Abstract
Glioblastoma (GB) is by far the most hostile type of malignant tumor that primarily affects the brain and spine, derived from star-shaped glial cells that are astrocytes and oligodendrocytes. Despite of significant efforts in recent years in glioblastoma research, the clinical efficacy of existing medical intervention is still limited and very few potential diagnostic markers are available. Long non-coding RNAs (lncRNAs) that lacks protein-coding capabilities were previously thought to be “junk sequences” in mammalian genomes are quite indispensible epigenetic regulators that can positively or negatively regulate gene expression and nuclear architecture, with significant roles in the initiation and development of tumors. Nevertheless, the precise mechanism of these distortedly expressed lncRNAs in glioblastoma pathogenesis is not yet fully understood. Since the advent of high-throughput sequencing technologies, more and more research have elucidated that lncRNAs are one of the most promising prognostic biomarkers and therapeutic targets for glioblastoma. In this paper, I briefly outlined the existing findings of lncRNAs. And also summarizes the profiles of different lncRNAs that have been broadly classified in glioblastoma research, with emphasis on both their prognostic and therapeutic values.
Keywords: Glioblastoma, Long-non coding RNA, Biomarker, MALAT1, H19, HOXA11-AS, TUG1, HOTAIR, CASC2, CRNDE, NEAT1, GAS5, XIST, SOX2OT, ZFAS1
Glioblastoma; Long-non coding RNA; Biomarker; MALAT1; H19; HOXA11-AS; TUG1; HOTAIR; CASC2; CRNDE; NEAT1; GAS5; XIST; SOX2OT; ZFAS1
1. Introduction
In 2001, the main goal of the genome sequencing project is to systematically identify and characterize the complete map of human transcriptome. In recent years, due to the advancement of several new technologies, such as deep RNA sequencing and DNA tiling arrays, catalyzed by improved epigenomic technology and computational prediction techniques [1] increased our database of genetic elements with massive RNA transcripts. Despite the fact that nearly 70% of the mammalian genome is known to be transcribed, however, coding genes account for just 2% of the total genome transcript [2]. Non-coding RNAs (ncRNAs) previously thought to be “junk”, account for the vast majority of genome transcripts. Non-coding RNA (ncRNA) is a term used to describe a type of ribonucleic acid (RNA) molecule that does not encode proteins that include long non-coding RNA (lncRNA), small interfering RNA (siRNA), microRNA (miRNA), small nucleolar RNA (snoRNA) among others (Table 1) [3]. Non-coding RNAs have been shown to have sequence-specific recognition of another nucleic acid in several cases, though not always. They mediate gene expression at both the gene transcriptional and post-transcriptional levels such as splicing, RNA editing, translational inhibition, messenger RNA (mRNA) degradation, and chromosome maintenance and segregation, serves as an essential regulators of several genomic processes and diseases [4]. There is a plethora of evidence which suggests that ncRNAs play a pivotal role in multiple normal biological and disease processes. Therefore, identification of ncRNAs is important for further understanding their function and their presence in complex human diseases (Table 2)
Table 1.
List of small and long non-coding RNAs.
| Type | Subtype | Signature roles | References |
|---|---|---|---|
| Small non-coding RNA (sncRNA) | Transfer RNA (tRNA) | Decode mRNA sequence into a protein | [14] |
| Micro RNA (miRNA) | RNA silencing and post-transcriptional regulation | [15, 16] | |
| Ribosomal 5S and 5.8s RNA (rRNA) | Translation of mRNA | [17] | |
| Piwi interacting RNA (piRNA) | RNA silencing, promote heterochromatin assembly and DNA methylation | [18, 19] | |
| Small interfering RNA (siRNA) | RNA interference | [20] | |
| Small nuclear RNA (snRNA) | Pre-mRNA splicing | [21] | |
| Small nucleolar RNA (snoRNA) | Modification, maturation, and stabilization of rRNA | [22, 23] | |
| Vault RNA (vtRNA) | Intracellular and nucleocytoplasmic transport | [24] | |
| Antisense termini associated short RNA (aTASR) | Increasing transcript copy numbers of their host gene | [25, 26] | |
| Promoter-associated short RNA (PASR) | Act as scaffolds for antisense transcripts, regulate gene expression | [27, 28] | |
| Tiny transcription initiation RNA (tiRNA) | Modulate local epigenetic structure, regulates CTCF localization | [29, 30] | |
| Human Y RNA (hY RNA) | Initiation of chromosomal DNA replication | [31] | |
| Retrotransposon-derived RNA (RE-RNA) | Act as a promoter for transcription initiation | [32, 33] | |
| 3′ UTR-derived RNA (UaRNA) | Post-transcriptional regulation | [34] | |
| Long non-coding RNA (lncRNA) | Large intergenic ncRNA (lincRNA) | Chromatin remodeling, RNA stabilization, transcription regulation | [35, 36, 37] |
| Transcribed ultraconserved regions (T-UCR) | Homeostatic maintenance of splicing factors, regulation of transcription, epigenetic modification | [38, 39] | |
| Long intronic ncRNA | Regulation of alternative splicing, RNA stabilization | [40, 41] | |
| Antisense RNA (aRNA) | Genomic imprinting and X-chromosome inactivation | [42] | |
| Promoter-associated long RNA (PALR) | Serve as scaffolds for antisense transcripts, regulate gene expression | [2, 28] | |
| Pseudogenes | Regulators of multiple biological process, in cancer development | [43, 44] | |
| GAA-repeat containing RNA (GRC-RNA) | Maintenance of nuclear matrix and regulate gene expression | [45] | |
| Promoter upstream transcripts (PROMPTs) | Regulation of gene expression | [46] | |
| Ribosomal 18s and 28s RNA (rRNA) | Translation of mRNA | [17] | |
| Long stress-induced non-coding transcripts (LSINCTs) | Stabilizing and promoting the oncogenic function | [47] |
Table 2.
Functional characteristics of various long non-coding RNAs in glioblastoma.
| lncRNA Type | Biological activity | Expression in GB | Role in GB | Mechanism/Targets | References |
|---|---|---|---|---|---|
| MALAT1 | Proliferation, migration, invasion and apoptosis | Downregulated | Tumor suppression | Inactivation of ERK/MAPK pathway; enhance expression of FBXW7 | [69, 70, 71, 216] |
| HOTAIR | Proliferation, migration, invasion, apoptosis and angiogenesis | Overexpressed | Promote proliferation and tumorigenesis potential of GB cells | Interact with PRC2, regulate Wnt/β-catenin signaling; Interact with miR-148-3p and miR-326/FGF1 pathway | [217, 218, 219, 220] |
| HOXA11-AS | Proliferation, migration, invasion and apoptosis | Overexpressed | Promote tumorigenesis of GB cells | Interact with miR-214-3p/EZH2 axis; sponge miR-124-3p and miR-140-5p | [96, 98] |
| NEAT1 | Proliferation, migration, invasion and apoptosis | Overexpressed | Promote glioblastoma pathogenesis | Interact with miR-let-7e, miR-132/SOX2 and miR-449b-5p/c-Met axis; associate with EGFR/NEAT1/EZH2/β-catenin | [105, 108, 114, 221] |
| GAS5 | Proliferation, migration, invasion and apoptosis | Downregulated | Inhibit GSC proliferation, migration and invasion | Interact with miR-196a-5p/FOXO1 feedback loop; reduce miR-222 and increase bmf and plexin C1; repress miR-18a-5p | [116, 121, 222] |
| CRNDE | Proliferation, migration, invasion and apoptosis | Overexpressed | Promote glioma cell proliferation and migration and tumorigenesis | Interact with P70S6K-mediated mTOR signaling; attenuate miR-384/PIWIL4/STAT3 axis; negatively regulate miR-186 and miR-136-5p | [129, 132, 133, 134] |
| HOTTIP | Proliferation, cell-cycle progression, invasion and apoptosis | Downregulated | Inhibit cell proliferation and promote apoptosis | Down-regulate BRE expression; critical factor in the HIF-1a/miR-101/ZEB1 axis | [139, 143] |
| SOX2OT | Proliferation, migration, invasion and apoptosis | Overexpressed | Promotes growth, migration, invasion, and tumorigenesis of GSCs | Interact with SOX2OT/miR-194-5p/miR-122/SOX3/TDGF-1 axis; regulate CDK2, CDK2AP2, ACTR3, INCENP, SMC4 and GNL3L |
[150, 151, 152] |
| XIST | Proliferation, migration, invasion and angiogenesis | Overexpressed | Promote tumorigenicity and angiogenesis | CeRNA for miR-152, miR-429, miR-29c, miR-92b and miR-137; regulate miR-133a/SOX4 axis and miR-137/Rac1 axis | [156, 157, 158, 160] |
| H19 | Proliferation, angiogenesis and temozolomide resistance | Overexpressed | Promote angiogenesis, invasion, stemness and tumorigenicity of GB cell | Interact with miR-675/CDK6, miR-29a/VASH2 and miR-140/iASPP (inhibitor of apoptosis stimulating protein of p53) axis; reduce miR-152 | [179, 180, 182] |
| TUG1 | Proliferation, apoptosis | Overexpressed | Maintains stemness and tumorigenic properties of GSCs; modulates BTB; and increases glioma-induced angiogenesis | CeRNAs for miR-26a, miR-144, miR-299, and miR-145; interact with PRC2 and YY1 | [193, 196, 197, 199] |
| ZFAS1 | Proliferation, migration, invasion, disease prognosis | Overexpressed | Promote cell proliferation, invasion and tumorigenicity of glioma | Activates the Notch signaling pathway; regulating miR-150-5p/PLP2 axis | [206, 215] |
Glioblastoma (GB), also referred as Grade IV astrocytoma, is an extremely severe form of primary intracranial tumor that affects the brain and spine, with a variety of malignancies and histological subtypes [5]. GB is an exceedingly fast growing glioma that can arise in the brain “de novo” or evolves from lower grade oligodendrogliomas or astrocytomas. In adults, GB occurs most frequently in the cerebral hemisphere region, chiefly in the temporal and frontal lobes [6]. The median survival rate of patients with GB was just 12–15 months, with less than 5% of patients surviving more than 5 years after diagnosis despite the use of multiple treatment strategies, such as chemotherapy and radiotherapy even after maximal resection was performed [7]. Cancer is a multistage process encompassing the acquisition of several genetic and epigenetic changes. In recent years, non-coding RNAs, especially miRNAs and lncRNAs, have received a great deal of interest in cancer biology [8, 9, 10, 11]. Dysregulation of lncRNA expression may lead to the instigation and progression of GB, and these changes may prompt the potential of lncRNA as diagnostic, prognostic and predictive markers [12, 13]. However, it is still in its initial phase to assess the apt ways to address such improvements for therapeutic interventions. Therefore, future research should be focused on further characterization of these lncRNAs, especially in terms of structure and mechanism, prior to their implementation as drug targets. In this review, I summarized the existing knowledge of lncRNAs and their role in glioblastoma research that has been under intensive investigation in recent years, with a focus on their prognostic and therapeutic implications.
2. Long non-coding RNA (lncRNA)
Long non-coding RNAs (lncRNAs) exhibit the largest class of ncRNAs with approximately 10,000 lncRNA genes. LncRNAs are linear mRNAs like transcripts, the length of which ranges from 200 nucleotides (nt) to approximately 100 kilobase (kb) lacking significant open reading frames (ORFs) [48, 49]. However, some lncRNAs contain short ORFs and go through translation, though only a limited percentage of such translation procedures result in functional and stable peptides [50]. The majority of eukaryotic lncRNAs are constructed by RNA polymerase II (RNAPII). There have been few examples, however, such as B2-short interspersed elements (B2-SINE) RNA synthesized through RNA polymerase III (RNAPIII).
Since lncRNAs are more often found in the nucleus than mRNAs, they are prone to improper splicing and polyadenylation, as well as resistance to exosome-mediated chromatin degradation. However, once in the nucleus they serve as effective regulators of the nuclear organization and function [51, 52]. LncRNAs exhibits poor sequence conservation across species. However, the structure of RNA is heavily conserved than its sequence, which shows that alignment of conventional sequences across species may truly not reveal the functionally conserved patterns of the RNA. Thus, poorly conserved lncRNAs can still be functionally active [53, 54].
The number of lncRNA genes in the human genome is estimated to be approximately 7000 to 23,000, suggesting that this class of ncRNAs represents an enormous yet undiscovered portion of normal cellular pathways that may be impaired in cancer [55]. H19 was the very first eukaryotic lncRNA gene to be identified, and was soon accompanied by the discovery of the silencing X-inactive-specific transcript (XIST) lncRNA gene, which portrays a significant role in X-chromosome inactivation [56, 57]. H19 expression in transgenic mice has been shown to be lethal at the prenatal stage, suggesting that the dosage of the lncRNA is tightly regulated and contributes in embryonic development [58].
