Summary
We developed a modified protocol, based on differential ultracentrifugation (dUC), to isolate extracellular vesicles and particles (specifically exomeres) (EVPs) from various human and murine sources, including cell lines, surgically resected tumors and adjacent tissues, and bodily fluids, such as blood, lymphatic fluid, and bile. The diversity of these samples requires robust and highly reproducible protocols and refined isolation technology, such as asymmetric-flow field-flow fractionation (AF4). Our isolation protocol allows for preparation of EVPs for various downstream applications, including proteomic profiling.
For complete details on the use and execution of this protocol, please refer to Hoshino et al. (2020).
Subject Areas: Cell culture, Cell isolation, Cell Membrane, Exosomes, Exomeres, Extracellular vesicles and particles, Isolation
Graphical abstract

Highlights
-
•
EVP isolation from human cell lines, tissues, and bodily fluids
-
•
First protocol describing EVP isolation from human tumor tissues and matched adjacent tissues
-
•
Improved EVP isolation and total EVP protein yield by dissecting tissues into pieces
-
•
AF4 fractionation to further analyze EVP samples
We developed a modified protocol, based on differential ultracentrifugation (dUC), to isolate extracellular vesicles and particles (specifically exomeres) (EVPs) from various human and murine sources, including cell lines, surgically resected tumors and adjacent tissues, and bodily fluids, such as blood, lymphatic fluid, and bile. The diversity of these samples necessitates robust and highly reproducible protocols and refined isolation technology, such as asymmetric-flow field-flow fractionation (AF4). Our isolation protocol allows for preparation of EVPs for various downstream applications, including proteomic profiling.
Before you begin
This protocol describes the isolation of small and heterogeneous EVPs from tissue culture supernatants (conditioned media) derived from cell lines or tissue explants as well as bodily fluids, such as blood, lymphatic fluid, and bile. Below are listed the different sample types, their generation, and pre-processing before EVP isolation.
Cell culture conditions
Timing: 3–4 days
All cell lines are mycoplasma-tested prior to EVP isolation.
-
1.Prepare appropriate cell culture media, i.e., RPMI1640 or DMEM supplemented with penicillin (100 U/mL), streptomycin (100 mg/mL) and 10% EVP-depleted fetal bovine serum (FBS).
-
a.We routinely prepare our own EVP-depleted fetal bovine serum (FBS). FBS is depleted of EVPs by ultracentrifugation at 100,000 × g for 70 min at 10°C and sterile-filtered through a 0.2 μm low protein-binding membrane. The EVP-depleted FBS can be stored in aliquots at −20°C (short term) and −80°C (long term).
-
a.
-
2.
Culture cells at 37°C, 5% CO2 in EVP-depleted media for 3–4 days until 80% confluence.
-
3.
Collect conditioned culture media for EVP isolation. See Table 1 for typical cell culture conditions and EVP yields.
CRITICAL: EVP-depleted FBS is also commercially available (Gibco #A2720801). We recommend using the EVP-depleted FBS prepared using exactly the same method or from the same commercial provider for any comparison study and to test each lot for cell growth and EVP production in a cell culture system.
CRITICAL: Avoid fully confluent cell cultures and maintain consistent culture conditions. For each cell line seeding number/density and culture time (48 to 72 h) need to be empirically determined depending on growth rate. If using trypsin to harvest adherent cells, remember to wash it off before seeding. Assess cell viability using a suitable method, i.e., manual cell counting and trypan blue exclusion, automatic cell counting with dead cell discrimination or flow cytometry with annexin V or propidium iodide, at the time of media collection.
Table 1.
Examples of typical cell culture conditions and EVP yields
| Cell line | Type of culture dish, growth area (cm2) | Cultured media volume (mL) | Cell number at seeding | Total EVP protein yield |
|---|---|---|---|---|
| MDA-MB-231 | T175 flask, 175 cm2 | 25–30 mL | 2.0 × 106 cells | 14.0 μg |
| MDA231-LM2-4175 | T175 flask, 175 cm2 | 25–30 mL | 1.8 × 106 cells | 17.0 μg |
| 4T1 | 150 mm plate, 148 cm2 | 25–30 mL | 2.0 × 106 cells | 50.0 μg |
| PANC-1 | 150 mm plate, 148 cm2 | 25–30 mL | 8.3 × 106 cells | 20.4 μg |
| AsPC-1 | 150 mm plate, 148 cm2 | 25–30 mL | 9.0 × 106 cells | 26.0 μg |
Abbreviation: EVP, extracellular vesicle and particle
Tissue explant culture reagents and tools
Timing: 30 min
-
4.Prepare explant culture media: RPMI1640 supplemented with penicillin (100 U/mL) and streptomycin (100 mg/mL).
-
a.Media needs to be warmed to ~15°C–20°C prior to use.
-
a.
-
5.
Sterilize surgical instruments (forceps, scalpel holder and scissors) by autoclaving (120°C, 40 min, dry cycle), otherwise use disposable instruments.
CRITICAL: For short-term (<16–18 h) tissue explant cultures use serum-free media to avoid introducing bovine proteins and EVPs into the preparations. EVP-depleted media can also be used depending on downstream applications and experimental aim, especially for long-term cell cultures, such as explant outgrowth or organoid culture.
Harvest of murine organs for EVP yield optimization
Timing: approximately 20–30 min per mouse
-
6.Mouse organ harvesting.
-
a.Mice should be anesthetized with isoflurane (3.0% isoflurane at 2.0 L/min oxygen) delivered through a nose cone and placed in a supine position before perfusing.
-
b.Perfuse mouse organs and flush blood out by injecting a bolus of 30 mL PBS/5 mM EDTA into the left ventricle using a 26 G × 1/2 needle and a 30 mL syringe.
-
c.Remove the organs of interest under aseptic conditions and immerse them in ice-cold sterile PBS; place on ice until further dissection. Dissect organs in the biosafety cabinet.
-
a.
Optional: Organ perfusion and harvesting can be performed after CO2 euthanasia, or other IACUC approved methods, such as terminal anesthesia (pentobarbital), if an efficient workflow can be maintained.
CRITICAL: A minimum of 5–10 mice is required to isolate EVPs from normal organs. Tumor-bearing organs have higher EVP yields and 2–3 animals could be enough, depending on tumor type and size. Matched organs should be pooled to obtain sufficient amounts of EVPs for further characterization, such as nanoparticle tracking analysis (NTA), electron microscopy (EM), and protein quantification (Bicinchoninic Acid Assay - BCA). Timely processing of tissues within 30 min of harvest is required to ensure maximal EVP yield.
Collection of blood and other bodily fluids
Timing: 10 min (blood samples), sample-dependent for other bodily fluids
Blood collection from patients and healthy subjects should typically be performed in the morning, or after fasting. Phlebotomy training is required to perform the peripheral blood draw.
Bile collection can be performed by needle cannulation of the common bile duct at the time of surgery. Bile is snap-frozen and stored at −80°C until analysis.
Lymphatic fluid can be collected after radical lymphadenectomy from routinely used collection drainage. To ensure that the sample of lymph fluid does not contain any surgical debris, only the fluid released between 24 and 48 h should be collected (the fluid collected in the first 24 h is discarded).
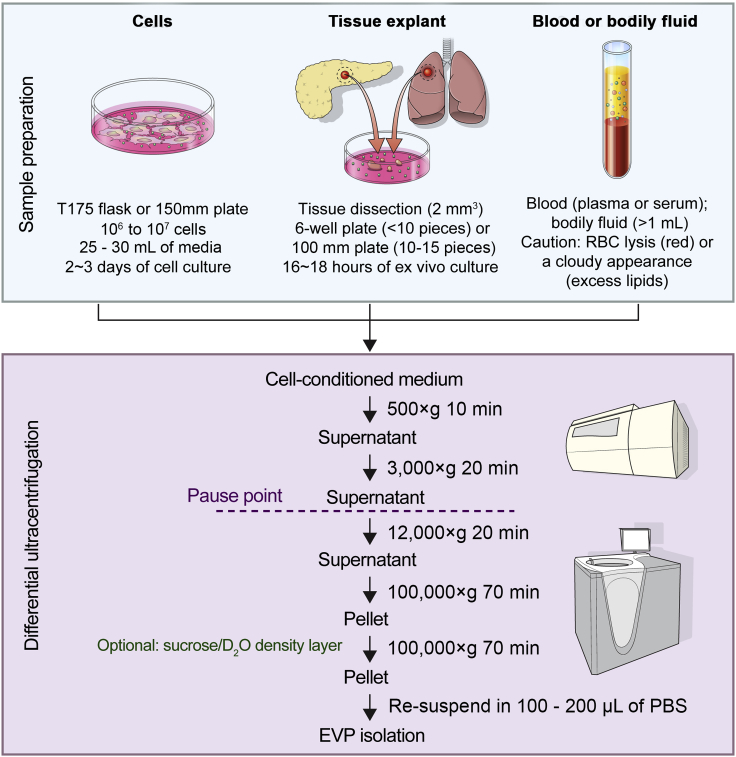
Downstream analysis of bodily fluid-derived EVPs follows the same isolation steps (Figure 1).
-
7.Collect the venous blood sample in EDTA (purple top) blood collection tube(s).
-
a.Invert the tube slowly, several times, immediately upon collection, to mix the blood with the anticoagulant and prevent clot formation.Note: One collection tube of 10 mL of whole blood typically yields approximately 5 mL of plasma, which is usually sufficient for downstream applications, such as BCA, NTA, EM, proteomic mass spectrometry (see further details in “Expected outcomes” section). We recommend a minimum starting volume of no less than 1 mL of bodily fluid to ensure sufficient EVP yield.
-
b.Place the tube of blood on ice to keep it cold during eventual transport.
