Abstract
Cytokines and chemokines are important regulators of airway hyper-responsiveness, immune cell infiltration, and inflammation and are produced when mast cells are stimulated with interleukin-33 (IL-33). Here, we establish that the salt-inducible kinases (SIKs) are required for the IL-33-stimulated transcription of il13, gm-csf and tnf and hence the production of these cytokines. The IL-33–stimulated secretion of IL-13, granulocyte-macrophage colony stimulating factor, and tumor necrosis factor was strongly reduced in fetal liver–derived mast cells from mice expressing a kinase-inactive mutant of SIK3 and abolished in cells expressing kinase-inactive mutants of SIK2 and SIK3. The IL-33–dependent secretion of these cytokines and several chemokines was also abolished in SIK2/3 double knock-out bone marrow–derived mast cells (BMMC), reduced in SIK3 KO cells but little affected in BMMC expressing kinase-inactive mutants of SIK1 and SIK2 or lacking SIK2 expression. In SIK2 knock-out BMMC, the expression of SIK3 was greatly increased. Our studies identify essential roles for SIK2 and SIK3 in producing inflammatory mediators that trigger airway inflammation. The effects of SIKs were independent of IκB kinase β, IκB kinase β-mediated NF-κB-dependent gene transcription, and activation of the mitogen-activated protein kinase family members p38α and c-jun N-terminal kinases. Our results suggest that dual inhibitors of SIK2 and SIK3 may have therapeutic potential for the treatment of mast cell–driven diseases.
Keywords: asthma, chemokine, granulocyte-macrophage colony stimulating, innate immunity, interleukin-13, interleukin-33, mast cells, protein kinase, salt-inducible kinase, tumor necrosis factor
Abbreviations: BMMC, bone marrow-derived mast cell; CCL, chemokine (C-C motif) ligand; CREB, cAMP response element-binding protein; FLMC, fetal liver–derived mast cell; GM-CSF, granulocyte-macrophage colony stimulating factor; IKK, IκB kinase; IL, interleukin; JNK, c-jun N-terminal kinase; KO, knock-out; MAP, mitogen-activated protein; SIK, salt-inducible kinase; TNF, tumor necrosis factor
According to the World Health Organization, the number of asthma patients worldwide exceeds 200 million. Airway inflammation is a feature of many asthma subtypes, so that understanding the interplay between the immune system and the cell types lining the airways is important. Mast cells have a significant role in asthma because they migrate into the airway smooth muscle, epithelium, and mucous glands of asthma patients where they release mediators of asthma (1, 2, 3). Mast cells, which are present in higher numbers in the airways of asthmatics (4), drive the disease by releasing cytokines that trigger airway inflammation, as well as histamine, prostaglandin D2, and leukotriene C4, which promote smooth muscle constriction and mucus secretion by the lungs (5). Other immune cells, such as eosinophils, are also important in late phase or chronic asthma (5, 6, 7).
Interleukin-33 (IL-33) is upregulated in and correlates with disease severity in asthma patients (8, 9). It stimulates the secretion of other cytokines, such as interleukin-13 (IL-13), granulocyte-macrophage colony stimulating factor (GM-CSF), and tumor necrosis factor (TNF) in mast cells (10, 11). IL-13 promotes airway obstruction through the overproduction of mucus (12, 13), as well as airway hyper-responsiveness and the infiltration of eosinophils into the lungs (12, 13, 14). GM-CSF has a critical role in promoting the proliferation and maturation of antigen-presenting cells involved in the presentation of allergens to T cells (15). Indeed, airway administration of GM-CSF in vivo was found to lower the threshold for the secretion of Th2 cytokines, such as IL-13, and eosinophil infiltration into the lungs in the house dust mite model of asthma and to increase IL-33 levels in this model (16). GM-CSF can also act together with IL-33 to promote the secretion of IL-13 in GM-CSF–derived macrophages and dendritic cells (17, 18). TNF has been implicated in many aspects of airway pathology in asthma (2, 19), whereas a number of chemokines promote the recruitment of additional immune cells to the lungs (20, 21). For example, the chemokine (C-C motif) ligand 24 (CCL24, also called eotaxin-2) stimulates the recruitment of eosinophils (20), which are associated with more severe forms of asthma. These findings have generated interest in developing therapies aimed at neutralising these cytokines or suppressing their production in asthma patients.
IL-33 is an interleukin-1 (IL-1) family member that exerts its effects on cells by binding to the ST2 component of the IL-33 receptor (22). Similar to IL-1, the interaction of IL-33 with its receptor induces the recruitment of myeloid differentiation factor 88 and IL-1 receptor–associated kinases 1, 2 and 4, forming oligomeric complexes, termed myddosomes (23). This leads to the activation of mitogen-activated protein (MAP) kinases, such as p38α and c-jun N-terminal kinase 1 (JNK1) and JNK2, and the IκB kinases (IKKs) which activate transcription factors, such as NF-κB. The p38α MAP kinase is required for the IL-33–stimulated secretion of IL-13, GM-CSF, TNF, and the chemokines CCL3 and CCL4 (11, 24).
The salt-inducible kinases (SIKs) are constitutively active, but their activity can be decreased by phosphorylation catalyzed by cyclic AMP-dependent protein kinase, in response to ligands that elevate intracellular cyclic AMP, such as prostaglandin E2 (25). The SIKs regulate cytokine production in macrophages and dendritic cells by impinging on myddosome-dependent processes (26, 27, 28). For example, SIK2 and SIK3 phosphorylate and inactivate cAMP response element-binding protein (CREB)-regulated transcription co-activator 3 in macrophages, preventing activation of the transcription factor CREB that drives the production of antiinflammatory molecules, such as IL-10 (25, 26, 29). In this way the SIKs antagonise a p38α-dependent pathway involving the activation of mitogen- and stress-activated kinases 1/2, the kinases that activate CREB (30). The SIKs also promote the production of TNF and other cytokines in macrophages and dendritic cells by an as yet unidentified mechanism (26, 27, 28, 29).
In this article, we have identified new and essential roles for the SIK family members, SIK2 and SIK3, in the production of cytokines and chemokines by mast cells, which suggests that inhibitors of these protein kinases may have therapeutic potential for the treatment of mast cell–driven diseases, such as asthma.
Results
SIK inhibition suppresses IL-33–dependent cytokine secretion from mast cells
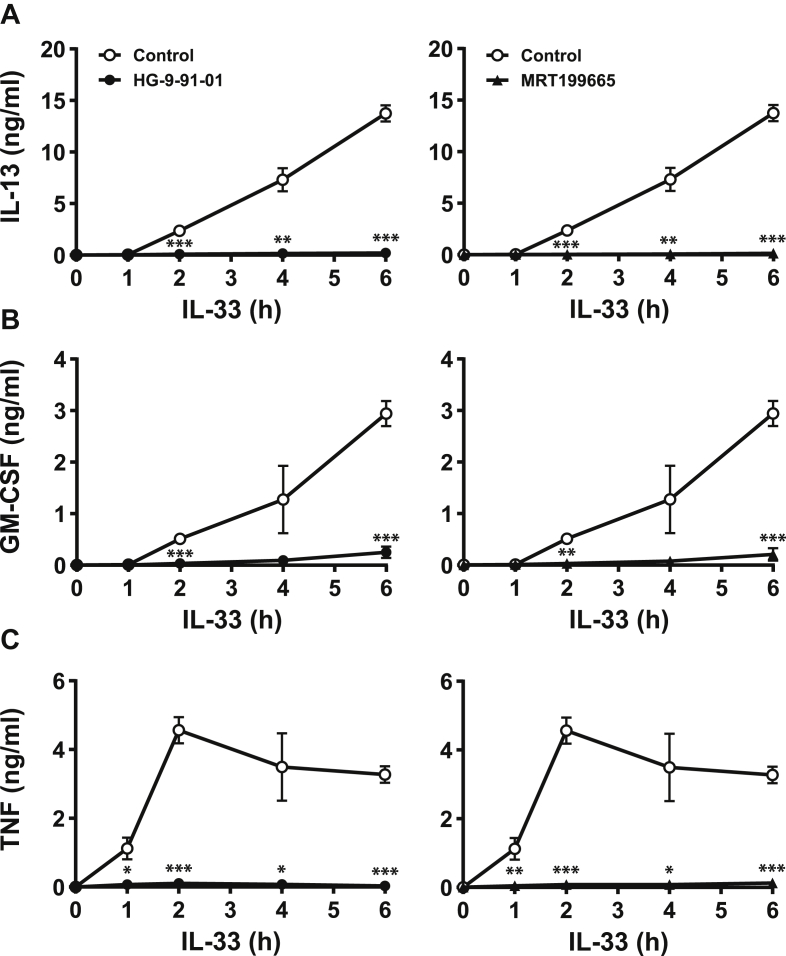
We initially studied the IL-33–dependent secretion of IL-13, GM-CSF, and TNF from WT bone marrow–derived mast cells (BMMCs) (Fig. 1). The secretion of IL-13 and GM-CSF was elevated 2 h after stimulation with IL-33 and continued to accumulate in the culture medium up to 6 h after stimulation (Fig. 1, A and B). In contrast, TNF secretion was detectable after 1 h and peaked after 2 h (Fig. 1C). The IL-33–dependent secretion of all three cytokines was drastically decreased by treatment with HG-9-91-01 or MRT199665 (Fig. 1), which are structurally unrelated small molecule inhibitors of all three SIK isoforms (26).
