Abstract
The prostate develops by epithelial budding and branching processes that occur during fetal and postnatal stages. The adult prostate demonstrates remarkable regenerative capacity, with the ability to regrow to its original size over multiple cycles of castration and androgen administration. This capacity for controlled regeneration prompted the search for an androgen-independent epithelial progenitor in benign prostatic hyperplasia (BPH) and prostate cancer (PCa). BPH is hypothesized to be a reawakening of ductal branching, resulting in the formation of new proximal glands, all while androgen levels are decreasing in the aging male. Advanced prostate cancer can be slowed with androgen deprivation, but resistance eventually occurs, suggesting the existence of an androgen-independent progenitor. Recent studies indicate that there are multiple castration-insensitive epithelial cell types in the proximal area of the prostate, but not all act as progenitors during prostate development or regeneration. This review highlights how recent cellular and anatomical studies are changing our perspective on the identity of the prostate progenitor.
Introduction
The human prostate is a walnut shaped organ situated at the base of the bladder. The glandular tissue of the prostate surrounds the portion of the urethra called the ‘prostatic urethra’. During in utero growth, epithelial outgrowths from the urethral wall extend outward into the surrounding mesenchyme to form prostate rudiments (Timms 2008). During the course of development, prostate rudiments elongate, branch and canalize to form a network of ducts that end in acini. The glands of the prostate drain into the urethra through ducts that open into the urethral wall.
Early studies using wax reconstructions of preserved human fetal samples identified clusters of prostate ducts emerging from different locations around the urethra (Lowsley 1912, Johnson 1920). In the adult, these locational distinctions are less obvious, but McNeal’s description of the zonal anatomy of the prostate (McNeal 1981) closely agreed with the earlier observations from the fetal prostate. The region surrounding the urethra was called the transition zone while the outer regions of the prostate were named the peripheral zone. The central zone was described as the collection of glands situated around the ejaculatory ducts. It was observed that BPH growth was confined to the peri-urethral transition zone of the prostate while prostate cancer was predominantly found in the peripheral zone (McNeal 1981, McNeal 1983). This finding raised the intriguing possibility that BPH and PCa have distinct cells of origin.
Rapid growth and differentiation of the prostate occurs during early development. Benign prostate enlargement is thought to be a recapitulation of this early embryonic growth (McNeal 1978, Brennen and Isaacs 2018). In BPH patients, nodules composed of stroma, epithelium or a mixture of both are found adjacent to the urethra (Strand et al. 2017). During embryonic prostate development, emerging prostate rudiments bud from the urethral epithelium and extend into mesenchymal condensations of fibroblasts in response to androgens (Cunha and Lung 1978). A similar process has been proposed for the development of BPH glandular nodules, where reawakening of embryonic growth signals from the mesenchyme induces new epithelial growth (McNeal 1978, Timms et al. 1994, Bierhoff et al. 1997). The mechanisms responsible for the susceptibility of peri-urethral glands to proliferative signals during the onset of BPH at middle age remain unknown. This could be explained by cell composition differences between the inner ducts and the outer glands. Recent single cell RNA sequencing (scRNA-seq) studies have shown that outgrowths from the prostatic urethra extend into the transition zone and have a distinct cellular composition compared to peripheral prostate glands, which could explain the regional incidence of disease (Henry et al. 2018, Joseph et al. 2020).
Prostate cancer is predominantly found in the peripheral zone and displays a luminal phenotype (De Marzo et al. 2007). Prostate cancer is the second leading cause of cancer mortality in the US and has been treated with surgical and chemical castration for decades (Huggins and Hodges 2002). Most men with advanced prostate cancer initially respond to first- and second-generation androgen deprivation, but castration-resistant prostate cancer (CRPC) almost universally recurs (Leuprolide Study 1984). To determine the cellular origin of CRPC, commonly found genetic losses and mutations have been driven by promoters of genes expressed in basal or luminal epithelia (Wang et al. 2009, Choi et al. 2012). With the advent of single cell RNA sequencing and the molecular identification of new epithelial cell types resistant to castration, the search for a cell of origin for prostate cancer has broadened (Guo et al. 2020). However, cell type-specific analyses of the prostate under castration and regeneration shows that not all castration-insensitive cell types are involved in prostate regeneration (Crowley et al. 2020, Guo et al. 2020, Karthaus et al. 2020, Mevel et al. 2020), which highlights the need to understand the molecular identification and anatomical context of each cell type in development and disease.
Prostate progenitors during fetal development
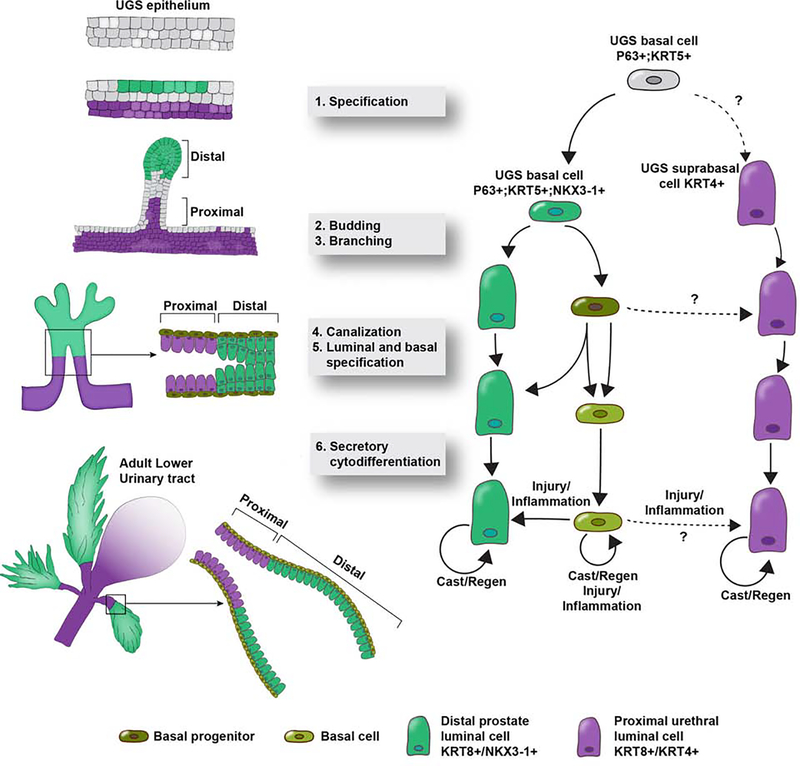
The urogenital system, of which the prostate is a part, develops from a transient endoderm-derived structure called the cloaca. The cloaca undergoes septation to form the urogenital sinus (UGS) and anorectal tracts (Gupta et al. 2014). Androgen action on the mesenchyme (Cunha 1984) surrounding the UGS epithelium induces the outgrowth of prostate buds starting from 10 weeks of gestation in humans and between embryonic day (E) 16–18 in mice (Timms 2008, Cunha et al. 2018). Prostate development has been studied in the greatest detail in mouse and rat and closely parallels human prostate development. Prostate development can be divided into six stages: 1) specification, 2) emergence of solid prostatic epithelial buds from UGS epithelium, 3) bud elongation and branching, 4) canalization of solid epithelial cords, 5) differentiation of luminal and basal epithelial cells, and 6) secretory cytodifferentiation (Cunha et al., 2018). The last two stages, differentiation and secretory cytodifferentiation, are completed at puberty in response to a surge in androgen production.
