Abstract
Soil fungi link above‐ and belowground carbon (C) fluxes through their interactions with plants and contribute to C and nutrient dynamics through the production, turnover, and activity of fungal hyphae. Despite their importance to ecosystem processes, estimates of hyphal production and turnover rates are relatively uncommon, especially in temperate hardwood forests. We sequentially harvested hyphal ingrowth bags to quantify the rates of Dikarya (Ascomycota and Basidiomycota) hyphal production and turnover in three hardwood forests in the Midwestern United States, where plots differed in their abundance of arbuscular (AM)‐ vs. ectomycorrhizal (ECM)‐associated trees. Hyphal production rates increased linearly with the percentage of ECM trees and annual production rates were 66% higher in ECM‐ than AM‐dominated plots. Hyphal turnover rates did not differ across the mycorrhizal gradient (plots varying in their abundance of AM vs. ECM trees), suggesting that the greater fungal biomass in ECM‐dominated plots relates to greater fungal production rather than slower fungal turnover. Differences in hyphal production across the gradient aligned with distinctly different fungal communities and activities. As ECM trees increased in dominance, fungi inside ingrowth bags produced more extracellular enzymes involved in degrading nitrogen (N)‐bearing relative to C‐bearing compounds, suggesting greater fungal (and possibly plant) N demand in ECM‐dominated soils. Collectively, our results demonstrate that shifts in temperate tree species composition that result in changes in the dominant type of mycorrhizal association may have strong impacts on Dikarya hyphal production, fungal community composition and extracellular enzyme activity, with important consequences for soil C and N cycling.
Keywords: Dikarya fungal biomass, ergosterol, hyphal ingrowth bags, hyphal production, hyphal turnover, mycelium, mycorrhizal type, temperate forest
Introduction
Forests mitigate the effects of climate change by sequestering carbon (C) in woody biomass and soils (Pan et al. 2011), and many of the factors that control C and nitrogen (N) cycling in ecosystems are driven by plant–microbe interactions occurring belowground (Phillips et al. 2013, Frey 2019, Lu and Hedin 2019). Free‐living saprotrophic fungi form the base of the detrital food web and perform key ecosystem services by recycling growth‐limiting nutrients such as N and stabilizing soil organic matter (SOM). Likewise, root‐associated mycorrhizal fungi can be the dominant pathway for C inputs to soil (Godbold et al. 2006, Clemmensen et al. 2013) and control C and N cycling through their effects on SOM decomposition (Talbot et al. 2008). Despite the importance of fungi to ecosystems, only a handful of studies have quantified the rates of hyphal production and turnover (e.g., Ekblad et al. 2016, Hendricks et al. 2016, Hagenbo et al. 2017), and even fewer studies have examined how fungal communities influence hyphal production and turnover across space and time (Cairney 2012, Hagenbo et al. 2018, Averill et al. 2019b ). A better understanding of how hyphal production and turnover rates vary in different forests will improve our ability to predict the magnitude and direction of feedbacks between tree species composition and soil C dynamics, both of which are essential for informing Earth System Models aimed at predicting future forest C sink.
Nearly all tree species associate exclusively with either arbuscular mycorrhizal (AM) or ectomycorrhizal (ECM) fungi (Brundrett and Tedersoo 2018), and both mycorrhizal types are sympatric across most of the temperate zone (Read 1991, Jo et al. 2019, Steidinger et al. 2019). Soils dominated by either AM‐ or ECM‐associated tree species often possess different communities of fungi (Rosling et al. 2016) and bacteria (Bahram et al. 2020, Beidler et al. 2020), which may be partly explained by mycorrhizal group variation in plant nutrient use traits (Averill et al. 2019a , b ), leaf litter quality (Cornelissen et al. 2001, Sun et al. 2018, Keller and Phillips 2019) and root litter quality (See et al. 2019). However, the ecosystem consequences of microbial community variation are poorly understood. Cheeke et al. (2017) reported that as the percentage of ECM trees in a temperate forest increase, standing fungal biomass of Dikarya (Basidiomycota, Ascomycota) in soil increases, suggesting that soil fungi may be the primary drivers of the observed biogeochemical variation. Whether the greater fungal biomass detected in ECM soils is driven by ectomycorrhizal or saprotrophic fungi is not clear. In a recent trenching experiment in a temperate hardwood forest (Indiana, USA) in which hyphal ingrowth bags were inserted into PVC tubes to create a mycorrhizal and root‐free environment, phospholipid fatty acid analysis (PLFA) showed that saprotrophic fungi produced almost twice as much mycelia in ECM stands relative to AM stands (Midgley and Phillips 2019). However, another study (Quebec, Canada) showed that saprotrophic taxa were more abundant in temperate forest plots dominated by AM‐associated tree species relative to ECM‐associated tree species (Carteron et al. 2020).
It is also unclear whether the greater standing fungal biomass reported in ECM soils (Cheeke et al. 2017) results from higher rates of hyphal production or slower hyphal turnover (i.e., rate at which hyphal biomass is replaced per unit time; Finlay and Clemmensen 2017). Studies of mycorrhizal fungi suggest a relatively slow turnover rate (months to years) for ECM fungal biomass (Treseder et al. 2004, 2005, Pritchard et al. 2008), while turnover rates for AM fungal biomass are presumed to be much shorter (days to weeks; Staddon et al. 2003, Vargas and Allen 2008), although a recent study suggests AM turnover rates on the order of weeks to months (Schäfer et al. 2019). Moreover, differences in fungal biomass may relate to saprotrophic fungi, as mycorrhizal fungi represent only part of the total fungal community in soil. Turnover for saprotrophic fungal biomass appears to overlap with mycorrhizal fungi, and may range from weeks to months (Moorhead and Reynolds 1992, Rousk and Bååth 2007). However to date, there have been no comparisons of hyphal production and turnover rates across stands varying in relative abundance of AM‐ vs. ECM‐associated tree species in the same forest.
Differences in the relative dominance of different fungal guilds may be responsible, in part, for the observed differences in C, N, and P cycling between AM‐ and ECM‐dominated stands (Phillips et al. 2013, Lin et al. 2016, Rosling et al. 2016, Craig et al. 2018). For example, AM, ECM, and saprotrophic fungi differ in a number of traits (Tedersoo and Bahram 2019) including hyphal exploration strategy (Agerer 2001), which could have consequences for biomass and necromass build‐up and turnover in soil. Ectomycorrhizal fungi have a variety of hyphal growth strategies, including both short‐distance and long‐distance explorers (Agerer 2001), while AM fungi are thought to explore shorter distances (Churchland and Grayston, 2014). Saprotrophic fungi vary in their hyphal extension rate, e.g., ranging from 0.2 to 10.8 mm/d (Maynard et al. 2019). This variation in hyphal exploration type and/or extension rate may lead to variation in fungal biomass production in ECM relative to AM forests. Moreover, melanized cell walls in ECM species such as Cenococcum geophilum are resistant to decay (Fernandez and Koide 2014), allowing buildup of ECM necromass in soil, which could increase soil C (Fernandez et al. 2019, Beidler et al. 2020), at least in the short term. Saprotrophic fungi and many ECM fungi also produce extracellular enzymes that release nutrients from SOM (Talbot et al. 2013), a process that reduces soil C and recycles N (Lindahl and Tunlid 2015). Given the shifts in dominance of AM‐ vs. ECM‐associated tree species that are currently occurring across the temperate forest region due to changing climate, N deposition, and fire suppression (Averill et al. 2018, Jo et al. 2019), it is critical to understand how hyphal production and turnover rates vary in forests that differ in tree species composition (Frey 2019). However, investigations of how shifts in fungal communities influence hyphal production and activity in forests that contain both ECM‐ and AM‐associated tree species are rare, and most previous studies have been conducted in pine forests, where ECM dominate (Cairney 2012).
In this study, we utilized sequentially harvested hyphal ingrowth bags to quantify Dikarya biomass and variation in hyphal production and turnover rates across “mycorrhizal gradients” of plots differing in abundance of AM‐ and ECM‐associated tree species in Indiana, USA, to assess drivers of standing fungal biomass in soil. To explore links between fungal communities and C and nutrient cycling, and to identify key taxonomical and functional groups contributing to hyphal production and turnover, we analyzed fungal community composition and quantified extracellular enzyme activity in both ingrowth bags and in soil. We hypothesized that (1) plots dominated by ECM‐associated tree species would have greater Dikarya hyphal production but similar turnover rates compared to plots dominated by AM‐associated trees, (2) Dikarya hyphal production in ingrowth bags would be dominated by ECM fungi in plots dominated by ECM‐associated trees, and would be dominated by saprotrophic fungi in plots dominated by AM‐associated trees (e.g., Hagenbo et al. 2019), (3) mycorrhizal communities detected in ingrowth bags would be similar to those found in soil, and that (4) the enzymatic activities of the fungi growing inside the bags would follow the patterns observed in bulk soil: greater N‐degrading enzymes in ECM‐dominated plots and C‐degrading enzymes in AM‐dominated plots (Cheeke et al. 2017).
Methods
Site description
This study was conducted across mycorrhizal forest gradients in three temperate hardwood forests in central Indiana, USA. Griffy Woods (GW) is a 75‐ha research forest within the Griffy Nature Preserve in Monroe County (39.19441 N 86.50923 W), with trees approximately 70–80 yr old. Morgan Monroe State Forest (MMSF) is an AmeriFlux site located in south‐central Indiana (39.32325 N 86.41318 W), with trees that are approximately 80 yr old. Lilly‐Dickey Woods (LDW) is a 223‐ha forest in Brown County (39.23982 N 86.21858 W), with trees that are > 150 yr old (Johnson et al. 2018). Although previous work showed that standing fungal biomass based on ergosterol, PLFA, and qPCR was greatest in the oldest forest (LDW), all three forests showed similar patterns of fungal biomass in soil across their respective mycorrhizal gradients (Cheeke et al. 2017). Soils in these forests are unglaciated, silty‐loams derived from sandstone, shale, and limestone. Central Indiana has hot, humid summers and cold, dry winters with mean annual precipitation of 1,200 mm and mean annual temperature of 11.6°C.