2.1. Classification of long non-coding RNAs (lncRNAs)
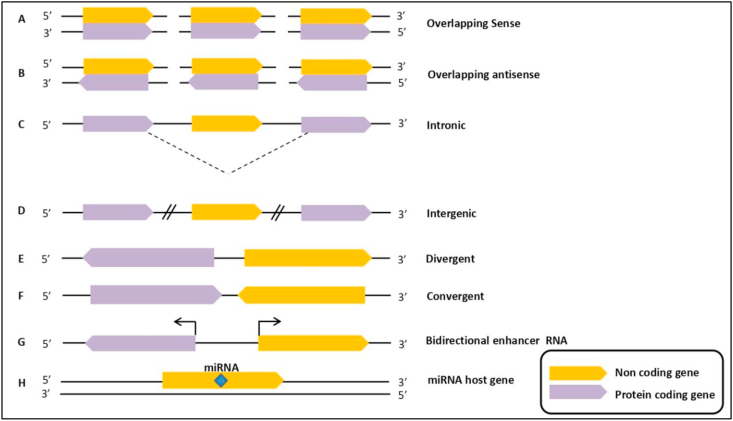
LncRNAs represents the major class of non-coding RNAs and accounts for a greater proportion of the transcribed genome. The traditional classification of lncRNAs based on their locations, including five main subtypes: (i) Antisense RNA refer to those RNA which have a complementary sequence to another RNA transcript. This group comprises natural antisense transcripts (NATs), which may occur in cis-NATs (affects the expression level of the corresponding sense transcript) or in trans-NATs (regulate expression level of non-paired genes from other genomic locations); (ii) long or large intergenic non-coding RNA (lincRNA) located between protein-coding genes, but it can also be found in introns; (iii) overlapping sense RNA, encompassing exons or whole protein coding genes (PCGs) within their introns and without any sense exon overlapping, they are transcribed in the same sense direction; (iv) sense intronic RNA, an RNA transcript that is encoded within the intron of the coding gene but does not overlap with other exons on the same strand; (v) bidirectional lncRNAs, transcribed on the opposite strand from the nearest PCG (Figure 1) [36,41,49]. Moreover, as various new types of lncRNAs were progressively being investigated, the standard classification of lncRNA did not encompass the entire domain and needed to be explained in greater detail. Sandre et al., categorized lncRNAs into eight groups: (i) intronic, transcribed from an intron of another gene; (ii) divergent (pancRNA), derived from the opposite strand of the same promoter region of protein coding gene; (ii) convergent, encoded on the opposite strands; (iv) intergenic, located distant from other genes; (v) overlapping sense, overlapped with other genes on the same strand; (vi) overlapping antisense, overlapped with other genes on the opposite strand; (vii) miRNA host gene; (viii) enhancer RNA, expressed as uni- or bi-directional transcripts [59].
Figure 1.
Classification of long non-coding RNAs. LncRNAs can be grouped into the following groups based on their location in the genome: (A) Overlapping sense; (B) Overlapping antisense; (C) Intronic; (D) Intergenic; (E) Divergent; (F) Convergent; (G) Bidirectional enhancer RNA; (H) miRNA host gene. Protein coding genes are represented by purple color, while long non-coding genes are represented by orange color.
To date, lncRNAs primarily act as mRNA decay mediators, structural scaffolds, signaling molecules, histone modification and chromatin remodeling regulators and as miRNA sponges. They also have been involved in pluripotency, cell reprogramming and recruiting RNA polymerase to promoter regions [60, 61, 62]. Nevertheless, the functional roles of the majority of lncRNAs still lack strong supporting evidence compared to other ncRNAs for instance miRNAs, which offers numerous opportunities for understanding and analyzing the functions of lncRNAs in cancer.
3. Overview of long non-coding RNAs in glioblastoma
3.1. MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1)
LncRNA MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1) is also known as NEAT2 (Non-coding Nuclear Enriched Abundant Transcript 2), and is dominantly expressed in the nucleus of the cell. It is about 7 kb in length and spliced and polyadenylated long non-coding RNA at chromosome 11q13.1 (Chr11q13.1) locus [63]. MALAT1 is majorly enriched in the paraspeckle, an integral nuclear structure that contains large quantities of various different proteins and regulates the pre-mRNA alternative splicing mechanism by controlling the amounts of serine and arginine nuclear factors, and also performs the transport and storage of RNA [63, 64]. MALAT1 is one the most abundant lncRNA in benign tissues and acts as a decoy, functioning as a link in ribonucleoprotein (RNP) complexes [65, 66].
According to Liao et al. MALAT1 knockdown prevents the spread of GB proliferation and its progression in vitro, as well as reducing tumor volume in an orthotopic GB murine model. They also showed a novel regulatory mechanism between MALAT1 and ZHX1 (Zinc-fingers and homeoboxes 1), whereby MALAT1 functions as a competitive endogenous RNA (ceRNA) against miR-199a. Blockage of the MALAT1/ZHX1/miR-199a axis inhibited GB proliferation and progression [67]. This suggested that MALAT1 plays an oncogenic role in the growth of glioma tumors. Xiong et al. reported that ablation of MALAT1 suppressed the proliferation of glioma stem cells by increasing miR-129, resulting in decreased glioma stem cell development by blocking SRY-box transcription factor 2 (SOX2) expression [68].
Han et al., on the other hand, ascertained that knockdown of MALAT1 facilitates glioma invasion and proliferation via the downstream effects on ERK/MAPK (extracellular signal-regulated kinase/mitogen-activated protein kinase) signaling pathway and expression of matrix metalloproteinase 2 (MMP2) [69].
Additionally, Fu et al. also detected increased MALAT1 expression and autophagic activity in glial tumor tissues. They reported an inverse correlation exists between miR-101 expression and MALAT1 in GB tissues. MALAT1 has been shown to stimulate cell proliferation and autophagy; as well as target miR-101, which, in turn, regulates the expression of stathmin 1 (STMN1), Ras-related protein Rab-5A (RAB5A) and autophagy related 4D cysteine peptidase (ATG4D). The expression of autophagy genes is decreased when miR-101 is overexpressed or MALAT1 is underexpressed [70].
In another study, MALAT1 appeared as a tumor suppressor in glioblastoma. Cao et al. demonstrated that MALAT1 negatively regulates miR-155 expression. MiR-155 is located on chromosome location 21q-21.3 (Chr21q-21.3). And it suppresses the tumor suppressor genes such as adenomatous polyposis coli (APC) and suppressor of cytokine signaling 1 (SOCS1), and the tyrosine kinase WEE1, which initially inhibits Cdc2 (cell division cycle protein 2) and hinders their entry into mitosis phase. MiR-155-induced oncogenesis is linked with the downregulation of FBXW7 (F-box and WD repeat domain containing 7) protein. FBXW7 acts as a substrate adapter for the SCF (SKP1-Cullin1-F-box protein)-type ubiquitin (E3) ligases complex and suppresses various types of oncoproteins. Based on their results, they stated that MALAT1 suppresses the growth of glioma cells via the down-regulation of miR-155 expression and promotes the activity of FBXW7 protein [71].
Though mutations in isocitrate dehydrogenase (IDH) genes take place much frequently in secondary glioblastoma (>80%) and are associated with better prognosis outcomes, the impact of IDH gene mutations on the prognosis of primary glioblastoma (<5%) remains uncertain [72, 73]. In a recent study, Argadal et al. determined the prognostic importance of MALAT1 expression in primary glioblastoma in general and in relation to IDH1/2 mutation profile. In their study, they found that upregulation of MALAT1 expression was associated with shorter survival rate in IDH1/2 wild-type primary glioblastoma patients, and overexpressed MALAT1 was associated with younger age and left-sided glioblastoma localization. In addition, they also reported that MALAT1 expression substantially increased in the insular lobe relative to other lobes [74]. This indicates that MALAT1 might be regard as a potential biomarker and therapeutic target in primary glioblastoma patients. However, the prognostic magnitude of MALAT1 in primary glioblastoma is far from certain. Further clinical studies are therefore needed to determine the functional role of MALAT1 in primary GB.
Temozolomide is a first-line chemotherapeutic drug commonly used for the management of glioblastoma. However, a large number of glioblastoma cells have inherent or developed resistance towards temozolomide-based therapy, which adversely affects therapeutic outcomes [75, 76]. As a result, a comprehensive analysis of the molecular characteristic that strengthens temozolomide resistance is needed. The study revealed that expression of MALAT1 was increased in patients with glioblastoma and promoted telozolomide resistance by suppressing miR-203 signaling and stimulating the expression of thymidylate synthase (TS) [77]. It was also revealed that knockdown of MALAT1 can significantly reduce temozolomide resistance in GB cells and influence cell cycle progression via enhancing miR-203 and disrupting TS expression. This suggest that overexpression of MALAT1 exerts a poor chemotherapeutic efficiency [77]. Cai and his colleagues have also confirmed that the upregulation of MALAT1 has been substantially associated with temozolomide resistance in GB cells while knockdown of MALAT1 resulted in decreased resistance of temozolomide in GB cells both in vitro and in vivo by inhibiting proliferation and promoting apoptosis [78]. They further demonstrated that MALAT1 promotes temozolomide resistance by repressing miR-101 through direct binding of it in GB cells. Conversely, the overexpression of miR-101 decreased the temozolomide resistance of GB cells and acted as an antagonist. Furthermore, their study also found that MALAT1 knockdown and overexpression of miR-101 inhibited several enzymes, such as methyl-guanine transferase (MGMT) and glycogen synthase kinase 3β (GSK3β), which are involved in chemotherapy and apoptotic resistance of GB cells [78].
All these different findings implied that MALAT1 may be either act as a positive regulator or a negative regulator in glioma tumorigenesis which all depends on different cellular context. At present, all these data in regard to MALAT1 actions seems to be quite conflicting. Therefore, a precise analysis of glioma specimens at the molecular level needed to obtain which may possibly resolve this controversy.
3.2. HOTAIR (HOX Transcript Antisense Intergenic RNA)
HOTAIR (HOX Transcript Antisense Intergenic RNA) is a trans-acting lncRNA involved in chromatin modulation and regulation. It is a spliced and polyadenylated RNA comprising 6 exons and 2,158 nucleotides. HOTAIR results from the transcription of the antisense strand of HoxC gene, which is located between HoxC 11 and HoxC 12 on chromosome location 12q13.13 (Chr12q13.13) [79]. HOTAIR acts as a cellular scaffold and chromatin regulator by binding, with its 5′ terminal, the Polycomb Repressor Complex 2 (PRC2), and with its 3’ terminal, the lysine specific demethylase 1/REST corepressor/RE-1 silencing transcription factor (LSD1/CoREST/REST) complex [62, 79].
Alves et al. explored the characteristics of HOTAIR in the epithelial-mesenchymal transition (EMT) and its function in the development and management of cancer stem cells (CSCs). They discovered that HOTAIR plays a critical role in the tumorigenic process by activating EMT and acquiring stemness properties [80]. As a result, researchers can diagnose the stage of cancer progression and predict the patient's chances of survival by measuring the level of HOTAIR expression.
Aberrant HOTAIR expression has been associated to cancer metastasis and has been described as a negative prognosis factor. Expression of HOTAIR in glioblastoma was reported to be substantially higher than normal brain tissues and involved in cell proliferation, metastasis, and invasion [81, 82, 83, 84].
Gupta et al. first found that the loss of HOTAIR decreases the invasive properties of cancer cells, adding to the growing body of evidence that HOTAIR plays a key role in a number of cancers, including glioblastoma [85].
Wang et al. have shown that HOTAIR promotes malignancy-related factors whereas miR-148-3p suppresses those malignant factors. They demonstrated that miR-148-3P has a tumor suppressive role by down-regulating the regulation of HOTAIR in glioma. Their data also showed that miR-148-3p inhibits cell proliferation, blocks cell cycle regulation and reduces cell invasion [86].
As per research, inhibition of HOTAIR expression provokes apoptotic pathways. Pastori et al. showed that BET (Bromodomain and extraterminal) family protein BRD4 (Bromodomain containing 4) controls the HOTAIR expression. In their study, they successfully demonstrated that the treatment of GB cells with the BET bromodomain inhibitor I-BET151 reduced levels of HOTAIR expression and thus effectively downregulated the proliferation of GB cells in vivo and in vitro. This suggests that BET proteins may also directly control the regulation and expression of HOTAIR [87].