The early prostate transcription factor NK3 Homeobox 1 (Nkx3.1) marks prostate outgrowths into the budding, branching and canalization stage, and ultimately becomes largely restricted to secretory luminal cells in the mature prostate (Bhatia-Gaur et al. 1999). p63 expression marks the basal layer of the UGS epithelium. Proliferative activity is concentrated in the basal layer of the UGS and in the developing prostate buds, which appear to be evaginations from the basal layer (Cunha et al. 2018). p63 knockout mice fail to develop prostate buds suggesting that the fetal prostate progenitor is basal in origin (Signoretti et al. 2000, Signoretti et al. 2005). Mouse models that indelibly mark basal (Keratin 5/Keratin 14) and luminal (Keratin 8) lineages confirm that basal epithelia are multipotent during prostate budding and branching until puberty, at which point basal and luminal lineages are self-sustained (Choi et al. 2012, Ousset et al. 2012, Tika et al. 2019).
The establishment of proximal and distal compartments occurs during early development, with the proximal region being derived from non-Nkx3.1 expressing urethral epithelial cells marked by Krt4 and Runx1 (Mevel et al. 2020). Differences in progenitor cell potential in the proximal and distal prostate epithelial compartments have been described in the adult prostate (Goto et al. 2006). Recent evidence indicates that the proximal and distal compartments are established concurrently with prostate budding. The proximal region appears to be an extension of urethral epithelium following prostate bud outgrowth. Outgrowths of proximal ducts do not express the Nkx3.1, but instead express markers specific to urethral epithelium (Mevel et al. 2020).
We previously defined the cell type composition of the human prostatic urethra, which includes p63+ basal cells as well as club and hillock cells that are named after similar cell types found in the lung (Henry et al. 2018). Club cells were named after lung club cells which function as progenitors, contribute to airway fluid and metabolize xenobiotics (Zuo et al. 2018). In the lower urinary tract, club cells are mostly confined to the prostatic urethra and proximal ducts (Henry et al. 2018). Club cells express secretoglobins (SCGB3A1 and SCGB1A1) in addition to several genes involved in innate immunity including LCN2, PIGR, PI3 and SLPI. Hillock cells were named after unique stratified structures with high turnover in the lung (Montoro et al. 2018), which are comprised of stacked KRT13-expressing cells. Hillock cells expressing the squamous epithelial markers KRT13 and KRT4 are found in the suprabasal layers of the prostatic urethra and are also found in small structures of contiguous stacked cells in prostate glands. In the developing human prostate, club and hillock cells of the urogenital sinus epithelium extend into the proximal ducts. The distribution of these cells becomes sparse within elongating buds (Joseph et al. 2020).
A recent study shows that a castration-resistant luminal lineage marked by Runx1 in the mouse is established at the onset of prostate development and specifically gives rise to the proximal region of prostate ducts. Runx1 expression and other urethral luminal markers such as Krt4 are localized to the inner luminal layers of the urethral epithelium at the time of bud outgrowth (Mevel et al. 2020). The proximal urethral luminal compartment, marked by Runx1 and Krt4 expression, is maintained as a distinct entity from the Nkx3.1+ prostate secretory luminal compartment into postnatal and adult stages (Joseph et al. 2020). Lineage tracing demonstrates that early Nkx3.1+ urogenital sinus cells contribute to the Nkx3.1− proximal urethral compartment in the adult (Crowley et al. 2020). Although facultative assays suggest that proximal luminal cells of the urethra can give rise to Nkx3.1+ prostate luminal cells (and p63+ basal cells) ex vivo, lineage tracing under physiological conditions in vivo shows little evidence that proximal Krt4+/Sca-1+/Trop2+/Psca+ urethral luminal cells are progenitors for prostate in development or adult castration/regeneration (Guo et al. 2020, Karthaus et al. 2020, Kwon et al. 2020).
Regional niches of epithelial progenitors in the adult prostate
Progenitor cells participate in organ morphogenesis and become less numerous by adulthood. These cells are often confined to specialized niches surrounded by inductive stroma and supporting cells. In the lung, stem/progenitor cells reside in the proximal airway and proliferate in response to injury. These progenitors can differentiate into multiple cell types in the lung (Chen et al. 2012, Salwig et al. 2019). Studies in the mouse bladder have demonstrated transient embryonic progenitors that contribute to early bladder development are replaced by a different set of progenitors in the adult bladder (Gandhi et al. 2013). In mouse prostate development, multipotent basal progenitors are replaced by lineage-committed, unipotent basal and luminal progenitors that contribute to adult growth and maintenance of the organ (Ousset et al. 2012, Tika et al. 2019, Kwon et al. 2020, Wang et al. 2020).
The adult mouse prostate shrinks at the distal axis in response to castration due to the loss of androgen-dependent secretory luminal cells. However, the prostate only regrows to its original size pointing to a growth-restricting mechanism (Coffey et al. 1968, English et al. 1985, English et al. 1987, English et al. 1989). Most of the DNA synthesis activity during regeneration occurs in the distal prostate. This led to the proposal that an androgen-independent progenitor exists in the distal adult prostate and can repopulate the castrated prostate when testosterone is re-administered (Sugimura et al. 1986, Kinbara et al. 1996).
In contrast, studies over the last 20 years have opened up the possibility that luminal cells in the proximal prostate could serve as androgen-independent prostate progenitors. A series of facultative, colony formation experiments using dissected tissue from the urethra, proximal ducts and distal prostate, suggested that the highest regenerative capacity resided in the urethra and proximal prostate (Tsujimura et al. 2002, Goto et al. 2006). The interpretation was that the enhanced ability to form colonies ex vivo was indicative of high proliferative potential and stemness. This group went on to show that dissected tissue from the mouse urethra and proximal ducts was more efficient in tissue regeneration and branching assays, and expressed high levels of Sca-1, a cell surface marker commonly expressed by stem cells in other tissues (Burger et al. 2005). Sca-1+ luminal cells in the proximal region have low levels of androgen signaling and do not express the androgen receptor target gene Nkx3.1 (Kwon et al. 2016). This made them ideal candidates for the androgen-independent proximal prostate progenitor. However, despite Sca-1+ positive cells being capable of differentiating into Sca-1−/Nkx3.1 + prostate luminal cells and p63+ basal cells ex vivo (Xin et al. 2005), lineage tracing of Sca-1 + luminal cells shows that they are largely self-sustaining, only regenerating more Nkx3.1−/Sca-1 + luminal cells (Kwon et al. 2020). These results were recently confirmed by lineage tracing proximal androgen-independent cells marked by Runx1, Psca or Krt4 (Guo et al. 2020, Mevel et al. 2020).
We recently demonstrated by scRNA-seq the molecular identity of Sca-1+ luminal cells in the mouse. Sca-1+ luminal epithelia in the mouse proximal prostate represent extensions of the urethra into the proximal ducts that drain the prostate of its secretions (Joseph et al. 2020). Mouse urethral luminal epithelia express markers previously suggested to represent proximal progenitors including Sca-1 (Shi et al. 2014, Kwon et al. 2016), Trop2 (Goldstein et al. 2008), Psca (Crowell et al. 2019), and Krt4 (Sackmann Sala et al. 2017, Karthaus et al. 2020). The Krt4+ proximal luminal cell compartment is derived from the suprabasal layers of the UGS and is established early in prostate development (Joseph et al. 2020, Mevel et al. 2020). Although the Sca-1 gene, Ly6a, does not have a direct human homolog, Sca-1 is one of several markers that are expressed in urethral luminal cells residing in the urethral epithelium and proximal ducts. Results from studies on Sca-1+ proximal luminal cells can be extended to the entire proximal urethral cell population. It is important to note that Sca-1 is also expressed in proximal basal cells and stromal cells, so special care has to be taken to isolate luminal Sca-1 expressing cells for experimental analyses. The transcriptome of mouse urethral luminal cells shows high correlation with human urethral cells (Joseph et al. 2020). Experimental studies on Sca-1 in the mouse can be used to gain insights into human urethral luminal cells, especially where such studies would be unworkable in humans.
