Abstract
The three domains of life employ various strategies to organize their genomes. Archaea utilize features similar to those found in both eukaryotic and bacterial chromatin to organize their DNA. In this review, we discuss the current state of research regarding the structure-function relationships of several archaeal chromatin proteins (histones, Alba, Cren7, and Sul7d). We address individual structures as well as inferred models for higher-order chromatin formation. Each protein introduces a unique phenotype to chromatin organization, and these structures are put into the context of in vivo and in vitro data. We close by discussing the present gaps in knowledge that are preventing further studies of the organization of archaeal chromatin, on both the organismal and domain level.
2. Introduction
All living things utilize scaffolding proteins to package their DNA as chromatin in order to protect their genetic code and regulate transcription. In eukaryotes, between 10 and 10,000 mega base pairs (Mbp) of DNA are packaged into a nucleus with a diameter of ~6 μm, with the metaphase chromosomes as the ultimate condensed form of the genome. The fundamental unit for this extreme level of compaction is the nucleosome, which consists of a histone octamer (composed of two copies each of histones H2A, H2B, H3, and H4) wrapping ~147 bp of DNA. Multiple nucleosomes are connected by linker DNA to form the classical “beads-on-a-string” arrangement.1,2 Numerous chromatin architectural proteins, ATP-dependent chromatin remodellers, histone chaperones, and pioneer transcription factors work in conjunction with histones to manipulate the accessibility of nucleosomes and eukaryotic chromatin, both locally and globally, during transcription and replication.3–5 In contrast, archaea and bacteria, which do not have a nucleus, store only 0.1–10 Mbp of DNA in the nucleoid, a compacted circular plasmid in the cytosol. Bacteria utilize a plethora of small cationic proteins, called nucleoid associated proteins (or NAPs),6 to scaffold and protect their chromatin. Archaea, on the other hand, appear to combine the two strategies, and the use of NAPs and/or histones varies significantly within the domain and between species.
A unique feature of many archaea is their ability to grow in extreme conditions. Common extremophilic organisms include thermophiles (which grow at high temperatures, above 60°C), halophiles (which grow at high salt conditions, even above 2 M), and acidophiles (which are able to grow at acidic pH). Each of these environments brings unique challenges to the task of organizing and structuring DNA. In thermophiles, proteins must allow for DNA flexibility, so as to permit transcription and replication, but they must simultaneously protect DNA from melting and heat-denaturation. In halophiles, elevated salt levels within the cell can cause denaturation of scaffolding proteins without proper sequence adaptations.7 Similarly, extremely low pH environments can induce charge-reversal in DNA molecules and cause the DNA double helix to melt, as well as dramatically alter the ability of cations to condense DNA.8 Even under these extreme conditions, archaea have found ways to maintain structured chromatin, highlighting the importance of ordering their genomes with the aid of proteins. It stands to reason that the proteins from each group of archaea may have separately evolved to handle specific environments, but no direct correlation or pattern between specific architectural protein sequences and type of extreme environment has been determined. This indicates that the choice of scaffolding protein may be somewhat stochastic and highlights the adaptability of protein sequences to accommodate unique conditions.
Here, we focus on four key proteins that are utilized by various archaea to organize their genomes: histones, Alba, Cren7, and Sul7d. We discuss available structures of the proteins and their unique interactions with DNA. We also discuss how these structures can be used to infer models of higher-order chromatin organization in archaea, as well as how these models can be interpreted in light of current in vivo and in vitro data. We close by discussing several key areas where advances are needed to further our understanding of archaeal chromatin structure and function.
3. Chromatin proteins
3.1. Histones
Histones are perhaps the most widely studied chromatin architectural protein. They are defined by their ability to bind DNA through their core fold motif of three α-helices connected by short flexible loops (α1-L1-α2-L2-α3; Figure 2A, C). For several decades, histones were thought to be exclusive to eukaryotes, where histones form an octamer to bind ~147 bp of DNA in the “beads on a string” arrangement.1,2 However, the Reeve lab shifted this paradigm in 1990 when they discovered the first archaeal histone in Methanothermus fervidus.11 In a follow up study, they showed that M. fervidus actually encodes two histone genes, denoted as “HMfA” and “HMfB”, and they further noted that these histones can form both homo- and heterodimers,12 in contrast to the strictly heterodimeric arrangements found in eukaryotic nucleosomes. Furthermore, eukaryotic histone sequences have additional features, such as disordered and cationic N-terminal tail extensions. They also contain unique secondary structure elements (including the H3 αN helix, the H2A “docking domain”, and the H2B αC helix) that further stabilize histone-histone and histone-DNA interactions. In contrast, HMfA and HMfB are rather short sequences (~65 amino acids) and lack cationic tails as well as any additional structural features beyond the histone fold motif. With the identification and sequencing of more archaea across the domain, it was found that the majority of archaeal genomes encode putative histone sequences with features similar to those of M. fervidus.13
Figure 2: Archaeal histones wrap DNA into a superhelix.
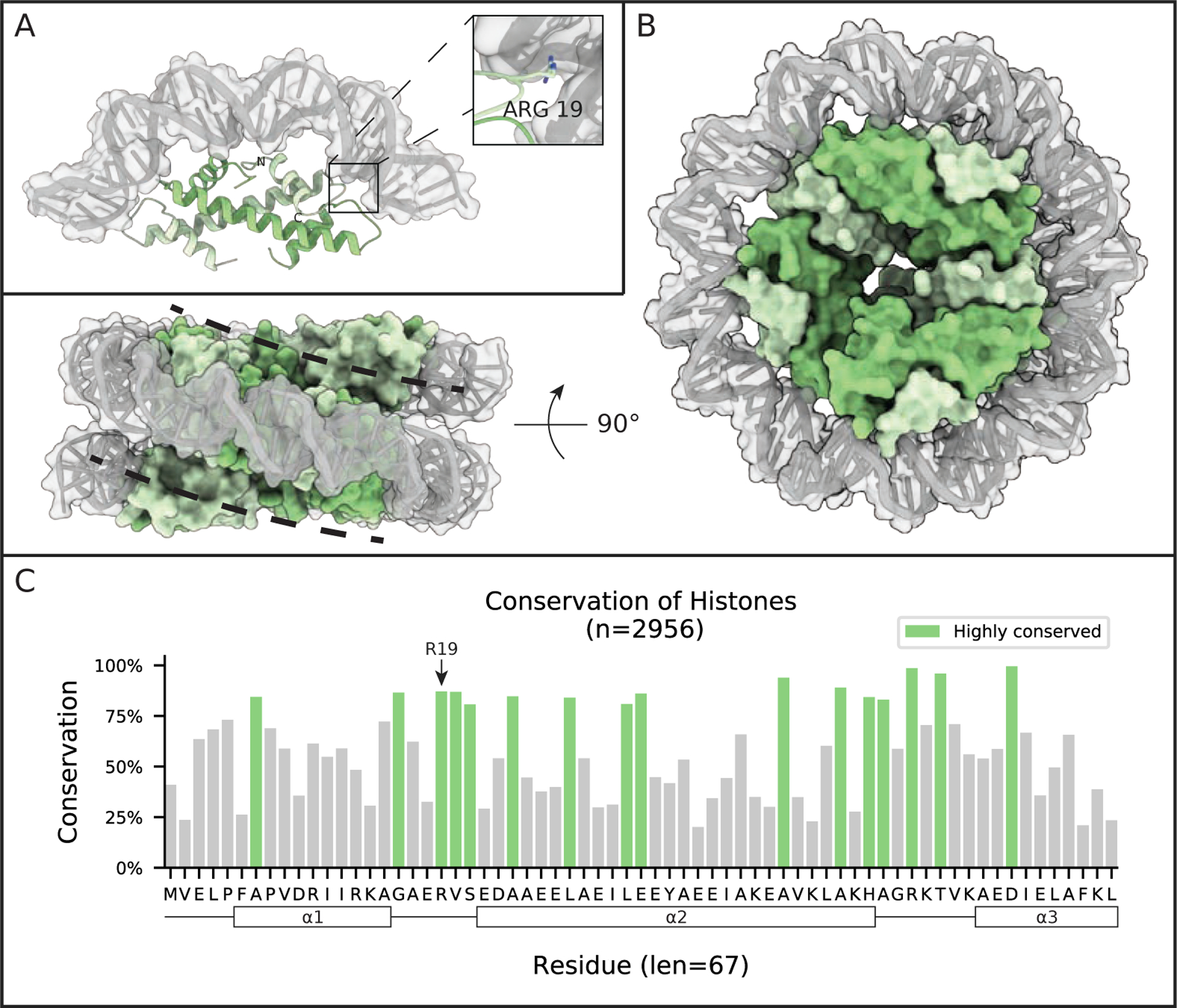
(A) The crystal structure of HMfB dimers bound to 90 bp of DNA (only 30 bp of DNA is shown with one dimer bound) shows the arching of DNA induced upon binding by histones (PDB 5T5K). The RMSD between this histone arrangement and DNA-bound eukaryotic dimers is only 1.7 Å.10 (B) Crystal contacts in the 5T5K structure suggests an organization for higher-order chromatin compaction, where repeated stacking of dimers wraps DNA into a nucleosome-like helical ramp. Shown from both face and side views, is a model of 4 sets of HMfB dimers wrapping 120 bps of DNA. (C) Conservation of putative histone homologs found in Archaea. Highlighted regions are residues conserved at least one standard deviation more than the mean conservation across the alignment. Black frames in the sequence identify predicted α-helical structures, based on homology with HMfB. The short loops connecting the α-helices are involved in DNA binding (see A).