 CRITICAL: Occasionally invert the blood collection tube to avoid red blood cell lysis or clotting during transport.Note: Blood collection from mice can be performed from the retro-orbital capillary sinus by using a heparinized capillary tube. Alternatively, blood can be collected via cardiac puncture in syringes containing anti-coagulants at the time of sacrifice. Approximately 50% of the blood volume will be represented by plasma and murine blood should be processed in the same manner as described for human samples. Cardiac puncture allows collection of 0.5–1.2 mL of blood per animal. Retro-orbital collection allows for around 0.5 mL. We recommend to use a minimum 1 mL of plasma which yields approximately 5 μg of EVPs. It is thus necessary to combine the plasma of multiple mice to obtain enough plasma EVPs for analysis.
CRITICAL: Occasionally invert the blood collection tube to avoid red blood cell lysis or clotting during transport.Note: Blood collection from mice can be performed from the retro-orbital capillary sinus by using a heparinized capillary tube. Alternatively, blood can be collected via cardiac puncture in syringes containing anti-coagulants at the time of sacrifice. Approximately 50% of the blood volume will be represented by plasma and murine blood should be processed in the same manner as described for human samples. Cardiac puncture allows collection of 0.5–1.2 mL of blood per animal. Retro-orbital collection allows for around 0.5 mL. We recommend to use a minimum 1 mL of plasma which yields approximately 5 μg of EVPs. It is thus necessary to combine the plasma of multiple mice to obtain enough plasma EVPs for analysis.
-
a.
Figure 1.
Workflow for EVP isolation from cell culture, tissue explants, and bodily fluids
Preparation of the AF4 instrument
Timing: 30 min
EVPs can be further separated into exomeres and distinct extracellular vesicle subpopulations by utilizing asymmetric-flow field-flow fractionation (AF4) technology (Zhang et al., 2018). A detailed protocol encompassing the working principles, AF4 method development and troubleshooting, comparison with other technologies, limitations, and applications has been recently reported (Zhang and Lyden, 2019). Here, we will focus our method description on preparation of the instruments prior to fractionation and sample processing and recovery. In brief, we use the Eclipse AF4 (Wyatt Technology) system to fractionate samples in a short channel (144 mm length, Wyatt Technology). The channel is equipped with a 10 kDa MWCO Regenerated Cellulose membrane (Millipore) on the accumulation bottom wall and a 490 mm spacer. An Agilent Iso pump, autosampler, and fraction collector are included for running buffer, loading samples, and collecting fractions, respectively. Online real-time monitors include the Agilent Multiple Wavelength Detector (280 nm) and Wyatt Technology DAWN HELEOS II (multi-angle light scattering-MALS detector, at 664 nm) with QELS (dynamic light scattering-DLS detector) installed at detector 12 (100°).
Optional: Assemble a new membrane for the short channel.
-
8.
Equilibrate the system by running PBS at 1 mL/min for a minimum of 30 min.
-
9.
Switch on lasers for all monitors ~15 min prior to sample loading.
-
10.
Switch on thermostats for both the autosampler and fraction collector and set temperature at 4°C, keeping samples and fractions at 4°C throughout the whole fractionation process.
Optional: Coat the membrane with BSA by running 30–40 μg BSA 2–3 times using the following method (this step is only needed when a new membrane is installed).
- Forward channel flow: 1 mL/min
- Elution, 2 min, Vx (cross flow) 1.5 mL/min;
- Focus, 1 min, Vx 1.5 mL/min;
- Focus + Injection, 2 min, Vx 1.5 mL/min, injection flow 0.2 mL/min;
- Elution, 15 min, Vx 3 mL/min;
- Elution, 5 min, Vx 0 mL/min;
- Elution + Injection, 5 min, Vx 0 mL/min;
-
11.
Pre-chill 96-well fraction collection plates by storing them in a cold room before use. Before starting the fractionation process, install two plates in the designated positions (pre-set in the Chemstation operating program).
CRITICAL: For long-term storage (>1-2 weeks), the system should be maintained in 20% ethanol. Flushing the system first with a large volume of water is essential before switching to PBS buffer to avoid precipitation. The PBS buffer should be filtered through 0.1 or 0.2 μm filter units. An inline filter can be used as well. For short-term storage (<1-2 weeks), the system can be stored in water (preferred) or PBS and run in a Night Rinse mode (0.2 mL/min).
Note: The short channel membrane can perform well without a notable difference in particle separation resolution for multiple runs. A new membrane should be installed when obvious shifting of the elution peaks and/or noisy baseline occurs.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human cancer and adjacent normal tissues, blood, bile, and lymphatic fluid | Memorial Sloan Kettering Cancer Center and Yonsei Cancer Center | n/a |
| Chemicals, peptides, and recombinant proteins | ||
| Sterile PBS | VWR | Cat#45000-446 |
| BSA | Sigma | Cat#A1900 |
| Premium-grade FBS | VWR | Cat#97068-085 |
| Gibco fetal bovine serum, exosome-depleted | Fisher Scientific | Cat#A2720801 |
| RPMI | Corning | Cat#10-040-CV |
| DMEM | Corning | Cat#10-013-CV |
| L-Glutamine, 100× | Corning | Cat#25-005-CI |
| Penicillin–streptomycin solution, 50× | Corning | Cat#30-001-CI |
| TrypLE | Thermo Fisher Scientific | Cat#12604-039P |
| UltraPure 0.5 M EDTA, pH 8.0 | Corning | Cat#15575020 |
| Isoflurane solution (formerly IsoThesia) | Covetrus | Cat#11695067772 |
| Deuterium oxide | Sigma | Cat# 1133660500 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| Experimental models: cell lines | ||
| Human: MDA-MB-231 | ATCC | HTB-26 |
| Human: MDA-MB-4175 | Dr. J. Massague (Minn et al., 2005) | n/a |
| Human: PANC-1 | ATCC | CRL-1469 |
| Human: AsPC-1 | ATCC | CRL-1682 |
| Mouse: B16-F10 | ATCC | CRL-6475 |
| Mouse: 4T1 | ATCC | CRL-2539 |
| Experimental models: organisms/strains | ||
| Mouse: wild-type BALB/c | Jackson Laboratory | Cat#000651 |
| Mouse: wild-type C57BL/6 | Jackson Laboratory | Cat#000664 |
| Software and algorithms | ||
| Astra 6 with an Eclipse module integrated to operate flow | Wyatt Technology | n/a |
| ChemStation | Agilent Technologies | n/a |
| Other | ||
| Ultracentrifuge | Beckman Coulter https://www.beckmancoulter.com/ | Optima XE/XPE, Cat#A94469 |
| Fixed-angle rotor 50.4Ti | Beckman Coulter | Cat# 347299 |
| Fixed-angle rotor 70Ti | Beckman Coulter | Cat# 337922 |
| Fixed-angle rotor 45Ti | Beckman Coulter | Cat# 339160 |
| Ultracentrifugation tube (45Ti rotor) | Beckman Coulter | Cat#355628 |
| Ultracentrifugation tube (70Ti rotor) | Beckman Coulter | Cat#355631 |
| Ultracentrifugation tube (50.4Ti rotor) | Beckman Coulter | Cat#355645 |
| 500 mL Supor MachV PES filter units | VWR | Cat#73520-984 |
| Integra disposable scalpel blades no. 10 | Miltex | REF 4-410 |
| Mouse Surgical Kit | Kent Scientific | INMOUSEKIT |
| 26 G × 1/2 in PrecisionGlide needle | Becton Dickinson | Cat#305111 |
| Cell strainer, 40 μm | Corning | Cat#CLS431750 |
| Millex-GV Filter, 0.22 μm | Millipore | Cat# SLGVV255F |
| 96-well plate | VWR | Cat#62406-081 |
| Blue screw caps | Agilent | Cat#5182-0717 |
| Screw cap vials | Agilent | Cat#5182-0714 |
| Vial insert, 250 μL pulled point glass | Agilent | Cat#5183-2085 |
| 96-well plate, 1.0 mL, polypropylene | Agilent | Cat#8010-0534 |
| Sealing tape, clear polyolefin | Thermo Fisher Scientific | Cat#232701 |
| Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-30 membrane | Millipore | Cat#UFC903024 |
| Amicon Ultra-4 Centrifugal Filter Unit with Ultracel-30 membrane | Millipore | Cat#UFC803024 |
| Millipore Reg. Cellulose membrane 10KD SC | Wyatt technology | Cat#4057 |
| Inline filter membrane (0.1 μm) | Wyatt Technology | Cat#1871 |
| 1260 Infinity analytical- and preparative-scale fraction collectors | Agilent | Cat# G1364C |
| 1290 thermostat | Agilent | Cat#G1330B |
| 1260 Infinity standard autosampler | Agilent | Cat#G1329B |
| 1260 Infinity multiple wavelength detector | Agilent | Cat#G1365D |
| 1260 Infinity isocratic pump | Agilent | Cat#G1310B |
| TG-14 HPLC vacuum degasser | GASTORR | Cat# TG-14 |
| DAWN HELEOS II with QELS installed at detector 12 (100°) | Wyatt Technology | n/a |
| Eclipse AF4 | Wyatt Technology | n/a |
| Short channel | Wyatt Technology | n/a |
| Tabletop Heraeus Multifuge x3R centrifuge | Thermo Fisher Scientific | Cat#75004501 |
| Microcentrifuge | Eppendorf | Cat#5424R |
| AccuScan GO UV/vis microplate spectrophotometer | Fisher Scientific | Cat#14-377-579 |
| BD microtainer, capillary blood collection tube hematology K2 EDTA additive | Becton Dickinson | Cat#365974 |
| Micro-hematocrit capillary tubes | Kimble Chase | Cat#41B2501 |
| BD Vacutainer Red Top blood collection tube | Becton Dickinson | Cat#366434G |
Materials and equipment
Sucrose/deuterium oxide mixture for density purification
A sucrose/deuterium oxide mixture can be used to further purify EVP samples by a density cushion.
| Reagent | Final concentration | Amount |
|---|---|---|
| Sucrose | 30% | 15 g |
| Tris (Trizma base, Sigma) | 20 mM | 1.2 g |
| D2O | n/a | Fill to 48 mL |
| HCl | n/a | 1–2 mL |
| Total | n/a | 50 mL |
To prepare the sucrose/deuterium oxide mixture (density 1.210 g/cm3), weigh sucrose and Tris and add D2O. Shake until completely dissolved and measure the pH of the mixture. Adjust the pH to 7.4 with HCl and correct the volume to 50 mL. Filter the mixture through a 0.22 μm filter into a sterile tube. The prepared sucrose mixture will be stable for one month at 4°C. After this period, measure the pH and adjust to 7.4.
