Abstract
Inhibitors of the immunoproteasome (i-20S) have shown promise in mouse models of autoimmune diseases and allograft rejection. In this study, we used a novel inhibitor of the immunoproteasome, PKS3053, that is reversible, noncovalent, tight-binding and highly selective for the β5i subunit of the i-20S to evaluate the role that i-20S plays in regulating immune responses in vitro and in vivo. In contrast to irreversible, less selective inhibitors, PKS3053 did not kill any of the primary human cell types tested, including plasmacytoid dendritic cells (pDCs), conventional dendritic cells (cDCs), macrophages and T cells, all of which expressed genes encoding both the constitutive proteasome (c-20S) and i-20S. PKS3053 reduced TLR-dependent activation of pDCs, decreasing their maturation and IFN-α response and reducing their ability to activate allogenic T cells. In addition, PKS3053 reduced T cell proliferation directly and inhibited TLR-mediated activation of cDCs and macrophages. In a mouse model of skin injury that shares some features of cutaneous lupus erythematosus (CLE), blocking i-20S decreased inflammation, cellular infiltration, and tissue damage. We conclude that the immunoproteasome is involved in the activation of innate and adaptive immune cells, that their activation can be suppressed with an i-20S inhibitor without killing them, and that selective inhibition of β5i holds promise as a potential therapy for inflammatory skin diseases such as psoriasis, CLE and systemic sclerosis (SSc).
INTRODUCTION
The eukaryotic 26S proteasome is a specialized complex responsible for regulating many cellular signaling pathways and for quality control of the proteome via targeted degradation of poly-ubiquitinylated (polyUb) proteins (1). It is composed of the cylindrical 20S core particle (CP) capped on one or both ends by a 19S regulatory particle (RP) (2). The RP is composed of ubiquitin (Ub) receptors that bind and position substrates, deubiquitylating enzymes (DUBs) that remove the polyUb tag and an ATPase ring that unfolds the substrate and abuts the ends of the CP barrel. The CP is composed of four stacked rings with seven different α subunits constituting each of the two outer rings and seven different β subunits forming each of the two inner rings (3). Subunits β1c, β2c and β5c each have proteolytic active sites, with caspase-like, trypsin-like and chymotrypsin-like activities, respectively (3).
In immune cells, the catalytic subunits of the constitutive proteasome (β1c, β2c, β5c) can be replaced by the inducible subunits β1i (LMP2), β2i (MECL-1), and β5i (LMP7) to form the immunoproteasome (4). In T cells, immunoproteasome expression is upregulated in response to IFNγ (5) and upon allo-activation (6). Immunoproteasomes take part in antigen processing and presentation on MHC class I molecules (7, 8). DPLG3, a selective N,C-capped dipeptide inhibitor of β5i, suppressed T cell proliferation and decreased pDCs’ response to Toll-like receptor 9 (TLR9) without inducing cell death (6). In pDCs, the immunoproteasome controls the intracellular trafficking of TLR9 from the endoplasmic reticulum to endolysosomes and the nuclear translocation of IRF7 and NFκB (9). The immunoproteasome can be upregulated in other cells in response to injury or infection, such as cells in the central nervous system (10) and in cardiac and skeletal muscle (11), and cells can express proteasomes with mixtures of constitutive and inducible subunits (12).
There is extensive evidence for the beneficial effect of immunoproteasome inhibitors in various mouse models of inflammatory or autoimmune disorders (13–18), However, those studies involved inhibitors that react covalently and irreversibly with β5i. Such inhibitors react to a lesser extent with β5c, but over time, the proportion of inactivated β5c rises, leading to significant cellular toxicity (9, 19–21). This makes it hard to distinguish which pathways in a given cell are i-20S dependent as opposed to the results reflecting that all pathways in a cell are dependent on the cell’s viability. Blocking i-20S along with c-20S using bortezomib has proven effective in mouse models of lupus (22) but its clinical use is limited due to the neurotoxicity of bortezomib (23). Similarly, ONX0914 and carfilzomib prevented lupus nephritis in MRL/lpr mice in association with decreases in plasma cells and serum autoantibodies (19), but the toxicity of these compounds (19, 24) raises concerns about their use for the chronic treatment of non-malignant disease.
We have recently characterized a novel reversible and highly selective β5i inhibitor, PKS3053 (25), which we now report has little to no toxicity to the cells studied and which serves as a probe for identifying immune cell responses that depend on β5i. It also allowed us to determine the role of i-20S in vivo in skin injury caused by tape stripping, which has been used as a mouse model for human psoriasis, atopic dermatitis (26–29) and CLE (30, 31). The stripping provokes transient inflammation and tissue damage in association with the recruitment of multiple CD45-positive immune cell subsets, including pDCs, macrophages, neutrophils, other myeloid cells and T cells (28, 30, 31).
MATERIALS AND METHODS
Reagents
TLR ligands (TLR-L) used are as follows: TLR9-L (CpG ODN C274) from TriLink 5’-TCGTCGAACGTTCGAGATGAT-3’, TLR8-L (ORN8-L) from Chemgenes 5’-M2UGCUGCUUGUG-/glycerol/-GUGUUCGUCGUM2–3’ and TLR4 agonist lipopolysaccharide (LPS) from Invivogen. Immunoproteasome inhibitor PKS3053 was resuspended in DMSO for in vitro studies or in 0.1% methyl cellulose + 0.1% Tween 90, which was used as vehicle for in vivo experiments. Flow cytometry was performed on the BD Canto™ II. Antibodies or dye used for flow cytometry are as follows: CD83, CD86, CD11c, CD14, CD3, CD4, CD20 and CD123 (all from BD), BDCA4 and BDCA2 (Miltenyi), HLA-DR (Biolegend) and cell trace violet (Invitrogen). Flow cytometry analysis was conducted using FlowJo v10.6.2. ELISA kits for human cytokines IL-6, IFNɤ were purchased from Mabtech.
Preparation of peripheral blood mononuclear cells (PBMCs)
Enriched leukocytes were obtained from the New York Blood Center (Long Island City, NY) after informed consent of donors who were deemed healthy by the Blood Center’s criteria and used under a protocol approved by the Institutional Review Board of the Hospital for Special Surgery and the Institutional Biosafety Committee of WCM. Peripheral blood mononuclear cells (PBMCs) were prepared using Ficoll-Paque density gradient (GE Healthcare) as previously described (32). The clinical and demographic characteristics of the donors reflect the diverse population of New York City and are as follow. Mean age of donors is 42 (SD=2.7); sex is 53% male and 47% female; Ethnic distribution was self-reported as 54% unknown or of multiple ethnicity, 35% Caucasian, 4% Asian and 2% African American.
Immune cells viability assay
Freshly isolated PBMCs were resuspended at 0.5 × 106 cells/ 100 μl RPMI 1640 medium (Gibco) and cultured for 24 h either alone or with the immunoproteasome inhibitors PKS3053 or ONX0914 at 0.1, 0.3, 1 or 10 μM. The following day, PBMCs were resuspended in FACS buffer and incubated with FcR blocking reagent (Miltenyi Biotec) for 10 min at 4 °C. The cells were then stained with the following monoclonal antibodies: CD14, HLA-DR, CD11c, CD123, BDCA4, CD3 and CD20 for 15 min at 4 °C and washed twice in FACS buffer. 100μl of 123count eBeads™ (Thermofischer) were added to each samples and cells+beads were acquired by a fluorescence activated cell sorter (FACS) Canto flow cytometer (BD Biosciences) and analysis was performed using FlowJo analysis software (TreeStar Inc.). 75,000 beads were acquired for every samples to normalize total number of cells acquired. The gating strategy involved progressively measuring total cells, viable cells (defined by SSC, FSC) and specific immune cell types (Fig. S2A and B). The monocytes were defined as CD14+; pDCs were identified as CD14−HLA-DR+CD11c−CD123+BDCA4+; and cDCs were gated as CD14−HLA-DR+ CD123−BDCA4−CD11c+. B cells and T cells were CD14−CD20+ and CD14−CD3+, respectively. Cell viability was expressed as the percentage of the count of viable immune cells for each condition relative to the number of viable cells in the medium alone condition.
Preparation and culture of immune cells
pDCs and monocytes, were isolated from PBMCs by positive selection using BDCA4- (33), and CD14-conjugated microbeads (Miltenyi) respectively. Purity of isolated pDCs (identified as CD123+BDCA2+ by flow cytometry) were more than 95% for each donor (Fig. S4A). Macrophages and cDCs were differentiated from monocytes by culturing with 20 ng/ml M-CSF for 5 days or with 50 ng/ml GM-CSF and 100 ng/ml IL-4 for 7 days respectively. After differentiation, the purity of macrophages (CD14+) and cDCs (CD11c+) was analyzed by flow cytometry and was more than 99% and 97% respectively for each donor (Fig. S4B & C). For functional assay, freshly isolated pDCs were resuspended at 0.5 × 105 cells/ 100 μl RPMI 1640 medium (Gibco) and cultured for 6 or 24h either alone, with the TLR9-L (C274 at 0.1 μM) or with C274+ PKS3053 at 1, 3 or 10 μM. After differentiation of macrophages and cDCs, the cells were cultured at 1 × 105 cells/ 100 μl RPMI 1640 medium for 6 or 24h either alone, with either the TLR4-L (LPS at 50 ng/ml) or the TLR8-L (ORN8-L at 60 μg/ml ) or with TLR4/8-L + PKS3053 at 0.02, 0.06, 0.2, 0.6, 2 or 6 μM. After 6h, all immune cells were lysed in RLT buffer (Qiagen) prior to RNA preparation. After 24h, supernatant was collected for cytokine analysis by ELISA and the expression of costimulatory markers (CD83 and CD86) was evaluated by flow cytometry.