Recently, Magalhães et al. have demonstrated that HOTAIR is overexpressed in glioblastoma. In their study, they found that the co-expression of HOTAIR and HOXA9 (Homeobox A9) occurs frequently in glioblastoma condition that is regulated by the Phosphoinositide 3-kinases (PI3K) signaling pathway via inhibition of EZH2 (enhancer of zeste homolog 2)-mediated H3K27 trimethylation. In sum, they concluded that HOXA9 was a novel direct regulator of HOTAIR and established HOTAIR as an independent prognostic marker [88]. All of these findings suggest that HOTAIR expression initiates and facilitates GB growth and development via a variety of regulatory signals. However, the underlying molecular mechanism of HOTAIR and its aberrant activation in glioblastoma are still awaiting additional studies to specifically identify their regulatory roles.
3.3. HOXA11-AS (HOXA11 antisense RNA)
The homeobox (HOX) gene family is a strongly conserved homeodomain that allows HOX to contract with particular DNA regions and govern gene transcription during the embryonic and tumorigenic development. In humans, HOX genes are categorized into four different clusters (A, B, C and D) which are clustered at four different chromosome locations (7p15, 17q21, 12q13 and 2q31, respectively) [89, 90]. There are 39 HOX genes in the HOX gene family, based on their position within the cluster and the homeobox sequence similarity they are assigned into 13 paralog groups. And each cluster contains 9 to 11 genes. There are three lncRNAs (i) HOXA11-AS; (ii) HOX10-AS and (iii) HOXA, transcribed from the 5’ region of the HOXA transcript [91]. To date, few studies have documented interaction between HOXA11-AS and glioblastoma.
Long non-coding RNA HOXA11-Antisense RNA (HOXA11-AS) is also known as NCRNA00076, which was originally discovered in a mouse embryonic cDNA library [92]. The HOXA11-AS is about 3,885 nucleotides long and is mapped to the chromosome 7p15.2 locus [93].
HOXA11-AS was greatly overexpressed in surgically excised and also in glioma cell lines, such as U251 and SHG44 relative to normal human astrocytes. This elevated HOXA11-AS level suggests a low survival rate and poor prognosis in glioma patients [94].
Wang et al. first defined the prognostic importance of HOXA11-AS in gliomas. They found that the HOXA11-AS is closely associated with cell cycle progression and facilitates cell proliferation [95]. In glioma cell lines, overexpression of HOXA11-AS culminated in increased S-phase rate, colony formation and expression of proteins needed for G1/S transition for example cyclin D1, CDK2, CDK4, and cyclin E. In addition, the up-regulation in HOXA11-AS expression reduced the expression of cyclin-dependent kinase (CDK) inhibitors such as p16, p21, and p27 and the expression of retinoblastoma (Rb) protein [96]. Furthermore, HOXA11-AS also facilitates tumorigenesis by sponging miR-140-5p, miR-124-3p and miR-214-3p [96–98]. Sun et al. found that miR-124-3p mimics significantly suppressed expression of HOXA11-AS in glioma cells, which resulted in increased proliferation, invasion and decreased apoptosis in glioma cells [97].
C. Xu et al. recently discovered that HOXA11-AS acts as a ceRNA for miR-214-3p, which specifically influences the enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2). In glioblastoma, expression of EZH2 is significantly correlated with HOXA11-AS expression and inversely associated with miR-214-3P expression. Their findings indicated that HOXA11-AS knockdown prevented the proliferation, migration and invasion of glioma cells via targeting miR-214-3p axis [98]. Even so, the specific fundamental mechanism has not yet been elucidated. Nonetheless, all the above studies and results propose that HOXA11-AS may serve as a potential prognostic marker for the evaluation of glioblastoma pathogenesis and also as a possible therapeutic approach in the future.
3.4. NEAT-1 (Nuclear Enriched Abundant Transcript 1)
NEAT-1 (Nuclear Enriched Abundant Transcript 1) is an intranuclear, 4 kb long lncRNA, enriched in the nucleus and also present in cytoplasm of the cell. It is a crucial structural constituent for the development of paraspeckles. Paraspeckles are located in the interchromatin region of the nucleus and participates in gene regulation by a number of mechanisms, such as mRNA retention, mRNA breakage and protein seizing [99]. NEAT-1 is transcribed from the familial tumor syndrome multiple endocrine neoplasia (MEN) type 1 location on chromosome 11q13.1, which encodes the two most abundant isoforms; (i) NEAT1-S (short NEAT1), 3735 base pairs (bp) long and (ii) NEAT1-L (long NEAT1), 22,743 bp [100]. NEAT1-L is required for the development of paraspeckles. Although its expression levels are much lower in most tissues relative to other isoforms [101, 102, 103].
West et al. have shown that NEAT1 binds to various genomic sites in cells, primarily near active transcription sites [104]. This postulated that NEAT1 is a regulatory factor for several genes and signaling pathways.
Expression of NEAT1 was found to be elevated in several solid tumors, including glioblastoma [105], osteosarcoma [106], colorectal cancer [107] and many more, supporting NEAT1 as a potential prognostic biomarker.
Zhen et al. discovered that NEAT1 was upregulated in glioma tissues. Via the regulation of the miR-449b-5p/c-Met axis, NEAT1 acts as an oncogene, facilitating cell proliferation, migration, and invasion while also deregulating apoptotic activities [105].
In another study, the expression of NEAT1 and miR-let-7e were found to be negatively correlated. NEAT1 was first discovered to be highly expressed in glioma stem cells (GSCs) by Gong et al. MiR-let-7e expression, conversely, decreased in glioma tissues and GSCs. The findings showed that NEAT1 knockdown or the restitution of miR-let-7e suppressed the cellular proliferation, migration and invasion, promoted apoptosis and dysregulated the malignant characteristics of GSCs [108]. Therefore, therapies that target the NEAT1/miR-let-7e/NRAS axis may be a promising therapeutic approach for glioma treatment. MiR-92b has also been shown to inhibit glioma cell apoptosis by downregulating the expression of Dickkopf Homolog 3 (DKK3) [109]. Liu et al. discovered that overexpression of NEAT1 prevents glioma cell proliferation and stimulates apoptosis by downregulating miR-92b and then upregulating DKK3, implying that the NEAT1/miR-92b/DKK3 axis may have been a potential target in glioma [110]. Previous studies indicated that knockdown of DKK3 expression facilitated the migration capabilities of glioma cells [111] and that overexpression of DKK3 improved temozolomide sensitivity in glioma [112]. However, it remains to be determined if NEAT1 influences migration and chemosensitivity properties via the miR-92b/DKK3 pathway.
Moreover, in another study it was found that NEAT1 enhanced tumorigenesis and growth of glioma, partially via miR-152-3p and chaperonin containing TCP1 subunit 6A (CCT6A) pathway. In this study, the authors revealed that levels of NEAT1 and CCT6A were highly expressed, but miR-152-3p was reduced and NEAT1 modulated the CCT6A expression to accelerate the progression of glioma by sponging miR-152-3p [113]. Conversely, they also discovered that overexpression of miR-152-3p suppressed cell viability, migratory, and invasive abilities whereas enhancing apoptotic mechanism in glioma cells (A172 and U251) [113]. This demonstrated that NEAT1 interacted negatively with miR-152-3p. The MiR-152-3p/CC6TA axis was thus represented as an explicit target of NEAT1 and could provide beneficial approach for glioma patients.
It was also found that NEAT1 could also be up-regulated by the oncogenic epidermal growth factor receptor (EGFR) pathway, which then promotes progression and invasion of GB cells by working as a molecular scaffold for the recruitment of the histone modification enzyme EZH2, leading to the stimulation of the Wnt/β-catenin cellular pathway. Therefore, the EGFR/NEAT1/EZH2/-catenin axis is a key player in the development and progression of glioma tumors [114].
Furthermore, Yang and colleagues hypothesized that NEAT1 is essential for the stemness of GSCs to be preserved. In their study, they discovered that NEAT1 increased expression of CDK6 via regulating miR-107 in the U87 cell line that ultimately mediated the cell cycle of GSCs [115].
3.5. GAS5 (Growth arrest-specific 5)
A snoRNA host gene, growth arrest-specific 5 (GAS5), encodes the lncRNA GAS5. It belongs to the 5′ terminal oligopyrimidine (5′TOP) gene family [116, 117]. GAS5 is a tumor suppressive gene, located at chromosome location 1q25.11 and comprised of total 12 exons, which constitutes a short ORFs that does not transcribe any protein [116]. GAS5 regulates proliferation, invasion and metastasis and apoptosis of cancer cells which may have diagnostic and prognostic significance in a multitude of interventional settings. GAS5 expression has been shown to be downregulated in a variety of cancers, including glioblastoma [116, 118, 119].
Kino et al. found that GAS5 functions as a decoy for glucocorticoid receptor (GR). They demonstrated that GAS5 attaches to the DNA-binding domain of ligand activated GR via the decoy RNA glucocorticoid response element (GRE) and suppressed GR-mediated transcriptional activity of endogenous glucocorticoid-responsive genes by inhibiting binding of GRs to target genes [120].
GAS5 also functions as a ceRNA. And like every other lncRNA, it also acts to sequester miRNAs. Zhao et al. reported that GAS5 would suppress the tumor by down-regulating the miR-222-3p. The researchers discovered that plasmid transfection of GAS5 increased the expression of the tumor suppressor Bcl-2 modifying factor (bmf) and plexin C1 by specifically targeting miR-222-3p and reducing its expression. Downregulated miR-222-3p expression resulted in decreased proliferation and invasion, and also promoted apoptosis in glioma [116]. MiR-196a-5p is another GAS5 target. MiR-196a-5p is an oncogenic miRNA that promotes GSCs proliferation, migration and invasion while suppressing apoptosis by down-regulating the expression of Forkhead box protein O1 (FOXO1). According to Zhao et al., up-regulation of FOXO1 intensified the expression of the phophotyrosine interaction domain containing 1 (PID1) and the migration and invasion inhibitory proteins (MIIP) which eventually resulted in the inhibition of GSCs tumorigenicity and development. They also found that FOXO1 facilitates the transcriptional activity of GAS5. Thus, forming a miR-196a-5p/FOXO1 positive regulatory feedback loop which inhibited GSCs proliferation, migration and invasion and induces apoptosis [121]. In a recent study, Li et al. have investigated the effects of GAS5 on the proliferation, migration, invasion and apoptosis of glioma cell lines (U251 and U87) by targeting the expression of glutathione-S-transferase M3 (GSTM3), a significant member of the glutathione-S-transferase (GST) family. Their study discovered that GAS5 overexpression exerts tumor suppressive roles via downregulating the expression of GSTM3 [122]. In addition, previous studies have also shown that knockdown of GSTM3 expression affected the apoptosis pathway and significantly inhibited the growth of cervical cancer cells [123]. Therefore, this new finding of the GAS5/GSTM3 axis might contribute in the advancement of alternative potential therapeutic targets for glioma. The detailed function of the GAS5 and GSTM3 regulatory pathways and their underlying mechanisms, however, still required to be addressed in a significant length.
All of the above studies thus concluded that considerably higher GAS5 levels were linked to a relatively low risk of mortality, relapse, and advancement rates in GB patients. Therefore, it could be used as a prognostic marker for both survival and disease progression in GB.
3.6. CRNDE (Colorectal Neoplasia Differentially Expressed)
The CRNDE (Colorectal Neoplasia Differentially Expressed) gene expresses multiple splice variants and exhibits tissue-specific expression. It is located on chromosome location 16q12.2 and about 84,001 bp in length [124]. CRNDE gene expression was initially identified as substantially higher in colorectal cancer [125, 126]. However, it is also found up-regulated in various other solid tumors, such as astrocytomas and glioblastomas [127, 128]. A significant number of research have exhibited that CRNDE works through multiple epigenetic mechanisms to regulate cell differentiation and pluripotency [129]. By influencing multiple target genes and signaling pathways, CRNDE, as an oncogene, assists in a myriad of cancer-related cellular mechanisms, like cell growth, invasion, and metastasis, triggers EMT, regulates the tumor's immune microenvironment, and inhibits apoptosis. This aberrant upsurge of CRNDE directs the poor prognosis and a lower survival rate. In addition, an elevated level of CRNDE is also capable of making cancerous cells particularly robust to multiple anti-cancer agents and sabotaging the active chemotherapeutic strategies. A detailed overview of the specific biological functions of CRNDE and its diverse molecular mechanisms in cancers has been published by Lu et al. [130].