Recent studies in the mouse have demonstrated that Krt4+ luminal epithelia are resistant to castration (Sackmann Sala et al. 2017). Ly6d+ luminal cells, which are resistant to castration and co-express Krt4, were found to be enriched in the proximal prostate and likely represent the same androgen-independent luminal population (Barros-Silva et al. 2018). Using scRNA-seq of urethra and prostate, we demonstrated that Krt4+ cells are a luminal cell of the urethra and express most of the previously identified markers of facultative progenitors (Joseph et al. 2020). The low levels of androgen signaling and castration resistance of proximal luminal cells can be explained by their urethral origin.
Two other groups also identified a distinct luminal cell type in the mouse proximal prostate by scRNA-seq. Karthaus et al. identified a Krt8+/Sca-1+/Krt4+/Psca+/Trop2+ ‘proximal luminal’ cell type that displayed enhanced survival in facultative assays, but only regenerated locally after castration and subsequent androgen re-administration according to clonal analysis of a Krt8-driven lineage tracing model. Notably, Krt8 marks Krt4+ urethral luminal epithelia in the proximal prostate ducts as well as Nkx3.1+ prostate luminal epithelia distally (Karthaus et al. 2020). Guo et al. identified a Krt8+/Sca-1+/Krt4+/Psca+/Trop2+ luminal epithelial cell type in the proximal prostate ducts and also found them in distal prostate invaginations. Lineage tracing these luminal cells using either Psca or Krt4 promoters produced similar results to Karthaus et al. In essence, Krt4+ luminal epithelia are castration-resistant, but they do not regenerate Nkx3.1 + prostate luminal epithelia proximally. This is in direct contrast to facultative spheroid and tissue regeneration assays that show the ability of proximal Krt4+ urethral luminal epithelia to differentiate into Nkx3.1+ prostate luminal epithelia (as well as p63+ basal epithelia) (Guo et al. 2020). These results highlight the widespread discordance seen in facultative assays compared to in situ lineage tracing experiments. While there is evidence that a small population of distal Krt4+ luminal epithelia can give rise to Nkx3.1+ prostate epithelia in castration/regeneration experiments in vivo (Guo et al. 2020), the contribution to prostate regeneration is a fraction of the contribution of castration-resistant prostate luminal epithelia (Karthaus et al. 2020).
Facultative stem cell assays vs. lineage tracing
The discordant results from facultative stem cell assays and lineage tracing experiments beg an important question: do the traditional facultative assays that are used to identify progenitors such as spheroid formation, tissue regeneration, and label retention accurately reflect the lineage hierarchy in vivo? Facultative stem cell assays have been widely used as a surrogate for multipotency and suggest that a progenitor niche exists in the proximal region of the prostate, but the recent scRNA-seq data and lineage tracing studies detailed above suggest that surviving castration does not necessitate that a cell type is a progenitor for Nkx3.1+ prostate luminal epithelia during androgen-stimulated prostate regeneration.
Spheroid formation in vitro has been widely used to assess self-renewal and regenerative capacity. Basal cells have superior sphere formation capacity compared to luminal cells (Lawson et al. 2007), but, as noted above, rarely give rise to luminal cells in the adult (except under injury as discussed below). A Trop2+ basal cell population, localized in the proximal region of the mouse prostate, had enhanced sphere formation capacity in vitro compared to Trop2− basal cells (Goldstein et al. 2008). We now understand that Trop2+ basal epithelia are preferentially located in the prostatic urethra, but as noted above, the cells of the prostatic urethra appear to be lineage-restricted in the adult. In addition to Trop2, Sca-1 is expressed predominantly by epithelial cells of the urethra and proximal ducts (Kwon et al. 2016, Joseph et al. 2020). A Sca-1+ basal population also showed increased susceptibility to transformation and greater sphere formation capacity compared to prostate luminal cells (Lawson et al. 2010), but failed to differentiate into Nkx3.1+ prostate luminal cells (Kwon et al. 2020).
Tissue regeneration assays determine the progenitor capacity of a cell type by assessing its ability to reconstitute differentiated prostate epithelium. A cell population of interest is recombined with inductive fetal urogenital sinus mesenchyme (UGM) under the kidney capsule of immunocompromised mice. Although tissue recombination and grafting studies have provided important insights into stromal-epithelial interactions in the prostate (Chung and Cunha 1983, Kinbara et al. 1996), the inductive capacity of UGM is so strong that it can induce artificial prostate differentiation in other epithelia such as the bladder (Cunha et al. 1983). Basal cells from both mouse and human prostate can be transduced with oncogenes and initiate luminal prostate cancer in tissue regeneration assays (Goldstein et al. 2008, Goldstein et al. 2010, Lawson et al. 2010), but it has yet to be demonstrated that basal epithelia harbor mutations in human prostate cancer. Given the discordant results of facultative stem cell assays and tissue regeneration assays described above, the higher regenerative capacity of basal compared to luminal epithelia and proximal urethral luminal compared to distal prostate luminal cells in tissue regeneration and organoid assays could be due to the relative survival of these cell types.
Label retention experiments have been used in other organ systems like the skin to identify slow cycling progenitors (Braun and Watt 2004). Rodents are injected with a chemical, like BrdU or EdU, which is incorporated into cells that are synthesizing DNA. The intensity of BrdU or EdU staining is assessed in the cells after a wash-out period. Fast cycling cells will dilute out the label during the wash out period while slow cycling cells will retain the label. Progenitor cells that are actively dividing during embryonic prostate growth take up the label during the pulse period. Proliferating cells labeled in embryonic day 16.5 urogenital sinus retained the label into adulthood and were found in the proximal region of the prostate, suggesting that progenitors from early development were maintained in the adult proximal prostate (Ceder et al. 2017, Wei et al. 2019). However, we now understand that the Krt4+/Trop2+/Sca-1+/Ly6d+/Psca+ cells of the proximal prostate are extensions of the prostatic urethra (Joseph et al. 2020). In addition, luminal LRCs enriched in the mouse proximal prostate that were resistant to castration highly expressed Krt19, an intermediate cell marker, which is also highly expressed in the urethral luminal cell compartment (Zhang et al. 2018, Joseph et al. 2020).
There is increasing consensus that the proximal and distal epithelial compartments are maintained independently. In the lung, distinct progenitor populations exist for the proximal and distal regions (Danopoulos et al. 2018). This appears to be true for the prostate as well, with different pools of progenitors maintaining the proximal ducts and distal tips of the prostate. In summary, the androgen-independent, slow cycling luminal cells of the prostatic urethra extend into the proximal prostate, are resistant to castration, and form prostate glands in facultative assays, but there is little evidence to date that these cells contribute to prostate formation during development or adult regenerative growth (Figure 1, Table 1).
Figure 1.
Establishment of proximal and distal compartments during prostate budding. The proximal luminal compartment is comprised of urethral luminal cells marked by KRT8 and KRT4 expression. The distal luminal compartment is comprised of KRT8 and NKX3-1 expressing luminal cells. The proximal and distal compartments are maintained separately during budding, branching, canalization, specification and differentiation. During androgen mediated regenerative growth after castration, the proximal and distal luminal lineages are largely self-sustaining. Under injury or inflammation, basal cells have been shown to reconstitute the luminal compartment.
Table 1.