T cell proliferation and stimulation assay
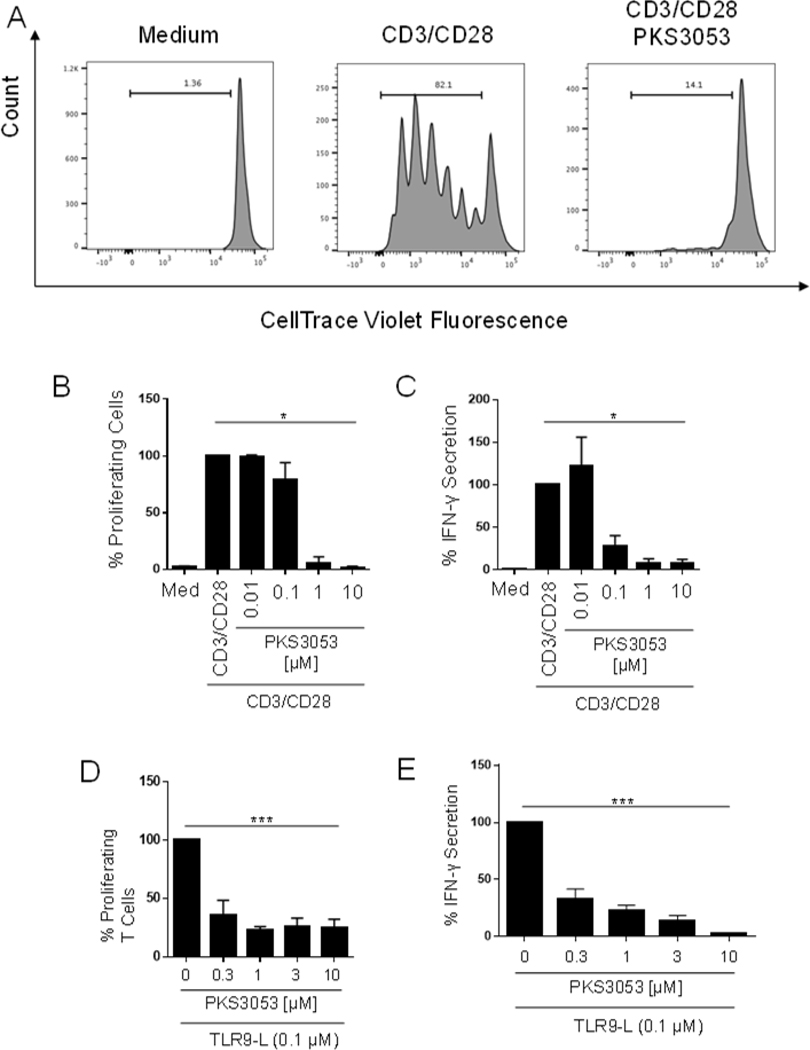
Naïve T cells were isolated by negative selection with CD4 microbeads and labelled with the fluorescent dye, cell trace violet (CTV) at 5 μM according to manufacturer protocol. 5 μg/ml anti-CD3 Ab were coated on wells of a 96-well U-bottom plate and CTV-labelled CD4 T cells were added at a concentration of 2 × 105 cells/ 200 μl RPMI in the presence of soluble anti-CD28.2 (1 μg/mL; BD Biosciences) and in the presence of PKS3053 at 0.01, 0.1, 1, or 10 μM. After 120h, supernatant was collected for cytokine analysis by ELISA and T-cell proliferation was evaluated by measuring the CTV dilution using flow cytometry.
Allogeneic T cell stimulation by pDCs
For co-culture experiments, freshly isolated pDCs resuspended at 1 × 105 cells/ 100 μl RPMI was cultured for 48h in 96-well U-bottom plate either alone, with C274 at 0.1 μM or with C274+ PKS3053 at 0.3, 1, 3 or 10 μM. After 48h, pDCs were washed and CTV-labelled allogenic CD4+ T cells were added at 2 × 105 cells/ 200 μl RPMI for an additional 96h. Supernatant was then collected, and T-cell proliferation was analyzed.
Animals and in vivo treatments
C57BL/6 female mice were either purchased from the Jackson lab or bred within the WCM Research Animal Resource Center (RARC) animal facility and used at 10–16 weeks of age. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC). Tape stripping was performed after shaving the dorsal area (3×3 cm) and applying the tape and stripping it from the skin using 10 successive, freshly cut 2–3 cm pieces of duct tape on the back of the mice as described (30). No depilatory creams or lotions were used, as their use might impact the inflammatory response in the skin. Shaved mice were used as control. PKS3053 and vehicle was administered intraperitoneally just prior to tape stripping. Skin was collected for histological analysis and RNA extraction after 24 hours.
Histological analysis of skin inflammation and tissue pathology
The biopsy specimens were fixed in paraformaldehyde 4%, dehydrated successively and embedded in paraffin. Five μm sections were cut, rehydrated and stained with hematoxylin and eosin. Multiple skin sections of 7–12 mice per group were evaluated in a blinded fashion. The following histological features: 1) dermal blister, 2) dermal-adipose tissue inflammation and 3) ulceration were assessed and scored 0 when absent. For dermal blisters, observation of focal subepidermal and/or intradermal microblistering was scored 1. Score 2 was given when multiple areas of subepidermal and intradermal microblistering and clefts in the dermo-adipose tissue junction were observed. Presence of numerous subepidermal and intradermal blisters and broad clefts between dermis and adipose tissue was assigned a score of 3. Inflammation was given score 1 when inflammatory cells gathered in small groups within the superficial dermis or the dermal-adipose junction; score 2 was assigned to a band-like inflammation structure along the dermal-adipose tissue interface, with scattering through the adipose tissue; score 3 corresponded to thick band of inflammatory cells, within the dermis and/or along the dermal-adipose junction involving extensively the deeper adipose tissue. Ulceration was scored 1 when few areas of partial loss of epidermal layers was observed, scored 2 when moderate areas of the whole epidermis were missing leading to exposure of the dermis and scored 3 for large areas of partial or total loss of epidermis layers. The individual parameters were scored separately and summed to a total disease score.
Immunohistochemical analysis
For immunohistochemical analyses, sections 5 μm thick were cut from 5 formalin-fixed and paraffin-embedded (FFPE) mouse skin samples for each condition. For visualization of CD45 and F4/80 by immunohistochemistry, the 5-μm sections were rehydrated before antigen retrieval and incubation with anti-CD45 or anti-F4/80 antibodies. Detection of anti-CD45 and anti-F4/80 antibodies was done using horseradish peroxidase (hrp)-conjugated secondary antibodies and DAB substrate. The slides were counterstained with hematoxylin and analyzed with a Zeiss microscope using the Zen2 software. For quantitation of CD45 and F4/80-immunostained cells in mouse skin samples, positive cells were counted in 5 non-overlapping microscopic fields (x10).
