TMC can be removed without causing a functional defect in vivo despite its strict conservation [25], although the β-jellyroll of LptC remains essential. The role of TMC remains to be elucidated, but in vitro evidence suggests that it may regulate the ATPase activity of LptB2FGC [15,24,26]. Below, we discuss the proposed placement of TMC during the transport cycle.
A Model for LPS Extraction from the IM
To understand LPS transport, we must know how LptB2FGC interacts with LPS, and how the energy derived from ATP is used by the transporter. A combination of structural, genetic, and biochemical studies has recently shed light on these issues, leading to a refinement of the model for LPS extraction by LptB2FGC (Fig. 2B). A fundamental feature of the model is that the cavity formed by LptFGC is the substrate-binding site of the transporter. Genetic evidence and subsequent cryo-EM structures and crosslinking data described below demonstrated that indeed LPS interacts with this cavity during transport. Details about how LPS enters the cavity and is loaded onto the periplasmic bridge have also been recently unveiled.
Figure 2: A model for LPS extraction from the IM by the LptB2FGC transporter.
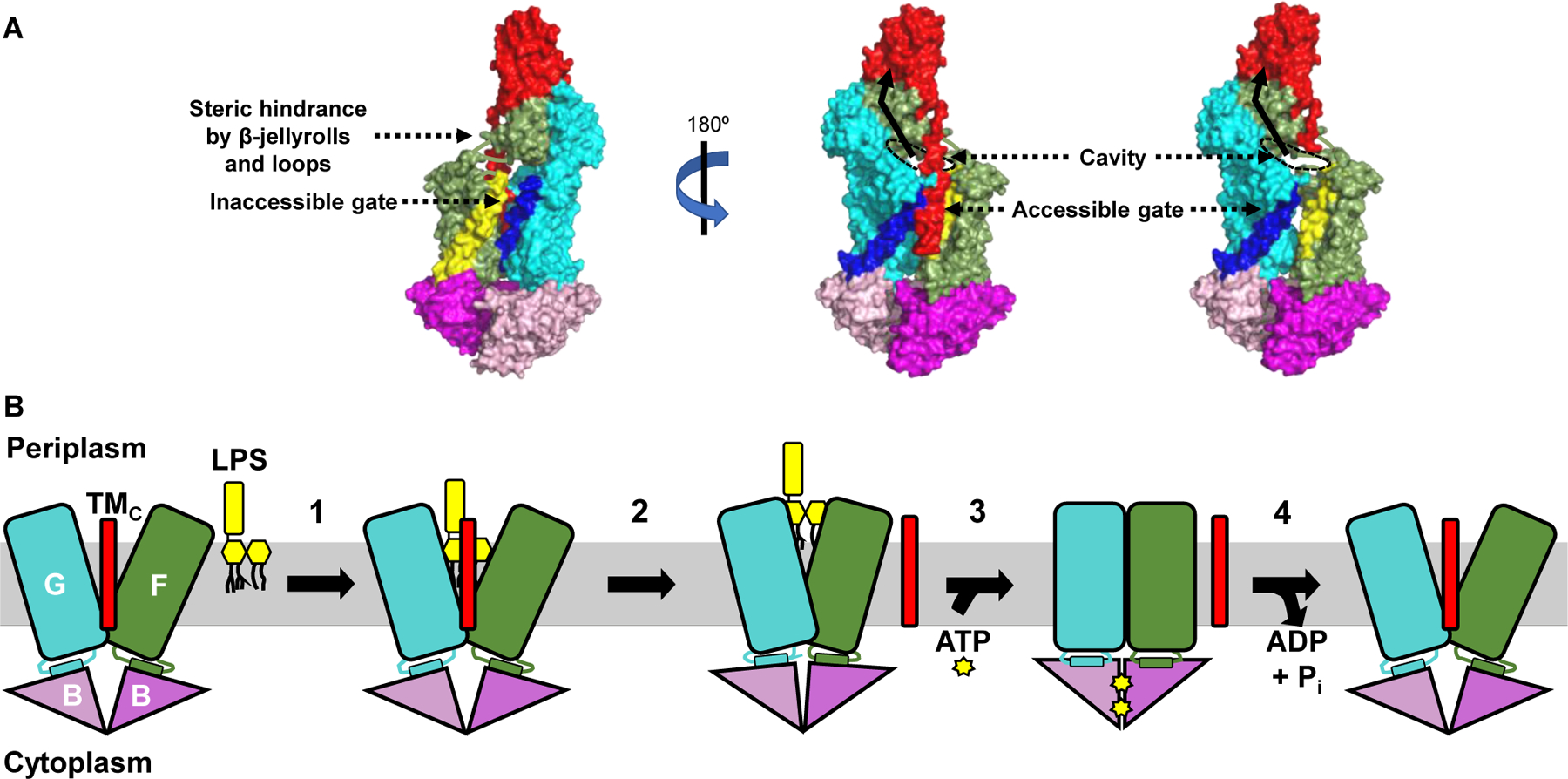
A) Crystal structure (PDB ID: 6MJP) of LptB2FGC derived from Vibrio cholerae is shown with surface rendition. LPS is thought to enter the cavity formed by LptF and LptG through one gate of the transporter only, although two putative gates exist. The far-left image shows the first gate referenced in the text, with LptF TM1 in yellow and LptG TM5 in dark blue. Steric hindrance imposed by β-jellyroll domains of LptFG and the loops connecting the transmembrane and periplasmic regions of LptF/LptG is thought to impede entry of LPS through this gate. The central and far-right images show the second gate referenced in the text, with LptF TM5 in yellow, LptG TM1 in dark blue, and LptC in red. The TMC has been omitted from the far-right image to