Within each forest, nine plots (15 × 15 m) were selected to create three replicate mycorrhizal gradients driven by variation in the dominant type of mycorrhizal association formed by tree species. There was one full mycorrhizal gradient in each forest for a total of 27 plots in the study. Plots were identified as ECM (≥70% ECM‐associated tree species by basal area), AM (≥70% AM‐associated tree species), or mixed plots (containing approximately 40–60% mixture of AM and ECM tree species). Tree species in these forests that form associations with AM fungi include sugar maple (Acer saccharum), white ash (Fraxinus americana), tulip poplar (Liriodendron tulipifera), black walnut (Juglans nigra), black cherry (Prunus serotina), and sassafras (Sassafras albidum). ECM‐associated tree species include bitternut hickory (Carya cordiformis), pignut hickory (Carya glabra), shagbark hickory (Carya ovata), American beech (Fagus grandifolia), white oak (Quercus alba), chestnut oak (Quercus prinus), northern red oak (Quercus rubra), and black oak (Quercus velutina; for more information see Table S1 in Cheeke et al. 2017).
Methods for estimating fungal biomass and characterizing fungal communities
Since no single method can accurately estimate the biomass or characterize the composition of entire soil fungal communities simultaneously (AM, ECM, and saprotrophic fungi), we focus on estimating rates of hyphal production and turnover of Dikarya (Basidiomycota and Ascomycota) and characterizing the community composition of the predominantly non‐AM fungal community across the temperate forest gradient. We do this for several reasons: (1) Cheeke et al. (2017) showed greater standing biomass of Dikarya in ECM, relative to AM plots across the forest gradient, however, it was not clear whether this pattern was driven by higher rates of hyphal production or slower rates of hyphal turnover; (2) esterified ergosterol is a fungal biomarker commonly used to quantify Dikarya biomass, however, ergosterol is known to be variable, and even absent in some AM fungi (Olsson et al. 2003); (3) the timescale at which we harvested our hyphal ingrowth bags (2‐month intervals) may have been too long to capture the turnover rates of AM fungi (previous estimates of AM turnover times range from days to months); and (4) most primers are not well suited to capture the both AM and non‐AM fungal communities simultaneously. We focus this study on Dikarya using ITS2 amplicons (AM fungal communities are often characterized using the SSU rRNA gene or portions of the LSU rRNA gene; Hart et al. 2015). Although AM fungi, a key component of the fungal community, are missing from the results presented here, we aim for this study to be a first step in elucidating potential drivers of higher standing Dikarya biomass previously reported in ECM relative to AM plots across this temperate hardwood forest gradient (Cheeke et al. 2017). For a more detailed discussion of methodological considerations, see Appendix S1.
Hyphal ingrowth bags
Hyphal ingrowth bags (2 × 10 cm, cylindrical design) were constructed using 50‐μm nylon mesh (Sefar, Buffalo, New York, USA) allowing for ingrowth of fungal hyphae but not plant roots or larger soil organisms (Wallander et al. 2001). Ingrowth bags were filled with 40 g of washed and dried silica sand (Fairmount Handy Sand, Sugarland, Texas, USA) heated to 500°C in a muffle furnace for 5 h to remove organic matter. Sand‐filled ingrowth bags have been used in a number of studies to selectively capture ECM hyphae (e.g., Hagenbo et al. 2017). The low organic matter content of the bags is thought to discriminate against saprotrophic fungi (Wallander et al. 2001, 2010, Parrent and Vilgalys 2007), although Hagenbo et al. (2019) showed that saprotrophic fungi can also colonize ingrowth bags depending on, e.g., incubation time and forest age. For further methodological considerations on the use of ingrowth bags for trapping fungal hyphae, including potential biases against short‐distance ECM hyphal exploration types, see Appendix S1.
Harvest and sampling of hyphal ingrowth bags
Ingrowth bags were installed in each plot across the replicated mycorrhizal gradients in May 2014 and sequentially harvested and replaced in 1‐ or 2‐month intervals to quantify variation in Dikarya hyphal biomass in each plot over a growing season (Fig. 1). One set of bags (i.e., “long‐term” bags) was installed in each plot in May and remained undisturbed in the soil until December (Fig. 1). Triplicate bags were buried just below the soil surface at a 45° angle (Wallander et al. 2013) in each plot, for a total of 405 ingrowth bags installed over the course of the experiment. At each harvest, new bags were inserted into the same holes to minimize disturbance and all sampling was performed in a 12 × 12 m internal plot to minimize edge effects. At each harvest, the three replicate bags in each plot were pooled into one composite sample per plot. Samples for ergosterol and DNA extraction were freeze‐dried, homogenized, and stored in the dark at room temperature and samples for enzyme assays were stored at −80°C.
Fig. 1.
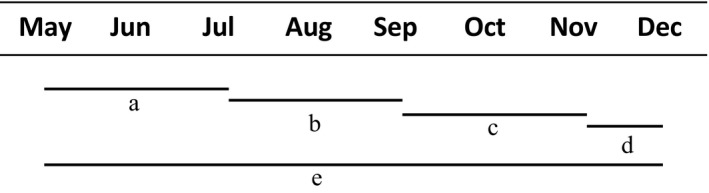
Incubation scheme for hyphal ingrowth bags installed and harvested between May and December 2014. Ingrowth bags were installed in each plot in May, harvested and replaced in July (a), September (b), and November (c), with a final harvest in December (d). Long‐term bags (e) remained in the soil from May to December. Three replicate bags were incubated in each plot and pooled before analysis to capture spatial heterogeneity within each of the 15 × 15 m forest plots. The study included three temperate hardwood forests in Indiana, USA, with replicated mycorrhizal gradients, and nine plots in each forest.
Given that sand‐filled bags may discriminate against certain fungal groups (Wallander et al. 2013), we also deployed ingrowth bags (7 × 12 cm) filled with 80% acid‐washed sand and 20% site‐specific soil (80 g of substrate per bag) in each plot in the previous year (2013) and harvested using the same harvesting scheme (Fig. 1). Correlations between fungal biomass in sand‐only and sand/soil bags, as well as fungal biomass data from the sand/soil ingrowth bags deployed over the 2013 growing season, are presented in the supplemental information (Appendix S2: Figs. S1, S2). Here, we focus on ingrowth bags with only sand to more easily compare our data with previous studies on hyphal production and turnover (e.g., Hagenbo et al. 2017).
Soil sampling
Soil samples collected in 2013 (0–5 cm) were used to compare fungal community composition in soils with the fungal community composition of ingrowth bags collected across the mycorrhizal gradients in 2013 (containing sand/soil mix) and 2014 (containing sand only). Soil samples were also collected from a subset of plots in September 2014 (0–7 cm, n = 18 plots in MMSF and LDW) to confirm that standing fungal biomass in soils collected in September 2013 was similar to those collected September 2014 (Appendix S2: Fig. S3). Data on soil properties and nutrients (pH, C, N), ergosterol concentrations, and enzyme activities were previously reported (Cheeke et al. 2017).
Quantification of fungal biomass
Dikarya fungal biomass was quantified from samples (5 g from ingrowth bags, 1 g from soil) using the fungal biomarker ergosterol, the most common sterol of Ascomycota and Basidiomycota and commonly used to estimate ECM fungal biomass in ingrowth bag studies (Wallander et al. 2013). Using established methods (Nylund and Wallander 1992), esterified ergosterol was extracted with 10% KOH in MeOH, filtered through a 0.45‐μm teflon filter, and 50 μL of each sample was analyzed using high performance liquid chromatography, with a C18 reverse‐phase column (Nova‐Pak; 3.9 × 150 mm; Waters, Milford, Connecticut, USA) preceded by a C18 reverse‐phase guard column (Nova‐Pak; 3.9 × 20 mm; Waters). The ergosterol peak was detected at 282 nm using an UV detector. Fungal biomass was determined from ergosterol concentrations using a conversion factor of 3 μg ergosterol/mg dry sample (Salmanowicz and Nylund 1988) and a correction factor (1/0.62) to compensate for unextracted ergosterol (Montgomery et al. 2000). Carbon content was estimated from fungal biomass data based on approx. 45% C concentration (Taylor et al. 2003). For more methodological considerations on the use of ergosterol for quantification of fungal biomass, see Appendix S1.
Rates of hyphal turnover and production
To determine rates of Dikarya hyphal turnover (μ), an exponential decay model was applied to hyphal biomass data from each plot across the gradient (see Ekblad et al. 2016 for model assumptions, and Hagenbo et al. 2017 for application in boreal forests). Briefly, ergosterol data was used to quantify Dikarya biomass in ingrowth bags at each harvest (bags a–d) and over the entire growing season (bag e; see Fig. 1 for harvesting scheme). We then used the Goal Seek function of Excel to estimate the rate of Dikarya hyphal turnover (μ) across the gradient, as Dikarya hyphal biomass from bags a + b + c + d is expected to be lower than the biomass in bag e due to loss by hyphal turnover (Hagenbo et al. 2017). Site‐specific correlations from a subset of nine ingrowth bags (three from across the gradient in each site) were used to estimate the turnover rate in the November–December incubation period (d), as ingrowth bags were not deployed in all plots after tree leaves had senesced. The mean residence time of Dikarya hyphae was calculated as 1/μ for each plot. Dikarya hyphal production rates (p) were determined from bags a–e using the following equation as described in Hagenbo et al., 2017: p = B(t)μ/(1 −e‐ μ t), where the change in hyphal biomass B(t) is a function of hyphal production (p), hyphal turnover (μ), and incubation time (t). Dikarya production and turnover rates from each plot (kg·ha−1·d−1) were then used to estimate annual Dikarya fungal biomass production in each plot (kg·ha−1·yr−1) assuming a 200‐d growing season (Brzostek et al. 2014).
