Abstract
In response to bone fracture, periosteal progenitor cells proliferate, expand, and differentiate to form cartilage and bone in the fracture callus. These cellular functions require the coordinated activation of multiple transcriptional programs, and the transcriptional regulators Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) regulate osteochondroprogenitor activation during endochondral bone development. However, recent observations raise important distinctions between the signaling mechanisms used to control bone morphogenesis and repair. Here, we tested the hypothesis that YAP and TAZ regulate osteochondroprogenitor activation during endochondral bone fracture healing in mice. Constitutive YAP and/or TAZ deletion from Osterix-expressing cells impaired both cartilage callus formation and subsequent mineralization. However, this could be explained either by direct defects in osteochondroprogenitor differentiation after fracture, or by developmental deficiencies in the progenitor cell pool prior to fracture. Consistent with the second possibility, we found that developmental YAP/TAZ deletion produced long bones with impaired periosteal thickness and cellularity. Therefore, to remove the contributions of developmental history, we next generated adult onset-inducible knockout mice (using Osx-CretetOff) in which YAP and TAZ were deleted prior to fracture, but after normal development. Adult onset-induced YAP/TAZ deletion had no effect on cartilaginous callus formation, but impaired bone formation at 14 days post-fracture (dpf). Earlier, at 4 dpf, adult onset-induced YAP/TAZ deletion impaired the proliferation and expansion of osteoblast precursor cells located in the shoulder of the callus. Further, activated periosteal cells isolated from this region at 4 dpf exhibited impaired osteogenic differentiation in vitro upon YAP/TAZ deletion. Finally, confirming the effects on osteoblast function in vivo, adult onset-induced YAP/TAZ deletion impaired bone formation in the callus shoulder at 7 dpf, prior to the initiation of endochondral ossification. Together, these data show that YAP and TAZ promote the expansion and differentiation of periosteal osteoblast precursors to accelerate bone fracture healing.
Keywords: fracture healing, osteoblasts, transcription factors, genetic animal models
INTRODUCTION
Bone is a remarkable tissue in its capacity to heal without forming a scar, and most bone injuries heal readily, with bone fractures healing at success rates of 90–95%(1). This is due, in part, to the maintenance of a skeletal stem cell population capable of recapitulating many aspects of embryological programs for adult tissue regeneration(2). As in development, bone formation during fracture repair can occur through both direct intramembranous ossification and endochondral ossification via a cartilage intermediate(3–6). However, the source, niche, and molecular regulation of the progenitor cells responsible for bone fracture repair are distinct from those that produce the skeleton during development(2). In the embryo, mesodermal mesenchymal progenitors in the limb bud form a template of the skeletal elements, while bone fracture healing initiates by expansion and differentiation of osteochondroprogenitor cells resident in the bone-lining periosteum(3,4,7). Understanding the mechanisms that regulate the proliferation and differentiation of these cells will be critical to develop new therapeutic strategies for accelerating fracture repair and regenerating challenging bone injuries that cannot heal on their own.
To define the roles of the molecular mediators that regulate adult periosteal progenitor cell function, we must decouple the developmental history of the osteochondroprogenitor cells that come to reside in the periosteum from the regenerative function of these cells after fracture to accurately evaluate their contribution to responding to injury in the adult. For example, a recent study in which PDGFRβ was deleted from Osterix-expressing cells found no notable defects in skeletal development, but severe impairment of fracture healing(8), demonstrating contextual distinctions between development and fracture repair. Further, skeletal cell-specific gene deletion during development may alter the number, location, or niche of progenitor cells that, during injury, are activated for skeletal regeneration(9). In this study, we assessed the effects of conditional gene deletion from osteochondroprogenitors on endochondral bone fracture repair, with deletion performed either constitutively during development or inducibly after normal development at skeletal maturity prior to fracture. Several types of inducible Cre-loxP systems exist to temporally regulate Cre-mediated gene recombination, including the interferon-responsive Mx1 promoter(10), tamoxifen-inducible mutated estrogen- and progesterone-receptors(11), and tetracycline-controlled systems(12). Here, we used the Osterix-CretetOff (Osx-Cre) mouse in which Cre-recombinase is driven by the Osterix promoter and temporally controlled by tetracycline (or its more stable derivative, doxycycline)(13). Osx-Cre is expressed in hypertrophic chondrocytes and osteoprogenitors, including those of the primary ossification center in the embryo as well as the periosteum in the adult(13,14).
Periosteal cell expansion and differentiation require the coordinated activation of multiple transcriptional programs, and the transcriptional regulators, Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) have recently emerged as critical mediators of osteoblast progenitor proliferation and differentiation during bone development(15–17). Previously, we found that constitutive homozygous deletion of both YAP and TAZ from Osterix-expressing cells caused severe skeletal fragility and neonatal lethality(15). Mice with a single allele of either YAP or TAZ survived, but sustained spontaneous long bone fractures due to both reduced bone mass and defects in bone matrix collagen that caused weaker bone mechanical properties(15). Despite fracture, the neonatal limbs exhibited natural reduction(18) and eventually healed through callus formation(15). However, the roles of YAP and TAZ in periosteal progenitor cell function and their contributions to bone fracture healing are unknown.
Here, we conditionally deleted YAP and/or TAZ from Osterix-expressing cells using either constitutive or tetOff-inducible deletion and evaluated adult endochondral bone fracture healing. We found that constitutive YAP and/or TAZ deletion impaired both callus formation and subsequent mineralization, due to developmental deficiencies in the progenitor cell pool prior to fracture. In contrast, adult onset-induced YAP/TAZ deletion had no effect on cartilaginous callus formation, but impaired both the proliferation of osteoblast precursor cells located in the shoulder of the callus and their osteogenic differentiation, both, in vitro and in vivo. Together, these data show that YAP and TAZ promote the expansion and differentiation of periosteal osteoblast precursors to accelerate bone fracture healing.
MATERIALS AND METHODS
Animals
Mice harboring loxP-flanked exon 3 alleles in both YAP and TAZ on a mixed C57BL/6J genetic background were kindly provided by Dr. Eric Olson (University of Texas Southwestern Medical Center). Tetracycline responsive B6.Cg-Tg(Sp/7-tTA,tetO-EGFP/Cre)1AMc/J (Osx-CretetOff) mice from The Jackson Laboratory (Bar Harbor, MA, USA) were used to generate two mouse models in which we conditionally deleted YAP and/or TAZ from Osterix-expressing cells (Table 1). In both mouse models, tetracycline (or its more stable derivative, doxycycline) administration prevents tetracycline-controlled transactivator protein (tTA) binding to the tetracycline-responsive promoter element (TRE) in the promoter of the Cre transgene, allowing Cre expression only in the absence of doxycycline(13) for temporal control of Osx-Cre-mediated gene deletion.
Table 1:
Experimental fracture healing models, genotypes, and abbreviations.
| Deletion approach | Genotype | Abbreviation |
|---|---|---|
| Constitutive | Yapfl/fl;Tazfl/fl | YAPWT;TAZWT |
| Yapfl/+;Tazfl/+;Osx-Cre | YAPcHET;TAZcHET | |
| Yapfl/fl;Tazfl/+;Osx-Cre | YAPcKO;TAZcHET | |
| Yapfl/+;Tazfl/fl;Osx-Cre | YAPcHET;TAZcKO | |
| Yapfl/fl;Tazfl/fl;Osx-Cre | YAPcKO;TAZcKO | |
| Inducible | Osx-CretetOff | Osx:Cre |
| Yapfl/fl;Tazfl/fl | YAPWT;TAZWT | |
| Yapfl/fl;Tazfl/fl;Osx-CretetOff | YApcKOi;TAZcKOi |
In our first study, we evaluated constitutive allele dose-dependent deletion of YAP and/or TAZ in skeletally mature mice 16–21 weeks of age (Table 1). Mice with homozygous floxed alleles for both YAP and TAZ (YAPfl/fl;TAZfl/fl) were mated with double heterozygous conditional knockout mice (YAPfl/+;TAZfl/+;Osx-Cre) to produce eight possible genotypes in each litter, but only Cre-positive and YAPfl/fl;TAZfl/fl animals were compared (Table 1). As, constitutive, dual homozygous YAP/TAZ deletion using Osx-Cre is perinatal lethal(15), we could not evaluate this genotype in the constitutive deletion fracture healing model. Here, the littermate YAPfl/fl;TAZfl/fl mice were considered the wild-type control genotype. All of these mice were bred, raised, and evaluated without tetracycline administration to induce gene recombination in Osx-Cre-expressing cells during embryonic development, for the duration of the analyses in the constitutive deletion model. In the constitutive deletion fracture healing model, 5 – 8 mice per genotype were evaluated at 14- and 42- days post fracture (dpf) using microCT and mechanical testing.
In our second study, we allowed mice to develop to skeletal maturity and induced homozygous YAP/TAZ deletion two weeks prior to fracture at 16–18 weeks of age (Table 1). In the inducible deletion model, both littermate YAPfl/fl;TAZfl/fl (YAPWT;TAZWT) mice and separately-bred Osx-CretetOff mice were evaluated as wild type control genotypes (Table 1). All mice were bred and raised until skeletal maturity with doxycycline in their drinking water to prevent Cre-mediated gene recombination. For all in vivo fracture healing assessments, doxycycline was removed two weeks prior to fracture surgery and normal drinking water was provided for the remainder of the study. For periosteal progenitor cell isolations from fractured limbs, doxycycline was provided for both YAPWT;TAZWTand YAPcKOi;TAZcKOi mice throughout skeletal development and the duration of the fracture healing experiment. In the inducible deletion fracture healing model, 3–9 mice per genotype were evaluated at 4, 7, and 14 dpf using qPCR, histology, and microCT.
In both studies, mice were tail or ear clipped after weaning or prior to euthanasia and genotyped by an external service (Transnetyx, Inc.). All mice were fed regular chow (PicoLab Rodent Diet, Cat#: 0007688, LabDiet) ad libitum and housed in cages containing 2–5 animals each. Mice were maintained at constant 25°C on a 12-hour light/dark cycle. Both male and female mice were evaluated with the same fracture healing procedure for both the constitutive and inducible deletion models of fracture healing (Table 1). All protocols were approved by the Institutional Animal Care and Use Committees at the University of Notre Dame and the University of Pennsylvania. All animal procedures were performed in adherence to federal guidelines for animal care and conform to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
Open femoral fracture model and timepoints
An open, unilateral, and intramedullary pin-stabilized femoral fracture model was used to study bone repair in both the inducible and constitutive deletion studies. Femora were surgically exposed and manually fractured by applying a bending moment with a scalpel, but stabilized with an intramedullary pin(19). For the surgical procedure, animals were anesthetized using isoflurane (1–5%), all hair was removed from the surgical site, and the area was cleansed with sterile water followed by betadine. A 25-gauge needle was inserted in a retrograde manner into the intramedullary canal of the right femur(20). Subsequently, the muscle surrounding the same femur was blunt dissected to expose the femoral midshaft and a reproducible fracture was created by applying a three-point bending moment using a scalpel in the femur containing the intramedullary pin. The contralateral leg was left intact. Any animals that displayed intramedullary pin displacement or fractures that were comminuted or too oblique were removed. The mice were allowed to recover under a heating lamp and after awakening returned to their cages and allowed to ambulate freely. In the constitutive deletion model, mice were euthanized at 14 days post-fracture (dpf) and 42 dpf. In the inducible deletion model, mice were euthanized at 4 dpf, 7 dpf, and 14 dpf. At 4 dpf, mice were injected intraperitoneally with 5-ethynyl-2’-deoxyuridine (EdU; E10187; Invitrogen) at 10 mg/kg 3 hours prior to euthanasia to assay cellular proliferation.
Microcomputed tomography
Micro-computed tomography (microCT) was performed according to published guidelines(21) on two separate systems correlating with the constitutive and inducible model. In accordance to published microCT guidelines(21), the same threshold was applied across all genotypes for a given system. In the constitutive deletion study, 14 and 42 dpf fractured limbs were dissected free from surrounding musculature and the intramedullary pins were removed. Samples from 14 and 42 dpf limbs were wrapped in PBS-soaked gauze and frozen at −20°C. When removed from the freezer, bones were allowed to thaw while being imaged using a μCT 35 system (Scanco Medical). Samples from 14 and 42 dpf bone were imaged with an X-ray intensity of 114 μA, energy of 70 kVp, integration time of 200 ms, and resolution of 15μm. As described previously(20), we defined the fracture callus mineralization threshold as 50% of the mineral density that we used to segment intact cortical bone under these conditions on this system. 2D tomograms of the entire fracture callus both excluding and including the intact cortical bone were manually contoured, stacked and binarized by applying a Gaussian filter (sigma =0.8, support =1) at a threshold of 345 mg HA/cm3. 3D quantification of the callus was conducted in two ways. First, to determine the callus volume and mineralization percentage of the callus, microCT slices were contoured pre-segmentation to produce a region of interest that excluded the intact cortical bone, but included the non-mineralized callus. Total callus volume, percent callus mineralization, and volumetric bone mineral density of the mineralized tissue were evaluated using the segmentation and thresholding above. Second, to evaluate the contours of the outer boundary of the callus, including both non-mineralized and mineralized tissues, microCT slices were contoured pre-segmentation and the polar moment of inertia (pMOI) was computed on a transverse slice-by-slice basis as described in detail below. Five to eight mice were analyzed per group. Investigators were blinded to animal genotype during scan quantification.
