Abstract
The mechanisms that regulate platelet biogenesis remain unclear; factors that trigger megakaryocytes (MKs) to initiate platelet production are poorly understood. Platelet formation begins with proplatelets which are cellular extensions that originate from the MK cell body. Proplatelet formation is a highly dynamic and asynchronous process which poses unique challenges for researchers to accurately capture and analyze. We have designed an open-source, high-content, high-throughput, label-free analysis platform. Phase-contrast images of live, primary MKs are captured over a 24-hour period. Pixel-based machine-learning classification done by ilastik generates probability maps of key cellular features (circular MKs and branching proplatelets), which are then processed by a customized CellProfiler pipeline to identify and filter structures of interest based on morphologic parameters. CellProfiler Analyst, provides a final, supervised, machine learning classification to bolster accurate identification of cellular structures. This entire workflow yields the percent of proplatelet production, area, and count of proplatelets and MKs, as well as other statistics including skeletonization information for measuring proplatelet branching and length. We propose using a combination of these analyzed metrics, in particular the area measurements of MKs and proplatelets, when assessing in-vitro proplatelet production. Accuracy was validated against manually counted images and an existing algorithm. We then used the new platform to test compounds known to cause thrombocytopenia, including bromodomain inhibitors, and uncovered previously unrecognized effects of drugs on proplatelet formation, thus demonstrating the utility of our analysis platform. This advance in creating unbiased data analysis will increase the scale and scope of proplatelet production studies and potentially serve as a valuable resource for investigating molecular mechanisms of thrombocytopenia.
Introduction
Platelets and their precursor cells, megakaryocytes (MKs), are essential for hemostasis. MKs, which reside mainly in the bone marrow, generate platelets by remodeling their cytoplasm into long, beaded proplatelet extensions, which function as the assembly lines for platelet production1. However, the mechanisms that regulate platelet biogenesis remain unclear due to a limited understanding of factors that trigger MKs to initiate proplatelet production. A better understanding of the signaling pathways that power platelet production may yield new therapeutic strategies for the treatment of thrombocytopenia (low platelet counts), a major clinical problem encountered across a number of conditions including sepsis, cancer, and autoimmune diseases. Thrombocytopenia can be caused due to a wide variety of medications, and can range from mild to life-threatening in severity. Drug-induced thrombocytopenia can occur due to the destruction of circulating platelets, and/or defects in MKs which render them unable to replenish platelet counts2. While platelet survival and clearance rates can be readily measured via flow cytometry, quantification of platelet production from MKs, such as the rate and extent of proplatelet production, currently requires direct visualization and low throughput analysis. As such, it is often difficult to distinguish what stage of thrombopoiesis is affected in thrombocytopenic patients and consequently it is hard to optimize the best therapeutic strategy.
Platelet formation from MKs in vitro is a highly dynamic and asynchronous process which poses unique challenges for researchers to accurately capture and analyze. Culturing primary MKs has allowed examination of platelet production in controlled, in vitro settings. Platelet production begins with the extension of large pseudopodia from the MK cell body using microtubule-dependent forces3. Microtubules then band together into bundles lining proplatelet shafts and form looped structures at their tips3. Powered by dynein-dependent microtubule sliding, proplatelets undergo extensive elongation and can reach millimeters in length4–6. During the final stages of maturation, MKs release proplatelets which reorganize into platelets. This process occurs spontaneously in vitro and has been directly visualized in vivo4, 7, 8. To date, practical limitations associated with culturing primary MKs9, live-cell imaging, and data analysis have impeded discoveries of molecular mechanisms capable of regulating proplatelet production.
Live-cell, label free analysis of MKs over time is optimal for quantifying proplatelet production and necessary for high-throughput applications. To date, manual counting of live proplatelet-producing MKs is the gold standard for quantification of proplatelet production. However, manual cell counts of proplatelet-producing cells vary widely, and in addition, only a small percentage of MKs undergo proplatelet production in vitro such that large numbers must be tediously assessed. While fluorescent labeling is a valuable tool for imaging other cell types, MKs present challenges because they are non-adherent; the fragile nature of proplatelets causes them to disconnect from the cell body after manipulation, and it can be difficult to image the thin bridges connecting beaded proplatelets. These practical limitations of imaging proplatelet production have made thresholding individual cells and high-throughput quantitation challenging. In fact, most studies have used static snapshots from a limited number of fixed cells that yield predominantly qualitative results8, 10.
Another limitation in current analysis techniques is the ability to classify and score proplatelet morphology. A true ‘high content’ analysis provides information on several morphological or phenotypical metrics11. To date, fluorescent labels on fixed cells during different stages of development have provided most of the qualitative information we have on proplatelet morphology. However, the ability to perform qualitative analysis in a live-cell, label-free system, with a high-throughput capacity, has not been previously described. In addition, proplatelet formation can be a rare event in different classes of megakaryocytes (i.e. murine bone marrow-derived versus fetal liver derived). Primary human megakaryocytes also produce significantly smaller and less elaborate proplatelet formations in culture. These rare events can often render the metric of ‘percent proplatelet production’ uninformative and are often over/under inflated by current analysis methods. As such, an unbiased analysis platform capable of delivering raw outputs (i.e. percentage of proplatelet-making MKs) with morphological scoring (normal extensions, diminished extensions, etc) would be invaluable for examining proplatelet formation.
We previously described a high-content, high-throughput assay, utilizing automated live-cell captures from the Incucyte Zoom (Essen BioScience) to quantitate and measure proplatelet production12. While data analysis using our custom ImageJ pipeline enabled real-time detection of the rate and extent of proplatelet production, accurate image segmentation and identification of proplatelet branches belonging to individual cells remained a key barrier to analyzing data. Furthermore, artificial inflation of proplatelet production due to high background as well as difficulties in batch processing have limited the ability to obtain high-throughput data. Here, we describe an improved workflow to analyze the biological mechanisms of platelet production using supervised machine learning algorithms in place of traditional, rule-based image analysis. We demonstrate that this image analysis approach can be used to capture and quantify the complex morphological features of MKs undergoing proplatelet formation. Further, we demonstrate that this workflow can be applied to clinically-relevant cases of drug-induced thrombocytopenia to help elucidate the cause of platelet production defects.