DNA extraction, PCR amplification, and sequencing
DNA was extracted from 0.30 g (approximate dry mass) of soil and 0.50 g of material from ingrowth bags using a CTAB extraction process (Clemmensen et al. 2016). Briefly, samples were homogenized in a Precellys bead‐beating machine and 1,000 μL CTAB buffer (3% cetyltrimethylammonium bromide, 2mmol/L EDTA, 150 mmol/L Tris‐HCL, 2.5 mol/L NaCl, pH 8) was added to each tube. Samples were incubated in CTAB at 65°C for 1 h. DNA was extracted with 500 μL of chloroform and precipitated by 500 μL of isopropanol. The precipitated DNA was washed with ethanol, resuspended in double deionized water, and quantified using a NanoDrop. The internal transcribed spacer (ITS) region is the universal barcode for fungi, and ITS2 amplicons were produced using the forward primer gITS7 (GTGARTCATCGARTCTTTG; Ihrmark et al. 2012) and the two mixed reverse primers ITS4 (75%; 5′‐TCCTCCGCTTATTGATATGC‐3′; White et al. 1990) and ITS4arch (25%; 5′‐CACACGCTGTCCTCGCCTTATTGATATGC‐3′; Rosling et al. 2016), elongated with unique identification tags (Clemmensen et al. 2016). PCR was run in 50‐mL reactions with 25 μL DNA template (diluted to 0.5 ng/μL), 0.2 mmol/L of each nucleotide, 0.75 mmol/L MgCl2, forward primer at 0.5 μmol/L, reverse primer at 0.3 μmol/L and 0.5 U polymerase (DreamTaq, Thermo Fisher Scientific, Waltham, MA, USA) in PCR buffer on a 2720 Thermal Cycler (Thermo Fisher Scientific). PCR conditions were 5 minutes at 94°C, 25–35 cycles of [30 s at 95°C, 30 s at 56°C, 30 s at 72°C] and 7 minutes at 72°C. The number of cycles (25–35) was adapted for each sample to give weak to moderately strong bands on the agarose gel with approximately the same strength for all samples to avoid oversaturation and distortion of the PCR pool. Three pooled PCR replicates from each sample were purified using the AMPure XP (Beckman Coulter, Agencourt, Indianapolis, IN, USA) according to the manufacturer’s instructions, and quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific). The products were mixed in equal amounts into four pools, and cleaned using the E.Z.N.A. Cycle Pure Kit (Omega bio‐tek, Norcross, Georgia, USA). Adaptor ligation and Pacific Biosciences RSII sequencing (Pacific Biosciences, Menlo Park, California, USA) were performed by SciLifeLab (National Genomics Infrastructure, Uppsala, Sweden) using 6 SMRT cells.
Sequence analyses
Raw sequence reads from all samples together were analyzed using the bioinformatics pipeline SCATA (Ihrmark et al. 2012; available online).7 Sequences were quality filtered and screened for primers and identification tags as described in Kyaschenko et al. (2017) with some adjustments. After removal of sequences with mean quality of 20 bases and lower or containing bases with quality of 3, sequences (complementary reversed, if needed) were searched for primers and identification tags. Only sequences containing matching tags at both ends were retained. All sequences were clustered into study‐level species hypotheses (SHs; Kõljalg et al. 2013) using a 1.2% threshold distance for sequences to enter an SH. Sequence data are stored at NCBI’s Sequence Read Archive (see Data Availability). To remove non‐fungal sequences, the representative sequences from each SH were compared against GenBank nucleotide database using BLASTn, after which we used MEGAN (Huson et al. 2011) for the BLAST results and fasta file to assign the lowest common ancestor and identify sequences that were not fungal (Balint et al. 2014). The representative sequences from each study‐level SH were then compared to all global SHs using the massBLASTer through the PlutoF platform in UNITE (Abarenkov et al. 2010) and assigned to appropriate taxonomic level (at least 97% similarity was required for species‐level identification, 90% for genus, 85% for family, 80% for order, 75% for class, and 70% to division/phylum), in order of decreasing global relative abundance until 70% of the sequences were covered. Study‐level SHs were assigned to ecological functions using FUNGuild software (Nguyen et al. 2016; see Appendix S2). See Data S1 for functional groups and exploration type assignments for the 834 most common identified OTUs (Agerer 2001), and Data S2 for relative abundance in soil (2013) and ingrowth bag samples (2014).
Extracellular enzyme activity
The potential activity of five extracellular enzymes, acid phosphatase (AP), associated with phosphate mobilization; β‐N‐Acetylglucosaminidase (NAG), involved in depolymerizing organic N; β‐glucosidase (BG), associated with cellulose degradation; polyphenol oxidase (Phenox) and peroxidase (Perox), both associated with lignin degradation, was measured using established methods (Saiya‐Cork et al. 2002) from ingrowth bags harvested across the mycorrhizal gradients in July 2014. Enzyme stoichiometry (BG activity/NAG or AP activity) was used to determine the relative investment in the decomposition of C‐, N‐, or P‐containing compounds across the mycorrhizal gradients. Potential extracellular enzyme activity rates were calculated as μmol reacted substrate·g−1 soil·h−1.
Calculations and statistical analysis
Rates of Dikarya hyphal turnover (μ) were determined using the Goal Seek function of Excel (as in Hagenbo et al. 2017) and used to estimate Dikarya hyphal production rates and mean Dikarya hyphal residence times as described in the Methods. Statistical analyses were performed using Rv.3.1.2 (R Core Team, 2014). To test for significant differences in Dikarya hyphal production, turnover, and residence time, and extracellular enzyme activity across the mycorrhizal gradient, single linear regressions were performed with percentage of ECM‐associated tree species in each plot (by basal area) as the independent variable and dependent variables were Dikarya hyphal turnover rate, mean residence time, hyphal production rate, and enzyme activities of BG, AP, NAG, Phenox, and Perox. Differences in enzyme stoichiometry across the gradient were evaluated similarly using BG activity divided by NAG or AP activity in each plot (to obtain a ratio of C to nutrient‐degrading activity) as the dependent variables. Pearson’s correlations were used to determine the relationship between concentration of ergosterol and extracellular enzyme activity in the ingrowth bags. Fungal communities in hyphal ingrowth bags and soil were analyzed using multivariate methods in CANOCO 5 (Microcomputer Power, Ithaca, New York, USA). Variation of fungal community (total fungal community; 807 SHs, and ECM fungal community; 177 SHs, see Data S1 and Data S2) was visualized using detrended correspondence analysis (DCA). Separate multivariate analyses were done for hyphal ingrowth bags, soil, and ingrowth bags and soil samples together, based on the individual samples. Correlations between site, mycorrhizal type (AM, ECM, and mixed), harvest time, ergosterol concentration, and enzyme activities (soil samples only), and both the total and ECM fungal community were tested using canonical correspondence analysis (CCA) with Monte Carlo permutations. Additionally, one CCA was done for a subset of ingrowth bags (July samples) where the available enzyme activity data were included. Treatment level communities were calculated as ergosterol‐weighted averages of relative abundances in all samples within the treatment according to
where Ei is the ergosterol concentration in treatment i, Rxi relative abundance of species X in treatment i, and n is the total number of samples in a treatment, to account for different fungal biomass in different ingrowth bags. For hyphal ingrowth bags and soil separately, treatment‐level communities were calculated per plot, i.e., integrating fungal communities from all sampling times. Correlations between either mycelial production (average production per year, n = 23) or mycelial turnover (annual turnover rate, n = 26) with site and mycorrhizal type, and their relationship with both the total and ECM fungal community, was tested using redundancy analysis (RDA) with Monte Carlo permutations (total fungal community) and CCA (ECM community). The DCAs, CCAs, and RDA reported were performed with identified communities (with the exception for an overall comparison of the total communities in all ingrowth bags and the soil samples), but additional analyses (not reported) were done with total fungal communities including all study‐level SHs to confirm that they produced the same pattern as the subset of identified SHs. To compare overall distribution of fungal functional groups in soil with ingrowth bags harvested at different times and for different mycorrhizal treatments, ergosterol‐weighted averages of relative abundances of functional groups in ingrowth bags (separately for harvest times July, September, November and long‐term across all forest sites and mycorrhizal types, and separately for the mycorrhizal types AM, mixed and ECM across forest sites and harvest times) and soil (one composite value across all sites, harvest times and mycorrhizal types) were calculated using Eq. 1. Species data were arcsine transformed to achieve normal distribution before all multivariate analyses, with the exception for the RDAs where species data was log‐transformed. Rare SHs were down‐weighted in DCA and CCA of the total fungal community analysis using default settings in Canoco. Diversity measurements (SHs richness and Shannon Wiener H index) based on all study‐level SHs are reported as averages (± SE) per site × mycorrhizal type × harvest time.