CRITICAL: HCl is highly corrosive and irritant and can cause serious eye and skin damage and respiratory irritation. Handle HCl in a chemical hood and use protective clothing, goggles, and gloves.
Note: We recommend keeping ultracentrifuge rotors in 4°C cold rooms and setting the temperature of the ultracentrifuge to 10°C to extend the life expectancy of the machine.
Alternatives: Alternatives to ultracentrifugation with sucrose/deuterium oxide cushion for further clean-up and purification of EVP samples are ultrafiltration, size-exclusive chromatography, two-phase isolation, magnetic capture, immunoprecipitation, density gradient (such as Optiprep) and microfluidic devices. The outcome of these alternatives has to be determined by the end user. As an alternative for fixed-angle rotors mentioned in this protocol, swing buckets could also be used but specific handling, such as supernatant removal and pellet re-suspension will have to be optimized by the user.
Step-by-step method details
Isolation of EVPs from cell culture
Timing: 3.5–4 h (time increases with number of samples)
Perform serial centrifugation and ultracentrifugation steps to isolate EVPs from conditioned cell culture media.
-
1.Harvest culture media and remove cells, debris, apoptotic bodies, and contaminants through serial centrifugation.
-
a.Centrifuge sample at 500 × g for 10 min at 4°C to pellet cells and debris.
-
b.Collect the supernatant without disturbing the cell/debris pellet. This can be done by quickly pouring off the supernatant into a new container/tube or carefully pipetting off the supernatant. It is essential to work as quickly as possible during these steps, as the pellet will become dislodged over time.Note: For this and all subsequent steps, supernatants and re-suspended EVP pellets should be kept on ice between isolation steps.Note: A step of 3,000 × g centrifugation for 20 min at 4°C should be added for cell culture supernatants before short-term storage (<24 h) at 4°C or before freezing to remove additional cell debris contaminants. This step has to be performed if further analysis of the post-12,000 × g microvesicle sample is planned.
 Pause Point: Conditioned media supernatants can be stored at −80°C for long-term storage or at 4 °C for short-term (<24 h) storage before ultracentrifugation. Minor differences have been observed between EVPs isolated from fresh versus frozen samples in terms of yield and purity. However, frozen samples are often used to allow for retrospective studies, collaborative studies between different institutions and batch processing of test samples/subjects and controls.
Pause Point: Conditioned media supernatants can be stored at −80°C for long-term storage or at 4 °C for short-term (<24 h) storage before ultracentrifugation. Minor differences have been observed between EVPs isolated from fresh versus frozen samples in terms of yield and purity. However, frozen samples are often used to allow for retrospective studies, collaborative studies between different institutions and batch processing of test samples/subjects and controls. -
c.Centrifuge the supernatant at 12,000 × g for 20 min at 4°C to remove apoptotic bodies and aggregates and larger vesicles that could otherwise be co-isolated with EVPs and “contaminate” the EVP preparation.Note: This step can be performed in the ultracentrifuge or tabletop centrifuge depending on volume and availability of the ultracentrifuge. We recommend performing the 12,000 × g spin right before ultracentrifugation since contaminating material could develop by freeze-thaw cycles or extended storage times.
-
d.Collect supernatant without disturbing the pellet and transfer to new/clean ultracentrifuge tube by pipetting, leaving a small volume behind to avoid contamination from pellet.
 CRITICAL: Wash newly purchased ultracentrifugation tubes with soap and water and rinse them extensively in ddH2O before the first EVP isolation, since the factory coating on the inside of the tube will prevent the EVP pellet from adhering to the tube. After use, the tubes should be washed extensively and sterilized by autoclaving if required. Cleaning of ultracentrifugation tubes was performed as per standard cleaning procedure for laboratory glassware and utensils used for molecular and cellular biology applications. We extensively wash the tubes in soap and hot water while scrubbing with a brush. The tubes are then thoroughly rinsed with tap water to remove any traces of soap, followed by additional rinses in ddH2O. If EVPs are isolated for in vitro or in vivo work rather than molecular biology, the tubes can be autoclaved, this however shortens the life of the tubes. No contamination between samples has ever been observed using these procedures. The tube used depends on the volume of starting material rather than the type of specimen.Note: It is important to avoid disturbing the pellet with the pipette tip in these centrifugation steps since the aim is to remove pelleted contamination; instead, leave a minimal amount of media above the pellet to avoid dissociating the pellet. Remember to work as quickly as possible during these steps as the pellet will become dislodged over time.
CRITICAL: Wash newly purchased ultracentrifugation tubes with soap and water and rinse them extensively in ddH2O before the first EVP isolation, since the factory coating on the inside of the tube will prevent the EVP pellet from adhering to the tube. After use, the tubes should be washed extensively and sterilized by autoclaving if required. Cleaning of ultracentrifugation tubes was performed as per standard cleaning procedure for laboratory glassware and utensils used for molecular and cellular biology applications. We extensively wash the tubes in soap and hot water while scrubbing with a brush. The tubes are then thoroughly rinsed with tap water to remove any traces of soap, followed by additional rinses in ddH2O. If EVPs are isolated for in vitro or in vivo work rather than molecular biology, the tubes can be autoclaved, this however shortens the life of the tubes. No contamination between samples has ever been observed using these procedures. The tube used depends on the volume of starting material rather than the type of specimen.Note: It is important to avoid disturbing the pellet with the pipette tip in these centrifugation steps since the aim is to remove pelleted contamination; instead, leave a minimal amount of media above the pellet to avoid dissociating the pellet. Remember to work as quickly as possible during these steps as the pellet will become dislodged over time.
-
a.
-
2.Isolate EVPs by ultracentrifugation
-
a.Centrifuge supernatant at 100,000 × g for 70 min at 10°C in a fixed-angle rotor in a Beckman Coulter ultracentrifuge (see Materials and equipment) to collect EVPs.
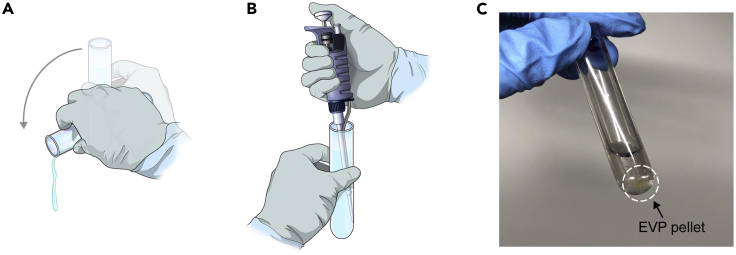
 CRITICAL: It is very important to accurately balance the tubes in the ultracentrifuge. An unbalanced rotor can cause spillage, contamination, run failure, or even damage the rotor/ultracentrifuge.Note: Before inserting tubes into the rotor, we recommend marking an area on the bottom of the tube where the pellet is expected to form, since it may not be easily visible. Avoid disturbing the pelleted area when discarding the supernatant and pay careful attention to it for successful retrieval of your EVP pellet (Figure 2).
CRITICAL: It is very important to accurately balance the tubes in the ultracentrifuge. An unbalanced rotor can cause spillage, contamination, run failure, or even damage the rotor/ultracentrifuge.Note: Before inserting tubes into the rotor, we recommend marking an area on the bottom of the tube where the pellet is expected to form, since it may not be easily visible. Avoid disturbing the pelleted area when discarding the supernatant and pay careful attention to it for successful retrieval of your EVP pellet (Figure 2). -
b.Discard the supernatant by decanting or carefully pipetting without disturbing the pellet. If pipetting off the supernatant, keep the tip of the pipet on the side of the tube opposite from the pellet. If decanting the supernatant, be sure to pour the supernatant off the side of the tube opposite the pellet and use one quick, careful motion to invert the tube.Optional: Store supernatant if interested in EVP-depleted media for measurement of other secreted factors, such as cytokines.Note: In this and subsequent steps it is important to work quickly and remove the supernatant as fast as possible after the ultracentrifuge has stopped to prevent dissociation of the pellet. After removing the supernatant, leave the pellet untouched and place the inverted tube in a rack for 1–3 min to avoid backflow of liquid. This ensures minimal carryover of contaminants and prevents dissociation of the pellet. Remove drops of liquid from tube walls by vacuum aspiration or using a Kimwipe, if necessary. Troubleshooting 1.
-
c.To wash the pellet, re-suspend it in 500 μL of cold PBS by pipetting and flushing the marked area at the bottom of the ultracentrifugation tube. Ensure the pellet is re-suspended well so contaminants trapped in the pellet (free, abundant proteins) are washed away. Transfer the re-suspended pellet to a new ultracentrifuge tube and add 3 mL (tube #355645), 20 mL (tube #355631) or 44 mL (tube #355628) of cold PBS. We recommend using the same size tube throughout the protocol (i.e., depending on starting volume) since the starting volume affects the yield. Thus, small volumes, 1–3 mL should be spun in a small tube where these fewer EVPs can more readily be collected and concentrated. Troubleshooting 2.
-
d.Centrifuge the re-suspended pellet at 100,000 × g for 70 min at 10°C to collect the washed EVP pellet.
