Abstract
The outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts contain β-barrel integral membrane proteins. The five-protein β-barrel assembly machine (Bam) accelerates the folding and membrane integration of these proteins. The central component of the machine, BamA, contains a β-barrel domain that can adopt a lateral-open state with its N- and C-terminal β-strands unpaired. Recently, strategies have been developed to capture β-barrel folding intermediates on the Bam complex. Biochemical and structural studies provide support for a model in which substrates assemble at the lateral opening of BamA. In this model, the N-terminal β-strand of BamA captures the C-terminal β-strand of substrates by hydrogen bonding to allow their directional folding and subsequent release into the membrane.
Keywords: Bam complex, outer membrane protein, β-barrel, protein folding, Gram-negative bacteria
Introduction
A characteristic of mitochondria, chloroplasts, and the Gram-negative group of bacteria is that they contain a double layer of membranes. The outer membrane contains proteins of β-barrel structure. Conserved multi-subunit machines assemble these β-barrel proteins into the outer membrane. In Escherichia coli, this machine is the β-barrel assembly machine (Bam) complex [1]. Homologous machines are present in the outer membranes of mitochondria (Sam, sorting and assembly machinery) [2,3] and in the outer membranes of chloroplasts (OEP80, outer envelope protein) [4]. How the Bam complex performs rapid folding and membrane integration of β-barrel membrane proteins, repeatedly and without an input of energy (e.g., from ATP), has been a longstanding question since the discovery of the first components [1,5,6].
To learn how the Bam complex catalyzes folding, several related aspects of the process must be understood: how substrates are held by the Bam components during folding, how the machine promotes folding, and how folded substrates are released to allow catalyst turnover. Observing folding intermediates in complex with the machine is required to answer these questions. In this review, we highlight major milestones in our understanding of β-barrel assembly, emphasizing work that has been done in the past few years.
Structure and function of the Bam complex
The multi-subunit machines that assemble β-barrel membrane proteins in Gram-negative bacteria, mitochondria, and chloroplasts each contain a β-barrel component. It has long been believed that this protein plays the central role in folding β-barrel substrates. In Gram-negative bacteria, the central component is BamA, an essential protein that belongs to the Omp85 superfamily of outer membrane proteins [6]. The Bam complex from E. coli also includes four lipoproteins: BamB, BamC, BamD, and BamE [1,7]. BamA is the only transmembrane component and catalyzes the folding process [8-12]. BamA contains five N-terminal soluble periplasmic polypeptide transport-associated (POTRA) domains and a C-terminal β-barrel transmembrane domain. The POTRA domains act as a scaffold that mediates interaction with the lipoproteins [13-17]. Although all four lipoproteins are required for maximal folding efficiency [18], BamD is the only essential lipoprotein [19]. Several reports have shown that BamA alone catalyzes the chemistry required for cell survival [11,20,21], providing support for the longstanding belief that the roles of the lipoproteins must be regulatory [20-25].