Early crosslinking studies on M. fervidus chromatin suggested that its histones exist as dimers in solution, similar to the eukaryotic H2A-H2B system, but they tetramerize in the presence of DNA, as do eukaryotic H3–H4 heterodimers.14 Crystal structures of HMfA and HMfB confirm that the homodimers are nearly identical to the core histone folds of the eukaryotic H3–H4 heterodimers.15 Low resolution structural studies showed that at sufficient concentration, homodimers are able to self-arrange on DNA to form beads-on-a-string-like assemblies that resemble eukaryotic nucleosome arrays.2,11,16 In 2017, we determined the crystal structure of HMfB homodimers bound to DNA (Figure 2A), revealing a tertiary structure that is nearly identical to DNA-bound eukaryotic (H3–H4)2 “tetrasomes”. This structure shows a conserved four-helix bundle interaction between dimers and DNA binding interactions that were highly conserved through contacts, such as arginine sidechains inserting to the DNA minor groove (Figure 2A, inset).10 The crystal lattice also suggests the ability of archaeal histones to form continuous wraps of DNA beyond the tetrasome (Figure 2B), and micrococcal nuclease digestions of archaeal chromatin from Thermococcus kodakarensis suggested that these long chromatin “slinkies” could span anywhere from 200 to 500 bp in the cell. For further comparisons of eukaryotic vs archaeal histone-based chromatin, we direct to prior literature.10,17
In a recent follow up study on the closely related HTkA histone from T. kodakarensis, analytical ultracentrifugation and cryoEM showed that archaeal histones indeed wrap long stretches of DNA (> 200 bp), but the level of compaction inferred from the crystal structure requires free Mg2+ ions in solution.18 Analysis of cryoEM data further suggests that T. kodakarensis chromatin might be organized as a series of “nucleosome-like” wrappings, each containing three to five histone dimers binding 90 to 150 base pairs of DNA, and connected by a near zero-length linker DNA. Loss of the four-helix bundle interactions across the bridging DNA sequence results in a perpendicular orientation between neighboring subunits. This stands in contrast to the characteristic wrapping of ~147 bp in particles connected by variable-length linker DNA, as observed in the 10 nm fiber of eukaryotic nucleosomes.2,19 Therefore, naming these similar yet different structures as “archaeal nucleosomes” may be misleading, as nucleosomes are of a defined size and occupy distinctly distant sites on linearized DNA, whereas archaeal histone-based chromatin can form complexes of varying size in close proximity to one another. We suggest the term “archaeasome” in place of “archaeal nucleosome”, where the defining feature for archaeasome nomenclature is the net length of DNA bound by the construct (i.e., a shorthand of “Arc210” defines an archaeasome with 210 bp of DNA). In the context of our solution data,18 Arc210 (and larger) complexes may sporadically transition between complete 210 base pair wrappings and (Arc150 + Arc60) and (Arc120 + Arc90) complexes, as well as exist as compressed or extended “slinkies” as a result of local salt conditions.
Since the discovery of histones in M. fervidus, and with the deluge of new archaeal species isolated and sequenced from a wide range of habitats, putative histones or histone-like proteins have been identified in a wide variety of archaea.13 Some of these organisms live in extreme habitats ranging from high temperatures to low pH to salt-rich environments. In general, their histone sequences most closely resemble the H4 eukaryotic family, and they are typically 68 amino acids in length. In the recently discovered Asgard superphylum, histone sequences containing positively charged and disordered N-terminal tails have been identified,13 but little is known about the chromatin structure of those species, in a large part due to the difficulties in isolating and cultivating model Asgards.20 Histone doublets (i.e. two diverse histone sequences fused head-to-tail by a linker sequence, typically 10–20 amino acids in length) have been identified in the Halobacterium family. A doublet histone gene is also present in a distantly related halophile, Methanopyrus kandleri. Its crystal structure has been solved in the absence of DNA and was shown to be nearly identical to the dimer fold of M. fervidus histones, despite a large divergence in amino acid sequence. These histones also appear to have a four-helix bundle tetramerization interface.21 In these fused doublets, each histone fold is only 35% conserved with respect to the other, and therefore these form an obligate fused heterodimer, rather than the mix of homo- and heterodimers found in M. fervidus.12 Many of these doublet histones are characterized by a low isoelectric point (pI ~4.5), which one would expect to be not conducive to binding DNA and forming archaeasomes. Nevertheless, DNA-binding has been inferred for doublets in Haloferix volcanii, due to the similarity to nucleosome occupancy apparent in micrococcal nuclease digestion of its chromatin.22 The acidity of these histones may enhance solubility of the proteins in high intracellular salt concentrations, a typical adaptation for halophilic organisms.23 Furthermore, the peptide linker connecting the two histone fold units might enforce the proper handedness for DNA wrapping, as the fused dimers from M. kandleri introduce strictly negative supercoiling to chromatin, while M. fervidus histones show the ability to supercoil in either handedness, depending on environmental conditions.24,25
While we have shown that archaeasomes in T. kodakarensis can adopt varying sizes in solution, there is little data regarding the generality of archaeasomes across species and the maximum archaeasome size in vivo. Micrococcal nuclease digestion has repeatedly shown maximum protection footprints between 150 and ~300 bp across a variety of organisms, including T. kodakarensis, M. fervidus, Methanothermobacter thermautotrophicus and H. volcanii.10,22,26,27 However, how the cell regulates the extent of continuously wrapped DNA is not understood. Inherent DNA flexibility, topological problems from wrapping extended segments of DNA in the same handedness, histone variant effects, or competing interactions with other DNA-binding proteins may each be responsible for size regulation. While ATP-dependent remodellers are largely responsible for spacing eukaryotic nucleosomes, orthologous proteins have yet to be found in archaea. A recent preprint of a combined in silico and in vivo study by Stevens et al. has suggested that histone variants may play a direct role in regulating archaeasome length.28 Molecular dynamics simulations were conducted for DNA-bound tetramers composed of homo- and heterodimers constructed from varying Methanobrevibacter stadtmanae histone sequences. Specific sequences were found to reduce dimer-dimer interaction strengths, thereby disrupting the four-helix bundle interface and potentially acting as “capstones” to limit archaeasome size. Interestingly, computational evaluation of the tetramerization strength inversely correlated with protein abundance in the exponential phase (weaker tetramer-forming sequences were less abundant during this phase). Future studies on the effects of variant sequences on in vitro and in vivo structures across a wide range of species, as well as variant-specific localization along individual genomes, may provide the information needed to elucidate a generalized variant-based mechanism in archaea.
Because histone chaperones, assembly factors, and ATP-dependent remodellers have yet to be identified in any archaeal genome, DNA sequence is thought to be an alternative driver of archaeasome occupancy in vivo,27 especially given the absence of known ATP-dependent remodellers and histone chaperones in archaea. In vitro studies of HMf histones showed a clear preference for adenosine-containing sequences, as well as little overlap with eukaryotic nucleosome positioning sequences,29,30 and the preference of HMf histones for AT-rich sequence is consistent with in vivo mapping.26 However, histones from M. thermautotrophicus and T. kodakarensis have been shown to preferentially occupy eukaryote-like positioning sequences that are rich in GC pentamers.27 A generalized mechanism for sequence-driven histone occupancy is further obscured by the wide range of GC content in histone-containing archaeal genomes (24–74%, according to the NCBI genome database). Moreover, there is a wide disparity even among model organisms from each family (M. fervidus = 31.6%; T. kodakarensis = 52%; M. thermautotrophicus = 48.5%; Halobacterium salinarum = 65.9%). While histones in M. fervidus display a preference for AT-rich sequences in vivo, Rojec et al. recently showed that these same HMf histones adopt a preference for GC-rich sequences when expressed in E. coli (50.8% GC), which highlights the impact of host genome content on positioning.26 Nevertheless, significantly more research must be conducted in order to understand the structural mechanisms at play between genome sequence and histone occupancy in archaeal chromatin.
