Abstract
Synthesis of the bacterial cell envelope requires a regulated partitioning of resources from central metabolism. Here, we consider the key metabolic junctions that provide the precursors needed to assemble the cell envelope. Peptidoglycan synthesis requires redirection of a glycolytic intermediate, fructose-6-phosphate, into aminosugar biosynthesis by the highly regulated branchpoint enzyme GlmS. MurA directs the downstream product, UDP-GlcNAc, specifically into peptidoglycan synthesis. Other shared resources required for cell envelope synthesis include the isoprenoid carrier lipid undecaprenyl phosphate and amino acids required for peptidoglycan cross-bridges. Assembly of the envelope requires a sharing of limited resources between competing cellular pathways and may additionally benefit from scavenging of metabolites released from neighboring cells or the formation of symbiotic relationships with a host.
Keywords: UDP-GlcNAc, peptidoglycan, teichoic acid, lipid II, capsule, GlmS, MurA, branch point regulation, PASTA kinase, UPD-GlcNAc
Graphical abstract
Introduction
“There is no delight in owning anything unshared.” ~Seneca (1st century A.D.)
The metabolic pathways that sustain life are a deeply entangled network of reactions that allow the efficient conversion of nutrients into energy and biomass [1]. Detailed reconstructions of metabolism in Escherichia coli include more than 2700 reactions that link ~1200 metabolites, with a small subset serving as key nodes for metabolism or as global regulatory signals [2]. Building a cell envelope is a resource-intensive process and imposes a substantial metabolic burden. Gram-positive peptidoglycan (PG) is multi-layered and can represent >20% of cell dry weight [3]. Although the PG layer in Gram-negative bacteria is thinner, the cell envelope additionally includes an outer membrane. Many Gram-positive bacteria also elaborate wall-linked capsular polysaccharide (CPS) and extracellular polysaccharides (EPS). The substantial metabolic flux associated with PG synthesis is unmasked when this process is blocked by antibiotics. Cell wall deficient L-forms of B. subtilis (which emerge upon interruption of PG synthesis) have increased carbon flux into lower glycolysis and the TCA cycle, which enhances flux through the electron transport chain and triggers oxidative stress [4].
Here, we highlight the regulation of the key branchpoint enzymes GlmS and MurA. GlmS controls the entry of fructose-6-phosphate (F6P) into envelope synthesis, and MurA directs UDP-GlcNAc into PG synthesis. PG also relies on other shared metabolites (glutamine, acetyl-CoA, UTP) and a shared anchor lipid, undecaprenyl phosphate (UP). Cells can also acquire metabolites for cell wall synthesis from their neighbors, and in extreme cases symbiotic partners may share the burden of synthesizing the enzymes required for PG synthesis.
Balancing glycolysis with synthesis of aminosugars.
PG is a glycopolymer comprised of two aminosugars, N-acetyl-glucosamine (GlcNAc) and N-acetyl-muramic acid (MurNAc). Aminosugar biosynthesis initiates when the transaminase GlmS converts F6P to glucosamine 6-phosphate (GlcN6P) using glutamine as the amino donor. Subsequent steps catalyzed by GlmM (mutase) and GlmU (bifunctional acetyl/uridine transferase) generate the key branchpoint intermediate, UDP-GlcNAc (Fig. 1A).
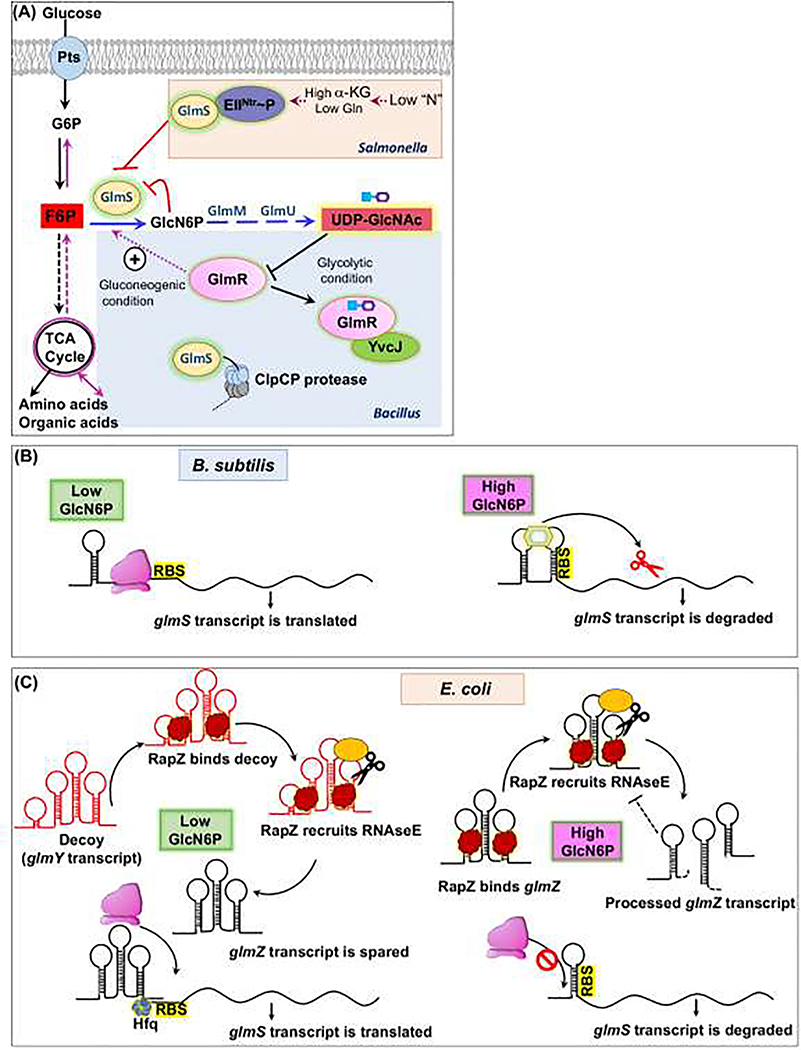
Figure 1. Regulation of the GlmS branchpoint enzyme.
A) Post-translational regulatory mechanisms. Glucose (Glc) is imported through a PTS and enters glycolysis where the branchpoint metabolite fructose-6-phosphate (F6P) is partitioned to aminosugar biosynthesis by GlmS. In Salmonella Typhimurium, GlmS is inhibited by the phosphorylated EII protein of the PTSNTR system under conditions of nitrogen deficiency (sensed by a high α-ketoglutarate to glutamine ratio). In B. subtilis, GlmS is stimulated the regulator GlmR(YvcK), and this activation is inhibited by binding to the next key branchpoint metabolite UDP-GlcNAc (UDP=blue square; GlcNAc is purple hexagon), GlmR:UDP-GlcNAc binds to YvcJ, which reduced GlmS activity/ GlmS is also subject to ClpCP-dependent proteolysis. (B) Feedback regulation of GlmS translation. In B. subtilis, the glmS transcript includes a 5’-regulatory ribozyme that binds GlcN6P, resulting in cleavage and inactivation of the mRNA. (C) Translation of the E. coli glmS mRNA is activated for recognition by the ribosome (pink) when bound to the regulatory RNA glmZ. However, when GlcN6P is high the glmZ transcript is degraded by RNase E, which is recruited by RapZ. When GlcN6P is low, RapZ triggers expression of a decoy transcript (GlmY) that sequesters RapZ, and the GlmZ sRNA is spared to activate glmS translation.