better show the substrate-binding cavity. The rim of the cavity that would be occupied by LPS is outlined with a black dashed oval, and the pathway LPS would take upon extraction from the IM is shown as a black solid arrow. B) The LptB2FGC complex is shown embedded in the IM but lacking the periplasmic β-jellyroll domains of LptCFG for clarity. Numbers indicate each step in the transport cycle. 1) LPS enters a cavity formed by LptFGC and makes initial, weak contacts with LptF and LptG. 2) The TMC is ejected from the cavity, which partially collapses to stabilize LPS through numerous contacts. The position of the TMC after its exit from the cavity is unknown. 3) Two ATP molecules bind the LptB dimer, causing its closure and facilitating complete collapse of the LptFG cavity. 4) ATP is hydrolyzed by the LptB dimer and ADP + Pi are released to reset the transporter.
First, LPS must enter the cavity formed by LptFGC (Fig. 2). Based on the earlier LptB2FG structures, two possible entry gates were proposed: lateral openings into the cavity between LptF TM1 and LptG TM5, or LptF TM5 and LptG TM1 [22] (Fig. 2A). LptB2FGC structures showing the LptF-LptC connection suggested that the first gate is inaccessible due to steric hindrance imposed by the LptFGC β-jellyrolls [15,24] (Fig. 2A, left image). Curiously, the opening (or second gate) between LptF TM5 and LptG TM1 is where the TMC resides (Fig. 2A, center image). Nonetheless, LPS is thought to pass through this lateral gate, likely by transiently breaking hydrophobic interactions between LptG TM1 and TMC without displacing TMC [15]. Supporting this model, LPS can be crosslinked to residues in this (but not the other) gate, and cryo-EM structures of the transporter have been obtained with LPS inside the cavity that is still aligned by TMC [15,24]. Importantly, entry of LPS through the gate does not require ATP, since LPS can be cross-linked to residues within the LptFG cavity in the absence of the nucleotide [15].