In most eukaryotes, loss of a single histone carries catastrophic consequences.31–33 In contrast, archaea seem to be able to adapt to the removal of histone genes but with varying phenotypes. In the halophile H. salinarum, deletion of the HpyA histone gene causes changes in cell morphology (an increase in cell width to length ratio), but viability is maintained under both nominal and stress conditions.34 In T. kodakarensis, two histone sequences are encoded (HTkA and HTkB), and while both are necessary for cell viability, loss of only one sequence does not result in cell death.35 Interestingly, deletion of HTkA renders cells impervious to transformation, yet this phenotype is not recaptured by the loss of HTkB despite the histones being 83% identical. Additionally, mutating a single residue in HTkA (G17), which was shown to preclude the formation of archaeasome assemblies beyond a stack of three dimers, does not slow growth under normal conditions but results in a weakened stress response.10 To understand the opposite extreme (introducing histones to a system not evolved to handle their presence), Rojec et al. modified E. coli cells to express HMfA or HMfB histones.26 Quite surprisingly, cells expressing these histones had no change in viability even though these histone assemble archaeasomes on the E. coli genome. However, some differences in cell morphology were observed, and stress responses in modified cells show a significant growth reduction, mimicking the effect of archaeasome-destabilizing mutations in T. kodakarensis.10
3.2. Alba
The Alba (“Aceytlation Lowers Binding Affinity”) protein family is possibly the most widespread archaeal chromatin architecture protein identified to-date (Figure 3). Unlike histones, which have only been found in eukaryotes and archaea, Alba or Alba-like proteins exist in all three domains of life.36 There are two main Alba families, and their sequences are more divergent across the eukaryotic domain than what is observed for archaea. Eukaryotic Alba sequences are frequently found attached to additional domains with expanded cellular roles including RNA metabolism, sugar transport, and even protein ubiquitination.36 In archaea, Alba proteins were discovered in the 1980’s under the names like Sac10b, Sso10b, and Ssh10b,37–39 and genes encoding Alba family proteins have been identified in nearly all archaeal lineages over the last 40 years, with the notable exceptions of Halobacterium and Methanosarcina genera.40–42
Figure 3: Alba dimers encase DNA.
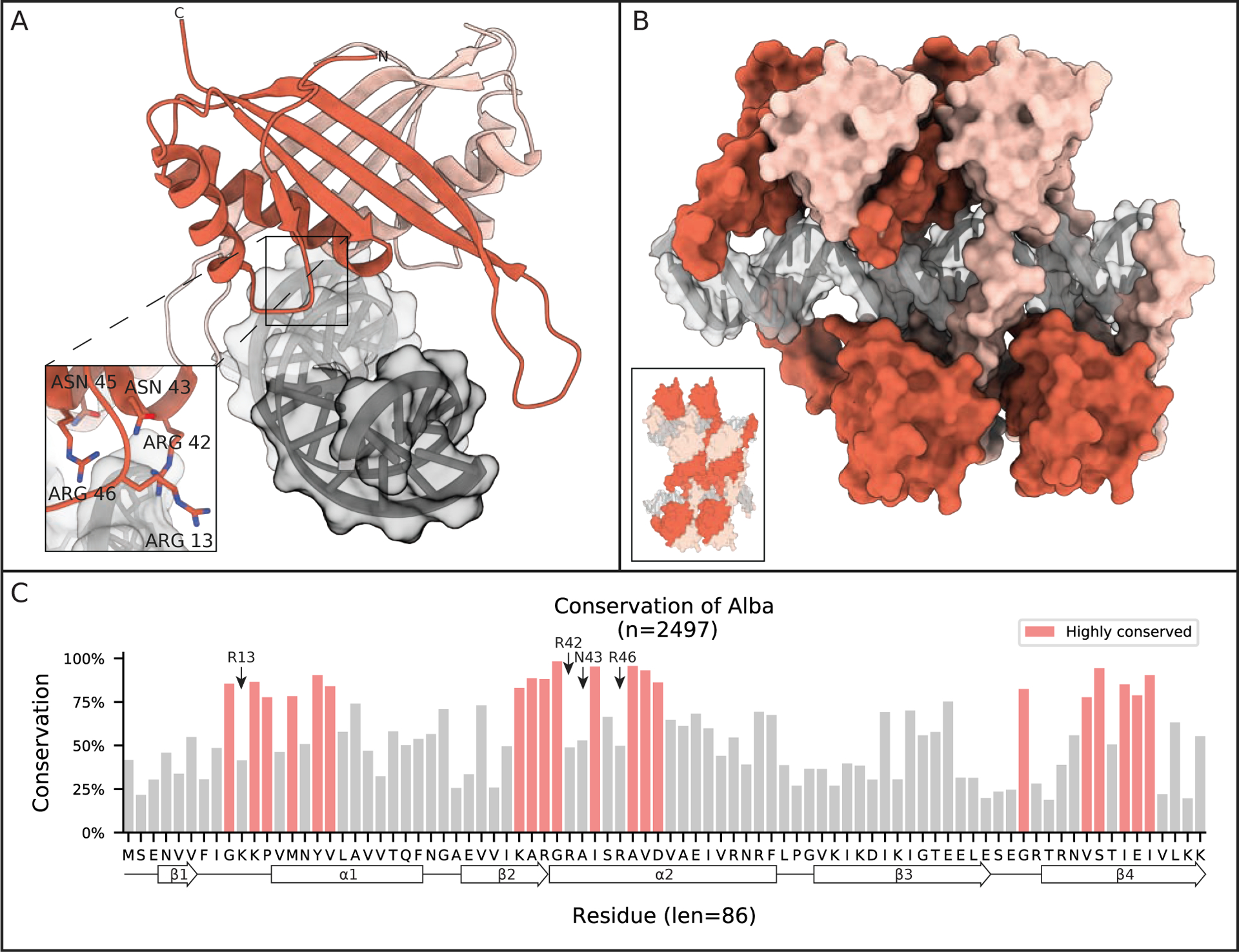
(A) Crystal structure of an Alba dimer from Aeropyrum pernix K1 bound to 16 bp of DNA (PDB 3U6Y). Only the first 4 bp, shown in dark grey, were resolved in the asymmetric unit, the rest are modeled in based on adjacent asymmetric units. Inset shows important DNA binding residues. (B) A model suggested by Tanaka et al. of higher order chromatin filament induced by continuous Alba dimer binding (based on PDB 3U6Y). The inset shows model of adjacent antiparallel Alba-DNA filaments. (C) Conservation of Alba family proteins found in Archaea. Highlighted regions are residues conserved at least one standard deviation more than the mean conservation across the alignment. Secondary structural elements are projected along the residue consensus sequence on the bottom of the plot.
Nearly 20 years after its initial classification, the crystal structure of an archaeal Alba was determined by Wardleworth et al.43 The protein forms a dimer and has a mixed α-helix and β-strand structure with high similarity to the N-terminal domain of DNase I (Cα RMSD of 2.1 Å). This structure is maintained when binding to DNA, as shown by Kumarevel et al. 10 years later (Figure 3A).44 The only conformational change upon DNA binding is observed in the loops connecting the β3-β4 hairpin of each monomer bending ~9 Å towards the complex center of the DNA-bound state.44 Dimerization in both the apo and DNA-bound state occurs symmetrically through the second α-helix and the third and fourth β-strand of each monomer, and this 690 Å2 interface is well conserved (Figure 3C). The primary interactions between the Alba dimer and DNA consist of cationic sidechains contacting the phosphates along the minor groove of DNA (Figure 3A, inset).
Many archaea encode two different families of Alba, labeled “Alba1” and “Alba2”, and the sequences between the two families can be quite divergent.45 For example, there is only 36% identity between the two proteins in Sulfolobus solfataricus.46 NMR spectroscopy done with mixtures of Alba1 and Alba2 from S. solfataricus shows that they form heterodimers at a 1:1 stoichiometric ratio, indicating that heterodimers are preferred over homodimers.45 Crystal structures of these heterodimers, in the absence of DNA, show an identical binding mode as homodimers.45 At low protein:DNA ratios, both EM and atomic force microscopy (AFM) images show that Alba1 homodimers and Alba1-Alba2 heterodimers form similar filaments on DNA.45,46 However, differences in packaging phenotype are observed at higher ratios, where heterodimers form condensed forms of chromatin while Alba1 homodimers form thick filaments bridging the DNA. Laurens et al. used optical tweezer measurements to further show that these changes in packaging phenotype have mechanical consequences.46 Alba1 homodimer filaments shift the measured persistence length - a measure of polymer “stiffness”, where larger lengths correspond to stiffer segments - of DNA from 49 nm (~150 bp) to 250 nm (~735 bp). In contrast, the heterodimer phenotype only increases the persistence length to 75nm (~220 bp) in the same concentration range, indicating reduced DNA coverage by the heterodimers when compared to Alba1 homodimers.46 It was further observed that Alba1 homodimers bound cooperatively to DNA fragments, but Alba1-Alba2 heterodimers and F60A mutant Alba1 homodimers, with a weakened dimer-dimer interface, do not exhibit the same binding cooperativity. This suggests that the more efficient coating of DNA by the homodimers is a direct result of cooperative binding.