Methods
MK culture and purification
Primary mouse MKs were derived from fetal liver cultures as previously described9. Briefly, fetal livers were extracted from CD-1 pregnant mice at day 13.5 of gestation. Homogenized fetal liver cells were then cultured in Dulbecco’s Modified Eagle Media (DMEM) in the presence of recombinant murine thrombopoietin (TPO; 70ng/mL) for 4 days. On day 4, mature MKs were purified by a bovine serum albumin (BSA) density gradient. Primary human MKs were generated from cord blood CD34+ cells cultured in serum free stem cell medium containing 70ng/mL TPO13. Mature human MKs were purified by magnetic bead separation on day 11.
Live cell imaging
Live cell imaging was performed using the IncuCyte® System (Essen BioScience, Ann Arbor, MI, USA). MK cultures were adjusted to a cell density of 250-500 cells/well and plated in 96 well plates (Opticlear, Greiner Bio-One, Monroe, NC, USA). Plates were maintained in an XL-3 incubation chamber at 37°C incubator with 5% CO2. Wells were imaged using 10x and 20x objectives in phase, for mouse and human MKs, respectively. Phase-contrast image frames were captured at 1-hour intervals from multiple 950 × 760–μm2 regions per well.
Inhibitor studies
After maturation, MKs were treated with one of the following compounds, before undergoing live-cell imaging for 24 hours, as described above: Panobinostat (1-10 μM; Selleckchem, Houston, TX, USA), Valproic acid (1-10μM; Sigma-Aldrich, St Louis, MO, USA), OTX015 (1-10μM; Selleckchem), I-BET762 (1-10μM; Sigma-Aldrich), Puromycin (100ng/mL; Sigma-Aldrich), Taxol (10μM; Sigma-Aldrich), or vehicle control (0.1% DMSO). Technical and biological replicates for each condition were performed, as indicated in the corresponding figure legends.
Analysis of MKs and proplatelet formation
Phase images were exported as 8-bit tiff stacks (frames over time) that captured stages of MK differentiation and proplatelet formation (Figure 1A), ilastik (version 1.3.0+) is an open source tool for segmentation and classification of images based on pixel intensities, ilastik’s autocontext workflow was used to expertly annotate and segment training images (Figure 1B)14, 15. The first stage of training classified pixels as either background, cell background, white, black, and grey. A second layer of classifiers was then annotated to identify higher level objects as background, cell boundary, MK, and proplatelet. To avoid image overtraining, ilastik’s probability maps were checked at each step, for every training image, to improve upon areas of ambiguity. We found that approximately 15-20 labels of each classifier, per image, at each stage, with a focus on variety and areas of ambiguity, gave optimal results.
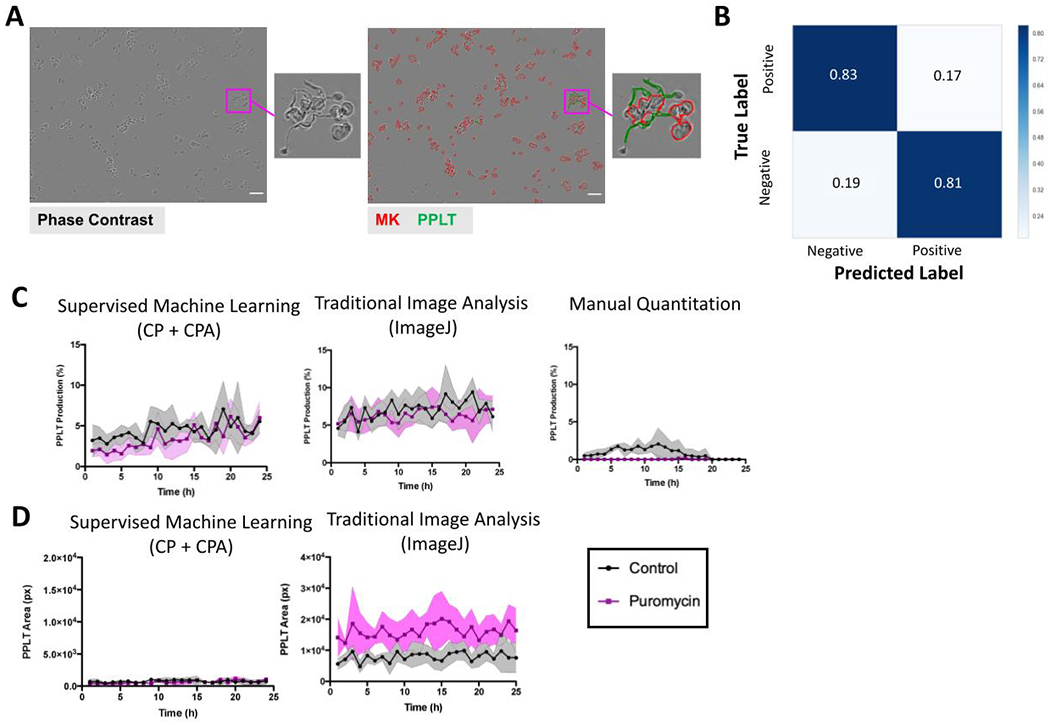
Figure 1. Development of a label-free proplatelet analysis platform using supervised machine learning.
