Abstract
Here we present a plasma proteomics dataset that was generated to understand the importance of self-reported race for biomarker discovery in Alzheimer's disease. This dataset is related to the article “Why inclusion matters for Alzheimer's disease biomarker discovery in plasma” [1]. Plasma samples were obtained from clinically diagnosed Alzheimer's disease and cognitively normal adults of African American/Black and non-Hispanic White racial and ethnic backgrounds. Plasma was immunodepleted, digested, and isobarically tagged with commercial reagents. Tagged peptides were fractionated using high pH fractionation and resulting fractions analysed by liquid chromatography – mass spectrometry (LC-MS/MS & MS3) analysis on an Orbitrap Fusion Lumos mass spectrometer. The resulting data was processed using Proteome Discoverer to produce a list of identified proteins with corresponding tandem mass tag (TMT) intensity information.
Keywords: Alzheimer's disease, Plasma, Proteomics, Biomarker, African American, Black, disparities
Specifications Table
| Subject | Chemistry, Biology, Neuroscience |
| Specific subject area | Alzheimer's disease, quantitative proteomics, health disparities |
| Type of data | Table Figure Mass spec raw files Proteome Discoverer msf files |
| How data were acquired | Liquid chromatography separation coupled to high-resolution mass spectrometer (LC-MS/MS), MS3 quantification using Tandem Mass Tagging (TMT) strategies. LC parameters: Nano UHPLC (Thermo Scientific) coupled with autosampler, 105 min gradient, self-packed trap column (100 µM ID × 2.5 cm) and analytical column (100 µM ID × 25 cm). MS parameters: Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific), data dependent acquisition (top speed precursor selection), synchronous precursor selection (top 10) for MS3 quantification. |
| Data format | Raw mass spectrometry files, PD read out files |
| Parameters for data collection | Plasma samples obtained from African American probable Alzheimer's disease (AD) (N=30) and cognitively normal (CN) (N=26) individuals, non-Hispanic White probable Alzheimer's disease (N=29) and cognitively normal (N=28) individuals. The patients had an average age of 71.8 (CN) and 75.4 (AD), average MMSE score of 27 (CN) and 15 (AD). No significant differences among the groups in terms of age, sex and presence of comorbidities. |
| Description of data collection | Samples were randomly divided into two experimental sets (Set 1 N=73 sample and Set 2 N=40 samples). Further, for Set 1, samples were divided into 8 batches, whereas there were 4 batches for Set 2 samples, with representation of all four groups in all batches. Plasma samples were immunodepleted of the top 6 most abundant proteins and digested using trypsin-LysC mix. Resulting peptides were labelled using TMT 10/11-plex tags. Labelled peptides were further fractionated using high pH fractionation. Finally, each fraction was randomly injected in duplicates into the mass spectrometer and subject to LC-MS/MS, MS3 analysis. |
| Data source location | Vanderbilt University Nashville, TN United States of America (USA) |
| Data accessibility | Proteomics Identification Database (PRIDE) ProteomeXchange Consortium: Dataset identifier: PXD022265 http://www.ebi.ac.uk/pride/archive/projects/PXD022265 |
| Related research article | M.J. Khan, H. Desaire, O.L. Lopez, M.I. Kamboh, R.A.S. Robinson, Why inclusion matters for Alzheimer's disease biomarker discovery in plasma Journal of Alzheimer's Disease (2021) Jan 5. doi: 10.3233/JAD-201318. [1] |
Value of the Data
-
•
The data provides information about proteins identified in plasma samples collected from Alzheimer's disease patients.
-
•
This data could be useful for individuals interested in the plasma proteome of Alzheimer's disease patients and also proteins changing due to AD.
-
•
This data could be a valuable source of potential peripheral protein biomarkers in Alzheimer's disease.
-
•
Proteomics biomarker discovery efforts that focus on diverse patient groups are few and this dataset includes African American/Black adults.
-
•
Plasma proteomics coupled with machine learning offers a powerful approach to identify potential diagnostic biomarker candidates in Alzheimer's disease and related dementias research.