Zhang et al. analyzed microarray data of glioma samples and confirmed that CRNDE was highly upregulated by up to 32-fold in glioma tissues [131]. CRNDE function as an oncogene through the negative regulation of miR-186 expression. Zheng et al. reported that the expression of CRNDE was up-regulated whereas the expression of miR-186 was downregulated in GSCs. Overexpression of CRNDE curtailed the expression of XIAP (X-linked inhibitor of apoptosis protein) and PAK7 (p21 protein (Cdc42/Rac)-activated kinase 7) by binding to miR-186 and negatively regulating it [132]. Additionally, both miR-384 and miR-136-5p have been identified as potential CRNDE targets [129, 133]. In their study, Zheng et al. determined the expressions of CRNDE, miR-384 and PIWIL4 (piwi like RNA-mediated gene silencing 4) in cell lines and glioma tissues. CRNDE facilitates the development of malignant gliomas by inhibiting the miR-384/PIWIL4/STAT3 signaling pathway. In glioma cell lines, miR-384 has been shown to function as a tumor suppressor by reducing the protein PIWIL4. Furthermore, they reported that knockdown of CRNDE along with the overexpression of miR-384 contributed to suppression of tumors in vivo [129].
CRNDE may also function as a ceRNA, which can facilitate glioma malignancy by mitigating the expression of Bcl-2 and Wnt2 from being downregulated by miR-136-5p [133]. This represents an essential role of the CRNDE/miR-136-5p/Bcl2/Wnt regulatory pathway in the progression and growth of glioma.
In another study, Wang et al. reported that upregulation of CRNDE is also regulated via P70S6K facilitated mammalian Target of Rapamycin (mTOR) signaling pathway. In their study, they blocked the mTOR pathway with rapamycin, which reduced the rate of proliferation and development of glioma cells. In addition, the expression of other cell proliferation genes, including c-Myc and cyclin D1, has been down-regulated in rapamycin treated cells, whereas tumor suppressor genes such as PTEN (Phosphatase and tensin homolog) and p53 have been up-regulated [134].
According to the National Center for Biotechnology Information (NCBI), a total of five CRNDE transcript variants (TV1-5) with possibly different functional roles have been identified [135]. Data suggest that these different transcript isoforms might vary in expression levels and functions in glioblastoma [136]. In preliminary studies of glioblastoma specimens and cell lines, the expression levels of both transcript variants, TV-1 and TV-5, were found strongly upregulated compared to normal astrocytes. Conversely, transcript variants TV-2, TV-3 and TV-4 were found to be expressed only in small amounts in both healthy and cancerous brain tissues [135]. Kiang et al. further assessed the therapeutic significance of these CRNDE transcripts in glioblastoma through the Kaplan-Meier survival test. Their preliminary results revealed that a low TV-1:TV-5 ratio was correlated with a more resilient disease result, suggesting that the oncogenic characteristics of TV-1 and TV-5 contradict each other [135]. In that regard, this finding indicates a potential use of CRNDE transcripts expression in predicting patient's survival outcomes. Therefore, further investigation needed of these differentially expressed CRNDE transcript isoforms, which may provide evidence on how it can be used as potential prognostic markers in glioblastoma.
3.7. HOTTIP (HOXA transcript at the Distal Tip)
The lncRNA HOTTIP (HOXA Transcript at the Distal Tip) is also known as HOXA-AS6 and HOXA13-AS1. It is about 3,764 nucleotide long, situated at the 5′end of the HOXA gene cluster on chromosome location 7p15.2 [137]. A plethora of studies have revealed that HOTTIP is associated with polycomb repressive complex 2 (PRC2) and WDR5/MLL1 chromatin–modifying complexes and is directly binds to WDR5 (WD repeat domain 5). Thus, direct binding of HOTTIP with the WDR5 protein and the subsequent recruitment of the WDR5/MLL complexes across HOXA locus leads to H3K4 methylation and initiation of transcription of the HOXA gene [137]. HOTTIP exhibited tissue-specific expression patterns in hepatocellular carcinoma (HCC) and Hirschsprun (HSCR) disease. However, HOTTIP expression levels and their underlying mechanism remain unclear in glioma tissues. Moreover, HOTTIP functions as an independent biomarker in various types of cancers [93, 95, 138]. HOTTIP has been seen to play a significant role in a number of conditions, such as differentiation, proliferation, cell-cycle transition and invasion, and apoptosis. In high-grade glioma tissue samples and GB cell lines, HOTTIP expression is significantly down-regulated [139]. Overexpression of HOTTIP, on the other hand, prevents cell proliferation, cell cycle progression, and apoptosis processes.
Brain and reproductive expression (BRE) gene (one of the BRCA1-A complex subunits) is expressed in various types of tissues, such as brain, testis, ovary, adrenal glands and kidneys [140]. It is strongly expressed in the nervous and reproductive systems. And it also modulates the activity of various hormones and cytokines in response to stress, cell survival and in cancer-like pathologies [141]. Overexpression of BRE stimulates cell growth and attenuates apoptotic cell death [142]. Xu et al. showed that HOTTIP act as a tumor suppressor in glioma cells in vivo and in vitro. Mechanistic study has shown that overexpression of HOTTIP inhibited the expression of CDK2 and cyclin A proteins and subsequent enhancement via downregulation of BRE gene [139].
In another study, Zhang et al. showed a significant association between HOTTIP and hypoxia inducible factor 1-α (HIF-1α) in glioma metastasis and poor prognosis. They found that in hypoxia-treated glioma cells HOTTIP is significantly up-regulated leading to EMT and glioma metastasis through modulation of miR-101/ZEB1 (zinc finger E box binding homeobox 1) axis. Furthermore, they found that decreased HOTTIP levels inhibited hypoxia-induced ZEB1 expression in glioma [143].
3.8. SOX2OT (SOX2 Overlapping Transcript)
LncRNA SOX2OT (SOX2 Overlapping Transcript) is mapped to the chromosome 3q26.3 locus in humans, contains at least eight transcript variants and is abundantly expressed in embryonic stem cells [144, 145]. SOX2 is embedded in the intronic region of the SOX2OT [146]. The SOX2OT gene is comprised of ten exons and more than two transcription start sites that exhibited complicated transcriptional activities. Some SOX2OT transcript variants are related to cancer and stem cell related pathways [147]. SOX2OT is a transcription factor that is essential for the development and maintenance of the pluripotency of CSCs [148]. Recent research has shown that SOX2OT is up- or down-regulated in different cancer cell lines and tissues, affecting the development and growth of tumors. Dysregulation of SOX2OT is seen in a multitude of cancers, including glioblastoma, osteosarcoma, lung cancer, breast cancer, gastric cancer, and many others, where it typically act as an oncogene and likely as a tumor suppressor [149]. SOX2OT regulates the expression of SOX2 to influence glioblastoma cell proliferation, migration and invasion in addition to the expression of some CSCs markers, like miR-194-5p, miR-122, SOX-3, TDGF1 (teratocarcinoma-derived growth factor 1), JAK/STAT (Janus kinase-signal transducer and activator of transcription), NSPc1 (nervous system polycomb 1), CDK2, INCENP (Inner centromere protein), GNL3L (G Protein Nucleolar 3 Like), SMC-4 (Structural maintenance of chromosomes protein 4), CDK2AP2 (cyclin dependent kinase 2 associated protein 2) [150–152].
Su et al. reported that silencing of SOX2OT increased miR194-5p and miR-122 expression, reduced SOX3 expression and decreased TDGF-1 expression, and also inhibited the JAK/STAT signaling pathway. It consequently prevented GSCs from proliferation, migration, and invasion, as well as prompted the death of GSCs [150]. This suggested that the SOX2OT/miR-194-5p/miR-122/SOX3/TDGF-1 pathway establishes a positive feedback loop that regulates the biological activities of GSCs.
Saghaeian et al. have also validated that knockdown of SOX2OT has an effect on cellular proliferation and growth processes. Particularly, SOX2OT controls the expression of cell cycle and activity of mitotic regulatory genes, such as CDK2, CDK2AP2, and ACTR3 (actin related protein 3), as well as the chromosome structure of certain genes like INCENP (inner centromere protein), GNL3L (guanine nucleotide-binding protein-like 3) and SMC4 (structural maintenance of chromosomes protein 4). Altogether, the findings revealed that knockdown of SOX2OT can significantly alter the gene expression of cancer cell lines targeting the cell cycle proliferation and cytoskeleton-related genes [152]. These results indicate that SOX2OT is a possible diagnostic and prognostic prediction marker for glioblastoma. However, its chemical stability and expression in body fluids are not clear. Therefore, additional study is warranted before it can be used as a viable prognostic marker.
3.9. XIST (X – Inactive Specific Target)
The lncRNA XIST (X – Inactive Specific Target) is a key modulator for the X-chromosome inactivation (XCI) process in mammals during embryonic development. XCI mechanism ensures dosage compensation between male (XY) and female (XX) individuals by random inactivation of one of the X-chromosomes [153, 154]. Long non-coding RNA XIST is transcribed from the XIST gene, located on the chromosome location Xq13 [155]. It plays a critical role in cell differentiation, proliferation and genomic maintenance. Several studies have shown that the lncRNA XIST is up-regulated in glioma tissues and GSCs, and that it facilitates tumorigenicity and angiogenesis processes [156].
Yao et al. have demonstrated that expression of XIST has been up-regulated in glioma tissues and GSCs. They observed that suppression of XIST had tumor-suppressive properties. Furthermore, a reciprocal suppression feedback loop between miR-152 and XIST was discovered by mechanistic analysis. They confirmed that the repression of XIST expression prevented the GSCs proliferation, migration and invasion and prompted apoptosis via the up-regulation of miR-152 [157].
Wang et al. further reported that XIST was an oncogenic lncRNA that facilitated glioma tumorigenesis by up-regulating the expression of miR-137 targeted gene Rac1 (ras-related C3 botulinum toxin substrate 1). In addition, overexpression of miR-137 or inhibition of Rac1 has been found to prevent the rate of proliferation, migration and invasion of glioma cells [156].
In another study, Peng et al. demonstrated that XIST/miR-29c regulates the chemo-resistance activities of glioma cells to temozolomide by regulating the function of DNA repair protein O6-methylguanine-DNA methyltransferase and transcription factor specificity protein 1 (SP1) expression. Moreover, they found that knocking down XIST alone was enough to inhibit glioma cell proliferation and improve temozolomide-mediated cell proliferation inhibition [158].
Recently, Luo et al. ascertained that XIST up-regulates proliferation and migration of glioma cells by regulating the miR-133a/SOX4 axis. They observed negative regulation between XIST and miR-133a in glioma cell lines. Their study reported that overexpression of miR-133a prevented glioma cell proliferation, metastasis and EMT by reducing the expression of SOX4 [159]. Consistently, other miRNAs, such as miR-429 [160], miR-210 [161], miR-92b [162] and miR-204-5p [163] may also modulates the expression of XIST in gliomas.
These findings have shown that XIST acts as an oncogene in glioma tumorigenesis; however, the concrete cellular mechanisms are still not quite clear. Therefore, in glioblastoma research, more consideration should be given to the XIST-miRNA axis.
3.10. CASC2 (Cancer Susceptibility Candidate 2)
LncRNA CASC2 (Cancer Susceptibility Candidate 2; formerly C10orf5) is first transcribed from an allelic loss region at chromosome location 10q26.11 in endometrial cancer [164]. Genomic and cDNA sequence analysis inferred the occurrence of three alternatively spliced CASC2 transcripts, CASC2a, CASC2b and CASC2c. Despite the fact that they all originate from CASC2, they only share the first three exons but comprised of different downstream exons [164]. In multiple cancers, the lncRNA CASC2a acted as a tumor suppressor [165], including in glioma [166] and inhibited proliferation, migration and invasion as well as the development of the cellular colonies. Conversely, CASC2c functions as an oncogenic RNA. In glioblastoma, expression of CASC2c was found upregulated and promotes tumorigenesis [167].
Wang et al. confirmed that overexpression of CASC2a suppressed the glioma cells malignancy, including proliferation, migration and invasion and mediated apoptosis via negative regulation of miR-21. They stated that miR-21 act as an oncogene in glioma cells. It was also discovered that CASC2 and miR-21 had a reciprocal suppression in an argonaute 2-dependent manner [166]. Liao et al. have also showed that CASC2 negatively regulates glioma. Their data indicate that overexpression of CASC2 could sensitize temozolomide by up-regulating PTEN protein and down-regulating p-AKT protein via miR-181a regulation, while knockdown of CASC2 has had opposite effects [168]. In another study, Liu et al. used a miR-101 sponge, which act as a decoy, to demonstrate that a high degree of CASC2c was positively associated with astrocytoma tumorigenesis. Their study confirmed that CASC2c is regulated directly by miR-101. Thus, knockdown of CASC2c negatively regulates the expression of miR-101 [167]. This suggested that miR-101 would act as a tumor suppressor. Therefore, combination of CASC2 and miR-101 represents a novel approach for the prognosis and treatment strategies.