Published markers of proposed prostate stem and progenitor populations
| Reference | Proposed population | Luminal/Basal | Location | Species | Selected markers |
|---|---|---|---|---|---|
| (Wang et al. 2001) | Adult progenitor/stem cells | Embryonic like basal cells | Human | KRT5, KRT14, KRT8, KRT18, KRT19, TP63, GSTP1 | |
| (Burger et al. 2005) | Adult prostatic stem cells | Proximal | Mouse | Ly6a (Sca-1), Itga6 (CD49f), Bcl2 | |
| (Xin et al. 2005) | Androgen independent prostate regenerating cells | Proximal | Ly6a (Sca-1) | ||
| (Goto et al. 2006) | Androgen independent stem cells | Proximal, urethra | Mouse | Ly6a (Sca-1), Itga6 (CD49f), Bcl2 | |
| (Goldstein et al. 2008) | Adult prostatic stem cells | Basal | Proximal | Human, Mouse | TACSTD2 (Trop2), ITGA6 (CD49f) |
| (Leong et al. 2008) | Adult prostatic stem cells | Basal | Proximal | Mouse | Kit (GD117), Ly6a (Sca-1), Prom1 (CD133), Cd44 |
| (Wang et al. 2009) | Prostate luminal stem cell | Luminal | Mouse | Nkx3-1, Krt18, Ar | |
| (Goldstein et al. 2010) | Prostate cancer cell of origin | Basal | Human | TACSTD2 (Trop2), ITGA6 (CD49f) | |
| (Lawson et al. 2010) | Prostate stem cells | Basal | Mouse | Ly6a (Sca-1), Itga6 (CD49f) | |
| (Liu et al. 2011) | Androgen independent luminal cells that can regenerate prostate | Luminal | Mouse | PSA-creErt2 lineage cells | |
| (Shi et al. 2014) | Label retaining prostate progenitors | Basal | Proximal | Mouse | Kit (GD117), Ly6a (Sca-1), Prom1 (GD133), Cd44 |
| (Kwon et al. 2016) | Bipotent luminal cells | Luminal | Proximal | Mouse | Ly6a (Sca-1), Krt8 |
| (Liu et al. 2016) | Stem-like tubule initiating cells | Basal | Human | KRT13 | |
| (Liu et al. 2016) | Progenitor-like Inflammation- Associated Luminal Cells (CD38-IOW cells) | Luminal | Human | PIGR, PSCA, CD74, BCL2, SCGB1A1, SCGB3A1, LCN2 | |
| (Yoo et al. 2016) | Castration-resistant luminal progenitor cells | Luminal | Proximal | Mouse | BMI1 |
| (Ceder et al. 2017) | Label retaining stem-like cells | Luminal | Proximal | Mouse | Ly6a(Sca-1), Tacstd2(Trop2), Prom1 (CD133), Cd44, Kit (CD117), Krt7 |
| (Moad et al. 2017) | Multipotent Basal Stem Cells | Basal | Proximal | Human | DLK1, KLF4, KCNQ3 |
| (Sackmann Sala et al. 2017) | Castration-resistant prostate progenitor cell | Luminal | Proximal | Mouse | Psca, Reg3b, Cxcl17, Edn1, Krt4, Pglyrp1 |
| (Barros-Silva et al. 2018) | Castration-resistant prostate progenitor cell | Luminal | Proximal | Mouse | Tacstd2 (Trop2), Ly6a (Sca-1), Ly6d, Alcam (CD166) |
| (Zhang et al. 2018) | Label retaining prostate luminal cell | Luminal | Proximal | Mouse | Krt19, Ly6a (Sca-1) |
| (McCray et al. 2019) | Prostate epithelial stem cells | Human | KRT13, LY6D, PSCA | ||
| (Wang et al. 2020) | Multipotent prostate basal stem cells | Basal | Proximal | Mouse | Zeb1, Snai1, Prrx1 and Prrx2 |
| (Karthaus et al. 2020) | Androgen independent proximal luminal cells | Luminal | Proximal | Mouse | Psca, Ly6a (Sca-1), Krt4 |
| (Joseph et al. 2020) | Androgen independent urethral luminal cells | Luminal | Proximal | Human, Mouse | KRT13, SCGB1A1 Krt4, Ly6d, Psca, Tacstd2 (Trop2) |
| (Crowley et al. 2020) | Proximal luminal cells | Luminal | Proximal | Mouse | Ppplrib |
| (Mevel et al. 2020) | Proximal luminal castration resistant cells | Luminal | Proximal | Mouse | Runx1, Krt4, Tacstd2 (Trop2) |
Influence of aging and inflammation on progenitors in prostate disease
It is important to note that different progenitors could exist in development, regeneration, and injury. Bipotent basal progenitors are present until postnatal day 30 in the mouse after which unipotent progenitors that maintain basal and luminal lineages separately take over (Ousset et al. 2012, Wuidart et al. 2016). The normal lineage hierarchy during development and homeostasis can be disrupted during injury. In an experimental mouse model, luminal cell anoikis prompts the conversion of basal cells to luminal cells, showing that basal cells in injury differ from that during regeneration of the prostate (Toivanen et al. 2016). In addition, environmental conditions such as inflammation and high fat diet can drive basal to luminal differentiation (Kwon et al. 2014, Kwon et al. 2016). These data suggest that distinct cell populations can act as progenitors during normal development, injury, and disease.
BPH and prostate cancer are diseases of aging and inflammation (De Marzo et al. 2007, Nickel et al. 2008, Platz et al. 2012). Initiation of growth in the transition zone is thought take place in the fourth decade of life when the prevalence of BPH is only 8%. The prevalence increases to 50% in men between 51–60 years (Berry et al. 1984). Aging expands the proximal luminal compartment (Trop2+, Psca+, Krt4+) (Crowell et al. 2019), and these cells are resistant to 5 alpha reductase inhibitor treatment (Joseph et al. 2020). However, it is still unclear whether urethral luminal epithelia are acting as progenitors for prostate growth or simply expanding in response to inflammation.
BPH is associated with aging and a pro-inflammatory state (Berry et al. 1984, Untergasser et al. 2005, Nickel et al. 2008). Club and hillock cells of the prostatic urethra extend into the proximal region of the prostate ducts and express immunomodulatory genes, including anti-bacterial proteins and chemokines, in the normal adult (Henry et al. 2018), and these genes are further increased in BPH (Joseph et al. 2020). Based on their location in the apical and supra-basal layers of the urethral and ductal epithelium, these cells could act as the first line of defense against bacterial infection and other foreign agents, similar to the protective role of club and hillock epithelia in the trachea and proximal lung.
Inflammation in the context of the prostate has been widely studied (Chughtai et al. 2011). Prostatitis due to bacterial or non-bacterial causes is a common health issue in men. Prostate secretions are missing until puberty, which could make the prostate susceptible to autoimmune attacks (Kramer et al. 2007). Ascending bacteria from the urethra also makes the prostate prone to infection. Like the lung, the urethra is regularly exposed to environmental antigens that create an inflammatory microenvironment. Recruitment of immune cells and bone marrow derived mesenchymal stem cells could create an environment conducive to the generation of inductive stromal nodules (Brennen and Isaacs 2018). Inductive stroma is suggested to play a role in the development of BPH by triggering new epithelial growth around the transition zone.
Bacterial instillation in mice produces a rapid inflammatory response resulting in reversible stromal cell activation and collagen deposition (Wong et al. 2014, Wong et al. 2015). Chronic bacterial inflammation can induce the formation of neoplasia in mouse prostate and a reduction of androgen receptor expression (Elkahwaji et al. 2009). Androgen receptor expression is anti-inflammatory (Zhang et al. 2016) and both hypogonadism and anti-androgen treatment are associated with increased inflammation (Sorrentino et al. 2011, Vignozzi et al. 2012).