These interpretations are in agreement with high-resolution models for Alba-coated filaments that have been inferred from the Alba-DNA crystal structure.44 Along the DNA length, the β3 strand of the central unit forms a dimerization interface with β1 of the neighboring molecule, and this head-to-tail interaction is thereby repeated many times over (Figure 3B). In this arrangement, bending of the DNA would be heavily constrained by the high density of Alba coating the DNA surface, thereby increasing the persistence length of the filament. The overlap of neighboring units further shows the additional role of Alba as a DNA bridging molecule that helps to hold two non-ligated DNA fragments together, end-to-end. Moreover, the crystal packing suggests that neighboring filaments can be further reinforced by interdimer interactions through antiparallel alignment of their α1 helices (Figure 3B, inset).44 This interaction was independently observed through NMR classification of Alba oligomers.47
Alba proteins across different archaeal species display varying levels of DNA sequence specificity. It is generally understood that Alba homologs in hyperthermophilic species display little to no sequence specificity.48,49 In contrast, Mma10b, an Alba homolog in the mesophilic Methanococcus maripaludis, preferentially binds to specific sites throughout the genome.50 Additionally, the expression level of Mma10b is two orders of magnitude lower than Ssh10b, the Alba protein of Sulfolobus shibatae, and this reduced abundance, in conjunction with a particular DNA sequence motif, is more in-line with the behavior of a transcription factor than a simple architectural protein.49
Many archaea encode genes for histones in addition to Alba (Figure 1). While histones drastically bend and wrap DNA into archaeasomes, Alba coats the DNA and yields rigidified chromatin filaments. This raises the question: how do these opposing architectural principles interface? This was one focus of a recent study where AFM was used to observe the changes in reconstituted T. kodakarensis chromatin structure in relation to Alba-to-histone ratio.51 It was found that increasing Alba concentration does not remove the presence of “archaeasome beads” on the DNA fragment, but rather increases the length of DNA between clusters of archaeasomes, albeit at HTkA-to-Alba ratios an order of magnitude above estimated in vivo ratios.52 Nevertheless, this shows that Alba and histones competitively bind to DNA, and local compaction of chromatin by archaeasomes may be reduced by the presence of Alba.
Figure 1: Groups of archaea utilize different proteins to structure their genomes.
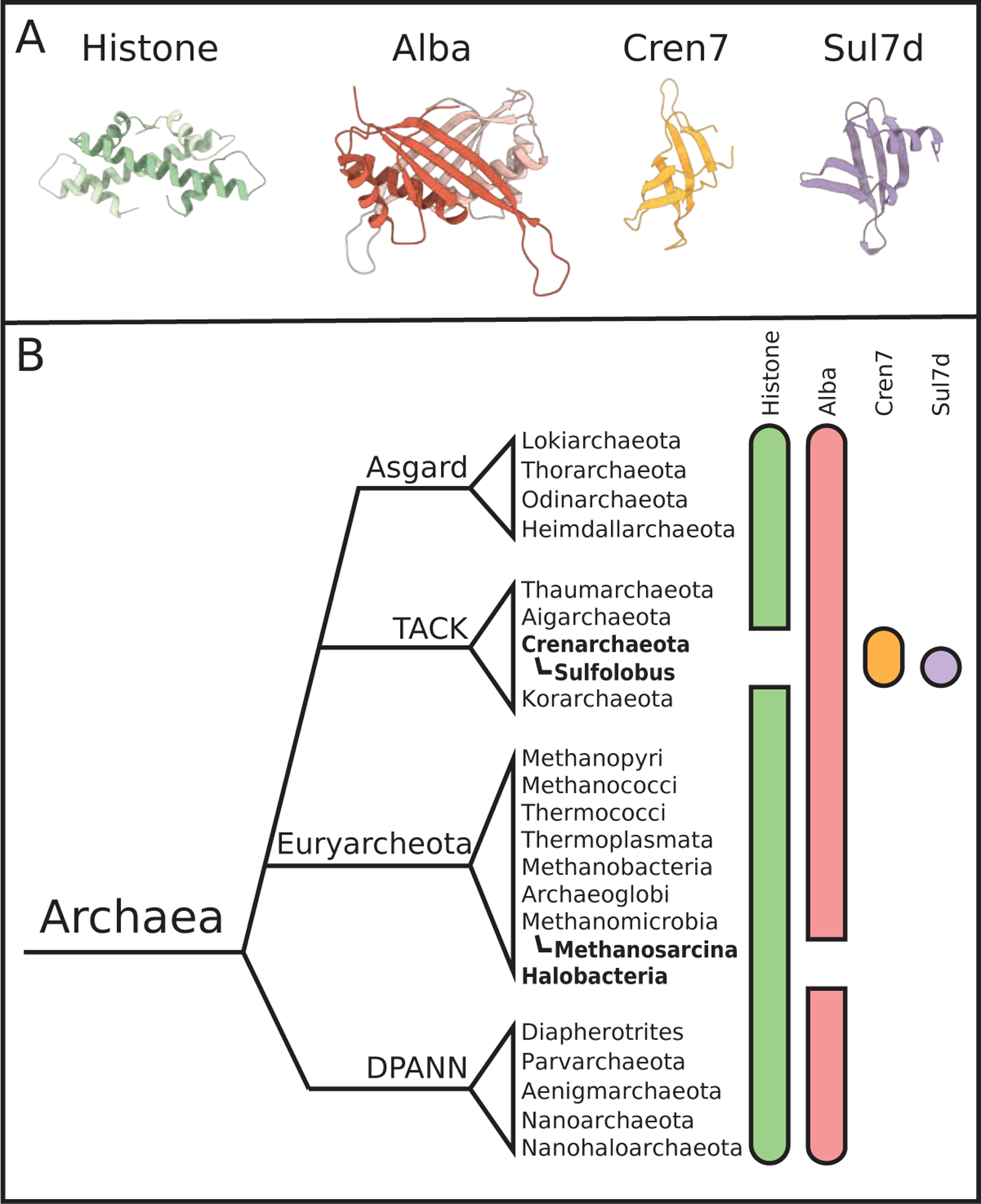
(A) Crystal structures of the four archaeal DNA-structuring proteins summarized in this review. (B) A simplified phylogeny, based on Williams et al.,9 showing the distribution of chromatin structure-related proteins among archaea. Although debate over the true taxonomy of Archaea is ongoing, we have placed four widely discussed branches on this tree: Asgard, TACK, Euryarcheota, and DPANN. Colored bars beside each phylum denotes the type of chromatin-organizing protein present in those species. Many archaea encode both histones and Alba, but bold text highlights taxa without identified histone or Alba sequences, as shown by gaps in the green and red bars, respectively. Cren7 and Sul7d exist only in Crenarcheota, which do not typically contain histones.
Until now, we have been discussing Alba as a DNA-organizing protein in the archaeal nucleoid. However, Alba is unique among the other proteins discussed in this review in that it also binds well to RNA duplexes,53 and there is evidence that archaeal Alba proteins may even facilitate RNA metabolism.54 Treatment of insoluble chromatin fractions extracted from S. solfataricus separately by DNase I and RNase A each release Alba, suggesting further that Alba is associated with both DNA and RNA in vivo.55 This complicates investigation of their in vivo functions. Guo et al. previously showed through crystallography that Alba tetramers can bind to an RNA duplex and perturb the local A-form arrangement of the RNA,53 but the physiological function of this alteration is still an open question. One possible role is that Alba serves as an archaeal member of the translation pathway for leaderless mRNA,53,56,57 but an alternative interpretation may be that Alba can serve as a chaperone for RNA molecules.58
3.3. Cren7
Aside from widespread chromatin structuring proteins such as histones and Alba, some Archaea have also evolved other proteins to help structure their genomes. Two of these, Cren7 and Sul7d (Figures 4 and 5, respectively), exist only in Crenarchaeota, which typically encode Alba but not histones. Cren7 is phylo-genetically sparse, being restricted only to Crenarchaeota, but it is encoded within almost every sequenced Crenarchaeota genome. The notable exception is the genome of Thermofilum pendens Hrk5, which is thought to lack a Cren7 homolog. Thermofilum is one of the only representatives of Crenarchaeota (along with species from Vulcanisaeta and Caldivirga) to encode a histone homolog. This limited dispersion of histone homologs in Cren7 containing species suggests that histones and Cren7 may serve redundant roles.49,59
Figure 4: Cren7 kinks DNA into S-shaped filaments.