Figure 1.
SIK inhibition suppresses IL-33–dependent secretion of IL-13, GM-CSF and TNF in BMMC.A–C, BMMCs from WT mice were incubated for 1 h without (open circles, control) or with 0.5 μM HG-9-91-01 (closed circles, left hand panels) or 3 μM MRT199665 (closed triangles, right hand panels) and then stimulated with 10 ng/ml IL-33 for the times indicated. IL-13 (A), GM-CSF (B), and TNF (C) secreted into the culture medium was measured as in Experimental procedures. The results using HG-9-91-01 and MRT199665 are from the same experiment but plotted in separate panels for clarity and are presented as the mean and standard deviation of four biological replicates. Similar results were obtained in three independent experiments. Statistical analysis is represented by repeated measures two-way ANOVA with Dunnett’s multiple comparison test, comparing each inhibitor to control; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001. BMMC, bone marrow-derived mast cell; GM-CSF, granulocyte-macrophage colony stimulating factor; IL, interleukin; SIK, salt-inducible kinase; TNF, tumor necrosis factor.
Complete suppression of cytokine secretion was observed when HG-9-91-01 and MRT199665 were used at 0.5 μM and 3 μM, respectively (Fig. S1, A–C). These concentrations were used for all subsequent studies, unless indicated otherwise and are similar to those previously found to inhibit SIKs in macrophages (26). Stimulation of mast cells for 6 h with IL-33 in the absence of inhibitors decreased the number of mast cells by about one third compared with mast cells incubated for 6 h in the absence of IL-33 (Fig. S1D). The inclusion of HG-9-91-01 and MRT199665 improved cell viability in IL-33–stimulated cells, indicating that these compounds are not toxic and that the suppression of IL-13, GM-CSF, and TNF secretion does not result from the inhibitors causing cell death (Fig. S1D). These results suggested that the catalytic activity of one or more SIK isoforms was required for the secretion of IL-13, GM-CSF, and TNF from BMMCs.
SIK inhibition suppresses IL-33–dependent transcription of il13, gm-csf, and tnf
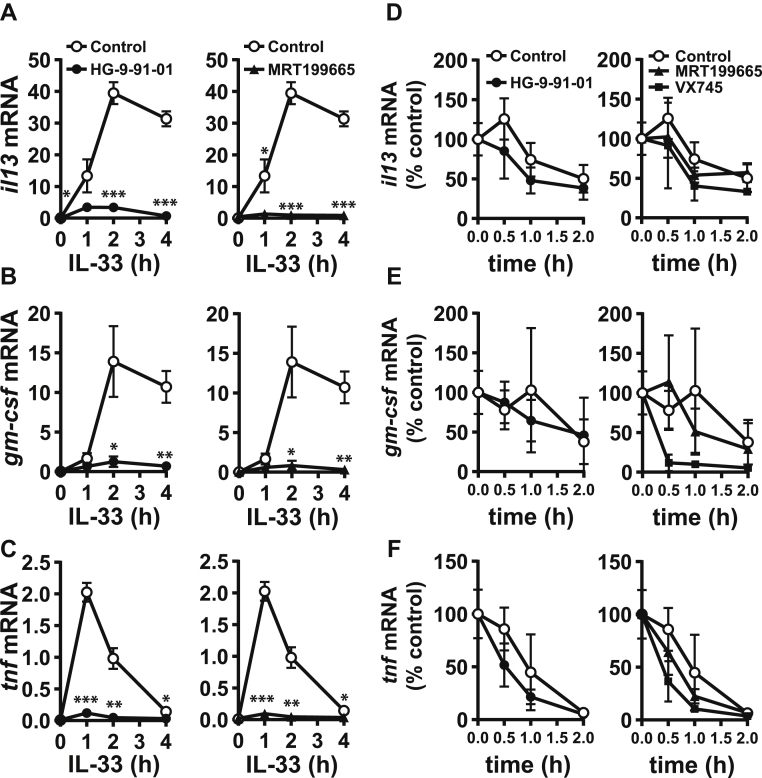
To investigate how IL-33 induced the production of IL-13, GM-CSF, and TNF, we studied the formation of the mRNAs encoding these cytokines (Fig. 2). IL-33 stimulated an increase in il13 and gm-csf mRNA, which occurred after 1 h, peaked after 2 h and was sustained for at least a further 2 h (Fig. 2, A and B). In contrast, tnf mRNA production peaked after 1 h and then declined rapidly (Fig. 2C). These results are consistent with the effects of IL-33 on cytokine secretion (Fig. 1).
Figure 2.
SIK inhibition suppresses IL-33–dependent production of il13, gm-csf, and tnf mRNA in BMMC.A–C, The experiment was performed as in Figure 1 except that, at the times indicated, cells were lysed and the mRNA encoding IL-13 (A), GM-CSF (B), and TNF (C) measured as described in Experimental procedures. The figures show the mRNA level relative to gapdh mRNA and are presented as the mean and standard deviation of four biological replicates. The results using HG-9-91-01 and MRT199665 are from the same experiment but plotted in separate panels for clarity, and similar results were obtained in two independent experiments. Statistical analysis is represented by repeated measures two-way ANOVA with Dunnett’s multiple comparison test, comparing each inhibitor to control; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001. D–F, WT BMMCs were incubated for 2 h with 10 ng/ml IL-33. Cells were then treated with 50 μM DRB and 5 μg/ml actinomycin D to block transcription without (control, open circles) or with 0.5 μM HG-9-91-01 (closed circles, left panel), 3 μM MRT199665 (closed triangles, right panel), or 1 μM VX745 (closed squares, right panel). At the times indicated, cells were lysed, and the il13 (D), gm-csf (E), and tnf (F) mRNA levels were measured, normalized to gapdh mRNA, and plotted relative to mRNA levels measured at the point of transcription inhibition. The results are presented as the mean and standard deviation of three replicates. Similar results were obtained in three independent experiments. The results using HG-9-91-01, MRT199665 and VX745 are from the same experiment, but plotted in separate panels for clarity. DRB, 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside; BMMC, bone marrow-derived mast cell; GM-CSF, granulocyte-macrophage colony stimulating factor; IL, interleukin; SIK, salt-inducible kinase; TNF, tumor necrosis factor.
We next investigated the rate at which these mRNAs declined when gene transcription was arrested by the addition of actinomycin D and 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside. The SIK inhibitors induced only a modest increase in the rate of mRNA decay that was not statistically significant (Fig. 2, D–F). In contrast, and as expected from previous studies (11), the p38α MAP kinase inhibitor VX745 increased the rate of tnf mRNA decay and strikingly increased the rate of decay of gm-csf mRNA but had no effect on il13 mRNA decay (Fig. 2, D–F). Taken together, the results suggested that SIKs regulate transcription of the genes encoding these cytokines with little effect on mRNA stability.