Inflammation alters the normal lineage hierarchy in the prostate by accelerating the conversion of basal to luminal cells (Kwon et al. 2014). Studies in human prostate tissue have shown that a CD38-low (CD38lo) luminal cell population expands in response to inflammation. CD38lo cells are concentrated around regions of inflammatory infiltrate and show increased NF-KB signaling activity. CD38lo cells around sites of inflammation have reduced androgen signaling and lower androgen receptor expression. Based on gene expression comparison, CD38lo cells express several characteristics of urethral luminal cells including expression of SCGB1A1, SCGB3A1, LCN2, PIGR, PSCA and KRT4 (Liu et al. 2016, Henry et al. 2018). Although club and hillock epithelial cells are concentrated in the urethra and proximal ducts, small patches of these cells can be observed in the distal regions of normal prostates (Henry et al. 2018). Considering that club and hillock cells are also present in developing prostate buds (Joseph et al. 2020), it is possible that patches of these cells seen in the adult prostate are remnants from embryonic growth. Androgen-independent club and hillock cells survive the castration-like effect of finasteride treatment in BPH (Joseph et al. 2020) or androgen deprivation therapy in prostate cancer (Karthaus et al. 2020). Inflammation in the prostate could trigger growth and expansion of the club and hillock cells from growth signals like IL-1 released by infiltrating macrophages.
Pro-inflammatory signaling from the expanding club and hillock cells could further exacerbate the immune response. More evidence is needed to determine whether urethral luminal club and hillock epithelia are progenitors for Nkx3.1+ prostate luminal epithelia during BPH or prostate cancer using in situ lineage tracing techniques in human tissue (Moad et al. 2017).
Identity and function of the stroma in prostate development and disease
Hormonal action and epithelial-stromal signaling direct the development and growth of the prostate. In the bid to uncover the epithelial lineage hierarchy, the contribution of the stroma is often overlooked (Cunha 1976). The urogenital sinus mesenchyme (UGM) is a specialized mesenchymal cell layer surrounding the urogenital sinus epithelium during embryonic development. Reciprocal signaling between the epithelium and mesenchyme induces the outgrowth of prostate buds. The UGM is extremely inductive and is used for recombination experiments with epithelial cells from different sources. Androgen signaling from the UGM is sufficient to induce prostate outgrowth and differentiation even when the epithelium is deficient for androgen receptor expression (Donjacour and Cunha 1993). Considering the critical role of UGM in prostate development, the prostate stroma is expected to play an important role in the development of BPH. McNeal’s embryonic reawakening theory hypothesizes that a reactivation of early growth signaling activates BPH (McNeal 1978). Direct implantation of fetal UGS tissue into the ventral prostate of immunocompromised mice results in prostate overgrowth, indicating that the adult prostate can be induced to proliferate in response to embryonic growth signals (Chung et al. 1984). This lends support to the embryonic reawakening theory. Yet, the stroma remains relatively understudied compared to the prostate epithelium.
Reciprocal stroma-epithelial interactions are involved in prostate development and homeostasis. During development, growth signaling from the stroma mediated by andromedins induce prostate bud outgrowths from the urogenital sinus epithelium. Sonic hedgehog signaling (Shh) from the epithelium signals to the stromal layer by inducing Gli1 expression. In the mouse, a Gli1+ stromal cell sub-type located directly adjacent the prostate epithelium exhibits long-term self-renewal capacity over multiple rounds of castration and regeneration (Peng et al. 2013). Recent studies have used advanced techniques to assess the stromal composition of the prostate. The prostate stroma was found to contain two specialized fibroblast populations (Kwon et al. 2019). Of particular interest is the proximal fibroblast population enriched in Wnt signaling which is present around the proximal prostate ducts in mice and human. This specialized stromal cell type contributes to a proximal niche that suppresses growth and maintains the quiescence of proximal progenitor-like cells (Kwon et al. 2019, Wei et al. 2019). Stromal TGF-β signaling activity is also concentrated in the proximal region and suppresses apoptosis in quiescent proximal cells (Salm et al. 2005).
McNeal proposed that the distinct composition of the peri-urethral stroma was responsible for the inductive epithelial-stromal interactions leading to prostate hyperplasia (McNeal 1981). In humans, the urogenital sinus mesenchyme is a condensation of immature mesenchymal cells up to week 17 of gestation, when most of the budding has occurred. The mesenchyme then progressively displays fibroblastic, fibromuscular and smooth muscle characteristics as development progresses. The natural progression of stromal nodules in BPH patients resembles that of the developing urogenital sinus mesenchyme. Different compositions of stromal nodules are observed, immature mesenchymal, fibroblastic, fibromuscular, and smooth-muscular, mirroring the developmental trajectory of the embryonic mesenchyme (Bierhoff et al. 1997). Stromal dysregulation with aging or inflammation could trigger changes in the proximal stromal niche leading to prostate hyperplasia.
Identifying the human prostate progenitor
A significant impediment to progress in the identification of the prostate progenitor has been the lack of studies using human tissue. Rodent models to study prostate growth and development were implemented to work around the hurdles posed by human tissue research. The early development of the rodent prostate is remarkably similar to the human (Timms et al. 1994). However, significant differences emerge when considering the adult prostate. The rodent prostate has a lobular architecture, being comprised of paired anterior, ventral and dorso-lateral lobes. Lobes are not evident in the human prostate, which is enclosed by a fibromuscular capsule. Dense stroma surrounds human prostate glands while rodent prostate glands are surrounded by thin stroma.
Comparative analysis of human and mouse prostate cell types reveal a mouse equivalent of human hillock cells that resides in the luminal layer of the urethra and proximal prostate ducts (Crowley et al. 2020, Guo et al. 2020, Joseph et al. 2020, Karthaus et al. 2020). These cells express several progenitor markers and display lower androgen signaling activity and dense heterochromatin, suggesting a quiescent state that has been corroborated by label retention studies. Lineage tracing in mice has revealed several key insights into the hierarchy of cells in the prostate and prostatic urethra. Performing similar studies in the human prostate is challenging. A recent study used mitochondrial DNA mutations to follow clonal expansion of cells in the prostate. This technique revealed that proximal multipotent basal cells give rise to prostate luminal cells in the distal prostate (Moad et al. 2017). Understanding whether these were urethral or prostate basal cells will require the discovery of anchor genes that can distinguish the two types of basal epithelia.
A critical question in human prostate research is the identification of androgen independent progenitors that play a role in prostate disease. Club and hillock cells can survive androgen deprivation therapies (Karthaus et al. 2020) and the castration-like effect of finasteride treatment (Joseph et al. 2020). Further research is required to see if these cells directly serve as prostate epithelial progenitors in BPH and prostate cancer or whether they act as modulators of the environment.
Some evidence suggests that hillock cells marked by KRT13 could play a role in prostate cancer. Benign human prostate tubule-initiating cells display several characteristics of prostate cancer including androgen independence. KRT13, the hillock cell marker, is highly enriched in tubule initiating cells and fetal prostate cells (Liu et al. 2016). Prostate organoids derived from primary human prostate cells express high levels of KRT13, a hillock cell marker, (Plasschaert et al. 2018) as well as LY6D and PSCA (McCray et al. 2019). Club and hillock markers are enriched in 3D spheroids derived from primary prostate cells (McCray et al. 2019). Further research is required to determine whether the enrichment of these cells in organoids might be attributed to their progenitor capacity, androgen independence or their intrinsic resistance to apoptosis.