Quantitative PCR analysis
PCR was performed as described previously (34). RNA was extracted from human cells and mice skin tissue using RNeasy® Mini Kit (QIAGEN) per manufacturer instructions. Quantity of RNA was measured by Nanodrop and cDNA (20–50 ng) was synthesized using the Multiscribe® High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Reactions were performed in StepOnePlus™ Real-Time PCR System with Power SYBR™ Green Master Mix (Applied Biosystems). Gene expression levels were calculated based on relative cycle threshold (CT) values. This was done using the formula: Relative CT = 100 × 1.8 (UBI-GENE), where UBI is the mean CT of the housekeeping gene ubiquitin in duplicate, GENE is the mean CT of the target gene in duplicate, and 100 is arbitrarily chosen as a factor to bring all values above 0. Primer sequences used were as follows: human PSMB6 forward 5’-GATTCAGGCGGGTGGTAAA-3’, reverse 5’-TTCACTGCCAATGCTCTCGC-3’, human PSMB7 forward 5’- TGCCAGTTTTCCGGACCTTT-3’, reverse 5’- CTGTGTCGGTGTATGCTCCA-3’, human PSMB5 forward 5’- TACAGCGGGTGCTTACATTG-3’, reverse 5’- ATGGTGCCTAGCAGGTATGG-3’, human PSMB9 forward 5’- TGCTGACTCGACAGCCTTTT-3’, reverse 5’- GGTACCCTTCCACTTGGCTG-3, human PSMB10 forward 5’- GTGGCTAAGGCTTGTCGGAG-3’, reverse 5’- CGTGCAGGCTTTGGTATTGG-3’, human PSMB8 forward 5’- AAGCTGCGCCTTTAGATGAC-3’, reverse 5’- TGGGCCATCTCAATCTGAAC-3, human IL-6 forward 5’-TACCCCCAGGAGAAGATTCC-3’, reverse 5’-GCCATCTTTGGAAGGTTCAG-3’, human IFN-α forward 5’-CCCAGGAGGAGTTTGGCAA-3’, reverse 5’-TGCTGGATCATCTCATGGAGG-3’, human TNF forward 5’-CTTCTGCCTGCTGCACTTTG-3’ reverse 5’-CTGGGCCAGAGGGCTGAT-3’, human IRF7 forward 5’- CTGTTTCCGCGTGCCCT-3’, reverse 5’- GCCACAGCCCAGGCCTT-3’, human ISG54 forward 5’- CTGGACTGGCAATAGCAAGCT-3’, reverse 5’- AGAGGGTCAATGGCGTTCTG-3’, mouse IRF7 forward 5’- ACAGGGCGTTTTATCTTGCG-3’, reverse 5’- TCCAAGCTCCCGGCTAAGT-3’ mouse IP10 forward 5’- GACGGTCCGCTGCAACTG-3’, reverse 5’- GCTTCCCTATGGCCCTCATT-3’, mouse ISG20 forward 5’- GTCACGCCTCAGCACATGGT-3’, reverse 5’- CCACCAGCTTGCCTTTCAGAA-3’, ISG15 forward 5’- ACGGTCTTACCCTTTCCAGTC-3’, reverse 5’- CCCCTTTCGTTCCTCACCAG-3’ mouse TNF forward 5’- GCCACCACGCTCTTCTGTCT-3’, reverse 5’- GGTCTGGGCCATAGAACTGATG-3’, mouse IL1β forward 5’- GACGGCACACCCACCCT-3’, reverse 5’- AAACCGTTTTTCCATCTTCTTCTTT-3’.
Disassociation rate constant Koff determination.
Human immunoproteasome (E-370) and mouse immunoproteasome (E-376) were purchased from Boston Biochem, Cambridge, MA, USA. Suc-LLVY-AMC was purchased from Bachem, Torrance, California, USA. Human i20S (40 nM) or mouse i20S (60 nM) and PKS3053 (100 nM) or DMSO in assay buffer (25 mM HEPES, pH7.5, 0.5 mM EDTA, 0.01% BSA and 0.02% SDS) were incubated at room temperature for 30 minutes. DMSO concentration remained constant at 1% in all reaction mixtures. The mixtures were then diluted 100-fold into 200 μL assay buffer containing 25 μM suc-LLVY-AMC at 37˚C. The fluorescence of hydrolyzed AMC (Ex 360 nm / Em 460 nm) was immediately recorded on a plate reader (Molecular Devices SpectraMax Spectrofluorometers M5) for 1 hour.
Statistical analysis
Data are expressed as means ± SEM unless otherwise indicated. Non-parametric Mann-Whitney tests were performed using Prism 8 (Graphpad Software, San Diego, CA), and a p value of ≤ 0.05 was considered significant.
RESULTS
Both immunoproteasome and constitutive proteasome subunits are expressed in human immune cells; selective inhibition of the β5i subunit using PKS3053 is not toxic to immune cells
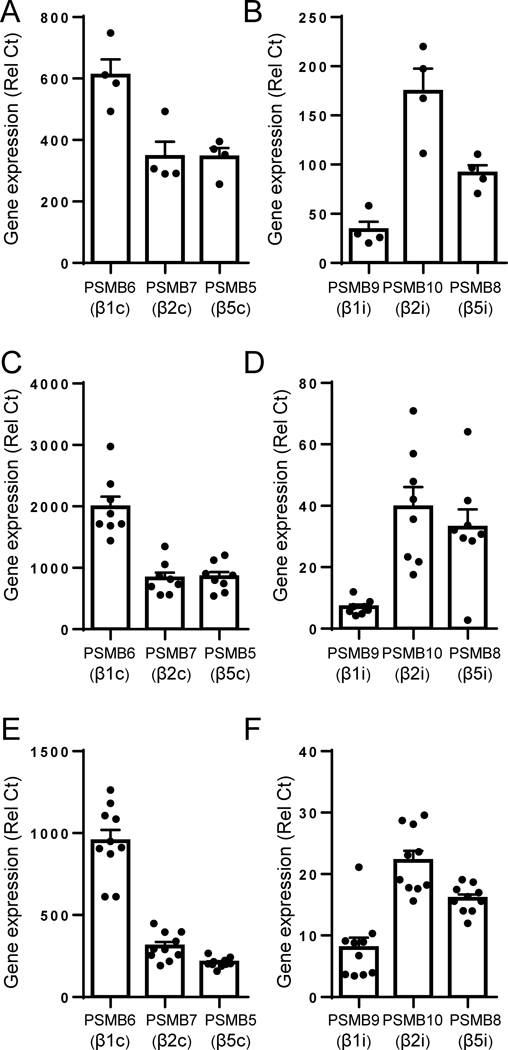
We first quantified the expression levels of transcripts for both c-20S and i-20S subunits in purified human immune cells including pDCs (Fig. 1A, B), cDCs (Fig. 1C, D) and macrophages (Fig. 1E, F). Consistent with published reports (35, 36), we observed that i-20S is expressed in all the cell types tested. However, transcripts for the subunits of the i-20S seem to be less abundant than those of c-20S, which reflects on the key contribution by c-20S to essential cellular functions. We have recently generated and characterized a novel, highly specific, non-covalent, reversible inhibitor of the β5i subunit of i-20S, called PKS3053 (25). PKS3053 is a derivative of our previously described inhibitor DPLG3 (6) with better drug-like properties and higher potency against β5i (IC50 of 0.5 nM) compared to DPLG3 (IC50 4.5 nM) when tested against the isolated enzyme and even further improved potency for proteasome inhibition within cells of the i-20S-expressing cell line Karpas 1106P (4.7 nM vs 106 nM)(25). PKS3053 is a β5i-specific inhibitor with a time-dependent modality of inhibition (Fig. S1 and (25)).
Figure 1. Relative expression of the immunoproteasome and constitutive proteasome subunits in purified human pDCs, cDCs and macrophages.
(A, B) Purified human pDCs, (C, D) monocyte-derived cDCs or (E, F) macrophages were purified from human PBMC and the expression of subunits of the (A, C, E) constitutive proteasome PMSB6, PMSB7 and PMSB5 or of the (B, D, F) immunoproteasome PMSB9, PMSB10 and PMSB8 were quantified by qPCR relative to the housekeeping gene UBI. All results are represented as a mean ± SEM and individual donors are shown.
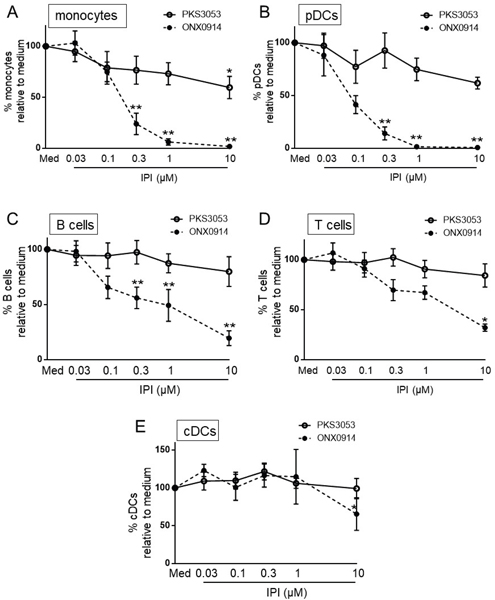
We evaluated the effect of PKS3053 on the viability of various immune cells compared to ONX0914 (previously known as PR-957), an irreversible inhibitor that is relatively selective for β5i (14). Even at low concentrations, ONX0914 was toxic to most cell types tested, except cDC. In contrast, we observed little to no toxicity with PKS3053, except at the highest concentration of 10 μM (Fig. 2A-E), and no cell death was induced by PKS3053 in pDCs responding to agonists of TLR9 (Fig. S3A) or in cDCs responding to TLR8 (Fig. S3B).
Figure 2. Inhibition of β5i with PKS3053 has no apparent toxicity for immune cells.