The budding model for β-barrel assembly
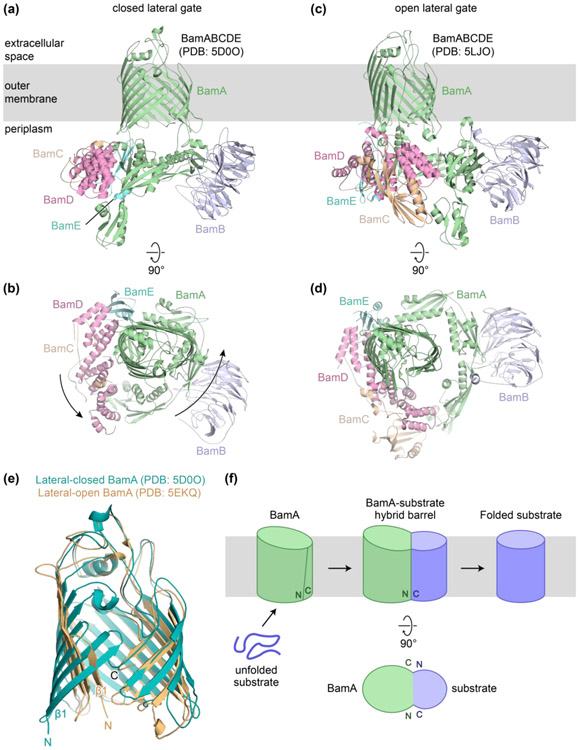
Structural studies of the Bam complex from bacteria provided the first clear ideas about the mechanism of substrate folding [8-10,14-17]. These structures showed that BamA can exist either in a closed state or an open state based on the pairing of the N- and C-terminal β-strands (Figure 1a-e). From the structures it was proposed that this open seam pairs by hydrogen-bonding with substrate β-strands to nucleate β-sheet formation [8,26]. A model was proposed in which β-strands of the substrate assemble at the seam and folded portions of the substrate bud through this opening to enter the membrane (Figure 1f). This “budding model” addresses a key aspect of the folding mechanism, which is the pathway by which substrates enter the membrane. As we discuss below, several recent discoveries support the proposal that the β-barrel of BamA opens and β-strands of nascent substrates pass through this opening into the membrane. Recently, structures of the Sam complex were reported that showed the central component, Sam50, in a lateral-open state [27]. These structures suggest that a similar mechanism for β-barrel folding might exist in Gram-negative bacteria and mitochondria.
Figure 1. Structures of the Bam complex and the budding model for folding.
(a) Structure of the BamABCDE complex in which the lateral gate of BamA is closed (PDB: 5D0O). (b) Top-down view of complex in (a). (c) Structure of the BamABCDE complex in which the lateral gate of BamA is open (PDB: 5LJO). (d) Top-down view of complex in (c). (e) Overlay of the β-barrel domain of BamA from a structure with a closed lateral gate (teal; PDB: 5D0O) and a structure with an open lateral gate (orange; PDB: 5EKQ). In the lateral-open structure, the initial β-strands are flipped outward approximately 60 degrees. (f) The budding model for β-barrel assembly into the outer membrane.
Capturing β-barrel folding intermediates
The early structural studies of BamA and the Bam complex identified a pathway for how substrates enter the membrane once the β-strands are assembled into a β-sheet but did not explain how the β-sheet itself forms. More recently, several groups have developed strategies to capture β-barrel folding intermediates by crosslinking them to their assembly machines [12,28-30].
One crosslinking study on Sam50, the BamA homolog in mitochondria, used a series of radiolabeled C-terminal fragments of substrates that were designed to mimic sequential β-hairpin intermediates [28]. These substrates all formed strong crosslinks from the C-terminal β-strand in the substrate to the N-terminal β-strand of the Sam50 β-barrel; however, residues in the N-terminal β-strand of a substrate fragment formed crosslinks to both the N- and C-terminal edges of the Sam50 β-barrel. Because crosslinking was stronger to the C-terminal β-strand of the substrate, it was suggested that the N-terminal β-strand of Sam50 formed a more stable interaction with the C-terminal β-strand of the substrate during folding. Interactions between N-terminal β-strands of the growing substrate with the C-terminal edge of the Sam50 β-barrel were proposed to be transient, forming as new β-strands inserted at the C-terminal edge of the lateral gate and then breaking to allow the additional β-strands to insert. In this model, the N- and C-terminal edge of the lateral gate templates β-sheet formation.