Future directions
The identification of new epithelial and stromal cell types in the prostate demands a reworking of existing models of lineage hierarchy and niche composition. The advent of new technologies has accelerated research in prostate biology. Technologies like single cell transcriptomics have enabled the unbiased comparison of cell types across different models based on thousands of genes and removed the reliance on a handful of markers. This has led to the realization that the cell types described as castration-resistant luminal progenitors in multiple studies were indeed the same cell type, found in the luminal layer of the urethra and proximal prostate ducts. Although this cell type shows androgen independence and high regenerative potential ex vivo, recent lineage tracing studies have shown that these cells do not contribute significantly to the regeneration of secretory prostate luminal cells during castration-regeneration studies. Instead, the proximal luminal compartment is maintained independently of the distal secretory luminal compartment.
Urethral luminal cells are resistant to castration and expand in mouse models of prostate cancer. It is important to assess the contribution of this cell type in the human prostate. Rodent prostate research is replete with genetic tools that are missing from human prostate research. Although the rodent and human prostates have several similarities, we cannot conclude that the lineage hierarchy is the same. To bridge the gap with our knowledge on the rodent prostate, continued advances in human prostate research are required.
Highlights.
Review of human and mouse prostate development
Review of latest developments in cell identities of the prostate and urethra
Comparison of ex vivo culture experiments vs. in vivo lineage tracing experiments
Acknowledgments
Financial support came from AUA Research Scholar Award 659333 (D.B.J.), R01 DK115477 (D.W.S.); U54DK104310 (D.W.S. and C.M.V.), U01DK110807 and R01DK099328 (C.M.V.).
Footnotes
Disclosure / Conflict of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Uncategorized References
- Barros-Silva JD, Linn DE, Steiner I, Guo G, Ali A, Pakula H, Ashton G, Peset I, Brown M, Clarke NW, Bronson RT, Yuan GC, Orkin SH, Li Z, Baena E, 2018. “Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer.” Cell Rep 25, 3504–3518 e3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SJ, Coffey DS, Walsh PC, Ewing LL, 1984. “The development of human benign prostatic hyperplasia with age.” J Urol 132, 474–479. [DOI] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C, Shen MM, 1999. “Roles for Nkx3.1 in prostate development and cancer.” Genes Dev 13, 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff E, Walljasper U, Hofmann D, Vogel J, Wernert N, Pfeifer U, 1997. “Morphological analogies of fetal prostate stroma and stromal nodules in BPH.” Prostate 31, 234–240. [DOI] [PubMed] [Google Scholar]
- Braun KM, Watt FM, 2004. “Epidermal label-retaining cells: background and recent applications.” J Investig Dermatol Symp Proc 9, 196–201. [DOI] [PubMed] [Google Scholar]
- Brennen WN, Isaacs JT, 2018. “Mesenchymal stem cells and the embryonic reawakening theory of BPH.” Nat Rev Urol 15, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, Wilson EL, 2005. “Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue.” Proc Natl Acad Sci U S A 102, 7180–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceder JA, Aalders TW, Schalken JA, 2017. “Label retention and stem cell marker expression in the developing and adult prostate identifies basal and luminal epithelial stem cell subpopulations.” Stem Cell Res Ther 8, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Matsumoto K, Brockway BL, Rackley CR, Liang J, Lee JH, Jiang D, Noble PW, Randell SH, Kim CF, Stripp BR, 2012. “Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury.” Stem Cells 30, 1948–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N, Zhang B, Zhang L, Ittmann M, Xin L, 2012. “Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation.” Cancer Cell 21,253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai B, Lee R, Te A, Kaplan S, 2011. “Role of inflammation in benign prostatic hyperplasia.” Rev Urol 13, 147–150. [PMC free article] [PubMed] [Google Scholar]
- Chung LW, Cunha GR, 1983. “Stromal-epithelial interactions: II. Regulation of prostatic growth by embryonic urogenital sinus mesenchyme.” Prostate 4, 503–511. [DOI] [PubMed] [Google Scholar]
- Chung LW, Matsuura J, Runner MN, 1984. “Tissue interactions and prostatic growth. I. Induction of adult mouse prostatic hyperplasia by fetal urogenital sinus implants.” Biol Reprod 31, 155–163. [DOI] [PubMed] [Google Scholar]
- Coffey DS, Shimazaki J, Williams-Ashman HG, 1968. “Polymerization of deoxyribonucleotides in relation to androgen-induced prostatic growth.” Arch Biochem Biophys 124, 184–198. [DOI] [PubMed] [Google Scholar]
- Crowell PD, Fox JJ, Hashimoto T, Diaz JA, Navarro HI, Henry GH, Feldmar BA, Lowe MG, Garcia AJ, Wu YE, Sajed DP, Strand DW, Goldstein AS, 2019. “Expansion of Luminal Progenitor Cells in the Aging Mouse and Human Prostate.” Cell Rep 28, 1499–1510 e1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, Li W, Hibshoosh H, Loda M, Rabadan R, Shen MM, 2020. “A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors.” Elife, DOI: 10.7554/eLife.59465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha G, Lung B, 1978. “The possible influences of temporal factors in androgenic responsiveness of urogenital tissue recombinants from wild-type and androgen-insensitive (Tfm) mice.” J. Exp. Zool. 205, 181–194. [DOI] [PubMed] [Google Scholar]
- Cunha GR, 1976. “Epithelial-stromal interactions in development of the urogenital tract.” Int Rev Cytol 47, 137–194. [DOI] [PubMed] [Google Scholar]
- Cunha GR, 1984. “Androgenic effects upon prostatic epithelium are mediated via trophic influences from stroma.” Prog Clin Biol Res 145, 81–102. [PubMed] [Google Scholar]
- Cunha GR, Sekkingstad M, Meloy BA, 1983. “Heterospecific induction of prostatic development in tissue recombinants prepared with mouse, rat, rabbit and human tissues.” Differentiation 24, 174–180. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Vezina CM, Isaacson D, Ricke WA, Timms BG, Cao M, Franco O, Baskin LS, 2018. “Development of the human prostate.” Differentiation 103, 24–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danopoulos S, Alonso I, Thornton ME, Grubbs BH, Bellusci S, Warburton D, Al Alam D, 2018. “Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9.” Am J Physiol Lung Cell Mol Physiol 314, L144–L149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG, 2007. “Inflammation in prostate carcinogenesis.” Nat Rev Cancer 7, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donjacour AA, Cunha GR, 1993. “Assessment of Prostatic Protein Secretion in Tissue Recombinants Made of Urogenital Sinus Mesenchyme and Urothelium from Normal or Androgen-Insensitive Mice.” Endocrinology 132, 2342–2350. [DOI] [PubMed] [Google Scholar]
- Elkahwaji JE, Hauke RJ, Brawner CM, 2009. “Chronic bacterial inflammation induces prostatic intraepithelial neoplasia in mouse prostate.” Br J Cancer 101, 1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English HF, Drago JR, Santen RJ, 1985. “Cellular response to androgen depletion and repletion in the rat ventral prostate: autoradiography and morphometric analysis.” Prostate 7, 41–51. [DOI] [PubMed] [Google Scholar]
- English HF, Kyprianou N, Isaacs JT, 1989. “Relationship between DNA fragmentation and apoptosis in the programmed cell death in the rat prostate following castration.” Prostate 15, 233–250. [DOI] [PubMed] [Google Scholar]