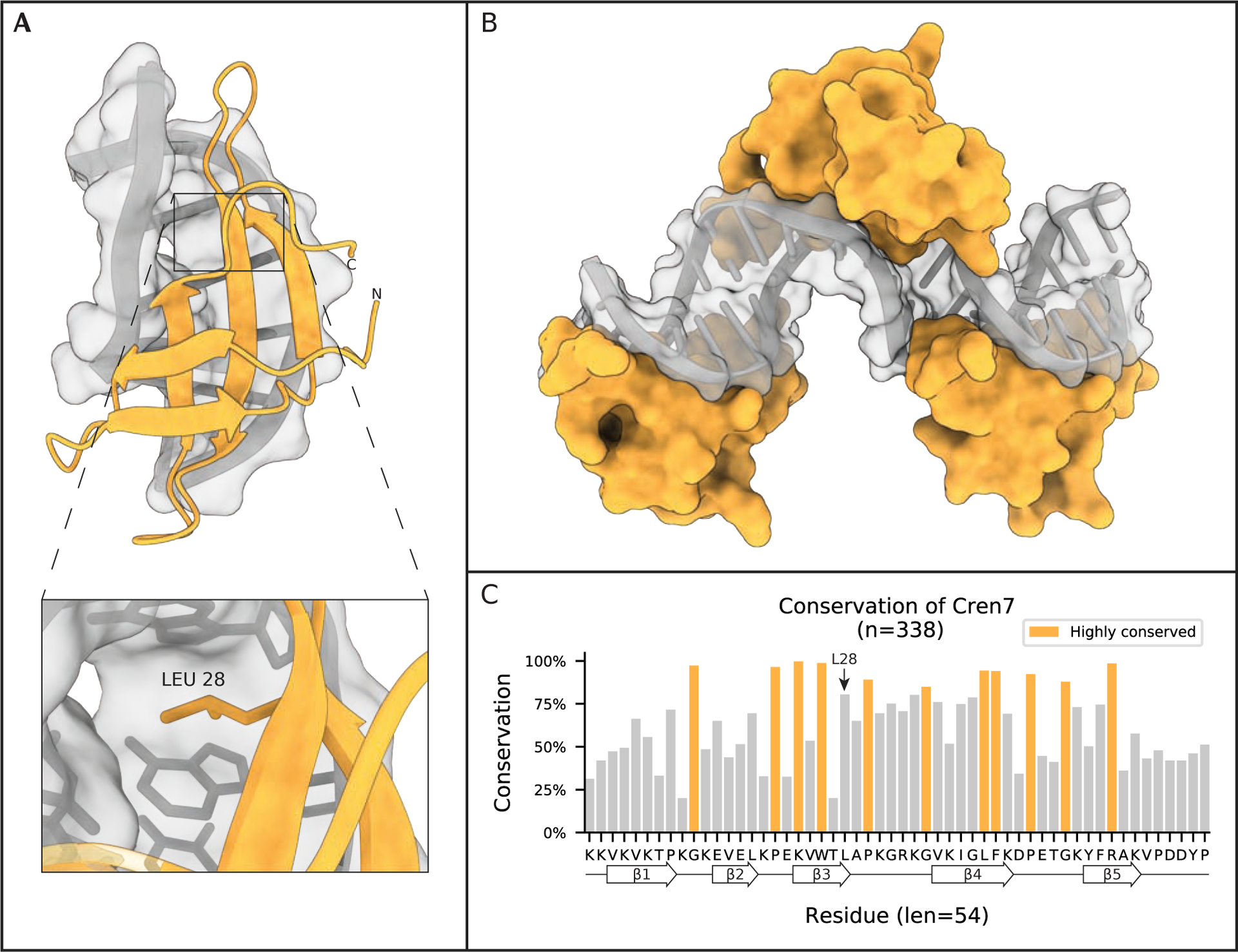
(A) Crystal structure of Cren7 from S. solfataricus P2 bound to 8 bp of dsDNA (PDB 4R56). Inset highlights incalation of L28 into the DNA ladder, an interaction that has been shown to stabilize the kinking of DNA strands by ~50° by Cren7. (B) Model of chromatin induced by Cren7 based on PDB 6A2I, in which monomers of Cren7 bind DNA in a head-to-tail fashion while structuring the DNA into an S-shaped filament. (C) Conservation of Cren7 homologs found in Archaea. Highlighted regions are residues conserved at least one standard deviation more than the mean conservation across the alignment. Secondary structural elements are projected along the residue consensus sequence on the bottom of the plot.
Figure 5: Sul7d kinks DNA.
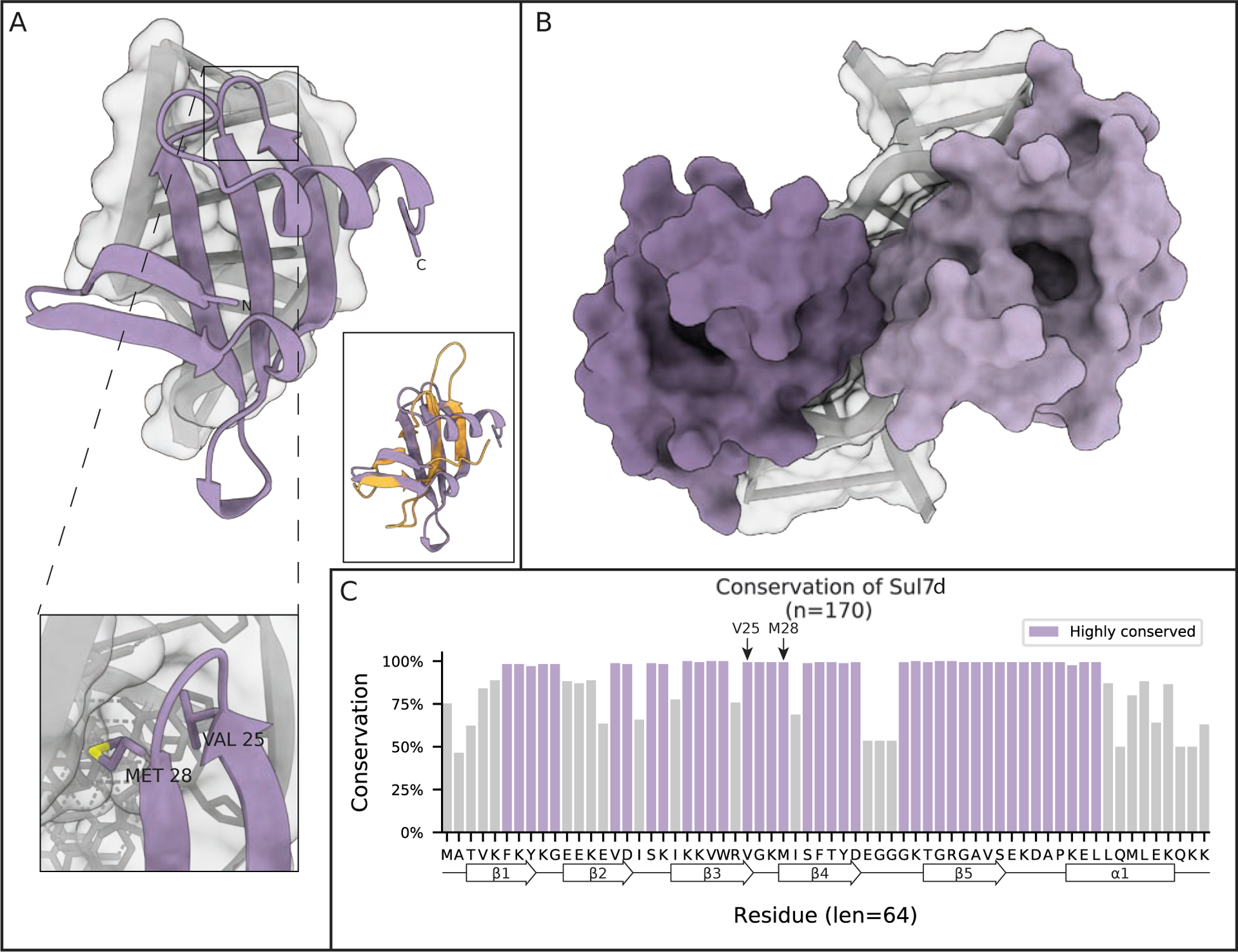
(A) Crystal structure of the Sul7d-DNA complex showing the kinking of DNA induced by Sul7d from S. acidocaldarius (PDB 1BF4). One inset shows two important residues that intercalate in the DNA double helix in a similar manner as L28 in Cren7 to stabilize the DNA kink. The other inset shows the structure of Cren7 (orange) overlayed onto that of Sul7d (purple). (B) Solution NMR structure of S. solfataricus Sul7d bound to 12 bp of DNA showing the head to head binding mode of Sul7d on DNA. (C) Conservation of Sul7d homologs found in Archaea. Highlighted regions are residues conserved at least one standard deviation more than the mean conservation across the alignment. Secondary structural elements are projected along the residue consensus sequence on the bottom of the plot.
Cren7 was first identified in 2008 through a pulldown in S. solfataricus as an association partner of Sul7d. Cren7 is a small, basic, highly conserved protein that comprises almost 1% of the total protein expressed in S. solfataricus60 (Figure 4C). Guo et al. explored the nucleic acid binding activity of Cren7 and showed that it prefers binding double stranded DNA over single stranded DNA, can stabilize double stranded DNA against thermal denaturation, and constrains DNA into negative supercoils as demonstrated by nick closure assays.60 The group went on to solve the first NMR structure of Cren7, which clearly defined it as an SH3-domain protein consisting of 5 β-strands connected by 4 loops. In 2010, two groups published crystal structures of Cren7 bound to short pieces of DNA61,62 (Figure 4A). These structures demonstrated the ability of Cren7 to kink DNA at ~50°, while twisting it 29°, enough to induce negative supercoiling. The structure also highlighted the importance of the interaction between the β3–4 loop and DNA through contacts with the minor groove. Deletion of this β3–4 loop led to a 60-fold reduction in DNA binding.61 Intercalation of a leucine (L28) residue into the DNA ladder, which was later shown to stabilize, although not induce, the kinking of DNA by Cren763 (Figure 4A inset).