IL-33–dependent cytokine production in mast cells from mice deficient in the activity or expression of SIKs
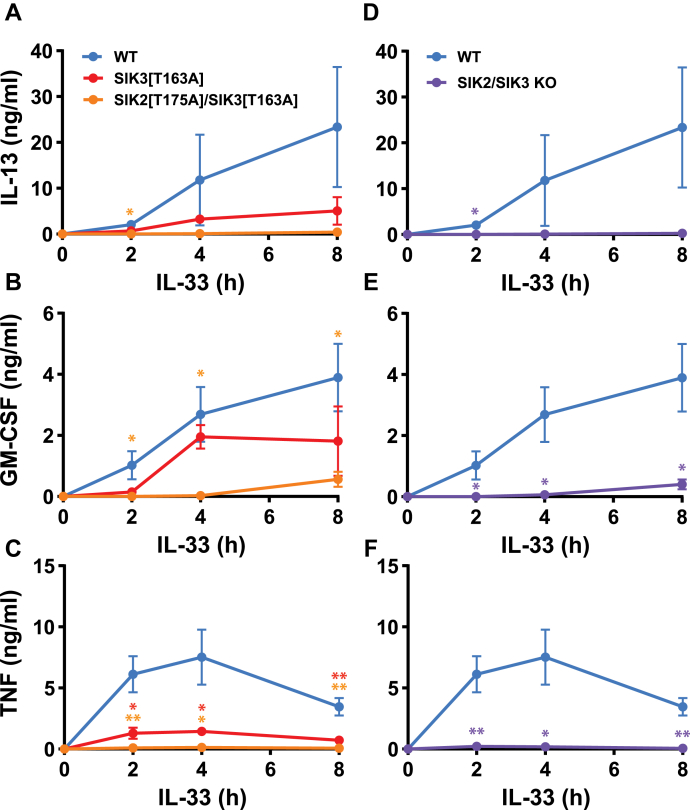
The compounds HG-9-91-01 and MRT199665 inhibit all three SIK isoforms with similar potency (26). To identify the SIK isoform(s) relevant to IL-33–dependent cytokine production, we studied mast cells from mice expressing kinase-inactive mutants of SIKs (29) and mast cells from SIK knock-out (KO) mice. The kinase-inactive SIK2[T175A]/SIK3[T163A] double knock-in mouse dies at a late stage of embryonic development (29), and so, studies with this double knock-in were performed in mast cells derived from fetal livers, in which the SIK inhibitors have a similar effect to that observed in BMMC (compare Fig. S2 with Fig. 1). We found that the IL-33–dependent secretion of IL-13, GM-CSF, and TNF was virtually abolished in SIK2[T175A]/SIK3[T163A] double knock-in fetal liver–derived mast cells (FLMCs) (Fig. 3, A–C), and similar results were obtained in FLMC from SIK2/SIK3 double KO mice (Fig. 3, D–F). The IL-33–stimulated secretion of IL-13 and TNF was strongly reduced (Fig. 3, A and C), and the secretion of GM-CSF partially reduced (Fig. 3B) in FLMC from SIK3[T163A] mice.
Figure 3.
SIK3 catalytic activity is critical for IL-13, GM-CSF, and TNF secretion in FLMC.A–F, FLMC from WT embryos, embryos expressing the kinase-inactive mutants of SIK3 (SIK3[T163A]) or both SIK2 and SIK3 (SIK2[T175A]/SIK3[T163A]) as well as SIK2/SIK3 double KO embryos were stimulated for the times indicated with 10 ng/ml IL-33. The concentrations of IL-13 (A, D), GM-CSF (B, E), and TNF (C, F) in the cell culture medium were then analyzed. The results in A–F are from a single experiment using FLMC from four WT, four SIK3[T163A], six SIK2[T175A]/SIK3[T163A], and four SIK2/3 KO mice and are plotted as the mean and standard deviation. Similar results were obtained in two independent experiments. The statistical analysis is represented by repeated measures two-way ANOVA with Dunnett’s multiple comparison test comparing all mutants to WT mast cells; ∗ p < 0.05, ∗∗ p < 0.01. BMMC, bone marrow-derived mast cell; FLMC, fetal liver–derived mast cell; GM-CSF, granulocyte-macrophage colony stimulating factor; IL, interleukin; KO, knock-out; SIK, salt-inducible kinase; TNF, tumor necrosis factor.
These results indicate that SIK1 is unable to compensate for the combined loss of SIK2/3 expression or catalytic activity in FLMC and is consistent with quantitative proteomic experiments that we have carried out in BMMC. These studies showed that mast cells express 5973 ± 109 copies of SIK3 and 675 ± 29 copies of SIK2 per cell, but SIK1-specific peptides were not detected in any of these experiments. Moreover, we were unable to detect a protein with the molecular mass of SIK1 by immunoblotting of BMMC using two different commercially available SIK1 antibodies (see Experimental procedures). This suggests that either SIK1 is not expressed in mast cells or that the level of expression is very low. Consistent with this conclusion, the loss of SIK1 catalytic activity in BMMC had no effect on the secretion of IL-13, GM-CSF, or TNF (Fig. S3).
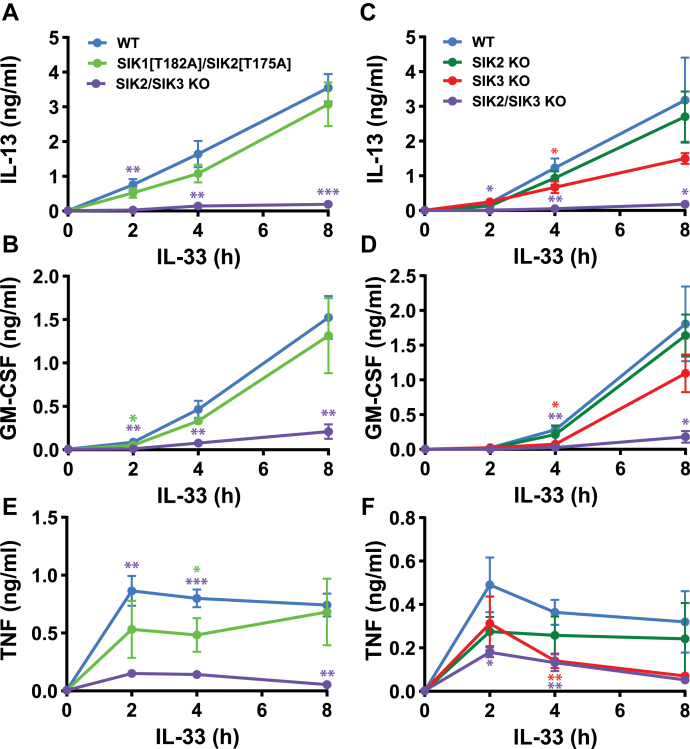
The SIK1[T182A]/SIK2[T175A] double knock-in mice are viable, and we therefore studied their contribution to IL-33–dependent cytokine production in BMMC from adult mice. These experiments showed that the loss of both SIK1 and SIK2 catalytic activity did not have a statistically significant effect on IL-13 or GM-CSF secretion (Fig. 4, A and B) or on the level of il13 and gm-csf mRNA (Fig. S4, A and B), indicating that cytokine production in these cells is controlled by SIK3. The contribution of SIK3 may be enhanced by the modest (50 %) increase in its expression that occurs in BMMC from SIK1[T182A]/SIK2[T175A] double knock-in mice (Fig. S5A). This is probably caused by the loss of SIK2 catalytic activity because there was no increase in SIK3 in extracts from SIK1[T182A] single knock-in BMMC (Fig. S5B). In SIK3 KO BMMC, the secretion of IL-13 and GM-CSF was reduced partially (Fig. 4, C and D), suggesting that another SIK isoform is compensating for the loss of SIK3. This is presumably SIK2, as the KO of both SIK2 and SIK3 abolished cytokine secretion. Similar to BMMC from SIK1[T182A]/SIK2[T175A] double knock-in mice, the KO of SIK2 alone did not have a statistically significant effect on the secretion of IL-13 or GM-CSF (Fig. 4, C and D). This appears to be because of compensation by SIK3, whose expression is greatly increased in SIK2 KO BMMC (Fig. S5C). In contrast, the expression of SIK2 was little affected in BMMC from SIK3 KO mice, but quantitation was not possible because another protein migrating very close to but slightly faster than SIK2 was recognized nonspecifically by the antibody (Fig. S5C).
Figure 4.