Once LPS enters the LptFGC cavity, it makes preliminary contacts with a ring of positively charged residues located at the rim of the cavity [24,27]. These interactions stabilize the negatively charged phosphates of LPS and orient the glycolipid for extraction. The importance of some of these interactions was first demonstrated through genetic experiments showing that mutations that change the positive charge of some of the stabilizing residues in LptG TM1 result in severe transport defects [27]. Importantly, these defects were shown to be suppressed by changing LPS structure so that its phosphates were modified with positive moieties. These results therefore suggest direct interactions between LptG TM1 and LPS in vivo. In addition, structural studies show that only a few residues in the LptFGC cavity are involved in the initial binding of LPS, and suggest that the next step in transport involves the formation of more contacts after TMC is removed from the cavity in an ATP-independent manner (Fig. 2B) [24]. It is therefore possible that the presence of LPS in the cavity weakens the association of TMC with LptFG, aiding in the movement of the TMC away from the cavity. While it is unclear exactly when and how the TMC moves away from the cavity, all structural studies suggest that this is a step that occurs prior to complete cavity closure and LPS extraction [15,24,26]. Cryo-EM structures lacking TMC also illustrate both a partial collapse of the cavity and a slight elevation of LPS, which allows its phosphates to interact with a larger ring of positively charged residues in the LptFG cavity [24]. The formation of these numerous substrate-cavity contacts likely primes the transporter for LPS extraction from the cavity and its transit to the β-jellyroll of LptF.
Extrusion of LPS from the cavity requires the complete collapse of the LptFG cavity (Fig. 2B). Several cryo-EM structures in this closed state also showed the cytoplasmic LptB2 ATPase in the closed-dimer conformation [24,26]. Furthermore, these conformations were only seen in the presence of either the non-hydrolyzable ATP analog AMP-PNP, or ATP and vanadate, which traps ATPases in a nucleotide-bound state mimicking the transition state. These findings suggested that LPS extraction might be induced by the binding of LptB to ATP, which would then cause the concomitant inward movement of the LptFG TMs to collapse the cavity. Indeed, genetic and biochemical evidence supports that, in vivo, ATP binding causes the closure of the LptB dimer [28]. The movement that results from closing the LptB dimer is proposed to be directly transduced to the TMs of LptFG through rigid-body coupling that relies on the direct physical contact of each LptB monomer to short “coupling helices” in the cytoplasmic loop connecting TM2–3 of LptF and LptG [24,29]. This event is thought to generate the squeezing force to vertically push LPS out of the LptFG cavity. Genetic evidence also suggests that a conserved glutamate in LptF’s coupling helix is required for coupling the closure of the LptB dimer to that of the LptFG cavity, and that interactions between the LptFG cavity and acyl chains of LPS can stimulate LptB’s activity [23,29]. Thus, coupling between LptB and LptFG is bidirectional and affected by both binding of ATP to LptB and binding of LPS to LptFG.
If ATP binding causes LptB2FGC to transport LPS, what is the role of ATP hydrolysis? As shown in other ABC transporters, ATP hydrolysis and the subsequent release of ADP and Pi are required to re-open the LptB dimer and thereby LptFG cavity, resetting the transporter to its initial state in the transport cycle (Fig. 2B) [28]. Interestingly, a unique novel region at the C-terminus of LptB is essential for proper ATP hydrolysis and opening of the LptB dimer [28]. It still remains unknown when and how TMC re-associates with LptFG and whether LPS moves to the β-jellyroll of LptF or beyond in each ATP cycle. If LPS only transits to LptF, a proposed valve at the base of LptF’s β-jellyroll might prevent the extracted LPS molecule from falling back into the cavity once it re-opens [15]. Thus, although the last three years have been very fruitful in Lpt studies, there are still many questions that need to be addressed.
Phospholipid Transport to the OM
Phospholipids are transported both to and from the OM [30]. Anterograde transport to the OM has been proposed to occur both through sites of IM-OM hemi-fusion that allow passive phospholipid transport, as well as through protein-mediated systems (Fig. 3) [31–34]. The take-home message is that we still do not understand how this transport occurs. The bi-directionality of phospholipid transport and possible redundancy between systems might have limited progress in understanding the essential transport of phospholipids to the OM.
Figure 3: Proposed mechanisms of phospholipid transport to the OM.