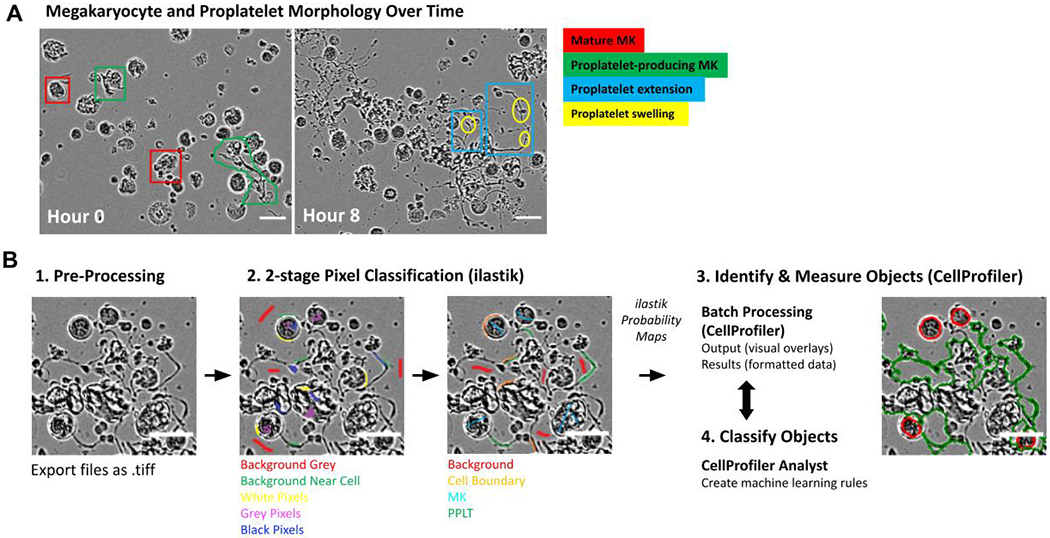
(A) Phase contrast images show megakaryocytes (MKs) undergoing different stages of maturation and proplatelet formation over time. Round, mature MKs (red) undergo cytoskeletal transformation to begin producing proplatelets (green). Notable features of proplatelets include long cytoskeletal extensions (blue) and terminal swellings thought to be early platelets (yellow). (B) Schematic of ilastik training (2-stage pixel classification) to segment MK classes. The software was trained using ilastik where pixels were classified as background gray, background near cell, white pixels, grey pixels or black pixels. In a second stage, pixels were classified as background, cell boundary, cell and protrusion. The probability maps generated were fed into CellProfiler, where overlays can be checked and undergo further CellProfiler Analyst rule creation. Scale bars are 50 μm.
After the 2-stage pixel classification training in ilastik, probability maps of features were generated for each image (background, cell boundary, MK, and proplatelet, Figure 1B, 2A). These feature maps were then processed in a CellProfiler (version 3.0.0+) pipeline which identified all cellular objects within an image. After running this pipeline, segmented images underwent a final round of training in CellProfiler Analyst (CPA - version 2.2.1) to create machine learning rules (in the form of a text file) for proplatelet structures. At this stage, 50 cells from each category (MK or proplatelet) were labeled by an expert as either positive or negative to improve recognition of complex proplatelet structures (Figure 2B). We classed proplatelet-forming cells as any MK lacking a round cell body (early stages of proplatelet formation) and/or displaying long extended protrusions (Figure 2B). By forming this preliminary understanding of the cellular objects within the phase-contrast images, a final, filtering CellProfiler pipeline, bolstered by the CPA-created text rules, was run to accurately segment genuine MKs and proplatelets. This pipeline screened the images by solidity (< 0.8), and size (diameter/major axis length/Zernike) exclusion metrics. Finally, the workflow was automated via Python programming, with the code, ilastik files, and CellProfiler pipelines located at https://github.com/broadinstitute/Italiano_MK_Analysis.
Figure 2. Supervised machine learning algorithms accurately segment stages of MK maturation.
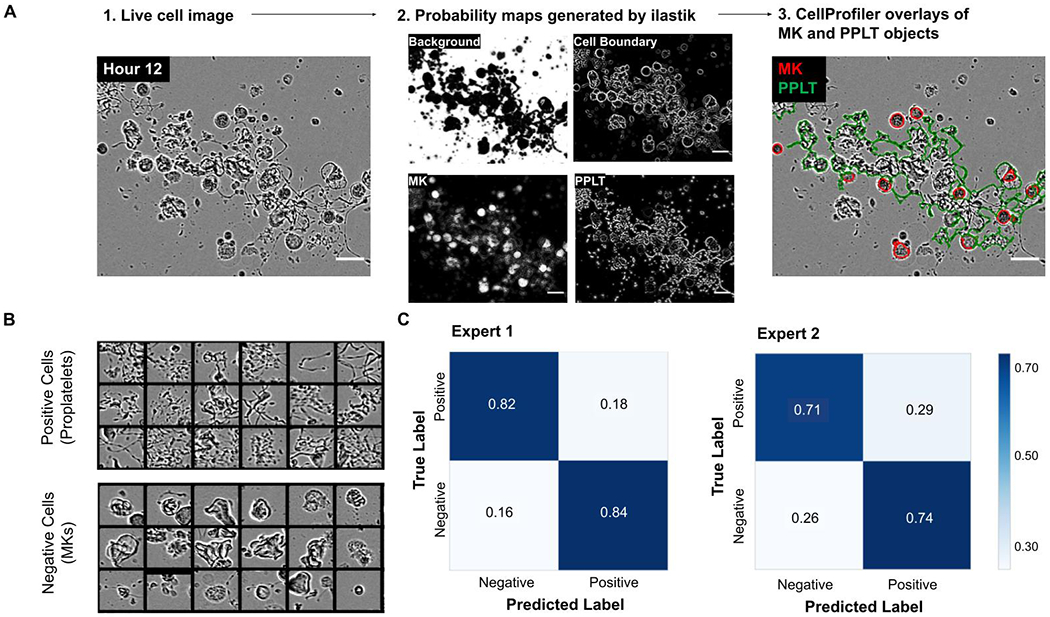
(A) 1: Representative input of an 8-bit single image, 2: The 4 probability maps generated by ilastik, and 3: The overlays generated by CellProfiler, showing accurate segmentation of round MKs (red) and proplatelet structures (green). Scale bars are 50 μm. (B) The machine learning algorithms were internally validated within CellProfiler. Events were binned as positive (proplatelet-forming) or negative (non-proplatelet forming MKs) in CellProfiler Analyst. 200 cells were then manually classified/annotated and compared to the CellProfiler predictions. (C) Confusion matrices were generated based on two independent expert annotators, showing percentages of the predicted versus true label.
A skeletonization pipeline was also developed to gain more in-depth information of proplatelet structures. 16-bit images of masked proplatelets (emanating protrusions from MK bodies which are identified by the MK pipeline), were analyzed through an additional CellProfiler pipeline that converted identified mouse proplatelet objects into skeletonized structures (Figure 5A). The skeletonized image was made up of nodes (vertices) linked by line segments (edges). The output provided tabulated information of statistical features of each skeletonized object, allowing quantitation of proplatelet lengths and in-depth visualization of spatial distributions over the differentiation timeline, within the well/image.
Figure 5. Supervised machine learning pipeline combined with skeletonization modules provides high content proplatelet profiling.