Crosslinking studies of the Bam complex were also reported using substrates accumulated in vivo. One study looked at the folding of an autotransporter containing a C-terminal β-barrel domain. This family of outer membrane proteins also contain an N-terminal passenger (extracellular) domain, which is translocated through the β-barrel during its folding by the Bam complex [31]. This secretion step is required to complete folding of the β-barrel domain [32]. Therefore, to stall the β-barrel on the Bam complex during folding, a full-length autotransporter substrate was fused to a soluble maltose-binding protein at the N-terminus, which prevented translocation of the passenger domain [29]. Crosslinking studies suggested that this substrate interacted asymmetrically with the BamA β-barrel, with the N-terminus of the BamA β-barrel forming strong interactions to the C-terminus of the autotransporter substrate, and the C-terminus of the BamA β-barrel forming two different interaction patterns with the N-terminus of the substrate. These two interaction patterns were proposed to reflect a folding state before and a folding state after membrane integration. The authors proposed a model in which the β-sheet of the substrate forms in the periplasm [33], and then BamA mediates the membrane integration of the largely folded β-barrel through a swinging motion into the membrane.
Another study of folding intermediates accumulated on the Bam complex examined the assembly of LptD, which forms a β-barrel around a lipoprotein plug, LptE [12]. This two-protein complex forms the translocon that inserts lipopolysaccharide into the outer membrane (see review by Ruiz and colleagues in this issue), and LptD assembles orders of magnitude more slowly than other β-barrel substrates [34]. Variants of LptD that fold even more slowly had previously allowed extensive crosslinking experiments to probe interactions between the β-barrel of BamA and the folding substrate [35]. This more recent study showed that the C-terminus of LptD is held at the N-terminal edge of the BamA β-barrel [12]. However, the N-terminal region of LptD was found to form a β-sheet along the concave interior wall of the BamA β-barrel. Changes to residues in strands of the LptD β-sheet that contact the BamA interior, or changes to residues on the interior surface of BamA itself, can increase rate of folding of a substrate that would otherwise accumulate. It was proposed that the interior wall of the BamA β-barrel forms a confined, cage-like environment that serves as an active site for β-sheet formation by reducing the loss of entropy upon folding. Once sufficient nascent structure forms, the substrate could exit the interior of BamA and enter the membrane through the lateral gate.
Structure of a folding intermediate on the Bam complex reveals how turnover can occur
All experimental studies to characterize how substrates interacted with the Bam complex during folding showed a strong interaction between the C-terminal β-strand of the substrate and the N-terminal β-strand of the BamA β-barrel. It makes sense that a β-barrel assembly machine would hold onto a substrate tightly until folding is complete. To complete folding, the N- and C-terminal β-strands of the substrate must pair with one another so the β-barrel can close. Therefore, the C-terminal β-strand of the substrate must somehow dissociate from the N-terminal edge of BamA. Proposed folding models did not provide a mechanism for how release occurs. It was observed that the folded β-barrel is stable in the membrane, but a thermodynamic driving force cannot explain how the Bam complex catalyzes folding, which depends on kinetic barriers. This machine carries out rapid, repeated assembly of β-barrel substrates, and operates without an exogenous energy source [18,36]. How the Bam complex can hold substrates stably at its N-terminus during folding without releasing them prematurely, but then release them rapidly to complete folding, remained a question.
The structure of a nascent β-barrel as it folds on the Bam complex was reported recently and suggested a mechanism for release that was supported experimentally [30] (Figure 2). BamA itself is folded by the Bam complex and a series of BamA substrates was generated, each lacking one of the eight extracellular loops. Loop deletions were expected to slow folding of the substrate to allow accumulation and capture on the machine. Experiments with these mutants in cells established that folding proceeds from the C-terminus of the substrate towards the N-terminus. A structure of the Bam complex bound to a loop-deleted substrate showed a snapshot of a very late-stage folding intermediate. At this stage, the C-terminal β-strand of the substrate is paired with the N-terminal β-strand of the machine via a series of six hydrogen bonds. The nascent β-barrel protrudes into the membrane. The N-terminal edge of the substrate is not hydrogen-bonded to the C-terminal edge of the BamA β-barrel, but instead curves inward and points toward its own C-terminal β-strand with the two β-barrels contacting each other on their exterior surfaces. This closed interface ensures that the unpaired edges of each β-barrel face into an aqueous lumen where they are solvated by water, while also ensuring that membrane lipids do not fill the interior.