To further explore how Cren7 interacts with DNA, molecular dynamics simulations were performed on crystal structures of Cren7 bound to DNA. These simulations showed that upon binding to DNA, the β3–4 loop of Cren7 is greatly stabilized. Much of the binding free energy contribution comes from one strand of DNA. These simulations also highlight residues important to DNA binding, in agreement with previous published biochemical data.64 In another study of Cren7, molecular dynamics simulations were used to look at how each of these DNA binding residues contributed to binding through free energy decomposition, as well as mutation. It was shown that most of the binding energy of Cren7 and DNA come from nonpolar solvation interactions.65 A third molecular dynamics simulation study further verified that Cren7 kinks DNA at an angle of ~50°.66
To understand how Cren7 might structure longer stretches of DNA than the 8 bp used in previous structural studies, Zhang et al. determined the crystal structure of Cren7 bound to 18 and 20 bp DNA fragments.67 These structures revealed two intriguing aspects of Cren7 chromatin. First, Cren7 monomers bind DNA in a head-to-tail fashion on opposites sides of the DNA, preventing them from contacting each other. Second, Cren7 creates a continuous S-shaped filament of DNA by inducing kinks in opposite directions (Figure 4B). Although this model of Cren7 chromatin seems to structure DNA, it does not compact it. In addition, Cren7 shows a preference for AT-rich DNA.67 Using a combination of SM-TIRF (single-molecule level by total internal reflection fluorescence) microscopy and AFM it was shown that Cren7 is only able to compact DNA to a small degree through the direct binding and DNA bending events that occur at low protein concentrations. However, at high protein concentrations, Cren7 is able to greatly compact DNA (by >13-fold) via bridging interactions in which DNA bound molecules of Cren7 interact with distal DNA bound Cren7 monomers to create highly condensed protein-DNA cores.68
3.4. Sul7d
The second well studied Crenarchaeotal specific chromatin architectural protein is Sul7d (Figure 5). It was identified in the 1980’s in a study that aimed to identify ribosomal proteins in Sulfolobus acidocaldarius, but also discovered a number of small DNA binding proteins, then referred to as the Sac7 proteins because of their 7 kDa size (Sul7d was originally called Sac7d).69 Sul7d is confined to a limited taxonomy of the Sulfolobus family where it coexists with Cren7 as the dominant chromatin architectural protein. In these organisms, Sul7d constitutes as much as 5% of the total protein in cells and thus classifies as an abundant chromatin architectural protein.70 Like Cren7, Sul7d is an SH3-domain protein. The two proteins share remarkable structural conservation, despite rather poor sequence conservation, perhaps suggesting an ancient common lineage.71 These two proteins also share specific structural characteristics, such as residues in the loop between β3 and 4, which are important for DNA binding. Within this loop, there seems to be conservation of a small hydrophobic residue (L28 in Cren7 and V25 in Sul7d) that helps stabilize DNA kinking. The kink induced by Sul7d on dsDNA is similar to that of Cren7, ~60° (compared to Cren7’s 50°)72 (Figure 5A and 5C). In Sul7d, both V25 and M28 are able to intercalate in the minor groove of the DNA, causing it to widen.73 Although these structural similarities would suggest a redundant role for the two proteins, both proteins appear to be essential and are thought to serve non-redundant roles.67
Differences between Cren7 and Sul7d become apparent in the structures of multiple molecules bound to DNA. While Cren7 binds DNA in a head-to-tail fashion, Sul7d adopts a head-to-head conformation (Figure 5B). Another structural difference is that the length of the important β3–4 loop, is much longer and more flexible in Cren7 than Sul7d. Additionally, Sul7D has a C-terminal alpha helix that is not present in Cren7 (Figure 5A).74 These structural differences seem to lower the ability of Sul7D to compact DNA compared to Cren7, and abolish the preference for AT-rich DNA of Cren7. Compared to Cren7, Sul7d does not significantly compact DNA through DNA bending at low protein concentrations. However, as is the case with Cren7, Sul7d is able to compact DNA at high protein concentrations through bridging interactions, as demonstrated by AFM experiments. These interactions do not seem to lead to the highly compacted cores found in Cren7 chromatin, but instead result in large DNA loops at high Sul7d concentrations.68
4. Concluding Remarks and Future Perspective
One of the fundamental challenges an organism faces is the requirement to package its genome in a manner that protects DNA from damage, but also makes it available for transcription and replication without generating tangles and knots. While the organizational strategies of eukaryotes and bacteria are fairly well conserved within each domain of life, archaea employ a wide variety of DNA packaging principles across the domain. Euryarcheaota use minimalist histones to construct archaeasomes of varying sizes to superhelically wrap DNA,10,13 but the extent to which DNA sequence and variant sequences regulate archaeasome size is not well known. In contrast, Crenarchaeota utilize small NAPs to coat DNA and create chromatin filaments with sharp kinks.67,72 Alba proteins span both of these archaeal phyla (and indeed all domains of life), and their role in DNA compaction shows a competitive interplay with other DNA bending or wrapping proteins.51 However, their potential role as RNA chaperones or active members in translation muddles interpretation of their in vivo roles.54 In order to answer these questions, many gaps in our knowledge must be filled.
The most glaring gap in the structural biochemistry of archaeal chromatin is the lack of high- and intermediate-resolution in vitro and in situ structures of higher-order chromatin organization. AFM and negative-stain EM provide important descriptions of architectural phenotypes, but a full description of scaffolding mechanisms requires a significant increase in structural resolution. Indeed, a crystal structure of the nucleosome array fiber was solved within a decade of the original nucleosome structure,1,75 with a cryoEM-derived structure of higher-order fibers published in the following decade.76 In situ cryo-electron tomography has further probed the structure of eukarotic oligonucleosomes in a cellular context.77 Analogous data for archaea would provide valuable insight regarding the organization of their genomes in cells.
In addition to missing information regarding higher-order chromatin structures reconstituted purely from one architectural protein, knowledge of the interplay and crosstalk between chromatin-organizing and chromatin-regulating proteins is lacking. In eukaryotes, complex communication pathways exist between nucleosomes and extra-nucleosomal factors, such as other chromatin architectural proteins,78 transcription factors,79 chromatin remodelers,4,5 and post translational modification readers and writers.80,81 Furthermore, inclusion of additional proteins, such as linker “histone” H1, results in additional structural rearrangements, such as the protection of additional DNA per nucleosome and tighter compaction of nucleosome arrays.82,83 For a complete picture of DNA organization in archaea, similar information must be sought for archaeal chromatin.
Another outstanding question is how the transcription and replication machinery interacts with chromatin-organizing proteins and their higher-order structures. To date, homologs of the histone chaperones or chromatin remodellers present in eukaryotes have not yet been identified in archaea. If these external factors are indeed absent from archaeal genomes, archaeal cells may utilize interesting (yet unidentified) mechanisms for chromatin dissassembly. One alternative influence may be the intrinsic flexibility of various codons.84 In this way, particular sequences with high rigidity may bias against archaeasome formation, or they could be poised for easy release from archaeasome complexes or kinked Cren7/Sul7d conformations. The replisome or transcription machinery may also be able to dispense and reorganize architectural proteins without the need for additional co-factors.
A key issue in our understanding of chromatin architecture in the archaeal domain of life is the genome sampling of underrepresented taxonomies, as well as the fluidity of archaeal taxonomy itself. This poses a problem for identifying trends in archaeal chromatin organization that might exist in nature that are overlooked because we have not sampled enough organisms to be able to make strong conclusions. This is linked to the problem of finding reliable means to infer relations throughout the domain that would allow us to speculate how these architectural proteins have evolved over time in complex cohabitation networks, such as those found in the human body,85–87 as well as how they have adapted to specific and often extreme environments. With advances in sequencing technology and availability, in addition to the growing number of genomic scaffolds upon which to build genomes, the scientific community is poised to vastly expand our understanding of how archaea fit into the tree of life. Not only will advances in sequencing help us define new species, but this will also provide new information on how genomes are organized. For example, a recent Hi-C study of two members in the Sulfolobus family showed that these genomes use SMC-like proteins to compartmentalize their genomes in a manner similar to that of eukaryotes and even proposed spatial organizations of those genomes.88 More complete genome coverages may also identify the chaperones and chromatin remodellers currently absent in archaea, or future bioinformatic analysis may uncover chromatin assembly factors with previously unknown sequences.
However, perhaps the biggest roadblock for archaeal research is simply our inability to efficiently isolate and grow enough model organisms in the lab. Although a few species have been cultured, isolating new cultures can require specialized equipment and resources that are beyond most labs, such as custom engineered methane-fed bioreactors.20 In part due to this restriction, current model species come from a vary narrow distribution of taxonomic and ecological niches that leave much of the Archaeal domain un- or underrepresented. Without a wider selection of genetically tractable model organisms that represent the diversity we see within this vast and diverse domain of live, and without the ability to investigate different modes of chromatin organization, it will be hard to verify many of the general, domain-wide chromatin models that have been proposed, and insights that are only possible through in vivo experiments are beyond reach for many organisms.
The future of archaea research and the structure-function mechanisms governing genome organization in this domain promises to be exciting, as there are many unanswered questions. Do archaea utilize chromatin-organizing proteins purely to package chromatin, or do they participate in transcription regulation? What role, if any, do post-translational modifications play in regulating archaeal chromatin structure? What unknown assembly mechanisms remain to be uncovered by studying archaeal systems? Technological advances such as the ability to identify and rapidly assemble genomes, revolutions in structure determination (including in situ cryo-electron tomography), and the possibility of conducting molecular dynamics simulations en masse will accelerate the rate and efficiency with which discoveries are made. This will hopefully be paralleled by increased diversity of model organisms that can be cultivated and genetically manipulated in the lab.