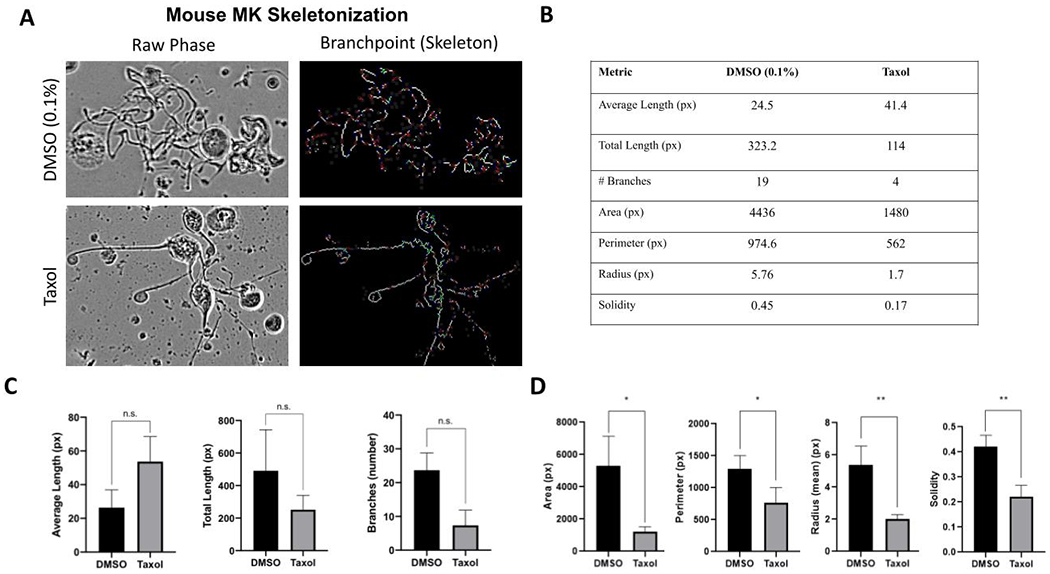
A skeletonization module was developed to allow deeper analysis of individual mouse proplatelet structures with different morphologies. (A) representative images of a typical proplatelet structure versus Taxol-treated cells, including their corresponding skeletonized image. The skeletonized image is made of trunks (red), branches (green), and endpoints (or vertices, blue), linked by “edges” or line structures (white). (B) A table comparing single cell metrics of the DMSO and Taxol images in 5A obtained from the skeleton analysis and metadata from the proplatelet analysis pipeline. (C) Quantitation of proplatelet skeleton from replicates of DMSO versus Taxol treated structures. The Taxol structures have increased average length of segments, reflecting the simplified skeletonized structure. Number of branches/masked protrusions is decreased with Taxol treatment, suggesting this is a more appropriate metric to detect complexity (n=3). (D) Quantitation of values that correlate with masked protrusions. Note area, perimeter, radius and solidity are all decreased in the Taxol-treated structure (n=3).
Results
Supervised machine learning algorithms accurately segment stages of MK maturation
We designed a supervised machine learning approach to accurately identify complex proplatelet objects in images. Mature MKs were imaged over a 24-hour period, during which they underwent several morphological changes as they transition to proplatelet production1 (Figure 1A). Mature MKs are large, round cells, which elongate in an asymmetrical, irregular fashion, indicating the initiation of proplatelet production. Within the proplatelet structure, extensions/protrusions form a complex web, with smaller beads or swellings also identifiable. The image analysis pipeline (Figure 1B) begins with phase contrast images (single, 8-bit); two rounds of supervised machine learning in ilastik generated probability maps of cellular features. These are then processed in CellProfiler to identify a potential pool of MKs and proplatelet structures. To identify true MK and proplatelet objects, images underwent a second round of machine learning, at the object rather than pixel level, using CellProfiler Analyst. The workflow produced segmented and overlaid images of the original raw phase image, allowing the monitoring of object classification accuracy throughout the stages of analysis. The workflow accurately identified structures of interest; an example processed test image is shown in Figure 2A. We quantitatively validated the workflow by testing its ability to distinguish 200 cells from non-training images as positive (MKs with proplatelets) and negative (round MKs, Figure 2B). Accuracy was ~70-80% when compared to annotation by 2 independent, blinded experts (Figure 2C). Of note, expert annotation was also within this range, suggesting that the workflow accuracy is within the range of human variability.
Supervised machine learning algorithms offer improved accuracy of detecting proplatelet structures compared to traditional data analysis methods
The supervised machine learning analysis platform was validated against two alternative methods of proplatelet quantification – traditional rule-based image analysis using a previously developed ImageJ macro12, and manual counting, which is the current gold standard of proplatelet analysis (Figure 3). Mature murine MKs were treated with either puromycin (100ng/mL), a protein synthesis inhibitor known to reduce proplatelet production16, or vehicle control, and imaged over 24 hours. In untreated control wells, proplatelet-making MKs accounted for approximately 30% of MKs, often peaking between 10 and 15 hours. As expected, cultures treated with puromycin had a reduced number of proplatelet structures. Images were quantified using the new analysis pipeline, ImageJ, and manual counting by 3 independent, blinded experts (Figure 3A). The proposed machine-learning based pipeline matched manual counts of percent proplatelet production most closely, indicating higher accuracy than the ImageJ macro that is based on circularity and size exclusion rules.
Figure 3. Supervised machine learning offers improved accuracy of proplatelet quantitation compared to alternative image analysis methods.