1. Data Description
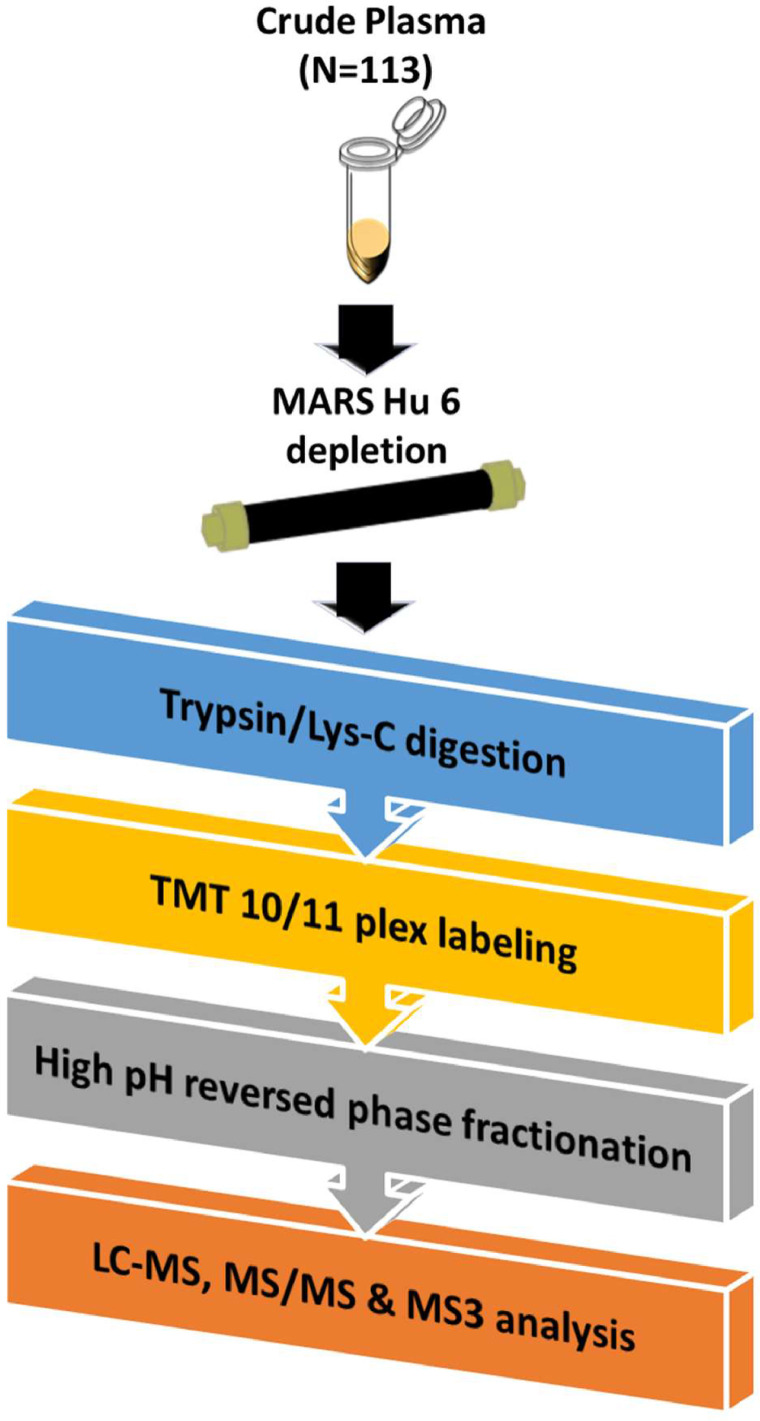
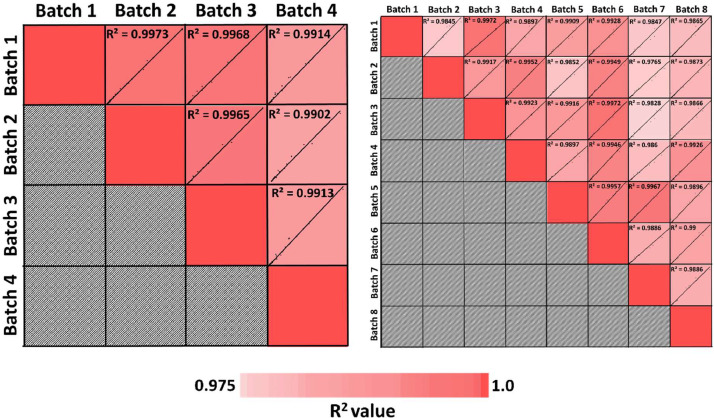
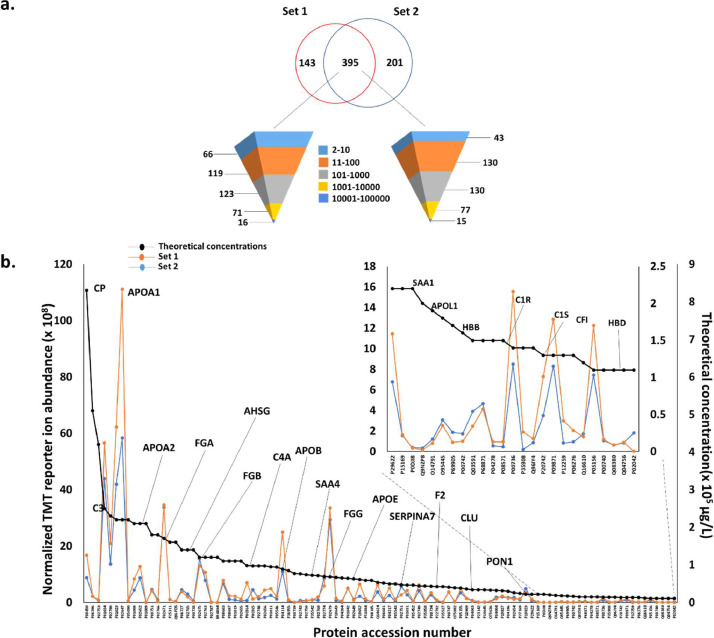
The data presented here include an in-depth plasma proteomics analysis of samples from participants from a national Alzheimer's Disease Research Center. A total of 113 patient samples from African American/Black and non-Hispanic White adults that were clinically diagnosed with probable Alzheimer's disease or were cognitively normal [1] are a part of this study. Samples were subject to quantitative plasma proteomics analysis. In order to accommodate the 113 samples and a quality control (QC) for data normalization, a total of 12 randomized TMT 10/11-plex experiments were performed. Fig. 1 displays the overall proteomics workflow employed in this experiment. The plasma samples were divided into two sets of N=73 (Set 1) and N=40 (Set 2) samples and taken through the entire workflow separately. Plasma samples were first immunodepleted, digested using trypsin/Lys-C mix, and labelled with TMT 10/11 plex tags (with one QC sample added into each batch), fractionated into 12 fractions and finally analysed using LC-MS, MS/MS, and MS3 analysis. Lists of proteins identified in each fraction have been provided in the Supplemental Information. Fig. 2 is a correlation plot of average normalized TMT reporter ion intensities for all proteins between different batches for both sets demonstrating high reproducibility among the batches in each experiment. The average R2 values for Set 1 and Set 2 were 0.99 and 0.9939, respectively. Fig. 3a