5. Acknowledgments
This work was funded by the Howard Hughes Medical Institute, and SB was supported by the National Institutes of General Medical Sciences of the National Institutes of Health under award number F32GM137496. The content of this article are the sole responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Luger K; Mäder AW; Richmond RK; Sargent DF; Richmond TJ Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- [2].Olins AL; Olins DE Spheroid Chromatin Units (ν Bodies). Science 1974, 183, 330–332. [DOI] [PubMed] [Google Scholar]
- [3].Hammond CM; Str?mme CB; Huang H; Patel DJ; Groth Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol 2017, 18, 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clapier CR; Iwasa J; Cairns BR; Peterson CL Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol 2017, 18, 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Markert J; Luger K Nucleosomes Meet Their Remodeler Match. Trends in Biochemical Sciences 2020, [DOI] [PubMed] [Google Scholar]
- [6].Dillon SC; Dorman CJ Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol 2010, 8, 185–195. [DOI] [PubMed] [Google Scholar]
- [7].DasSarma S; DasSarma P Halophiles and their enzymes: negativity put to good use. Curr. Opin. Microbiol 2015, 25, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guo Z; Wang Y; Yang A; Yang G The effect of pH on charge inversion and condensation of DNA. Soft Matter 2016, 12, 6669–6674. [DOI] [PubMed] [Google Scholar]
- [9].Williams TA; Szöllősi GJ; Spang A; Foster PG; Heaps SE; Boussau B; Ettema TJG; Embley TM Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proceedings of the National Academy of Sciences 2017, 114, E4602–E4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mattiroli F; Bhattacharyya S; Dyer PN; White AE; Sandman K; Burkhart BW; Byrne KR; Lee T; Ahn NG; Santangelo TJ et al. Structure of histone-based chromatin in Archaea. Science (New York, N.Y.) 2017, 357, 609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sandman K; Krzycki JA; Dobrinski B; Lurz R; Reeve JN HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. U.S.A 1990, 87, 5788–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sandman K; Grayling RA; Dobrinski B; Lurz R; Reeve JN Growth-phase-dependent synthesis of histones in the archaeon Methanothermus fervidus. Proc. Natl. Acad. Sci. U.S.A 1994, 91, 12624–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Henneman B; Emmerik C. v.; Ingen H. v.; Dame RT Structure and function of archaeal histones. PLOS Genetics 2018, 14, e1007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pereira SL; Grayling RA; Lurz R; Reeve JN Archaeal nucleosomes. Proceedings of the National Academy of Sciences 1997, 94, 12633–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Starich MR; Sandman K; Reeve JN; Summers MF NMR Structure of HMfB from the Hyperthermophile,Methanothermus fervidus, Confirms that this Archaeal Protein is a Histone. Journal of Molecular Biology 1996, 255, 187–203. [DOI] [PubMed] [Google Scholar]
- [16].Tomschik M; Karymov MA; Zlatanova J; Leuba SH The Archaeal Histone-Fold Protein HMf Organizes DNA into Bona Fide Chromatin Fibers. Structure 2001, 9, 1201–1211. [DOI] [PubMed] [Google Scholar]
- [17].Bhattacharyya S; Mattiroli F; Luger K Archaeal DNA on the histone merry-go-round. The FEBS Journal 2018, 285, 3168–3174. [DOI] [PubMed] [Google Scholar]
- [18].Bowerman S; Wereszczynski J; Luger K Archaeal chromatin ‘slinkies’ are inherently dynamic complexes with deflected DNA wrapping pathways. bioRxiv 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Olins DE; Olins AL Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol 2003, 4, 809–814. [DOI] [PubMed] [Google Scholar]
- [20].Imachi H; Nobu MK; Nakahara N; Morono Y; Ogawara M; Takaki Y; Takano Y; Uematsu K; Ikuta T; Ito M et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 2020, 577, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fahrner RL; Cascio D; Lake JA; Slesarev A An ancestral nuclear protein assembly: Crystal structure of the Methanopyrus kandleri histone. Protein Science 2001, 10, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ammar R; Torti D; Tsui K; Gebbia M; Durbic T; Bader GD; Giaever G; Nislow C Chromatin is an ancient innovation conserved between Archaea and Eukarya. eLife 2012, 1, e00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tadeo X; López-Méndez B; Trigueros T; Laín A; Castaño D; Millet O Structural Basis for the Aminoacid Composition of Proteins from Halophilic Archea. PLOS Biology 2009, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Musgrave DR; Sandman KM; Reeve JN DNA binding by the archaeal histone HMf results in positive supercoiling. Proceedings of the National Academy of Sciences 1991, 88, 10397–10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Musgrave D; Forterre P; Slesarev A Negative constrained DNA supercoiling in archaeal nucleosomes. Mol. Microbiol 2000, 35, 341–349. [DOI] [PubMed] [Google Scholar]
- [26].Rojec M; Hocher A; Stevens KM; Merkenschlager M; Warnecke T Chromatinization of Es-cherichia coli with archaeal histones. eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nalabothula N; Xi L; Bhattacharyya S; Widom J; Wang JP; Reeve JN; Santangelo TJ; Fondufe-Mittendorf YN Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs. BMC Genomics 2013, 14, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stevens KM; Swadling JB; Hocher A; Bang C; Gribaldo S; Schmitz RA; Warnecke T Histone variants in archaea and the evolution of combinatorial chromatin complexity. Proceedings of the National Academy of Sciences 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bailey KA; Pereira SL; Widom J; Reeve JN Archaeal histone selection of nucleosome positioning sequences and the procaryotic origin of histone-dependent genome evolution11Edited by T. Richmond. Journal of Molecular Biology 2000, 303, 25–34. [DOI] [PubMed] [Google Scholar]
- [30].Pereira SL; Reeve JN Archaeal nucleosome positioning sequence from Methanothermus fervidus. J. Mol. Biol 1999, 289, 675–681. [DOI] [PubMed] [Google Scholar]
- [31].Günesdogan U; Jäckle H; Herzig A Histone supply regulates S phase timing and cell cycle progression. eLife 2014, 3, e02443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Smith MM; Stirling VB Histone H3 and H4 gene deletions in Saccharomyces cerevisiae. J Cell Biol 1988, 106, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McKay DJ; Klusza S; Penke TJ; Meers MP; Curry KP; McDaniel SL; Malek PY; Cooper SW; Tatomer DC; Lieb JD et al. Interrogating the Function of Metazoan Histones using Engineered Gene Clusters. Developmental Cell 2015, 32, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dulmage KA; Todor H; Schmid AK Growth-Phase-Specific Modulation of Cell Morphology and Gene Expression by an Archaeal Histone Protein. mBio 2015, 6, e00649–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Čuboňová L; Katano M; Kanai T; Atomi H; Reeve JN; Santangelo TJ An Archaeal Histone Is Required for Transformation of Thermococcus kodakarensis. Journal of Bacteriology 2012, 194, 6864–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Goyal M; Banerjee C; Nag S; Bandyopadhyay U The Alba protein family: Structure and function. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2016, 1864, 570–583. [DOI] [PubMed] [Google Scholar]
- [37].Reddy TR; Suryanarayana T Novel histone-like DNA-binding proteins in the nucleoid from the acidothermophillic archaebacterium Sulfolobus acidocaldarius that protect DNA against thermal denaturation. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1988, 949, 87–96. [Google Scholar]
- [38].Dijk J; Reinhardt R The Structure of DNA-Binding Proteins from Eu- and Archaebacteria. In Bacterial Chromatin, Gualerzi CO, Pon CL (eds). 1986, 185–218. [Google Scholar]
- [39].Sandman K; Reeve JN Archaeal chromatin proteins: different structures but common function? Current Opinion in Microbiology 2005, 8, 656–661. [DOI] [PubMed] [Google Scholar]
- [40].Green GR; Searcy DG; DeLange RJ Histone-like protein in the Archaebacterium Sulfolobus acidocaldarius. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1983, 741, 251–257. [DOI] [PubMed] [Google Scholar]
- [41].Forterre P; Confalonieri F; Knapp S Identification of the gene encoding archeal-specific DNA-binding proteins of the Sac10b family. Molecular Microbiology 1999, 32, 669–670. [DOI] [PubMed] [Google Scholar]
- [42].White MF; Bell SD Holding it together: chromatin in the Archaea. Trends Genet. 2002, 18, 621–626. [DOI] [PubMed] [Google Scholar]
- [43].Wardleworth BN; Russell RJ; Bell SD; Taylor GL; White MF Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002, 21, 4654–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tanaka T; Padavattan S; Kumarevel T Crystal structure of archaeal chromatin protein Alba2-double-stranded DNA complex from Aeropyrum pernix K1. J. Biol. Chem 2012, 287, 10394–10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jelinska C; Conroy MJ; Craven CJ; Hounslow AM; Bullough PA; Waltho JP; Taylor GL; White MF Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure 2005, 13, 963–971. [DOI] [PubMed] [Google Scholar]