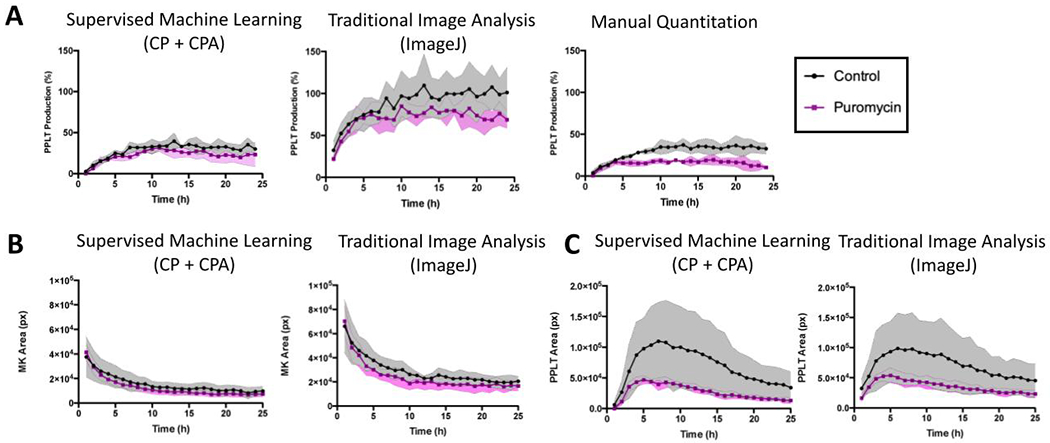
(A) Comparative analysis of proplatelet (PPLT) production between CellProfiler, ImageJ, and manually counted images (n=3 people) was performed to validate the platform against existing methods of quantification. Fetal-liver derived mouse megakaryocytes (MKs) were imaged live in culture over a period of 24 hours while they underwent proplatelet formation. Note both CellProfiler and manual counting show the number of proplatelet objects beginning at 0% at time 0, and peaking at under 50%, with a similar degree of variation. In contrast, analysis via a previously published investigator-coded ImageJ macro shows the same trend, however with an inflated estimation of the amount of proplatelet objects and a higher degree of variation. (B) Area (pixels) of MK and (C) proplatelet objects were also compared to the values obtained via analysis with ImageJ. These data show ImageJ analysis estimated a higher MK area, suggesting CellProfiler may more accurately segment single cells. Data are mean ± SD of n=3 independent experiments.
In addition to the percentage of MKs making proplatelets, other values such as total area of MKs and proplatelets per image were generated using both CellProfiler and ImageJ analysis platforms. These values are useful in conjunction with percentages, as they reveal more detail about the structures being analyzed. Images analyzed by ImageJ/traditional analysis showed a higher estimate of MK area and a lower estimate of proplatelet area than supervised machine learning algorithms (Figure 3B–C). This suggests that the source of the ImageJ macros over-estimation of percentage of proplatelets stems from difficulties in accurately segmenting whole proplatelet objects.
Shape and area metrics are more appropriate to assess human proplatelet production
Human, CD34+ cell-derived MKs were also validated using the supervised machine learning analysis platform. Of note, human MKs are typically smaller and produce significantly fewer proplatelets with less elaborate extensions in culture than their murine fetal liver-derived counterparts9 (Figure 4A). The difference in morphology and rare occurrence of proplatelet production events poses particular challenges for automated data analysis techniques. Due to these differences in morphology, we created a separate pipeline where workflow training was performed on human MKs. This yielded a similar degree of accuracy to the murine system (Figure 4B). Human MKs were then treated with either vehicle or puromycin (100 ng/mL) and imaged under the same conditions as the murine MKs, as described above. Supervised machine learning-based analysis estimated a similar percentage of proplatelet-producing MKs as ImageJ (approximately 5%), with the manual quantitation estimation being significantly lower than both (<5%) (Figure 4C). However, when area parameters were analyzed, CellProfiler estimated an almost negligible number of pixels, consistent with the true small number of human proplatelet structures (Figure 4D). In contrast, ImageJ analysis provided a much higher area of proplatelets, further suggesting inaccuracies in identifying bonafide extensions (Figure 4D). We therefore suggest that proplatelet area, or more detailed shape metrics, are more relevant metrics for analyzing proplatelet formation from human MKs.
Figure 4. Supervised machine learning allows for analysis of human MKs.
(A) Representative phase contrast and CellProfiler overlaid images of human CD34+ derived MKs (MKs, red) and proplatelet (PPLT, green) structures. Note the smaller size of proplatelet branches formed from human MKs compared to murine fetal-liver derived MKs, which has posed challenges for accurate detection using image analysis software. (B) A confusion matrix showing validation of the human MK pipeline. (C) Comparison of proplatelet quantification overtime between CellProfiler, ImageJ and manual counting (n=3 people). (D) Area (pixels) of MK and proplatelet objects were also compared to the values obtained via analysis with ImageJ. Scale bar is 50 μm. Data are mean ± SD (shaded regions) of n=3 independent experiments.
Skeletonization pipeline allows high content analysis of proplatelet structures
To analyze proplatelet structures in greater detail, we developed an additional skeletonization pipeline. This extension of the proposed workflow creates skeletonized images of the proplatelets, thus allowing us to monitor the biological significance of different mouse proplatelet morphologies and structures (Figure 5A). These ‘skeletonized’ images are made of trunks (red), branches (green), and endpoint nodes (or vertices, blue), linked by “edges” or line structures (white). The edge and vertex information (combined with location values) generated from the skeletonization pipeline allowed quantification of the number and length (in pixels) of proplatelet protrusions within a single proplatelet structure of interest (Figure 5C).
To identify which metrics would best reveal changes in the morphological structure of proplatelet objects, we performed an in-depth analysis of two previously determined, morphologically distinct proplatelet structures. Taxol, an anti-cancer drug and known to stabilize microtubules, causes a distinct, reproducible proplatelet phenotype which is characterized by long, unbranched protrusions as opposed to multiple long protrusions that tangle together to form a complex web17. Using the skeleton analysis, we identified several features that can be used to create a profile of complex vs. non-complex objects. Taxol-treated proplatelets had, on average, longer segmented branches, indicative of less complex, retracted protrusions. In contrast, normal proplatelets were characterized by a larger number of segmented branches, total proplatelet length within the objects, greater area, perimeter, radius, and higher solidity value (Figures 5B–D). Two-tailed t-tests were performed on these 7 metrics, with area (p = 0.0193), perimeter (p = 0.0424), radius (p = 0.0079), and solidity (p = 0.0055) showing significant differences between DMSO and Taxol.