As befits its role as an essential branchpoint enzyme, GlmS is highly regulated. In Salmonella Typhimurium, GlmS activity is regulated by nitrogen status, consistent with the requirement of glutamine (Gln) as amino donor. Regulation involves a nitrogen-metabolic phosphotransferase system, PTSNtr, analogous to PTS systems involved in sugar import. The PTSNtr EI enzyme is phosphorylated under low nitrogen conditions (signaled by a high α-ketoglutarate/Gln ratio), and this phosphoryl group is transferred through NPr (an HPr paralog) to EIIANtr. The resulting EIINtr~P sequesters GlmS in an inactive complex, thereby shutting down aminosugar synthesis (Fig. 1A). When Gln availability is high, EIIA(Ntr) is dephosphorylated and degraded by Lon protease, thereby relieving GlmS inhibition [5].
GlmS production may also be feedback regulated by its product, GlcN6P (Fig 1B,1C). In E. coli, the mRNA of the bicistronic glmUS operon is processed to yield an unstable glmS mRNA, which in turn is stabilized and translationally activated by the action of a small regulatory RNA (sRNA), GlmZ [6] (Fig. 1C). GlmZ is unstable, being targeted for degradation by RNase E by the adaptor protein RapZ [7]. The induction of GlmS expression relies on a decoy sRNA, GlmY, which also binds RapZ. In addition to its role as an RNase E adaptor, RapZ serves as the sensor GlcN6P [8] (Fig. 1C). When GlmS activity is low, GlcN6P-free RapZ indirectly activates transcription of the glmY decoy sRNA. The net result is that GlmS translation responds sensitively to changes in the GlmS product, GlcN6P.
GlmS regulation in Bacillus subtilis is also complex. The first level of feedback regulation occurs when the glmS ribozyme cleaves and inactivates the glmS mRNA in response to the GlmS product, GlcN6P [9] (Fig. 1B). Additional control is provided by the GlmR(YvcK) regulator. GlmR stimulates GlmS activity when not bound to the downstream metabolite UDP-GlcNAc [10,11]. GlmR is required for growth on gluconeogenic carbon sources [12], where low F6P levels limit GlmS activity [13] (Fig. 1A). This requirement for GlmR can be bypassed by mutations that inactivate the glmS ribozyme or by exogenous aminosugars [14]. When bound to UDP-GlcNAc, GlmR associates instead with a RapZ homolog, YvcJ (Fig 1A) [10,11]. GlmS may also be regulated by ClpCP-dependent proteolysis [15]. Collectively, these mechanisms ensure that GlmS partitions sufficient carbon from glycolysis/gluconeogenesis into aminosugar biosynthesis for cell wall synthesis. A glmR/yvcK homolog is essential in Staphylococcus aureus [16], and mutants in Listeria monocytogenes (yvcK) and Mycobacterium tuberculosis (cuvA) are defective in cell wall biosynthesis [17,18]. However, it is unclear if all GlmR family members have this same mechanism.
UDP-GlcNAc is a precious cargo.
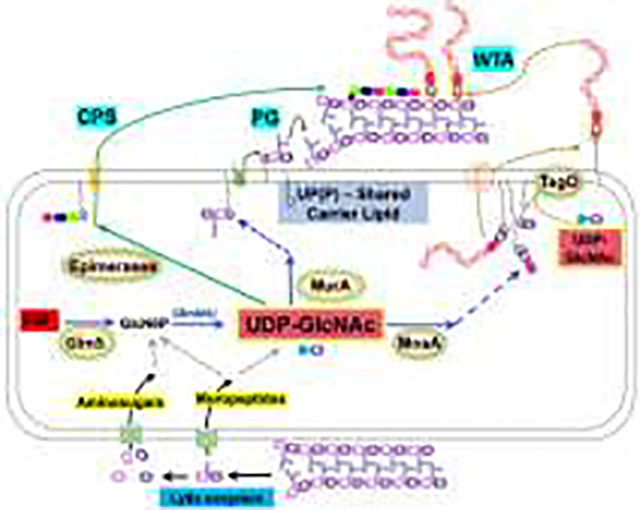
The next key branchpoint intermediate is UDP-GlcNAc, a shared metabolite used by MurA in the first committed step of PG biosynthesis (Fig. 2). In Gram-positive bacteria, UDP-GlcNAc is also used for synthesis of the cell surface glycopolymers wall teichoic acid (WTA) and CPS [19,20]. WTA is a long copolymer consisting of glycerol-P (or ribitol-P) units and is covalently linked to PG by a short linker often comprised of GlcNAc and N-acetylmannosamine (ManNAc) [21–23]. UDP-GlcNAc can also serve as a donor for WTA glycosylation [24,25]. UDP-GlcNAc is also the sugar donor for the synthesis of bacilllithiol (BSH), comprised of cysteine, D-glucosamine, and malic acid. BSH is the major low molecular weight thiol in many Gram-positive bacteria and can be present at millimolar levels [26].