Wnt/β-catenin signaling pathway is a highly conserved pathway that has been reported to be aberrantly activated in glioblastoma due to the accumulation of different mutations and/or deregulation of several proteins [169]. Activation of Wnt signaling in tumors is associated with perturbations of key components of this pathway, such as β-catenin, APC, and TCF-4 (Transcription factor 4) [170]. In a recent study, it was exhibited that overexpression of CASC2 extensively repressed the rate of proliferation, migration, and invasion of glioma cells by suppressing the Wnt/β-catenin signaling pathway. Thus, in order to investigate the interaction between Wnt/β-catenin pathway and CASC2, authors measured the mRNA level of β-catenin, cyclin D1 and c-Myc in glioma tissues and found that their levels were elevated relative to adjacent normal brain tissue. This indicated a reciprocal relationship between the CASC2 and Wnt/β-catenin pathway. Furthermore, they stated that the overexpression of CASC2 reduced nuclear β-catenin levels and its downstream targets [171]. By using the Cox regression analysis, they also reported that CASC2 could be reflected as an independent indicator of poor prognosis [171]. Taken together, this implies that CASC2 may possibly assist as an effective diagnostic marker as well as a promising therapeutic target in glioblastoma.
To assess the function of CASC2 in chemo-resistant glioma cells, Jiang et al. have established two temozolomide-resistant glioma cell lines (U257TR and U87TR). After treatment of glioma cells with temozolomide, the expression levels of CASC2, miR-193a-5p and mTOR were assessed. Their results revealed that CASC2 negatively controls miR-193a-5p expression through direct interaction in glioma cells [172]. Inhibition of miR-193a-5p or overexpression of CASC2 made glioma cells sensitive to temozolomide treatment by reducing autophagy via regulating mTOR expression. In addition, they found that the reduction of autophagy was due to the miR-193a-5p which directly binds to the 3’ untranslated region (UTR) of the mTOR and inhibited its expression [172]. Taken together, they came to the conclusion that restricting autophagy might help to improve the efficiency of temozolomide. This suggests that the combination of autophagy inhibitor and temozolomide drug can advance the prognosis of cancer patients and might be helpful in the management of glioblastoma patients who are resistant to temozolomide therapy.
3.11. H19 (H19 Imprinted Maternally Expressed Transcript)
H19 (H19 Imprinted Maternally Expressed Transcript) is a paternally imprinted gene, located on chromosome 11p15.5 in humans and on chromosome 7 in the mouse and is only transcribed from the maternally inherited allele [173]. The H19 gene constructs a 2.3 kb of spliced, capped and polyadenylated lncRNA. It also directs the expression of several genes within the imprinted gene network (IGN) and plays a notable role in embryonic development [58]. Several studies have reported up-regulation of H19 levels in various cancer tissue samples and cell lines, including glioblastoma. In addition, functional studies have verified that H19 could potentially serve as an oncogenic lncRNA [127, 174]. Mechanistically, the overexpression of H19 promotes cancer by triggering autophagy, inhibiting apoptosis, and enhancing EMT process, which in turn facilitates invasion, angiogenesis and other invasive properties of cancer cells [175, 176, 177].
Barsyte – Lovejoy et al. have first demonstrated that the product of MYC oncogene, c-Myc targets the expression of H19 in an allele–specific manner, thereby potentiating glioma tumorigenesis. This suggests that the c-Myc activation of the H19 gene plays a critical role in the transformation process [178]. Furthermore, H19 also acts as a precursor of miRNA and modulates the progression of glioma by generating miR-675. MiR-675/H19 regulates proliferation and migration of glioma cells via CDK6 and promotes glioma cell invasion by targeting the expression of cancer – associated cadherin 13 (CDH13), a particular target of miR-675 [175,179]. Various other miRNAs, such as miR-152 [180], miR-29a [181], miR-140 [182], miR-130a-3p [183], miR-326 [184] and miR-200a [185] were also shown to regulate the expression of H19 in glioma tumorigenesis.
Recently, according to Barbara et al. expression of H19 is inversely related to NKD1 (naked cuticle homolog 1), a negative regulator of the Wnt pathway, indicating that H19 may regulate the transcription of NKD1 via EZH2 – induced H3K27 trimethylation of its promoter. Moreover, it has also been showed that the knockdown of H19 impaired the H19 binding to EZH2 and EZH2 binding to NKD1 and other promoters in glioblastoma cells [186].
Furthermore, Li W et al. stated that H19 overexpression could also maintain the stemness properties of GB cells. CSC markers including CD133, Oct4 (octamer-binding transcription factor 4), NANOG (Nanog homeobox) and SOX2 were found to be substantially down-regulated in H19 deficient cells [187]. This was further validated by Jiang et al. who showed that the concentration of H19 was extensively higher in CD133-positive GB cells, and that increased expression of H19 facilitated invasion, migration, angiogenesis and stemness of GB cells [188].
3.12. TUG1 (Taurine Upregulated Gene 1)
Taurine Upregulated Gene 1 (TUG1), with a length of 7,598 nucleotides, is located at chromosome location 22q12.2 [189]. TUG1 was first acknowledged as a transcript regulated by taurine in mouse retinal cells, where it participates in retinal growth and photoreceptor formation [190]. Recent studies have shown that TUG1 can regulate the expression of genes through a number of mechanisms, such as by functioning as a miRNA sponge and via interacting with the PRC2 or the PRC1. The binding of TUG1 to PRC2 results in epigenetic silencing [191, 192]. TUG1 has also been reported to act as ceRNA to target miRNAs, thereby inhibiting their biological functions [193, 194, 195]. In GB tissues, TUG1 is ubiquitously expressed than in healthy brain tissues [196].
Keisuke et al. revealed that activation of Notch 1 signaling in GSCs explicitly stimulates the expression of TUG1. TUG1 initially maintains the tumorigenic and stemness characteristics of GSCs via sponging of miR-145 in the cytoplasm, maintaining stemness associated genes such as SOX2 and Myc, and by recruiting PRC2 to suppress differentiation genes by methylation of histone H32K7 via YY1 (Yin Yang 1) binding activity. This study emphasizes the important roles of the Notch/TUG1 axis in the regulation of the self-renewal of glioma cells [197].
Li et al. revealed that TUG1 regulated the apoptosis of glioma cells by up-regulating PTEN, partly sponging miR-26a leading to proliferation and promotion of apoptosis. It was found that TUG1 acts as a tumor suppressor in glioma cells, and that decreased TUG1 expression is linked to miR-26a up-regulation [196]. This was further confirmed by another study which showed that up-regulation of TUG1 prompts apoptosis of glioma cells by triggering caspase-3 and 9 mediated anti-apoptotic pathway, acting as a tumor suppressor in glioma cells [198]. In addition, serving as ceRNA for miR-144 in gastric epithelial cells (GECs) and miR-299 in GSCs, TUG1 is also able to modulate blood tumor barrier (BTB) and enhance glioma-induced angiogenesis, respectively [193, 199]. Cai et al. showed that knockdown of TUG1 expression decreased the expression of tight endothelial junction proteins via down-regulating heat shock transcription factor 2 (HSF2), and promoting therapeutic agents BTB permeability. Therefore, there may be a possible role of TUG1 in BTB function and in glioma therapy [193].
3.13. ZFAS1 (Zinc finger nuclear transcription factor, X-box binding 1-type containing 1 antisense RNA 1)
The ZFAS1 (Zinc finger nuclear transcription factor, X-box binding 1-type containing 1 antisense RNA 1) gene is located on chromosome location 20q13.13 and is transcribed from the antisense strand near the 5’ terminal of the protein-encoding gene ZNFX1 (Zinc Finger NFX1-Type Containing 1) and the hosts three C/D box snoRNAs (Snord 12, -12b and -12c) [200]. It was originally identified in the mammary gland as a regulator of epithelial cell differentiation and alveolar growth [200]. Previous findings have shown that ZFAS1 has been overexpressed in various cancers and acts as a cancerous oncogene, including in glioma, ovarian cancer, gastric cancer, colorectal cancer, melanoma and in many more [201, 202, 203, 204]. However, Lee et al. observed a down-expression of ZFAS1 in HER-2 (human epidermal growth factor receptor 2) positive breast cancer cells. According to Fan et al., overexpression of ZFAS1 significantly repressed the proliferation, migration and invasion [205].
ZFAS1 was shown to be up-regulated in glioma tissues and cell lines. Gao et al. stated that ZFAS1 exhibited an oncogenic role in the growth of glioma by regulating the EMT process and Notch signaling pathways. They also discovered that knocking down ZFAS1 in glioma cells blocked EMT and notch signaling, as well as cell proliferation, migration, and invasion [206]. The EMT process has been shown to play a potential role in the cancer metastasis [207]. Lv et al. have confirmed that the expression of ZFAS1 promotes EMT process. In their study, it was discovered that knocking down ZFAS1 reduced the expression of MMP2, MMP9, N-cadherin, Integrin β1, ZEB1, and markedly up-regulated the level of E-cadherin in glioma cells [208]. Moreover, Notch is the induction signal of EMT and the chief regulator of GSCs [209]. Upon activation by binding to the ligands, Notch produces an active fragment, termed Notch intracellular fragment (NIC), which translocate to the nucleus to replace the repressor and to recruit co-activators. Many genes which are involved in survival and differentiation activities, for example cyclin D1, Hes and c-Myc are then activated [210]. By means of activating the notch signaling pathway, ZFAS1 facilitates EMT of glioma cells.
The levels of the proteolipid protein 2 (PLP2) have also increased in glioma tissues and cell lines. It has also been implicated in the growth of various tumors [211, 212, 213]. PLP2 is an endoplasmic reticulum omentum protein with four membrane-spanning domains that are present mainly on the surface of the cells [214]. Li et al. reported that ZFAS1 functions as a sponge of miR-150-5p to regulate PLP2 and promotes the progression of glioma both in vivo and in vitro [215]. In sum, all of these findings suggested that ZFAS1 could serve as a useful marker and possible treatment option for glioblastoma. However, limited amount of research has been conducted on the detailed mechanism of ZFAS1 in glioma development.