PBMCs (n=6) were cultured for 24h with media alone or with either PKS3053 or ONX0914 at 0.03, 0.1, 0.3, 1 and 10 μM as indicated. The viability of (A) monocytes (CD14+ cells), (B) pDCs (CD14−CD11c− CD123+ BDCA4+ cells), (C) B cells (CD14−CD20+ cells), (D) T cells (CD14−CD3+ cells) and (E) cDCs (CD14−CD123−CD11c+ cells) was evaluated by flow cytometry. All results are represented as mean ± SEM. Statistical significance was evaluated using a Mann-Whitney U-test and *p≤0.05; **p≤0.01; ***p≤0.001
The immunoproteasome contributes to the response of pDCs to TLR9 activation
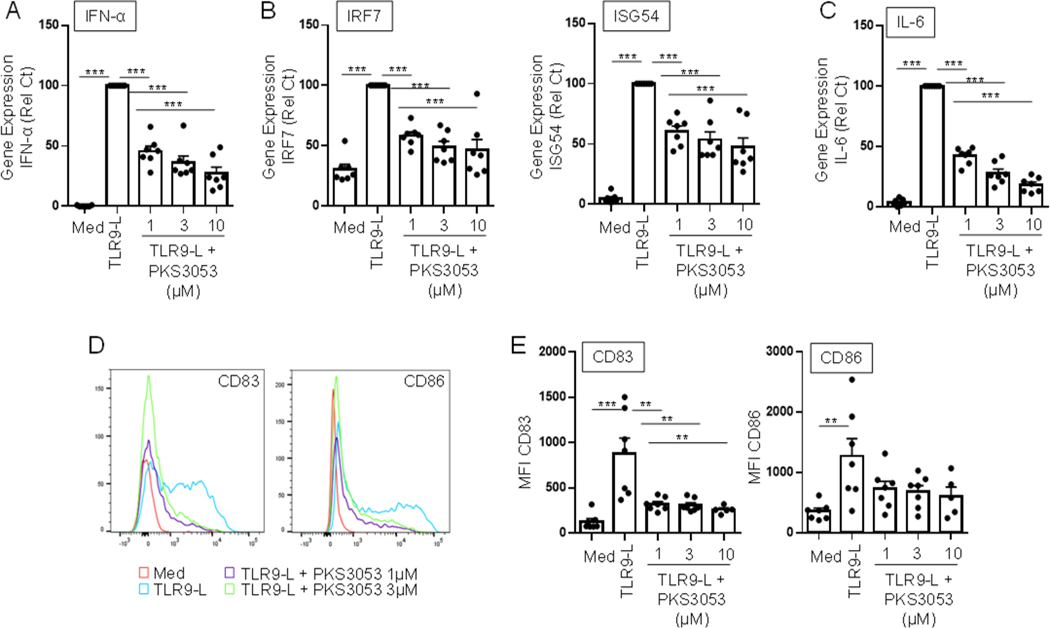
pDCs are a key cell type involved in anti-viral responses, but their chronic activation can lead to autoimmune diseases (37, 38). We and others have shown that TLR9 signaling can induce pDCs to secrete type I IFN as well as other proinflammatory cytokines such as IL-6 and can induce the maturation of pDCs into antigen presenting cells with the expression of costimulatory molecules such as CD80/83/86 (37, 38). DPLG3, a selective N,C-capped dipeptide inhibitor of the immunoproteasome subunit β5i, decreased pDCs’ response to TLR9 (6). PKS3053 significantly inhibited pDC response to TLR9 as seen with decreased expression of Type I IFN (Fig. 3A), of the IFN-regulated genes IRF7 and ISG54 (Fig. 3B) and of the pro-inflammatory cytokine IL-6 (Fig. 3C). PKS3053 also decreased the TLR9-induced expression of the maturation markers CD83 and CD86 in pDCs (Fig. 3D and E). These data demonstrate that the immunoproteasome controls both the production of several cytokines and maturation of pDCs and this control can be blocked without inducing cell death.
Figure 3. Blocking the immunoproteasome prevents activation of TLR9-induced pDCs without inducing toxicity.
Purified pDCs were cultured for 6h (A, B, C) or 24h (D, E) with media alone, with the TLR9 ligand CpG-C274 (0.1 μM) or with the TLR9-L and either PKS3053 or ONX0914 at the indicated concentration. Gene expression levels of (A) IFNα, (B) IRF7, ISG54, and (C) IL-6 relative to the housekeeping gene UBI were quantified via qPCR (n = 7). (D) Histograms and (E) mean fluorescence intensity (n = 5 to 7) of costimulatory molecules CD83 and CD86 were obtained by flow cytometry. All results are represented as a mean ± SEM and individual donors are shown. Statistical significance was evaluated using a Mann-Whitney U-test and *p≤0.05; **p≤0.01; ***p≤0.001
TLR-dependent activation of macrophages and cDCs is under the control of the immunoproteasome
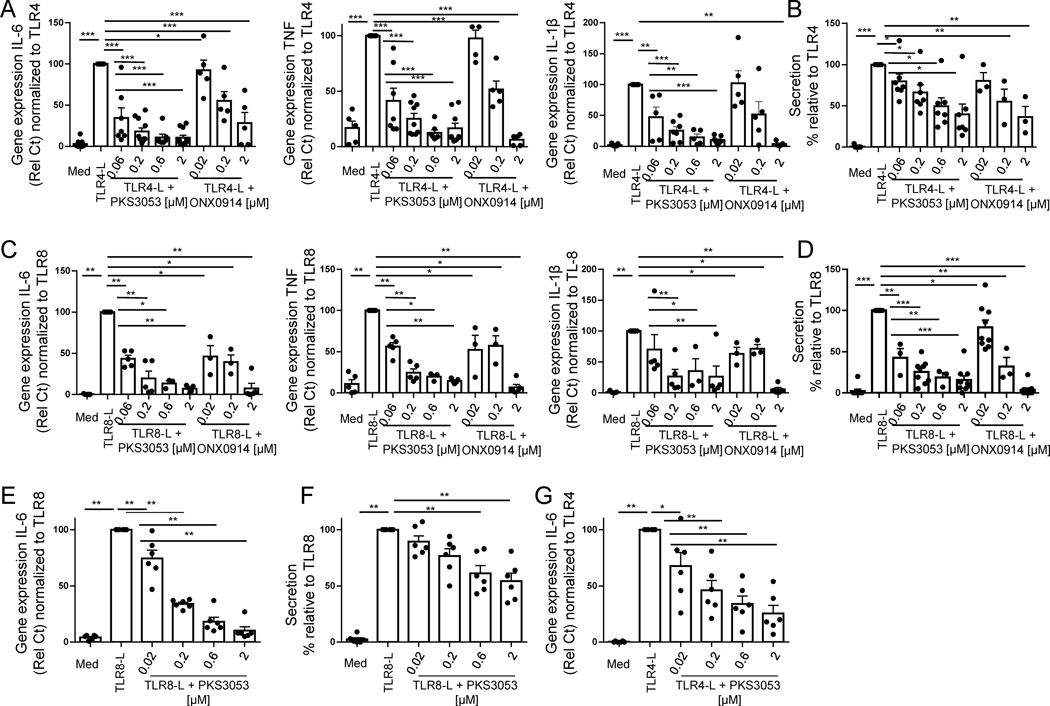
We next investigated whether the immunoproteasome controls TLR-induced pro-inflammatory responses in other innate cell types. Human macrophages and monocytes as well as other myeloid cells express little TLR9 (39) can but produce a massive proinflammatory response when triggered through TLR4 or TLR8 (39). Expression of IL-6, TNF and IL-1β by TLR4- (Fig.4A) or TLR8-activated (Fig. 4C) macrophages was markedly inhibited by both ONX0914 and PKS3053. Likewise, these inhibitors prevented the secretion of IL-6 by TLR4- (Fig. 4B) or TLR8-induced macrophages (Fig. 4D). Similarly, PKS3053 decreased IL-6 gene expression (Fig. 4E and G) and secretion (Fig. 4F) by TLR8-or TLR4-activated cDC. These data suggest that the immunoproteasome is an important regulator of the inflammatory response to TLR agonists by innate immune cells and that PKS3053 as efficient as ONX0914 at inhibiting TLR4- and TLR8-induced responses in macrophages and cDC without detectable toxicity at the concentrations used (Fig. 2).
Figure 4. Immunoproteasome inhibitor suppresses the pro-inflammatory response to TLR ligands in myeloid cells.
Macrophages and cDCs differentiated from monocytes of healthy donors were cultured with media alone, with the TLR4 agonist (LPS) or TLR8 agonist (ORN8-L) with either PKS3053 or ONX0914. (A and C) Gene expression levels of IL-6, TNF and IL-1β relative to UBI were quantified by qPCR in macrophages cultured for 6 hours (n = 5–11) with media alone or with either LPS (A) or ORN8-L (C) in addition to PKS3053 (0, 0.06, 0.2, 0.6, and 2μM) or ONX0914 (0.02, 0.2 and 2 μM). (B & D) IL-6 secretion by (B) TLR4- or (D) TLR8-induced macrophages was quantified by ELISA after 24 hours of culture. (E, F, G) cDCs (n=6) were cultured for (E, G) 6h or (F) 24h with media alone, with (E,F) LPS (50 ng/ml) or with (G) the ORN8-L (60 μg/ml) with PKS3053 (0, 0.02, 0.2, 2 & 6 μm). (E, G) IL-6 gene expression was quantified by qPCR in (E) TLR8- or (G) TLR4-induced cDCs. (F) IL-6 secretion was quantified by ELISA in TLR8-induced cDCs All results are represented as a mean ± SEM and individual donors are shown. Statistical significance was evaluated using a Mann-Whitney U-test and *p≤0.05; **p≤0.01.