In the inducible deletion study, 7 and 14 dpf limbs were dissected free from surrounding musculature and the intramedullary pins were removed. Bones from 7 and 14 dpf were then snap-frozen in liquid nitrogen-cooled isopentane for 1-minute, wrapped in gauze and imaged on a vivaCT 80 system (Scanco Medical). Samples from 7 and 14 dpf were imaged with an X-ray intensity of 114 μA, energy of 70 kVp, integration time of 200 ms, and resolution of 15μm. We again defined the fracture callus mineralization threshold as 50% of the mineral density that we used to segment intact cortical bone under these conditions on this system. 2D tomograms of the entire fracture calluses both excluding and including the intact cortical bone were manually contoured, stacked and binarized by applying a Gaussian filter (sigma =0.8, support =1) at a threshold of 254 mg HA/cm3. 3D quantification was applied as described above. Six to eight mice were analyzed per group. Investigators were blinded to animal genotype during scan quantification.
To quantify the contours of the outer boundary of the callus along the length of the bone axis, the “Bone Midshaft” evaluation script (Scanco Medical) was used to measure the polar moment of inertia (pMOI), also known as the second polar moment of area, a measure of radial distribution(22). Limbs from 14 dpf in the constitutive deletion model and 14 dpf from the inducible deletion model were analyzed using a negative, non-physiological threshold within the contours of the callus to include both mineralized and non-mineralized tissues in the “Bone Midshaft” evaluation. Limbs from 42 dpf were scanned on the μCT 35 system were analyzed using the same threshold for mineralized tissue above (345 mg HA/cm3) to include only the mineralized tissue in the “Bone Midshaft” evaluation. In both cases, pMOI values for all groups were binned into 25 equal distance bins from the center of the fracture using a custom MATLAB script. Data presented as the mean polar moment of inertia at each axial position, with error bars corresponding to the standard deviation (SD).
Mechanical testing
Following microCT scanning, 14 and 42 dpf limbs from the constitutive deletion model were tested in torsion to failure. For torsional testing, we used fixtures and a custom potting apparatus that allowed us to reproducibly align and pot the fractured limbs in polymethylmethacrylate bone cement. After the fractured limbs were potted, they were loaded in torsion at a rate of 1°/s until failure using a custom-designed micro-torsional testing system. Recorded torque-rotation data were normalized by gauge length on a per-sample basis(23). Torsional rigidity, maximum torque to failure, work to maximum torque and work to failure were quantified from the normalized torque-rotation data using a custom MATLAB script(15). Five to eight mice were analyzed per group per timepoint. Investigators were blinded to animal genotype during data quantification.
Histology, immunohistochemistry, and immunofluorescence
Limbs from 7 and 14 dpf were fixed with 10% neutral buffered formalin for 48 hours and decalcified for 4 weeks with 0.25M EDTA (pH 7.4) at 4°C. Paraffin sections (5 μm thickness) were processed for either immunohistochemistry or histology. Primary antibodies were compared to normal rabbit sera IgG control sections. For immunostaining, anti-OSX (1:500, ab22552; abcam), anti-YAP (1:500, 14074; Cell Signaling), and anti-TAZ (1:250 NB110–58359; Novus Biologicals) primary antibodies were applied overnight. Next, sections were incubated with corresponding biotinylated secondary antibody, avidin-conjugated peroxidase, and diaminobenzidine substrate chromogen system (329ANK-60; Innovex Biosciences), which allowed for immunohistochemical detection of positively stained cells. Hematoxylin and eosin stains (H&E), Safranin-O, and Picrosirius Red stains were used to stain for bone, cartilage, and collagen.
Limbs from 4 dpf and intact femora were fixed with 10% neutral buffered formalin for 48 hours at 4°C, transferred to 30% sucrose in PBS overnight at 4°C, and then embedded in O.C.T. compound (Tissue-Tek). Thin sections (7 μm thickness) were made from undecalcified fractured femurs using cryofilm IIC tape (Section Lab Co. Ltd.) as previously described(24) and processed for immunofluorescence and/or aqueous H&E staining. Taped sections were glued to microscope slides using a UV-adhesive glue, rehydrated and then decalcified with 0.25M EDTA (pH 7.4) for 3 minutes prior to staining. 5-ethynyl-2’-deoxyuridine (EdU)) staining were performed using the Click-iT Plus EdU Assay kit (C10339; Invitrogen) according to the manufacturer’s instructions.
Imaging and histomorphometric analysis
Histological and immunohistochemical sections were imaged on either on an Axio Observer Z1 (Zeiss) at the 10x and 25x objectives or using an Axioscan microscope (Zeiss) at the 10x and 20x objective. Histomorphometry quantification at 4, 7 and 14 dpf was performed using ImageJ (NIH). To evaluate bone and cartilage area parameters at 7 and 14 dpf, the entire callus area was evaluated using ImageJ (NIH) on at least 3 sections per sample with 3–4 mice per group per timepoint. To evaluate the number of positively immunostained cells at 7 and 14 dpf, individual cells were manually scored as either positive or negative, using ImageJ (NIH), for each given antibody. Data are reported as the percentage of positively-stained osteoblasts, osteocytes, and chondrocytes per total number of each cell type on each sample. For all samples, approximately 500 cells were evaluated where osteoblasts and osteocytes were primarily evaluated in the “shoulder” region of the callus while chondrocytes were evaluated in the cartilage near the fracture line. To evaluate periosteal parameters at 4 dpf, 4 regions of interest were outlined per sample 1–3 mm from the fracture on the periosteal cortical bone(25), which corresponded in this femoral fracture model to areas of the callus where intramembranous ossification occurred.
Samples from 7 and 14 dpf were stained with Picrosirius Red and imaged under polarized light using an Axioscan microscope (Zeiss) at the 20x objective and using second harmonic generated (SHG) microscopy. SHG images were taken on a TCS SP8 Multiphoton Confocal microscope (Leica) at a fundamental wavelength of 880 nm with the 10x and 40x objective on sections oriented in the same direction for all groups. All SHG images were quantified using ImageJ (NIH) and reported as mean pixel intensity within the cortical and callus region relative to WT bone. Mean pixel intensities across four separate regions of interests were averaged as technical replicates for a given sample within either the callus or cortex area with 3–4 mice per group.
Immunofluorescence sections of 4 dpf limbs were imaged on an Axio Observer Z1 (Zeiss) at the 5x, 10x, and 25x objectives. To evaluate the number of positively EdU-stained periosteal cells, 4 regions of interest were outlined per mouse 1–3 mm from the fracture on the periosteal cortical bone(25). Images were manually scored as either positive or negative, averaged together from each of the 4 regions and reported as percent positively stained per total number of periosteal cells in ImageJ (NIH). Periosteal area and average thickness from these same 4 regions of interested were outlined in ImageJ (NIH) using both immunofluorescence and aqueous H&E sections and averaged together with 6–9 mice per group.
Periosteal cell isolation and osteogenic differentiation
Mouse periosteal cells were isolated from either WT or Osterix-conditional YAP/TAZ deficient (YAPcKOi;TAZcKOi) femurs on 4 dpf and cultured at 37°C and 5% O2, as described previously(26). Briefly, mice were anesthetized by carbon dioxide inhalation and euthanized via cervical dislocation. Fractured limbs were carefully dissected of all non-osseous tissue, the epiphyses were then removed, and marrow cavities were flushed. The periosteum was scraped and enzymatically digested for 1 hour at 37°C on an orbital shaker (0.5 mg/ml collagenase P, 2mg/ml hyaluronidase in PBS). Following washing, 2 × 104 cells/cm2 were seeded in growth medium (α-MEM, 10% FBS, 1% penicillin-streptomycin, and 1ug/ml doxycycline) and cultured in 5% oxygen for the first 4 days. Half of the media was changed on day 4 and cultures were then incubated in 21% O2. Primary cells reached confluence by day 7 and were passaged once into osteogenic differentiation experiments.
Passage 1 periosteal cells were then seeded at 21% O2 into 24-well plates (15 × 103 cells/cm2) and cultured in growth medium. After reaching confluence, primary periosteal cell cultures were induced towards osteogenic differentiation (50 μg/mL ascorbic acid and 4mM β-glycerophosphate) for 21 days. Osteogenic media was changed every two days prior to RNA isolation and alizarin red staining for mineral deposition at 21 days.
RNA isolation and qPCR
Limbs from 7 and 14 dpf were carefully dissected and removed of all non-osseous tissue. The intramedullary pin was removed, the ends of the femur were cut, and marrow flushed before snap-freezing the entire callus in liquid nitrogen-cooled isopentane for 1 minute prior to microCT imaging and storage at −80°C until processing. Tissues were then homogenized via mortar and pestle and RNA from the sample was collected using Trizol Reagant (15596026; Life Technologies) followed by centrifugation in chloroform. RNA from fractured limb tissue and cell culture experiments were purified using the RNA Easy Kit (74106; Qiagen) and quantified by spectrophotometry using a NanoDrop 2000 (Thermo-Fisher Scientific). Reverse transcriptase polymerase chain reaction (RT-PCR) was performed on 0.5 μg/μl concentration of RNA using the High-Capacity cDNA Reverse Transcription Kit (4368814; Thermo-Fisher Scientific). Quantitative polymerase chain reaction (qPCR) assessed RNA amount using a StepOnePlus™ Real-Time PCR System (Thermo-Fisher Scientific) relative to the internal control of 18S ribosomal RNA (18S rRNA). Data are presented using the ΔΔCt method. Six mice per group were used. Specific mouse primer sequences are listed (Supplemental Table 1).
Statistics
Sample sizes were selected a priori by power analyses based on effect sizes and population standard deviations taken from published data on the developmental bone phenotype of Osterix-conditional YAP/TAZ deletion(15). As fracture healing outcomes tend to have more variability than developmental phenotyping outcomes, we assumed 1.5 times the standard deviation for outcomes from Osterix-conditional YAP/TAZ deletion(15), which resulted in an effect size of 1.94, a power of 80%, and α=0.05. From the power analysis, a sample size of 6 was selected. Some samples were lost during initial protocol optimization. All statistics and power analyses were performed in GraphPad Prism or R (Version 3.5.1). Comparisons between two groups were made using the independent t-test while comparisons between 3 or more groups were made using a one-way ANOVA with post-hoc Tukey’s multiple comparisons test, if the data were normally distributed according to D’Agostino-Pearson omnibus normality test and homoscedastic according to Bartlett’s test. When parametric test assumptions were not met, data were log-transformed, and residuals were evaluated. If necessary, either the non-parametric Kruskal-Wallis test with post-hoc Dunn’s multiple comparisons or the non-parametric Mann-Whitney test were used. A p-value < 0.05 (adjusted for multiple comparisons) was considered significant. On the graphs, exact p-values to 3 decimal places were indicated above the bracketed lines to indicate significant comparisons. Post-hoc comarisons were performed using adjusted post-hoc Tukey’s multiple comparisons test for comparisons between 3 or more groups or two-tailed t-tests when comparing 2 groups. If a p-value is not indicated between two groups the differences between those groups were not statistically significant. Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range.
RESULTS
Constitutive Osterix-conditional YAP and/or TAZ deletion impaired fracture healing
To evaluate the roles of YAP and TAZ in fracture repair, we used Cre-lox to delete YAP and/or TAZ from Osterix-Cre expressing cells during embryonic development(13). We selected a breeding strategy that generated YAP/TAZ allele dosage-dependent Osterix-conditional knockouts with four genotypes (see Table 1). Constitutive, Osterix-conditional, dual homozygous YAP/TAZ knockouts were not evaluated due to perinatal lethality(15). We then evaluated adult bone fracture repair at 14- and 42-days post fracture (dpf).
All genotypes exhibited callus formation by 14 dpf (Fig. 1A). However, constitutive, Osterix-conditional YAP and/or TAZ deletion reduced total callus volume and mineralized callus percentage (i.e. BV/TV) at 14 dpf in an allele dosage-dependent manner (Fig. 1B,C). Similarly, constitutive Osterix-conditional YAP and/or TAZ deletion also reduced mineralized tissue volume and volumetric mineral density at 14 dpf (Supplemental Fig. 1A,B). We then tested 14 dpf limbs in torsion to failure and observed a similar reduction in maximum torque to failure and torsional rigidity (Fig. 1D,E). However, work to max torque and work to failure did not reach statistically significant differences between genotypes (Supplemental Fig. 1C,D).
Figure 1. Constitutive, combinatorial YAP/TAZ deletion from Osterix-expressing cells impaired fracture healing.