-
e.Discard the PBS supernatant by decanting and remove all liquid by gently tapping the inverted tube to avoid backflow.
-
f.Re-suspend the EVP pellet in 100 μL (see note for range below) of cold PBS and store at −80°C. Reserve 10 μL aliquots for downstream quantification and characterization, such as protein quantification, NTA, and EM, to avoid freeze-thaw cycles.
-
a.
Note: The final re-suspension volume can be adjusted to create an appropriate concentration. As low as 50 μL or up to 500 μL can be used for high yields depending on the total EVP yield. Because cell lines vary in their EVP production, we recommend that users empirically establish their own EVP yield for each cell type. Final concentrations will depend on downstream applications but should be above the lower limit of detection of the BCA assay or other assay for protein quantification. If desired, the EVP pellet can be re-suspended directly in cell lysis buffer for downstream RNA or protein applications.
Optional: After thawing of stored EVP samples and before downstream applications, depending on the type of downstream analysis, an additional centrifugation at 12,000 × g for 5 min at 4°C can be performed to remove potential aggregates/precipitates formed during storage.
Figure 2.
Careful handling of the EVP pellet
(A) After ultracentrifugation, carefully decant supernatant and (B and C) re-suspend the pellet in a small volume of PBS (0.5–1 mL depending on tube size) by pipetting and flushing the marked area of the lower side of the ultracentrifugation tube. Avoid generating bubbles.
Isolation of EVPs from tissue samples
Timing: 2 days
This section describes culture of dissected tissue explants and serial centrifugation steps to isolate EVPs from conditioned media. Fresh human tumor tissues and peritumoral adjacent tissues were obtained from patients surgically treated at Memorial Sloan Kettering Cancer Center (MSKCC) and Yonsei Cancer Center (YCC). The ages of patients ranged from 3 to 88 years old. All individuals or legal guardians provided informed consent for tissue donation according to protocols approved by the institutional review board of MSKCC, WCM, and YCC (IRB 11-033A, 16-774, 16-1514, and 15-015, MSKCC; IRB 0604008488, WCM; IRB 4-2019-0811, YCC). The study is compliant with all relevant ethical regulations regarding research involving human participants. All mouse experiments were performed in accordance with Institutional Animal Care and Use Committees (IACUC) and American Association for Laboratory Animal Science (AAALAS) guidelines (Weill Cornell Medicine animal protocol 0709-666A). Six to eight weeks old wild-type C57BL/6 or BALB/c female mice were purchased from Jackson Laboratories.
-
1.
Collect the tumor tissue and adjacent tissue in ice-cold PBS at the time of surgery and transfer them on ice to the class II biosafety cabinet within 30 min.
Optional: Tissue weight can be used to normalize the EVP yield. Work quickly or in PBS to avoid drying out tissue and decreasing EVP yield.
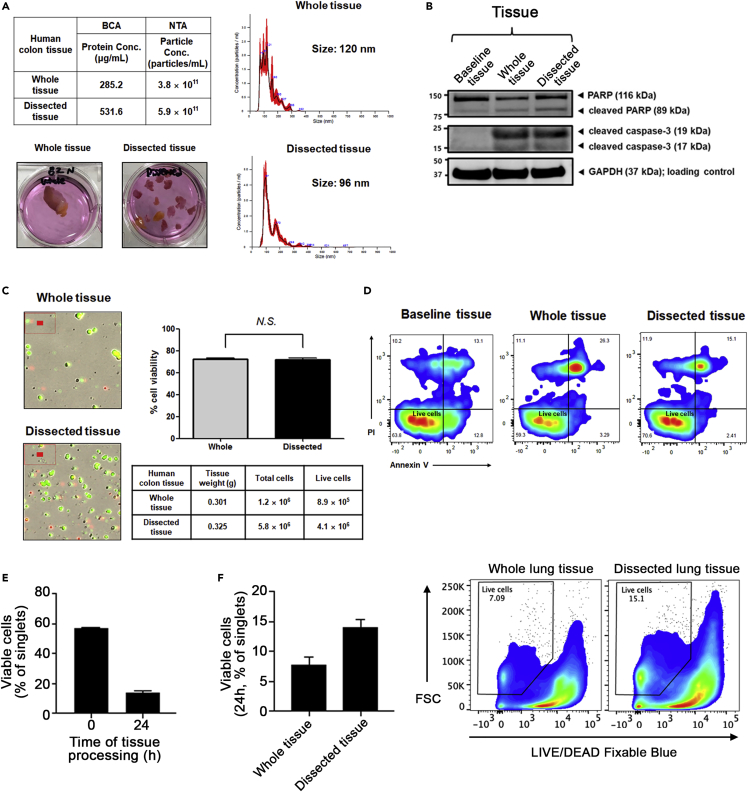
Note: Viability will vary between tissue explant cultures depending on a number of factors, such as surgical approach, time to culture, and tissue composition. Viability can be assessed using suitable methods such as manual cell counting and trypan blue exclusion, automatic cell counting with dead cell discrimination dyes or flow cytometry with annexin V or propidium iodide. The process of making single cell suspensions can also affect the viability. We have compared our dissected tissue explants to whole tissue explants and found similar, slightly enhanced, viability in the dissected cultures (Figure 3).
Figure 3.
Comparison of cell viability between whole tissue versus tissue dissected into smaller pieces
(A) Nanosight (NTA) profiles and protein concentration by bicinchoninic acid (BCA) assay for human colon that was cultured as whole tissue or dissected into smaller pieces.
(B) Immunoblotting of lysates (20 μg of proteins) from baseline tissue (directly after collection, timepoint 0 h) immediately placed into lysing buffer without culturing, whole tissue, and dissected tissue (after 12 h culture) derived from the human colon. PARP antibody (#9542, Cell Signaling Technology [CST]), cleaved caspase-3 antibody (#9661S, CST), GAPDH antibody (#2118S, CST) were used for immunoblotting.
(C) Comparison of human colon whole tissue and dissected tissue cell viability as determined by dual Acridine Orange / Propidium Iodide fluorescence and brightfield optics, using the LUNA automated cell counter. Live cells are green and dead cells are red.
(D) Comparison of human colon whole tissue and dissected tissue cell viability by apoptotic flow staining shows similar viability between baseline tissue directly after collection and whole and dissected tissues after 12 h tissue explant culture.
(E) Comparison of cell viability between baseline and 24 h cultures of mouse lung tissue explant.
(F) Cell viability, as determined by flow cytometry with LIVE/DEAD™ Fixable Blue Dead Cell Stain, shows increased viability in dissected tissue cultures compared to whole tissue culture from mice.
-
2.
Wash tissues in cold PBS × 3 and quickly cut the tissues into 2 mm3-sized pieces. Rinse tissue pieces again in PBS before plating if needed to remove debris and/or blood.
Note: Heterogeneous tumor and adjacent tissues might require dissection of specific areas of interest, such as removal of adipose tissue, tumor and dissection of specific structures (i.e. tumor margin), and thus, trained personnel is required.
-
3.
Place tissue pieces in a 6-well plate (<10 pieces) or a 10-cm dish (10–15 pieces).
CRITICAL: It is important to limit the number of dissected tissues per dish to no more than 15 pieces, which will minimize loss of tissue viability.
-
4.
If possible, allow tissue pieces to adhere to the plastic dishes for a few minutes before adding media. Pipet media slowly against the side of the dish to avoid disturbing the tissue pieces. Ensure enough media is added to cover the tissue pieces and try to minimize detachment from the plate.
Note: After initial media collection, consider adding EVP-depleted FBS and other essential growth factors to culture media if establishing long-term viable cultures is desired. Alternatively, organoids can be generated from the fresh tissue and EVPs can be continuously collected from these organoid cultures.
-
5.
Carefully transfer plates to 37°C incubator with 5% CO2.
Note: Keep in a separate incubator reserved for ex vivo work, away from routine mycoplasma-free cell cultures, if feasible.
-
6.
After 16–18 h, carefully collect culture media without disturbing tissue explants, especially if continuous culturing is planned. Keep the collected media on ice.
-
7.Centrifuge explant culture media according to the centrifugation steps described above for EVP isolation from cell cultures.
-
a.Centrifuge the culture media at 500 × g for 10 min at 4°C. Remove supernatant to a new tube.
-
b.Centrifuge the supernatant at 3,000 × g for 20 min at 4°C. Remove supernatant to a new tube.
-
c.Centrifuge the supernatant at 12,000 × g for 20 min at 4°C, in either a tabletop centrifuge or in the ultracentrifuge, depending on volume.
-
a.
CRITICAL: Given the complex nature of tissue samples and the presence of significant debris compared to cell lines, always perform all three pre-ultracentrifugation steps (500 × g, 3,000 × g, and 12,000 × g) mentioned above to remove insoluble material. Additional filtrations and purification steps can be added as described below.
-
8.
Isolate EVPs by ultracentrifugation at 100,000 × g for 70 min at 10°C.