The immunoproteasome regulates the pDC-driven proliferation and activation of T cells
Blocking the immunoproteasome with DPLG3 inhibited the proliferation of mouse CD4 and CD8 T cells (6). We investigated whether the immunoproteasome impacts human T cell activation directly or indirectly by preventing an optimal APC-T cell interaction. Naïve T cells were labelled and stimulated with anti-CD3/CD28 for 5 days and proliferation was quantified by flow cytometry. PKS3053 suppressed both the proliferation (Fig. 5A and B) and the secretion of IFNγ (Fig. 5C) from anti-CD3/CD28 activated T cells in a dose dependent manner. Since we showed that inhibiting the immunoproteasome decreased the activation of pDCs, we tested whether pDCs that are triggered through TLR9 in the presence of PKS3053 could activate allogeneic naïve T cells. The pDCs were first activated for 48h with a TLR9 agonist with or without PKS3053, then washed to remove PKS3053 from the culture and used to activate allogeneic T cells. Blocking pDCs’ function with PKS3053 led to a reduced allogeneic T cell response as assessed by decreased proliferation (Fig. 5D) and IFNγ production (Fig. 5E). This demonstrates that the immunoproteasome is a key regulator of T cell responses, both via a direct effect on the T cells and by interfering with APC-T cell interaction.
Figure 5. The immunoproteasome controls APC-driven activation of T cells.
T cells were stained with Cell Trace Violet and activated by anti-CD3 (5 μg/ml, coated) and anti-CD28.2 (1 μg/ml, soluble) antibodies and treated with media alone or PKS3053 (0.01, 0.1, 1, 3, 10 μM) for 5 days. (A) Histograms and (B) percentage of proliferating T cells (n = 5) were acquired via flow cytometry (C) IFNγ secretion was quantified by ELISA with the same conditions. (D) Histograms and percentage of proliferating (n = 8) T cells were acquired by flow cytometry after coculture for 96 hours with pDCs pre-activated with C274 (0.1 μM) with or without PKS3053 (0.3, 1, 3, 10 μM) for 48 hours. (E) IFNγ secretion was measured by ELISA with the same conditions. All results are represented as a mean ± SEM. Statistical significance was evaluated using a Mann-Whitney U-test and *p≤0.05; **p≤0.01; ***p≤0.001.
Inhibition of immunoproteasome protects from inflammation and reduces skin injury in a mild cutaneous injury model in mice
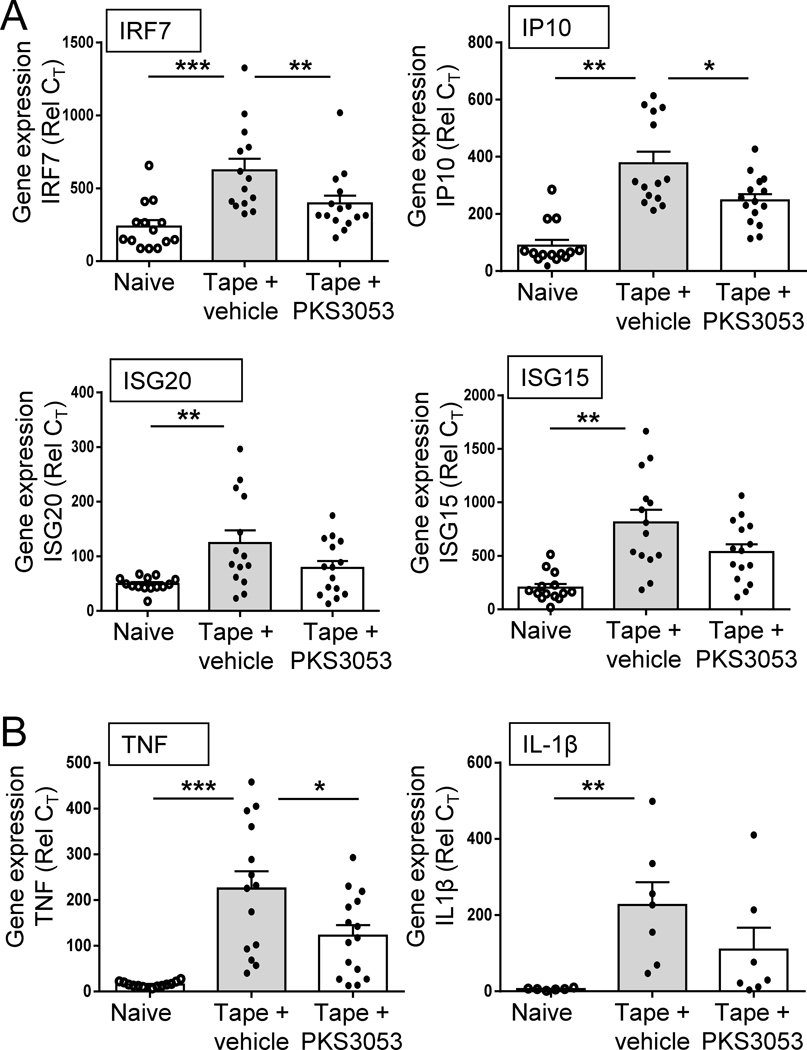
We and others have previously described a model of mild cutaneous injury in mice that resulted in a rapid and strong inflammatory response in the skin that involves multiple cell types, including pDCs that secrete IFN-I and myeloid cells that produce a large set of pro-inflammatory mediators (30, 31). This provides a way to evaluate the effect of PKS3053 in vivo in a model where the nature of the cytokines produced in the skin has been associated with specific cell types (30). The tape stripping method has been used to mimic psoriasis and atopic dermatitis in mice (26–28). At 24 h post tape stripping, we observed the induction in the skin of the mice of several IFN-regulated genes, including IRF7, IP10, ISG20 and ISG15 (Fig. 6A), as well as pro-inflammatory cytokines TNF and IL1β (Fig. 6B). A single injection of the immunoproteasome inhibitor PKS3053 immediately prior to the tape stripping prevented the induction of these genes (Fig. 6A and B).
Figure 6: Inhibition of immunoproteasome protects from inflammation in a mild cutaneous injury model in mice.
(A-B) Age matched C57BL/6 mice were either shaved only (naïve, open circle, n=14) or tape stripped (tape) to provoke inflammation and injected i.p either with vehicle (grey bar, n=13) or PKS3053 at 50 mg/kg (n=14). 24 h later, skin biopsies were collected, and gene expression were evaluated by quantitative PCR. (A) Relative expression of IFN regulated genes as indicated (B) Relative expression of pro-inflammatory genes as indicated. All results are represented as a mean ± SEM from three independent experiments for each group and statistical significance was evaluated using a Mann-Whitney U-test and *p≤0.05; **p≤0.01; ***p≤0.001
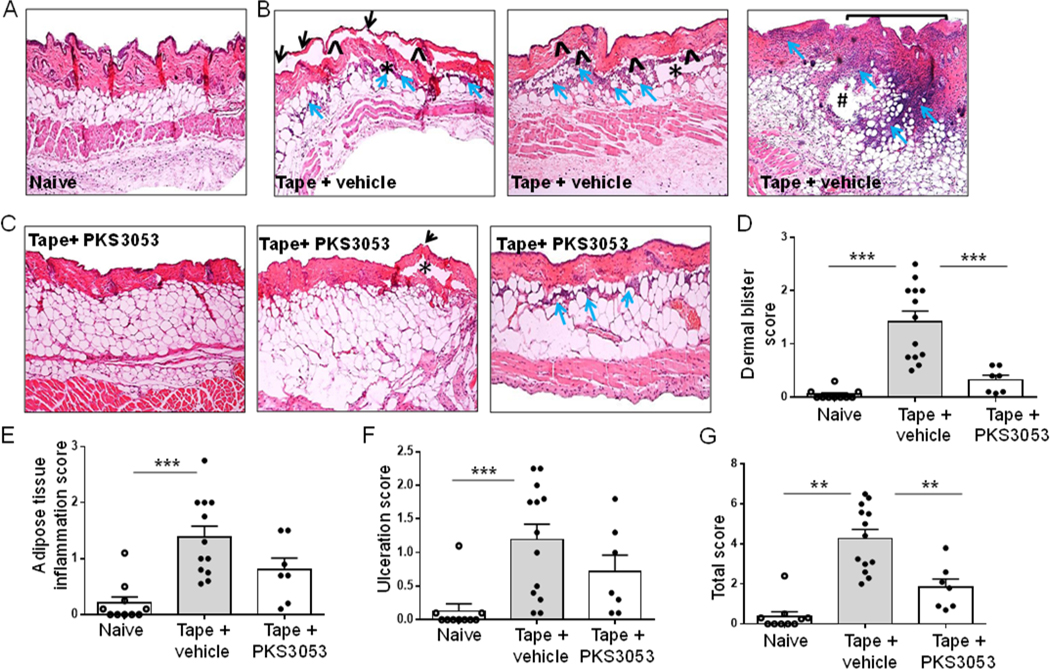
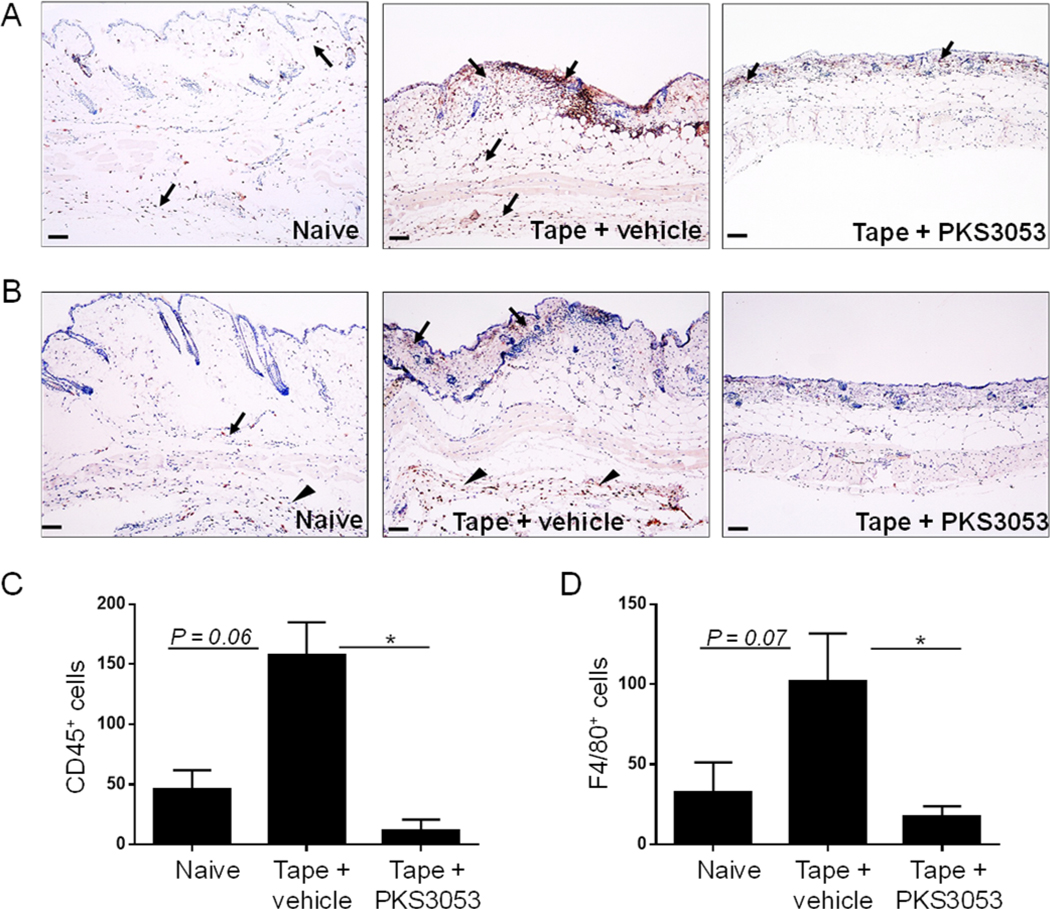
We have shown in a previous study that tape stripping in WT mice leads to a transient inflammation and injury after 1 day, which resolves spontaneously (30). In order to evaluate the role of the immunoproteasome in cutaneous injury as well as in inflammation, PKS3053 or vehicle was injected intraperitoneally in C57BL/6 mice just before tape stripping and skin biopsies on the back were collected and tissue injury evaluated at 24 h in terms of ulceration, dermal and adipose tissue inflammation and blistering. No inflammation and no epithelial damage were observed in control mice where the back skin was only shaved (Fig. 7A). In tape-stripped mice treated with vehicle, signs of skin damage included extensive ulcerations with exposed dermis (square bracket, Fig. 7B); multiple areas with partial or complete loss of epidermis (black arrow, Fig. 7B); and blistering in the dermoepidermal junction or along the dermal-adipose junction (*), with marked inflammation (blue arrows) (Fig. 7B). In some cases, the inflammation reached the subcutaneous adipose tissue and included polymorphonuclear cells (Fig. 7B, #). In the skin of tape-stripped mice treated with PKS3053, no (Fig. 7C, left panel) or minimal alterations (Fig. 7C, middle and right panel) were observed. While in some of tape-stripped mice receiving injection of PKS3053 there were no signs of skin damage (Fig. 7C, left panel), other mice (Fig. 7C, middle panel) showed small areas of thinned epidermis (black arrows) and a