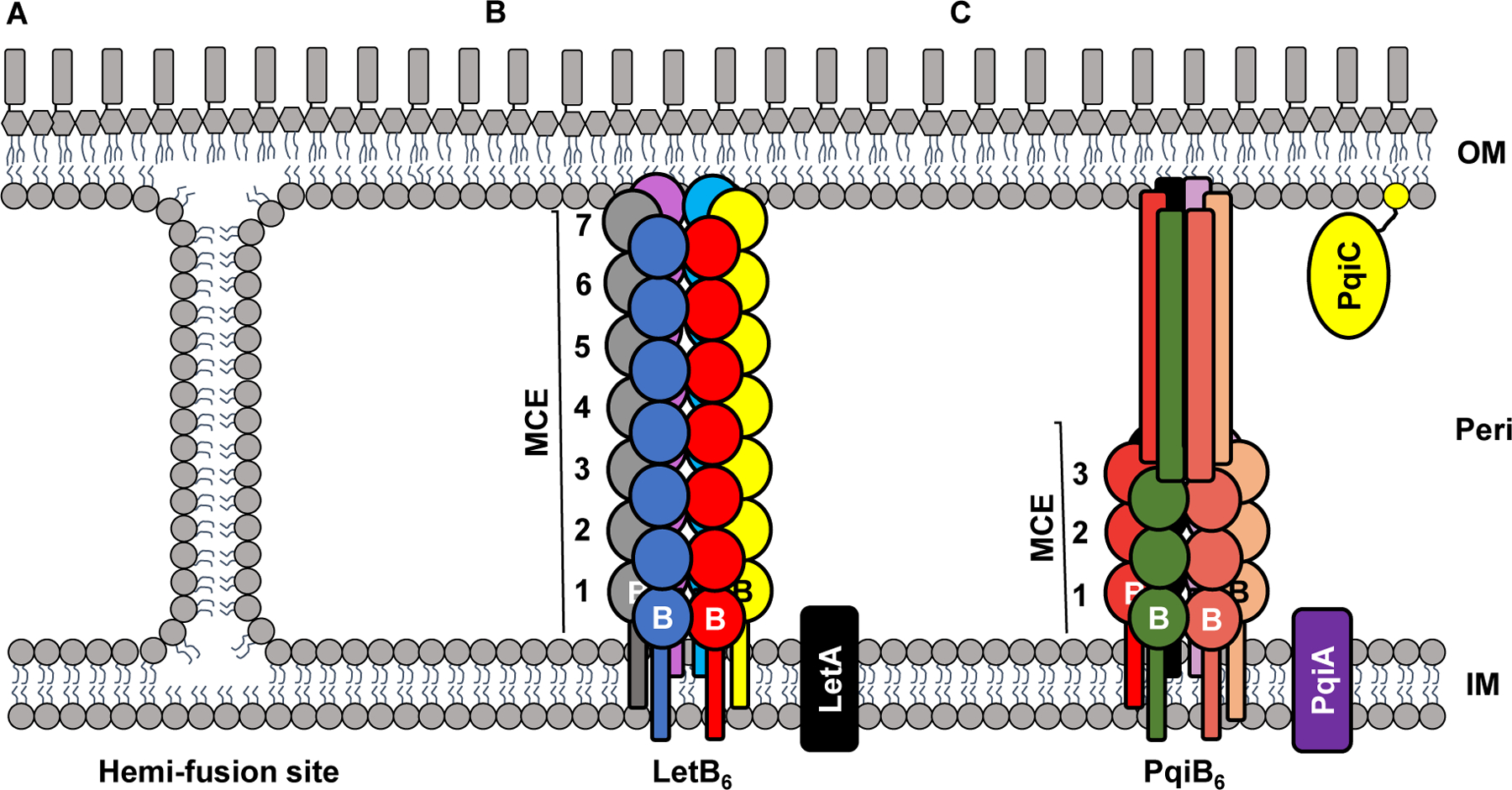
The peptidoglycan layer has been omitted for simplicity. A) IM-OM hemi-fusion sites, also called Bayer junctions. These sites would allow for the bi-directional diffusion of phospholipids. B) LetAB. LetA is an IM protein. A homohexamer of LetB (with each monomer labeled B and colored differently) forms a large structure containing seven MCE domains (numbered). Interactions between LetA and LetB have been proposed but not demonstrated. C) PqiABC. PqiA is an IM protein and PqiC is an OM lipoprotein. Both PqiA and PqiC are hypothesized to interact with PqiB. PqiB forms a homohexamer (with each monomer labeled B and colored differently) containing three MCE domains (numbered). The C-terminus of PqiB monomers (shown as tubes) form a long, needle-like extension. OM, outer membrane. IM, inner membrane. Peri, periplasm.