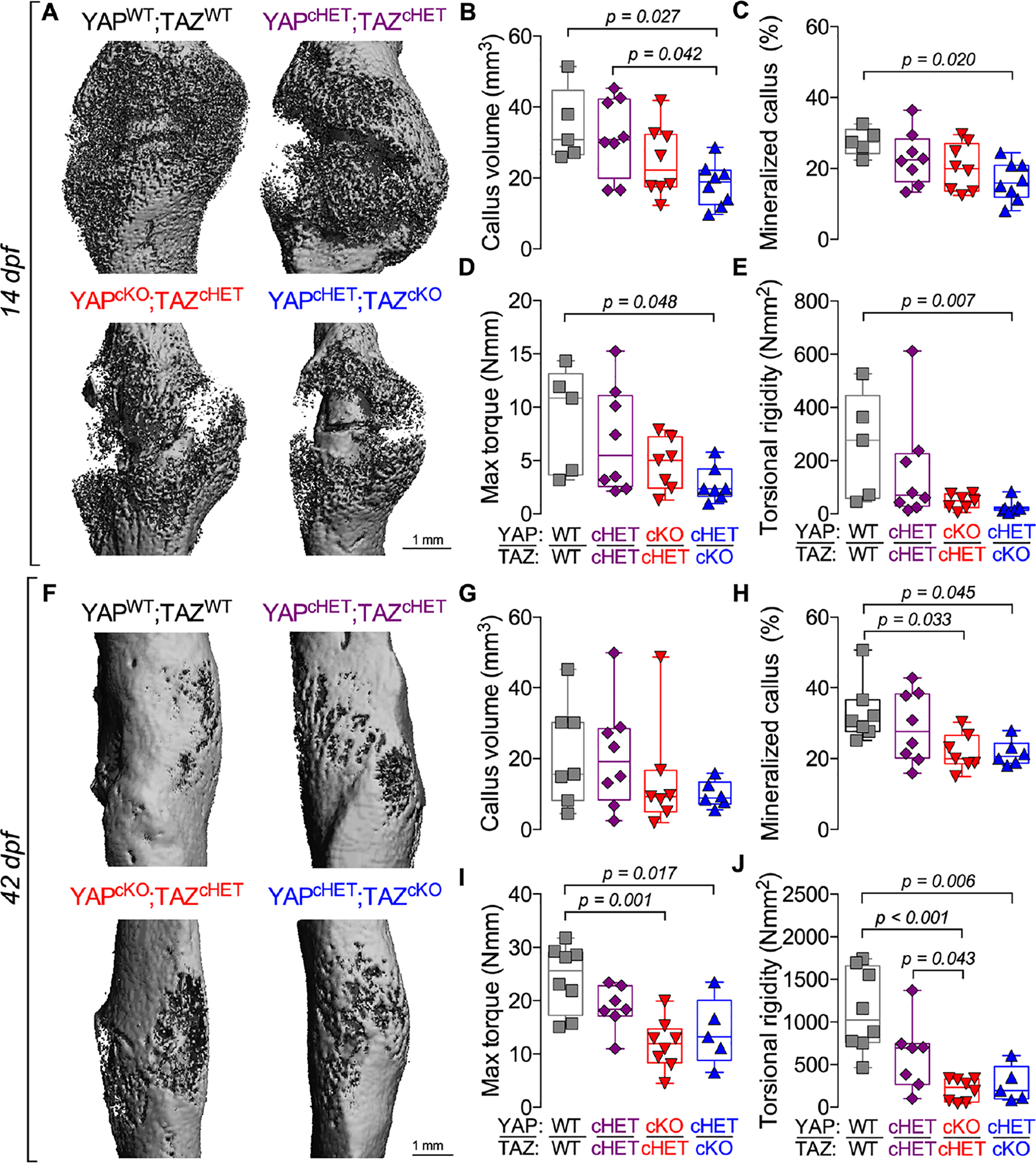
A) MicroCT reconstructions at 14 days post-fracture (dpf). Quantification of 14 dpf callus architecture: (B) total callus volume and (C) mineralized callus percentage. Quantification of 14 dpf callus mechanical testing in torsion to failure: (D) maximum torque and (E) torsional rigidity. F) MicroCT reconstructions at 42 dpf. Quantification of 42 dpf callus architecture: (G) total callus volume and (H) mineralized callus percentage. Quantification of 42 dpf callus mechanical testing in torsion to failure: (I) maximum torque and (J) torsional rigidity. Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. Data were evaluated by one-way ANOVA with post-hoc Tukey’s multiple comparisons tests. Groups with significant pairwise comparisons are indicated by bracketed lines p-values, adjusted for multiple comparisons. Sample sizes, N = 5–8. Scale bars indicate 1 mm for microCT reconstructions.
At 42 dpf, all genotypes underwent hard callus formation and remodeling (Fig. 1F). However, constitutive Osterix-conditional YAP and/or TAZ deletion again delayed healing with reduced mineralized callus percentage, maximum torque to failure, and torsional rigidity, but at this timepoint differences in total callus volume did not reach statistical significance (Fig 1G–J). At 42 dpf, constitutive, Osterix-conditional YAP and/or TAZ deletion reduced volumetric mineral density, but did not significantly reduce mineralized tissue volume, work to maximum torque, or work to failure (Supplemental Fig. 1E–H).
Constitutive Osterix-conditional YAP and/or TAZ deletion reduced callus size
To evaluate the radial distribution of the total callus at 14 dpf, we quantified the polar moment of inertia of the entire callus, including bone, cartilage, and fibrous tissue. Independent of mineralized tissue formation, constitutive, Osterix-conditional YAP/TAZ deletion reduced total callus size, particularly in the YAPcHET;TAZcKO mice, which are homozygous for TAZ deletion and heterozygous for YAP (Fig. 2A,B). At 42 dpf, similar results were observed where the polar moment of inertia distribution of mineralized tissue within the callus were reduced in the YAPcHET;TAZcKO mice (Supplemental Fig. 2A,B). At 42 dpf, we performed an analysis of covariance (ANCOVA) using linear regression to decouple the contributions of callus mineralization and geometric distribution from the mechanical behavior(15,27), since constitutive, Osterix-conditional YAP/TAZ deletion reduced both callus mineralization and geometry. We found that individual regression lines for each genotype best predicted maximum torque to failure and torsional rigidity, suggesting that differences in connectivity or composition also contribute to mechanical behavior (Supplemental Fig. 2C,D).
Figure 2. Constitutive, combinatorial YAP/TAZ deletion from Osterix-expressing cells reduced callus size.
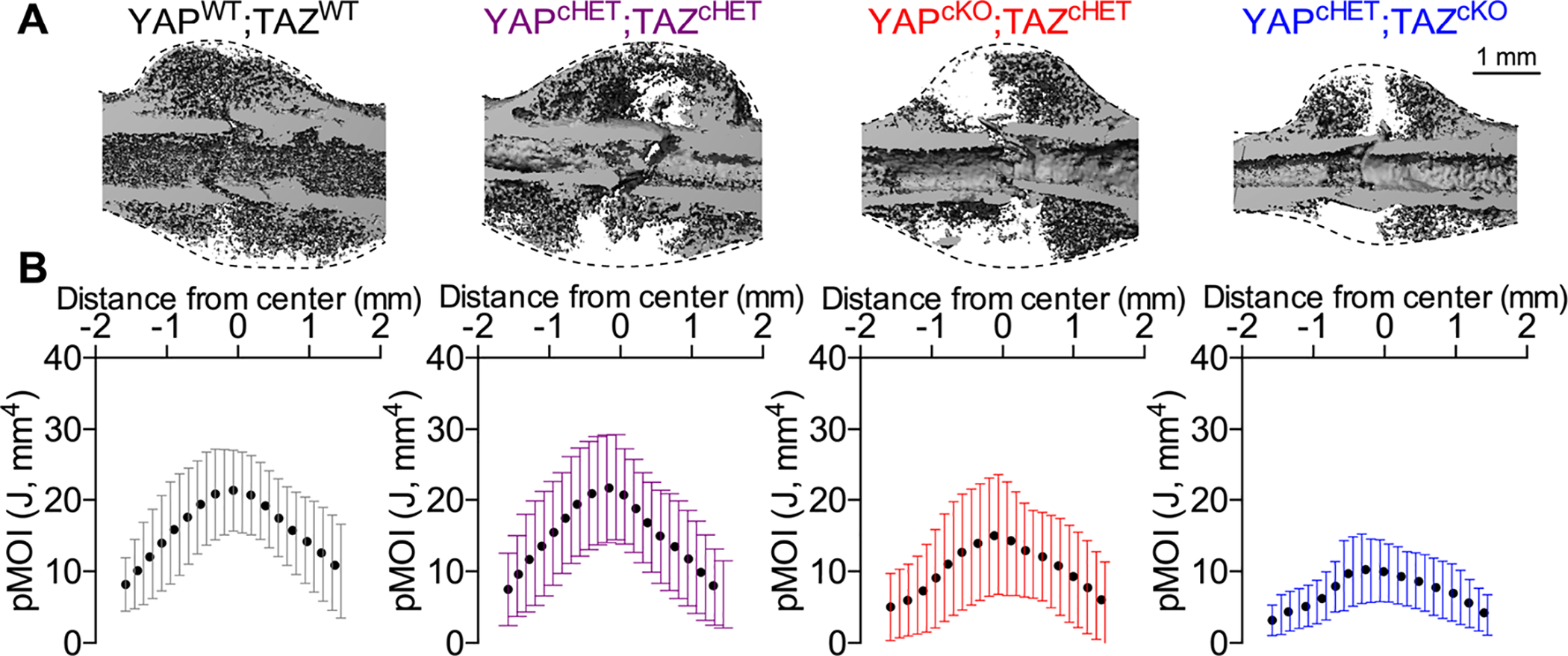
A) MicroCT reconstructions at 14 dpf showing longitudinal cut-planes within the callus. Dotted lines indicate the callus boundary, the region within which the total callus moment of inertia was quantified. B) Polar moment of inertia distributions of the entire callus for each of the YAP/TAZ allele dose-dependent knockout genotypes. Data were binned into 25 equal distance bins from the center of the callus and presented as dots representing the mean and error bars corresponding to the standard deviation (SD). Sample sizes, N = 5–8. Scale bar indicates 1 mm for microCT reconstructions.
Constitutive Osterix-conditional YAP and/or TAZ deletion impaired periosteal development
To evaluate whether developmental defects in the periosteum contributed to the reduced callus size, we evaluated the periosteal thickness and cellularity of intact femurs from constitutive knockout mice. Constitutive, Osterix-conditional YAP and/or TAZ deletion significantly reduced periosteal thickness and area (Fig. 3A–C). Similar to younger mice(15), constitutive YAP and/or TAZ deletion reduced periosteal cell number per bone surface in an allele dose-dependent manner (Fig 3. D), suggesting defective periosteal development.
Figure. 3. Constitutive, combinatorial YAP/TAZ deletion from Osterix-expressing cells impaired periosteal development in intact bone.
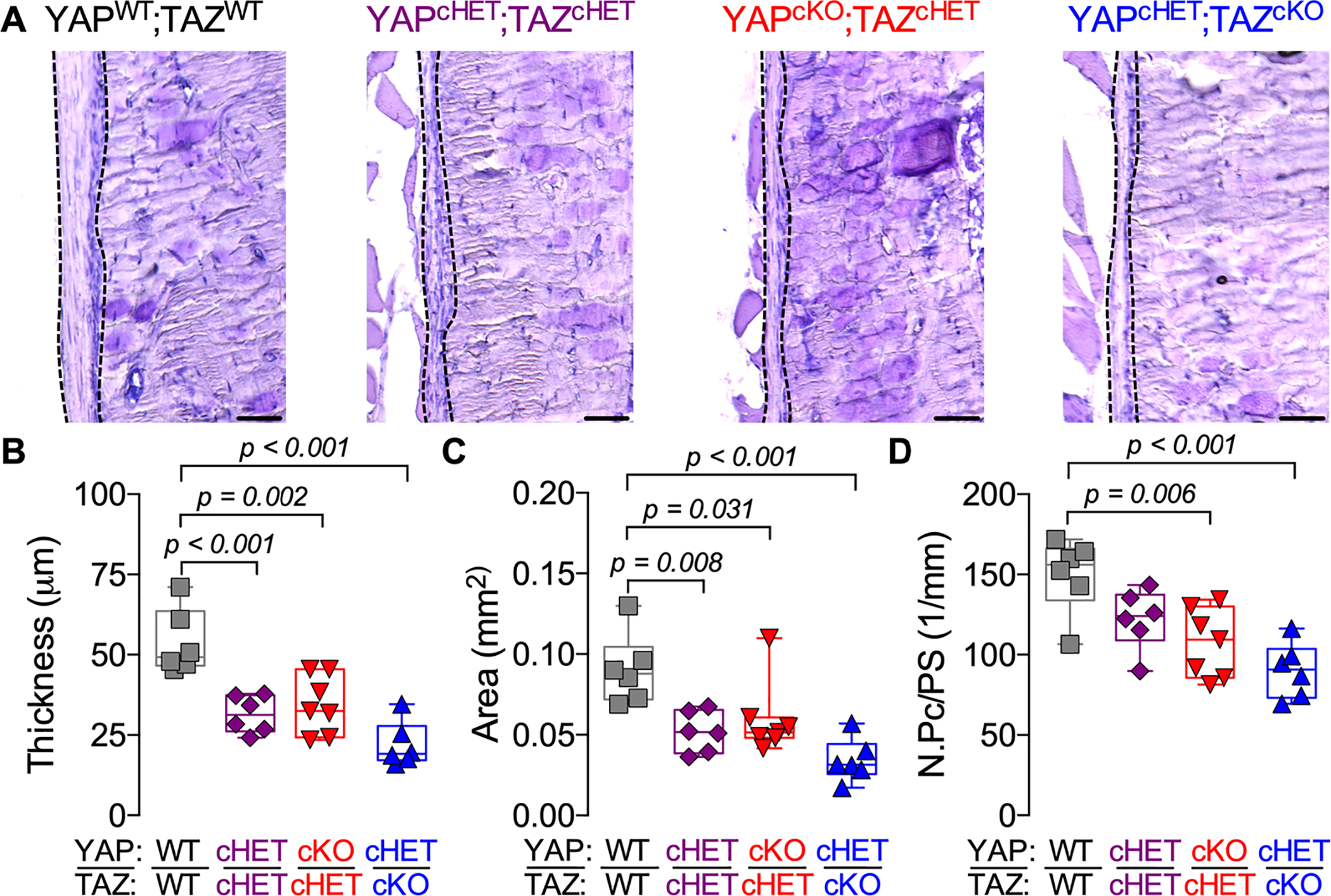
A) Representative micrographs of 18–21 weeks-old distal femur cortical bone stained by aqueous H&E. Dotted lines indicate the periosteum. Quantification of (B) periosteal thickness, (C) periosteal area and (D) periosteal cell number per bone surface (N.Pc /PS). Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. Data were evaluated by one-way ANOVA with post-hoc Tukey’s multiple comparisons tests. Groups with significant pairwise comparisons are indicated by bracketed lines p-values, adjusted for multiple comparisons. Sample sizes, N = 6–7. Scale bars indicate 50 μm for all images.