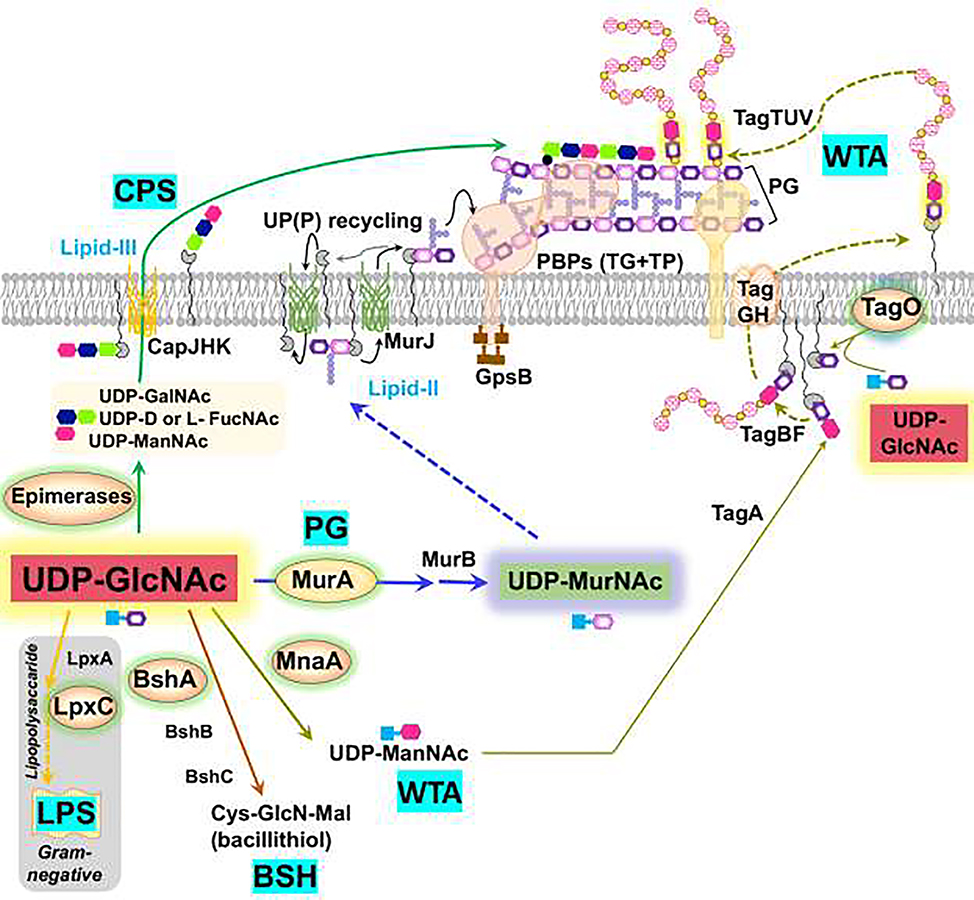
Figure 2. UDP-GlcNAc is a key branchpoint metabolite.
UDP-GlcNAc (red) is a shared precursor directed to peptidoglycan (PG) synthesis by MurA. PG synthesis relies on a UP-linked disaccharide-pentapeptide (lipid II), which is exported by MurJ so that penicillin-binding proteins (PBPs) and the Rod complex (elongasome) can catalyze the transglycosylation (TG) and transpeptidation (TP) reactions for PG synthesis. In Gram-positive bacteria, UDP-GlcNAc is additionally directed to wall teichoic acid (WTA) synthesis by MnaA (for UDP-ManNAc synthesis) and TagO (which couples GlcNAc to UP). WTA polymers are synthesized in the cytosol, flipped across the membrane by the TagGH complex, and covalently linked to PG (TagTUV enzymes). UDP-GlcNAc is also required for bacillithiol (BSH) synthesis, CPS synthesis and, in Gram-negative bacteria, for LPS biosynthesis. Branchpoint enzymes are highlighted with a yellow background.
The mechanisms that balance the flux of UDP-GlcNAc between PG synthesis and competing pathways have been studied in detail in L. monocytogenes. The conserved, multimeric GpsB protein functions as a cell cycle regulator by interacting with penicillin-binding proteins (PBPs) and scaffolding the assembly of multi-protein complexes [27–29] (Fig. 2). Genetic analysis revealed that gpsB mutations are suppressed by increasing the level of MurA through loss of the ClpCP protease, or decreasing activity of other enzymes that compete for UDP-GlcNAc [30]. These competing enzymes include GtcA and LMO2550 (involved in GlcNAc decoration of WTA) and MnaA (generates UDP-N-acetylmannosamine in support of WTA synthesis). In B. subtilis, the major MurA isozyme (MurAA) is also degraded by ClpCP, but whether this serves a regulatory role is not yet resolved [31].
Capsular polysaccharide (CPS) helps pathogens evade complement fixation, opsonization, and phagocytosis [32]. CPS production initiates with UDP-GlcNAc, a parent compound for capsule building blocks such as UDP-NAc-fucosamine and UDP-D-N-mannosaminuronic acid (Fig. 2). CPS is often subject to nutritional regulation. For example, S. pneumoniae grown in galactose (abundant in respiratory mucus) or GlcNAc have the highest level of capsule, followed by glucose or sucrose. The lowest levels of CPS were observed during growth on fructose due to decreased pools of the UDP-sugar precursor [33]. Mutations that affect metabolism often have pleiotropic effects. In the case of S. pneumoniae lysine decarboxylase (cadA), deletion indirectly affected glycolysis and in turn caused reduced UDP-sugars and impaired CPS biosynthesis [32].
In E. coli, UDP-GlcNAc is a shared substrate for MurA and LpxA. LpxA directs UDP-GlcNAc into LPS synthesis by conjugation to an R-3-hydroxyacyl chain. However, the next reaction (LpxC deacetylase) is the committed step for the LPS pathway. LpxC activity is reduced by FtsH-dependent proteolysis when LPS is in excess [34–36]. Proteolysis is also central to the regulation of LPS synthesis in Francisella tularensis. In this microbe, RipA is a conserved membrane protein that stabilizes LpxA to direct entry of GlcNAc into LPS biosynthesis [37].
Catch and release: PASTA kinases coordinate envelope biogenesis.
Many Gram-positive bacteria contain eukaryotic-like serine/threonine kinases (eSTK) with extracellular PBP and serine/threonine kinase associated (PASTA) domains [38]. These signaling kinases may respond to peptidoglycan-associated muropeptides [39,40] and, in at least some systems, directly regulate PG synthesis. In Listeria monocytogenes, the PrkA kinase regulates a protease adaptor protein, ReoM, that activates ClpCP-dependent degradation of MurA, thereby reducing PG synthesis. Activation of PrkA by muropeptides leads to ReoM phosphorylation, thereby decreased MurA degradation [41]. PASTA kinases are also critical for cell envelope homeostasis in M. tuberculosis [42], where PknB phosphorylates proteins related to PG synthesis including GlmU [43] and CwlM, an essential intracellular cell wall amidase homolog. CwlM localizes to the membrane and regulates the lipid II flippase MurJ. However, upon phosphorylation CwlM~P localizes to the cytoplasm where it activates MurA [44,45] (Fig. 3). PknB also regulates outer membrane mycolic acid synthesis [46,47].