4. Concluding remarks
Glioblastoma remains one of the most aggressive primary brain tumors, despite advancements in radiotherapy, chemotherapy, and surgical treatments. Finding both new therapeutic targets and prognostic biomarkers could supplement to unravel the mystery of glioblastoma mechanism at the molecular level. Previously considered “junk sequences” in human genomes, lncRNAs have now been shown to be an important cellular regulatory network for various types of cancers, including glioblastoma. A significant proportion of lncRNAs have been identified with dysregulated gene expression and imbalanced biological processes, including proliferation, metastasis, migration, invasion, angiogenesis, stemness and EMT in all facets of GB cells. LncRNAs are likely to allow researchers to discover molecular signatures that help to fine-tune prognosis and discern between from one type of cancer to another. Focusing on lineage and disease-specific lncRNAs appears to be the best approach for investigating possible biomarkers or therapeutic targets. Since lncRNAs are often expressed at lower levels than protein-coding genes, it is critical to find certain candidates that can be easily identified. Thus, it appears that expression levels are also a pivotal factor in the production of lncRNA-based therapeutics and diagnostics. However, only a hand full of lncRNAs was identified in glioblastoma research so far, and our understanding of the domain of lncRNA biology is still limited and lacks understanding of the dynamic mechanism of these regulatory molecules. It is also inappropriate to use lncRNAs for diagnostic or therapeutic purposes without a thorough understanding of their functions. A significant number of studies and basic experimental work are therefore needed to fully assess their potential in cancer biology.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
No data was used for the research described in the article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Clark M.B., Mercer T.R., Bussotti G., Leonardi T., Haynes K.R., Crawford J. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat. Methods. 2015;12(4):339–342. doi: 10.1038/nmeth.3321. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (80- ) 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Vidigal J.A., Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25:137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 6.Hanif F., Muzaffar K., Perveen K., Malhi S.M., Simjee S.U. Glioblastoma multiforme: a review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac. J. Cancer Prev. APJCP. 2017;18:3–9. doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E. The epidemiology of glioma in adults: a state of the science review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu C., Li D., Zhang X., Liu N., Chi G., Jin X. LncRNA PVT1 facilitates tumorigenesis and progression of glioma via regulation of MiR-128-3p/GREM1 Axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–1157. doi: 10.1007/s13311-018-0649-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Chen G., Cao Y., Zhang L., Ma H., Shen C., Zhao J. Analysis of long non-coding RNA expression profiles identifies novel lncRNA biomarkers in the tumorigenesis and malignant progression of gliomas. Oncotarget. 2017;8(40):67744–67753. doi: 10.18632/oncotarget.18832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toraih E.A., Aly N.M., Abdallah H.Y., Al-Qahtani S.A., Shaalan A.A.M., Hussein M.H. MicroRNA–target cross-talks: key players in glioblastoma multiforme. Tumor Biol. 2017;39(11):1–22. doi: 10.1177/1010428317726842. [DOI] [PubMed] [Google Scholar]
- 11.Godlewski J., Krichevsky A.M., Johnson M.D., Chiocca E.A., Bronisz A. Belonging to a network-microRNAs, extracellular vesicles, and the glioblastoma microenvironment. Neuro Oncol. 2015;17:652–662. doi: 10.1093/neuonc/nou292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10 doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 14.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stults D.M., Killen M.W., Pierce H.H., Pierce A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18(1):13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 19.Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- 20.Kawaji H., Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:3–8. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipowicz W., Pogači V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 24.Stadler P.F., Chen J.J.L., Hackermüller J., Hoffmann S., Horn F., Khaitovich P. Evolution of vault RNAs. Mol. Biol. Evol. 2009;26(9):1975–1991. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 25.Ma X., Han N., Shao C., Meng Y. Transcriptome-wide discovery of PASRs (promoter-associated small RNAs) and TASRs (terminus-associated small RNAs) in Arabidopsis thaliana. PloS One. 2017;12(1) doi: 10.1371/journal.pone.0169212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapranov P., Ozsolak F., Kim S.W., Foissac S., Lipson D., Hart C. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466(7306):642–646. doi: 10.1038/nature09190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonen H., Kol N., Shomron N., Leibowitz-Amit R., Quagliata L., Lorber T. Promoter-associated RNAs regulate HSPC152 gene expression in malignant melanoma. Non-coding RNA. 2016;2(3) doi: 10.3390/ncrna2030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fejes-Toth K., Sotirova V., Sachidanandam R., Assaf G., Hannon G.J., Kapranov P. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457(7232):1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taft R.J., Glazov E.A., Cloonan N., Simons C., Stephen S., Faulkner G.J. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 30.Taft R.J., Simons C., Nahkuri S., Oey H., Korbie D.J., Mercer T.R. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010;17(8):1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 31.Christov C.P., Gardiner T.J., Szüts D., Krude T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol. Cell Biol. 2006;26(18):6993–7004. doi: 10.1128/MCB.01060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faulkner G.J., Carninci P. Altruistic functions for selfish DNA. Cell Cycle. 2009;8:2895–2900. doi: 10.4161/cc.8.18.9536. [DOI] [PubMed] [Google Scholar]
- 33.Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41(5):563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 34.Mercer T.R., Wilhelm D., Dinger M.E., Soldà G., Korbie D.J., Glazov E.A. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 2011;39(6):2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Vol. 19. Nat. Rev. Mol. Cell Biol. 2018:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Morales D.R. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scaruffi P. The transcribed-ultraconserved regions: a novel class of long noncoding RNAs involved in cancer susceptibility. TheScientificWorldJournal. 2011;11:340–352. doi: 10.1100/tsw.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaruffi P., Stigliani S., Moretti S., Coco S., De Vecchi C., Valdora F. Transcribed-ultra conserved region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer. 2009;9:1–9. doi: 10.1186/1471-2407-9-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louro R., Smirnova A.S., Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Rearick D., Prakash A., McSweeny A., Shepard S.S., Fedorova L., Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res. 2011;39(6):2357–2366. doi: 10.1093/nar/gkq1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M. Molecular biology: antisense transcription in the mammalian transcriptome. Science (80- ) 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 43.Zheng D., Frankish A., Baertsch R., Kapranov P., Reymond A., Siew W.C. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res. 2007;17(6):839–851. doi: 10.1101/gr.5586307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z.D., Frankish A., Hunt T., Harrow J., Gerstein M. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 2010;11(3) doi: 10.1186/gb-2010-11-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng R., Shen Z., Tripathi V., Xuan Z., Freier S.M., Bennett C.F. Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J. Cell Sci. 2010;123(21):3734–3744. doi: 10.1242/jcs.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M.S., Mapendano C.K. RNA exosome depletion reveals transcription upstream of active human promoters. Science (80- ) 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 47.Silva J.M., Perez D.S., Pritchett J.R., Halling M.L., Tang H., Smith D.I. Identification of Long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95(6):355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N. Molecular biology: the transcriptional landscape of the mammalian genome. Science (80- ) 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 50.Andrews S.J., Rothnagel J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 51.Melé M., Mattioli K., Mallard W., Shechner D.M., Gerhardinger C., Rinn J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27(1):27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlackow M., Nojima T., Gomes T., Dhir A., Carmo-Fonseca M., Proudfoot N.J. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell. 2017;65(1):25–38. doi: 10.1016/j.molcel.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taft R.J., Pheasant M., Mattick J.S. The relationship between Non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 54.Mathews D.H., Moss W.N., Turner D.H. Folding and finding RNA secondary structure. Cold Spring Harbor Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipovich L., Johnson R., Lin C.Y. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochimica et Biophysica Acta - Gene Regul. Mech. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Bartolomei M.S., Zemel S., Tilghman S.M. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 57.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 58.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K.C., Chang H.Y. molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin. Cell Dev. Biol. 2011;22:343–350. doi: 10.1016/j.semcdb.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 62.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F. Long noncoding RNA as modular scaffold of histone modification complexes. Science (80- ) 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han Y., Zhou L., Wu T., Huang Y., Cheng Z., Li X. Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell. Mol. Neurobiol. 2016;36(7):1097–1107. doi: 10.1007/s10571-015-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Y., Ma L. New insights into long non-coding rna malat1 in cancer and metastasis. Cancers. 2019;11 doi: 10.3390/cancers11020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baspinar Y., Elmaci I., Ozpinar A., Altinoz M.A. Long non-coding RNA MALAT1 as a key target in pathogenesis of glioblastoma. Janus faces or Achilles’ heal? Gene. 2020;739 doi: 10.1016/j.gene.2020.144518. [DOI] [PubMed] [Google Scholar]
- 67.Liao K., Lin Y., Gao W., Xiao Z., Medina R., Dmitriev P. Blocking lncRNA MALAT1/miR-199a/ZHX1 Axis inhibits glioblastoma proliferation and progression. Mol. Ther. Nucleic Acids. 2019;18:388–399. doi: 10.1016/j.omtn.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong Z., Wang L., Wang Q., Yuan Y. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J. Cell Mol. Med. 2018;22(8):3929–3940. doi: 10.1111/jcmm.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han Y., Wu Z., Wu T., Huang Y., Cheng Z., Li X. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu Z., Luo W., Wang J., Peng T., Sun G., Shi J. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem. Biophys. Res. Commun. 2017;492(3):480–486. doi: 10.1016/j.bbrc.2017.08.070. [DOI] [PubMed] [Google Scholar]
- 71.Cao S., Wang Y., Li J., Lv M., Niu H., Tian Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J Cancer Res. 2016;6(11):2561–2574. [PMC free article] [PubMed] [Google Scholar]
- 72.Cohen A.L., Holmen S.L., Colman H. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 2013;13(5) doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen J.R., Yao Y., Xu H.Z., Qin Z.Y. Isocitrate dehydrogenase (IDH)1/2 mutations as prognostic markers in patients with glioblastomas. Medicine (United States) 2016;95(9) doi: 10.1097/MD.0000000000002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Argadal O.G., Mutlu M., Aksoy S.A., Kocaeli H., Tunca B., Civan M.N. Long noncoding rna malat1 may be a prognostic biomarker in idh1/2 wild-type primary glioblastomas. Bosn. J. Basic Med. Sci. 2020;20(1):63–69. doi: 10.17305/bjbms.2019.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goellner E.M., Grimme B., Brown A.R., Lin Y.C., Wang X.H., Sugrue K.F. Overcoming temozolomide resistance in glioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Res. 2011;71(6):2308–2317. doi: 10.1158/0008-5472.CAN-10-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yip S., Miao J., Cahill D.P., Iafrate A.J., Aldape K., Nutt C.L. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin. Cancer Res. 2009;15(14):4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen W., Xu X.K., Li J.L., Kong K.K., Li H., Chen C. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8(14):22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai T., Liu Y., Xiao J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018;7(4):1404–1415. doi: 10.1002/cam4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alves C.P., Fonseca A.S., Muys B.R., Bueno R.D.B.E.L., Burger M.C., De Souza J.E.S. Brief report: the lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cell. 2013;31(12):2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 81.Wu Y., Zhang L., Wang Y., Li H., Ren X., Wei F. Long noncoding RNA HOTAIR involvement in cancer. Tumor Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 82.Huang K., Sun J., Yang C., Wang Y., Zhou B., Kang C. HOTAIR upregulates an 18-gene cell cycle-related mRNA network in glioma. Int. J. Oncol. 2017;50(4):1271–1278. doi: 10.3892/ijo.2017.3901. [DOI] [PubMed] [Google Scholar]
- 83.Shen J., Hodges T.R., Song R., Gong Y., Calin G.A., Heimberger A.B. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol. Carcinog. 2018;57(1):137–141. doi: 10.1002/mc.22739. [DOI] [PubMed] [Google Scholar]