few intradermal blisters (asterix) with no inflammation. Some mice showed just a mild (Fig. 7C, right panel) inflammation (blue arrows) in the dermal-adipose junction with no ulceration or blistering. Blistering (Fig. 7D), inflammation (Fig. 7E) and ulceration (Fig. 7F) were scored as described in Materials and Methods. They were significantly increased in tape-stripped mice receiving vehicle alone as compared to shaved mice and reduced in tape-stripped mice treated with the immunoproteasome inhibitor (Fig. 7D, E and F). The total score of skin damage was significantly reduced in tape stripped mice receiving PKS3053 compared to vehicle (Fig. 7G). We further quantified inflammation by analyzing infiltration of CD45+ cells and macrophages, identified by the F4/80 marker. Inflammation induced by tape-stripping resulted in infiltration of CD45+ cells in the dermis and adipose tissue, but mice treated with PKS3053 had a significant decrease in the number of these cells (Fig. 8A and C). CD45+ cells were scattered all across the skin as well as in the deeper subcutaneous soft tissue in tape-stripped mice injected with vehicle, while PKS3053 reduced the number of CD45+ cells, which were mostly concentrated in the superficial dermis. Infiltration of macrophages in tape-stripped mice receiving vehicle injection was increased and mostly concentrated in the deep tissues (arrow head) (Fig. 8B, middle panel), while few to no F4/80+ cells were observed in skin from mice treated with PKS3053 (Fig. 8B & D). These data support the conclusion that PKS3053 reduced the overall inflammatory status in the skin of the tape-stripped mice.
Figure 7: Blocking the immunoproteasome reduces skin injury in mice.
(A-G) Age matched C57BL/6 mice were either shaved only (naïve) or tape stripped (tape) to provoke injury and injected i.p either with vehicle or PKS3053 at 50 mg/kg. 24 h later, skin biopsies were collected, stained and histology scores were evaluated (A-C) Representative images of Hematoxylin & Eosin-stained skin sections at a magnification of x10. Presence of blisters is indicated by ^ for intradermal bullae or by * for subepidermal cracks, of inflammation by blue arrows, spreading in the deeper adipose tissue forming abscesses (#) and of ulcerations is represented by square bracket for large areas of disepithelization or by black arrow for micro regions of epidermal loss in (A) control mice that have been shaven only, (B) tape stripped mice injected with vehicle, (C) tape stripped mice injected with PKS3053, (D-G) Average histology parameters scored from 0– 3 in 4 skin sections of naïve (open circle, n=10), tape + vehicle (grey bar, n=12) or tape+ PKS3053 (n=7) for (D) Dermal blister score (E) Degree of adipose tissue inflammation (F) Ulcer score (G) Total score represented as the average sum of the three histology parameters. Histology scores are represented as a mean ± SEM from two independent experiments for each group and statistical significance was evaluated using a Mann-Whitney U-test and *p≤0.05; **p≤0.01; ***p≤0.001.
Figure 8: The immunoproteasome inhibitor decreases infiltration of inflammatory cells in tape-stripped skin of mice.
(A-D) Age matched C57BL/6 mice were either shaved only (naïve) or tape stripped (tape) to provoke injury and injected i.p either with vehicle or PKS3053 at 50 mg/kg. 24 h later, skin biopsies were collected, stained with CD45 or f4/80 antibodies and number of positive cells were counted (A, B) Representative images (Scale bar = 50 μm) of (A) CD45 and (B) F4/80 immunohistochemistry of skin sections at 10X magnification. Presence of positively stained cells are represented by arrows and arrow heads (C, D) averages of the number of (C) CD45+ and (D) F4/80+ cells. All values represent mean ± SEM; n=5 mice for each group. Data are cumulative from two independent experiments. Statistical significance was evaluated using a Mann-Whitney U-test. *p≤0.05; **p≤0.01; ***p≤0.001.
DISCUSSION
This study makes new three points. First, a β5i-selective IPI is effective at suppressing innate immune cell function, T cell function and dermal inflammation without co-inhibition of additional active subunits of proteasomes. Second, an IPI can exert these effects without dependence on a cytotoxic effect. Third, a noncovalent IPI is effective in vitro and in vivo in these settings.
The proteasome shares with autophagy the responsibility for the degradation of most proteins in the cell. However, the proteasome plays an equally important, additional role in pre- and post-transcriptional cell signaling through the degradation of certain proteins that control the function of others. Both sets of processes are largely controlled by selective ubiquitinylation of proteins targeted for degradation, although this requirement can be bypassed for heavily oxidized proteins. This proteasome’s role in cell signaling extends to cellular survival, activation, differentiation and proliferation and has thus generated significant scientific and therapeutic interest. Bortezomib was approved by the FDA in 2003 for the treatment of myeloma and later for mantle cell lymphoma. Mechanistic studies suggested that its ability to inhibit the proteasome in cells with a high load of misfolded proteins and dependence on NF-kB for avoidance of apoptosis contribute to its lethality for these tumor cells. However, its toxicity extends to other cells as well, including in the nervous, hematopoietic and cardiovascular systems. These toxicities limit its usefulness for the chronic treatment of autoimmune diseases and have directed attention to the development of more selective inhibitors of immunoproteasomes.