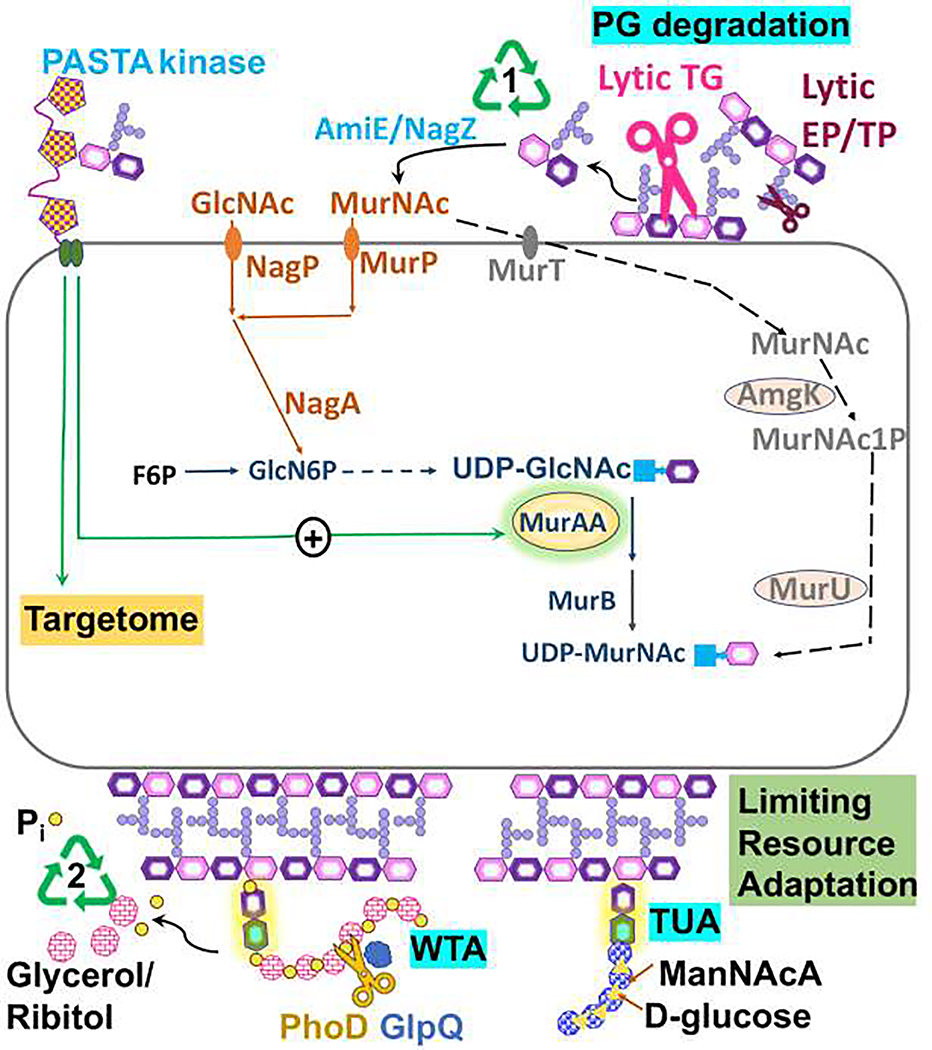
Figure 3. Degradation and recycling of the cell envelope.
Cell growth is accompanied by release of muropeptides generated by lytic transglycosylases (lytic TG) and endopeptidases/transpeptidases (lytic EP.TP) (1), which may be recycled. Recycling pathways involve degradation to release GlcNAc and MurNAc sugars and amino acids. Gram-negative bacteria may import muropeptodes and recycle intracellularly (not shown). In B. subtilis (orange), MurNAc and GlcNAc, are imported by PTS transporters MurP and NagP. The oral pathogen Tannerella forsythia is auxotrophic for MurNAc, which is imported by MurT and processed to generate UDPMurNAc (gray). Muropeptides can also activate some PASTA kinases, which phosphorylate many target proteins (targetome). In some systems, this can regulate the PG branchpoint enzyme MurA. (2) During phosphate limitation, Gram-positive bacteria may recycle WTA (glycerol-P copolymer) aided by the GlpQ and PhoD proteins. Secreted teichoicases may also cleave WTA off of nearby cells to scavenge phosphate. Phosphate limitation in B. subtilis triggers a switch from synthesis of a glycerol-P-based WTA to a functionally similar, but phosphate-free teichuronic acid polymer.
Walls and bridges: diversion of amino acids to PG.
In addition to aminosugars, PG synthesis requires amino acids for the peptide crosslinks. A typical pentapeptide, such as in E. coli or B. subtilis, consists of L-Ala, D-Glu, meso-diaminopimelate (mDAP), and two D-Ala residues. However, there is some variation in other organisms [48]. Both Ala and Glu are abundant amino acids, and racemases facilitate the interconversion of the L- and D-isomers. Meso-diaminopimelate (mDAP) is a precursor on the pathway to lysine.
Mutations that perturb pentapeptide synthesis can trigger morphological abnormalities or cell lysis. In Caulobacter crescentus mutants lacking the RNA chaperone Hfq are slow growing and morphologically altered [49]. Genetic studies traced this defect to decreased activity of TCA cycle enzymes. The resulting increase in α-ketoglutarate (KG) inhibited succinyldiaminopimelate aminotransferase (KG is a product of the reaction), an enzyme required for mDAP biosynthesis [49]. A similar stalling of PG synthesis was observed in a B. subtilis strain lacking aspartate transaminase (AspB), the first enzyme in Asp biosynthesis, when grown in rich medium. Asp is a precursor for mDAP biosynthesis, and limitation therefore compromises PG synthesis [50].
Crossing the border: undecaprenol phosphate as a carrier lipid.
Undecaprenyl phosphate (UP) is a C55 isoprenoid lipid needed to shuttle hydrophilic envelope precursors across the membrane [51] (Fig. 2). UP serves as a lipid anchor during assembly of PG, LPS O-antigen, and WTA [20]. UP is synthesized in its pyrophosphate form (UPP), and PG transglycosylases also release UPP. Therefore, phosphatases are required for UP synthesis and recycling. The total number of UP(P) carrier molecules in the cell is limited, with an estimated ~1.5 × 105 copies per cell in E. coli and S. aureus [52]. The UP lipid carrier is coupled to PG precursors to generate the lipid II donor for PG synthesis, and then released and dephosphorylated after transglycosylation. This cycle is estimated to take ~90 seconds, with the flipping of UP(P) back to the cytosol likely rate-limiting [53].
Depletion of the UP(P) carrier lipid disrupts PG synthesis. Indeed, this is the mechanism of action of bacitracin, which sequesters UPP. Depletion can also result from mutations or antibiotics that block other UP-dependent pathways, including lipo/oligosaccharides, WTA, and CPS synthesis. The impact of UP sequestration first emerged in studies of WTA. Mutations that block late stages of WTA synthesis led to cell death, whereas loss of the first enzyme in the pathway (TagO, Fig 2) leads to cells that are misshapen, but viable [54]. This insight led to the development of Targocil, an antibiotic that inhibits WTA export thereby depleting UP and triggering a shutdown of PG synthesis [55]. In E. coli, PG synthesis is also highly sensitive to conditions that lead to UP limitation [56,57].
Reuse it or lose it: PG recycling.