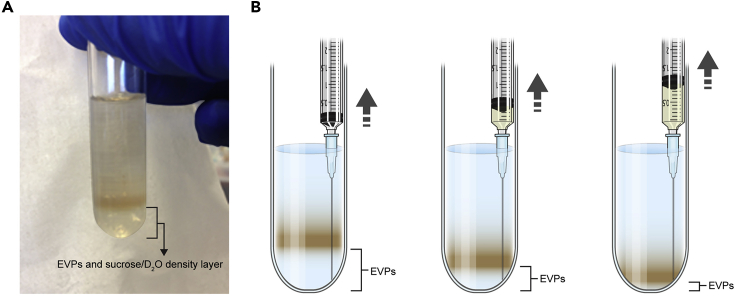
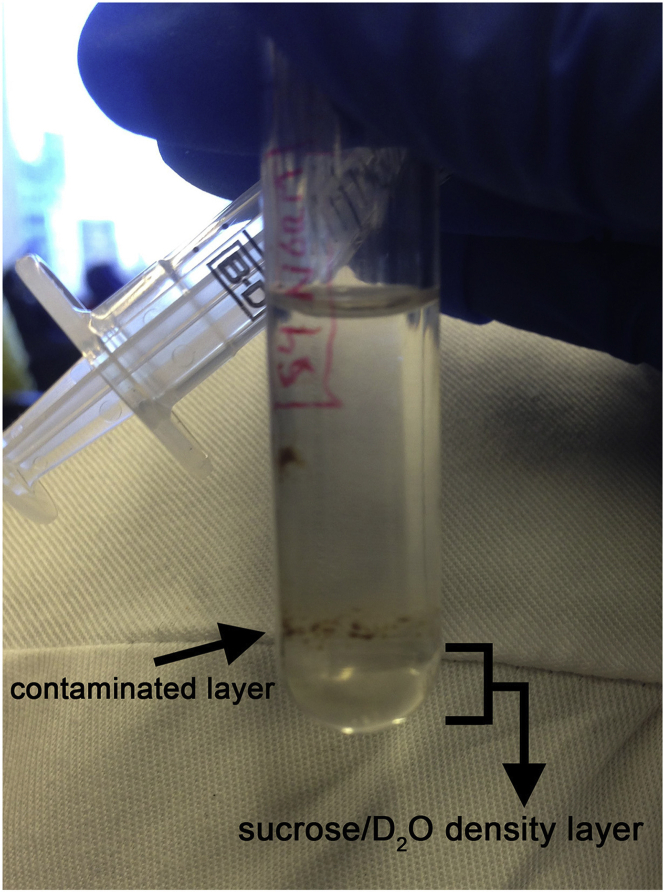
Optional: If further purification of a tissue sample EVP pellet is desired, re-suspend the EVP pellet in 2.5 mL of PBS and slowly overlay onto a 0.5 mL sucrose/deuterium oxide cushion layer by tilting the tube to 45°; then centrifuge at 100,000 × g for 70 min at 10°C (Figure 4). Aspirate the lower fraction using an 18 G needle and 3 mL syringe. Ensure not to disturb the layers or eventual pellet and collect a volume similar to the amount of sucrose/deuterium oxide mixture added (0.5 mL here). Note: the amount of sucrose/deuterium oxide mixture will depend on the volume of the sample/ultracentrifugation tube size used and should be used in a ratio of 1:5 sucrose/D2O:EVP/PBS suspension (Lamparski et. al, 2002). The end user will need to confirm that sucrose density-cushion purified EVPs retain their functional characteristics. Troubleshooting 3.
Figure 4.
Ultracentrifugation of tissue sample EVPs over a sucrose/deuterium oxide density layer
(A) EVPs re-suspended in PBS could be further purified by overlaying onto a sucrose/deuterium oxide cushion and centrifuging at 100,000 × g, resulting in a lower EVP-enriched layer that could be aspirated from the upper layer containing any potential contaminants.
(B) Aspiration of the lower fraction using an 18G needle and 3 mL syringe.
-
9.
Re-suspend EVP pellet in 3 mL of PBS to wash and collect by ultracentrifugation at 100,000 × g for 70 min at 10°C.
-
10.
Discard the supernatant and re-suspend collected EVP pellet in 100 μL of PBS.
-
11.
Store at −80°C. Save 10 μL aliquots for protein quantification, NTA, and EM to avoid freeze-thaw cycles.
Isolation of EVPs from blood samples and other bodily fluids
Timing: 4 h
This section describes differential centrifugation steps to isolate EVPs from whole blood or bodily fluids, such as lymphatic fluid, and bile. Human blood and bodily fluids were obtained from patients treated at Memorial Sloan Kettering Cancer Center (MSKCC). The ages of patients ranged from 1 to 83 years old. All individuals or legal guardians provided informed consent for blood/fluid donation according to protocols approved by the institutional review board of MSKCC (IRB 11-033A, 16-774, 16-1514, and 15-015; MSKCC, IRB 0604008488, WCM). The study is compliant with all relevant ethical regulations regarding research involving human participants. All mouse experiments were performed in accordance with Institutional Animal Care and Use Committees (IACUC) and American Association for Laboratory Animal Science (AAALAS) guidelines (Weill Cornell Medicine animal protocol 0709-666A). Six to eight weeks old wild-type C57BL/6 or BALB/c female mice were purchased from Jackson Laboratories.
-
1.Remove cells, debris, apoptotic bodies, and other contaminants by serial centrifugation steps.
-
a.Centrifuge the EDTA tube containing the blood at 500 × g for 10 min at 4°C. The blood will separate into a lower layer containing red blood cells (RBC) and white blood cells (WBC) and an upper layer of plasmaNote: The plasma should be yellow and clear but not red, which is a sign of RBC lysis at collection that could potentially affect and contaminate the EVP preparation. Following blood collection in anti-coagulant-coated tubes, we follow the manufacturer’s protocol, and roll/invert tubes several times, to ensure the blood is mixed with the anti-coagulant, thus minimizing clotting and hemolysis. However, if visual observation of hemolysis is not sufficient and the user would like to confirm by quantitative measurement, we recommend the Nature protocol by Dr. Olivier Wever and Dr. An Hendrix using a spectrophotometer to confirm the absence of hemolysis by the lack of a spectrophotometric absorbance peak for free hemoglobin at 414 nm (Tulkens et al., 2020). Additionally, the presence of large amounts of lipids, as indicated by a cloudy appearance, might indicate the failure of fasting in subjects prior to blood collection. Consider excluding such samples.Note: For bile samples, and other very viscous samples, such as pancreatic duct fluid, add ice-cold PBS at a ratio of 1:1 and homogenize the mixture by repeated pipetting. Follow the same isolation steps as for blood.
 CRITICAL: It is important not to disturb the pellet after each centrifugation step. Instead, collect most of the plasma while leaving a small residual volume above the pellet. We recommend handling a maximum of 10 plasma samples at one time since additional samples will increase the delay and the risk of EVP loss.
CRITICAL: It is important not to disturb the pellet after each centrifugation step. Instead, collect most of the plasma while leaving a small residual volume above the pellet. We recommend handling a maximum of 10 plasma samples at one time since additional samples will increase the delay and the risk of EVP loss. -
b.Collect the plasma by pipetting the top layer without touching the interphase layer and transfer into a new tube.
-
c.Centrifuge the plasma at 3,000 × g for 20 min at 4°C. This step will remove platelets that could otherwise be co-isolated with EVPs and contaminate the EVP preparation.Note: Platelet numbers can be assessed by classic hemocytometer counting, cell counting with size exclusion or more precisely through flow cytometry of the platelet suspension stained for CD61 (platelet glycoprotein IIb, broadly expressed on resting and activated platelets).
-
d.Collect the plasma and discard the platelet-rich pellet if not needed.
 Pause Point: The plasma can be frozen at −80°C for long-term storage.
Pause Point: The plasma can be frozen at −80°C for long-term storage. -
e.Centrifuge the plasma at 12,000 × g for 20 min at 4°C in either a tabletop microcentrifuge or in an ultracentrifuge.Note: This step will remove apoptotic bodies and larger vesicles that could otherwise be co-isolated with EVPs and contaminate the EVP preparation.Optional: The sample can be further processed to remove contaminants, such as lipids, by filtering the cleared post-12,000 × g plasma supernatant through a 1.2 μm membrane.Note: We recommend performing the 12,000 × g spin right before the ultracentrifugation steps, especially for plasma and other bodily fluids as well as tissue-derived media, since protein aggregates and precipitates could develop through freeze-thaw cycles or over extended waiting times.Optional: After the 12,000 × g spin, save 100 μL aliquots of cleared plasma for EVP quantification and characterization by NTA.
-
a.
-
2.Isolate EVPs by ultracentrifugation.
-
a.Transfer the plasma supernatant to an ultracentrifuge tube (see Materials and equipment). For plasma, we recommend the 50.4Ti rotor with the #355645 tube for volumes up to 4 mL.
-
b.Centrifuge sample at 100,000 × g for 70 min at 10°C in a fixed angle rotor in a Beckman Coulter ultracentrifuge (see Materials and equipment).
-
c.Decant or carefully pipet off the plasma, leaving the pellet untouched; place the inverted tube in a tube rack to drain and to avoid liquid backflow. Note the volume of plasma for calculating EVP numbers and protein per milliliter plasma.
-
a.
-
3.
Re-suspend the EVP pellet in 500 μL of ice-cold PBS and transfer to a second ultracentifuge tube.
-
4.
Add 3.5 mL of PBS to wash the pellet.
-
5.
Centrifuge the sample at 100,000 × g for 70 min at 10°C.
-
6.
Discard the supernatant by decanting and ensure there is no liquid backflow onto the pellet.
-
7.
Re-suspend the pellet in 100 μL of PBS. Store at −80°C. Save aliquots for protein quantification, NTA, and EM to avoid freeze-thaw cycles.
CRITICAL: EVPs can also be isolated from serum (in a red top blood collection tube without anticoagulant, Cat#366434G) using the same procedure; however, serum EVPs and plasma EVPs are not equivalent.
Note: The complex nature of plasma and heterogeneity between patients makes EVP isolation challenging. Additional filtering and PBS dilutions can be made but could also introduce loss or bias. Optimization protocols are being developed to decrease the inter-sample variation in EVP isolation from blood. See further discussion under “Limitations” section.
EVP sample preparation for AF4
Timing: 10 min
EVPs isolated from different sources via ultracentrifugation are pre-processed for AF4 analysis.
-
1.
Adjust the protein concentration of freshly isolated or thawed frozen EVPs to 0.5–1 μg/μL with PBS.
-
2.
Pre-clear the EVP suspension by centrifuging in a tabletop microcentrifuge at 12,000 × g at 4°C for 5 min to remove large protein and nanoparticle aggregates.
-
3.