Inducible Osterix-conditional YAP/TAZ deletion impaired callus mineralization, but not formation
The impairment of bone fracture healing observed in the constitutive, Osterix-conditional YAP/TAZ deletion knockout model could be a consequence of defective periosteal stem cell supply, expansion, and/or differentiation. To address this question, we generated adult-inducible, dual homozygous YAP/TAZ knockout mice in which the periosteal progenitor population was allowed to develop normally prior to fracture. Here, we induced homozygous Osterix-conditional YAP/TAZ deletion two weeks prior to fracture. During those two weeks prior to fracture, inducible Osterix-conditional YAP/TAZ deletion did not significantly affect periosteal cell thickness, area, or cell number (Fig. 4A–D).
Figure 4. Periosteal thickness and cellularity developed normally in adult onset-induced, Osterix-conditional, homozygous YAP/TAZ knockout mice.
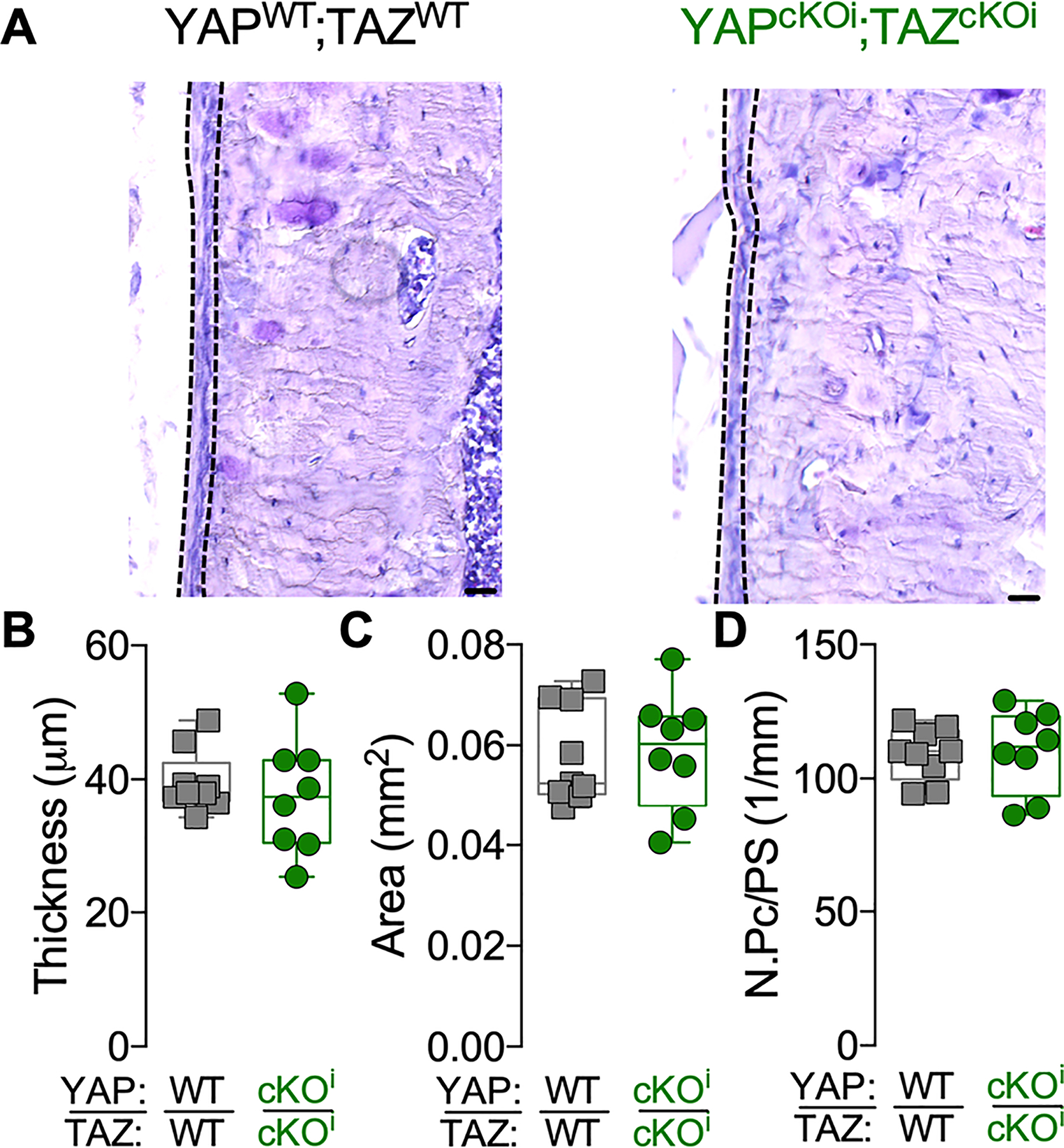
A) Representative micrographs of 16–18 weeks-old cortical bone stained by aqueous H&E. Dotted lines indicate the periosteum. Quantification of (B) periosteal thickness, (C) periosteal area and (D) periosteal cell number per bone surface (N.Pc/PS). Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. Groups were compared by two-tailed Student’s t-test. There were no significant differences between groups. Sample sizes, N = 8–9. Scale bars indicate 30 μm for all images.
To analyze the recombination efficiency of inducible Osterix-Cre-mediated YAP/TAZ deletion following fracture, we evaluated YAP/TAZ expression in chondrocytes, osteoblasts, and osteocytes within the callus at 14 dpf by immunohistochemistry and qPCR (Supplemental Fig. 3A–C). The percent of YAP-positive cells were significantly reduced for chondrocytes (23% reduction), osteoblasts (23% reduction), and osteocytes (26% reduction) (Supplemental Fig. 3D–F). YAP mRNA expression was reduced by 59% in full callus lysate preparations (Supplemental Fig. 3G). The percent of TAZ-positive cells were moderately reduced for chondrocytes (11% reduction; p = 0.106) and significantly reduced for osteoblasts (18% reduction), and osteocytes (12% reduction) (Supplemental Fig. 3H–J). TAZ mRNA expression was reduced by 49% in full callus lysate preparations (Supplemental Fig. 3K).
All genotypes underwent initial callus formation by 14 dpf (Fig. 5A). Inducible Osterix-Cre-mediated YAP/TAZ deletion reduced mineralized callus percentage and volumetric bone mineral density, but differences in total callus volume between groups were not observed (Fig. 5B–D). Further, inducible Osterix-Cre-mediated YAP/TAZ deletion qualitatively increased variability in the polar moment of inertia distribution of the total callus size in comparison to YAPWT;TAZWT and Osx:Cre mice (Fig. 5B; Supplemental Fig. 4A,B).
Figure 5. Adult onset-inducible, homozygous YAP/TAZ deletion from Osterix-expressing cells impaired callus mineralization, but not size.
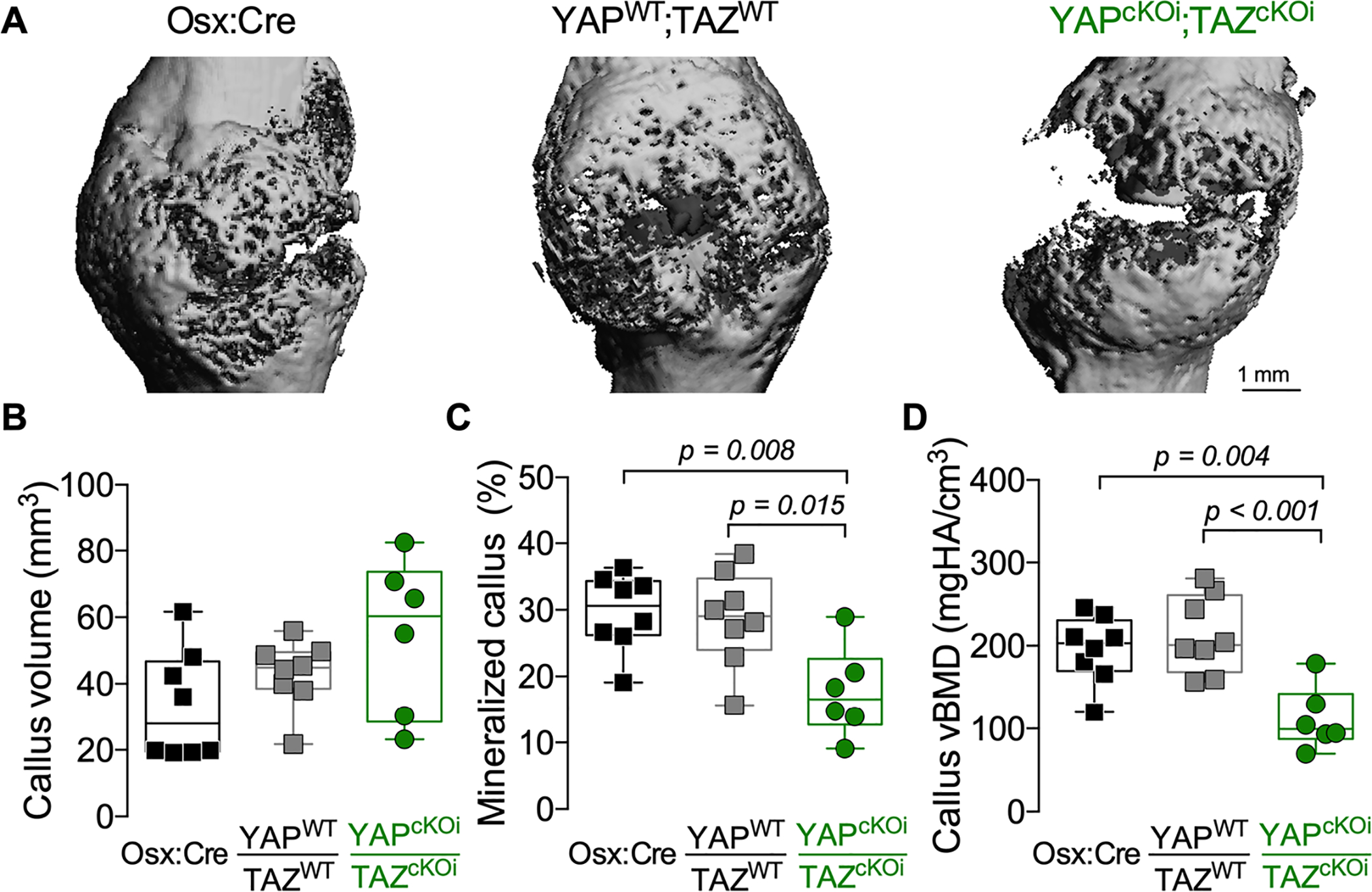
A) MicroCT reconstructions at 14 dpf. Quantification of 14 dpf callus architecture: (B) total callus volume, (C) mineralized callus percentage, and (D) volumetric mineral density. Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. Data were evaluated by one-way ANOVA with post-hoc Tukey’s multiple comparisons tests. Groups with significant pairwise comparisons are indicated by bracketed lines and p-values, adjusted for multiple comparisons. Sample sizes, N = 6–8. Scale bars indicate 1 mm for microCT reconstructions.