PG synthesis relies on the ordered insertion of newly synthesized glycan strands into the existing wall structure. While the details are still debated, active PG synthesis requires endopeptidases to cleave peptide crossbridges to make room for new strand insertion [58]. In addition, lytic transglycosylases degrade some of the existing PG structure to release muropeptides [58]. The generation and release of muropeptides during growth is substantial (Fig. 3), with up to ~50% of PG recycled each generation. In Gram-negative bacteria, where muropeptides are recaptured from the periplasm, recycling is efficient [59]. In Gram-positive bacteria there is significant loss of muropeptides to the environment. Studies in S. aureus, B. subtilis, and Streptomyces coelicolor suggest that aminosugars released from the cell wall may be recycled upon entry into stationary phase. However, the efficiency of recycling is low, with only 5–10% MurNAc recovery [60].
The Gram-positive cell wall additionally contains WTA, which can comprise up to 60% of the wall mass [61]. Synthesis of WTA, a sugar-phosphate copolymer, places a high demand on cellular phosphate reserves. In B. subtilis, the PhoPR regulatory system responds to phosphate limitation by repressing WTA synthesis and activating the synthesis of a substitute phosphate-free polymer, teichuronic acid (Fig 3) [62]. The cell may also recycle phosphate from existing WTA. PhoR activates expression of a WTA catabolic teichoicase (GlpQ) and phosphohydrolase (PhoD) (Fig 3) [63]. This phosphate scavenging mechanism can be used to selectively degrade the WTA of neighboring bacteria. For example, S. aureus produces a ribitol-phosphate WTA, and phosphate-limited cells express a teichoicase that releases glycerol-phosphate from the WTA of neighboring bacteria [64].
The virtue of charity: interspecies resource sharing.
The large metabolic demands associated with envelope synthesis may be reduced if precursors can be obtained from neighboring organisms. For example, the obligate intracellular pathogen Rickettsia relies on >50 host-derived metabolites for growth [65]. Among these, isoprenoid precursors are required for synthesis of the UP(P) carrier lipid [66]. As a result, treatment of mammalian cells with statins that reduce isoprenoid synthesis lead to a decreased proliferation and morphological defects in resident Rickettsia (Fig 4) [66].
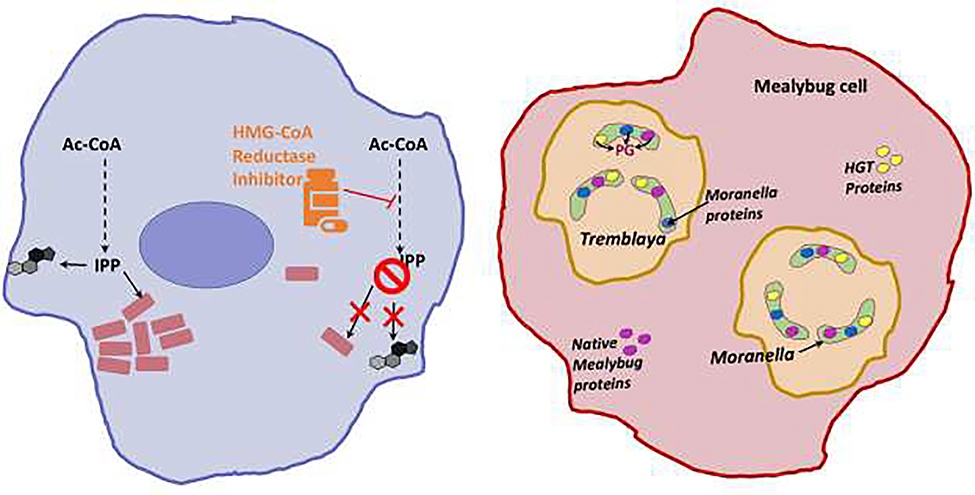
Figure 4. Resource sharing beyond kin.
Left: Rickettsia (pink rectangles) obtains isoprenoid precursors from its host. The inhibition of host HMG-CoA reductase restricts this metabolic crossfeeding and ultimately affects Rickettsia survival. Right: Moranella is snugly housed in Tremblaya, which in turn lives as an endosymbiont in its insect host, the mealybug Planoccous citri cells. These nested cells share the burden of PG synthesis for the diminutive Moranella cell. The initial enzymes for PG biosynthesis are imported from the Mealybug and include both housekeeping enzymes (Glm-S,U, and M-depicted in pink) and enzymes acquired by horizontal gene transfer from Bacteria (MurA-F, DdlB, DapF, MltB, AmiD- depicted in yellow). The cytosolic PG intermediate synthesized by these imported enzymes can then be assembled by later stage enzymes retained by Moranella (MraY-Pbp’s- depicted in blue).
Interspecies cross-feeding is also essential for the survival of Tannerella forsythia. This organism lacks GlmS, MurA, and MurB synthetic enzymes, and relies instead on scavenging of PG turnover products from its periodontal neighbors [59]. Co-habiting Fusobacterium nucleatum may act as a patron to provide muropeptides and perhaps LPS-derived sugars, and sialic acid may be scavenged from salivary mucins and glycoproteins in the oral mucosa [67]. It is likely that further analysis of complex microbial communities will reveal other examples of metabolically interdependent cells that share the burden of synthesizing cell envelope precursors.
Organisms in long-term associations may instead share the burden of enzyme production. The ability of an endosymbiont to transfer the metabolic burden of gene maintenance, transcription, and protein synthesis to its host is evolutionarily advantageous, assuming that the enzyme can be imported to fulfill its necessary function. The amoeba Paulinella chromatophora, for example, grows photosynthetically by virtue of a symbiotic association with a cyanobacterium that resides as a plastid (chromatophore) in the host cytosol [68]. This plastid retains a 1 Mb genome, but several metabolic pathways, including PG synthesis, require enzymes whose genes now reside in the amoeba. In the case of PG, the bacterial MurF enzyme for attaching D-Ala-D-Ala to the stem peptide is host encoded and must be imported for PG synthesis [68].
This type of gene transfer has been taken even further in a bacterium (Moranella) residing within another bacterium (Tremblaya), which in turn resides as a symbiont in an insect (mealybug) (Fig 4) [69]. Both bacterial symbionts have greatly reduced genomes, with only 400 protein-coding genes in Moranella, and 120 in Tremblaya. Although Moranella retains genes for most of the enzymes needed to assemble PG in the periplasm, it lacks those needed for the cytosolic steps leading to the lipid I precursor. These genes are in the insect genome, having been imported from multiple bacteria. Remarkably, to function in Moranella, these insect-synthesized proteins must cross five lipid bilayers (Tremblaya has three and Moranella two). The ability of Moranella to synthesize PG therefore reflects a shared burden of enzyme synthesis, with enzymes for the late stages of synthesis encoded by the endosymbiont and those for early stages imported from the insect host [69]. Given the ubiquity of reductive genome evolution in symbionts, it is likely that cell envelope synthesis is a shared burden in many other systems.