Results
Effect of dominant mycorrhizal association on hyphal production, turnover, standing fungal biomass, and hyphal residence time
Dikarya hyphal biomass production increased linearly with increasing abundance of ECM‐associated tree species across the mycorrhizal gradient (Fig. 2a; Appendix S2: Table S1). Annual Dikarya hyphal production rates (based on a 200‐d growing season) were 1.7 times higher in ECM plots relative to AM plots (ECM plot mean 73.3 ± 7.4 kg·ha−1·yr−1; AM plot mean 44.2 ± 7.2 kg·ha−1·yr−1). Dikarya hyphal production rates ranged from an average of 0.22 ± 0.04 kg·ha−1·d−1 in AM plots, to 0.28 ± 0.03 kg·ha−1·d−1 in mixed plots, and 0.37 ± 0.04 kg·ha−1·d−1 in ECM plots (Appendix S2: Table S1), with an average hyphal production rate of 58.6 ± 4.5 kg·ha−1·yr−1. Estimates of C contributions from Dikarya mycelia range from an average of 1.99 ± 0.33 g C·m−2·yr−1 in AM plots, 2.56 ± 0.29 C·m−2·yr−1 in mixed plots, and 3.30 ± 0.33 g C·m−2·yr−1 in ECM plots (Appendix S2: Table S1). Estimates of standing Dikarya biomass in soil collected in September 2014 were similar to soil samples collected in September 2013 (Appendix S2: Fig. S3).
Fig. 2.
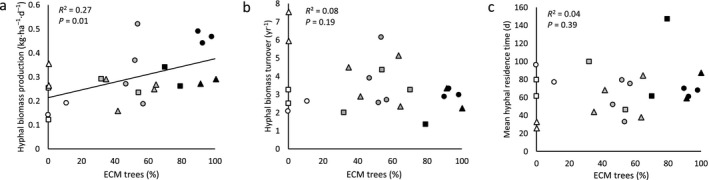
Hyphal production, turnover, and residence time in mycorrhizal gradients in the three temperate hardwood forests Griffy Woods (GW, circles), Lilly‐Dickey Woods (LDW, squares), and Morgan Monroe State Forest (MMSF, triangles) in central Indiana, USA. (a) Hyphal biomass production increased linearly with increasing abundance of ectomycorrhizal (ECM)‐associated tree species (R 2 = 0.27, P = 0.01, n = 23 plots) but there were no differences in (b) hyphal turnover rate (R 2 = 0.08, P = 0.19, n = 23 plots) or (c) hyphal residence time (R 2 = 0.04, P = 0.39, n = 23 plots) across the mycorrhizal gradients. ECM abundance was measured as a percentage of basal area. Symbols are color coded according to plot type (black symbols indicate forest plots containing ≥ 70% ECM‐associated tree species, white symbols indicate plots that contain ≥ 70% AM‐associated tree species, and gray symbols indicate mixed AM/ECM plots). Hyphal production data were collected from hyphal ingrowth bags (sand‐only) harvested sequentially across the gradient from May to December 2014. Four plots were excluded from the final analysis due to outliers or missing values at one or more of the sampling times over the growing season. Annual turnover rates are based on a 200‐d growing season.
Dikarya hyphal turnover rates did not differ across the mycorrhizal gradient (Fig. 2b; Appendix S2: Table S1) or by plot type (AM, ECM, or mixed plots; F 2,14 = 1.01, P = 0.39). Dikarya hyphal turnover rates ranged from one to eight times per year, with an average turnover rate of 3.5 ± 0.3 times per year (Fig. 2b, Appendix S2: Table S1), based on a 200‐d growing season. The mean residence time of Dikarya hyphae ranged from 26 to 147 d and did not differ significantly across the mycorrhizal gradient (Fig. 2c).
Sequencing output
Sequencing generated a total of 628,270 reads, of which 259,646 reads passed quality control (QC). After removal of non‐fungal sequences (5,679 reads, corresponding to 2.2% of reads that passed QC) and singletons the remaining 246,945 reads clustered into 5,616 study‐level SHs, out of which the most common 834 SHs (76% of the sequences in the ingrowth bags from 2014 and 70% of the sequences in soil samples from 2013) were subjected to taxonomic and functional identification (Data S1, Data S2). Each sample had an average of 281 reads (971 maximum, 200 minimum). The sequence data showed distinct differences in fungal community composition among the different forest plot types (plots dominated by AM‐associated tree species, ECM‐associated tree species, and plots containing a mixture of AM/ECM trees). See Data S3 for relative abundance for the most common identified OTUs in sand/soil ingrowth bag samples from 2013.
Fungal community composition
Ingrowth bags incubated across the mycorrhizal gradients were colonized by distinct fungal communities, with the largest difference found between the AM‐ and ECM‐dominated plots (Fig. 3). Phyla in the sequence data, hereafter referred to as the “total fungal community” (Data S1), included primarily Ascomycota and Basidiomycota (Dikarya) but also some Mucoromycota (comprises Glomeromycotina, Mortierellomycotina, and Mucoromycotina; sister to Dikarya; Spatafora et al. 2016). Total fungal communities in ingrowth bags were significantly related to mycelial production (kg−1·ha−1·d−1) for the total fungal community in each plot (RDA; not shown, Table 1; P = 0.028), but this correlation did not hold for the ECM community (CCA; not shown, Table 1). Mycelial production, site and mycorrhizal type collectively explained 28.5% of the variation in total fungal community, and 25.5% of variation in ECM community. Mycelial turnover was not related to either total fungal or ECM community composition in ingrowth bags (RDA and CCA for plot‐level community; not shown). Community composition in ingrowth bags based on all individual samples was significantly related to mycorrhizal type, harvest time, and site (CCA; Appendix S2: Fig. S4, Table S2). In AM plots Mortierella sp., Cladosporium, Ochrocladosporium, Hypocreales, Xylariales, Dothideomycetidae, and Sordariomycetes were common (Fig. 3). In ECM plots, Laccaria bicolor, Tomentella ellisii, and putative ECM Meliniomyces sp. were commonly occurring (Fig. 3), together with Amanita sp., C. geophilum, and Thelephora friulana (Appendix S2: Fig. S4). The distribution of functional groups differed among mycorrhizal treatments; ingrowth bags from ECM plots had the highest proportion of ECM fungi and also more ECM fungi compared to soil, while ingrowth bags from AM plots had more saprotrophic fungi, putative endophytes, and plant pathogens (Fig. 4). In ingrowth bags across the gradient, 177 SHs were classified as ECM. ECM community composition showed a less clear overall separation between AM and ECM plots, but when considering each site separately, the different mycorrhizal types separated and especially ECM plots differed from mixed and AM plots (Appendix S2: Fig. S5). No pattern was found regarding ECM exploration types. When ECM sequences were removed from the analysis, we saw similar patterns for ingrowth bags (Appendix S2: Fig. S6a) and soil (Appendix S2: Fig. S6b).
Fig. 3.
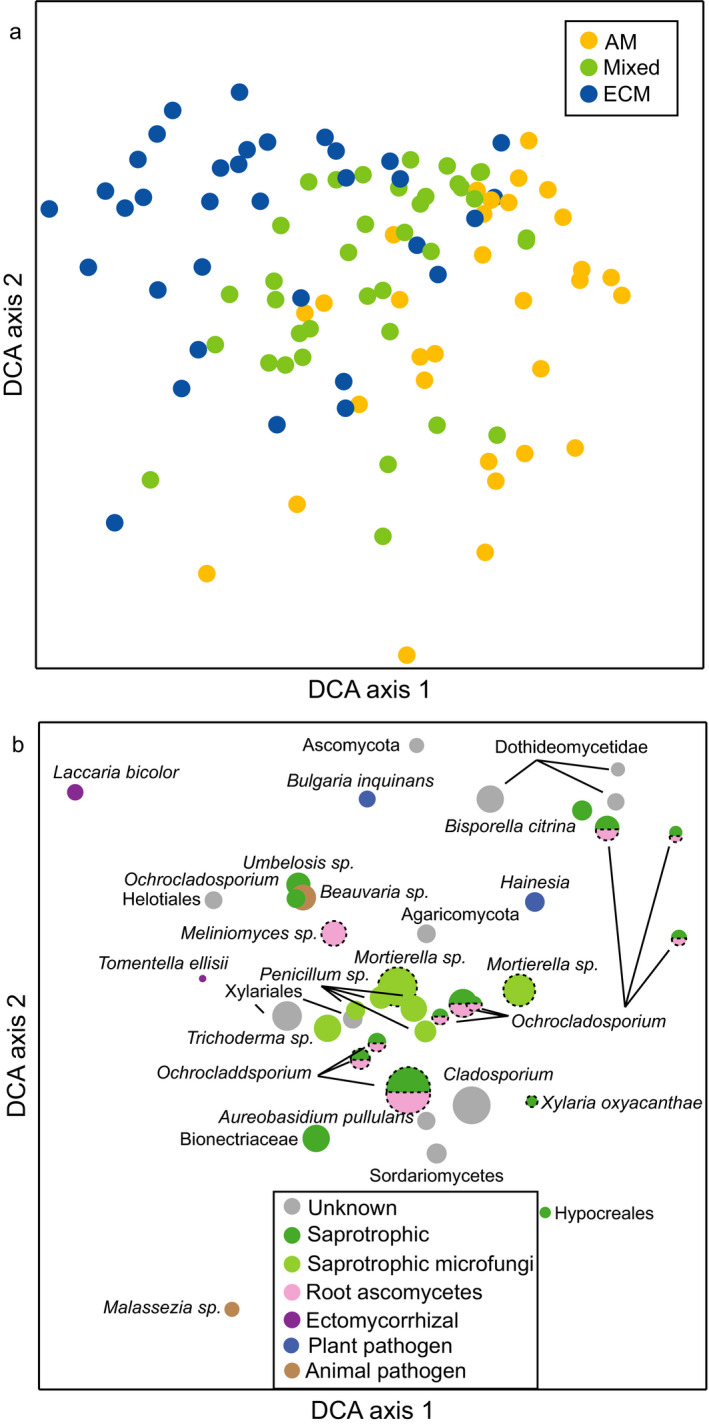
Variation in fungal community composition in ingrowth bags (sand‐only) incubated in mycorrhizal gradients with increasing abundance of ectomycorrhizal (ECM) tree species in three forests (GW, LDW, MMSF), as visualized by (a) a sample plot and (b) species plot of a detrended correspondence analysis (DCA) based on PacBio sequencing of amplified ITS2 markers. DCAs were based on 807 fungal taxa. Circles are color coded according to (a) plot type (yellow symbols represent fungal community composition in forest plots dominated by arbuscular mycorrhizal [AM]‐associated tree species, blue symbols indicate samples from plots dominated by ectomycorrhizal [ECM]‐associated tree species, and green symbols indicate plots that contain a mixture of AM and ECM associated tree species) and (b) functional groups with area indicating relative abundance of the 40 most abundant orders/genera. Circles are color coded according to functional group: gray indicates unknown functional group, dark green indicates saprotrophic fungi, light green indicates saprotrophic microfungi, pink indicates root ascomycetes, purple indicates ECM fungi, blue indicates plant pathogens, brown indicates animal pathogens, and dotted lines indicate unsure functional group classifications. Axes 1 and 2 explained 4.9% and 3.6%, respectively, of the total inertia of 6.72.