Figure 2. Structure of a late-stage folding intermediate on the Bam complex.
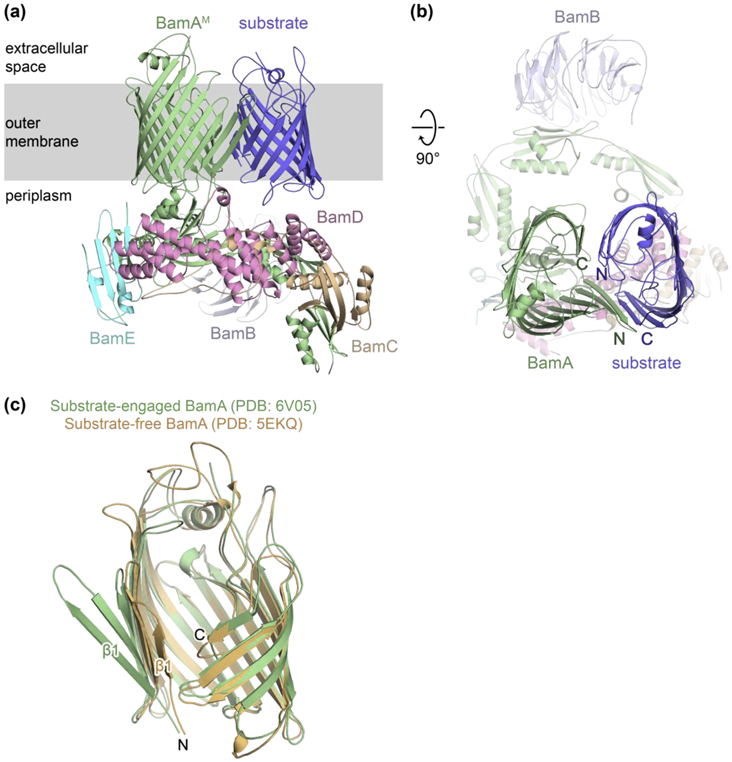
(a) Side view of the cryo-EM structure of the Bam complex bound to a late-stage BamA folding intermediate. (b) Top-down view of complex in (a). (c) Overlay of the β-barrel domain of BamA from the substrate-engaged complex (green; PDB: 6V05) and a substrate-free, lateral-open complex (orange; PDB: 5EKQ).
In order for the substrate to release from the machine, a total of six hydrogen bonds must be broken so that the N- and C-terminal strands of the substrate can pair. In a membrane where the dielectric is low, these bonds are worth ~25 kcal/mol, and breaking them simultaneously would have a huge kinetic barrier. Given that turnover occurs in the absence of ATP, there must be a mechanism for release that occurs through a pathway that does not require simultaneous rupture of all of these bonds.
The structure showed an overhang at the very C-terminus of the stalled substrate that did not pair with the N-terminal edge of the BamA β-barrel. It was proposed that this region of the substrate can initiate pairing with the free N-terminal β-strand of the substrate, allowing a stepwise exchange of hydrogen bonds to the machine for hydrogen bonds between the two terminal edges of the substrate. Multiple lines of evidence showed the importance of the C-terminal overhang in release. Dramatically, single amino acid changes at the C-terminus of the substrate were found to stall an otherwise completely intact substrate on the machine.