Inducible Osterix-conditional YAP/TAZ deletion did not alter callus size, but reduced bone formation
As the formation of a cartilaginous callus template is a critical step during endochondral fracture healing(3–5), we histologically evaluated cartilage, bone, and fibrous tissue formation at 14 dpf (Fig. 6A,E–G; Supplemental Fig. 5). Consistent with microCT, inducible Osterix-Cre-mediated YAP/TAZ deletion did not affect total callus area at 14 dpf (Fig. 6E). Differences in total cartilage area, percent cartilage area, and the number of Osterix (OSX)-positive hypertrophic chondrocytes were not detected between groups (Fig. 6F–H). Fibrous tissue composition was evaluated by qualitative analysis of H&E- and picrosirius red-stained adjacent sections to differentiate between fibrotic tissue and bone, but differences in fibrous tissue were not observed between groups (Supplemental Fig. 5A; Fig. 6B). At 14 dpf, inducible Osterix-Cre-mediated YAP/TAZ deletion did not significantly alter mRNA expression of markers for chondrogenesis, including SRY-Box Transcription Factor 9 (Sox9), Aggrecan (Acan), and collagen, type II, alpha 1 (Col2a1) (Supplemental Fig. 5D).
Figure. 6. Adult onset-inducible, homozygous YAP/TAZ deletion from Osterix-expressing cells reduced callus bone formation.
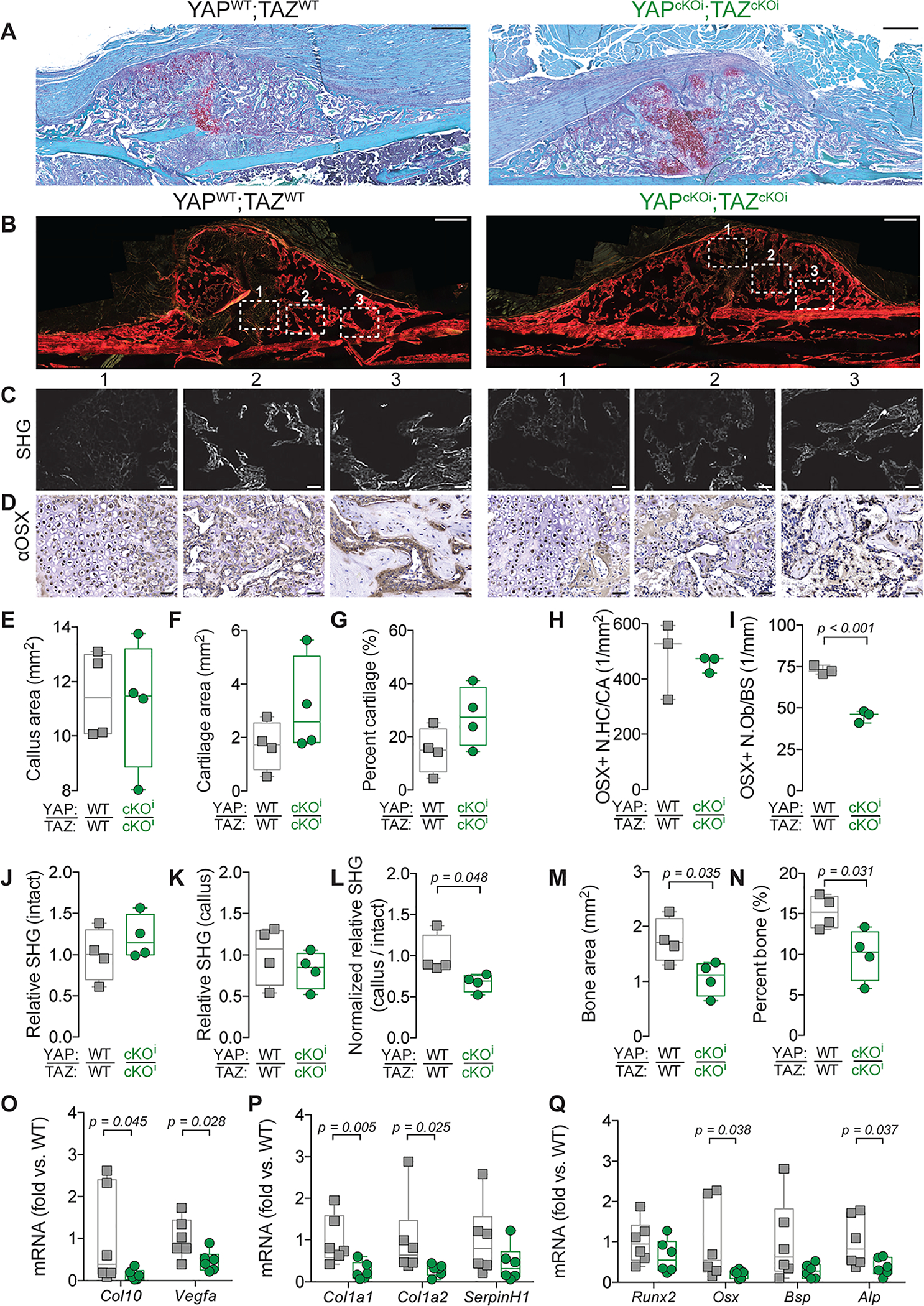
A) Representative micrographs of Safranin-O-stained calluses at 14 dpf. B) Representative micrographs of Picrosirius Red-stained calluses at 14 dpf. White boxes outline three regions of interest in which second harmonic generated imaging (SHG) (C) and anti-Osterix (αOSX) immunostaining (D) are highlighted. E-G) Quantification of cartilage callus histomorphometry at 14 dpf. (E) Callus area, (D) cartilage area, and (E) percent cartilage area. H, I) Quantification of the number of Osterix-positive hypertrophic chondrocytes per cartilage area (OSX+ N.HC/CA) (H) and Osterix-positive osteoblasts per bone surface (OSX+ N.Ob/BS) (I). J-L) Quantification of relative SHG intensity per bone area at 14 dpf. (J) Intact cortical bone, (K) the newly formed callus bone, and (L) normalized callus-to-intact SHG intensity. M, N) Histomorphometry of bone formation. (M) Bone area and (N) percent bone area. O-Q) Messenger RNA was extracted from callus lysate preparations at 14 dpf and gene expression was normalized to 18S rRNA and quantified as fold-change relative to wild type. O) Col10 and Vegfa mRNA expression. P) Col1a1, Col1a2, and SerpinH1 mRNA expression. Q) Runx2, Osx, Bsp, and Alp mRNA expression. Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. Data were evaluated either by one way ANOVA with post-hoc Tukey’s multiple comparisons tests, or by two-tailed Student’s t-tests, as appropriate. Groups with significant pairwise comparisons are indicated by bracketed lines and p-values, adjusted for multiple comparisons where necessary. Sample sizes N = 3–4 per group for histomorphometry and N= 6 for gene expression. Scale bars indicate 50 μm for all high-power images and 500 μm for callus images.
Following formation of the cartilaginous callus, bone formation occurs through both intramembranous and endochondral ossification(4). Therefore, we next evaluated endochondral ossification and bone formation at 14 dpf (Fig. 6; Supplemental Fig. 5A). At 14 dpf, inducible Osterix-Cre-mediated YAP/TAZ deletion reduced the number of OSX-positive osteoblasts per bone surface, relative SHG measurement of collagen organization, and histomorphometric bone area and percent bone area (Fig. 6I–N). Further, inducible Osterix-Cre-mediated YAP/TAZ deletion reduced mRNA expression of markers for hypertrophic chondrocytes, including collagen, type X, alpha 1 (Col10) and vascular endothelial growth factor (Vegfa) (Fig. 6O). Similarly, inducible Osterix-Cre-mediated YAP/TAZ deletion reduced mRNA expression of markers of collagen, including collagen type I, alpha 1 (Col1a1) and collagen type I, alpha II (Col1a2), but not serpin family H member 1 (SerpinH1) (Fig. 6P). Lastly, inducible Osterix-Cre-mediated YAP/TAZ deletion reduced mRNA expression markers of osteogenesis, including osteoblast-specific transcription factor Osterix (Osx) and alkaline phosphatase (Alp), while reductions in runt-related transcription factor 2 (Runx2) or bone sialoprotein (Bsp) did not reach statistical significance (Fig. 6Q).
Inducible Osterix-conditional YAP/TAZ deletion reduced periosteal osteoblast precursor expansion in vivo and osteogenic differentiation in vitro
Given the osteogenic defect resulting from inducible Osterix-Cre-mediated YAP/TAZ deletion, we evaluated activated periosteal progenitors at 4 dpf in four regions distal and proximal to the fracture line, in which periosteal osteoblast precursors are primarily fated to form bone through direction intramembranous ossification(6) (Fig. 7A). At 4 dpf, inducible YAP/TAZ deletion reduced periosteal osteoprogenitor cell expansion in terms of total area and average thickness (Fig. 7B–D). However, inducible YAP/TAZ deletion did not reduce the number of periosteal osteoprogenitor cells per expanded periosteal area, but significantly reduced the percentage of proliferating, EdU-positive periosteal osteoprogenitor cells (Fig. 7E,F).
Figure 7. Adult onset-inducible, homozygous YAP/TAZ deletion from Osterix-expressing cells impaired periosteal osteoblast precursor cell expansion and osteogenic differentiation.
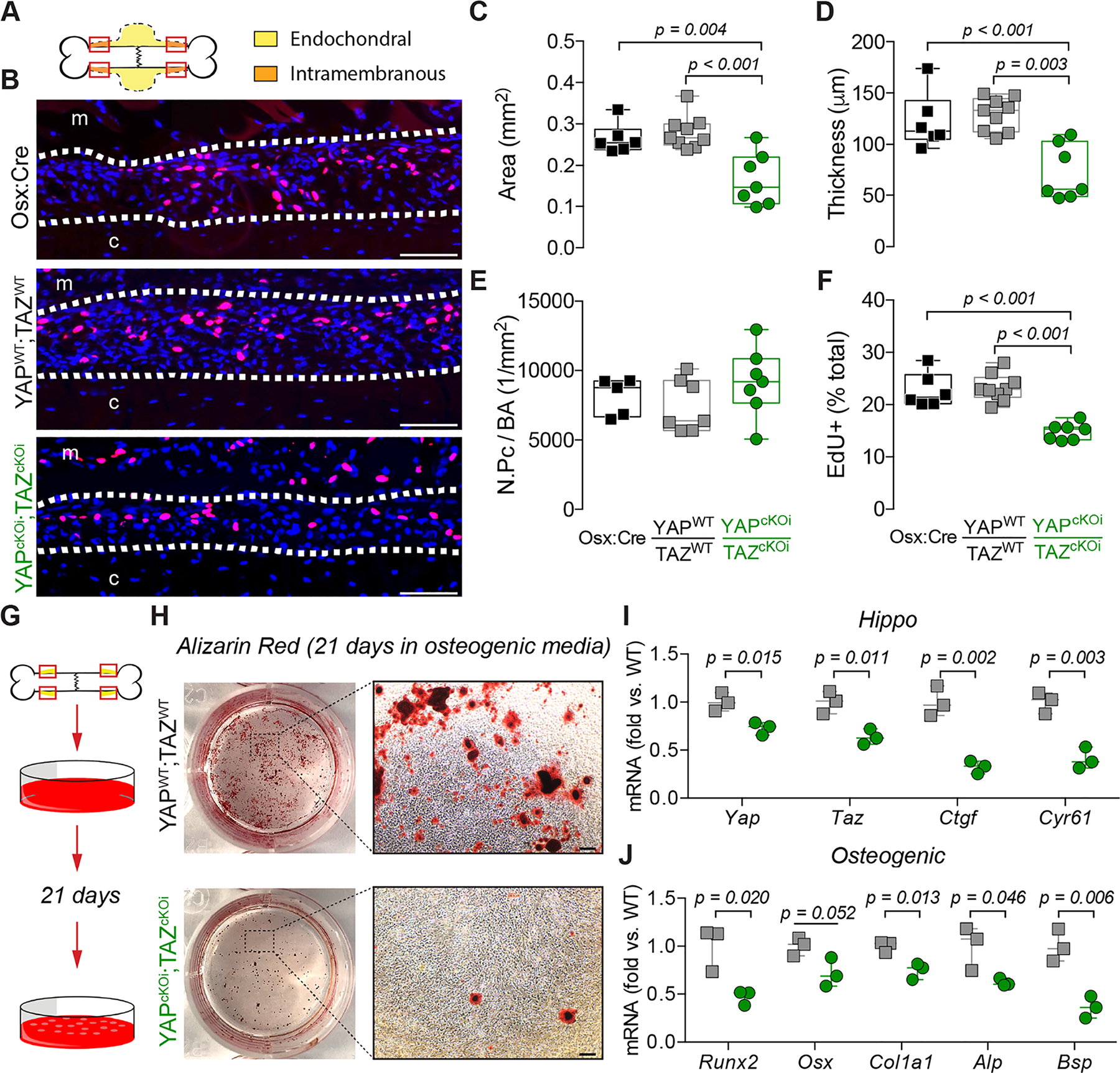
A) Representation of the regions of interest in the callus “shoulder,” where bone formation initiates by intramembranous ossification, and where we evaluated periosteal osteoblast precursor expansion and proliferation. B) Representative micrographs of EdU+ periosteal cells (red) at 4 dpf, with all nuclei counterstained by DAPI (blue). White dotted lines indicate periosteal cell expansion zone; “m” indicates muscle, “c” indicates cortical bone. Quantification of the expanded (C) periosteal area, (D) average thickness, (E) number of periosteal cells per bone area (N.Pc/BA), and (F) percentage of EdU-positive periosteal cells. G) Activated periosteal cells isolated from 4 dpf limbs were cultured in osteogenic media for 21 days. H) Representative Alizarin Red staining of mineral deposition following osteogenic induction. I) Yap, Taz, Ctgf, and Cyr61 and J) Runx2, Osx, Alp, and Bsp mRNA expression, relative to 18S rRNA, from periosteal progenitor cell cultures following 21 days of osteogenic induction. Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. Data were evaluated either by one-way ANOVA with post-hoc Tukey’s multiple comparisons tests, or by two-tailed Student’s t-tests. Groups with significant pairwise comparisons are indicated by bracketed lines and p-values, adjusted for multiple comparisons. N = 6–9 per group for in vivo data and N = 3 for in vitro data. Scale bars indicate 100 μm for all high-power EdU images and 1 mm for high-power Alizarin Red images.