is a Venn diagram of identified proteins that are in common between the two experimental sets. The pyramid plot shows the distribution of proteins commonly identified between the two experimental sets based on the total number of peptide spectral matches (PSMs) identified per protein across all the batches. The distribution of proteins was similar for both experimental sets with a similar number of proteins identified with the same number of PSMs. For example, there were 16 proteins with over 10,000 PSMs in Set 1, while Set 2 had 15 proteins meeting the same criteria. On the other hand, there were 66 and 43 proteins having at least 2-10 PSMs for Set 1 and Set 2, respectively. For both Set 1 and Set 2, Complement C3 and α-2 macroglobulin were identified with the most PSMs. Fig. 3b displays a comparison between the theoretical concentrations of proteins [2] and the corresponding average normalized TMT abundances for the top 100 most abundant proteins in plasma. For both sets, apolipoprotein A1 had the highest TMT abundance, whereas ceruloplasmin had the highest theoretical concentration [2]. Other example proteins having high TMT intensities included complement C3, α-2 macroglobulin, fibrinogen α chain, fibrinogen γ chain, fibrinogen β chain, hemopexin, and apolipoprotein B100. Lower abundant proteins are also highlighted in the inset of Fig. 3b, and include serum amyloid A1, α-1antitrypsin, complement C1r subcomponent, and immunoglobulin λ-like polypeptide 5. For a majority of the proteins, similar TMT abundances were measured in both Set 1 and Set 2 samples.