Current model for β-barrel assembly
The recent work described here leads to a model for β-barrel assembly by the Bam complex (Figure 3). Initially, the substrate associates with Bam components at the outer membrane (step a). Interactions with BamD may trigger opening of the BamA β-barrel at its seam [24]. The C-terminal β-strand of the substrate is then captured by the N-terminal β-strand of BamA to form a continuous β-sheet (step b). The substrate enters the interior of the BamA β-barrel, where folding of the β-sheet, catalyzed by stabilizing interactions with the interior wall of the BamA β-barrel, results in the addition of β-strands from the C-terminus of the substrate towards the N-terminus (step c). Once a significant amount of folding occurs, the substrate passes into the membrane through the open seam of BamA (step d). Features at each end of the substrate promote release by a stepwise exchange of hydrogen bonds that result in the closure of the substrate β-barrel (step e).
Figure 3. Current model for β-barrel assembly.
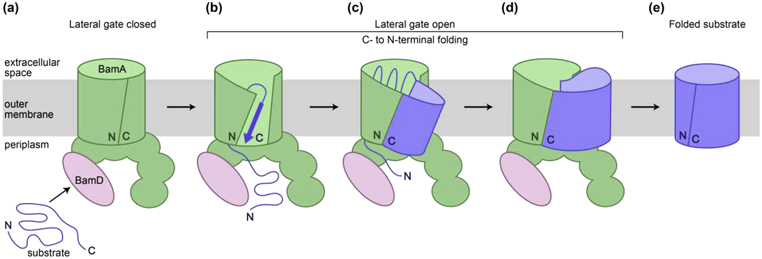
(a) The substrate is recruited to the Bam complex. (b) Opening of the seam of the BamA β-barrel allows interactions with the unfolded substrate. (c) Folding proceeds in a C- to N-terminal direction as β-strands are added within the interior of BamA to form a β-barrel. (d) Full membrane integration occurs at a late stage of folding, and the remaining β-strands are added to complete the β-barrel. (e) Release of the substrate occurs by a hydrogen bond exchange mechanism. For simplicity, only BamA (containing N-terminal soluble domains and a C-terminal β-barrel transmembrane domain) and BamD are shown.
Conclusions and Future Directions
Important advances in our understanding of β-barrel assembly have been made over the past few years. One surprising discovery was that BamA captures only one end of the substrate by hydrogen bonding, while the other end remains unpaired. One remaining question is how folding is initiated to form the hybrid β-barrel. It has been proposed that BamD or the POTRA domains of BamA can recruit substrates [12,13,24,37,38], possibly by interactions with chaperones [39,40]; however, it is unclear how substrates are then passed to the open seam of BamA. Another interesting question is how β-barrels that form oligomers assemble in the membrane. It has been shown that multiple Bam complexes can co-localize in outer membrane “precincts” mediated by interactions between BamB within adjacent complexes, and these precincts were shown to be important in the assembly of β-barrel trimers [41]. It has also been assumed that substrates traverse the periplasm via chaperones that deliver them to the outer membrane [39]. Recently, however, it has been proposed that a super-complex consisting of the Sec translocon, the chaperone SurA, and the Bam complex exists to connect the inner and outer membrane for protein delivery [42]. Additional work will confirm the relevance of these protein bridges to β-barrel assembly in vivo. The Bam complex assembles a variety of substrates, some of which have a plug-and-barrel architecture [43-45]. A recent structure showed that the interior of the BamA β-barrel can accommodate a folded lipoprotein, RcsF (see review by Collet and colleagues in this issue) [46]. If the Bam complex is functioning properly, RcsF is believed to be transferred from BamA to the interior of some substrate β-barrels. The recent structural work described above provides a picture for how lipoprotein transfer could occur—when substrates are folding, RcsF can move from the interior of BamA to that of the substrate, since the interiors of the two β-barrels are connected. Future work will address whether other lipoproteins, such as LptE, which is involved in assembly of the β-barrel LptD, can also be held initially within the interior of BamA prior to transfer to LptD. Finally, future studies should be aimed at capturing different substrates at various stages of folding to construct a complete physical picture of the process.