Though unexplored in periosteal osteoprogenitor cells, YAP and TAZ are known to mediate osteogenic differentiation in the MSCs(28–30), which originate from a common mesenchymal embryonic lineage(31). To elucidate if inducible YAP/TAZ deletion regulated periosteal osteoprogenitor differentiation, we isolated activated periosteal progenitor cells at 4 dpf (Fig. 7G)(25,26). Following culture for 21 days in osteogenic media, inducible, Osterix-conditional YAP/TAZ deletion reduced mineral deposition stained with Alizarin Red (Fig. 7G–H). As expected, Osterix-conditional inducible YAP/TAZ deletion in vitro reduced mRNA expression of Yap and Taz as well as their canonical downstream target, Ctgf and Cyr61 in periosteal progenitor cells from YAPcKOi;TAZcKOi mice (Fig. 7I). Lastly, Osterix-conditional inducible YAP/TAZ deletion in vitro reduced mRNA expression of osteogenic differentiation genes including, Runx2, Col1a1, Alp, and Bsp while reductions in Osx (p = 0.052) did not reach statistical significance (Fig. 7J).
Given the defective periosteal osteoblast precursor expansion and osteogenic differentiation, we histologically evaluated the callus at 7 dpf. All genotypes underwent periosteal expansion and mineralization (Fig. 8A). Further, inducible Osterix-Cre-mediated YAP/TAZ deletion did not alter total callus volume but reduced mineralized callus percentage and volumetric mineral density at 7 dpf (Fig. 8B–D). Significant differences in cartilaginous callus formation, or fibrous tissue were not detected at 7 dpf (Fig. 8 E–G; Supplemental Fig. 6). Similarly, inducible Osterix-Cre-mediated YAP/TAZ deletion did not significantly alter endochondral ossification, matrix collagen, or osteogenic gene expression at 7 dpf (Supplemental Fig. 7). At 7 dpf, differences in bone area did not reach statistical significance (Fig. 8H), yet inducible Osterix-Cre-mediated YAP/TAZ deletion reduced the percentage of bone within the total callus area (Fig. 8H,I). Lastly, inducible Osterix-Cre-mediated YAP/TAZ deletion reduced OSX-positive osteoblasts per bone surface within regions of the callus undergoing intramembranous bone formation (Fig. 8J–L), consistent with our observations at 4 dpf in vivo and in vitro.
Figure 8. Adult onset-inducible, homozygous YAP/TAZ deletion from Osterix-expressing cells impaired periosteal osteoblast precursor bone formation.
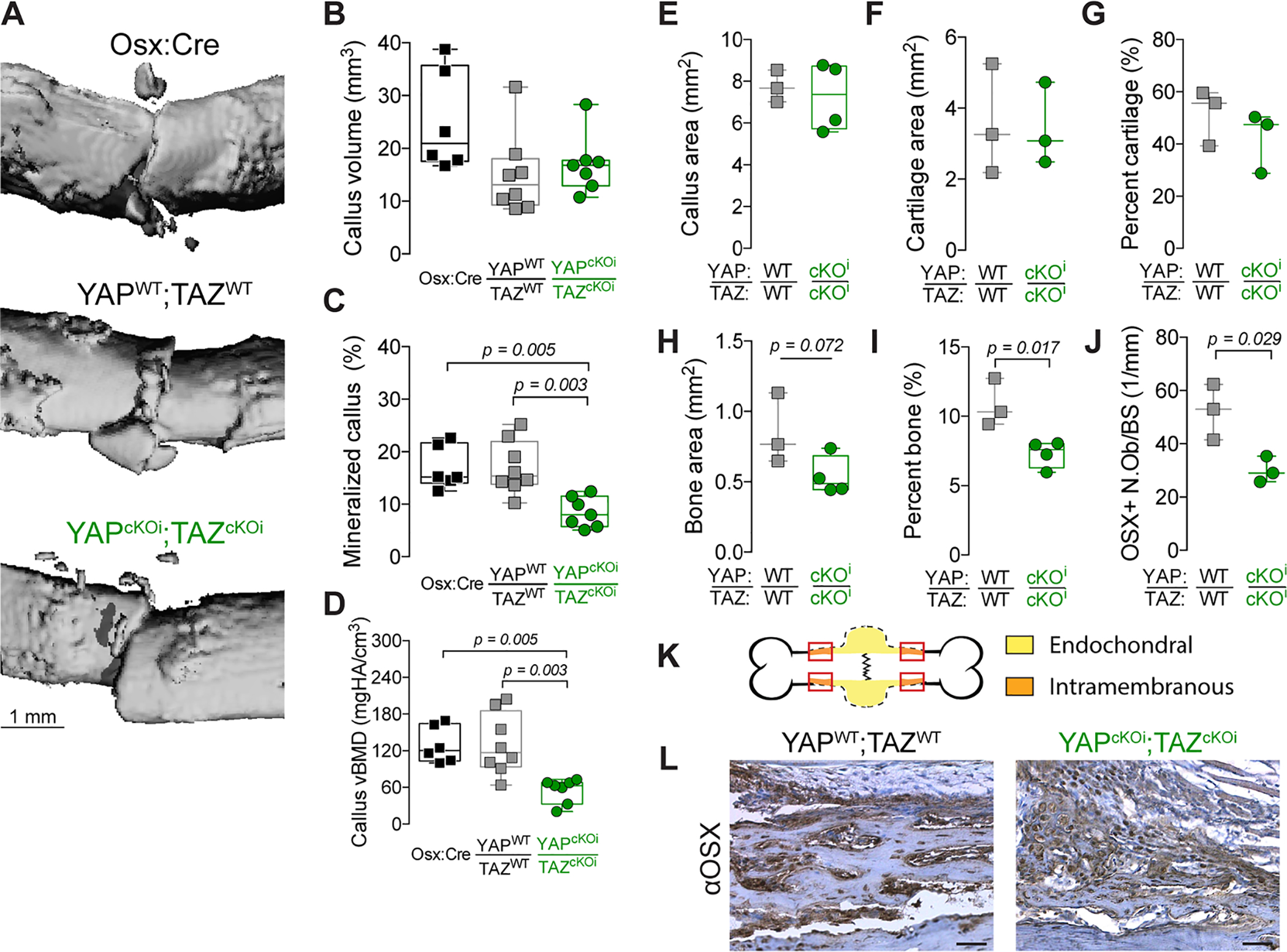
A) MicroCT reconstructions at 7 days post-fracture (dpf). Quantification of 7 dpf callus architecture: (B) callus volume, (C) mineralized callus percentage and (D) volumetric bone mineral density. Quantification of total callus histomorphometry at 7 dpf of (E) total callus area, (F) cartilage area, (G) percent cartilage, and (H) bone area, and (I) percent bone area, and (J) Osterix-positive osteoblasts per bone surface (OSX+ N.Ob/BS). K) Representation of the callus “shoulder,” where bone formation initiates by intramembranous ossification, and where we evaluated (L) anti-Osterix (αOSX) immunostaining at 7 dpf. Data are presented as individual samples in scatterplots and boxplots corresponding to the median and interquartile range. When appropriate, an independent t-test was used and p values are shown with bracketed lines. If not shown, p value for comparison > 0.05. Sample sizes, N = 3–8. Scale bars indicate 1 mm for microCT reconstructions and 50 μm for micrographs.
DISCUSSION
This study identifies new roles for the transcriptional regulators, YAP and TAZ, in bone fracture healing, adding to our understanding of periosteal osteoblast precursor cell regulation. Here, we show that YAP and TAZ promote expansion and osteoblastic differentiation of periosteal osteoblast precursors to promote bone fracture healing. Constitutive YAP and/or TAZ deletion from Osterix-expressing cells impaired bone fracture healing by reducing both callus size and subsequent mineralization, due in part developmental defects in the periosteal progenitor supply. When we allowed for the development of a normal periosteal progenitor population prior to fracture, adult onset-induced YAP/TAZ deletion did not impair callus size, but delayed mineralization as a result of impaired osteoblast precursor cell differentiation. Together, these data demonstrate that the transcriptional co-activators, YAP and TAZ, promote the expansion and differentiation of periosteal osteoblast precursors to accelerate bone fracture healing.
Fracture healing recapitulates many aspects of embryonic skeletal development, but features a unique post-natal environment, resulting in contextual differences(3). We previously found that Osterix-conditional YAP/TAZ deletion in the embryo caused a severe skeletal fragility phenotype(15), while Xiong et al. induced Osterix-conditional YAP/TAZ deletion at post-natal day 21 (P21) and performed skeletal phenotyping at P84, observing increased osteoblast numbers but no measurable effect on whole bone microarchitecture(16). Our present data resolve the differences between these two studies, establishing a critical role for YAP and TAZ in the development of the postnatal osteoprogenitor niche and demonstrating critical roles for YAP and :TAZ in osteoblast precursor proliferation and differentiation in a context of rapid bone formation, similar to that which occurs during bone development, in contrast to postnatal growth and homeostasis. The present data are further consistent with other reports. For example, deletion of the YAP/TAZ-regulated transcription factors, Snail and Slug, from Osterix-expressing cells reduced both the proliferative potential of bone surface-associated osteoprogenitors and osteogenic differentiation capacity of adult skeletal stem cells(32). Similarly, conditional deletion of YAP from Osteocalcin-expressing cells reduced osteoblast progenitor cell proliferation and differentiation, further supporting a role for YAP and TAZ in promoting osteoblast progenitor cell function(17).
Endochondral bone fracture repair includes both the formation of a cartilage template as well as subsequent osteoblast-mediated mineralization(3–5). Here, we found that while constitutive deletion impaired callus size, adult onset-inducible YAP/TAZ deletion did not significantly affect cartilage formation during fracture healing. A previous study found that YAP overexpression in developing chondrocytes, using the Col2a1 promoter, as well as deletion of MST1/2 using Dermo-Cre impaired endochondral fracture healing(33), which appears to contradict the results described here. However, both models exhibit a developmental skeletal phenotype prior to fracture(33), and in particular, observed that YAP/TAZ negatively regulate chondrogenesis. Thus, inducible targeting models are needed to decouple the developmental history from the process of fracture repair(34,35). Here, we selected the tetOFF Osterix-Cre inducible system instead of the tamoxifen-inducible Osterix-CreERT2 (36), as tamoxifen is rapidly cleared(37), resulting in transient Cre-activity(38) in newly-generated Osterix-positive cells during fracture repair. Furthermore, targeting conditional gene inactivation in chondrocytes versus osteoblast-lineage cells can result in drastically different phenotypes. For example, conditional deletion of Runx2 in chondrocytes using Col2a1-Cre phenocopied global Runx2 gene inactivation with perinatal lethality and a lack of mineralization while conditional deletion of Runx2 in osteoblasts using 2.3kb-Col1a1-Cre resulted in a moderate low bone mass phenotype(39,40). Nonetheless, hypertrophic chondrocytes are known to express Osterix during endochondral ossification(41,42) and we observed moderate Osterix-Cre-mediated YAP/TAZ recombination in hypertrophic chondrocytes, suggesting that the relative contributions of YAP and TAZ during endochondral ossification are potentially stage-dependent(15–17,33). Future studies will identify the temporal and cell-specific contributions of YAP and TAZ to bone development and repair.