Transfer the supernatant into a pre-chilled screw cap vial. A pulled point glass vial insert can be used for samples with small volume.
-
4.
Place the sample vial into the designated position in the pre-chilled autosampler.
CRITICAL: To achieve the best resolution and yield, we have determined empirically that 40–100 μg of EVP protein is required for EVPs isolated from conditioned media of cell culture for AF4 processing (with our instrument settings). Due to the size of the sample injection loop, which determines the maximum sample volume to be loaded, a sample concentration range of 0.4–1 μg/μL is required for AF4. However, for EVPs derived from other sources, these parameters need to be optimized empirically due to different EVP complexity.
AF4 of EVPs and data collection
Timing: 70 min
EVPs are separated based on their hydrodynamic sizes via AF4, and real-time data collection is carried out for concentration and size determination.
-
1.
Astra 6 (Wyatt Technology) is used for data acquisition and analysis, including MALS, DLS, and UV signals. Open a new experiment with the current configuration of the instrument, and set parameters for data collection: MALS data collection interval is 1 s and QELS interval is 2 s; total data collection time is 60 min. Save the file and click “RUN.” Data collection automatically starts once triggered by the autosampler sample loading.
-
2.Chemstation software (Agilent Technologies) with an integrated Eclipse module (Wyatt Technology) is used to operate the AF4. First, program a new method or load the desired method from the method library. The running method we have developed and optimized for EVP separation is the following:
- Forward channel flow: 1 mL/min
- Elution, 2 min, Vx 0.5 mL/min;
- Focus, 1 min, Vx 0.5 mL/min;
- Focus + Injection, 2 min, Vx 0.5 mL/min, injection flow 0.2 mL/min;
- Elution, 45 min, Vx a linear gradient decreasing from 0.5 to 0 mL/min;
- Elution, 5 min, Vx 0 mL/min;
- Elution + Injection, 5 min, Vx 0 mL/min;
-
3.
In Chemstation, set the input sample volume in the autosampler module (<=100 μL), and set the parameters for fraction collection in the fraction collector module. Here, we collect fractions by time slice (0.5 min/fraction).
-
4.
Click “Run a single sample.” The sample will be automatically loaded and fractionated accordingly, and data collection by Astra 6 will start once the sample loading is done.
-
5.
Fractions are collected into 96-well plates based on time slice by the Agilent Fraction Collector.
-
6.
After all samples have been processed for the day, switch off all lasers and thermostats for both autosamplers and fraction collector. Switch on the COMET of the Dawn Heleos II detector for ~30 min to clean the flow cell. Change the buffer to water and run the Night Rinse mode ~16–18 h. For long-term storage, the system can be stored in 20% ethanol after running a large volume of water through the system.
CRITICAL: A blank control of the running buffer (i.e., PBS) can be run using the same running method in parallel to the EVP samples. This control can assist in evaluating instrument performance, such as the background noise level and baseline drifting.
Note: Samples more than 100 μg or of a volume larger than 100 μL can be sequentially fractionated via AF4 using the same running method and fractions can be pooled together according to their hydrodynamic sizes.
Pause Point: Collected fraction plates can be sealed with adhesive tapes and kept on ice in 4°C for 2–3 days. However, we recommend proceeding to the next step of fraction concentration and characterization shortly after fraction collection if time allows.
Fraction concentration and characterization
Timing: 30 min or longer (timeframe varies based on the total amount of input samples to be analyzed)
The size of EVP fractions is analyzed using the Astra 6 software and fractions of similar size are pooled together for further concentration and downstream characterization.
-
1.
In Astra 6, adjust the baseline of data collected from each monitor in the experiment just completed and save the changes.
-
2.
Open a new window of EASI Graph in Astra 6, and plot the hydrodynamic radius (Rh) of particles vs Time. Please refer to Zhang and Lyden (2019) for the underlying principle of hydrodynamic radius determination and fractionation efficiency evaluation using the Autocorrelation function. Both QELS and UV absorbance are displayed on a relative scale to illustrate the relative abundance of each subpopulation in the sample. A single peak or multiple peak regions can be selected for analysis simultaneously.
Optional: Representative individual fractions can be first concentrated as described in steps 34–36 and a small aliquot can be subjected to negative staining TEM analysis for quick morphology examination.
-
3.
Guided by the particle size, abundance, and morphology, adjacent fractions with similar properties are then pooled together into 50 mL Falcon tubes and placed on ice.
-
4.
Pre-wet the Millipore centrifugal filter columns with Ultracel-30 membrane (30 kDa cutoff) with cold PBS (fill to full) and spin in tabletop clinical centrifuge at 3,700 × g at 4°C for 5 min. Discard the residual liquid in the filter unit and flow-through in the collection tube and place the filter column back into the collection tube.
-
5.
Transfer the pooled fractions into the filter column (top part) and spin at 3,700 × g at 4°C for 7–8 min. Discard the flow-through but keep the samples retained in upper chamber of the filter column (the concentrated sample).
-
6.
Repeat step 34 until each pooled set is concentrated to the desired volume using a single filter column.
-
7.
Mix the concentrated samples gently and transfer to Eppendorf tubes on ice. For better recovery, an additional volume of PBS can be added to rinse the filter column and combined with the sample.
-
8.
Record the volume of the final product and remove an aliquot for BCA assay and/or NTA analysis to determine the protein concentration and/or particle concentration. A small aliquot (~1 μg) can be saved to verify particle integrity and morphology by TEM.
-
9.
Use the EVP subsets immediately for downstream analysis or keep on ice for short-term storage. Aliquots can be kept frozen at −80°C for long-term storage.
CRITICAL: For EVP samples from a specific type of source material that has never been analyzed previously by AF4, it is critical to examine the morphology of EVPs present in representative individual fractions by TEM first, before pooling fractions together for further processing and analysis. NTA analysis is not applicable for exomeres, which, due to their size, are below the detection limit of NTA technology (minimum size detected is 50 nm).
Note: An alternative approach to concentrate a large volume of pooled fractions is ultracentrifugation at 100,000 × g at 10°C for 70 min. The pelleted EVP subsets can then be re-suspended in the desired volume of PBS or other types of solution as required by downstream analysis.
Note: This AF4 fractionation protocol is developed and optimized for EVPs isolated via dUC method from conditioned media of cell culture. For complex samples, such as plasma-derived EVPs, further preparation of samples prior to AF4 fractionation may be required.
Expected outcomes
Due to the recent interest in understanding the role of EVPs in inter-organ communication and their value as biomarkers, the development of methods that could improve and optimize EVP isolation protocols from different sources, such as blood and tissues, is critical.
Cell lines, as well as their EVPs, remain most valuable tools for functional studies, including cancer research. However, we have shown that cell line EVPs do not always recapture the complexity and heterogeneity of tissue samples, such as tumor explants (Hoshino et al., 2020). While a cell line will produce EVPs specific for that tumor cell type, tumor explants contain a mixture of EVPs derived from the tumor cells, tumor stroma, and infiltrating immune cells. Similarly, bodily fluids also contain EVPs derived from distant organs, such as the liver and bone marrow. The lower complexity of cell line-derived EVPs, compared to tissue explants or bodily fluids, makes them suitable for targeted approaches, such as immunoaffinity-based isolation methods. Cultured cell lines lack the tissue organization and microenvironment, which may alter cell physiology and lead to differences in EVP content and secretion, but they allow the study of EVPs derived from a single cell type source. To overcome these shortcomings, patient-derived organoids can be used for EVP isolation, allowing us to partially recapitulate the tissue architecture while limiting the sources of EVPs to one or two cell types. Another advantage of using cell lines is the possibility to scale up cultures to obtain large amounts of EVPs for downstream applications. For example, with our protocol, the commonly used, commercially available human breast cancer cell line MDA-MB-231 (2 × 106 cells, 4 days of culture) yields around 15 μg of EVP protein per 25 mL of media, and the well-known murine melanoma cell line B16-F10 yields 35 μg of EVP protein per 15 mL of media after three days of a culture, initially seeded with 1.5 × 106 cells. The amounts of EVPs will vary largely between cell lines and culture conditions and therefore the scale of the culture will have to be empirically determined for each experiment depending on the amount of EVPs required for downstream applications.
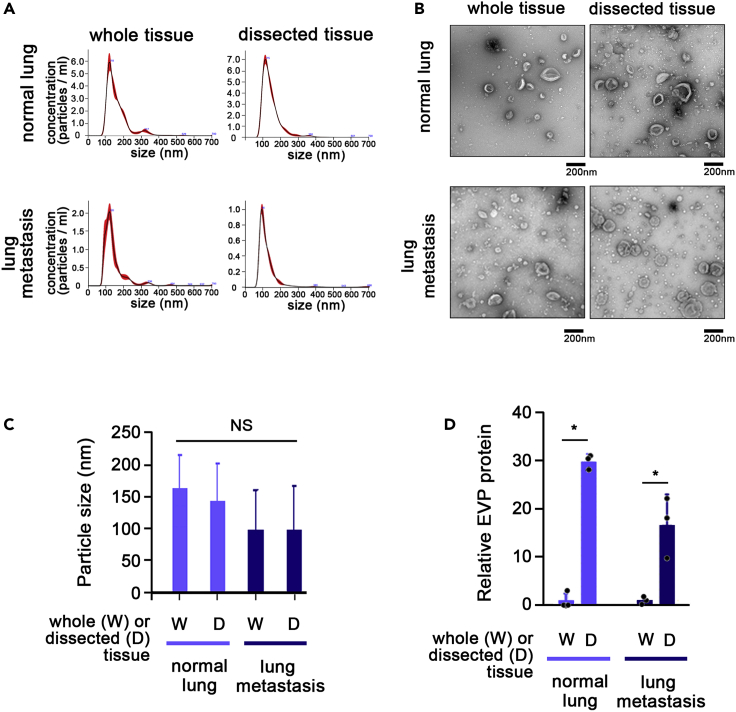
For optimization of EVP isolation from tissue samples, we used organs harvested from healthy wild-type mice, murine metastatic lung tumor tissue, and human adjacent colon tissue (Figure 5). We compared EVP yields from whole tissue/organs to dissected (2 mm3) pieces and performed the following downstream characterization: total EVP protein amounts were measured by BCA; particle number, size and concentration were measured by NTA; and morphology was determined by TEM. Combining 5 BALB/c mice per group, we found that dissecting tissue explants into smaller pieces resulted in an increase in EVP protein yields measured by BCA assay in all tested tissues (Table 2). Similar results were observed when EVP number per μg of tissue was measured by NTA, showing an increase in the number of EVPs in tissue explants that had been dissected into pieces. The more pronounced increase in EVPs detected by BCA, compared to NTA suggests that the increase in exomeres, or small protein particles, which are below the detection limit of NTA, is higher relative to the increase in larger extracellular vesicles/exosomes (this has to be further confirmed by AF4 experiments). Since downstream analysis protocols including proteomics and functional characterization require at least 10 μg of EVP protein, and NTA and TEM require at least 1 μg, increasing the yield of the tissue explant protocol ensures the feasibility of further experiments, while decreasing the number of animals required (meeting the 3 Rs principle - replacement, reduction, refinement), and reducing the cost of animal acquisition and maintenance. Taken together, these results demonstrate that dissecting tissue explants into smaller pieces before culture for EVP isolation improves EVP yield. However, it remains unclear whether dissected tissues versus whole tissue explants, and the tissue explant culture process itself, can alter tissue biology, promoting changes in the EVP cargo.