IL-33–dependent secretion of cytokines in BMMC from SIK1/2 double knock-in, SIK3 knock-out, and SIK2/3 double knock-out mice.A and B, The experiments were performed as in Figure 1 using BMMCs differentiated from WT, SIK1[T182A]/SIK2[T175A] double knock-in mice and SIK2/SIK3 double KO mice. After stimulation for the times indicated with 10 ng/ml IL-33, IL-13 (A), and GM-CSF (B) secreted into the culture medium was analyzed. C and D, BMMCs from SIK2 KO, SIK3 KO, SIK2/3 double KO, and WT mice were stimulated with IL-33 and cytokine secretion measured as in A and B. E and F, As for A-D except that TNF secretion was measured. The results shown are the mean and standard deviation from four mice per genotype. Similar results were obtained in two independent experiments. The statistical analysis is represented by repeated measures two-way ANOVA with Dunnett’s multiple comparison test, comparing SIK1[T182A]/SIK2[T175A] or SIK2/3 KO BMMC to WT BMMC (A, B, E) or comparing SIK2 KO, SIK3 KO or SIK2/3 KO BMMC to WT BMMC (C, D, F); ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001. BMMC, bone marrow-derived mast cell; GM-CSF, granulocyte-macrophage colony stimulating factor; IL, interleukin; KO, knock-out; SIK, salt-inducible kinase; TNF, tumor necrosis factor.
In contrast to IL-13 and GM-CSF, the secretion of TNF (Fig. 4, E and F) was reduced significantly in SIK1[T182A]/SIK2[T175A] double knock-in BMMC, as well as in SIK3 KO BMMC, indicating the involvement of more than one SIK isoform. These are presumably SIK2 and SIK3 because the KO of both SIK2 and SIK3 abolished cytokine secretion. The partial suppression of TNF secretion in BMMC from SIK2/SIK3 double KO mice and apparent absence of SIK1 in these cells suggest that another protein kinase may compensate for the absence of SIK2 and SIK3 when the expression of both of these isoforms is ablated. However, such compensation does not happen in FLMC expressing the kinase-inactive SIK2[T175A] and SIK3[T163A] mutants or in FLMC lacking any expression of SIK2 and SIK3 (Fig. 3, C and F).
SIK2/3-mediated, IL-33–dependent cytokine secretion occurs independently of p38α or JNK MAP kinases, IKKβ, or NF-κB
The cell surface levels of ST2 were unaffected by SIK inhibitors or in SIK2/3 double KO cells (Fig. S6, A and B), suggesting the SIKs control cytokine secretion “downstream” of the receptor. The p38α MAP kinase is required for cytokine and chemokine secretion in BMMC (11, 24). We found that SIK inhibitors did not affect the IL-33–stimulated phosphorylation (activation) of the MAP kinase family members p38α or JNK, indicating that the suppression of cytokine production by these compounds was not explained by inhibition of the pathway leading to the activation of these MAP kinases (Fig. S6, C and D). Similar results were obtained in BMMC from SIK2/3 double KO mice (Fig. S6, E and F). In addition, JNK-IN-8, a JNK inhibitor, suppressed the phosphorylation of its substrate c-Jun without affecting the IL-33–dependent secretion of IL-13 and GM-CSF, although TNF secretion was reduced by 70 % after 2 h and 35 % after 6 h (Fig. S7). Taken together, our results indicate that the SIKs exert their effects on cytokine secretion in mast cells by a mechanism that is independent of these MAP kinases. It should be noted that JNK-IN-8 does not affect the IL-33–stimulated phosphorylation of JNKs but nevertheless decreases their electrophoretic mobility because of its covalent mechanism of inhibition (31).
The IL-33–stimulated phosphorylation of p105/NF-κB1, a substrate of IKKβ (32, 33), was not reduced by the SIK inhibitors or in SIK2/3 double KO BMMC (Fig. S6, C and E), indicating that SIKs are not required at any step in the pathway leading to activation of the IKKβ component of the canonical IKK complex. Moreover, SIK inhibitors had little effect on IKKβ-mediated NF-κB–dependent gene transcription (Fig. S8A) at concentrations that completely prevented IL-33–stimulated cytokine secretion (Figs. 1 and S1). In contrast, two structurally unrelated small molecule inhibitors of IKKβ blocked IL-33–stimulated NF-κB–dependent transcription of a luciferase reporter gene, as expected (Fig. S8A). We also investigated how IKKβ inhibitors affected IL-33–stimulated cytokine secretion. GM-CSF and TNF secretions were reduced by about 50 %, whereas IL-13 secretion was unaffected (Fig. S8, B–D). In summary, our results indicate that the activation of IKKβ, IKKβ activity, and IKKβ-mediated, NF-κB–dependent gene transcription is not required for IL-33–stimulated IL-13 production in mast cells but may contribute to GM-CSF and TNF induction by an SIK-independent mechanism. Taken together, our results indicate that the SIKs exert their effects on cytokine secretion in mast cells by mechanisms that are largely independent of IKKβ and IKKβ-mediated, NF-κB-dependent gene transcription.
SIK activity is required for the secretion of several chemokines from mast cells
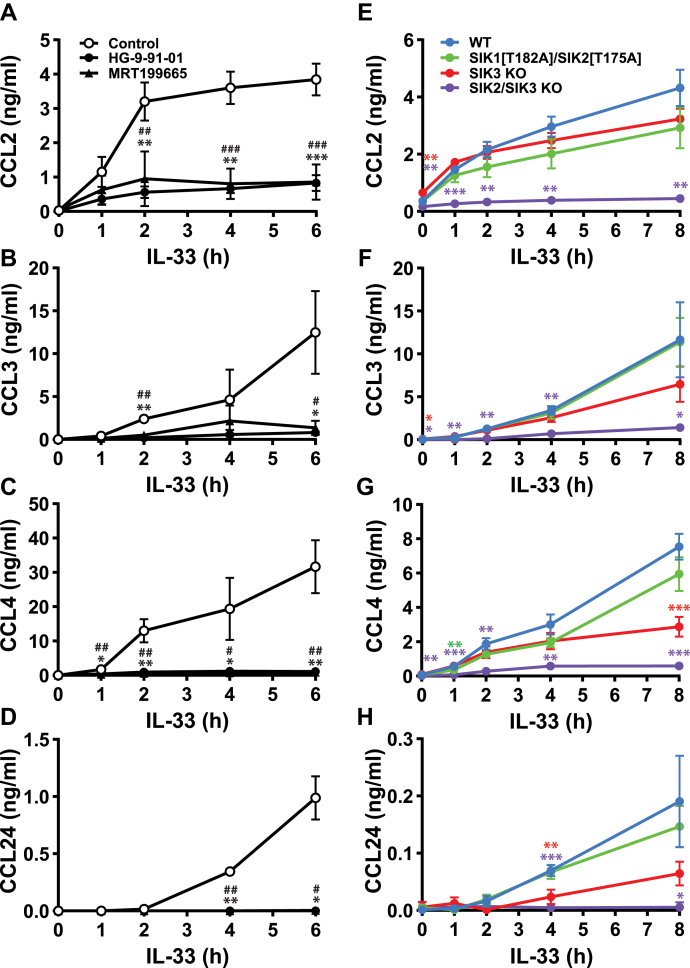
Mast cells also secrete chemokines, which have important roles in promoting the recruitment of immune cells to the lungs and therefore in establishing lung inflammation and bronchial hyper-responsiveness (20, 21). The chemokines CCL2 (also known as monocyte chemoattractant protein-1), CCL3 (also known as macrophage inflammatory protein-1α), and CCL4 (macrophage inflammatory protein-1β) were detected in the culture medium of WT BMMCs after stimulation for 1 h with IL-33, whereas CCL24 was detected after 2 h (Fig. 5, A–D). CCL3, CCL4, and CCL24 continued to accumulate for up to 8 h after stimulation (Fig. 5, B–D), but CCL2 secretion became near maximal after 2 h (Fig. 5A). The secretion of all four chemokines was strongly suppressed by SIK inhibitors (Fig. 5, A–D) or in SIK2/3 double KO BMMC (Fig. 5, E–H). We were unable to detect any CCL11 (eotaxin-1) in the culture medium after stimulation with IL-33.
Figure 5.