Live-cell microscopy has recently suggested that phospholipids may diffuse from the IM to the OM through sites of membrane fusion (Fig. 3A) in a process that involves the YhdP protein [35]. Whether this diffusion occurs by fusing the outer leaflet of the IM and the inner leaflet of the OM remains unknown. If it does, it will be important to understand how fusion sites form and allow for selective diffusion of phospholipids.
Recently, the proteins YebT (now called LetB for lipophilic envelope-spanning tunnel B) and PqiB, which are anchored to the IM through an N-terminal transmembrane α-helix and possess large periplasmic regions capable of spanning the periplasm, have been proposed to mediate phospholipid transport from the IM to the OM [36]. These proteins contain periplasmic mammalian cell entry (MCE) domains, which have been implicated in cell envelope maintenance and import of hydrophobic substrates [37]. Proteins with this domain can form higher-order structures. Cryo-EM images show that LetB hexamerizes into a structure with seven rings formed by MCE domains that form a tunnel long enough to span the periplasm. This tunnel has a central hydrophobic pore hypothesized to transport phospholipids (Fig. 3B) [38,39]. PqiB also forms a hexamer, but the structure has three MCE rings and a long α-helical C-terminal domain (Fig. 3C), resembling a barrel topped by a needle-like extension [34]. A long hydrophobic core runs the length of the barrel and needle, creating a tunnel which may allow for the transport of phospholipids. It has also been suggested, based on genomic co-localization into putative operons, that LetB and PqiB interact with putative partners LetA and PqiAC, respectively [36]. Although the LetB and PqiB structures are suggestive of a transport function, we await for experimental evidence supporting such function in cells and note that mutants lacking both of these proteins do not exhibit growth defects under standard growth conditions [37]. It is therefore unclear if these proteins are involved in phospholipid transport.
We should finally mention that a recent controversy has ensued regarding the Mla system, a multi-protein transporter that was initially proposed to transport phospholipids in retrograde fashion from the OM to the IM [40], and later proposed to mediate anterograde phospholipid transport [41,42]. A recent study has found no evidence to support anterograde phospholipid transport and has challenged the validity of earlier claims [43]. We encourage readers to judge for themselves by reading the primary literature on both sides of this controversy, but suggest that they first refer to an excellent perspective commentary on this debate [44].
Conclusion
Our understanding of LPS transport to the OM has significantly moved forward over the past few years. Key questions and mechanistic details remain to be elucidated, but the framework and experimental tools needed to address them exist. We therefore predict steady progress regarding the Lpt system. In contrast, the answer to the most fundamental question about phospholipid transport to the OM remains elusive: what mediates it? We hope that recent developments lead to the answer soon.