Antibiotic resistant infections, especially ones caused by Gram-negative bacteria, are an emerging problem. Recently, an antibody targeting BamA has been shown to perturb outer membrane integrity when added to E. coli cells and can prevent proper folding of outer membrane proteins by binding extracellular loops of BamA [47]. Furthermore, small molecules that interact with the Bam complex have been identified that lead to cell death by preventing efficient function of this essential molecular machine, likely by stabilizing the closed state of BamA (see review by Walker and colleagues in this issue) [48-50]. Recent structural and functional insights into the folding of β-barrels may enable the design of new antibiotics, as proper assembly of these proteins is critical for survival of Gram-negative bacteria.
Highlights.
The BamA β-barrel adopts an open state with its first and last β-strands unpaired.
BamA β-strand 1 captures the C-terminal β-strand of substrates by hydrogen bonding.
Folding proceeds from the C-terminus of the substrate towards the N-terminus.
Folding occurs in the interior of the BamA β-barrel prior to membrane integration.
Interactions between the N- and C-termini of the nascent β-barrel allow release.
Acknowledgements
This work was supported by NIH grants R01GM066174 and R01AI081059 (to D.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest:
** of outstanding interest:
- 1.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D: Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 2005, 121:235–245. [DOI] [PubMed] [Google Scholar]
- 2.Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W: Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 2003, 426:862–866. [DOI] [PubMed] [Google Scholar]
- 3.Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C: Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 2003, 424:565–571. [DOI] [PubMed] [Google Scholar]
- 4.Töpel M, Ling Q, Jarvis P: Neofunctionalization within the Omp85 protein superfamily during chloroplast evolution. Plant Signal Behm 2012, doi: 10.4161/psb.18677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggert US, Ruiz N, Falcone B V, Branstrom AA, Goldman RC, Silhavy TJ, Kahne D: Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science 2001, 294:361–4. [DOI] [PubMed] [Google Scholar]
- 6.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J: Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 2003, 299:262–5. [DOI] [PubMed] [Google Scholar]
- 7.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ: Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 2007, 104:6400–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK: Structural insight into the biogenesis of β-barrel membrane proteins. Nature 2013, doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, Huang Y: Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J 2014, doi: 10.1096/fj. 13–248450. [DOI] [PubMed] [Google Scholar]
- 10.Albrecht R, Schütz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, Zeth K: Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr Sect D Biol Crystallogr 2014, doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 11.Plummer AM, Fleming KG: BamA Alone Accelerates Outer Membrane Protein Folding in Vitro through a Catalytic Mechanism. Biochemistry 2015, doi: 10.1021/acs.biochem.5b00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Tomasek D, Santos T, May MD, Meuskens I, Kahne D: Formation of a β-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. Elife 2019, 8.** The authors trap assembly-defective forms of LptD on the Bam complex and characterize direct interactions between the machine and this substrate. Biochemical experiments show that the substrate forms extensive interactions with the interior of the BamA β-barrel, and a model for folding is proposed in which this region of BamA catalyzes β-sheet formation.
- 13.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D: Structure and function of an essential component of the outer membrane protein assembly machine. Science 2007, 317:961–964. [DOI] [PubMed] [Google Scholar]
- 14.Bakelar J, Buchanan SK, Noinaj N: The structure of the β-barrel assembly machinery complex. Science (80- ) 2016, 351:180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, et al. : Structural basis of outer membrane protein insertion by the BAM complex. Nature 2016, doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 16.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, et al. : Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 2016, doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 17.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA: Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commxm 2016, 7:12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagan CL, Kim S, Kahne D: Reconstitution of outer membrane protein assembly from purified components. Science 2010, 328:890–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ: YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 2006, 61:151–164. [DOI] [PubMed] [Google Scholar]
- 20.Hart EM, Gupta M, Wühr M, Silhavy TJ: The gain-of-function allele bamAE470K bypasses the essential requirement for BamD in β-barrel outer membrane protein assembly. Proc Natl Acad Sci U S A 2020, doi: 10.1073/pnas.2007696117.* This paper reports a bamA mutant that can survive in the absence of the otherwise essential lipoprotein BamD. The authors show that BamD is not directly involved in catalyzing β-barrel assembly but functions to regulate BamA.