- English HF, Santen RJ, Isaacs JT, 1987. “Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement.” Prostate 11, 229–242. [DOI] [PubMed] [Google Scholar]
- Gandhi D, Molotkov A, Batourina E, Schneider K, Dan H, Reiley M, Laufer E, Metzger D, Liang F, Liao Y, Sun TT, Aronow B, Rosen R, Mauney J, Adam R, Rosselot C, Van Batavia J, McMahon A, McMahon J, Guo JJ, Mendelsohn C, 2013. “Retinoid signaling in progenitors controls specification and regeneration of the urothelium.” Dev Cell 26, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON, 2010. “Identification of a cell of origin for human prostate cancer.” Science 329, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON, 2008. “Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics.” Proc Natl Acad Sci U S A 105, 20882–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Salm SN, Coetzee S, Xiong X, Burger PE, Shapiro E, Lepor H, Moscatelli D, Wilson EL, 2006. “Proximal prostatic stem cells are programmed to regenerate a proximal-distal ductal axis.” Stem Cells 24, 1859–1868. [DOI] [PubMed] [Google Scholar]
- Guo W, Li L, He J, Liu Z, Han M, Li F, Xia X, Zhang X, Zhu Y, Wei Y, Li Y, Aji R, Dai H, Wei H, Li C, Chen Y, Chen L, Gao D, 2020. “Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips.” Nat Genet 52, 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bischoff A, Pena A, Runck LA, Guasch G, 2014. “The great divide: septation and malformation of the cloaca, and its implications for surgeons.” Pediatr Surg Int 30, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW, 2018. “A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra.” Cell Rep 25, 3530–3542 e3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins C, Hodges CV, 2002. “Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941.” J Urol 167, 948–951; discussion 952. [PubMed] [Google Scholar]
- Johnson FP, 1920. “The later development of the urethra in the male.” J. Urol. 4, 447–501. [Google Scholar]
- Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, Abler LL, Sandhu SK, Cadena MT, Malladi VS, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Roehrborn CG, Baker LA, Vezina CM, Strand DW, 2020. “Urethral luminal epithelia are castration-insensitive cells of the proximal prostate.” Prostate 80, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, Carver B, Gopalan A, Abida W, Laudone V, Biton M, Chaudhary O, Xu T, Masilionis I, Manova K, Mazutis L, Pe’er D, Regev A, Sawyers CL, 2020. “Regenerative potential of prostate luminal cells revealed by single-cell analysis.” Science 368, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinbara H, Cunha GR, Boutin E, Hayashi N, Kawamura J, 1996. “Evidence of stem cells in the adult prostatic epithelium based upon responsiveness to mesenchymal inductors.” Prostate 29, 107–116. [DOI] [PubMed] [Google Scholar]
- Kramer G, Mitteregger D, Marberger M, 2007. “Is benign prostatic hyperplasia (BPH) an immune inflammatory disease?” Eur Urol 51, 1202–1216. [DOI] [PubMed] [Google Scholar]
- Kwon OJ, Choi JM, Zhang L, Jia D, Li Z, Zhang Y, Jung SY, Creighton CJ, Xin L, 2020. “The Sca-1(+) and Sca-1(−) mouse prostatic luminal cell lineages are independently sustained.” Stem Cells 38, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OJ, Zhang B, Zhang L, Xin L, 2016. “High fat diet promotes prostatic basal-to-luminal differentiation and accelerates initiation of prostate epithelial hyperplasia originated from basal cells.” Stem Cell Res 16, 682–691. [DOI] [PubMed] [Google Scholar]
- Kwon OJ, Zhang L, Ittmann MM, Xin L, 2014. “Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin.” Proc Natl Acad Sci U S A 111, E592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OJ, Zhang L, Xin L, 2016. “Stem Cell Antigen-1 Identifies a Distinct Androgen-Independent Murine Prostatic Luminal Cell Lineage with Bipotent Potential.” Stem Cells 34, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OJ, Zhang Y, Li Y, Wei X, Zhang L, Chen R, Creighton CJ, Xin L, 2019. “Functional Heterogeneity of Mouse Prostate Stromal Cells Revealed by Single-Cell RNA-Seq.” iScience 13, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON, 2007. “Isolation and functional characterization of murine prostate stem cells.” Proc Natl Acad Sci U S A 104, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON, 2010. “Basal epithelial stem cells are efficient targets for prostate cancer initiation.” Proc Natl Acad Sci U S A 107, 2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong KG, Wang BE, Johnson L, Gao WQ, 2008. “Generation of a prostate from a single adult stem cell.” Nature 456, 804–808. [DOI] [PubMed] [Google Scholar]
- Leuprolide Study G, 1984. “Leuprolide versus diethylstilbestrol for metastatic prostate cancer.” N Engl J Med 311, 1281–1286. [DOI] [PubMed] [Google Scholar]
- Liu J, Pascal LE, Isharwal S, Metzger D, Ramos Garcia R, Pilch J, Kasper S, Williams K, Basse PH, Nelson JB, Chambon P, Wang Z, 2011. “Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate.” Mol Endocrinol 25, 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cadaneanu RM, Zhang B, Huo L, Lai K, Li X, Galet C, Grogan TR, Elashoff D, Freedland SJ, Rettig M, Aronson WJ, Knudsen BS, Lewis MS, Garraway IP, 2016. “Keratin 13 Is Enriched in Prostate Tubule-Initiating Cells and May Identify Primary Prostate Tumors that Metastasize to the Bone.” PLoS One 11, e0163232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Grogan TR, Hieronymus H, Hashimoto T, Mottahedeh J, Cheng D, Zhang L, Huang K, Stoyanova T, Park JW, Shkhyan RO, Nowroozizadeh B, Rettig MB, Sawyers CL, Elashoff D, Horvath S, Huang J, Witte ON, Goldstein AS, 2016. “Low CD38 Identifies Progenitor-like Inflammation-Associated Luminal Cells that Can Initiate Human Prostate Cancer and Predict Poor Outcome.” Cell Rep 17, 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowsley OS, 1912. “The development of the human prostate gland with reference to the development of other structures at the neck of the urinary bladder.” Am. J. Anat. 13, 299–349. [Google Scholar]
- McCray T, Moline D, Baumann B, Vander Griend DJ, Nonn L, 2019. “Single-cell RNA-Seq analysis identifies a putative epithelial stem cell population in human primary prostate cells in monolayer and organoid culture conditions.” Am J Clin Exp Urol 7, 123–138. [PMC free article] [PubMed] [Google Scholar]
- McNeal JE, 1978. “Origin and evolution of benign prostatic enlargement.” Invest Urol 15, 340–345. [PubMed] [Google Scholar]
- McNeal JE, 1981. “The zonal anatomy of the prostate.” Prostate 2, 35–49. [DOI] [PubMed] [Google Scholar]
- McNeal JE, 1983. Relationship of the origin of benign prostatic hypertrophy to prostatic structure of man and other mammals. Benign Prostatic Hypertrophy. Hinman FJ. New York, Springer-Verlag, 152–166. [Google Scholar]
- Mevel R, Steiner I, Mason S, Galbraith LC, Patel R, Fadlullah MZ, Ahmad I, Leung HY, Oliveira P, Blyth K, Baena E, Lacaud G, 2020 “RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development.” Elife, DOI: 10.7554/eLife.60225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moad M, Hannezo E, Buczacki SJ, Wilson L, El-Sherif A, Sims D, Pickard R, Wright NA, Williamson SC, Turnbull DM, Taylor RW, Greaves L, Robson CN, Simons BD, Heer R, 2017. “Multipotent Basal Stem Cells, Maintained in Localized Proximal Niches, Support Directed Long-Ranging Epithelial Flows in Human Prostates.” Cell Rep 20, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, Rogel N, Burgin G, Tsankov AM, Waghray A, Slyper M, Waldman J, Nguyen L, Dionne D, Rozenblatt-Rosen O, Tata PR, Mou H, Shivaraju M, Bihler H, Mense M, Tearney GJ, Rowe SM, Engelhardt JF, Regev A, Rajagopal J, 2018. “A revised airway epithelial hierarchy includes CFTR-expressing ionocytes.” Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel JC, Roehrborn CG, O’Leary MP, Bostwick DG, Somerville MC, Rittmaster RS, 2008. “The relationship between prostate inflammation and lower urinary tract symptoms: examination of baseline data from the REDUCE trial.” Eur Urol 54, 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousset M, Van Keymeulen A, Bouvencourt G, Sharma N, Achouri Y, Simons BD, Blanpain C, 2012. “Multipotent and unipotent progenitors contribute to prostate postnatal development.” Nat Cell Biol 14, 1131–1138. [DOI] [PubMed] [Google Scholar]