Acknowledgements
This work was supported by the National Institutes of Health awards T32-GM086252 to A.W. and R01-GM100951 to N.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Silhavy TJ, Kahne D, Walker S: The bacterial cell envelope. Cold Spring Harb Perspect Biol 2010, 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamio Y, Nikaido H: Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 1976, 15:2561. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H: Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 2003, 67:593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delcour AH: Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 2009, 1794:808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani B, Ruiz N: Function and biogenesis of lipopolysaccharides. EcoSal Plus 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raetz C: Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol Rev 1978, 42:614–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson BW, May JM, Sherman DJ, Kahne D, Ruiz N: Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos Trans R Soc Lond B Biol Sci 2015, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.May JM, Sherman DJ, Simpson BW, Ruiz N, Kahne D: Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos Trans R Soc Lond B Biol Sci 2015, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperandeo P, Pozzi C, Deho G, Polissi A: Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol 2006, 157:547–558. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ: Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 2008, 105:5537–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D: Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 2006, 103:11754–11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun M, Silhavy TJ: Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol 2002, 45:1289–1302. [DOI] [PubMed] [Google Scholar]
- 13.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D: Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 2016, 14:337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freinkman E, Okuda S, Ruiz N, Kahne D: Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 2012, 51:4800–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.**Owens TW, Taylor RJ, Pahil KS, Bertani BR, Ruiz N, Kruse AC, Kahne D: Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 2019, 567:550–553.First LptB2FGC crystal structures showing the interdigitation of the LptC transmembrane α-helix with LptFG. This study reveals that LPS is transported through the β-jellyroll fold of LptF (and not LptG) to LptC.
- 16.Tomasek D, Kahne D: The assembly of β-barrel outer membrane proteins. Curr Opin Microbiol 2021, 60:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundquist KP, Gumbart JC: Presence of substrate aids lateral gate separation in LptD. Biochim Biophys Acta Biomembr 2019, 1862:183025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.**Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D: Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 2018, 359:798–801.This study reported the first in vitro full reconstitution of LPS transport from purified components. The work demonstrates that Lpt is necessary and sufficient to transport LPS unidirectionally through a bridge structure by utilizing ATP.
- 19.Xie R, Taylor RJ, Kahne D: Outer Membrane translocon communicates with inner membrane ATPase to stop lipopolysaccharide transport. J Am Chem Soc 2018, 140:12691–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong H, Zhang Z, Tang X, Paterson NG, Dong C: Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat Commun 2017, 8:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Q, Yang X, Yu S, Shi H, Wang K, Xiao L, Zhu G, Sun C, Li T, Li D, et al. : Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat Struct Mol Biol 2017, 24:469–474. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, Tang X, Zhang Z, Dong C: Structural insight into lipopolysaccharide transport from the Gram-negative bacterial inner membrane to the outer membrane. Biochim Biophys Acta Mol Cell Biol Lipids 2017, 1862:1461–1467. [DOI] [PubMed] [Google Scholar]
- 23.Simpson BW, Owens TW, Orabella MJ, Davis RM, May JM, Trauger SA, Kahne D, Ruiz N: Identification of residues in the lipopolysaccharide ABC transporter that coordinate ATPase activity with extractor function. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.**Li Y, Orlando BJ, Liao M: Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 2019, 567:486–490.Cryo-EM study revealing various states of LptB2FGC during the transport cycle, including LPS-bound and ADP-vanadate-bound. Additionally, this study highlights the numerous contacts LPS makes with the LptFG cavity
- 25.Villa R, Martorana AM, Okuda S, Gourlay LJ, Nardini M, Sperandeo P, Deho G, Bolognesi M, Kahne D, Polissi A: The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J Bacteriol 2013, 195:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.*Tang X, Chang S, Luo Q, Zhang Z, Qiao W, Xu C, Zhang C, Niu Y, Yang W, Wang T, et al. : Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat Commun 2019, 10:4175.Cryo-EM study showing the AMP-PNP-bound state of LptB2FGC in a closed or collapsed configuration. This study supports the model in which ATP binding facilitates LptFG cavity collapse and LPS extraction.
- 27.Bertani BR, Taylor RJ, Nagy E, Kahne D, Ruiz N: A cluster of residues in the lipopolysaccharide exporter that selects substrate variants for transport to the outer membrane. Mol Microbiol 2018, 109:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.**Simpson BW, Pahil KS, Owens TW, Lundstedt EA, Davis RM, Kahne D, Ruiz N: Combining mutations that inhibit two distinct steps of the ATP hydrolysis cycle restores wild-type function in the lipopolysaccharide transporter and shows that ATP binding triggers transport. mBio 2019, 10.The authors identified an essential region at the C-terminus of LptB with a functional link to the ATP-binding domain of LptB and proposed based on genetic and biochemical data that ATP binding to LptB2 triggers LPS extraction, while ATP hydrolysis resets the transporter.