In pioneering studies, the peptide epoxyketone ONX0914 was shown to suppress immune activation in various mouse models of inflammatory diseases with less toxicity than Bortezomib (14, 16, 17, 19, 40). Similar or improved versions of ONX0914 are being developed, but for the most part, they are covalent, irreversible binders of i-20S (41) with relatively modest selectivity for i-20S compared to c-20S. Over time, this can lead to cumulative inhibition of c-20S. Once both i-20S and c-20S are inhibited, non-malignant immune cells die, as seen not only with Bortezomib (9) but also with ONX0914 (Fig. 2). It has not been clear whether the immune suppressing effects of such agents are separable from their cytotoxic effects on immune cells. Our findings show that the immune-suppressing effects of an immunoproteasome-selective inhibitor do not have to be accompanied by or result from cytotoxicity. GANG PLEASE CHECK: In a study with covalently reactive proteasome inhibitors, impairment of immune cell function required combined inhibition of β5i and β1i (20, 42). However, this need not be the case, as PKS3053 is inactive against β1i. We do not know why the beneficial effects of PKS3053 are seen without the need to inhibit β1i along with β5i. Perhaps covalent attachment of some inhibitors to β5i alters the conformation of β1i in such a way as to increase its ability to compensate for inhibition of β5i, while PKS3053 does not have that effect.
Several novel classes of non-covalent, reversible inhibitors of i-20S have been reported that are highly β5i-selective (6, 43–45). Compared to DPLG3, PKS3053 is an improved N,C-capped dipeptide with 118-fold selectivity over β5c; its selectivity is maintained as the time of co-incubation is increased; and PKS3053 has markedly improved potency for in situ proteasome inhibition within cells. Because of its lack of cellular toxicity, even at high doses, we were able to demonstrate that the i-20S is essential for the optimal activation of immune cells under the conditions studied. The production of IFN-I, the induction of costimulatory molecules and the ability to activate T cells by TLR-activated pDCs were all dependent on i-20S as judged by the impact of PKS3053 (Fig. 2). Macrophages and cDCs required i-20S to produce proinflammatory cytokines in response to multiple TLR agonists (Fig. 3). These data suggest that innate immune cells require degradation of certain proteins in order for the stimulatory signals tested to fully activate these cells. It has been well established that the proteasome regulates NF-kB activation by degrading IKKα following its phosphorylation and subsequent ubiquitination (46). While i-20S is well known to degrade proteins, it was not appreciated that the immunoproteasome, rather than the constitutive proteasome, is involved in the cellular activation of multiple immune cells. Which proteins the immunoproteasome is responsible for degrading and what dictates selective dependence on the immunoproteasome are questions for further investigation. Blocking i-20S with PKS3053 also hindered T cell activation, whether triggered by anti-CD3/CD28 or by allogeneic reactions. Thus, non-cytotoxic inhibition of the immunoproteasome is sufficient to prevent the activation of both innate and adaptive responses. This highlights the potential of such an approach in many autoimmune diseases.
Tape stripping is a relevant model to study cutaneous inflammation in vivo as it recruits multiple immune cell types and pathways that are associated with IFN-I and other proinflammatory mediators (30). This model leads to moderate skin injury and the mechanism of inflammation is reminiscent of what is observed in cutaneous inflammatory diseases characterized by an interface dermatitis, such as CLE, dermatomyositis or lichen planus (47), as well as psoriasis, atopic dermatitis and systemic sclerosis (33). The infiltration of immune cells is not dependent on MyD88 or TLR7/9, but the cytokine response is, suggesting that the effect observed with PKS3053 is due to inhibition of cellular activation rather than a direct effect on cellular extravasation and homing. Because treating mice with PKS3053 reduced both the IFN-I-dependent and -independent responses, PKS3053 appears to have been effective at blocking not only pDCs, which we have shown to be the source of IFN-I in this model (30), but also myeloid cells responsible for the production of IL-1s and TNF. We observed that the overall reduction in the inflammatory response was associated with reduced cellular infiltration as the presence of CD45+ cells and of F4/80+ cells was attenuated in the PKS3053-treated animal, as compare to control. This inhibition of the cellular response in vivo by PKS3053 was associated with reduced tissue damage, including reduced ulceration, dermal blistering and inflammation of adipose tissues.
The contribution of pDCs to skin diseases that share a common pathological feature termed “interface dermatitis” (47) has been documented and their role in systemic disease is also clear (37, 48, 49). We recently showed that pDCs contribute to skin fibrosis (33) and a human trial of an antibody that attenuates the pDC response yielded promising results (50). Macrophages producing TNF contribute to arthritis and have been associated with fibrosis (51). Taken altogether, our data suggest that non-cytotoxic inhibition of i-20S might prove useful for the treatment of autoimmune diseases, including skin-related diseases such as CLE and scleroderma.
Supplementary Material
Key points :.
The immunoproteasome controls the activation of immune cells
A novel inhibitor of the immunoproteasome without cellular toxicity
PKS3053 decreases inflammation and tissue damage in a moderate cutaneous injury model
Acknowledgments
GRANT SUPPORT
FJB is supported by grants from the NIH 1R01AI132447, from the Scleroderma Research Foundation and the Scleroderma Foundation. GL acknowledges the support of the Alliance for Lupus Research, NIH R01AI143714 and the Daedalus Fund for Innovation at Weill Cornell Medicine. CN acknowledges the support of the Milstein Program in Chemical Biology and Translational Medicine. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Trust.
ABBREVIATIONS
- pDC
plasmacytoid dendritic cells
- cDC
conventional dendritic cells
- i-20S
immunoproteasome
- c-20S
constitutive proteasome
- CLE
cutaneous lupus erythematosus
- CTV
cell trace violet
References:
- 1.Goldberg AL 2007. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans 35: 12–17. [DOI] [PubMed] [Google Scholar]
- 2.Tomko RJ Jr., and Hochstrasser M. 2013. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem 82: 415–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumeister W, Walz J, Zuhl F, and Seemuller E. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K. 1994. Role of proteasomes modified by interferon-gamma in antigen processing. J Leukoc Biol 56: 571–575. [DOI] [PubMed] [Google Scholar]
- 5.Groettrup M, Kirk CJ, and Basler M. 2010. Proteasomes in immune cells: more than peptide producers? Nat Rev Immunol 10: 73–78. [DOI] [PubMed] [Google Scholar]
- 6.Sula Karreci E, Fan H, Uehara M, Mihali AB, Singh PK, Kurdi AT, Solhjou Z, Riella LV, Ghobrial I, Laragione T, Routray S, Assaker JP, Wang R, Sukenick G, Shi L, Barrat FJ, Nathan CF, Lin G, and Azzi J. 2016. Brief treatment with a highly selective immunoproteasome inhibitor promotes long-term cardiac allograft acceptance in mice. Proc Natl Acad Sci U S A 113: E8425–E8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehling HJ, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, and von Boehmer H. 1994. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science 265: 1234–1237. [DOI] [PubMed] [Google Scholar]
- 8.Ferrington DA, and Gregerson DS 2012. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci 109: 75–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirai M, Kadowaki N, Kitawaki T, Fujita H, Takaori-Kondo A, Fukui R, Miyake K, Maeda T, Kamihira S, Miyachi Y, and Uchiyama T. 2011. Bortezomib suppresses function and survival of plasmacytoid dendritic cells by targeting intracellular trafficking of Toll-like receptors and endoplasmic reticulum homeostasis. Blood 117: 500–509. [DOI] [PubMed] [Google Scholar]
- 10.Ferrington DA, Hussong SA, Roehrich H, Kapphahn RJ, Kavanaugh SM, Heuss ND, and Gregerson DS 2008. Immunoproteasome responds to injury in the retina and brain. J Neurochem 106: 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opitz E, Koch A, Klingel K, Schmidt F, Prokop S, Rahnefeld A, Sauter M, Heppner FL, Volker U, Kandolf R, Kuckelkorn U, Stangl K, Kruger E, Kloetzel PM, and Voigt A. 2011. Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog 7: e1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, and Van den Eynde BJ 2010. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci U S A 107: 18599–18604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basler M, Dajee M, Moll C, Groettrup M, and Kirk CJ 2010. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol 185: 634–641. [DOI] [PubMed] [Google Scholar]
- 14.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, and Groettrup M. 2009. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med 15: 781–787. [DOI] [PubMed] [Google Scholar]