New insights into mechanisms of drug-induced thrombocytopenia using high content MK analysis
Many drugs, especially chemotherapy agents, cause thrombocytopenia in patients; the mechanisms of which are largely unknown. To validate the clinical utility of our analysis platform, we tested several compounds, with known and unknown effects on proplatelet production18–20, to assess their impact on the ability of MKs to make platelets (Table 1). HDAC inhibitors, panobinostat and valproic acid, and the bromodomain inhibitors, OTX015 and I-BET172, were incubated with mature, cultured MKs and imaged over 24 hours, for a total of n=72 fields analyzed per inhibitor (over 3 independent/biological repeats). Then, a profile of several metrics including proplatelet production, area, and number of masked protrusions were compiled to reveal a detailed phenotype. We found that panobinostat decreased percent proplatelet formation (over time and at 20 hours), and the proplatelet structures that formed were smaller in area and had less masked protrusions (Figure 6A). In contrast, valproic acid did not significantly affect percent proplatelet production, but did show a decrease in the number of masked protrusions, suggesting the proplatelet quality could be affected by this compound (Figure 6B). In addition, two bromodomain inhibitors currently undergoing clinical trials were tested in the assay. The compound OTX150 had a subtle effect on proplatelet formation, with a more drastic effect on masked protrusions and proplatelet area (Figure 6C). Similarly, the compound IBET-762 affected all parameters (Figure 6D). Together, these data 1) corroborate previously reported effects of HDAC inhibitors on proplatelets, 2) suggest that bromodomain inhibitors may be associated with platelet production defects, and 3) support using the supervised-machine learning pipeline generated here to give a more comprehensive picture of the differential effects of drugs on proplatelet production.
Table 1.
Panel of inhibitors with known and unknown effects on MK development.
| Compound | Class | Associated with thrombocytopenia | Known MK defect |
|---|---|---|---|
| Panobinostat | HDAC inhibitor (broad spectrum) | Yes | Yes - platelet production (Bishton et al, Blood, 2011) |
| Valproic acid | HDAC inhibitor (HDAC1,2) | Yes | Yes – inhibition of MK differentiation (Bartles et al, PLoS One, 2018) |
| OTX015 | Bromodomain inhibitor (BET family) | Yes | Unknown |
| I-BET762 | Bromodomain inhibitor (BET family) | Unknown | Unknown |
Figure 6. Proplatelet profiling allows new insights into mechanisms of drug-induced thrombocytopenia.
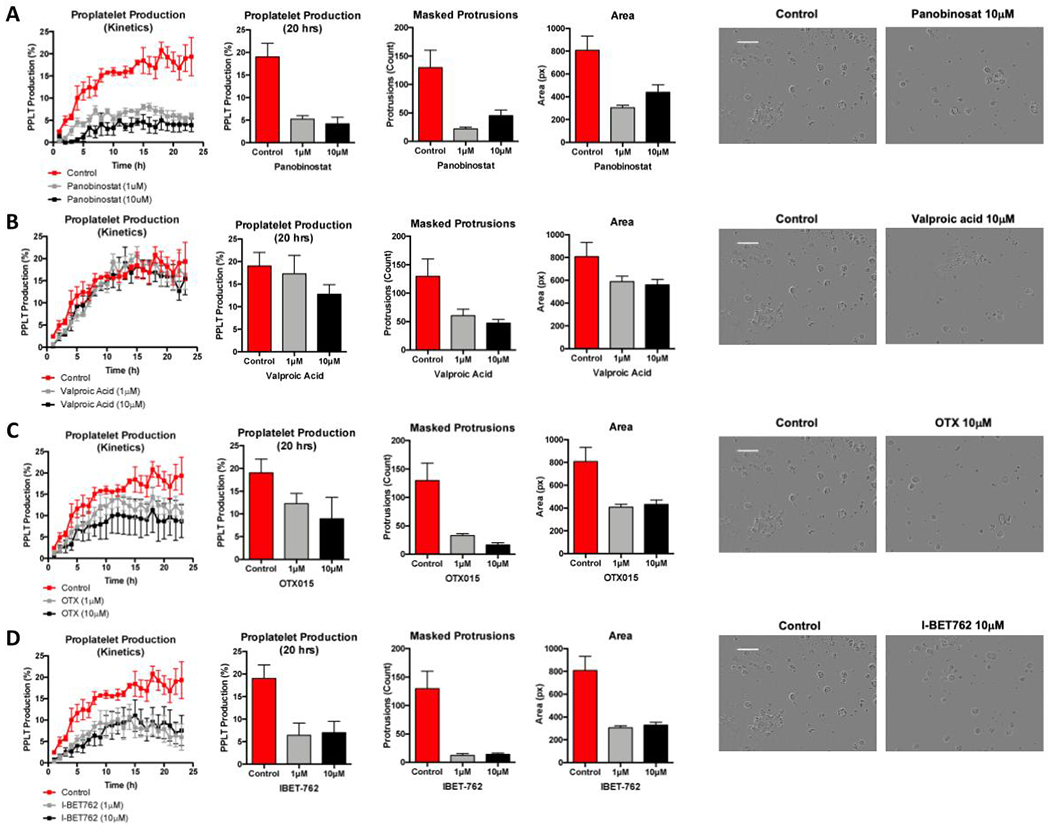
Mature fetal liver-derived megakaryocytes (MKs) were treated with a panel of inhibitors and were then screened for proplatelet (PPLT) production defects, using metrics from several analyses to create an individual profile for each inhibitor. Metrics assessed include PPLT production over time, percent PPLT production at 20 hours, number of masked protrusions within the PPLT objects, and average area of each PPLT object. Representative images for each condition at 20 hours are shown on the right. Scale bar is 100μm. (A) HDAC inhibitor panobinostat affects both the number and quality of PPLT extensions. (B) In contrast, valproic acid (also a HDAC inhibitor) was found to not significantly affect the number of MK-making PPLT, however did decrease the number of masked protrusions. The bromodomain inhibitors (C) OTX015 and (D) IBET-762 were also assessed and were found to impact both quality and quantity of PPLT formation. Data are mean ± SEM of n=3 independent (biological) experiments, all conditions were performed in duplicate in each experiment.
Discussion
We present an automated, high content framework for analyzing MKs and proplatelet production using supervised machine learning. In this unified platform, we have developed a novel workflow that identifies and quantitates MKs and proplatelets within label-free, phase-contrast images, based on distinguishing pixel-based features21. This is evidenced by improved recognition of proplatelet objects compared to traditional rule-based image analysis (ImageJ macro). Further, we have also developed a qualitative proplatelet analysis, utilizing CellProfiler’s skeletonization module combined with area and shape metrics. This advance in image analysis methodology for analyzing proplatelet production will accelerate new findings in the field.