Fig. 1.
Plasma proteomics workflow for Set 1 (N=73) and Set 2 (N=40) samples. Samples were randomized into eight batches for Set 1 and four batches for Set 2 with one QC pool sample and representative samples from each group contained in each batch. The samples were randomly assigned TMT channels for both experiments. The experimental workflow was maintained except for the digestion step, where ammonium bicarbonate based in solution digestion was used for Set 1, while urea-based filter-assisted sample preparation digestion was employed in Set 2.
Fig. 2.
Correlation plot of average normalized TMT reporter ion intensities for all proteins between different batches for both Set 1 and Set 2. Both displayed positive correlation among all the batches. In case of Set 1, batch 3 and batch 6 showed the best correlation with an R2 value of 0.9972, while batch 2 and batch 7 showed the lowest co-relation with an R2 value of 0.9765. On average Set 1 had an R2 value of 0.99. On the other hand, for Set 2, batch 1 and batch 2 had the best correlation with an R2 value of 0.9973, batch 2 and batch 4 had the lowest correlation, with an R2 value of 0.9902. On average Set 1 had an R2 value of 0.9939.
Fig. 3.
a) Venn diagram of identified proteins common between Set 1 and Set 2. The pyramid plots highlight the distribution of proteins identified with a given range of peptide spectral matches (PSMs). b) Line plot of theoretical protein concentration's for plasma [2] against the experimentally obtained relative abundances for these proteins using normalized TMT reporter ion intensities of the top 100 most abundant proteins in plasma. The insert is a zoomed-in version of proteins in the lower concentration range of the line plot. The black line represents the theoretical protein concentrations, while the orange and blue line represent corresponding TMT reporter ion abundances for Set 1 and Set 2 proteins, respectively. Abbreviation: CP- Ceruloplasmin, C3- Complement C3, APOA1-Apolipoprotein A1, APOA2- Apolipoprotein A2, FGA- Fibrinogen alpha chain, AHSG- Alpha-2-HS-glycoprotein, FGB- Fibrinogen beta chain, C4A- Complement C4-A, APOB- Apolipoprotein B, SAA4- Serum amyloid A4, FGG- Fibrinogen gamma chain, APOE- Apolipoprotein E, SERPINA7- Thyroxine-binding globulin, F2- Prothrombin, CLU- Clusterin, PON1- Serum paraoxonase/arylesterase 1, SAA1- Serum amyloid A1, APOL1- Apolipoprotein L1, HBB- Hemoglobin subunit beta, C1R- Complement C1r subcomponent, C1S- Complement C1s subcomponent, CFI- Complement factor I, HBD- Hemoglobin subunit delta.
2. Experimental Design, Materials and Methods
2.1. Plasma sample collection
Human plasma samples from four groups - African American/ Black cognitively normal (N=26) & Alzheimer's disease (N=30) non-Hispanic White cognitively normal (N=28) & Alzheimer's disease (N=29) were obtained from the University of Pittsburgh Alzheimer's Disease Research Center (ADRC). Detailed characteristics of the individuals have been described previously [1]. Approval for the participation of human subjects was obtained by the Institutional Review Boards of the University of Pittsburgh and Vanderbilt University. Informed consent was obtained for human subjects. The Mini-Mental State Examination was performed and disease individuals were clinically diagnosed with mild to moderate dementia at the time of draw according to the National Institute on Aging-Alzheimer's Association and National Alzheimer's Coordinating Center criteria. The samples were divided into two experimental sets (Set 1 N=73 samples, Set 2 N=40 samples)
2.2. Plasma depletion
Plasma samples were immunodepleted using Multiple Affinity Removal (MARS) Column Human 6 (Agilent, Santa Clara) of the 6 most abundant proteins (albumin, IgG, IgA, α1-antitrypsin, transferrin and haptoglobin). Samples (30 µL) were diluted 4 times using buffer A (Agilent, Santa Clara), and loaded onto a 0.22 µm spin filter, and centrifuged at 16,000 g for 1 min to remove any particulates from the sample. Samples were loaded onto a MARS 6 column for depletion of the top 6 most abundant proteins. The flow through fractions were collected and concentrated using a 5 kDa molecular weight cut off concentrator at 4695 g. Bicinchoninic acid (BCA) assay was used to determine protein concentration. A pooled sample containing equal amounts of protein from each of the plasma samples was generated and used as quality control (QC) sample.