- 21.Hart EM, Silhavy TJ: Functions of the BamBCDE Lipoproteins Revealed by Bypass Mutations in BamA. J Bacteriol 2020, doi: 10.1128/JB.00401-20.* The authors show that BamB, BamC, and BamE are involved in allowing proper coordination between BamD and BamA. Additionally, they identify a mutation in bamA that allows β-barrel assembly in vivo to proceed in the absence of the four lipoproteins, demonstrating that BamA alone can catalyze folding and membrane integration.
- 22.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ: BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol 2012, 194:1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricci DP, Hagan CL, Kahne D, Silhavy TJ: Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 2012, 109:3487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Sutterlin HA, Wzorek JS, Mandler MD, Hagan CL, Grabowicz M, Tomasek D, May MD, Hart EM, Silhavy TJ, et al. : Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc Natl Acad Sci U S A 2018, 115:2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCabe AL, Ricci D, Adetunji M, Silhavy TJ: Conformational changes that coordinate the activity of BamA and BamD allowing β-barrel assembly. J Bacteriol 2017, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK: Lateral opening and exit pore formation are required for BamA function. Structure 2014, doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diederichs KA, Ni X, Rollauer SE, Botos I, Tan X, King MS, Kunji ERS, Jiang J, Buchanan SK: Structural insight into mitochondrial β-barrel outer membrane protein biogenesis. Nat Commum 2020, doi: 10.1038/s41467-020-17144-1* The authors report the first structures of the Sam complex, the mitochondrial homolog of the Bam complex. These structures show that the β-barrel domain of Sam50 can open laterally, possibly allowing for interactions with substrates.
- 28.Höhr AIC, Lindau C, Wirth C, Qiu J, Stroud DA, Kutik S, Guiard B, Hunte C, Becker T, Pfanner N, et al. : Membrane protein insertion through a mitochondrial β-barrel gate. Science (80- ) 2018, 359:eaah6834.** The authors capture β-barrel substrates on the Sam complex and report interactions between the N- and C-terminal edges of Sam50 and the opposite edges of the trapped substrates. A model for folding is proposed in which substrates enter the interior of Sam50, interact with extracellular loop 6, and assemble at the lateral opening of Sam50.
- 29.Doyle MT, Bernstein HD: Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat Commim 2019, doi: 10.1038/s41467-01911230–9.** The authors generate a β-barrel substrate trapped on the Bam complex at a late stage of folding. Biochemical characterization of this trapped substrate shows that only the C-terminal end is stably captured by BamA, forming an asymmetric hybrid barrel consisting of BamA and the substrate. Membrane integration is proposed to occur via a swinging motion of the substrate into the membrane.
- 30.Tomasek D, Rawson S, Lee J, Wzorek JS, Harrison SC, Li Z, Kahne D: Structure of a nascent membrane protein as it folds on the BAM complex. Nature 2020, doi: 10.1038/s41586-020-2370-1.** The authors use extracellular loop deletions in BamA to trap this β-barrel as a substrate on the Bam complex. They present the first structure of a substrate-engaged machine, which shows how the N-terminal edge of BamA within the machine captures the C-terminal end of the substrate to allow for directional folding of the substrate and release from the machine once folding has finished.