Recent and ongoing studies have revealed remarkable diversity in both the cellular identity and regulatory signals that contribute to periosteal function. Gli1(43), Prx1(31), αSMA(44), cathepsin K(7), and Osterix(45) mark both overlapping and distinct periosteal progenitor cell populations, while markers previously thought to define bone marrow stromal cells, including CD73, CD90, CD105, PDGFRα, Gremlin 1, Cxcl12, and Nestin(31,46–49), also show high expression in the periosteal progenitors. Ongoing efforts continue to uncover new skeletal progenitor cell populations that contribute to fracture repair(48–52), and the intersection of this cellular diversity with YAP/TAZ signaling remains unclear. Here, we observed significant reductions in mRNA expression of endochondral-, collagen-, and osteogenesis-related gene expression signatures at 14 dpf, which can be explained either by indirect shifts in the cell populations that express these targets or by direct YAP/TAZ-mediated transcriptional regulation of those genes. Further research will be required to not only systematically identify the transcriptional co-effectors of YAP and TAZ in each cell type of interest, but also delineate the periosteal progenitor subpopulations affected by YAP/TAZ signaling.
YAP and TAZ may regulate osteoblast precursor cell proliferation and osteogenic differentiation through a variety of mechanisms. Conditional deletion of YAP in osteoblasts using Osteocalcin-Cre reduced osteoblast progenitor proliferation as well as osteogenic differentiation and proposed YAP stabilized β-catenin to promote β-catenin-mediated osteogenesis(17). However, evidence exists for YAP and TAZ playing both a positive and negative role in WNT/β-catenin signaling(53,54), suggesting that further investigating into YAP/TAZ-dependent regulation of this pathway in periosteal progenitors is needed. A similar study demonstrated that Snail and Slug form stable protein-protein complexes with both YAP and TAZ in tandem to promote osteoprogenitor proliferation and differentiation(32). In osteoprogenitors, the Snail/Slug-YAP/TAZ axis promotes proliferation by interacting with TEAD to enhance TEAD-dependent transcriptional activity and downstream expression of YAP/TAZ-TEAD target genes, such as Ctgf and Ankrd1(32). In contrast, the Snail/Slug-YAP/TAZ axis promotes osteogenic differentiation via Snail/Slug-TAZ interactions with Runx2 to promote Runx2-dependent transcriptional activity and downstream expression of osteogenic target genes, such as Osterix and Alp(32). Accordingly, evidence for TAZ interacting with Runx2 to promote downstream osteogenic gene expression in vitro is strong (30,55), but YAP has been observed to both inhibit and promote downstream osteogenic gene expression in vitro(56–59). Thus, future studies to identify the molecular mechanisms by which YAP and TAZ control periosteal progenitor expansion and differentiation are needed.
This study has several limitations. First, small sample sizes (N = 3–9) produced under-powered analyses for some fracture healing outcome measures, in particular histomorphometric analysis of cartilage. Nonetheless, we did observe statistically significant differences in osteogenic outcome measures of fracture healing in both the constitutive and inducible model, suggesting a larger effect size of Osterix-mediated YAP/TAZ deletion on osteogenesis compared to chondrogenesis during fracture healing. Regardless, not all the studies performed had sufficient power to compare sex as a variable, though we did not observe sexually dimorphic behavior for any outcome measure, and our prior assessment of YAP/TAZ regulation of bone development did not show an effect of sex(15). Second, we used an open fracture model, which may affect the kinetics and immunology of the fracture repair process(60,61). We initially began these experiments using a closed fracture model, following the Einhorn method(62), but this produced a high percentage of comminuted fractures in the constitutive, Osterix-conditional YAP and/or TAZ knockout genotypes. We therefore moved to an open fracture model in which the bending moment could be applied with lower kinetic energy. This observation suggests that YAP/TAZ deletion during development impaired the bone matrix fracture toughness, consistent with our prior report on the bone fragility phenotype(15). However, in the current study we did not observe differences in post-fracture work to maximum load or work to failure, suggesting potential differences between post-yield plastic deformation and elastic deformation of the calluses. Third, adult onset-inducible Osterix-conditional knockout increased variability in response to fracture, adding an additional layer of complexity to the already challenging study of endochondral bone fracture healing biology(5). The drug used to prevent Cre-mediated recombination, doxycycline, is a tetracycline derivative. Tetracycline exhibits high affinity for exposed mineral and is therefore commonly used as a label for dynamic bone histomorphometry(63). Potential embedding of doxycycline into the bone matrix during skeletal development and subsequent release following fracture could impair robust Cre-recombination and reduce the observed effect size for adult onset-inducible YAP/TAZ knockout mice. We recommend additional study to quantify the kinetics of tetOff inducible systems and efficiency of Cre-mediated inducible recombination in bone. Fourth, while callus size was equivalent in inducible knockout mice, we did not measure to ensure an equal length of cortical bone in samples prepared for PCR, which may contribute to the variability of observed outcomes. Lastly, the Osterix-Cre transgene is known to cause fracture callus formation(42) and defects in craniofacial development(64), depending on genetic background. However, on this background, we did not observe differences between Osx-Cre and YAPWT;TAZWT wild type mice, demonstrating phenotypic specificity for YAP/TAZ deletion.
In conclusion, this study identifies the transcriptional co-activators, YAP and TAZ, as regulators of bone fracture healing that promote periosteal osteoblast precursor proliferation and osteogenic differentiation to accelerate bone healing. Further elucidation of the mechanisms by which YAP and TAZ control the periosteal progenitor cell response to fracture may help guide the development of future targeted therapies to enhance bone fracture healing.
Supplementary Material
ACKNOWLEDGEMENTS
YAPfl/fl;TAZfl/fl mice were provided by Dr. Eric Olson (University of Texas Southwestern Medical Center). Theresa Sikorski (University of Notre Dame) provided initial mouse husbandry and maintenance. The authors declare no conflicts of interest. C.D.K., L.Q. and J.D.B. designed research; C.D.K., M.P.N., Y.M., and J.D.B. analyzed data; C.D.K., M.P.N., Y.M., H.B.P., J.H.D., and K.M.J. performed research; L.Q. contributed new reagents/analytic tools; C.D.K. and J.D.B. wrote the paper; All authors reviewed and approved the paper. C.D.K and J.D.B. take responsibility for integrity of data analysis. This work was supported by the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (NIAMS) of the National Institutes of Health through grants T32 AR007132 (to C.D.K.), P30 AR069619 (to J.D.B.), and R01 AR074948 (to J.D.B.).
Footnotes
DISCLOSURES: The authors have nothing to disclose.
REFERENCES
- 1.Musculoskeletal injuries report: incidence, risk factors and prevention. Am. Acad. Orthop. Surg 2000. [Google Scholar]
- 2.Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA. Fracture healing as a post-natal developmental process: Molecular, spatial, and temporal aspects of its regulation. J. Cell. Biochem 2003. April 1;88(5):873–84. [DOI] [PubMed] [Google Scholar]
- 3.Einhorn TA, Gerstenfeld LC. Fracture healing: Mechanisms and interventions. Nat. Rev. Rheumatol Nature Publishing Group; 2015. p. 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan EF, De Giacomo A, Gerstenfeld LC. Overview of skeletal repair (fracture healing and its assessment). Methods Mol. Biol. Humana Press Inc; 2014;1130:13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahney CS, Zondervan RL, Allison P, Theologis A, Ashley JW, Ahn J, Miclau T, Marcucio RS, Hankenson KD. Cellular biology of fracture healing [Internet]. J. Orthop. Res John Wiley and Sons Inc.; 2019. [cited 2020 Feb 23]. p. 35–50. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jor.24170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bragdon BC, Bahney CS. Origin of Reparative Stem Cells in Fracture Healing. Curr. Osteoporos. Rep Current Medicine Group LLC 1; 2018. August 1;16(4):490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debnath S, Yallowitz AR, McCormick J, Lalani S, Zhang T, Xu R, Li N, Liu Y, Yang YS, Eiseman M, Shim JH, Hameed M, Healey JH, Bostrom MP, Landau DA, Greenblatt MB. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature Nature Publishing Group; 2018. p. 133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhm AM, Dirckx N, Tower RJ, Peredo N, Vanuytven S, Theunis K, Nefyodova E, Cardoen R, Lindner V, Voet T, Van Hul M, Maes C. Activation of Skeletal Stem and Progenitor Cells for Bone Regeneration Is Driven by PDGFRβ Signaling. Dev. Cell. Cell Press; 2019. October 21;51(2):236–254. e12. [DOI] [PubMed] [Google Scholar]
- 9.Dallas SL, Xie Y, Shiflett LA, Ueki Y. Mouse Cre Models for the Study of Bone Diseases. Curr. Osteoporos. Rep Current Medicine Group LLC 1; 2018. p. 466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science (80-.). American Association for the Advancement of Science; 1995. September 8;269(5229):1427–9. [DOI] [PubMed] [Google Scholar]
- 11.Feil R, Brocard J, Mascrez B, Lemeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. U. S. A 1996. October 1;93(20):10887–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furth PA, Onge L St., Böger H, Gruss P, Gossen M, Kistner A, Bujard H, Hennighausen L. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc. Natl. Acad. Sci. U. S. A National Academy of Sciences; 1994. September 27;91(20):9302–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development [Internet]. 2006. August 15 [cited 2017 May 8];133(16):3231–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16854976 [DOI] [PubMed] [Google Scholar]
- 14.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell [Internet]. 2002. January 11 [cited 2017 May 8];108(1):17–29. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11792318 [DOI] [PubMed] [Google Scholar]
- 15.Kegelman CD, Mason DE, Dawahare JH, Horan DJ, Vigil GD, Howard SS, Robling AG, Bellido TM, Boerckel JD. Skeletal cell YAP and TAZ combinatorially promote bone development. FASEB J [Internet]. Federation of American Societies for Experimental BiologyBethesda, MD, USA; 2018. May 26 [cited 2018 May 10];32(5):2706–21. Available from: http://www.fasebj.org/doi/10.1096/fj.201700872R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong J, Almeida M, O’Brien CA. The YAP/TAZ transcriptional co-activators have opposing effects at different stages of osteoblast differentiation. Bone [Internet]. Elsevier; 2018. July 1 [cited 2018 May 10];112:1–9. Available from: https://www.sciencedirect.com/science/article/pii/S8756328218301479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan J-X, Xiong L, Zhao K, Zeng P, Wang B, Tang F-L, Sun D, Guo H, Yang X, Cui S, Xia W-F, Mei L, Xiong W-C. YAP promotes osteogenesis and suppresses adipogenic differentiation by regulating β-catenin signaling. Bone Res [Internet]. Nature Publishing Group; 2018. December 1 [cited 2019 Mar 3];6(1):18. Available from: http://www.nature.com/articles/s41413-018-0018-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rot C, Stern T, Blecher R, Friesem B, Zelzer E. A Mechanical Jack-like Mechanism Drives Spontaneous Fracture Healing in Neonatal Mice. Dev. Cell [Internet]. 2014. October 27 [cited 2017 May 10];31(2):159–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25373776 [DOI] [PubMed] [Google Scholar]
- 19.Holstein JH, Garcia P, Histing T, Kristen A, Scheuer C, Menger MD, Pohlemann T. Advances in the Establishment of Defined Mouse Models for the Study of Fracture Healing and Bone Regeneration. J. Orthop. Trauma [Internet]. 2009. May [cited 2020 Feb 23];23(SUPPL. 5):S31–8. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00005131-200905005-00008 [DOI] [PubMed] [Google Scholar]
- 20.Duvall CL, Taylor WR, Weiss D, Wojtowicz AM, Guldberg RE. Impaired Angiogenesis, Early Callus Formation, and Late Stage Remodeling in Fracture Healing of Osteopontin-Deficient Mice. J. Bone Miner. Res [Internet]. John Wiley & Sons, Ltd; 2006. November 6 [cited 2019 Sep 30];22(2):286–97. Available from: http://doi.wiley.com/10.1359/jbmr.061103 [DOI] [PubMed] [Google Scholar]
- 21.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res [Internet]. John Wiley & Sons, Ltd; 2010. June 7 [cited 2019 Jun 13];25(7):1468–86. Available from: http://doi.wiley.com/10.1002/jbmr.141 [DOI] [PubMed] [Google Scholar]