There have been several studies showcasing analysis platforms for quantifying proplatelet production, including rule-based, investigator-coded pipelines12, 10. A recent paper from Salzmann and colleagues10 describes a CellProfiler-based pipeline to examine proplatelet formation using fluorescent labels in fixed MKs spread on fibrinogen. While fluorescence imaging has been used with other cell types, it presents challenges in MKs. Notably, it is difficult to resolve the thin bridges connecting beaded proplatelets by fluorescence. Further, due to the presence of multiple nuclei22, MKs have a propensity to shed fluorescent labels as they undergo proplatelet production. Potential cytotoxicity and artifacts from overexpressing fluorescent proteins also compound issues with using fluorescent markers in proplatelet analysis. Furthermore, immunofluorescence approaches require cells to be fixed and adhered to a coverslip. Adherence of MKs on fibrinogen coated coverslips could bias results if there are differences in attachment of round versus proplatelet-producing MKs. Furthermore, adherent MKs tend to spread on substrates like fibrinogen and it can be difficult to differentiate between spreading and bona-fide proplatelet production. Another advantage of our platform is the acquisition of kinetic data in live cells over 24 hours. Given that proplatelet formation is asynchronous, analysis of fixed cells only provides a limited snapshot of this process. Our platform will enable investigators to more easily measure the effect of perturbations (e.g. small molecule inhibitors) on proplatelet formation over a time course. As such, live-cell label-free analysis of MKs over time is the optimal modality for examining and quantifying proplatelet production in vitro and has the potential to increase standardization between laboratories.
In this study, we performed a small-scale compound screen of inhibitors with known and unknown proplatelet defects. Drug-induced thrombocytopenia is a major clinical problem which often becomes apparent only when compounds are already in clinical trials. It is most often caused by either immune-mediated destruction and clearance of circulating platelets or defects at the MK/proplatelet formation level. Compounds commonly used for cancer treatments, such as HDAC inhibitors, are often associated with thrombocytopenia, presumably due to their global effects on blood cells from multiple lineages. Our analysis platform can be used to gain quantitative information on kinetics and percentage of proplatelet production, as well as qualitative information on proplatelet extensions and complexity. Morphological characterization of proplatelet structures might yield novel mechanistic insights into how platelets form, not just whether they form. An issue with the proplatelet structure analysis, in its current state, is the tendency for it to indiscriminately skeletonize all line-like structures that emanate from detected megakaryocyte bodies. However, even with the presence of additional noise/false-positives, the ability to quantitate major structure lengths is feasible, which affords the end user additional metrics to draw conclusions from. This could serve as a high-throughput, high-content preclinical tool, which will mitigate the risk of patients developing thrombocytopenia, as well as help to gain mechanistic insight into proplatelet production defects. We note that there are other methods for assessing the effects of drugs on proplatelet production that attempt to more faithfully recapitulate the in vivo microenvironment of platelet production23,24. While these microfluidic devices mimic physiological aspects of the bone marrow and blood vessel microenvironment, this is a low throughput method as one can only visualize a single chip at a time.
One of the main difficulties in creating an automated analysis of proplatelet structures is the complex morphological variations in MK shapes, and the complexity of branched, overlapping proplatelet structures. To define these features, we have used modular, machine-learning capable tools21. Modeled after a pre-existing ImageJ macro12 designed to quantify real-time differentiation, we have extended our analytical capabilities by leveraging open-source Python algorithms. This implementation provides a high level of flexibility that permits high-content and high-throughput quantification. However, despite many advances, there are still limitations which exist in our described method. Machine learning analyses are entirely reliant on an abundance of high quality training data. In terms of image analysis, this necessitates that images are captured in a consistent manner, exhibiting diverse and well defined pixel characteristics. Capturing rare, and anatomically convoluted events (in the case of human proplatelet formation, and mouse megakaryocyte shape variation, respectively) remains challenging for algorithms as well as distinguishing between single and multiple complex proplatelet objects. These limitations, however, provide further rationale for using shape and area metrics, in conjunction with raw percentages, when analyzing proplatelet formation. In addition, images acquired from different machines will need to be retrained at the pixel level, thus introducing a source of subjectivity and will require ongoing validation. However, both ilastik and CellProfiler offer multiple checkpoints for internal quality control - uncertainty values (ilastik), confusion matrices (CellProfiler Analyst), and masked overlays of all objects within the image (CellProfiler). Regardless, the only true validation is direct visualization, and a strength of this platform is that every stage can be manually validated and tailored to suit individual needs. Future directions of this work will therefore focus on standardizing this platform to provide an objective and transparent method of quantifying proplatelet production, and additionally reduce inconsistencies among different laboratories.
Conclusion
We have utilized open-source software to create a user-friendly, unbiased, accurate tool for measuring MK and proplatelet production in live-cell label free culture systems. The two-step supervised machine learning approach improves upon previously published methods of proplatelet quantification with improved accuracy, automation, flexibility and high content output. This advance in data analysis will help standardize proplatelet quantification in the field, increase researcher capability for analyzing proplatelet production, and potentially serve as a valuable clinical resource for investigating mechanisms of thrombocytopenia.
Essentials.
Platelet production is difficult to quantify due to limitations in live-cell image analysis methods.
We describe a novel, label-free analysis method that uses supervised machine learning.
We compared our analysis platform to manual counting by blinded experts to validate its accuracy.
We describe new insights into drug-induced thrombocytopenia using high content analysis.
Acknowledgements
The authors would like to thank Martha Sola-Visner and Zhi-Jian Liu for supplying cord blood derived CD34+ human cells. The authors would also like to thank Clarence Yapp and Beth Cimini for technical advice. This work was supported in part by grants from the National Heart, Lung, and Blood Institute (R01H168130 [J.E.I.], T32HL066987-16 [A.R.W]) the National Institute of Diabetes and Digestive and Kidney Diseases (K01DK111515 [K.R.M.]), and the National Institute of General Medical Sciences (R35GM122547 [AEC]), at the National Institutes of Health. J.E.I. and K.R.M. are American Society of Hematology Scholars. L.J.H. is supported by the National Science Foundation Graduate Research Fellowships Program (NSF GRFP, Grant no. DGE1745303) and the Ford Foundation Predoctoral Fellowship.