Conclusions.
Assembling a cell envelope imposes a large metabolic burden on cells. Cells have evolved a wide variety of mechanisms to economically regulate the flux of precursors needed for envelope biogenesis, often by controlling the activity of the enzymes at critical crossroads of metabolism. Situations that deplete required precursors, either through mutation, nutrient limitation, or the action of antimicrobial compounds, weaken the envelope and can lead to cell death. Cells may bypass precursor limitation by scavenging relevant nutrients from the environment or by cleaving cell surface structures from neighboring cells. Finally, we highlight examples where the metabolic burden is shared between intracellular bacteria and their hosts, either through provision of metabolic intermediates or by sharing the burden of enzyme synthesis.
Highlights.
Bacteria are protected and shaped by membranes and a peptidoglycan wall
Peptidoglycan is often linked to a polysaccharide capsule and teichoic acids
Synthesis of the cell envelope requires a partitioning of shared metabolites
The key branchpoint enzymes are regulated by multiple feedback mechanisms
Key metabolites may be obtained from other bacteria or a eukaryotic host
Acknowledgments:
We thank Anne Galinier, Kyu Rhee, and Tobias Dörr for critical reading of the manuscript.
Funding: Research reported in this publication was supported by the National Institutes of Health under award number R35GM122461 to JDH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest statement.
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Chubukov V, Gerosa L, Kochanowski K, Sauer U: Coordination of microbial metabolism. Nat Rev Microbiol 2014, 12:327–340. [DOI] [PubMed] [Google Scholar]
- 2.Monk JM, Lloyd CJ, Brunk E, Mih N, Sastry A, King Z, Takeuchi R, Nomura W, Zhang Z, Mori H, et al. : iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol 2017, 35:904–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reith J, Mayer C: Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol 2011, 92:1–11. [DOI] [PubMed] [Google Scholar]
- 4.Kawai Y, Mercier R, Mickiewicz K, Serafini A, Sorio de Carvalho LP, Errington J: Crucial role for central carbon metabolism in the bacterial L-form switch and killing by beta-lactam antibiotics. Nat Microbiol 2019, 4:1716–1726.• Blocking PG synthesis can trigger the emergence of L-forms, bacteria that lack a cell wall. However, the resultant increase in flux of carbon into lower glycolysis can ultimately the electron transfer chain triggers oxidative stress.
- 5.Yoo W, Yoon H, Seok YJ, Lee CR, Lee HH, Ryu S: Fine-tuning of amino sugar homeostasis by EIIA(Ntr) in Salmonella Typhimurium. Sci Rep 2016, 6:33055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopel Y, Khan MA, Gorke B: Menage a trois: post-transcriptional control of the key enzyme for cell envelope synthesis by a base-pairing small RNA, an RNase adaptor protein, and a small RNA mimic. RNA Biol 2014, 11:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durica-Mitic S, Gorke B: Feedback regulation of small RNA processing by the cleavage product. RNA Biol 2019, 16:1055–1065.•• The complex circuitry required for regulation of GlmS in E. coli is further resolved with the demonstration that RapZ is a GlcN6P sensor. GlcN6P-free RapZ stimulates the QseE/QseF TCS to activate expression of the decoy sRNA, GlmY, that sequesters RapZ and prevents GlmZ decay (Fig. 1C).
- 8.Khan MA, Durica-Mitic S, Gopel Y, Heermann R, Gorke B: Small RNA-binding protein RapZ mediates cell envelope precursor sensing and signaling in Escherichia coli. EMBO J 2020, 39:e103848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferre-D’Amare AR: The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA. Q Rev Biophys 2010, 43:423–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulquier E, Galinier A: YvcK, a protein required for cell wall integrity and optimal carbon source utilization, binds uridine diphosphate-sugars. Sci Rep 2017, 7:4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foulquier E, Pompeo F, Byrne D, Fierobe HP, Galinier A: Uridine diphosphate N-acetylglucosamine orchestrates the interaction of GlmR with either YvcJ or GlmS in Bacillus subtilis. Sci Rep 2020, 10:15938.• Biochemical studies demonstrate that B. subtilis GlmR(YvcK) activates GlmS. This activation is prevented by UDP-GlcNAc, and in the presence of UDP-GlcNAc GlmR instead forms a complex with the RapZ homolog YvcJ.
- 12.Gorke B, Foulquier E, Galinier A: YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology (Reading) 2005, 151:3777–3791. [DOI] [PubMed] [Google Scholar]
- 13.Kleijn RJ, Buescher JM, Le Chat L, Jules M, Aymerich S, Sauer U: Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J Biol Chem 2010, 285:1587–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel V, Wu Q, Chandrangsu P, Helmann JD: A metabolic checkpoint protein GlmR is important for diverting carbon into peptidoglycan biosynthesis in Bacillus subtilis. PLoS Genet 2018, 14:e1007689.• Genetic studies reveal that B. subtilis GlmR(YvcK) activates GlmS (see (11)), and this activation is critical for growth under nutrient conditions where the GlmS precursor, F6P, is present at low levels.