- 15.Inoue S, Nakase H, Matsuura M, Mikami S, Ueno S, Uza N, and Chiba T 2009. The effect of proteasome inhibitor MG132 on experimental inflammatory bowel disease. Clin Exp Immunol 156: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalim KW, Basler M, Kirk CJ, and Groettrup M. 2012. Immunoproteasome subunit LMP7 deficiency and inhibition suppresses Th1 and Th17 but enhances regulatory T cell differentiation. J Immunol 189: 4182–4193. [DOI] [PubMed] [Google Scholar]
- 17.Basler M, Mundt S, Muchamuel T, Moll C, Jiang J, Groettrup M, and Kirk CJ 2014. Inhibition of the immunoproteasome ameliorates experimental autoimmune encephalomyelitis. EMBO molecular medicine 6: 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mundt S, Engelhardt B, Kirk CJ, Groettrup M, and Basler M. 2016. Inhibition and deficiency of the immunoproteasome subunit LMP7 attenuates LCMV-induced meningitis. Eur J Immunol 46: 104–113. [DOI] [PubMed] [Google Scholar]
- 19.Ichikawa HT, Conley T, Muchamuel T, Jiang J, Lee S, Owen T, Barnard J, Nevarez S, Goldman BI, Kirk CJ, Looney RJ, and Anolik JH 2012. Beneficial effect of novel proteasome inhibitors in murine lupus via dual inhibition of type I interferon and autoantibody-secreting cells. Arthritis Rheum 64: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basler M, Claus M, Klawitter M, Goebel H, and Groettrup M. 2019. Immunoproteasome Inhibition Selectively Kills Human CD14(+) Monocytes and as a Result Dampens IL-23 Secretion. J Immunol 203: 1776–1785. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Koerner J, Basler M, Brunner T, Kirk CJ, and Groettrup M. 2019. Immunoproteasome inhibition induces plasma cell apoptosis and preserves kidney allografts by activating the unfolded protein response and suppressing plasma cell survival factors. Kidney Int 95: 611–623. [DOI] [PubMed] [Google Scholar]
- 22.Neubert K, Meister S, Moser K, Weisel F, Maseda D, Amann K, Wiethe C, Winkler TH, Kalden JR, Manz RA, and Voll RE 2008. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis. Nat Med 14: 748–755. [DOI] [PubMed] [Google Scholar]
- 23.Badros A, Goloubeva O, Dalal JS, Can I, Thompson J, Rapoport AP, Heyman M, Akpek G, and Fenton RG 2007. Neurotoxicity of bortezomib therapy in multiple myeloma: a single-center experience and review of the literature. Cancer 110: 1042–1049. [DOI] [PubMed] [Google Scholar]
- 24.Pletinckx K, Vassen S, Schlusche I, Nordhoff S, Bahrenberg G, and Dunkern TR 2019. Inhibiting the immunoproteasome’s beta5i catalytic activity affects human peripheral blood-derived immune cell viability. Pharmacol Res Perspect 7: e00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan W, Singh PK, Ban Y, Qing X, Ah Kioon MD, Fan H, Zhao Q, Wang R, Sukenick G, Salmon J, Warren JD, Ma X, Barrat FJ, Nathan CF, and Lin G. 2020. Structure-Activity Relationships of Noncovalent Immunoproteasome beta5i-Selective Dipeptides. J Med Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue J, Yotsumoto S, Sakamoto T, Tsuchiya S, and Aramaki Y. 2005. Changes in immune responses to antigen applied to tape-stripped skin with CpG-oligodeoxynucleotide in mice. J Control Release 108: 294–305. [DOI] [PubMed] [Google Scholar]
- 27.Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, Itami S, Nickoloff BJ, and DiGiovanni J. 2005. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med 11: 43–49. [DOI] [PubMed] [Google Scholar]
- 28.Jin H, Oyoshi MK, Le Y, Bianchi T, Koduru S, Mathias CB, Kumar L, Le Bras S, Young D, Collins M, Grusby MJ, Wenzel J, Bieber T, Boes M, Silberstein LE, Oettgen HC, and Geha RS 2009. IL-21R is essential for epicutaneous sensitization and allergic skin inflammation in humans and mice. J Clin Invest 119: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttman-Yassky E, Diaz A, Pavel AB, Fernandes M, Lefferdink R, Erickson T, Canter T, Rangel S, Peng X, Li R, Estrada Y, Xu H, Krueger JG, and Paller AS 2019. Use of Tape Strips to Detect Immune and Barrier Abnormalities in the Skin of Children With Early-Onset Atopic Dermatitis. JAMA Dermatol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guiducci C, Tripodo C, Gong M, Sangaletti S, Colombo MP, Coffman RL, and Barrat FJ 2010. Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. J Exp Med 207: 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregorio J, Meller S, Conrad C, Di Nardo A, Homey B, Lauerma A, Arai N, Gallo RL, Digiovanni J, and Gilliet M. 2010. Plasmacytoid dendritic cells sense skin injury and promote wound healing through type I interferons. J Exp Med 207: 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guiducci C, Ghirelli C, Marloie-Provost MA, Matray T, Coffman RL, Liu YJ, Barrat FJ, and Soumelis V. 2008. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J Exp Med 205: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ah Kioon MD, Tripodo C, Fernandez D, Kirou KA, Spiera RF, Crow MK, Gordon JK, and Barrat FJ 2018. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, and Coffman RL 2005. Nucleic Acids of Mammalian Origin Can Act as Endogenous Ligands for Toll-like Receptors and May Promote Systemic Lupus Erythematosus. J Exp Med 202: 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morel S, Levy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin AL, Monsarrat B, Van Velthoven R, Cerottini JC, Boon T, Gairin JE, and Van den Eynde BJ 2000. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity 12: 107–117. [DOI] [PubMed] [Google Scholar]
- 36.Haorah J, Heilman D, Diekmann C, Osna N, Donohue TM Jr., Ghorpade A, and Persidsky Y. 2004. Alcohol and HIV decrease proteasome and immunoproteasome function in macrophages: implications for impaired immune function during disease. Cell Immunol 229: 139–148. [DOI] [PubMed] [Google Scholar]
- 37.Barrat FJ, and Su L. 2019. A pathogenic role of plasmacytoid dendritic cells in autoimmunity and chronic viral infection. J Exp Med 216: 1974–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reizis B. 2019. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 50: 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guiducci C, Gong M, Cepika AM, Xu Z, Tripodo C, Bennett L, Crain C, Quartier P, Cush JJ, Pascual V, Coffman RL, and Barrat FJ 2013. RNA recognition by human TLR8 can lead to autoimmune inflammation. J Exp Med 210: 2903–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vachharajani N, Joeris T, Luu M, Hartmann S, Pautz S, Jenike E, Pantazis G, Prinz I, Hofer MJ, Steinhoff U, and Visekruna A. 2017. Prevention of colitis-associated cancer by selective targeting of immunoproteasome subunit LMP7. Oncotarget 8: 50447–50459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huber EM, de Bruin G, Heinemeyer W, Paniagua Soriano G, Overkleeft HS, and Groll M. 2015. Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors. J Am Chem Soc 137: 7835–7842. [DOI] [PubMed] [Google Scholar]
- 42.Basler M, Lindstrom MM, LaStant JJ, Bradshaw JM, Owens TD, Schmidt C, Maurits E, Tsu C, Overkleeft HS, Kirk CJ, Langrish CL, and Groettrup M. 2018. Co-inhibition of immunoproteasome subunits LMP2 and LMP7 is required to block autoimmunity. EMBO Rep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan H, Angelo NG, Warren JD, Nathan CF, and Lin G. 2014. Oxathiazolones Selectively Inhibit the Human Immunoproteasome over the Constitutive Proteasome. ACS Med Chem Lett 5: 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh PK, Fan H, Jiang X, Shi L, Nathan CF, and Lin G. 2016. Immunoproteasome beta5i-Selective Dipeptidomimetic Inhibitors. ChemMedChem 11: 2127–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos RLA, Bai L, Singh PK, Murakami N, Fan H, Zhan W, Zhu Y, Jiang X, Zhang K, Assker JP, Nathan CF, Li H, Azzi J, and Lin G. 2017. Structure of human immunoproteasome with a reversible and noncompetitive inhibitor that selectively inhibits activated lymphocytes. Nature communications 8: 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karin M, and Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol 18: 621–663. [DOI] [PubMed] [Google Scholar]
- 47.Wenzel J, and Tuting T. 2008. An IFN-Associated Cytotoxic Cellular Immune Response against Viral, Self-, or Tumor Antigens Is a Common Pathogenetic Feature in “Interface Dermatitis”. J Invest Dermatol 128: 2392–2402. [DOI] [PubMed] [Google Scholar]
- 48.Sisirak V, Ganguly D, Lewis KL, Couillault C, Tanaka L, Bolland S, D’Agati V, Elkon KB, and Reizis B. 2014. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J Exp Med 211: 1969–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowland SL, Riggs JM, Gilfillan S, Bugatti M, Vermi W, Kolbeck R, Unanue ER, Sanjuan MA, and Colonna M. 2014. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J Exp Med 211: 1977–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furie R, Werth VP, Merola JF, Stevenson L, Reynolds TL, Naik H, Wang W, Christmann R, Gardet A, Pellerin A, Hamann S, Auluck P, Barbey C, Gulati P, Rabah D, and Franchimont N. 2019. Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J Clin Invest 129: 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrat FJ, and Lu TT 2019. Role of type I interferons and innate immunity in systemic sclerosis: unbalanced activities on distinct cell types? Current opinion in rheumatology 31: 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.