Table 1.
Effect of mycelial production, mycorrhizal type, and site on plot‐level total fungal and ectomycorrhizal (ECM) community composition in ingrowth mesh bags incubated across three mycorrhizal gradients in temperate hardwood forests (Indiana, USA).
| Factor | Total fungal community | ECM community | ||
|---|---|---|---|---|
| P | Variation explained (%) | P | Variation explained (%) | |
| Mycelial production | 0.028 | 6.8 | 0.31 | 5.1 |
| Mycorrhizal type | ||||
| AM | 0.028 | 7.6 | 0.14 | 5.5 |
| ECM | 0.040 | 6.0 | 0.70 | 4.4 |
| Mixed | 0.26 | 4.8 | 0.40 | 4.8 |
| Site | ||||
| GW | 0.035 | 6.4 | 0.078 | 5.6 |
| LDW | 0.11 | 5.4 | 0.47 | 4.8 |
| MMSF | 0.035 | 6.0 | 0.34 | 5.0 |
The effect was evaluated by redundancy analysis (RDA) of the identified total fungal communities and canonical correspondence analysis (CCA) of the identified ECM communities, and conducted on species hypothesis level (n = 807 and n = 177 for total and ECM communities, respectively). Significant values (P < 0.05) are highlighted in boldface type. AM, arbuscular mycorrhizal associated communities. Sites are identified in Methods: Site description.
Fig. 4.
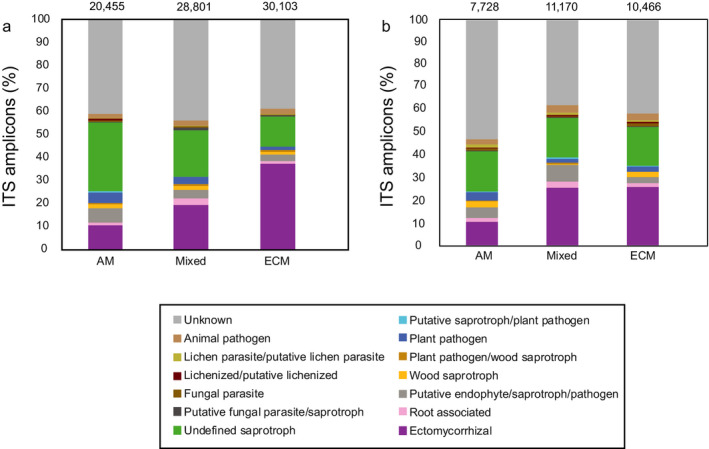
Distribution of fungal functional groups in (a) ingrowth bags (sand‐only) and (b) soil in stands dominated by arbuscular mycorrhizal (AM)‐associated tree species, mixed tree species, and ectomycorrhizal (ECM)‐associated tree species, as estimated by PacBio sequencing of amplified ITS2 markers. Abundances are given as the percentage of the identified amplicon sequences (accounting for 76% and 70% of total sequences for mesh bags and soil, respectively), and were ergosterol‐weighted averages of relative abundances per treatment, calculated across three replicate mycorrhizal gradients and different harvest times. Number of counts per treatment are given above bars.
The fungal community patterns in ingrowth bags largely mirrored those found in soil (Appendix S2: Fig. S7), with separated communities in AM‐ and ECM‐dominated plots. In soil, Russula spp. and Atheliaceae were common (Appendix S2: Fig. S7). There was higher variation in relative abundance for SHs among plots varying in mycorrhizal tree dominance (not shown), compared to ingrowth bags. Communities in soil were also significantly related to mycelial production for the total fungal community at the plot level (RDA; P = 0.019, not shown), and significantly related to the ECM community (CCA; P = 0.022, not shown). In soil, 145 of the identified SHs were classified as ECM. Turnover was not related to composition of either total soil fungal or ECM communities (RDA and CCA plot‐level community; not shown). Hyphal production, site and mycorrhizal type collectively explained 29.3% of the variation in total fungal community, and 24.6% of variation in ECM community. Community composition in soil based on all individual samples was significantly related to mycorrhizal type (AM, ECM, mixed), harvest time, site, enzyme activities (significant effects: AP; P = 0.002, Perox; P = 0.002) and fungal biomass (P = 0.002), explaining 27.8% of the variation. When comparing community composition in ingrowth bags (2014) directly with soil (2013), there was a clear separation between ingrowth bag and soil samples (Appendix S2: Fig. S8). When also including ingrowth sand/soil bags from 2013, the community composition of those mostly overlapped with soil samples (also from 2013), but with similar compositional patterns regarding mycorrhizal type (Appendix S2: Fig. S9). The separation between ingrowth bags and soil was partly explained by higher abundances in soil of, e.g., C. geophilum, SHs within Russulaceae and Atheliales (Piloderma sp.), and higher abundances in ingrowth bags of, e.g., L. bicolor, Cladosporium, Ochrocladosporium, and Dothideomycetidae (Appendix S2: Fig. S8), but not by any obvious differences in functional group distribution (Fig. 4). Functional group distribution in ingrowth bags from 2013 (sand/soil; Appendix S2: Fig. S10) were similar to the ingrowth bags containing only sand. Finally, some SHs were only found in ingrowth bags (Amanita rubescens, one Russula sp., Sebacina sp., Tomentella sp.), but were rare.
Seasonal variation in mycelial biomass in ingrowth bags and fungal community composition across the gradient
Mycelial biomass was highest in ingrowth bags harvested in July (1.05 ± 0.14 mg mycelia per bag, n = 27; Appendix S2: Fig. S11), followed by those harvested in September (0.48 ± 0.05 mg mycelia per bag, n = 27; Appendix S2: Fig. S11) and November (0.26 ± 0.05 mg mycelia per bag, n = 27; Appendix S2: Fig. S11), and was lowest in December (0.13 ± 0.03 mg mycelia per bag, n = 9). The long‐term bags that were incubated from May to December had an average of 0.53 ± 0.05 mg mycelia per bag (n = 26 plots). This corresponded to a higher proportion of ECM fungi in ingrowth bags in July compared to November (Appendix S2: Fig. S12), especially in ECM plots (Appendix S2: Fig. S13). At most harvest periods, fungal biomass (mg mycelia per bag) increased with increasing percentage of ECM‐associated tree species with the exception of bags harvested in November (Appendix S2: Fig. S11). Overall species richness (based on all study‐level SHs) in ingrowth bags increased over time (P = 0.009; July < November < September < long‐term), mostly showing the opposite pattern to mycelial biomass in ingrowth bags. In contrast, overall species richness in soil decreased over time (P < 0.001, May > July > September). When comparing average diversity between all treatment levels there were some significant effects of harvest time, mycorrhizal type, site, and for the interaction term site × harvest (Appendix S2: Fig. S14, Table S3). The distribution of functional groups in ingrowth bags varied over time; a higher proportion of ECM fungi (~30% of amplicons) was found in July and September bags compared to 12% in November and 20% in long‐term incubated bags (Appendix S2: Fig. S12).
Effect of dominant mycorrhizal association on enzyme activity and enzyme stoichiometry in ingrowth bags
Activities of NAG, AP, and BG in the ingrowth bags increased linearly with increasing abundance of ECM‐associated tree species across the mycorrhizal gradient (Fig. 5a–c). Phenox activity also increased linearly across the mycorrhizal gradient (Appendix S2: Fig. S15a) but there was no difference in Perox activity across the gradient (Appendix S2: Fig. S15b). Extracellular enzyme activities of NAG, AP, and BG were positively correlated with ergosterol concentrations in the ingrowth bags (Appendix S2: Fig. S16). Shifts in enzyme stoichiometry in hyphal ingrowth bags demonstrated that microbial investment in the degradation of C‐containing vs. N‐containing compounds decreased linearly with increasing abundance of ECM‐associated trees species across the mycorrhizal gradient (Fig. 5d). There was no difference in microbial investment in the degradation of C‐containing compounds vs. P‐containing compounds across the gradient (Fig. 5e).
Fig. 5.