Footnotes
Conflict-of-interest disclosure: J.E.I. has financial interest in and is a founder of Platelet BioGenesis, a company that aims to produce donor-independent human platelets from human-induced pluripotent stem cells at scale. J.E.I. is an inventor on this patent. The interests of J.E.I. were reviewed and are managed by the Brigham and Women’s Hospital and Partners Healthcare in accordance with their conflict-of-interest policies. The remaining authors declare no competing financial interests.
References
- 1.Machlus KR, Italiano JE Jr. The incredible journey: From megakaryocyte development to platelet formation. The Journal of cell biology. 2013;201(6):785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George JN, Aster RH. Drug-induced thrombocytopenia: pathogenesis, evaluation, and management. Hematology American Society of Hematology Education Program. 2009:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Italiano JE Jr., Patel-Hett S, Hartwig JH. Mechanics of proplatelet elaboration. Journal of thrombosis and haemostasis : JTH. 2007;5 Suppl 1:18–23. [DOI] [PubMed] [Google Scholar]
- 4.Italiano JE Jr., Lecine P, Shivdasani RA, Hartwig JH. Blood platelets are assembled principally at the ends of proplatelet processes produced by differentiated megakaryocytes. The Journal of cell biology. 1999; 147(6): 1299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender M, Thon JN, Ehrlicher AJ, Wu S, Mazutis L, Deschmann E, Sola-Visner M, Italiano JE, Hartwig JH. Microtubule sliding drives proplatelet elongation and is dependent on cytoplasmic dynein. Blood. 2015;125(5):860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thon JN, Montalvo A, Patel-Hett S, Devine MT, Richardson JL, Ehrlicher A, Larson MK, Hoffmeister K, Hartwig JH, Italiano JE Jr. Cytoskeletal mechanics of proplatelet maturation and platelet release. The Journal of cell biology. 2010;191(4):861–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junt T, Schulze H, Chen Z, Massberg S, Goerge T, Krueger A, Wagner DD, Graf T, Italiano JE Jr., Shivdasani RA, von Andrian UH. Dynamic visualization of thrombopoiesis within bone marrow. Science (New York, NY). 2007;317(5845): 1767–70. [DOI] [PubMed] [Google Scholar]
- 8.Ghalloussi D, Dhenge A, Bergmeier W. New insights into cytoskeletal remodeling during platelet production. Journal of thrombosis and haemostasis : JTH. 2019;17(9):1430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijey P, Posorske B, Machlus KR. In vitro culture of murine megakaryocytes from fetal liver-derived hematopoietic stem cells. Platelets. 2018;29(6):583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzmann M, Hoesel B, Haase M, Mussbacher M, Schrottmaier WC, Kral-Pointner JB, Finsterbusch M, Mazharian A, Assinger A, Schmid JA. A novel method for automated assessment of megakaryocyte differentiation and proplatelet formation. Platelets. 2018;29(4):357–64. [DOI] [PubMed] [Google Scholar]
- 11.Singh S, Carpenter AE, Genovesio A. Increasing the Content of High-Content Screening: An Overview. Journal of biomolecular screening. 2014;19(5):640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thon JN, Devine MT, Jurak Begonja A, Tibbitts J, Italiano JE Jr. High-content live-cell imaging assay used to establish mechanism of trastuzumab emtansine (T-DM1)--mediated inhibition of platelet production. Blood. 2012;120(10): 1975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu ZJ Jr., Italiano J, Ferrer-Marin F, Gutti R, Bailey M, Poterjoy B, Rimsza L, Sola-Visner M. Developmental differences in megakaryocytopoiesis are associated with up-regulated TPO signaling through mTOR and elevated GATA-1 levels in neonatal megakaryocytes. Blood. 2011;117(15):4106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer C, Strähle C, Köthe U & Hamprecht FA ilastik: Interactive Learning and Segmentation Toolkit. Proc Eighth IEEE International Symposium on Biomedical Imaging 2011:230–3. [Google Scholar]
- 15.Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, et al. ilastik: interactive machine learning for (bio)image analysis. Nature Methods. 2019. [DOI] [PubMed] [Google Scholar]
- 16.Machlus KR, Wu SK, Stumpo DJ, Soussou TS, Paul DS, Campbell RA, Kalwa H, Michel T, Bergmeier W, Weyrich AS, Blackshear PJ, Hartwig JH, Italiano JE, Jr. Synthesis and dephosphorylation of MARCKS in the late stages of megakaryocyte maturation drive proplatelet formation. Blood. 2016; 127(11): 1468–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tablin F, Castro M, Leven RM. Blood platelet formation in vitro. The role of the cytoskeleton in megakaryocyte fragmentation. Journal of cell science. 1990;97 (Pt 1):59–70. [DOI] [PubMed] [Google Scholar]
- 18.Bartels M, Govers A, Polak R, Vervoort S, van Boxtel R, Pals C, Bierings M, van Solinge W, Egberts T, Nieuwenhuis E, Mokry M, Coffer PJ. Megakaryocyte lineage development is controlled by modulation of protein acetylation. PloS one. 2018;13(4):e0196400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishton MJ, Harrison SJ, Martin BP, McLaughlin N, James C, Josefsson EC, Henley KJ, Kile BT, Prince HM, Johnstone RW. Deciphering the molecular and biologic processes that mediate histone deacetylase inhibitor-induced thrombocytopenia. Blood. 2011; 117(13):3658–68. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics. 2017;12(5):323–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan DJ, Shan J, Bhatia SN, Carpenter AE. Quantifying co-cultured cell phenotypes in high-throughput using pixel-based classification. Methods (San Diego, Calif). 2016;96:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravid K, Lu J, Zimmet JM, Jones MR. Roads to polyploidy: the megakaryocyte example. Journal of cellular physiology. 2002;190(1):7–20. [DOI] [PubMed] [Google Scholar]
- 23.Thon JN, Mazutis L, Wu S, Sylman JL, Ehrlicher A, Machlus KR, Feng Q, Lu S, Lanza R, Neeves KB, et al. Platelet bioreactor-on-a-chip. Blood 2014;124:1857–1867. doi: 10.1182/blood-2014-05-574913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito Y, et al. Turbulence Activates Platelet Biogenesis to Enable Clinical Scale Ex Vivo Production. Cell. 2018;174:636–648 e618. doi: 10.1016/j.cell.2018.06.011. [DOI] [PubMed] [Google Scholar]