2.3. Digestion
Two separate digestion protocols were used for the two experimental sets. In Set 1, in solution digestion was performed in 100 mM ammonium bicarbonate buffer. Proteins were reduced using 200 mM DTT for 45 mins at 55°C, alkylated using 200 mM IAM in the dark for 30 mins. Finally, proteins were digested overnight at 37°C using trypsin/Lys-C mix (Promega, Madison) (1:50 enzyme:protein ratio). For Set 2, a modified Filter Assisted Sample Preparation (FASP) [3] digestion was performed. In brief, 100 µg of proteins was loaded onto a 10 kDa molecular weight cut off filter (Sartorius, Gloucestershire, UK) followed by reduction with 20 mM dithiothreitol (DTT) in 100 mM Tris with 8 M urea for 15 min. This was followed by centrifugation at 14,000 rpm to remove the excess reagents from the sample. Proteins were then alkylated with 20 mM iodoacetamide (IAM) in the dark for 15 mins followed by centrifugation to remove the excess reagents. Samples were then centrifuged at 14,000 rpm using 100 mM Tris in 1 M urea to wash any remaining reagents. Finally, trypsin/Lys-C mix (Promega, Madison) was added to the sample at 1:50 enzyme:protein ratio and incubated at 37 °C. After 8 hours of digestion, the peptides were acidified with formic acid and desalted using HLB cartridges (Waters corporation, Milford) per manufacturer's instructions.
2.4. TMT tagging
Dried desalted peptides were labelled using either TMT 10-plex or 11-plex reagents (ThermoFisher Scientific, Waltham) following manufacturer's instructions. Briefly, 25 µg of peptide samples were reconstituted using 100 mM triethyl ammonium bicarbonate buffer. TMT reagents were activated by solubilizing with 41 µL of acetonitrile and added to the samples. After 1-hour incubation, the reaction was quenched using 5% hydroxylamine solution for 15 min. The samples were pooled into a single mixture and desalted to remove excess TMT reagents.
2.5. High pH reversed phase fractionation
Labelled peptides were fractionated using high pH (pH=10) fractionation into 12 fractions using an acetonitrile flow (ACN (%)- 3,5,8,10,13,18,22,30,45,60,80,95) with an HLB Oasis cartridge (Waters Corp). The fractions were dried down and reconstituted in water with 0.1% formic acid to a final concentration of 0.5 µg/µL.
2.6. LC-MS/MS and MS3 parameters
All sample were analysed using an Ultimate 3000 UHPLC system on the front end of an Orbitrap Fusion Lumos mass spectrometer. Peptides were loaded onto a self-packed C18 (5 µm, 200Ǻ, MICHROM Bioresources Inc.) trap column (100 µM ID × 2.5 cm, IntegraFrit Capillary), and separated using an in-house packed C18 (2.5 µm, 100Ǻ, XBridge BEH from Waters) capillary column (100 µM ID × 25 cm, Polymicro Technologies) using solvent A (water with 0.1 % formic acid) and solvent B (acetonitrile with 0.1% formic acid) buffers with the following gradient: 0-7 min, 10% B; 7-67 min, 10-30% B; 67-75 min, 30-60% B; 75-77 min, 60-90% B; 77-82 min, 90% B; 82-83 min, 90-10% B; 83-100 min, 10% B. Precursor scan parameters were as follows: mass range m/z 375–1500, resolution = 120,000, automatic gain control (AGC) target set at 4 × 105 ions and maximum ion injection (IT) time of 50 ms. The instrument was operated in data dependent acquisition (DDA) mode with a cycle time of 3 s. Fragmentation was performed with an NCE= 35% using collision-induced dissociation (CID). The AGC was set at 1 × 104 with an isolation width of 0.7 m/z, maximum injection time of 100 ms, and a dynamic exclusion of 20 s. Synchronous precursor selection (SPS-10) mode was used for collecting MS3 spectra, using higher-energy collisional dissociation (HCD) with the following Orbitrap parameters: NCE= 55%, scan range = 100-400 m/z, resolution = 60,000, AGC = 5 × 104, maximum injection time = 118 ms and isolation width = 2 m/z. Each fraction was injected in duplicate and the injection order was randomized for each batch.