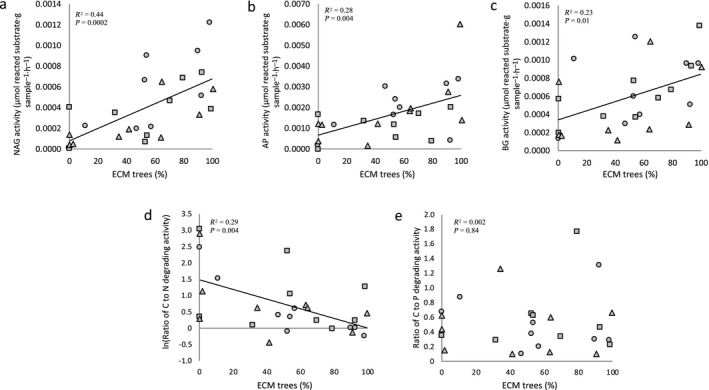
Extracellular enzyme activity of (a) β‐N‐acetylglucosaminidase (NAG, R 2 = 0.44, P = 0.0002), (b) acid phosphatase (AP, R 2 = 0.28, P = 0.004), and (c) β‐glucosidase (BG, R 2 = 0.23, P = 0.01) increased linearly with increasing dominance of ectomycorrhizal (ECM)‐associated tree species across the mycorrhizal forest gradient in Griffy Woods (circles), Lilly‐Dickey Woods (squares), and Morgan Monroe State Forest (triangles), Indiana, USA (n = 27 plots). The degradation of carbon (C)‐containing compounds vs. nitrogen (N)‐containing compounds decreased linearly with increasing ECM‐associated trees species across the mycorrhizal gradient as determined by (d) ln(BG/NAG) stoichiometry (R 2 = 0.29; P < 0.01, n = 27 plots) but there was no difference in (e) the degradation of carbon (C)‐containing compounds vs. phosphorus (P)‐containing compounds across the mycorrhizal gradient as determined by BG/AP stoichiometry (R 2 = 0.002, P = 0.84, n = 26 plots). Enzyme analyses were performed on samples from ingrowth bags (sand‐only) collected across the replicate mycorrhizal temperate forest gradients in July 2014.
Discussion
Shifts in the relative abundance of AM vs. ECM trees owing to global change (Jo et al. 2019, Steidinger et al. 2019) are likely to have profound consequences for ecosystem C and nutrient cycling given differences in how the two tree types influence soil processes (Phillips et al. 2013, Soudzilovskaia et al. 2019). Yet, while distinct biogeochemical syndromes are often apparent in AM vs. ECM‐dominated forest plots, the role of microbial communities, as opposed to variation in tree traits, is poorly understood. In this study, we investigated the degree to which variation in fungal communities (and fungal activity) can contribute to observed variation in soil C and N cycling. We found that the greater Dikarya biomass reported in ECM‐dominated forest soils relates to higher rates of Dikarya hyphal production (and not slower Dikarya hyphal turnover), and that variation in fungal communities across the landscape can have profound effects on soil C and N cycling. Of the fungi identified, we found a larger proportion of saprotrophic fungi in temperate forest stands dominated by AM‐associated tree species compared to stands dominated by ECM‐associated tree species; ECM stands were dominated by ECM fungi mixed with saprotrophic fungi. Taken together, our results provide some of the first evidence connecting the attributes of microbial communities to the observed biogeochemical differences between AM and ECM‐dominated forests.
Standing fungal biomass is determined by hyphal production, not turnover
Using hyphal ingrowth bags sequentially harvested over a growing season in three temperate hardwood forests in the Midwestern United States, we found that standing fungal biomass of Dikarya in temperate forest soils was driven by greater rates of Dikarya hyphal production, rather than slower Dikarya hyphal turnover (Fig. 2). Across a mycorrhizal gradient varying in the relative abundance of AM‐ and ECM‐associated tree species, rates of hyphal production were 66% higher in plots dominated by ECM‐associated tree species compared to plots dominated by AM‐associated tree species, confirming part of our first hypothesis. This finding is consistent with those from a trenching experiment conducted in a nearby forest, which reported greater fungal production in ECM‐dominated soils relative to AM‐dominated soils (Midgley and Phillips 2019). Despite the variation in tree species composition, dominant mycorrhizal association, and fungal community composition, we detected no differences in hyphal turnover rates or hyphal residence times across the mycorrhizal gradient. Taken together, our data show that the variation in standing fungal biomass in these temperate forests (Cheeke et al. 2017) was driven by higher rates of hyphal production, in contrast to a recent study in boreal forests in Sweden in which ECM hyphal turnover was the main driver (Hagenbo et al. 2017). One caveat of our study is that our methods did not specifically measure the effect of AM hyphal production on the variation in standing fungal biomass across the gradient or the impact of AM fungi on overall turnover rates. Thus, the absolute fungal production and/or turnover rates across the gradient may differ from those observed in the Dikarya community.
Hyphal production and turnover rates vary among forest types
Our estimates of Dikarya hyphal production (mean 58.6 ± 4.5 kg/ha per growing season) represent some of the first estimates of hyphal production rates in temperate hardwood forests. The annual hyphal production rate observed in our study was remarkably similar to that of an ECM‐dominated temperate Pinus palustris forest in Georgia, USA (hyphal production rate of 65 kg/ha per growing season; Hendricks et al. 2006) but was nearly three times lower than in ECM‐dominated boreal pine forests in Sweden (160 kg/ha per growing season; Hagenbo et al. 2017). However, hyphal turnover rates were similar between the temperate and boreal forests, with our temperate forests averaging hyphal turnover rates of 3.5 times per year (mean residence time of 67 d) and boreal pine forests averaging hyphal turnover rates of 3.2 times per year (residence time 55 d; Hagenbo et al. 2017). The turnover rates in our temperate hardwood forests and the boreal pine forests were lower than those reported in a subtropical longleaf pine forest in Georgia, USA (turnover 10 times per year, residence times of 28–52 d; Hendricks et al. 2016), and in a pine plantation in North Carolina, USA (turnover 13 times per year, residence time 28 d; Ekblad et al. 2016). However, because the latter two studies did not identify the fungi in hyphal ingrowth bags through sequencing, it is unclear if the data reported are exclusively ECM hyphae or a mixture of ECM and other fungi. Although we did not detect any differences in Dikarya hyphal turnover rates across the mycorrhizal gradient, it is possible that the saprotrophic and other fungi that grew into our mesh bags turned over at the same rate as ECM hyphae and/or that the lack of detection of AM hyphae in the mesh bags influenced our results. Because previous studies were conducted in systems dominated by ECM fungi (predominantly pines), it is unknown how the hyphal production and turnover rates we determined here compare to those of other mixed temperate hardwood forests. Future work should aim to determine how hyphal production and turnover rates vary in a greater diversity of forests, with differing tree species and understory composition, type of mycorrhizal association, geographic location, and environmental variables.
The absolute rates of Dikarya hyphal production, and their contribution to soil C inputs, are likely underestimated in our study (Appendix S2: Table S1), given the lack of organic matter and nutrients in the sand‐filled bags, and the potential bias against short‐distance and contact ECM exploration types (e.g., Russula). Values from ingrowth bags containing both sand and soil (Appendix S2: Table S4) were an order of magnitude greater (mean 20.4 ± 3.9 g C·m−2·yr−1 in AM plots and 32.6 ± 5.0 g C·m−2·yr−1 in ECM plots) than in the sand‐only ingrowth bags (mean 1.99 ± 0.33 g C·m−2·yr−1 in AM plots and 3.30 ± 0.33 g C·m−2·yr−1 in ECM plots; Appendix S2: Table S1), which is comparable to Swedish pine forests (where most ingrowth bag studies have been conducted) in which extraradical mycelia was estimated to contribute 16–42 g C·m−2·yr−1 (reviewed in Ekblad et al. 2013).
Shifts in ingrowth bag community composition correlate with hyphal production and extracellular enzyme activities
We found a significant correlation between fungal community composition and hyphal production in the ingrowth bags, suggesting that the shifting species composition underpin changes in mycelial biomass production across mycorrhizal gradients. Although we used a correlative approach to identify potential drivers of hyphal dynamics, the use of replicated mycorrhizal gradients enabled us to find clear and consistent differences in the fungal communities of Basidiomycota, Ascomycota, and Mucoromycota associated with AM‐ or ECM‐dominated hardwood forest plots. The replicated mycorrhizal gradients have previously shown consistent and strong patterns for key biogeochemical processes such as C cycling (Craig et al. 2018), N cycling (Phillips et al. 2013, Craig et al. 2019) and P cycling (Rosling et al. 2016), as well as enzyme stoichiometry (Cheeke et al. 2017). The distribution of functional groups also differed between the AM and ECM forest plots, providing support for our second hypothesis; ingrowth bags from the ECM‐dominated forest plots had the highest proportion of ECM fungi, while ingrowth bags from the AM‐dominated plots had more saprotrophic fungi, putative endophytes, and plant pathogens, reflecting the soil fungal community (supporting our third hypothesis). The higher proportion of ECM fungi may be also partly responsible for the greater biomass production we observed in plots dominated by ECM‐associated tree species. The higher proportion of Russula spp. in soil may reflect a bias of the ingrowth bags against short‐distance and contact exploration types, whereas higher Atheliales in soil than ingrowth bags may reflect their preference for organic‐rich soils.
Additionally, enzyme activities in ingrowth bags were positively correlated with increasing percentage of ECM‐associated tree species, and there was higher investment in the degradation of N‐containing compounds in ECM‐dominated plots, consistent with previous studies (Brzostek et al. 2015, Cheeke et al. 2017). The enzyme activity profiles inside the ingrowth bags appear to mirror those in soil (Cheeke et al. 2017), confirming our fourth hypothesis and suggesting that low N availability in ECM plots (Phillips et al. 2013, Craig et al. 2019) may constrain the activities of all microbes (free living and symbiotic) in ECM dominated plots. Because NAG activity is involved in the recycling of chitin, it could be associated with a shift in N source from inorganic to hyphal biomass. Increases in NAG activity across the gradient could also reflect increasing ECM fungal biomass in ECM‐ relative to AM‐dominated plots, as suggested by a recent girdling experiment. In a nearby forest (Moores Creek, Indiana, USA) when trees were girdled, NAG activity in the rhizosphere of ECM‐associated tree species decreased by 40% but there was no effect on NAG activity in the rhizosphere of AM‐associated trees (Brzostek et al. 2015). Finally, the higher abundance of certain ECM species, such as C. geophilum, in ECM plots compared to AM plots, may slow the recycling of C and N in soil, given that the melanized cell walls of this taxa are relatively resistant to decay (Fernandez and Koide 2014, Beidler et al. 2020). Thus, variation in the relative abundance of the dominant fungal taxa in AM vs. ECM forest plots could have important consequences for ecosystem C balance in temperate systems with co‐occurring tree species.