SIK activity is required for CCL2, CCL3, CCL4, and CCL24 secretion in BMMC.A–D, WT BMMCs were incubated for 1 h without (open circles, control) or with 0.5 μM HG-9-91-01 (closed circles) or 3 μM MRT199665 (closed triangles), then stimulated with 10 ng/ml IL-33. The concentrations of CCL2 (A), CCL3 (B), CCL4 (C), and CCL24 (D) in the cell culture medium were measured. The results shown are mean and standard deviation from four biological replicates (A–C) or three biological replicates (D) with similar results obtained in two independent experiments. Statistical analyses are shown as repeated measures two-way ANOVA with Dunnett’s multiple comparison test, comparing HG-9-91-01 (∗) or MRT199665 (#) to control; ∗,#p < 0.05, ∗∗,##p < 0.01, ∗∗∗,###p < 0.001. E–H, BMMCs generated from WT, SIK1[T182A]/SIK2[T175A] double knock-in, SIK3 KO and SIK2/SIK3 double KO mice were stimulated for the times indicated with 10 ng/ml IL-33. The concentrations of CCL2 (E), CCL3 (F), CCL4 (G), and CCL24 (H) in the cell culture medium were measured as described in Experimental procedures. The results shown are the mean and standard deviation from four biological replicates with similar results obtained in two independent experiments. Statistical analyses are shown as repeated measures two-way ANOVA with Dunnett’s multiple comparison test, comparing SIK1[T182A]/SIK2[T175A], SIK3 KO or SIK2/3 KO to WT BMMCs; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001. BMMC, bone marrow-derived mast cell; CCL, chemokine (C-C motif) ligand; GM-CSF, granulocyte-macrophage colony stimulating factor; IL, interleukin; KO, knock-out; SIK, salt-inducible kinase.
In contrast to the cytokines mentioned in the preceding paragraph, the SIK inhibitors did not block the secretion of chemokine (C-X-C motif) ligand 2 (also called MIP-2) (Fig. S9A), which is the mouse ortholog of human IL-8. Indeed, the secretion of chemokine (C-X-C motif) ligand 2 was actually elevated in the SIK3 KO or SIK2/SIK3 double KO mice (Fig. S9B).
Discussion
Our earlier studies in macrophages and dendritic cells established critical roles for the SIK subfamily of protein kinases, especially SIK2 and SIK3, in controlling the production of proinflammatory and antiinflammatory cytokines by TLR-activating ligands (26, 29). The inhibition of both SIK2 and SIK3 increased the production of the antiinflammatory cytokine IL-10 and drastically reduced the production of major proinflammatory cytokines, such as IL-6 and IL-12, (see the start of the text). These and other findings (34) suggested that drugs inhibiting both SIK2 and SIK3 might have therapeutic potential for the treatment of inflammatory and autoimmune diseases. Interestingly, the biotechnology company Galapagos disclosed on October 27th 2020 that compounds from their “Toledo” program target the SIK family of protein kinases and that a dual SIK2/SIK3 inhibitor GLPG3970 had passed Phase I trials and would soon be entering advanced clinical trials for psoriasis, rheumatoid arthritis, and ulcerative colitis. The present study which shows that the combined inhibition of SIK2/3 suppresses the production of a number of cytokines and chemokines that contribute to the pathogenesis of asthma now suggests that GLPG3970 and/or related compounds may be worth exploring as potential treatments for asthma patients who are refractory to the corticosteroid therapies that are in widespread use. The efficacy of antibodies that target a single cytokine has proved disappointing in advanced clinical trials for asthma, and several such trials have been stopped (35, 36, 37, 38). Drugs that suppress the production of many cytokines and chemokines that are important contributors to asthma pathology (such as SIK2/3 inhibitors) may have greater efficacy. Interestingly, genome-wide association studies have identified single nucleotide polymorphisms within the regions containing SIK genes that are associated with asthma or with allergic diseases such as hay fever and eczema (39).
In the present study, we found that SIK2/3 inhibition abolished transcription of the genes encoding IL-13, GM-CSF, and TNF, implying that a SIK2/3-catalyzed phosphorylation event(s) is essential for this process. The transcription factors NF-κB and IRF5, which are essential for the production of many proinflammatory cytokines in macrophages, are activated by IKKβ-catalyzed phosphorylation events (40, 41). We found that SIK inhibitors did not affect IKKβ activation or activity, indicating that SIKs are not required at any step between the IL-33 receptor and the activation of the IKKβ component of the canonical NF-κB complex (Fig. S6, C and E). The SIK inhibitors also had little effect on IL-33–stimulated NF-κB–dependent gene transcription (Fig. S8A) at concentrations that abolish IL-13, GM-CSF, and TNF production (Fig. S1, A–C). Conversely, IKKβ inhibitors blocked IL-33–stimulated NF-κB–dependent gene transcription (Fig. S8A) but had no effect on IL-33–stimulated IL-13 secretion (Fig. S8B) and only reduced GM-CSF and TNF secretion by 50 % in BMMCs (Fig. S8, C and D). Taken together, our results establish that SIKs do not regulate cytokine gene transcription via IKKβ or IKKβ-stimulated activation of NF-κB–dependent gene transcription. However, our results do not exclude the possibility that SIKs regulate IL-33–stimulated cytokine and chemokine production by modulating the noncanonical NF-κB pathway, which requires IKKα and not IKKβ activity.
It has been reported by others (42) that SIKs phosphorylate and inactivate histone deacetylase 4 and histone deacetylase 5, leading to increased acetylation of the p65 subunit of NF-κB, which is thought to stimulate its ability to bind to gene promoters and transcribe genes. Such genes include those encoding proinflammatory cytokines, including tnf (42). The results presented in this article (Figs. S6 and S8) indicate that this is not the mechanism by which SIKs mediate the transcription of genes encoding cytokines and chemokines in mast cells, otherwise the SIK inhibitors would have been expected to suppress IL-33–stimulated NF-κB–dependent gene transcription.
It is well established that the activation of p38α is required for the production of IL-13, GM-CSF, TNF, CCL3, and CCL4 in mast cells and that it stimulates GM-CSF and TNF production by stabilizing the mRNAs encoding these cytokines and not by regulating the transcription of their genes (11, 24). We found that SIK inhibition, or ablation of the expression of both SIK2 and SIK3, had no effect on the IL-33–stimulated activation of p38α (Fig. S6, C–F), and SIK inhibition did not accelerate the decay of GM-CSF or TNF significantly (Fig. 2, E and F). Our studies therefore suggest that SIKs are unlikely to control cytokine or chemokine gene transcription via p38α. Similarly, a JNK inhibitor blocked the phosphorylation (activation) of the transcription factor c-Jun (Fig. S7A) without affecting the secretion of IL-13 or GM-CSF (Fig. S7, B–D). Taken together, our results exclude the possibility that SIKs control il13 or gm-csf gene transcription in mast cells by suppressing the MAP kinase family members p38α and JNK. Identifying the transcription factor or transcriptional co-activator(s) whose phosphorylation by SIK2/3 is required for cytokine and chemokine gene transcription in mast cells is a key outstanding question that has yet to be solved.
Experimental procedures
Materials
Recombinant murine IL-3 and IL-33 were purchased from Peprotech, reconstituted in sterile water, and stored in aliquots at −80 °C. The structures of HG-9-91-01 and MRT199665 and their characterization as SIK inhibitors have been described previously (26). The IKKβ inhibitors BI605906 and PS1145, the p38α inhibitor VX745, and the JNK inhibitor JNK-IN-8 were synthesized by Natalia Shpiro in our unit. The protein kinase inhibitors used in this study can be obtained from MRC PPU Reagents and Services (https://mrcppureagents.dundee.ac.uk/). Actinomycin D was purchased from Cambridge Bioscience and 5,6-dichlorobenzimidazole 1-β-D-ribofuranoside from Sigma.
Generation of mice
SIK1[T182A] (Sik1tm1.1 Arte), SIK2[T175A] (Sik2tm1.1 Arte), and SIK3[T163A] (Sik3tm1.1 Arte) knock-in mice expressing catalytically inactive mutants of each SIK were generated by Taconic Biosciences and were characterized previously (29). The SIK2[T175A] mice contain LoxP sites flanking exons 5 to 7 of the sik2 gene, with recombination at this site resulting in loss of these exons (corresponding to amino acid residues 160–316) and a frame-shift mutation affecting the subsequent exons. After germline expression of a Cre transgene by crossing to the Cre Deleter strain (C57BL/6-Gt(ROSA)26Sortm16(cre)Arte, Taconic Biosciences), the Cre was bred out of the line, and mice testing negative for Cre expression for at least three generations were used as SIK2 KO mice.
To generate SIK3flox/flox mice, C57Bl/6N-Sik3tm1a(EUCOMM)Hmgu/Wtsi mice were purchased from the European Conditional Mouse Mutagenesis Program (EUCOMM). The targeted SIK3 allele contained a cassette expressing LacZ and a neomycin-resistance gene flanked by Frt sites (43). These mice were bred to those expressing a Flp transgene (Taconic Biosciences), resulting in loss of the LacZ-neomycin resistance cassette and generation of a sik3flox allele. To generate mice that do not express SIK3 within the hematopoietic lineages, SIK3flox/flox mice were bred to mice expressing a Cre transgene driven by the Vav promoter (44). This deletes exon 5 of sik3 (encoding amino acid residues 226–247) and causes frame-shift mutations affecting the subsequent exons and hence disrupts the gene.