2.7. Data Analysis
Raw files were searched against the Uniprot human reviewed protein database (07/17/2018, 20289 sequences) using SEQUEST-HT using Proteome Discoverer software (version 2.2). A maximum of two trypsin miscleavages were set with precursor mass tolerance of 10 ppm and fragment mass tolerance set at 0.6 Da. Dynamic modification of methionine oxidation (+15.995 Da), protein N-termini acetyl (42.011 Da), TMT 10 (229.163 Da)/11 plex (229.169 Da) on peptide N-termini and lysine residue were selected, while static modification of cysteine carbamidomethyl (+57.02 Da) was included. Decoy database searching was used to generate high confidence peptides (FDR < 1%). TMT reporter ions (i.e. m/z 126 – 129) were identified using the most confident centroid at a reporter ion mass tolerance of 20 ppm. Technical replicates and fractions from each batch were combined into a single file using a python script.
2.8. Data normalization and statistics
The final list of proteins was normalized using a two-step internal reference scaling (IRS) method [4] which involves within-batch and across-batch normalization. Briefly, the intensity of each TMT channel was summed, and a scaling factor (SF) was calculated by dividing the sum of intensity of the pooled channel by the sum of each TMT channel for each individual batch. Across-batch normalization involved the calculation of SF by calculating the geometric mean of TMT intensity of pooled samples for all the batches, and dividing that by the intensity of the individual pooled sample TMT intensity for each batch. This was followed by multiplying with the scaling factor to the in-batch normalized intensity for each protein. Once normalized, differentially-expressed proteins were determined by performing t-test between AD and CN samples groups from each race. A fold change cut-off of 1.23 (Set 1) and 1.33 (Set 2) was set based on prior power analysis [5].
Declaration of Competing Interest
The authors have declared no conflicts of interest.
Acknowledgments
The authors would like to thank the University of Pittsburgh ADRC and research participants for providing plasma samples. The authors acknowledge funding from the Alzheimer's Association (AARGD-17-533405), pilot funds from the University of Pittsburgh Alzheimer Disease Research Center funded by the National Institutes of Health and National Institute on Aging (P50AG005133, RASR), the National Institute on Aging (AG041718, AG030653, AG064877, AG066468 MIK) and the Vanderbilt Institute of Chemical Biology (T32-GM06508). The authors would also like to thank Dr. Lars Plate and Mr. Mahmud Reaz for providing the Python code for combining multi-batch data.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.106923.
Appendix. Supplementary materials
References
- 1.Khan M.J., Desaire H., Lopez O.L., Kamboh M.I., Robinson R.A.S. Why inclusion matters for Alzheimer's disease biomarker discovery in plasma. J. Alzheimer's Dis. 2021 doi: 10.3233/JAD-201318. Jan 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwenk J.M., Omenn G.S., Sun Z., Campbell D.S., Baker M.S., Overall C.M., Aebersold R., Moritz R.L., Deutsch E.W. The human plasma proteome draft of 2017: building on the human plasma peptideatlas from mass spectrometry and complementary assays. J. Proteome Res. 2017;16(12):4299–4310. doi: 10.1021/acs.jproteome.7b00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 4.Plubell D.L., Wilmarth P.A., Zhao Y., Fenton A.M., Minnier J., Reddy A.P., Klimek J., Yang X., David L.L., Pamir N. Extended multiplexing of tandem mass tags (TMT) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol. Cell Proteomics. 2017;16(5):873–890. doi: 10.1074/mcp.M116.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Z., Yende S., Kellum J.A., Robinson R.A.S. Additions to the human plasma proteome via a Tandem MARS depletion iTRAQ-based workflow. Int. J. Proteomics. 2013;2013 doi: 10.1155/2013/654356. 654356-654356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.