- 15.Gerth U, Kock H, Kusters I, Michalik S, Switzer RL, Hecker M: Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 2008, 190:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, et al. : Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 2009, 10:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pensinger DA, Boldon KM, Chen GY, Vincent WJ, Sherman K, Xiong M, Schaenzer AJ, Forster ER, Coers J, Striker R, et al. : The Listeria monocytogenes PASTA Kinase PrkA and Its Substrate YvcK Are Required for Cell Wall Homeostasis, Metabolism, and Virulence. PLoS Pathog 2016, 12:e1006001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir M, Prisic S, Kang CM, Lun S, Guo H, Murry JP, Rubin EJ, Husson RN: Mycobacterial gene cuvA is required for optimal nutrient utilization and virulence. Infect Immun 2014, 82:4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imperiali B: Bacterial carbohydrate diversity - a Brave New World. Curr Opin Chem Biol 2019, 53:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitfield C, Wear SS, Sande C: Assembly of Bacterial Capsular Polysaccharides and Exopolysaccharides. Annu Rev Microbiol 2020. [DOI] [PubMed] [Google Scholar]
- 21.Sumrall ET, Keller AP, Shen Y, Loessner MJ: Structure and function of Listeria teichoic acids and their implications. Mol Microbiol 2020, 113:627–637. [DOI] [PubMed] [Google Scholar]
- 22.Keinhorster D, George SE, Weidenmaier C, Wolz C: Function and regulation of Staphylococcus aureus wall teichoic acids and capsular polysaccharides. Int J Med Microbiol 2019, 309:151333. [DOI] [PubMed] [Google Scholar]
- 23.van Dalen R, Peschel A, van Sorge NM: Wall Teichoic Acid in Staphylococcus aureus Host Interaction. Trends Microbiol 2020. [DOI] [PubMed] [Google Scholar]
- 24.Sobhanifar S, Worrall LJ, Gruninger RJ, Wasney GA, Blaukopf M, Baumann L, Lameignere E, Solomonson M, Brown ED, Withers SG, et al. : Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid alpha-glycosyltransferase. Proc Natl Acad Sci U S A 2015, 112:E576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlach D, Guo Y, De Castro C, Kim SH, Schlatterer K, Xu FF, Pereira C, Seeberger PH, Ali S, Codee J, et al. : Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 2018, 563:705–709. [DOI] [PubMed] [Google Scholar]
- 26.Chandrangsu P, Loi VV, Antelmann H, Helmann JD: The Role of Bacillithiol in Gram-Positive Firmicutes. Antioxid Redox Signal 2018, 28:445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleverley RM, Rutter ZJ, Rismondo J, Corona F, Tsui HT, Alatawi FA, Daniel RA, Halbedel S, Massidda O, Winkler ME, et al. : The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes. Nat Commun 2019, 10:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halbedel S, Lewis RJ: Structural basis for interaction of DivIVA/GpsB proteins with their ligands. Mol Microbiol 2019, 111:1404–1415. [DOI] [PubMed] [Google Scholar]
- 29.Hammond LR, White ML, Eswara PJ: !vIVA la DivIVA! J Bacteriol 2019, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rismondo J, Bender JK, Halbedel S: Suppressor Mutations Linking gpsB with the First Committed Step of Peptidoglycan Biosynthesis in Listeria monocytogenes. J Bacteriol 2017, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kock H, Gerth U, Hecker M: MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol Microbiol 2004, 51:1087–1102. [DOI] [PubMed] [Google Scholar]
- 32.Ayoola MB, Shack LA, Nakamya MF, Thornton JA, Swiatlo E, Nanduri B: Polyamine Synthesis Effects Capsule Expression by Reduction of Precursors in Streptococcus pneumoniae. Front Microbiol 2019, 10:1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troxler LJ, Werren JP, Schaffner TO, Mostacci N, Vermathen P, Vermathen M, Wuthrich D, Simillion C, Brugger SD, Bruggmann R, et al. : Carbon source regulates polysaccharide capsule biosynthesis in Streptococcus pneumoniae. J Biol Chem 2019, 294:17224–17238.• An exploration of how different sugar sources affect the synthesis of capsule that illuminates why capsule levels are greatly reduced in cells grown on fructose.
- 34.Fivenson EM, Bernhardt TG: An Essential Membrane Protein Modulates the Proteolysis of LpxC to Control Lipopolysaccharide Synthesis in Escherichia coli. mBio 2020, 11.• Together with (35), this study further develops a model in which the inner membrane proteins YejM modulates LpxC turnover by FtsH-LapB to regulate LPS synthesis.
- 35.Guest RL, Same Guerra D, Wissler M, Grimm J, Silhavy TJ: YejM Modulates Activity of the YciM/FtsH Protease Complex To Prevent Lethal Accumulation of Lipopolysaccharide. mBio 2020, 11.Genetic studies here, and in (34), refine a model for the feedback regulation of LPS biosynthesis in which the YejM inner membrane protein regulates proteolysis of LpxC, possibly in response to accumulation of LPS in the inner membrane.
- 36.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CR, et al. : Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol 1999, 31:833–844. [DOI] [PubMed] [Google Scholar]
- 37.Miller CN, Steele SP, Brunton JC, Jenkins RJ, LoVullo ED, Taft-Benz SA, Romanchuk A, Jones CD, Dotson GD, Collins EJ, et al. : Extragenic suppressor mutations in Delta-ripA disrupt stability and function of LpxA. BMC Microbiol 2014, 14:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manuse S, Fleurie A, Zucchini L, Lesterlin C, Grangeasse C: Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol Rev 2016, 40:41–56. [DOI] [PubMed] [Google Scholar]
- 39.Shah IM, Laaberki MH, Popham DL, Dworkin J: A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 2008, 135:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardt P, Engels I, Rausch M, Gajdiss M, Ulm H, Sass P, Ohlsen K, Sahl HG, Bierbaum G, Schneider T, et al. : The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int J Med Microbiol 2017, 307:1–10. [DOI] [PubMed] [Google Scholar]
- 41.Wamp S, Rutter ZJ, Rismondo J, Jennings CE, Moller L, Lewis RJ, Halbedel S: PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM. Elife 2020, 9.•• This study reveals that the Listeria monocytogenes PrkA PASTA kinase regulates a conserved protein (ReoM) that is essential for mitigating ClpCP-dependent proteolytic degradation of MurA to enable PG synthesis.
- 42.Bellinzoni M, Wehenkel AM, Duran R, Alzari PM: Novel mechanistic insights into physiological signaling pathways mediated by mycobacterial Ser/Thr protein kinases. Microbes Infect 2019, 21:222–229. [DOI] [PubMed] [Google Scholar]
- 43.Jagtap PK, Soni V, Vithani N, Jhingan GD, Bais VS, Nandicoori VK, Prakash B: Substrate-bound crystal structures reveal features unique to Mycobacterium tuberculosis N-acetyl-glucosamine 1-phosphate uridyltransferase and a catalytic mechanism for acetyl transfer. J Biol Chem 2012, 287:39524–39537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turapov O, Forti F, Kadhim B, Ghisotti D, Sassine J, Straatman-Iwanowska A, Bottrill AR, Moynihan PJ, Wallis R, Barthe P, et al. : Two Faces of CwlM, an Essential PknB Substrate, in Mycobacterium tuberculosis. Cell Rep 2018, 25:57–67 e55.•• CwlM, a substrate of the PknB PASTA kinase, is a cytoplasmic regulator homologous to cell wall lytic enzymes. This study suggests that CwlB potentially regulates both the biosynthesis of PG precursors and their transport by MurJ across the cytoplasmic membrane.
- 45.Boutte CC, Baer CE, Papavinasasundaram K, Liu W, Chase MR, Meniche X, Fortune SM, Sassetti CM, Ioerger TR, Rubin EJ: A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le NH, Locard-Paulet M, Stella A, Tomas N, Molle V, Burlet-Schiltz O, Daffe M, Marrakchi H: The protein kinase PknB negatively regulates biosynthesis and trafficking of mycolic acids in mycobacteria. J Lipid Res 2020, 61:1180–1191.• An exploration of the role of the PknB PASTA kinase with a focus on its role in mycolic acid biosynthesis.