- Peng YC, Levine CM, Zahid S, Wilson EL, Joyner AL, 2013. “Sonic hedgehog signals to multiple prostate stromal stem cells that replenish distinct stromal subtypes during regeneration.” Proc Natl Acad Sci U S A 110, 20611–20616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, Jaffe AB, 2018. “A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte.” Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platz EA, Joshu CE, Mondul AM, Peskoe SB, Willett WC, Giovannucci E, 2012. “Incidence and progression of lower urinary tract symptoms in a large prospective cohort of United States men.” J Urol 188, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann Sala L, Boutillon F, Menara G, De Goyon-Pelard A, Leprevost M, Codzamanian J, Lister N, Pencik J, Clark A, Cagnard N, Bole-Feysot C, Moriggl R, Risbridger GP, Taylor RA, Kenner L, Guidotti JE, Goffin V, 2017. “A rare castration-resistant progenitor cell population is highly enriched in Pten-null prostate tumours.” J Pathol 243, 51–64. [DOI] [PubMed] [Google Scholar]
- Salm SN, Burger PE, Coetzee S, Goto K, Moscatelli D, Wilson EL, 2005. “TGF-{beta} maintains dormancy of prostatic stem cells in the proximal region of ducts.” J Cell Biol 170, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwig I, Spitznagel B, Vazquez-Armendariz AI, Khalooghi K, Guenther S, Herold S, Szibor M, Braun T, 2019. “Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo.” EMBO J, DOI: 10.15252/embj.2019102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Gipp J, Dries M, Bushman W, 2014. “Prostate progenitor cells proliferate in response to castration.” Stem Cell Res 13, 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoretti S, Pires MM, Lindauer M, Horner JW, Grisanzio C, Dhar S, Majumder P, McKeon F, Kantoff PW, Sellers WR, Loda M, 2005. “p63 regulates commitment to the prostate cell lineage.” Proc Natl Acad Sci U S A 102, 11355–11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M, 2000. “p63 is a prostate basal cell marker and is required for prostate development.” Am J Pathol 157, 1769–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino C, Musiani P, Pompa P, Cipollone G, Di Carlo E, 2011. “Androgen deprivation boosts prostatic infiltration of cytotoxic and regulatory T lymphocytes and has no effect on disease-free survival in prostate cancer patients.” Clin Cancer Res 17, 1571–1581. [DOI] [PubMed] [Google Scholar]
- Strand DW, Costa DN, Francis F, Ricke WA, Roehrborn CG, 2017. “Targeting phenotypic heterogeneity in benign prostatic hyperplasia.” Differentiation 96, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura Y, Cunha GR, Donjacour AA, Bigsby RM, Brody JR, 1986. “Whole-mount autoradiography study of DNA synthetic activity during postnatal development and androgen-induced regeneration in the mouse prostate.” Biol Reprod 34, 985–995. [DOI] [PubMed] [Google Scholar]
- Tika E, Ousset M, Dannau A, Blanpain C, 2019. “Spatiotemporal regulation of multipotency during prostate development.” Development, DOI: 10.1242/dev.180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms BG, 2008. “Prostate development: a historical perspective.” Differentiation 76, 565–577. [DOI] [PubMed] [Google Scholar]
- Timms BG, Mohs TJ, Didio LJ, 1994. “Ductal budding and branching patterns in the developing prostate.” J Urol 151, 1427–1432. [DOI] [PubMed] [Google Scholar]
- Toivanen R, Mohan A, Shen MM, 2016. “Basal Progenitors Contribute to Repair of the Prostate Epithelium Following Induced Luminal Anoikis.” Stem Cell Reports 6, 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura A, Koikawa Y, Salm S, Takao T, Coetzee S, Moscatelli D, Shapiro E, Lepor H, Sun TT, Wilson EL, 2002. “Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis.” J Cell Biol 157, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser G, Gander R, Lilg C, Lepperdinger G, Plas E, Berger P, 2005. “Profiling molecular targets of TGF-beta1 in prostate fibroblast-to-myofibroblast transdifferentiation.” Mech Ageing Dev 126, 59–69. [DOI] [PubMed] [Google Scholar]
- Vignozzi L, Morelli A, Sarchielli E, Comeglio P, Filippi S, Cellai I, Maneschi E, Serni S, Gacci M, Carini M, Piccinni MP, Saad F, Adorini L, Vannelli GB, Maggi M, 2012. “Testosterone protects from metabolic syndrome-associated prostate inflammation: an experimental study in rabbit.” J Endocrinol 212, 71–84. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM, 2009. “A luminal epithelial stem cell that is a cell of origin for prostate cancer.” Nature 461,495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, Fan L, Dong B, Xue W, Wai Chua C, Wu D, Gao WQ, He Zhu H, 2020. “Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate.” Nat Commun 11, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hayward S, Cao M, Thayer K, Cunha G, 2001. “Cell differentiation lineage in the prostate.” Differentiation 68, 270–279. [DOI] [PubMed] [Google Scholar]
- Wei X, Zhang L, Zhou Z, Kwon OJ, Zhang Y, Nguyen H, Dumpit R, True L, Nelson P, Dong B, Xue W, Birchmeier W, Taketo MM, Xu F, Creighton CJ, Ittmann MM, Xin L, 2019. “Spatially Restricted Stromal Wnt Signaling Restrains Prostate Epithelial Progenitor Growth through Direct and Indirect Mechanisms.” Cell Stem Cell 24, 753–768 e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, Hutson PR, Bushman W, 2014. “Prostatic inflammation induces fibrosis in a mouse model of chronic bacterial infection.” PLoS One 9, e100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, Hutson PR, Bushman W, 2015. “Resolution of chronic bacterial-induced prostatic inflammation reverses established fibrosis.” Prostate 75, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, Blanpain C, 2016. “Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells.” Genes Dev 30, 1261–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Lawson DA, Witte ON, 2005. “The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis.” Proc Natl Acad Sci U S A 102, 6942–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YA, Roh M, Naseem AF, Lysy B, Desouki MM, Unno K, Abdulkadir SA, 2016. “Bmi1 marks distinct castration-resistant luminal progenitor cells competent for prostate regeneration and tumour initiation.” Nat Commun, 12943 DOI: 10.1038/ncomms12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kwon OJ, Henry G, Malewska A, Wei X, Zhang L, Brinkley W, Zhang Y, Castro PD, Titus M, Chen R, Sayeeduddin M, Raj GV, Mauck R, Roehrborn C, Creighton CJ, Strand DW, Ittmann MM, Xin L, 2016. “Non-Cell-Autonomous Regulation of Prostate Epithelial Homeostasis by Androgen Receptor.” Mol Cell 63, 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jeter C, Gong S, Tracz A, Lu Y, Shen J, Tang DG, 2018. “Histone 2B-GFP Label-Retaining Prostate Luminal Cells Possess Progenitor Cell Properties and Are Intrinsically Resistant to Castration.” Stem Cell Reports 10, 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo WL, Shenoy SA, Li S, O’Beirne SL, Strulovici-Barel Y, Leopold PL, Wang G, Staudt MR, Walters MS, Mason C, Kaner RJ, Mezey JG, Crystal RG, 2018. “Ontogeny and Biology of Human Small Airway Epithelial Club Cells.” Am J Respir Crit Care Med 198, 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]