- 84.Tan S.K., Pastori C., Penas C., Komotar R.J., Ivan M.E., Wahlestedt C. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer. 2018;17 doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang G., Li Z., Tian N., Han L., Fu Y., Guo Z. MiR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol. Lett. 2016;12(2):879–886. doi: 10.3892/ol.2016.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pastori C., Kapranov P., Penas C., Peschansky V., Volmar C.H., Sarkaria J.N. The bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. U. S. A. 2015;112(27):8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xavier-Magalhães A., Gonçalves C.S., Fogli A., Lourenço T., Pojo M., Pereira B. The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget. 2018;9(21):15740–15756. doi: 10.18632/oncotarget.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhatlekar S., Fields J.Z., Boman B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 90.Lu S., Jiang X., Su Z., Cui Z., Fu W., Tai S. The role of the long non-coding RNA HOXA11-AS in promoting proliferation and metastasis of malignant tumors. Cell Biol. Int. 2018;42:1596–1601. doi: 10.1002/cbin.11045. [DOI] [PubMed] [Google Scholar]
- 91.Wei C., Zhao L., Liang H., Zhen Y., Han L. Recent advances in unraveling the molecular mechanisms and functions of HOXA11-AS in human cancers and other diseases (Review) Oncol. Rep. 2020;43:1737–1754. doi: 10.3892/or.2020.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chau Y.M., Pando S., Taylor H.S. HOXA11 silencing and endogenous HOXA11 antisense ribonucleic acid in the uterine endometrium. J. Clin. Endocrinol. Metab. 2002;87(6):2674–2680. doi: 10.1210/jcem.87.6.8527. [DOI] [PubMed] [Google Scholar]
- 93.Richards E.J., Permuth-Wey J., Li Y., Ann Chen Y., Coppola D., Reid B.M. A functional variant in HOXA 11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6(33):34745–34757. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nunes F.D., de Almeida F.C., Tucci R., de Sousa S.C. Homeobox genes: a molecular link between development and cancer. Pesquisa odontológica brasileira = Braz. Oral Res. 2003;17:94–98. doi: 10.1590/s1517-74912003000100018. [DOI] [PubMed] [Google Scholar]
- 95.Wang Q., Zhang J., Liu Y., Zhang W., Zhou J., Duan R. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016;373(2):251–259. doi: 10.1016/j.canlet.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 96.Cui Y., Yi L., Zhao J.Z., Jiang Y.G. Long noncoding RNA HOXA11-AS functions as miRNA sponge to promote the glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol. 2017;36(10):822–828. doi: 10.1089/dna.2017.3805. [DOI] [PubMed] [Google Scholar]
- 97.Sun Y., Yang J.X., Liu B., Yang B.Y., Meng Q. Long non-coding RNA homeobox (HOX) A11-AS promotes malignant progression of glioma by targeting miR-124-3p. Neoplasma. 2018;65(4):505–514. doi: 10.4149/neo_2018_170705N462. [DOI] [PubMed] [Google Scholar]
- 98.Xu C., He T., Li Z., Liu H., Ding B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed. Pharmacother. 2017;95:1504–1513. doi: 10.1016/j.biopha.2017.08.097. [DOI] [PubMed] [Google Scholar]
- 99.Lin Y., Schmidt B.F., Bruchez M.P., McManus C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018;46(7):3742–3752. doi: 10.1093/nar/gky046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bond C.S., Fox A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. Men ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sasaki Y.T.F., Ideue T., Sano M., Mituyama T., Hirose T. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U. S. A. 2009;106(8):2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakagawa S., Naganuma T., Shioi G., Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhen L., Yun-hui L., Hong-yu D., Jun M., Yi-long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumor Biol. 2016;37(1):673–683. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 106.Wang H., Yu Y., Fan S., Luo L. Knockdown of long non-coding RNA NEAT1 inhibits proliferation and invasion and induces apoptosis of osteosarcoma by inhibiting miR-194 expression. Yonsei Med. J. 2017;58(6):1092–1100. doi: 10.3349/ymj.2017.58.6.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Y., Li Y., Chen W., He F., Tan Z., Zheng J. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641–27650. doi: 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gong W., Zheng J., Liu X., Ma J., Liu Y., Xue Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget. 2016;7(38):62208–62223. doi: 10.18632/oncotarget.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Q., Shen K., Zhao Y., Ma C., Liu J., Ma J. MiR-92b inhibitor promoted glioma cell apoptosis via targeting DKK3 and blocking the Wnt/beta-catenin signaling pathway. J. Transl. Med. 2013;11(1) doi: 10.1186/1479-5876-11-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu D., Zou Z., Li G., Pan P., Liang G. Long noncoding RNA NEAT1 suppresses proliferation and promotes apoptosis of glioma cells via downregulating MiR-92b. Cancer Control. 2020;27(1) doi: 10.1177/1073274819897977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peng G., Yang C., Liu Y., Shen C. miR-25-3p promotes glioma cell proliferation and migration by targeting FBXW7 and DKK3. Exp. Ther Med. 2019;18(1):769–778. doi: 10.3892/etm.2019.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fujihara T., Mizobuchi Y., Nakajima K., Kageji T., Matsuzaki K., Kitazato K.T. Down-regulation of MDR1 by Ad-DKK3 via Akt/NFκB pathways augments the anti-tumor effect of temozolomide in glioblastoma cells and a murine xenograft model. J. Neuro Oncol. 2018;139(2):323–332. doi: 10.1007/s11060-018-2894-5. [DOI] [PubMed] [Google Scholar]
- 113.Li B., Lu X., Ma C., Sun S., Shu X., Wang Z. Long non-coding RNA NEAT1 promotes human glioma tumor progression via miR-152-3p/CCT6A pathway. Neurosci. Lett. 2020:732. doi: 10.1016/j.neulet.2020.135086. [DOI] [PubMed] [Google Scholar]
- 114.Chen Q., Cai J., Wang Q., Wang Y., Liu M., Yang J. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/b-catenin pathway by scaffolding EZH2. Clin. Cancer Res. 2018;24(3):684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 115.Yang X., Xiao Z., Du X., Huang L., Du G. Silencing of the long non-coding RNA NEAT1 suppresses glioma stem-like properties through modulation of the miR-107/CDK6 pathway. Oncol. Rep. 2017;37(1):555–562. doi: 10.3892/or.2016.5266. [DOI] [PubMed] [Google Scholar]
- 116.Zhao X., Wang P., Liu J., Zheng J., Liu Y., Chen J. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol. Ther. 2015;23(12):1899–1911. doi: 10.1038/mt.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Amaldi F., Pierandrei-Amaldi P. TOP genes: a translationally controlled class of genes including those coding for ribosomal proteins. Prog. Mol. Subcell. Biol. 1997;18:1–17. doi: 10.1007/978-3-642-60471-3_1. [DOI] [PubMed] [Google Scholar]
- 118.Ji J., Dai X., Yeung S.C.J., He X. The role of long non-coding RNA GAS5 in cancers. Canc. Manag. Res. 2019;11:2729–2737. doi: 10.2147/CMAR.S189052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen C.Y., Chen C.C., Shieh T.M., Hsueh C., Wang S.H., Leu Y.L. Corylin suppresses hepatocellular carcinoma progression via the inhibition of epithelial-mesenchymal transition, mediated by long noncoding RNA GAS5. Int. J. Mol. Sci. 2018;19(2) doi: 10.3390/ijms19020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3(107) doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao X., Liu Y., Zheng J., Liu X., Chen J., Liu L. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864(10):1605–1617. doi: 10.1016/j.bbamcr.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 122.Li G., Cai Y., Wang C., Huang M., Chen J. LncRNA GAS5 regulates the proliferation, migration, invasion and apoptosis of brain glioma cells through targeting GSTM3 expression. The effect of LncRNA GAS5 on glioma cells. J. Neuro Oncol. 2019;143(3):525–536. doi: 10.1007/s11060-019-03185-0. [DOI] [PubMed] [Google Scholar]
- 123.Checa-Rojas A., Delgadillo-Silva L.F., Velasco-Herrera M. del C., Andrade-Domínguez A., Gil J., Santillán O. GSTM3 and GSTP1: novel players driving tumor progression in cervical cancer. Oncotarget. 2018;9(31):21696–21714. doi: 10.18632/oncotarget.24796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cabili M., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Graham L.D., Pedersen S.K., Brown G.S., Ho T., Kassir Z., Moynihan A.T. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2(8):829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellis B.C., Molloy P.L., Graham L.D. CRNDE: a long non-coding RNA involved in CanceR Neurobiology, and DEvelopment. Front. Genet. 2012;3:1–15. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mei-Yee Kiang K., Zhang X.Q., Leung G.K.K. Long non-coding RNAs: the key players in glioma pathogenesis. Cancers. 2015;7:1406–1424. doi: 10.3390/cancers7030843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jing S.Y., Lu Y.Y., Yang J.K., Deng W.Y., Zhou Q., Jiao B.H. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur. Rev. Med. Pharmacol. Sci. 2016;20(19):3992–3996. [PubMed] [Google Scholar]
- 129.Zheng J., Liu X., Wang P., Xue Y., Ma J., Qu C. CRNDE promotes malignant progression of Glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol. Ther. 2016;24(7):1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Lu Y., Sha H., Sun X., Zhang Y., Wu Y., Zhang J. CRNDE: an oncogenic long non-coding RNA in cancers. Canc. Cell Int. 2020;20 doi: 10.1186/s12935-020-01246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang X., Sun S., Pu J.K.S., Tsang A.C.O., Lee D., Man V.O.Y. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012;48(1):1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 132.Zheng J., dong Li X., Wang P., bai Liu X., Xue Y.X., Hu Y. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6(28):25339–25355. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li D.X., Fei X.R., Dong Y.F., Cheng C.D., Yang Y., Deng X.F. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8(50):88163–88178. doi: 10.18632/oncotarget.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Y., Wang Y., Li J., Zhang Y., Yin H., Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367(2):122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 135.Kiang K.M.Y., Leung G.K.K. Clinical significance of CRNDE transcript variants in glioblastoma multiforme. Non-coding RNA Res. 2017;2(2):119–121. doi: 10.1016/j.ncrna.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Szafron L.M., Balcerak A., Grzybowska E.A., Pienkowska-Grela B., Felisiak-Golabek A., Podgorska A. The novel gene CRNDE encodes a nuclear peptide (CRNDEP) which is overexpressed in highly proliferating tissues. PloS One. 2015;10(5) doi: 10.1371/journal.pone.0127475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–126. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chui Y.L., Ma C.H., Li W., Xu Z., Yao Y., Lin F.K.Y. Anti-apoptotic protein BRE/BRCC45 attenuates apoptosis through maintaining the expression of caspase inhibitor XIAP in mouse Lewis lung carcinoma D122 cells. Apoptosis. 2014;19(5):829–840. doi: 10.1007/s10495-013-0963-y. [DOI] [PubMed] [Google Scholar]
- 139.Xu L.M., Chen L., Li F., Zhang R., Li Z.Y., Chen F.F. Over-expression of the long non-coding RNA HOTTIP inhibits glioma cell growth by BRE. J. Exp. Clin. Cancer Res. 2016;35(1):1–15. doi: 10.1186/s13046-016-0431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miao J., Panesar N.S., Chan K.T., Lai F.M.M., Xia N.S., Wang Y.B. Differential expression of a stress-modulating gene, BRE, in the adrenal gland, in adrenal neoplasia, and in abnormal adrenal tissues. J. Histochem. Cytochem. 2001;49(4):491–499. doi: 10.1177/002215540104900409. [DOI] [PubMed] [Google Scholar]
- 141.Mei K.T., Chun M.W., Sze W.S., Yiu L.C., Ching A.K.K., Pak H.C. Comparative proteomic analysis reveals a function of the novel death receptor-associated protein BRE in the regulation of prohibitin and p53 expression and proliferation. Proteomics. 2006;6(8):2376–2385. doi: 10.1002/pmic.200500603. [DOI] [PubMed] [Google Scholar]
- 142.Li Q., Ching A.K.K., Chan B.C.L., Chow S.K.Y., Lim P.L., Ho T.C.Y. A death receptor-associated anti-apoptotic protein, BRE, inhibits mitochondrial apoptotic pathway. J. Biol. Chem. 2004;279(50):52106–52116. doi: 10.1074/jbc.M408678200. [DOI] [PubMed] [Google Scholar]
- 143.Zhang S., Wang W., Liu G., Xie S., Li Q., Li Y. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed. Pharmacother. 2017;95:711–720. doi: 10.1016/j.biopha.2017.08.133. [DOI] [PubMed] [Google Scholar]
- 144.Amaral P.P., Neyt C., Wilkins S.J., Askarian-Amiri M.E., Sunkin S.M., Perkins A.C. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. Rna. 2009;15(11):2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shahryari A., Rafiee M.R., Fouani Y., Oliae N.A., Samaei N.M., Shafiee M. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cell. 2014;32(1):126–134. doi: 10.1002/stem.1542. [DOI] [PubMed] [Google Scholar]
- 146.Fantes J., Ragge N.K., Lynch S.A., McGill N.I., Collin J.R.O., Howard-Peebles P.N. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003;33(4):461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 147.Saghaeian Jazi M., Samaei N.M., Ghanei M., Shadmehr M.B., Mowla S.J. Identification of new SOX2OT transcript variants highly expressed in human cancer cell lines and down regulated in stem cell differentiation. Mol. Biol. Rep. 2016;43(2):65–72. doi: 10.1007/s11033-015-3939-x. [DOI] [PubMed] [Google Scholar]
- 148.Wang Z., Tan M., Chen G., Li Z., Lu X. LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2. IUBMB Life. 2017;69(11):867–876. doi: 10.1002/iub.1681. [DOI] [PubMed] [Google Scholar]
- 149.Wang Y., Wu N., Luo X., Zhang X., Liao Q., Wang J. SOX2OT, a novel tumor-related long non-coding RNA. Biomed. Pharmacother. 2020;123 doi: 10.1016/j.biopha.2019.109725. [DOI] [PubMed] [Google Scholar]
- 150.Su R., Cao S., Ma J., Liu Y., Liu X., Zheng J. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer. 2017;16(1) doi: 10.1186/s12943-017-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Y., Liang Z., Li H., Tao J., Sun Y., Gong Y. NSPc1 polycomb protein complex binds and cross-talks to lncRNAs in glioma H4 cells. Oncol. Rep. 2019;41(4):2575–2584. doi: 10.3892/or.2019.7000. [DOI] [PubMed] [Google Scholar]
- 152.Saghaeian Jazi M., Samaei N.M., Mowla S.J., Arefnezhad B., Kouhsar M. SOX2OT knockdown derived changes in mitotic regulatory gene network of cancer cells. Cancer Cell Int. 2018;18(1) doi: 10.1186/s12935-018-0618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kay G.F., Penny G.D., Patel D., Ashworth A., Brockdorff N., Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993;72(2):171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- 154.Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 155.Furlan G., Rougeulle C. Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wiley Interdiscip. Rev. RNA. 2016;7(5):702–722. doi: 10.1002/wrna.1359. [DOI] [PubMed] [Google Scholar]