SIK2 KO/SIK3flox/flox mice were generated by crossing SIK2ko/+ and SIK3flox/flox mice to produce a mouse carrying both the sik2ko and sik3flox alleles. Both sik2 and sik3 are located on chromosome 9 separated by approximately 4.6 Mb, and so a recombination event is required to link the sik2ko and sik3flox mutations. This required multiple rounds of breeding using SIK3flox/flox and SIK2ko/+SIK3flox/+ mice before a SIK2ko/+SIK3flox/flox mouse was detected. SIK2ko/+SIK3flox/flox mice were bred to mice expressing a Cre transgene driven by the Vav promoter (44). In this article, SIK3flox/floxVavcre+ and SIK2ko/koSIK3flox/floxVavcre+ mice are termed SIK3 KO and SIK2/3 double KO mice, respectively, and complete loss of SIK2 and SIK3 protein expression in BMMC was verified by immunoblotting (Fig. S5C).
SIK3flox/floxVavcre+ females are infertile, and the Vav promoter can result in Cre expression in the testes (44), therefore SIK3flox/+Vavcre+ or SIK2ko/koSIK3flox/+Vavcre+ females were used to maintain these lines.
WT mice from the relevant SIK line or the SIK3+/+Vavcre+ (WT Vavcre+) mice were used for control experiments as indicated in the figure legends. All mice were maintained on a C57Bl/6J background using mice obtained from Charles River Laboratories UK, and routine genotyping was carried out on genomic DNA from ear biopsy specimens. Primers used for genotyping SIK knock-in mice were published previously (29). The following primers were used for genotyping SIK KO mice; aagagagtgtgggactaacttgg, cttaaaagctgggcatagtgg and tgttctctaagcatgctaactactagg for SIK2 KO; gctgaagacgtggtgtggcag and gcaggtaacatttctgcttccagac for SIK3flox/flox; ctccaacctgctgactgtgc and caccagggacacagcattgg for Vavcre.
Animals were maintained under specific pathogen-free conditions consistent with EU and UK regulations. Mice were housed in individually ventilated cages at 21 °C, 45 to 55 % humidity, and a 12/12 h light/dark cycle, with free access to food (R&M3) and water. This work was performed under a U.K. Home Office Project Licence awarded after recommendation by the University of Dundee Ethical Review Committee.
Mast cell culture
BMMC were generated as described (45), using bone marrow collected from femurs and tibias of 3 to 6 months old mice. Fetal livers were differentiated into FLMCs. The fetal livers were dissected from embryos at E14.5-E16.5, dispersed to single cell suspensions in PBS using a 70 μm cell strainer, pelleted at 300g for 5 min at 21 °C, and re-suspended in 1.5 ml freezing media (90 % [v/v] fetal bovine serum, 10 % [v/v] dimethyl sulfoxide) per liver. Cells were then frozen at −80 °C for 24 h before transferring to liquid nitrogen. When sufficient fetal liver cells from each genotype had been collected, they were thawed rapidly in a 37 °C water bath and diluted with BMMC media (45) at 37 °C to reduce the dimethyl sulfoxide concentration to 1.5 % (v/v). Fetal liver cells were pelleted as described above, re-suspended in 15 ml BMMC media, and cultured as for BMMC.
Mast cells were seeded (typically at 106 cells/ml) in media containing 30 ng/ml IL-3 because this is required for their viability and for the expression of ST2 on the cell surface (46), and cells were rested for 4 h before experimentation.
Flow cytometry
Mast cell purity was monitored by flow cytometry (Figs. S10 and S11) using a FACSCanto (BD Biosciences). 4 x 105 cells were collected, pelleted, washed in 1 ml PBS containing 1 % (w/v) bovine serum albumin, and suspended in 50 μl of the same buffer containing 1 μl of anti-CD16/CD32 (BD Biosciences, clone 2.4G2). After 10 min at 4 °C, a further 50 μl of buffer containing the following stains was added and incubated for 30 min at 4 °C; 0.25 μl of anti-c-kit-PE (eBioscience, clone 2B8), 0.25 μl of anti-FcεR1-FITC (eBioscience, clone MAR-1) and 0.25 μl of anti-ST2-BV421 (BioLegend, clone DIH9). The cells were washed twice with PBS containing 1 % (w/v) bovine serum albumin and re-suspended in buffer containing 0.1 μM TO-PRO-3 Iodide (Invitrogen).
The gating strategy for mast cells is shown in Figure S10. As reported previously (47), the expression of the cell surface mast cell markers FcεR1 and c-kit was similar in BMMCs and FLMCs (Fig. S11A). In addition, BMMCs and FLMCs expressed identical levels of the IL-33 receptor, ST2 on their cell surface (Fig. S11, B and C). On the other hand, FLMCs appear more granular than BMMCs as determined by side scatter on the flow cytometer (Fig. S11D) (47). The purity of the mast cells used for experiments ranged from 80 to 95 % and averaged >85 % purity. For cell counting, mast cell cultures were diluted 1:10 in PBS containing 1 μg/ml DAPI (Thermo Fisher) and analyzed using FACSVerse (BD Biosciences) or NovoCyte (ACEA Biosciences). Data collected via flow cytometry were analyzed using FlowJo software version 10 (Tree Star).
Immunoblotting
Mast cells were lysed in 50 mM Tris/HCl pH 7.4, 1 mM EDTA, 1 mM EGTA, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 10 mM sodium β-glycerol 1-phosphate, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 0.27 M sucrose, 1 % (v/v) Triton X-100, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM phenylmethylsulphonyl fluoride, and cell lysates clarified by centrifugation at 17,000g for 10 min at 4 °C. The protein concentrations of the supernatants (cell extract) were determined using the Bradford assay, and 15 to 20 μg of cell extract was separated by SDS-PAGE. Where indicated in the figure legends, 10 mM MgCl2, 1 % (w/v) SDS and 0.05 % (v/v) Benzonase nuclease were added to the lysis buffer, and protein concentrations of the cell extracts were determined using BCA assay. In these experiments, 40 to 50 μg of cell extract was separated by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes, immunoblotted, and visualized by chemiluminescence using enhanced chemiluminescence substrate (Amersham) and the Chemidoc MP imaging system (Bio-Rad Laboratories). Densitometric analysis was performed using Image Lab 6.1 (Bio-Rad Laboratories). Antibodies were purchased from the following suppliers: anti-GAPDH (#2118), anti-SIK2 (#6919), anti-p38 (#9212), anti-phospho p105/NF-κB1 (Ser932) (#4806), anti-phospho p38 (Thr180/Tyr182) (#9211), anti-phospho c-Jun (Ser63) (#9261), anti-phospho JNK1/2 (Thr183/Tyr185) (#9251), and horseradish peroxidase-conjugated anti-rabbit antibodies from Cell Signaling Technology. Anti-SIK1 antibodies were obtained from Abcam (ab64428) and Santa Cruz (sc-83754). An anti-SIK3 antibody was raised in house against amino acid residues 926 to 1038 of SIK3 (sheep S373D, bleed 3) and can be obtained from MRC PPU Reagents and Services (https://mrcppureagents.dundee.ac.uk/). Horseradish peroxidase-conjugated anti-sheep secondary antibody was from Thermo Scientific.
RNA isolation and QPCR
RNA was isolated from mast cells using the MicroElute Total RNA kit (VWR) following the Manufacturers’ instructions. Reverse transcription to generate cDNA was performed using the iScript cDNA synthesis kit (Bio-Rad Laboratories) using 300 to 500 ng total RNA. SsoFast EvaGreen Supermix (Bio-Rad Laboratories) was used on a CFX384 RT-PCR machine (Bio-Rad Laboratories) to quantify cDNA products. The expression of each gene was normalized to mRNA levels of gapdh using 2ˆ(Ctgapdh-Ctgene of interest). The following primers were used for qPCR: gapdh F, tgcaccaccaactgcttag; gapdh R, gatgcagggatgatgttc; gm-csf F, gcagacaggagtgttgctct; gm-csf R, tgaaattgccccgtagaccc; il13 F, gcagcagcttgagcacattt; il13 R, gcagacaggagtgttgctct; tnf F, cagaccctcacactcagatcatc; tnf R, ggctacaggcttgtcactcg.