- 22.Boerckel JD, Kolambkar YM, Stevens HY, Lin ASP, Dupont KM, Guldberg RE. Effects of in vivo mechanical loading on large bone defect regeneration. J. Orthop. Res [Internet]. John Wiley & Sons, Ltd; 2012. July 1 [cited 2020 Feb 23];30(7):1067–75. Available from: http://doi.wiley.com/10.1002/jor.22042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodt MD, Ellis CB, Silva MJ. Growing C57Bl/6 Mice Increase Whole Bone Mechanical Properties by Increasing Geometric and Material Properties. J. Bone Miner. Res [Internet]. 1999. December 1 [cited 2019 Sep 30];14(12):2159–66. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10620076 [DOI] [PubMed] [Google Scholar]
- 24.Dyment NA, Jiang X, Chen L, Hong S-H, Adams DJ, Ackert-Bicknell C, Shin D-G, Rowe DW. High-Throughput, Multi-Image Cryohistology of Mineralized Tissues. J. Vis. Exp [Internet]. 2016. September 14 [cited 2018 Aug 13];(115):e54468–e54468. Available from: http://www.jove.com/video/54468/high-throughput-multi-image-cryohistology-of-mineralized-tissues [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Tower RJ, Chandra A, Yao L, Tong W, Xiong Z, Tang K, Zhang Y, Liu XS, Boerckel JD, Guo X, Ahn J, Qin L. Periosteal Mesenchymal Progenitor Dysfunction and Extraskeletally-Derived Fibrosis Contribute to Atrophic Fracture Nonunion. J. Bone Miner. Res [Internet]. John Wiley and Sons Inc.; 2019. March 1 [cited 2020 Feb 23];34(3):520–32. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jbmr.3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Matthews BG, Yu J, Novak S, Grcevic D, Sanjay A, Kalajzic I. PDGF Modulates BMP2-Induced Osteogenesis in Periosteal Progenitor Cells. JBMR Plus [Internet]. John Wiley & Sons, Ltd; 2019. May 1 [cited 2019 Sep 30];3(5):e10127. Available from: http://doi.wiley.com/10.1002/jbm4.10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guss JD, Horsfield MW, Fontenele FF, Sandoval TN, Luna M, Apoorva F, Lima SF, Bicalho RC, Singh A, Ley RE, van der Meulen MC, Goldring SR, Hernandez CJ. Alterations to the Gut Microbiome Impair Bone Strength and Tissue Material Properties. J. Bone Miner. Res [Internet]. 2017. March [cited 2017 May 8]; Available from: http://doi.wiley.com/10.1002/jbmr.3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature [Internet]. 2011. June 8 [cited 2017 May 8];474(7350):179–83. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21654799 [DOI] [PubMed] [Google Scholar]
- 29.Seo E, Basu-Roy U, Gunaratne PH, Coarfa C, Lim D-S, Basilico C, Mansukhani A. SOX2 Regulates YAP1 to Maintain Stemness and Determine Cell Fate in the Osteo-Adipo Lineage. Cell Rep [Internet]. 2013. June 27 [cited 2017 May 8];3(6):2075–87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23791527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong J-H, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a Transcriptional Modulator of Mesenchymal Stem Cell Differentiation. Science (80-.). [Internet]. 2005. [cited 2017 May 8];309(5737). Available from: http://science.sciencemag.org/content/309/5737/1074.long [DOI] [PubMed] [Google Scholar]
- 31.Duchamp De Lageneste O, Julien A, Abou-Khalil R, Frangi G, Carvalho C, Cagnard N, Cordier C, Conway SJ, Colnot C. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat. Commun Nature Publishing Group; 2018. December 1;9(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y, Feinberg T, Keller ET, Li XY, Weiss SJ. Snail/Slug binding interactions with YAP/TAZ control skeletal stem cell self-renewal and differentiation. Nat. Cell Biol Nature Publishing Group; 2016. September 1;18(9):917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Y, Wu A, Li P, Li G, Qin L, Song H, Mak KK. Yap1 Regulates Multiple Steps of Chondrocyte Differentiation during Skeletal Development and Bone Repair. Cell Rep [Internet]. 2016. March 8 [cited 2017 May 10];14(9):2224–37. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26923596 [DOI] [PubMed] [Google Scholar]
- 34.Garcia EL, Mills AA. Getting around lethality with inducible Cre-mediated excision. Semin. Cell Dev. Biol [Internet]. 2002. April [cited 2020 Mar 6];13(2):151–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12127267 [DOI] [PubMed] [Google Scholar]
- 35.Albanese C, Hulit J, Sakamaki T, Pestell RG. Recent advances in inducible expression in transgenic mice. Semin. Cell Dev. Biol Elsevier Ltd; 2002;13(2):129–41. [DOI] [PubMed] [Google Scholar]
- 36.Maes C, Kobayashi T, Kronenberg HM. A Novel Transgenic Mouse Model to Study the Osteoblast Lineage in Vivo. Ann. N. Y. Acad. Sci Blackwell Publishing Inc.; 2007. p. 149–64. [DOI] [PubMed] [Google Scholar]
- 37.Robinson SP, Langan-Fahey SM, Johnson DA, Jordan VC. Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab. Dispos 1991;19(1):36–43. [PubMed] [Google Scholar]
- 38.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. Current Biology Ltd; 1998. December 3;8(24):1323–6. [DOI] [PubMed] [Google Scholar]
- 39.Takarada T, Hinoi E, Nakazato R, Ochi H, Xu C, Tsuchikane A, Takeda S, Karsenty G, Abe T, Kiyonari H, Yoneda Y. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J. Bone Miner. Res [Internet]. John Wiley & Sons, Ltd; 2013. October 1 [cited 2020 Mar 5];28(10):2064–9. Available from: http://doi.wiley.com/10.1002/jbmr.1945 [DOI] [PubMed] [Google Scholar]
- 40.Adhami MD, Rashid H, Chen H, Clarke JC, Yang Y, Javed A. Loss of Runx2 in committed osteoblasts impairs postnatal skeletogenesis. J. Bone Miner. Res [Internet]. John Wiley and Sons Inc.; 2015. January 1 [cited 2020 Mar 5];30(1):71–82. Available from: http://doi.wiley.com/10.1002/jbmr.2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura R, Wakabayashi M, Hata K, Matsubara T, Honma S, Wakisaka S, Kiyonari H, Shioi G, Yamaguchi A, Tsumaki N, Akiyama H, Yoneda T. Osterix regulates calcification and degradation of chondrogenic matrices through matrix metalloproteinase 13 (MMP13) expression in association with transcription factor Runx2 during endochondral ossification. J. Biol. Chem [Internet]. 2012. September 28 [cited 2020 Feb 23];287(40):33179–90. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22869368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Shi Y, Regan J, Karuppaiah K, Ornitz DM, Long F. Osx-Cre Targets Multiple Cell Types besides Osteoblast Lineage in Postnatal Mice. Tjwa M, editor. PLoS One [Internet]. Public Library of Science; 2014. January 15 [cited 2017 May 10];9(1):e85161. Available from: http://dx.plos.org/10.1371/journal.pone.0085161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y, He G, Lee WC, McKenzie JA, Silva MJ, Long F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. Nature Research; 2017. December 1;8(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews BG, Grcevic D, Wang L, Hagiwara Y, Roguljic H, Joshi P, Shin D-G, Adams DJ, Kalajzic I. Analysis of αSMA-Labeled Progenitor Cell Commitment Identifies Notch Signaling as an Important Pathway in Fracture Healing. J. Bone Miner. Res [Internet]. John Wiley & Sons, Ltd; 2014. May 1 [cited 2019 Sep 30];29(5):1283–94. Available from: http://doi.wiley.com/10.1002/jbmr.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaback LA, Soung DY, Naik A, Smith N, Schwarz EM, O’Keefe RJ, Drissi H. Osterix/Sp7 regulates mesenchymal stem cell mediated endochondral ossification. J. Cell. Physiol [Internet]. 2008. January [cited 2020 Mar 1];214(1):173–82. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17579353 [DOI] [PubMed] [Google Scholar]
- 46.Méndez-Ferrer S, Michurina TV., Ferraro F, Mazloom AR, MacArthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature Nature Publishing Group; 2010. August 12;466(7308):829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenbaum A, Hsu YMS, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature [Internet]. 2013. March 14 [cited 2020 Feb 23];495(7440):227–30. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23434756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, Levin D, Schwartz MG, Uygur A, Hayakawa Y, Gross S, Renz BW, Setlik W, Martinez AN, Chen X, Nizami S, Lee HG, Kang HP, Caldwell JM, Asfaha S, Westphalen CB, Graham T, Jin G, Nagar K, Wang H, Kheirbek MA, Kolhe A, Carpenter J, Glaire M, Nair A, Renders S, Manieri N, Muthupalani S, Fox JG, Reichert M, Giraud AS, Schwabe RF, Pradere JP, Walton K, Prakash A, Gumucio D, Rustgi AK, Stappenbeck TS, Friedman RA, Gershon MD, Sims P, Grikscheit T, Lee FY, Karsenty G, Mukherjee S, Wang TC. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell [Internet]. Cell Press; 2015. January 15 [cited 2020 Feb 23];160(1–2):269–84. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25594183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morikawa S, Mabuchi Y, Kubota Y, Nagai Y, Niibe K, Hiratsu E, Suzuki S, Miyauchi-Hara C, Nagoshi N, Sunabori T, Shimmura S, Miyawaki A, Nakagawa T, Suda T, Okano H, Matsuzaki Y. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med [Internet]. 2009. October 26 [cited 2020 Feb 23];206(11):2483–96. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19841085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan CKF, Seo EY, Chen JY, Lo D, McArdle A, Sinha R, Tevlin R, Seita J, Vincent-Tompkins J, Wearda T, Lu WJ, Senarath-Yapa K, Chung MT, Marecic O, Tran M, Yan KS, Upton R, Walmsley GG, Lee AS, Sahoo D, Kuo CJ, Weissman IL, Longaker MT. Identification and specification of the mouse skeletal stem cell. Cell [Internet]. Cell Press; 2015. January 15 [cited 2020 Mar 2];160(1–2):285–98. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25594184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell [Internet]. 2010. August 17 [cited 2020 Mar 2];19(2):329–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20708594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marecic O, Tevlin R, McArdle A, Seo EY, Wearda T, Duldulao C, Walmsley GG, Nguyen A, Weissman IL, Chan CKF, Longaker MT. Identification and characterization of an injury-induced skeletal progenitor. Proc. Natl. Acad. Sci. U. S. A [Internet]. National Academy of Sciences; 2015. August 11 [cited 2020 Mar 2];112(32):9920–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26216955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 2011. April;332(6028):458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park HW, Kim YC, Yu B, Moroishi T, Mo J-S, Plouffe SW, Meng Z, Lin KC, Yu F-X, Alexander CM, Wang C-Y, Guan K-L. Alternative Wnt Signaling Activates YAP/TAZ. Cell [Internet]. 2015. [cited 2017 May 25];162(4):780–94. Available from: http://www.sciencedirect.com/science/article/pii/S0092867415008508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong J-H, Yaffe MB. TAZ: A β-Catenin-like Molecule that Regulates Mesenchymal Stem Cell Differentiation. Cell Cycle [Internet]. 2006. January 16 [cited 2017 May 8];5(2):176–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16397409 [DOI] [PubMed] [Google Scholar]
- 56.Yang B, Sun H, Chen P, Fan N, Zhong H, Liu X, Wu Y, Wang J. YAP1 influences differentiation of osteoblastic MC3T3-E1 cells through the regulation of ID1. J. Cell. Physiol John Wiley & Sons, Ltd; 2019. August;234(8):14007–18. [DOI] [PubMed] [Google Scholar]
- 57.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J [Internet]. European Molecular Biology Organization; 2004. February 25 [cited 2017 May 8];23(4):790–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14765127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan H, Xie Y, Zhang Z, Li K, Hu D, Zheng X, Fan Q, Tang T. YAP-mediated mechanotransduction regulates osteogenic and adipogenic differentiation of BMSCs on hierarchical structure. Colloids Surfaces B Biointerfaces. Elsevier; 2017. April;152:344–53. [DOI] [PubMed] [Google Scholar]
- 59.Murakami K, Kikugawa S, Kobayashi Y, Uehara S, Suzuki T, Kato H, Udagawa N, Nakamura Y. Olfactomedin-like protein OLFML1 inhibits Hippo signaling and mineralization in osteoblasts. Biochem. Biophys. Res. Commun Academic Press; 2018. October;505(2):419–25. [DOI] [PubMed] [Google Scholar]
- 60.Klein M, Stieger A, Stenger D, Scheuer C, Holstein JH, Pohlemann T, Menger MD, Histing T. Comparison of healing process in open osteotomy model and open fracture model: Delayed healing of osteotomies after intramedullary screw fixation. J. Orthop. Res [Internet]. John Wiley and Sons Inc.; 2015. July 1 [cited 2020 Mar 9];33(7):971–8. Available from: http://doi.wiley.com/10.1002/jor.22861 [DOI] [PubMed] [Google Scholar]
- 61.Haffner-Luntzer M, Kovtun A, Rapp AE, Ignatius A. Mouse Models in Bone Fracture Healing Research. Curr. Mol. Biol. Reports Springer Science and Business Media LLC; 2016. June 4;2(2):101–11. [Google Scholar]
- 62.Bonnarens F, Einhorn TA. Production of a standard closed fracture in laboratory animal bone. J. Orthop. Res [Internet]. John Wiley & Sons, Ltd; 1984. January 1 [cited 2020 Feb 23];2(1):97–101. Available from: http://doi.wiley.com/10.1002/jor.1100020115 [DOI] [PubMed] [Google Scholar]
- 63.Tam CS, Anderson W. Tetracycline labeling of bone in vivo. Calcif. Tissue Int [Internet]. 1980. [cited 2020 Mar 6];30(2):121–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/6155183 [DOI] [PubMed] [Google Scholar]
- 64.Wang L, Mishina Y, Liu F. Osterix-Cre Transgene Causes Craniofacial Bone Development Defect. Calcif. Tissue Int [Internet]. 2015. February [cited 2017 Nov 29];96(2):129–37. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25550101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