- 156.Wang Z., Yuan J., Li L., Yang Y., Xu X., Wang Y. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am. J. Transl. Res. 2017;9(4):1845–1855. [PMC free article] [PubMed] [Google Scholar]
- 157.Yao Y., Ma J., Xue Y., Wang P., Li Z., Liu J. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359(1):75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 158.Du P., Zhao H., Peng R., Liu Q., Yuan J., Peng G. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 2017;37(5) doi: 10.1042/BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Luo C., Quan Z., Zhong B., Zhang M., Zhou B., Wang S. lncRNA XIST promotes glioma proliferation and metastasis through miR-133a/SOX4. Exp. Ther. Med. 2020;19(3):1641–1648. doi: 10.3892/etm.2020.8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cheng Z., Li Z., Ma K., Li X., Tian N., Duan J. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J. Cancer. 2017;8(19):4106–4116. doi: 10.7150/jca.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fasanaro P., Greco S., Lorenzi M., Pescatori M., Brioschi M., Kulshreshtha R. An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 2009;284(50):35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhuang L.K., Yang Y.T., Ma X., Han B., Wang Z.S., Zhao Q.Y. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7(4) doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shen J., Xiong J., Shao X., Cheng H., Fang X., Sun Y. Knockdown of the long noncoding RNA XIST suppresses glioma progression by upregulating miR-204-5p. J. Cancer. 2020;11(15):4550–4559. doi: 10.7150/jca.45676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Baldinu P., Cossu A., Manca A., Satta M.P., Sini M.C., Rozzo C. Identification of a novel candidate gene, CASC2, in a region of common allelic loss at chromosome 10q26 in human endometrial cancer. Hum. Mutat. 2004;23(4):318–326. doi: 10.1002/humu.20015. [DOI] [PubMed] [Google Scholar]
- 165.Baldinu P., Cossu A., Manca A., Satta M.P., Sini M.C., Palomba G. CASC2a gene is down-regulated in endometrial cancer. Anticancer Res. 2007;27(1 A):235–243. [PubMed] [Google Scholar]
- 166.Wang P., Liu Y.H., Yao Y.L., Li Z., Li Z.Q., Ma J. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell. Signal. 2015;27(2):275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 167.Liu C., Sun Y., She X., Tu C., Cheng X., Wang L. CASC2c as an unfavorable prognosis factor interacts with miR-101 to mediate astrocytoma tumorigenesis. Cell Death Dis. 2017;8(3) doi: 10.1038/cddis.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liao Y., Shen L., Zhao H., Liu Q., Fu J., Guo Y. LncRNA CASC2 interacts with miR-181a to modulate glioma growth and resistance to TMZ through PTEN pathway. J. Cell. Biochem. 2017;118(7):1889–1899. doi: 10.1002/jcb.25910. [DOI] [PubMed] [Google Scholar]
- 169.Lee Y., Lee J.K., Ahn S.H., Lee J., Nam D.H. WNT signaling in glioblastoma and therapeutic opportunities. Lab. Invest. 2016;96(2):137–150. doi: 10.1038/labinvest.2015.140. [DOI] [PubMed] [Google Scholar]
- 170.Nager M., Bhardwaj D., Cantí C., Medina L., Nogués P., Herreros J. β -catenin signalling in glioblastoma multiforme and glioma-initiating cells. Chemother Res. Pract. 2012;2012:1–7. doi: 10.1155/2012/192362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang R., Li Y., Zhu G., Tian B., Zeng W., Yang Y. Long noncoding RNA CASC2 predicts the prognosis of glioma patients and functions as a suppressor for gliomas by suppressing Wnt/β-catenin signaling pathway. Neuropsychiatr. Dis. Treat. 2017;13:1805–1813. doi: 10.2147/NDT.S137171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jiang C., Shen F., Du J., Fang X., Li X., Su J. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed. Pharmacother. 2018;97:844–850. doi: 10.1016/j.biopha.2017.10.146. [DOI] [PubMed] [Google Scholar]
- 173.Gabory A., Ripoche M.A., Yoshimizu T., Dandolo L. Cytogenetic and Genome Research. 2006. The H19 gene: regulation and function of a non-coding RNA; pp. 188–193. [DOI] [PubMed] [Google Scholar]
- 174.Chen T., Yang P., He Z.Y. Long non-coding RNA H19 can predict a poor prognosis and lymph node metastasis: a meta-analysis in human cancer. Minerva Med. 2016;107:251–258. [PubMed] [Google Scholar]
- 175.Shi Y., Wang Y., Luan W., Wang P., Tao T., Zhang J. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PloS One. 2014;9(1) doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280(7):1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 177.Matouk I.J., Raveh E., Abu-lail R., Mezan S., Gilon M., Gershtain E. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843(7):1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 178.Barsyte-Lovejoy D., Lau S.K., Boutros P.C., Khosravi F., Jurisica I., Andrulis I.L. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 179.Li C., Lei B., Huang S., Zheng M., Liu Z., Li Z. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am. J. Transl. Res. 2015;7(10):1747–1764. [PMC free article] [PubMed] [Google Scholar]
- 180.Chen L., Wang Y., He J., Zhang C., Chen J., Shi D. Long noncoding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol. Res. 2018;26(9):1419–1428. doi: 10.3727/096504018X15178768577951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jia P., Cai H., Liu X., Chen J., Ma J., Wang P. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 182.Zhao H., Peng R., Liu Q., Liu D., Du P., Yuan J. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch. Biochem. Biophys. 2016;610:1–7. doi: 10.1016/j.abb.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 183.Hu Q., Yin J., Zeng A., Jin X., Zhang Z., Yan W. H19 functions as a competing endogenous RNA to regulate EMT by sponging miR-130a-3p in glioma. Cell. Physiol. Biochem. 2018;50(1):150–168. doi: 10.1159/000494002. [DOI] [PubMed] [Google Scholar]
- 184.Fawzy M.S., Ellawindy A., Hussein M.H., Khashana M.S., Darwish M.K., Abdel-Daim M.M. Long noncoding RNA H19, and not microRNA miR-326, is over-expressed and predicts survival in glioblastoma. Biochem. Cell. Biol. 2018;96(6):832–839. doi: 10.1139/bcb-2018-0122. [DOI] [PubMed] [Google Scholar]
- 185.Berthois Y., Delfino C., Metellus P., Fina F., Nanni-Metellus I., Al Aswy H. Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: evidence that miR200a-3p is regulated by O6-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol. Ther. 2014;15(7):938–950. doi: 10.4161/cbt.28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fazi B., Garbo S., Toschi N., Mangiola A., Lombari M., Sicari D. The lncRNA H19 positively affects the tumorigenic properties of glioblastoma cells and contributes to NKD1 repression through the recruitment of EZH2 on its promoter. Oncotarget. 2018;9(21):15512–15525. doi: 10.18632/oncotarget.24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Li W., Jiang P., Sun X., Xu S., Ma X., Zhan R. Suppressing H19 modulates tumorigenicity and stemness in U251 and U87MG glioma cells. Cell. Mol. Neurobiol. 2016;36(8):1219–1227. doi: 10.1007/s10571-015-0320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jiang X., Yan Y., Hu M., Chen X., Wang Y., Dai Y. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016;124(1):129–136. doi: 10.3171/2014.12.JNS1426. [DOI] [PubMed] [Google Scholar]
- 189.Young T.L., Matsuda T., Cepko C.L. The noncoding RNA Taurine Upregulated Gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 190.Zhang Q., Geng P.L., Yin P., Wang X.L., Jia J.P., Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac. J. Cancer Prev. 2013;14(4):2311–2315. doi: 10.7314/apjcp.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 191.Lin P.C., Huang H.D., Chang C.C., Chang Y.S., Yen J.C., Lee C.C. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16(1) doi: 10.1186/s12885-016-2569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D. NcRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cai H., Xue Y., Wang P., Wang Z., Li Z., Hu Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015;6(23):19759–19779. doi: 10.18632/oncotarget.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu L., Chen X., Zhang Y., Hu Y., Shen X., Zhu W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget. 2017;8(19):31386–31394. doi: 10.18632/oncotarget.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Long J., Menggen Q., Wuren Q., Shi Q., Pi X. Long noncoding rna taurine-upregulated gene1 (TUG1) promotes tumor growth and metastasis through tug1/mir-129-5p/astrocyte-elevated gene-1 (AEG-1) axis in malignant melanoma. Med. Sci. Monit. 2018;24:1547–1559. doi: 10.12659/MSM.906616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li J., An G., Zhang M., Ma Q. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem. Biophys. Res. Commun. 2016;477(4):743–748. doi: 10.1016/j.bbrc.2016.06.129. [DOI] [PubMed] [Google Scholar]
- 197.Katsushima K., Natsume A., Ohka F., Shinjo K., Hatanaka A., Ichimura N. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7 doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li J., Zhang M., An G., Ma Q. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp. Biol. Med. 2016;241(6):644–649. doi: 10.1177/1535370215622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cai H., Liu X., Zheng J., Xue Y., Ma J., Li Z. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36(3):318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 200.Askarian-Amiri M.E., Crawford J., French J.D., Smart C.E., Smith M.A., Clark M.B. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. Rna. 2011;17(5):878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang Y., Yu X., Chen L., Zhang Z., Feng S. EZH2 overexpression is associated with poor prognosis in patients with glioma. Oncotarget. 2017;8(1):565–573. doi: 10.18632/oncotarget.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.McTyre E., Lucas J.T., Helis C., Farris M., Soike M., Mott R. Outcomes for anaplastic glioma treated with radiation therapy with or without concurrent temozolomide. Am. J. Clin. Oncol. Cancer Clin. Trials. 2018;41(8):813–819. doi: 10.1097/COC.0000000000000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 204.Liu F., Gao H., Li S., Ni X., Zhu Z. Long non-coding RNA ZFAS1 correlates with clinical progression and prognosis in cancer patients. Oncotarget. 2017;8(37):61561–61569. doi: 10.18632/oncotarget.18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fan S., Fan C., Liu N., Huang K., Fang X., Wang K. Downregulation of the long non-coding RNA ZFAS1 is associated with cell proliferation, migration and invasion in breast cancer. Mol. Med. Rep. 2018;17(5):6405–6412. doi: 10.3892/mmr.2018.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gao K., Ji Z., She K., Yang Q., Shao L. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed. Pharmacother. 2017;87:555–560. doi: 10.1016/j.biopha.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 207.Imani S., Hosseinifard H., Cheng J., Wei C., Fu J. Prognostic value of EMT-inducing transcription factors (EMT-TFs) in metastatic breast cancer: a systematic review and meta-analysis. Sci. Rep. 2016;6 doi: 10.1038/srep28587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lv Q.L., Chen S.H., Zhang X., Sun B., Hu L., Qu Q. Upregulation of long noncoding RNA zinc finger antisense 1 enhances epithelial–mesenchymal transition in vitro and predicts poor prognosis in glioma. Tumor Biol. 2017;39(3) doi: 10.1177/1010428317695022. [DOI] [PubMed] [Google Scholar]
- 209.Nowell C.S., Radtke F. Notch as a tumour suppressor. Nat. Rev. Cancer. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 210.Espinoza I., Miele L. Deadly crosstalk: notch signaling at the intersection of EMT and cancer stem cells. Canc. Lett. 2013;341:41–45. doi: 10.1016/j.canlet.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 211.Zhu H., Miao M.H., Ji X.Q., Xue J., Shao X.J. MiR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia. Biochem. Biophys. Res. Commun. 2015;459(2):340–345. doi: 10.1016/j.bbrc.2015.02.116. [DOI] [PubMed] [Google Scholar]
- 212.Ding Z., Jian S., Peng X., Liu Y., Wang J., Zheng L. Loss of MiR-664 expression enhances cutaneous malignant melanoma proliferation by upregulating PLP2. Med (United States) 2015;94(33):e1327. doi: 10.1097/MD.0000000000001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zou Y., Chen Y., Yao S., Deng G., Liu D., Yuan X. MiR-422a weakened breast cancer stem cells properties by targeting PLP2. Cancer Biol. Ther. 2018;19(5):436–444. doi: 10.1080/15384047.2018.1433497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen Y.H., Hueng D.Y., Tsai W.C. Proteolipid protein 2 overexpression indicates aggressive tumor behavior and adverse prognosis in human gliomas. Int. J. Mol. Sci. 2018;19(11) doi: 10.3390/ijms19113353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Li X., Luo Y., Liu L., Cui S., Chen W., Zeng A. The long noncoding RNA ZFAS1 promotes the progression of glioma by regulating the miR-150-5p/PLP2 axis. J. Cell. Physiol. 2020;235(3):2937–2946. doi: 10.1002/jcp.29199. [DOI] [PubMed] [Google Scholar]
- 216.Li Z., Xu C., Ding B., Gao M., Wei X., Ji N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J. Neuro Oncol. 2017;134(1):19–28. doi: 10.1007/s11060-017-2498-5. [DOI] [PubMed] [Google Scholar]
- 217.Zhang K., Sun X., Zhou X., Han L., Chen L., Shi Z. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zhou X., Ren Y., Zhang J., Zhang C., Zhang K., Han L. HOTAIR is a therapeutic target in glioblastoma. Oncotarget. 2015;6(10):8353–8365. doi: 10.18632/oncotarget.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ke J., Yao Y.L., Zheng J., Wang P., Liu Y.H., Ma J. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6(26):21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sa L., Li Y., Zhao L., Liu Y., Wang P., Liu L. The role of HOTAIR/miR-148b-3p/USF1 on regulating the permeability of BTB. Front. Mol. Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 221.Zhou K., Zhang C., Yao H., Zhang X., Zhou Y., Che Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer. 2018;17(1) doi: 10.1186/s12943-018-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu Q., Yu W., Zhu S., Cheng K., Xu H., Lv Y. Long noncoding RNA GAS5 regulates the proliferation, migration, and invasion of glioma cells by negatively regulating miR-18a-5p. J. Cell. Physiol. 2018;234(1):757–768. doi: 10.1002/jcp.26889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.