Secretion assays
Cytokines in cell-free cell culture medium were analyzed using the Bio-Plex Pro Assay System (Bio-Rad Laboratories), Luminex 200 machine, and xPONENT 4.1 software (Luminex Corporation). The expression of Cre recombinase in WT Vavcre+ BMMCs did not affect the IL-33–stimulated secretion of cytokines from the cells (Fig. S12), and so WT mast cells not expressing Cre recombinase were used as controls, unless otherwise indicated.
Luciferase assays
To study NF-κB–dependent transcription, 4 million mast cells were transfected with 2 μg NF-κB–dependent reporter plasmid (PRDII elements from the IFNβ promoter cloned into the pLuc-MCS vector, a kind gift from Professor Katherine Fitzgerald, School of Medicine, University of Massachusetts) (48) and 0.4 μg pTK-renilla luciferase (Stratech) plasmid. Transfections were performed by electroporation using Cell Line Nucleofector kit V and programme T-030 on a Nucleofector II device (Lonza Bioscience). Cells were rested for 12 h posttransfection before experimentation. Luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega Corporation) and measured using PHERAstar FS (BMG Labtech Ltd).
Quantitative mass spectrometry
The number of molecules of each SIK isoform was determined by quantitative mass spectrometry following tandem mass tag labeling as described for T cells (49). The results for BMMC, with full details of how the experiments were performed and analyzed have been deposited on PRIDE project ID PXD020091.
Statistical analysis of the data
Results are presented as mean and standard deviation. Statistical differences between groups of experimental cells were assessed by one-way or two-way ANOVA with Dunnett’s or Sidak’s multiple comparison test as indicated in the figure legend. The results were considered significant if p < 0.05; n.s. not significant, ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001.
Data availability
All mass spectrometry data are available via PRIDE project ID PXD020091. Data used for generation of the figures in the paper are available upon request from the corresponding author (Philip Cohen, p.cohen@dundee.ac.uk) or the first author (Nicola Darling, n.j.darling@dundee.ac.uk).
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the content of this article.
Acknowledgments
This study was supported by a Programme Grant (MR/R021406/1) from the U.K. Medical Research Council (to P. C.).
Author contribution
N. J. D. and P. C. conceptualization; N. J. D. investigation; J. S. C. A. methodology; P. C. supervision; N. J. D. and P. C. writing (original draft); J. S. C. A writing (review and editing); P. C. funding acquisition.
Edited by Peter Cresswell
Supporting information
References
- 1.Carroll N.G., Mutavdzic S., James A.L. Increased mast cells and neutrophils in submucosal mucous glands and mucus plugging in patients with asthma. Thorax. 2002;57:677–682. doi: 10.1136/thorax.57.8.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradding P., Roberts J.A., Britten K.M., Montefort S., Djukanovic R., Mueller R., Heusser C.H., Howarth P.H., Holgate S.T. Interleukin-4, -5, and -6 and tumor necrosis factor-alpha in normal and asthmatic airways: Evidence for the human mast cell as a source of these cytokines. Am. J. Respir. Cell Mol. Biol. 1994;10:471–480. doi: 10.1165/ajrcmb.10.5.8179909. [DOI] [PubMed] [Google Scholar]
- 3.Brightling C.E., Bradding P., Symon F.A., Holgate S.T., Wardlaw A.J., Pavord I.D. Mast-cell infiltration of airway smooth muscle in asthma. N. Engl. J. Med. 2002;346:1699–1705. doi: 10.1056/NEJMoa012705. [DOI] [PubMed] [Google Scholar]
- 4.Balzar S., Chu H.W., Strand M., Wenzel S. Relationship of small airway chymase-positive mast cells and lung function in severe asthma. Am. J. Respir. Crit. Care Med. 2005;171:431–439. doi: 10.1164/rccm.200407-949OC. [DOI] [PubMed] [Google Scholar]
- 5.Bradding P. Asthma: Eosinophil disease, mast cell disease, or both? Allergy Asthma Clin. Immunol. 2008;4:84–90. doi: 10.1186/1710-1492-4-2-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cieslewicz G., Tomkinson A., Adler A., Duez C., Schwarze J., Takeda K., Larson K.A., Lee J.J., Irvin C.G., Gelfand E.W. The late, but not early, asthmatic response is dependent on IL-5 and correlates with eosinophil infiltration. J. Clin. Invest. 1999;104:301–308. doi: 10.1172/JCI7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein Wolterink R.G., Kleinjan A., van Nimwegen M., Bergen I., de Bruijn M., Levani Y., Hendriks R.W. Pulmonary innate lymphoid cells are major producers of IL-5 and IL-13 in murine models of allergic asthma. Eur. J. Immunol. 2012;42:1106–1116. doi: 10.1002/eji.201142018. [DOI] [PubMed] [Google Scholar]
- 8.Préfontaine D., Lajoie-Kadoch S., Foley S., Audusseau S., Olivenstein R., Halayko A.J., Lemière C., Martin J.G., Hamid Q. Increased expression of IL-33 in severe asthma: Evidence of expression by airway smooth muscle cells. J. Immunol. 2009;183:5094–5103. doi: 10.4049/jimmunol.0802387. [DOI] [PubMed] [Google Scholar]
- 9.Hamzaoui A., Berraies A., Kaabachi W., Haifa M., Ammar J., Kamel H. Induced sputum levels of IL-33 and soluble ST2 in young asthmatic children. J. Asthma. 2013;50:803–809. doi: 10.3109/02770903.2013.816317. [DOI] [PubMed] [Google Scholar]
- 10.Ho L.H., Ohno T., Oboki K., Kajiwara N., Suto H., Iikura M., Okayama Y., Akira S., Saito H., Galli S.J., Nakae S. IL-33 induces IL-13 production by mouse mast cells independently of IgE-FcεRI signals. J. Leukoc. Biol. 2007;82:1481–1490. doi: 10.1189/jlb.0407200. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy P.C., Phair I.R., Greger C., Pardali K., McGuire V.A., Clark A.R., Gaestel M., Arthur J.S.C. IL-33 regulates cytokine production and neutrophil recruitment via the p38 MAPK-activated kinases MK2/3. Immunol. Cell Biol. 2019;97:54–71. doi: 10.1111/imcb.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grünig G., Warnock M., Wakil A.E., Venkayya R., Brombacher F., Rennick D.M., Sheppard D., Mohrs M., Donaldson D.D., Locksley R.M., Corry D.B. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z., Homer R.J., Wang Z., Chen Q., Geba G.P., Wang J., Zhang Y., Elias J.A. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wills-Karp M., Luyimbazi J., Xu X., Schofield B., Neben T.Y., Karp C.L., Donaldson D.D. Interleukin-13: Central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 15.Shiomi A., Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm. 2015;2015:568543. doi: 10.1155/2015/568543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llop-Guevara A., Chu D.K., Walker T.D., Goncharova S., Fattouh R., Silver J.S., Moore C.L., Xie J.L., O'Byrne P.M., Coyle A.J., Kolbeck R., Humbles A.A., Stampfli M.R., Jordana M. A GM-CSF/IL-33 pathway facilitates allergic airway responses to sub-threshold house dust mite exposure. PLoS One. 2014;9:e88714. doi: 10.1371/journal.pone.0088714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Göpfert C., Andreas N., Weber F., Häfner N., Yakovleva T., Gaestel M., Kamradt T., Drube S. The p38-MK2/3 module is critical for IL-33-induced signaling and cytokine production in dendritic cells. J. Immunol. 2018;200:1198–1206. doi: 10.4049/jimmunol.1700727. [DOI] [PubMed] [Google Scholar]