- [46].Laurens N; Driessen RP; Heller I; Vorselen D; Noom MC; Hol FJ; White MF; Dame RT; Wuite GJ Alba shapes the archaeal genome using a delicate balance of bridging and sti ening the DNA. Nat Commun 2012, 3, 1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jelinska C; Petrovic-Stojanovska B; Ingledew WJ; White MF Dimer-dimer stacking interactions are important for nucleic acid binding by the archaeal chromatin protein Alba. Biochem J 2010, 427, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Črnigoj M; Podlesek Z; Zorko M; Jerala R; Anderluh G; Ulrih NP Interactions of Archaeal Chromatin Proteins Alba1 and Alba2 with Nucleic Acids. PLOS ONE 2013, 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Peeters E; Driessen RPC; Werner F; Dame RT The interplay between nucleoid organization and transcription in archaeal genomes. Nature Reviews. Microbiology 2015, 13, 333–341. [DOI] [PubMed] [Google Scholar]
- [50].Liu Y; Guo L; Guo R; Wong RL; Hernandez H; Hu J; Chu Y; Amster IJ; Whitman WB; Huang L The Sac10b homolog in Methanococcus maripaludis binds DNA at specific sites. J Bacteriol 2009, 191, 2315–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Maruyama H; Prieto EI; Nambu T; Mashimo C; Kashiwagi K; Okinaga T; Atomi H; Takeyasu K Different Proteins Mediate Step-Wise Chromosome Architectures in Thermoplasma acidophilum and Pyrobaculum calidifontis. Front Microbiol 2020, 11, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Maruyama H; Shin M; Oda T; Matsumi R; Ohniwa RL; Itoh T; Shirahige K; Imanaka T; Atomi H; Yoshimura SH et al. Histone and TK0471/TrmBL2 form a novel heterogeneous genome architecture in the hyperthermophilic archaeon Thermococcus kodakarensis. Mol. Biol. Cell 2011, 22, 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Guo L; Ding J; Guo R; Hou Y; Wang DC; Huang L Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea. J. Biol. Chem 2014, 289, 1478–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Aravind L; Iyer LM; Anantharaman V The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003, 4, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Marsh VL; Peak-Chew SY; Bell SD Sir2 and the Acetyltransferase, Pat, Regulate the Archaeal Chromatin Protein, Alba. Journal of Biological Chemistry 2005, 280, 21122–21128. [DOI] [PubMed] [Google Scholar]
- [56].Gissot M; Walker R; Delhaye S; Alayi TD; Huot L; Hot D; Callebaut I; Schaeffer-Reiss C; Dorsselaer AV; Tomavo S Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J. Mol. Biol 2013, 425, 1287–1301. [DOI] [PubMed] [Google Scholar]
- [57].Mani J; Gottinger A; Schimanski B; Heller M; Acosta-Serrano A; Pescher P; Sp?th, G.; Roditi, I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS ONE 2011, 6, e22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang N; Guo L; Huang L The Sac10b homolog from Sulfolobus islandicus is an RNA chaperone. Nucleic Acids Res. 2020, 48, 9273–9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Anderson I; Rodriguez J; Susanti D; Porat I; Reich C; Ulrich LE; Elkins JG; Mavromatis K; Lykidis A; Kim E et al. Genome sequence of Thermofilum pendens reveals an exceptional loss of biosynthetic pathways without genome reduction. Journal of Bacteriology 2008, 190, 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Guo L; Feng Y; Zhang Z; Yao H; Luo Y; Wang J; Huang L Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea. Nucleic Acids Research 2008, 36, 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Feng Y; Yao H; Wang J Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex. Protein Science : A Publication of the Protein Society 2010, 19, 1253–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang Z; Gong Y; Guo L; Jiang T; Huang L Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA. Molecular Microbiology 2010, 76, 749–759. [DOI] [PubMed] [Google Scholar]
- [63].Zhang Z; Zhao M; Wang L; Chen Y; Dong Y; Gong Y; Huang L Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA. Biochemical Journal 2017, 474, 1727–1739. [DOI] [PubMed] [Google Scholar]
- [64].Chen L; Zhang J-L; Yu L-Y; Zheng Q-C; Chu W-T; Xue Q; Zhang H-X; Sun C-C Influence of Hyperthermophilic Protein Cren7 on the Stability and Conformation of DNA: Insights from Molecular Dynamics Simulation and Free Energy Analysis. The Journal of Physical Chemistry B 2012, 116, 12415–12425. [DOI] [PubMed] [Google Scholar]
- [65].Chen L; Zheng Q-C; Zhang H-X Insights into the effects of mutations on Cren7–DNA binding using molecular dynamics simulations and free energy calculations. Physical Chemistry Chemical Physics 2015, 17, 5704–5711. [DOI] [PubMed] [Google Scholar]
- [66].Driessen RPC; Meng H; Suresh G; Shahapure R; Lanzani G; Priyakumar UD; White MF; Schiessel H; van Noort J; Dame RT Crenarchaeal chromatin proteins Cren7 and Sul7 compact DNA by inducing rigid bends. Nucleic Acids Research 2013, 41, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhang Z; Zhao M; Chen Y; Wang L; Liu Q; Dong Y; Gong Y; Huang L Architectural roles of Cren7 in folding crenarchaeal chromatin filament. Molecular Microbiology 2019, 111, 556–569. [DOI] [PubMed] [Google Scholar]
- [68].Zhang Z; Zhan Z; Wang B; Chen Y; Chen X; Wan C; Fu Y; Huang L Archaeal Chromatin Proteins Cren7 and Sul7d Compact DNA by Bending and Bridging. mBio 2020, 11, e00804–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Grote M; Dijk J; Reinhardt R Ribosomal and DNA binding proteins of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology 1986, 873, 405–413. [Google Scholar]
- [70].Wurtzel O; Sapra R; Chen F; Zhu Y; Simmons BA; Sorek R A single-base resolution map of an archaeal transcriptome. Genome Research 2010, 20, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kaur G; Iyer LM; Subramanian S; Aravind L Evolutionary convergence and divergence in archaeal chromosomal proteins and Chromo-like domains from bacteria and eukaryotes. Scientific Reports 2018, 8, 6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Robinson H; Gao Y-G; McCrary BS; Edmondson SP; Shriver JW; Wang AH-J The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 1998, 392, 202–205. [DOI] [PubMed] [Google Scholar]
- [73].Gao Y-G; Su S-Y; Robinson H; Padmanabhan S; Lim L; McCrary BS; Edmondson SP; Shriver JW; Wang AH-J The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nature Structural Biology 1998, 5, 782–786. [DOI] [PubMed] [Google Scholar]
- [74].Agback P; Baumann H; Knapp S; Ladenstein R; Härd T Architecture of nonspecific protein–DNA interactions in the Sso7d–DNA complex. Nature Structural Biology 1998, 5, 579–584. [DOI] [PubMed] [Google Scholar]
- [75].Schalch T; Duda S; Sargent DF; Richmond TJ X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 2005, 436, 138–141. [DOI] [PubMed] [Google Scholar]
- [76].Song F; Chen P; Sun D; Wang M; Dong L; Liang D; Xu RM; Zhu P; Li G Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 2014, 344, 376–380. [DOI] [PubMed] [Google Scholar]
- [77].Cai S; Böck D; Pilhofer M; Gan L The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin. Mol. Biol. Cell 2018, 29, 2450–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yan K; Yang J; Zhang Z; McLaughlin SH; Chang L; Fasci D; Ehrenhofer-Murray AE; Heck AJR; Barford D Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 2019, 574, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Makowski MM; Gaullier G; Luger K Picking a nucleosome lock: Sequence- and structure-specific recognition of the nucleosome. J Biosci 2020, 45. [PubMed] [Google Scholar]
- [80].Weaver TM; Morrison EA; Musselman CA Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 2018, 23, 2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chan JC; Maze I Nothing Is Yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends in Biochemical Sciences 2020, 45, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Arya G; Schlick T A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J Phys Chem A 2009, 113, 4045–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].White AE; Hieb AR; Luger K A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci Rep 2016, 6, 19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Basu A; Bobrovnikov DG; Qureshi Z; Kayikcioglu T; Ngo TTM; Ranjan A; Eustermann S; Cieza B; Morgan MT; Hejna M et al. Measuring DNA mechanics on the genome scale. Nature 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Probst AJ; Auerbach AK; Moissl-Eichinger C Archaea on Human Skin. PLOS ONE 2013, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Moissl-Eichinger C; Probst AJ; Birarda G; Auerbach A; Koskinen K; Wolf P; Holman HN Human age and skin physiology shape diversity and abundance of Archaea on skin. Sci Rep 2017, 7, 4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Koskinen K; Pausan MR; Perras AK; Beck M; Bang C; Mora M; Schilhabel A; Schmitz R; Moissl-Eichinger C First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 2017, 8, e00824–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Takemata N; Samson RY; Bell SD Physical and Functional Compartmentalization of Archaeal Chromosomes. Cell 2019, 179, 165–179.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]