Methodological considerations: Contributions of mycorrhizal hyphae to hyphal ingrowth
In these temperate hardwood forests our PacBio sequencing results showed that the ingrowth bags were colonized by a diverse fungal community, with a large proportion of saprotrophic fungi and other functional groups present, in contrast to previous studies of ECM‐dominated coniferous forests (e.g., Hagenbo et al. 2018). Although ECM fungi were overrepresented in the hyphal ingrowth bags incubated in the ECM‐dominated plots (37.5%) compared to soils (25.8%), they did not dominate in any of the ingrowth bags. In addition, AM fungal communities were not detected in our study, even in plots dominated by AM‐associated tree species, supporting the findings of Rosling et al. (2016), who used the same primer set to characterize fungal communities in a nearby forest (Moores Creek, Indiana, USA). The methods we used here (esterified ergosterol and ITS2 amplicons) were selected to target Dikarya and are not generally used for quantifying the biomass or community composition of AM fungi. However, the gITS7 primer has successfully captured AM fungi in at least one study (Osborne et al. 2018). Thus, it is also possible that AM fungal communities were not detected in our study because AM fungi are not abundant (in terms of DNA) relative to other fungal taxa in our plots (Maeda et al. 2018, Lofgren et al. 2019), AM fungi may not preferentially colonize sand‐filled mesh bags (Wallander et al. 2013), or because we excluded roots from ingrowth bags, which may contain higher concentrations of AM fungal DNA compared to soil or sand. An additional factor that may have limited detection of AM fungi is the placement of ingrowth bags near the soil surface, where AM fungi are less common (Carteron et al. 2020).
Future studies in mixed AM/ECM forests may benefit from using a second set of primers to specifically target the LSU or SSU rRNA gene to capture the AM fungi. However, when we used the AM fungal primer pair AML1/AML2 (Lee et al. 2008) to amplify the SSU rRNA gene in soil samples collected from these plots in a previous study, amplification of AM fungal DNA was also very low (Cheeke et al. 2017). Moreover, when AM fungal biomass was measured from hyphal ingrowth cores harvested from a nearby temperate forest (Moores Creek, Indiana, USA) using PLFA, M. G. Midgley and R. P. Phillips (unpublished data) found that although AM‐dominated stands had 44% more AM fungal biomass than ECM plots (16:1ω5; PLFA biomarker for AM fungi), the total amount of AM fungal biomass detected overall was very small. This supports findings from soil collected across the mycorrhizal gradients in two of the forests studied here (Griffy Woods and Lilly‐Dickey Woods, Indiana, USA), in which 16:1ω5 concentrations were also fairly low, even in the plots dominated by AM‐associated tree species (see Fig. S4 in Cheeke et al. 2017). Thus, even if our methods had measured the AM fungal biomass in the present study, it is likely that it would have been much lower than the Dikarya biomass detected in these plots.
By using the same analytical method (esterified ergosterol) to estimate standing fungal biomass in the soil of these plots (Cheeke et al. 2017) and in the hyphal ingrowth bags in the present study, we were able to determine that the variation in standing Dikarya biomass across the mycorrhizal forest gradient was driven by higher rates of Dikarya production rather than by slower rates of Dikarya hyphal turnover. However, because ergosterol is variable or absent in many AM fungi (Olsson et al. 2003), the relative contribution of AM fungi to hyphal production and turnover rates across the gradient remains unknown.
Conclusion
We show that variation in standing Dikarya biomass in the temperate hardwood forests of the Midwestern USA is driven by higher rates of Dikarya hyphal production in ECM‐dominated relative to AM‐dominated plots, rather than by differences in Dikarya hyphal turnover rates. Hyphal production was significantly related to total fungal community composition, which clearly differed between plots dominated by AM‐ and ECM‐associated tree species. Based on these and earlier results from the same forests, we hypothesize that ECM fungi contribute more to biomass production that the saprotrophic fungi. This is also in line with the results reported by Hagenbo et al.(2018) in a northern pine forest. Higher Dikarya biomass in ECM plots across the mycorrhizal forest gradient was correlated with increased activity of enzymes involved in N acquisition in ECM relative to AM‐dominated plots. Taken together, our results demonstrate that shifts in the dominant mycorrhizal association of temperate tree species can have strong impacts on soil fungal community structure and hyphal dynamics in soil, which may impact soil C storage in both the short‐term and long‐term. This work improves understanding of the complex linkages among tree species composition, fungal community composition, and activity that is increasingly important for accurately predicting the impact of range shifts on soil processes and soil C storage.
Supporting information
Appendix S1
Appendix S2
Metadata S1
Metadata S2
Metadata S3
Data S1
Data S2
Data S3
Acknowledgments
Funding for this work was provided to T. E. Cheeke by a Carl Tryggers postdoctoral fellowship, the Knut and Alice Wallander Foundation, the Indiana Academy of Sciences, a Forest Fungal Ecology Postdoctoral Research award from the Mycological Society of America, and a National Science Foundation (NSF) Postdoctoral Fellowship in Biology (#1401729). R. P. Phillips was funded by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research, Terrestrial Ecosystem Science Program, the U.S. National Science Foundation (Ecosystem Studies Program; #1153401) and the Indiana University’s Research and Teaching Preserve (RTP). We thank Michael Chitwood, who facilitates research at IU RTP. We thank Robin Johnson, Zach Brown, Mark Sheehan, Edward Brzostek, Meghan Midgley, Andreas Hagenbo, Katarina Ihrmark, Rena Gadjieva, Maria Jonsson, Tea Ammunet, and Laura Podzikowski for their contributions to this work. P. Fransson, R. P. Phillips, and T. E. Cheeke were responsible for the conceptual framework of the study. Author contributions: R. P. Phillips established the permanent research plots. T. E. Cheeke conducted the experiment, and performed the ergosterol assays and DNA extractions. A. Kuhn assisted with fieldwork and performed the enzyme assays. P. Fransson facilitated the preparation of samples for sequencing and analyzed the sequence data. Calculations and statistical analyses were performed by T. E. Cheeke, A. Rosling, and P. Fransson. T. E. Cheeke wrote the manuscript, with all authors contributing significantly to revisions. The authors declare no conflicts of interest.
Cheeke, T. E. , Phillips R. P., Kuhn A., Rosling A., and Fransson P..2021. Variation in hyphal production rather than turnover regulates standing fungal biomass in temperate hardwood forests. Ecology 102(3):e03260. 10.1002/ecy.3260
Corresponding Editor: Serita Frey.
Footnotes
Data Availability Statement
Sequencing data are available in the NCBI Sequence Read Archive under accession number PRJNA674497.