Figure 5.
Dissecting tissues into 2 mm3 pieces improves EVP yields without affecting EVP morphology
(A) Nanosight (NTA) profiles for whole organs and tissue dissected into smaller pieces for murine normal lung and metastasis-bearing lung.
(B) Transmission electron microscopy images of the same samples as in b, normal lung (upper row) and metastasis-bearing lung (lower row).
(C and D) (C) Particle size for EVPs from normal lung and metastasis-bearing lung and (D) EVP protein yields from whole tissue versus tissue dissected into smaller pieces. n = 3, mean and SD is shown; ∗p < 0.05.
Table 2.
Increased EVP yields from dissected tissue explant cultures versus whole tissue cultures
| Tissue | EVP concentration by BCA (ng/μL) |
EVP size by NTA (nm) |
The number of EVP particles by NTA (particles/mL) |
|||
|---|---|---|---|---|---|---|
| Whole | Dissected | Whole | Dissected | Whole | Dissected | |
| Human | ||||||
| Colon | 353.3 | 741.8 | 109 (±3) | 107 (±2) | 1.3 × 108 (±1.0 × 107) | 3.1 × 108 (±2.0 × 107) |
| Mouse | ||||||
| Brain | 29.1 | 329.4 | 134 (±6) | 144 (±4) | 3.2 × 108 (±2.9 × 107) | 4.9×108 (±1.8 × 107) |
| Kidney | 12.0 | 42.2 | 140 (±4) | 159 (±16) | 6.8 × 108 (±7.6×107) | 2.6 × 108 (±1.9×107) |
| Liver | 83.1 | 247.2 | 116 (±5) | 143 (±14) | 2.3 × 108 (±1.0 × 107) | 3.5 × 108 (±2.9 × 107) |
| Lung | 11.1 | 209.8 | 120 (±1) | 115 (±1) | 4.7 × 109 (±5.4 × 107) | 5.6 × 109 (±3.9 × 107) |
| Pancreas | 44.6 | 188.5 | 178 (±27) | 202 (±53) | 7.2 × 107 (±8.1 × 106) | 1.4 × 108 (±1.5 × 107) |
| Spleen | 7.7 | 92.0 | 145 (±14) | 163 (±27) | 6.1 × 108 (±2.9 × 107) | 5.3×108 (±2.0 × 107) |
| Thymus | 37.0 | 145.1 | 135 (±5) | 138 (±11) | 4.4 × 108 (±4.6 × 107) | 2.6 × 108 (±1.2 × 108) |
| White adipose tissue | 0.2 | 1,299.6 | 134 (±8) | 130 (±10) | 9.9 × 107 (±1.2 × 107) | 6.4 × 108 (±6.2 × 107) |
| Brown adipose tissue | 7.1 | 138.6 | 113 (±8) | 153 (±16) | 2.2 × 108 (±3.3 × 107) | 2.0 × 108 (±2.3 × 107) |
EVP size is similar between the two conditions. EVP protein concentration was measured by BCA (ng/μL, left), EVP size (nm, middle) and concentration (number of EVP/μg tissue) were measured by Nanosight (right) for human colon tissue and murine pancreas, white adipose tissue (WAT), brown adipose tissue (BAT), lung, brain, spleen, thymus, kidney, and liver.
Bodily fluids are often rich in EVPs and 1 mL of fluid is sufficient to isolate enough material for downstream characterization and proteomics. Blood, as the most easily accessible fluid, can yield from a few micrograms to 20 μg EVP protein per mL plasma whereas bile could be even richer and contain up to double the amount of EVP protein (and ~1 × 108 EVPs/mL) compared to plasma. The amounts will vary based on health status and other variables, such as cancer stage, treatment, etc.
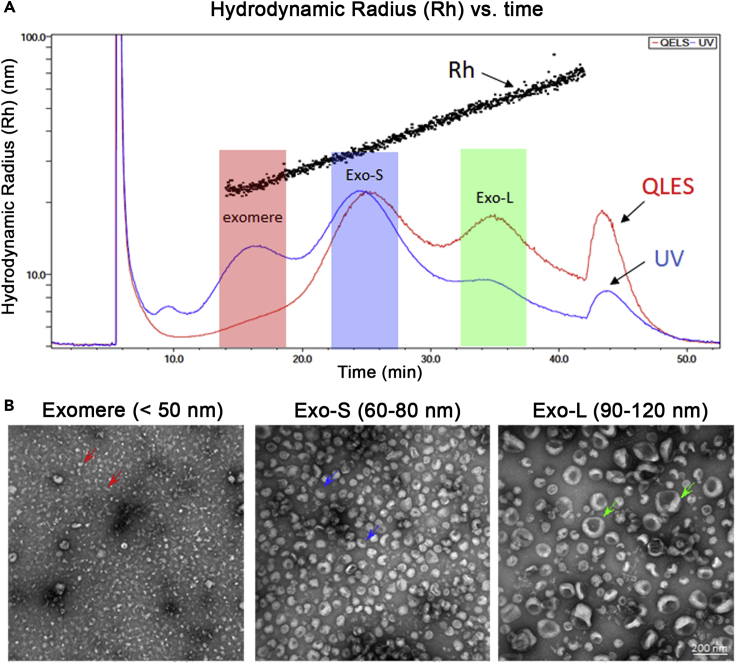
AF4 separates EVPs based on their hydrodynamic size within a large dynamic range. As reported in our recent work, AF4 facilitates the successful separation of distinct subsets of extracellular vesicles/exosomes and the identification of exomeres (Zhang et al., 2018). A representative AF4 fractionation profile of B16F10- derived EVPs and TEM images of pooled fractions corresponding to exomeres, Exo-S, and Exo-L are shown in Figure 6.
Figure 6.
Representative AF4 separation of EVPs prepared from B16F10 cells using differential ultracentrifugation
(A) Representative AF4 fractionation profile showing hydrodynamic radius (Rh) and fractograms of UV absorbance and QELS plotted against time.
(B) Representative negative staining TEM images of combined fractions resolved by AF4 for corresponding exomeres, Exo-S, and Exo-L subpopulations. Colored arrows: representative EVPs in each subpopulation (adapted from Zhang et al., 2018).
Limitations
Although the differential ultracentrifugation (dUC) protocol provides a robust methodology to process and analyze a large number of specimens from different sources and with a wide range of sample volumes, it presents several limitations. dUC separates particles based on their hydrodynamic size and density but the resolution and the resultant EVP purity is limited. Only several groups of large vesicles, microparticles, and small EVPs (relatively enriched for exomeres and exosomes) are isolated. Moreover, EVPs in each group will be heterogeneous and contamination by other non-EVP proteins will likely occur (Théry et al., 2006; Tauro et al., 2012). Furthermore, the composition of bodily fluids, such as blood plasma, is much more complex than most other types of specimens, such as conditioned media from cell culture. One major complication is the high abundance in plasma of albumin, lipoproteins and other soluble proteins. These proteins/complexes cannot be efficiently eliminated from the small EVP fraction by the dUC method. Moreover, some of the lipoproteins are within the size range of EVPs (e.g., LDL 18–28 nm in diameter; IDL, 25–50 nm; VLDL 30–80 nm; Chylomicrons 75–600 nm). The presence of high amounts of contaminants can interfere with specific downstream analysis, such as multi-omics profiling. Therefore, removal of these abundant contaminants by coupling dUC with other separation strategies, such as immunoaffinity capture or size-exclusion chromatography, based on distinct biophysical/biochemical properties of EVPs is necessary to further improve the quality of EVP preparations and expand the dynamic range of the analysis for EVP-based biomarker discovery. Accordingly, a larger volume of starting material is usually required if additional purification procedures are included to yield enough EVP for final analysis.