- 29.Lundstedt EA, Simpson BW, Ruiz N: LptB-LptF coupling mediates the closure of the substrate-binding cavity in the LptB2FGC transporter through a rigid-body mechanism to extract LPS. Mol Microbiol 2020, 114:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones NC, Osborn MJ: Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J Biol Chem 1977, 252:7405–7412. [PubMed] [Google Scholar]
- 31.Bayer ME: Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol 1968, 53:395–404. [DOI] [PubMed] [Google Scholar]
- 32.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ: Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 2016, 113:E1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrivastava R, Chng SS: Lipid trafficking across the Gram-negative cell envelope. J Biol Chem 2019, 294:14175–14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD: Architectures of lipid transport systems for the bacterial outer membrane. Cell 2017, 169:273–285 e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm J, Shi H, Wang W, Mitchell AM, Wingreen NS, Huang KC, Silhavy TJ: The inner membrane protein YhdP modulates the rate of anterograde phospholipid flow in Escherichia coli. Proc Natl Acad Sci U S A 2020, 117:26907–26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama T, Zhang-Akiyama QM: pqiABC and yebST, putative mce operons of Escherichia coli, encode transport pathways and contribute to membrane integrity. J Bacteriol 2017, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isom GL, Davies NJ, Chong ZS, Bryant JA, Jamshad M, Sharif M, Cunningham AF, Knowles TJ, Chng SS, Cole JA, et al. : MCE domain proteins: conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci Rep 2017, 7:8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.**Liu C, Ma J, Wang J, Wang H, Zhang L: Cryo-EM Structure of a bacterial lipid transporter YebT. J Mol Biol 2020, 432:1008–1019.The authors obtained cryo-EM images of LetB (YebT) and proposed its possible function as a phospholipid transporter. Structures contain densities identified as probable phospholipid molecules bound to LetB.
- 39.**Isom GL, Coudray N, MacRae MR, McManus CT, Ekiert DC, Bhabha G: LetB Structure reveals a tunnel for lipid transport across the bacterial envelope. Cell 2020, 181:653–664 e619.Cryo-EM study of LetB (YebT) revealing that LetB adopts a tunnelled structure with a hydrophobic core that is capable of spanning the periplasmic compartment. In vivo photo-crosslinking assays identified putative phospholipid binding sites within LetB.
- 40.Malinverni JC, Silhavy TJ: An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 2009, 106:8009–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.*Hughes GW, Hall SCL, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, et al. : Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol 2019, 4:1692–1705.In vitro assays showing transfer of phospholipids from MlaD to MlaC, but not vice versa, which would implicate the Mla system as a possible anterograde phospholipid transporter.
- 42.*Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, Kollman JM, Miller SI: The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 2019, 8.Study proposing that Mla transports phospholipids from the IM to the OM in Acinetobacter baumannii. Monitoring the localization of newly synthesized phospholipids appeared to show their accumulation in the IM of mla mutants. See reference 43.
- 43.**Powers MJ, Simpson BW, Trent MS: The Mla pathway in Acinetobacter baumannii has no demonstrable role in anterograde lipid transport. Elife 2020, 9:e56571.This study contradicts Kamischke et al. (reference 42) by demonstrating that Mla does not transport phospholipids in anterograde fashion in Acinetobacter baumannii. It also shows that the previous mla mutant strain used by Kamischke et al. (reference 42) contains an additional mutation that causes phenotypic abnormalities.
- 44.Powers MJ, Trent MS: Intermembrane transport: Glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc Natl Acad Sci U S A 2019, 116:17147–17155. [DOI] [PMC free article] [PubMed] [Google Scholar]