- 18.Aoki M., Yamaguchi R., Yamamoto T., Ishimaru Y., Ono T., Sakamoto A., Narahara S., Sugiuchi H., Hirose E., Yamaguchi Y. Granulocyte-macrophage colony-stimulating factor primes interleukin-13 production by macrophages via protease-activated receptor-2. Blood Cells Mol. Dis. 2015;54:353–359. doi: 10.1016/j.bcmd.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Howarth P.H., Babu K.S., Arshad H.S., Lau L., Buckley M., McConnell W., Beckett P., Al Ali M., Chauhan A., Wilson S.J., Reynolds A., Davies D.E., Holgate S.T. Tumour necrosis factor (TNFα) as a novel therapeutic target in symptomatic corticosteroid dependent asthma. Thorax. 2005;60:1012–1018. doi: 10.1136/thx.2005.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg H.F., Phipps S., Foster P.S. Eosinophil trafficking in allergy and asthma. J. Allergy Clin. Immunol. 2007;119:1303–1310. doi: 10.1016/j.jaci.2007.03.048. quiz 1311-1302. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalo J.A., Lloyd C.M., Wen D., Albar J.P., Wells T.N., Proudfoot A., Martinez-A C., Dorf M., Bjerke T., Coyle A.J., Gutierrez-Ramos J.C. The coordinated action of CC chemokines in the lung orchestrates allergic inflammation and airway hyperresponsiveness. J. Exp. Med. 1998;188:157–167. doi: 10.1084/jem.188.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chackerian A.A., Oldham E.R., Murphy E.E., Schmitz J., Pflanz S., Kastelein R.A. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J. Immunol. 2007;179:2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T.K., Zurawski G., Moshrefi M., Qin J., Li X., Gorman D.M., Bazan J.F., Kastelein R.A. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Drube S., Kraft F., Dudeck J., Muller A.L., Weber F., Göpfert C., Meininger I., Beyer M., Irmler I., Häfner N., Schütz D., Stumm R., Yakovleva T., Gaestel M., Dudeck A. MK2/3 are pivotal for IL-33-induced and mast cell-dependent leukocyte recruitment and the resulting skin inflammation. J. Immunol. 2016;197:3662–3668. doi: 10.4049/jimmunol.1600658. [DOI] [PubMed] [Google Scholar]
- 25.MacKenzie K.F., Clark K., Naqvi S., McGuire V.A., Noehren G., Kristariyanto Y., van den Bosch M., Mudaliar M., McCarthy P.C., Pattison M.J., Pedrioli P.G., Barton G.J., Toth R., Prescott A., Arthur J.S. PGE(2) induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A-SIK-CRTC3 pathway. J. Immunol. 2013;190:565–577. doi: 10.4049/jimmunol.1202462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark K., MacKenzie K.F., Petkevicius K., Kristariyanto Y., Zhang J., Choi H.G., Peggie M., Plater L., Pedrioli P.G., McIver E., Gray N.S., Arthur J.S., Cohen P. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16986–16991. doi: 10.1073/pnas.1215450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lombardi M.S., Gilliéron C., Dietrich D., Gabay C. SIK inhibition in human myeloid cells modulates TLR and IL-1R signaling and induces an anti-inflammatory phenotype. J. Leukoc. Biol. 2016;99:711–721. doi: 10.1189/jlb.2A0715-307R. [DOI] [PubMed] [Google Scholar]
- 28.Sundberg T.B., Choi H.G., Song J.H., Russell C.N., Hussain M.M., Graham D.B., Khor B., Gagnon J., O'Connell D.J., Narayan K., Dančík V., Perez J.R., Reinecker H.C., Gray N.S., Schreiber S.L. Small-molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12468–12473. doi: 10.1073/pnas.1412308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darling N.J., Toth R., Arthur J.S., Clark K. Inhibition of SIK2 and SIK3 during differentiation enhances the anti-inflammatory phenotype of macrophages. Biochem. J. 2017;474:521–537. doi: 10.1042/BCJ20160646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naqvi S., Martin K.J., Arthur J.S. CREB phosphorylation at Ser133 regulates transcription via distinct mechanisms downstream of cAMP and MAPK signalling. Biochem. J. 2014;458:469–479. doi: 10.1042/BJ20131115. [DOI] [PubMed] [Google Scholar]
- 31.Zhang T., Inesta-Vaquera F., Niepel M., Zhang J., Ficarro S.B., Machleidt T., Xie T., Marto J.A., Kim N., Sim T., Laughlin J.D., Park H., LoGrasso P.V., Patricelli M., Nomanbhoy T.K. Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol. 2012;19:140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterfield M., Jin W., Reiley W., Zhang M., Sun S.C. IkappaB kinase is an essential component of the Tpl2 signaling pathway. Mol. Cell Biol. 2004;24:6040–6048. doi: 10.1128/MCB.24.13.6040-6048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang V., Janzen J., Fischer G.Z., Soneji Y., Beinke S., Salmeron A., Allen H., Hay R.T., Ben-Neriah Y., Ley S.C. betaTrCP-mediated proteolysis of NF-kappaB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol. Cell Biol. 2003;23:402–413. doi: 10.1128/MCB.23.1.402-413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozanne J., Prescott A.R., Clark K. The clinically approved drugs dasatinib and bosutinib induce anti-inflammatory macrophages by inhibiting the salt-inducible kinases. Biochem. J. 2015;465:271–279. doi: 10.1042/BJ20141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molfino N.A., Kuna P., Leff J.A., Oh C.K., Singh D., Chernow M., Sutton B., Yarranton G. Phase 2, randomised placebo-controlled trial to evaluate the efficacy and safety of an anti-GM-CSF antibody (KB003) in patients with inadequately controlled asthma. BMJ Open. 2016;6:e007709. doi: 10.1136/bmjopen-2015-007709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matera M.G., Calzetta L., Cazzola M. TNF-alpha inhibitors in asthma and COPD: We must not throw the baby out with the bath water. Pulm. Pharmacol. Ther. 2010;23:121–128. doi: 10.1016/j.pupt.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Durham A.L., Caramori G., Chung K.F., Adcock I.M. Targeted anti-inflammatory therapeutics in asthma and chronic obstructive lung disease. Transl Res. 2016;167:192–203. doi: 10.1016/j.trsl.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipworth B., Jabbal S., Kuo C.R. Anti-interleukin 13 for asthma: Stick or twist? Lancet Respir. Med. 2018;6:e46–e47. doi: 10.1016/S2213-2600(18)30275-3. [DOI] [PubMed] [Google Scholar]
- 39.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., Suveges D., Vrousgou O., Whetzel P.L., Amode R., Guillen J.A. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Pelaez M., Lamont D.J., Peggie M., Shpiro N., Gray N.S., Cohen P. Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17432–17437. doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren J., Chen X., Chen Z.J. IKKβ is an IRF5 kinase that instigates inflammation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17438–17443. doi: 10.1073/pnas.1418516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luan B., Goodarzi M.O., Phillips N.G., Guo X., Chen Y.D., Yao J., Allison M., Rotter J.I., Shaw R., Montminy M. Leptin-mediated increases in catecholamine signaling reduce adipose tissue inflammation via activation of macrophage HDAC4. Cell Metab. 2014;19:1058–1065. doi: 10.1016/j.cmet.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yahara Y., Takemori H., Okada M., Kosai A., Yamashita A., Kobayashi T., Fujita K., Itoh Y., Nakamura M., Fuchino H., Kawahara N., Fukui N., Watanabe A., Kimura T., Tsumaki N. Pterosin B prevents chondrocyte hypertrophy and osteoarthritis in mice by inhibiting Sik3. Nat. Commun. 2016;7:10959. doi: 10.1038/ncomms10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 45.Jensen B.M., Swindle E.J., Iwaki S., Gilfillan A.M. Generation, isolation, and maintenance of rodent mast cells and mast cell lines. Curr. Protoc. Immunol. 2006;Chapter 3 doi: 10.1002/0471142735.im0323s74. Unit 3 23. [DOI] [PubMed] [Google Scholar]
- 46.Junttila I.S., Watson C., Kummola L., Chen X., Hu-Li J., Guo L., Yagi R., Paul W.E. Efficient cytokine-induced IL-13 production by mast cells requires both IL-33 and IL-3. J. Allergy Clin. Immunol. 2013;132:704–712.e710. doi: 10.1016/j.jaci.2013.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukuishi N., Igawa Y., Kunimi T., Hamano H., Toyota M., Takahashi H., Kenmoku H., Yagi Y., Matsui N., Akagi M. Generation of mast cells from mouse fetus: Analysis of differentiation and functionality, and transcriptome profiling using next generation sequencer. PLoS One. 2013;8:e60837. doi: 10.1371/journal.pone.0060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 49.Howden A.J.M., Hukelmann J.L., Brenes A., Spinelli L., Sinclair L.V., Lamond A.I., Cantrell D.A. Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol. 2019;20:1542–1554. doi: 10.1038/s41590-019-0495-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry data are available via PRIDE project ID PXD020091. Data used for generation of the figures in the paper are available upon request from the corresponding author (Philip Cohen, p.cohen@dundee.ac.uk) or the first author (Nicola Darling, n.j.darling@dundee.ac.uk).