In the past decade, a variety of strategies have been developed for EVP isolation. Besides the dUC method described here, examples include immunoaffinity capture (IAC), ultrafiltration (UF), size-exclusion chromatography (SEC), polymer-based precipitation, and microfluidics (Théry et al., 2006; Chen et al., 2010; Merchant et al., 2010; Lässer et al., 2012; Tauro et al., 2012; Jørgensen et al., 2013; Nakai et al., 2016; Ko et al., 2017; Reátegui et al., 2018; Brennan et al., 2020). We recently reported the application of AF4 for small EVP fractionation with improved resolution (Zhang et al., 2018; Zhang and Lyden, 2019). To choose the most suitable method for biomarker discovery using plasma samples, especially for potential clinical translation, several critical factors have to be taken into consideration, including the available specimen volume/size and the proposed downstream analysis. A compromise may have to be made between purity and yield. Labor- and time-efficiency are important factors to consider, as well. According to the MISEV2018 guidelines, dUC falls within the category of “intermediate recovery, intermediate specificity” (Théry et al., 2018). Given that dUC is reproducible and commonly used, can be quickly and easily mastered by researchers, and does not require specialized expertise or equipment, it is still the most accessible methodology for EVP isolation. This is very important for studies requiring cross-comparison and validation by multiple centers or studies involving sample collection and processing over a relatively long timespan. Another reason for dUC remaining the gold standard for EVP isolation is that no chemicals are used, thereby preserving the EV integrity, which is particularly important for downstream functional studies.
Troubleshooting
Problem 1
Variability in EVP yield.
Potential solution
Despite using the same ultracentrifuge manufacturer, model, and rotors, we have observed variability in EVP yield between different ultracentrifuges. Since each ultracentrifuge is unique, if possible, always use the same machine, or if it is necessary to use different or several machines, compare yields between machines and ensure test samples and controls are prepared using the same machine.
Problem 2
Low EVP yield.
Potential solution
One option is to use a smaller sized tube to concentrate the collection of the EVPs in a smaller target area at the bottom of the tube and avoid loss by washing in large amounts of PBS. Another possibility is that the EVP pellet may be lost by dissolving into supernatant during prolonged handling times between steps. Work quickly or handle fewer samples at a time to avoid this problem.
Problem 3
Especially when isolating EVPs from human samples, one can expect a variety of contaminants, such as tissue debris and protein aggregates, and therefore extensive processing is necessary to remove them and to achieve satisfactory quality of EVPs.
Potential solution
One approach for the purification of limited sample quantities is the use of a density separation step (see description in protocol and Figure 7). Other options are filtration and additional pre-ultracentrifugation spins, or extensive washes, all of which will lead to some loss of material. For sucrose/deuterium oxide cushion purification up to 50% loss in total EVP protein can be expected but depending on downstream applications, specific EVP populations can also be enriched. Purification steps should be included when the EVP product will be submitted for downstream “omics” analyses for which higher purity of the product is required. We recommend using the sucrose cushion for tissue explants where the collected EVP pellet still contains aggregates after the washing step, which are typically observed for certain organs, such as pancreas and liver. This also depends partly on disease condition and needs to be determined by the investigator.
Figure 7.
Example of “contaminated” samples purified over a sucrose/deuterium oxide density layer
Aggregates and insoluble material could contaminate the EVP pellet, especially in complex samples, such as tissue explants or bodily fluids. Purification methods, such as density cushion, could be performed to improve sample purity.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Lyden (dcl2001@med.cornell.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The protocol includes all datasets generated or analyzed during this study. The study did not generate new unique code.
Acknowledgments
We thank the Electron Microscopy & Histology services of the Weill Cornell Medicine Microscopy & Image Analysis Core for TEM with NIH, United States Shared Instrumentation Grant (S10RR027699) and Medical Illustration & Design, part of the Medical Research Support Services of Yonsei University College of Medicine, South Korea, for all artistic support related to this work. We are grateful for the great AF4 technical support from Wyatt Technology. The authors gratefully acknowledge support from: the National Cancer Institute, United States CA224175 (D.L.), CA210240 (D.L.), CA232093 (D.L.), CA163117 and CA207983 (D.L.), CA163120 (D.L.), CA169416 (D.L.), CA169538 (D.L.), CA218513 (D.L., H.Z.) and AI144301 (D.L., V.P.); the United States Department of Defense, United States W81XWH-13-1-0425 (D.L.), W81XWH-13-1-0427, W81XWH-13-1-0249 (D.L.), and W81XWH-14-1-0199 (D.L.); National Institutes of Health, United States/WCM CTSC, United States (NIH/NCATS (UL1TR00457) (H.Z.); NIH/NCATS (UL1TR002384) (D.L. and H.Z.); the Hartwell Foundation, United States (D.L.); the Thompson Family Foundation (D.L., D.K.); the STARR Consortium I9-A9-056 (D.L., H.Z.) and I8-A8-123 (D.L.); the Pediatric Oncology Experimental Therapeutics Investigator’s Consortium (D.L., T.M.T.); Alex’s Lemonade Stand Foundation, United Sates (D.L.); the Breast Cancer Research Foundation, United States (D.L.); the Feldstein Medical Foundation (D.L.); the Tortolani Foundation (D.L.); the Clinical & Translational Science Center (D.L., H.Z.); the Mary Kay Ash Charitable Foundation (D.L., I.M.); the Malcolm Hewitt Weiner Foundation (D.L.); the Manning Foundation (DL., A.H.); the Daniel P. and Nancy C. Paduano Family Foundation (D.L); the James Paduano Foundation (D.L.); the Sohn Foundation (D.L.); the AHEPA Vth District Cancer Research Foundation (D.L., L.B., S.L.); the Daedalus Fund Selma and Lawrence Ruben Science to Industry Bridge Award (D.L.); the Children’s Cancer and Blood Foundation, United States (D.L.); Susan G. Komen Postdoctoral Fellowship PDF15331556, JST PRESTO, Japan 30021, and JSPS KAKENHI, Japan JP19K23743 (A.H.); the National Research Foundation of Korea, South Korea (NRF) grant funded by the Korea government (MSIT) (2019R1C1C1006709, 2018R1A5A2025079, and 2020M3F7A1094093); a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, South Korea (KHIDIHI19C1015010020); "The Alchemist Project" through the Ministry of Trade, Industry and Energy (MOTIE, Korea) (20012443), and Severance Hospital Research fund for Clinical excellence (SHRC) (C-2020-0032 and C-2020-0025) (H.S.K.); The Swedish Cancer Society Pancreatic Cancer Fellowship, the Lions International Postdoctoral fellowship and the Sweden-America stipend (L.B.); the São Paulo Research Foundation (FAPESP) São Paulo, Brazil (2017/07117-7) (G.C.T); the Swiss National Science Foundation (SFNS), Switzerland Postdoc Mobility grant (P2SKP3_174785 and P400PB_186791) (F.A.V.); and the DoD PRCRP Horizon Award, United States (W81XWH-19-PRCRP-HA) (S.L).
Author contributions
L.B., A.H., and H.S.K designed the experimental approach, coordinated the project, interpreted the data, and wrote the manuscript. F.P.V., G.T., S.L., J.H., D.K., K.K. and Z.W. performed optimization experiments. T.E.H., M.P.Q., D.K., T.M.T., D.R.J., and W.R.J. provided human samples. K.E.G., C.M.K., V.P., I.M., and H.Z. contributed to data interpretation and wrote the manuscript. D.L. led the project, interpreted the data, and wrote the manuscript.
Declaration of interests
D.L., A.H., H.S.K., and L.B. have filed a US patent application related to this work.
Contributor Information
Haiying Zhang, Email: haz2005@med.cornell.edu.
Ayuko Hoshino, Email: ayukohoshino@bio.titech.ac.jp.
David Lyden, Email: dcl2001@med.cornell.edu.
References
- Brennan K., Martin K., FitzGerald S.P., O'Sullivan J., Wu Y., Blanco A., Richardson C., Mc Gee M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020;10:1039. doi: 10.1038/s41598-020-57497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Skog J., Hsu C.H., Lessard R.T., Balaj L., Wurdinger T., Carter B.S., Breakefield X.O., Toner M., Irimia D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 2020;182:1044–1061. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen M., Bæk R., Pedersen S., Søndergaard E.K., Kristensen S.R., Varming K. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J. Extracell. Vesicles. 2013;2:20920. doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparski H.G., Metha-Damani A., Yao J.Y., Patel S., Hsu D.H., Ruegg C., Le Pecq J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- Lässer C., Eldh M., Lötvall J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012;2012:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J., Bhagwat N., Black T., Yee S.S., Na Y.J., Fisher S., Kim J., Carpenter E.L., Stanger B.Z., Issadore D. miRNA profiling of magnetic nanopore–isolated extracellular vesicles for the diagnosis of pancreatic cancer. Cancer Res. 2017;78:3688–3697. doi: 10.1158/0008-5472.CAN-17-3703. [DOI] [PubMed] [Google Scholar]
- Merchant M.L., Powell D.W., Wilkey D.W., Cummins T.D., Deegens J.K., Rood I.M., McAfee K.J., Fleischer C., Klein E., Klein J.B. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin. Appl. 2010;4:84–96. doi: 10.1002/prca.200800093. [DOI] [PubMed] [Google Scholar]
- Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai W., Yoshida T., Diez D., Miyatake Y., Nishibu T., Imawaka N., Naruse K., Sadamura Y., Hanayama R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016;6:33935. doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reátegui E., van der Vos K.E., Lai C.P., Zeinali M., Atai N.A., Aldikacti B., Floyd F.P., Jr., Khankhel A.H., Thapar V., Hochberg F.H. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018;9:175. doi: 10.1038/s41467-017-02261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., Simpson R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Théry C., Witwer K.W., Aikawa E., Jose Alcaraz M., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulkens J., De Wever O., Hendrix A. Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat. Protoc. 2020;15:40–67. doi: 10.1038/s41596-019-0236-5. [DOI] [PubMed] [Google Scholar]
- Zhang H., Freitas D., Kim H.S., Fabijanic K., Li Z., Chen H., Mark M.T., Molina H., Martin A.B., Bojmar L. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018;20:332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lyden D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc. 2019;14:1027–1053. doi: 10.1038/s41596-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The protocol includes all datasets generated or analyzed during this study. The study did not generate new unique code.