- 47.Carette X, Platig J, Young DC, Helmel M, Young AT, Wang Z, Potluri LP, Moody CS, Zeng J, Prisic S, et al. : Multisystem Analysis of Mycobacterium tuberculosis Reveals Kinase-Dependent Remodeling of the Pathogen-Environment Interface. mBio 2018, 9.• A multi-omics analysis that documents the highly pleiotropic effects from inhibition of the PknA and PknB PASTA kinases, including effects on multiple pathways affecting the cell envelope.
- 48.Turner RD, Vollmer W, Foster SJ: Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 2014, 91:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irnov I, Wang Z, Jannetty ND, Bustamante JA, Rhee KY, Jacobs-Wagner C: Crosstalk between the tricarboxylic acid cycle and peptidoglycan synthesis in Caulobacter crescentus through the homeostatic control of alpha-ketoglutarate. PLoS Genet 2017, 13:e1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Roistacher DM, Helmann JD: Aspartate deficiency limits peptidoglycan synthesis and sensitizes cells to antibiotics targeting cell wall synthesis in Bacillus subtilis. Mol Microbiol 2018, 109:826–844.• This paper, together with (49), reveal how perturbations of central metabolism can restrict PG synthesis. In this case, exploration of a lysis phenotype in a transposon-generated mutant reveals that Asp limitation during growth triggers mDAP deficiency, whereas in (49) mDAP deficiency results from high alpha-ketoglutarate levels.
- 51.Workman SD, Strynadka NCJ: A Slippery Scaffold: Synthesis and Recycling of the Bacterial Cell Wall Carrier Lipid. J Mol Biol 2020, 432:4964–4982. [DOI] [PubMed] [Google Scholar]
- 52.Barreteau H, Magnet S, El Ghachi M, Touze T, Arthur M, Mengin-Lecreulx D, Blanot D: Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J Chromatogr B Analyt Technol Biomed Life Sci 2009, 877:213–220. [DOI] [PubMed] [Google Scholar]
- 53.Piepenbreier H, Diehl A, Fritz G: Minimal exposure of lipid II cycle intermediates triggers cell wall antibiotic resistance. Nat Commun 2019, 10:2733.• A comprehensive analysis of the lipid II cycle that integrates literature data, experimental results, and mathematical modeling to provide insights into the key steps that are limiting for PG synthesis.
- 54.D’Elia MA, Millar KE, Beveridge TJ, Brown ED: Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 2006, 188:8313–8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schirner K, Eun YJ, Dion M, Luo Y, Helmann JD, Garner EC, Walker S: Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat Chem Biol 2015, 11:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgenson MA, Young KD: Interrupting Biosynthesis of O Antigen or the Lipopolysaccharide Core Produces Morphological Defects in Escherichia coli by Sequestering Undecaprenyl Phosphate. J Bacteriol 2016, 198:3070–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jorgenson MA, MacCain WJ, Meberg BM, Kannan S, Bryant JC, Young KD: Simultaneously inhibiting undecaprenyl phosphate production and peptidoglycan synthases promotes rapid lysis in Escherichia coli. Mol Microbiol 2019, 112:233–248.• This paper, together with (56), explores the physiological consequences of conditions that trigger limitation of the UP carrier lipid.
- 58.Egan AJF, Errington J, Vollmer W: Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 2020, 18:446–460. [DOI] [PubMed] [Google Scholar]
- 59.Mayer C, Kluj RM, Muhleck M, Walter A, Unsleber S, Hottmann I, Borisova M: Bacteria’s different ways to recycle their own cell wall. Int J Med Microbiol 2019, 309:151326. [DOI] [PubMed] [Google Scholar]
- 60.Borisova M, Gaupp R, Duckworth A, Schneider A, Dalugge D, Muhleck M, Deubel D, Unsleber S, Yu W, Muth G, et al. : Peptidoglycan Recycling in Gram-Positive Bacteria Is Crucial for Survival in Stationary Phase. mBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown S, Santa Maria JP, Jr., Walker S: Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 2013, 67:313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Botella E, Devine SK, Hubner S, Salzberg LI, Gale RT, Brown ED, Link H, Sauer U, Codee JD, Noone D, et al. : PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol Microbiol 2014, 94:1242–1259. [DOI] [PubMed] [Google Scholar]
- 63.Myers CL, Li FK, Koo BM, El-Halfawy OM, French S, Gross CA, Strynadka NC, Brown ED: Identification of Two Phosphate Starvation-induced Wall Teichoic Acid Hydrolases Provides First Insights into the Degradative Pathway of a Key Bacterial Cell Wall Component. J Biol Chem 2016, 291:26066–26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorge AM, Schneider J, Unsleber S, Xia G, Mayer C, Peschel A: Staphylococcus aureus counters phosphate limitation by scavenging wall teichoic acids from other staphylococci via the teichoicase GlpQ. J Biol Chem 2018, 293:14916–14924.•• Phosphate is a limiting resource in many environments. S. aureus can take advantage of the abundant phosphate present in the WTA of neighboring bacteria. Secreted teichoicases can liberate phosphate from neighboring cells that have glycerol-P WTA, while sparing its own ribitol-P polymer.
- 65.Driscoll TP, Verhoeve VI, Guillotte ML, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF, Gillespie JJ: Wholly Rickettsia! Reconstructed Metabolic Profile of the Quintessential Bacterial Parasite of Eukaryotic Cells. mBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahyong V, Berdan CA, Burke TP, Nomura DK, Welch MD: A Metabolic Dependency for Host Isoprenoids in the Obligate Intracellular Pathogen Rickettsia parkeri Underlies a Sensitivity to the Statin Class of Host-Targeted Therapeutics. mSphere 2019, 4.•• Bacteria require isoprenoid lidids for synthesis of the C55 undecaprenyl phosphate lipid carrier and for the synthesis of quinones important for electron transport. The intracellular pathogen Rickettsia relies on a host isoprenoid precursor (isopentenyl-pyrophosphate).
- 67.Ruscitto A, Sharma A: Peptidoglycan synthesis in Tannerella forsythia: Scavenging is the modus operandi. Mol Oral Microbiol 2018, 33:125–132.• This review summarizes evidence that the oral pathogen Tannerella forsythia requires MurNAc released by cohabiting bacteria or host sialic acid (Neu5Ac) to support PG synthesis.
- 68.Nowack EC, Price DC, Bhattacharya D, Singer A, Melkonian M, Grossman AR: Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc Natl Acad Sci U S A 2016, 113:12214–12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bublitz DC, Chadwick GL, Magyar JS, Sandoz KM, Brooks DM, Mesnage S, Ladinsky MS, Garber AI, Bjorkman PJ, Orphan VJ, et al. : Peptidoglycan Production by an Insect-Bacterial Mosaic. Cell 2019, 179:703–712 e707.•• A fascinating example of endosymbiosis, genome reduction, and enzyme translocation to allow a reconstructed PG synthetic pathway to support the Moranella endosymbiont.