- 31.Albenne C, leva R: Job contenders: roles of the β-barrel assembly machinery and the translocation and assembly module in autotransporter secretion. Mol Microbiol 2017, doi: 10.1111/mmi.13832. [DOI] [PubMed] [Google Scholar]
- 32.leva R, Tian P, Peterson JH, Bernstein HD: Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci U S A 2011, 108:E383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikdar R, Peterson JH, Anderson DE, Bernstein HD: Folding of a bacterial integral outer membrane protein is initiated in the periplasm. Nat Commim 2017, doi: 10.1038/s41467-017-01246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chng S-S, Xue M, Garner RA, Kadokura H, Boyd D, Beckwith J, Kahne D: Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 2012, 337:1665–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, et al. : Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci 2016, 113:8717–8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan CL, Kahne D: The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of β-barrel assembly. Biochemistry 2011, 50:7444–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagan CL, Wzorek JS, Kahne D: Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci 2015, 112:2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wzorek JS, Lee J, Tomasek D, Hagan CL, Kahne DE: Membrane integration of an essential β-barrel protein prerequires burial of an extracellular loop. Proc Natl Acad Sci 2017, 114:2598–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sklar JG, Wu T, Kahne D, Silhavy TJ: Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Gems Dev 2007, 21:2473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daimon Y, Iwama-Masui C, Tanaka Y, Shiota T, Suzuki T, Miyazaki R, Sakurada H, Lithgow T, Dohmae N, Mori H, et al. : The TPR domain of BepA is required for productive interaction with substrate proteins and the β-barrel assembly machinery complex. Mol Microbiol 2017, 106:760–776. [DOI] [PubMed] [Google Scholar]
- 41.Gunasinghe SD, Shiota T, Stubenrauch CJ, Schulze KE, Webb CT, Fulcher AJ, Dunstan RA, Hay ID, Naderer T, Whelan DR, et al. : The WD40 Protein BamB Mediates Coupling of BAM Complexes into Assembly Precincts in the Bacterial Outer Membrane. Cell Rep 2018, doi: 10.1016/j.celrep.2018.04.093. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Wang R, Jin F, Liu Y, Yu J, Fu X, Chang Z: A supercomplex spanning the inner and outer membranes mediates the biogenesis of β-barrel outer membrane proteins in bacteria. J Biol Chem 2016, doi: 10.1074/jbc.M115.710715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freinkman E, Chng S-S, Kahne D: The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci U S A 2011, 108:2486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SH, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, Asmar A, Laloux G, Hov AK, Leverrier P, et al. : Detecting envelope stress by monitoring β-barrel assembly. Cell 2014, doi: 10.1016/jcell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 45.Konovalova A, Mitchell AM, Silhavy TJ: A lipoprotein/b-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife 2016, doi: 10.7554/eLife.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez-Alonso R, Létoquart J, Nguyen VS, Louis G, Calabrese AN, Iorga BI, Radford SE, Cho SH, Remaut H, Collet JF: Structural insight into the formation of lipoprotein-β-barrel complexes. Nat Chem Biol 2020, doi: 10.1038/s41589-020-0575-0.* This paper reports a structure of the lipoprotein RcsF within the interior of the BamA β-barrel. The structure provides an explanation for how RcsF can sense stress at the outer membrane and, more generally, how a lipoprotein can be inserted into the BamA β-barrel.
- 47.Storek KM, Auerbach MR, Shi H, Garcia NK, Sun D, Nickerson NN, Vij R, Lin Z, Chiang N, Schneider K, et al. : Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci U S A 2018, doi: 10.1073/pnas.1800043115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hart EM, Mitchell AM, Konovalova A, Grabowicz M, Sheng J, Han X, Rodriguez-Rivera FP, Schwaid AG, Malinverni JC, Balibar CJ, et al. : A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc Natl Acad Sci U S A 2019, doi: 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imai Y, Meyer KJ, Iinishi A, Favre-Godal Q, Green R, Manuse S, Caboni M, Mori M, Niles S, Ghiglieri M, et al. : A new antibiotic selectively kills Gram-negative pathogens. Nature 2019, doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luther A, Urfer M, Zahn M, Müller M, Wang SY, Mondal M, Vitale A, Hartmann JB, Sharpe T, Lo Monte F, et al. : Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 2019, doi: 10.1038/s41586-019-1665-6. [DOI] [PubMed] [Google Scholar]