Literature Cited
- Abarenkov, K., et al. 2010. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytologist 186:281–285. [DOI] [PubMed] [Google Scholar]
- Agerer, R. 2001. Exploration types of ectomycorrhizae—A proposal to classify ectomycorrhizal mycelial systems according to their patterns of differentiation and putative ecological importance. Mycorrhiza 11:107–114. [Google Scholar]
- Averill, C. , Bhatnagar J. M., Dietze M. C., Pearse W. D., and Kivlin S. N.. 2019a. Global imprint of mycorrhizal fungi on whole‐plant nutrient economics. Proceedings of the National Academy of Sciences USA 116:23163–23168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averill, C. , Cates L. L., Dietze M. C., and Bhatnagar J. M.. 2019b. Spatial vs. temporal controls over soil fungal community similarity at continental and global scales. ISME Journal 13:2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averill, C. , Dietze M. C., and Bhatnagar J. M.. 2018. Continental‐scale nitrogen pollution is shifting forest mycorrhizal associations and soil carbon stocks. Global Change Biology 24:4544–4553. [DOI] [PubMed] [Google Scholar]
- Bahram, M. , Netherway T., Hildebrand F., Pritsch K., Drenkhan R., Loit K., Anslan S., Bork P., and Tedersoo L.. 2020. Plant nutrient‐acquisition strategies drive topsoil microbiome structure and function. New Phytologist 227:1189–1199. [DOI] [PubMed] [Google Scholar]
- Balint, M. , Schmidt P. A., Sharma R., Thines M., and Schmitt I.. 2014. An Illumina metabarcoding pipeline for fungi. Ecology and Evolution 4:2642–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beidler, K. V. , Phillips R. P., Andrews E., Maillard F., Mushinski R. M., and Kennedy P. G.. 2020. Substrate quality drives fungal necromass decay and decomposer community structure under contrasting vegetation types. Journal of Ecology 108:1845–1859. [Google Scholar]
- Brundrett, M. C. , and Tedersoo L.. 2018. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytologist 220:1108–1115. [DOI] [PubMed] [Google Scholar]
- Brzostek, E. R. , Dragoni D., Brown Z. A., and Phillips R. P.. 2015. Mycorrhizal type determines the magnitude and direction of root‐induced changes in decomposition in a temperate forest. New Phytologist 206:1274–1282. [DOI] [PubMed] [Google Scholar]
- Brzostek, E. R. , Dragoni D., Schmid H. P., Rahman A. F., Sims D., Wayson C. A., Johnson D. J., and Phillips R. P.. 2014. Chronic water stress reduces tree growth and the carbon sink of deciduous hardwood forests. Global Change Biology 20:2531–2539. [DOI] [PubMed] [Google Scholar]
- Cairney, J. W. G. 2012. Extramatrical mycelia of ectomycorrhizal fungi as moderators of carbon dynamics in forest soil. Soil Biology & Biochemistry 47:198–208. [Google Scholar]
- Carteron, A. , Beigas M., Joly S., Turner B. L., and Laliberté E.. 2020. Temperate forests dominated by arbuscular or ectomycorrhizal fungi are characterized by strong shifts from saprotrophic to mycorrhizal fungi with increasing soil depth. Microbial Ecology. 10.1007/s00248-020-01540-7. [DOI] [PubMed] [Google Scholar]
- Cheeke, T. E. , Phillips R. P., Brzostek E. R., Rosling A., Bever J. D., and Fransson P.. 2017. Dominant mycorrhizal association of trees alters carbon and nutrient cycling by selecting for microbial groups with distinct enzyme function. New Phytologist 214:432–442. [DOI] [PubMed] [Google Scholar]
- Churchland, C. , and Grayston S. J.. 2014. Specificity of plant‐microbe interactions in the tree mycorrhizosphere biome and consequences for soil C cycling. Frontiers in Microbiology 5:222–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmensen, K. E. , Bahr A., Ovaskainen O., Dahlberg A., Ekblad A., Wallander H., Stenlid J., Finlay R. D., Wardle D. A., and Lindahl B. D.. 2013. Roots and associated fungi drive long‐term carbon sequestration in boreal forest. Science 339:1615–1618. [DOI] [PubMed] [Google Scholar]
- Clemmensen, K. E. , Ihrmark K., Durling M. B., and Lindahl B. D.. 2016. Sample preparation for fungal community analysis by high‐throughput sequencing of barcode amplicons. Pages 61–88. In Martin F. andUroz S., editors. Microbial environmental genomics (MEG), Methods in Molecular Biology, Vol. 1399. Humana Press, New York, New York, USA. [DOI] [PubMed] [Google Scholar]
- Cornelissen, J. H. C. , Aerts R., Cerabolini B., Werger M. J. A., and van der Heijden M. G. A.. 2001. Carbon cycling traits of plant species are linked with mycorrhizal strategy. Oecologia 129:611–619. [DOI] [PubMed] [Google Scholar]
- Craig, M. E. , Lovko N., Flory S. L., Wright J. P., and Phillips R. P.. 2019. Impacts of an invasive grass on soil organic matter pools vary across a tree‐mycorrhizal gradient. Biogeochemistry 144:149–164. [Google Scholar]
- Craig, M. E. , Turner B. L., Liang C., Clay K., Johnson D. J., and Phillips R. P.. 2018. Tree mycorrhizal type predicts within‐site variability in the storage and distribution of soil organic matter. Global Change Biology 24:3317–3330. [DOI] [PubMed] [Google Scholar]
- Ekblad, A. , Mikusinska A., Agren G. I., Menichetti L., Wallander H., Vilgalys R., Bahr A., and Eriksson U.. 2016. Production and turnover of ectomycorrhizal extramatrical mycelial biomass and necromass under elevated CO2 and nitrogen fertilization. New Phytologist 211:874–885. [DOI] [PubMed] [Google Scholar]
- Ekblad, A., et al. 2013. The production and turnover of extramatrical mycelium of ectomycorrhizal fungi in forest soils: role in carbon cycling. Plant and Soil 366:1–27. [Google Scholar]
- Fernandez, C. W. , Heckman K., Kolka R., and Kennedy P. G.. 2019. Melanin mitigates the accelerated decay of mycorrhizal necromass with peatland warming. Ecology Letters 22:498–505. [DOI] [PubMed] [Google Scholar]
- Fernandez, C. W. , and Koide R. T.. 2014. Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biology & Biochemistry 77:150–157. [Google Scholar]
- Finlay, R. D. , and Clemmensen K. E.. 2017. Immobilization of carbon in mycorrhizal mycelial biomass and secretions. Pages 413–440 in Johnson N. C., Gehring C., and Jansa J., editors. Mycorrhizal mediation of soil. Elsevier, Cambridge, Massachusetts, USA. [Google Scholar]
- Frey, S. D. 2019. Mycorrhizal fungi as mediators of soil organic matter dynamics. Annual Review of Ecology, Evolution, and Systematics 50:237–259. [Google Scholar]
- Godbold, D. L., et al. 2006. Mycorrhizal hyphal turnover as a dominant process for carbon input into soil organic matter. Plant and Soil 281:15–24. [Google Scholar]
- Hagenbo, A. , Clemmensen K. E., Finlay R. D., Kyaschenko J., Lindahl B. D., Fransson P., and Ekblad A.. 2017. Changes in turnover rather than production regulate biomass of ectomycorrhizal fungal mycelium across a Pinus sylvestris chronosequence. New Phytologist 214:424–431. [DOI] [PubMed] [Google Scholar]
- Hagenbo, A. , Hadden D., Clemmensen K. E., Grelle A., Manzoni S., Mölder M., Ekblad A., and Fransson P.. 2019. Carbon use efficiency of mycorrhizal fungal mycelium increases during the growing season but decreases with forest age across a Pinus sylvestris chronosequence. Journal of Ecology 107:2808–2822. [Google Scholar]
- Hagenbo, A. , Kyaschenko J., Clemmensen K. E., Lindahl B. D., and Fransson P.. 2018. Fungal community shifts underpin declining mycelial production and turnover across a Pinus sylvestris chronosequence. Journal of Ecology 106:490–501. [Google Scholar]
- Hart, M. M. , Aleklett K., Chagnon P. L., Egan C., Ghignone S., Helgason T., Lekberg Y., Opik M., Pickles B. J., and Waller L.. 2015. Navigating the labyrinth: a guide to sequence‐based, community ecology of arbuscular mycorrhizal fungi. New Phytologist 207:235–247. [DOI] [PubMed] [Google Scholar]
- Hendricks, J. J. , Mitchell R. J., Kuehn K. A., and Pecot S. D.. 2016. Ectomycorrhizal fungal mycelia turnover in a longleaf pine forest. New Phytologist 209:1693–1704. [DOI] [PubMed] [Google Scholar]
- Hendricks, J. J. , Mitchell R. J., Kuehn K. A., Pecot S. D., and Sims S. E.. 2006. Measuring external mycelia production of ectomycorrhizal fungi in the field: the soil matrix matters. New Phytologist 171:179–186. [DOI] [PubMed] [Google Scholar]
- Huson, D. H. , Mitra S., Ruscheweyh H. J., Weber N., and Schuster S. C.. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Research 21:1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrmark, K., et al . 2012. New primers to amplify the fungal ITS2 region—evaluation by 454‐sequencing of artificial and natural communities. FEMS Microbiology Ecology 82:666–677. [DOI] [PubMed] [Google Scholar]
- Jo, I. , Fei S., Oswalt C. M., Domke G. M., and Phillips R. P.. 2019. Shifts in dominant tree mycorrhizal associations in response to anthropogenic impacts. Science Advances 5:eaav6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. J. , Clay K., and Phillips R. P.. 2018. Mycorrhizal associations and the spatial structure of an old‐growth forest community. Oecologia 186:195–204. [DOI] [PubMed] [Google Scholar]
- Keller, A. B. , and Phillips R. P.. 2019. Leaf litter decay rates differ between mycorrhizal groups in temperate, but not tropical, forests. New Phytologist 222:556–564. [DOI] [PubMed] [Google Scholar]
- Kõljalg, U., et al . 2013. Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology 22:5271–5277. [DOI] [PubMed] [Google Scholar]
- Kyaschenko, J. , Clemmensen K. E., Hagenbo A., Karltun E., and Lindahl B. D.. 2017. Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. ISME Journal 11:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Lee S., and Young J. P. W.. 2008. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiology Ecology 65:339–349. [DOI] [PubMed] [Google Scholar]
- Lin, G. , McCormack M. L., Ma C., and Guo D.. 2016. Similar below‐ground carbon cycling dynamics but contrasting modes of nitrogen cycling between arbuscular mycorrhizal and ectomycorrhizal forests. New Phytologist 213:1440–1451. [DOI] [PubMed] [Google Scholar]
- Lindahl, B. D. , and Tunlid A.. 2015. Ectomycorrhizal fungi—potential organic matter decomposers, yet not saprotrophs. New Phytologist 205:1443–1447. [DOI] [PubMed] [Google Scholar]
- Lofgren, L. A. , Uehling J. K., Branco S., Bruns T. D., Martin F., and Kennedy P. G.. 2019. Genome‐based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Molecular Ecology 28:721–730. [DOI] [PubMed] [Google Scholar]
- Lu, M. , and Hedin L. O.. 2019. Global plant–symbiont organization and emergence of biogeochemical cycles resolved by evolution‐based trait modelling. Nature Ecology & Evolution 3:239–250. [DOI] [PubMed] [Google Scholar]
- Maeda, T. , Kobayashi Y., Kameoka H., Okuma N., Takeda N., Yamaguchi K., Bino T., Shigenobu S., and Kawaguchi M.. 2018. Evidence of non‐tandemly repeated rDNAs and their intragenomic heterogeneity in Rhizophagus irregularis . Communications Biology 1:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard, D. S. , Bradford M. A., Covey K. R., Lindner D., Glaeser J., Talbert D. A., Tinker P. J., Walker D. M., and Crowther T. W.. 2019. Consistent trade‐offs in fungal trait expression across broad spatial scales. Nature Microbiology 4:846–853. [DOI] [PubMed] [Google Scholar]
- Midgley, M. G. , and Phillips R. P.. 2019. Spatio‐temporal heterogeneity in extracellular enzyme activities tracks variation in saprotrophic fungal biomass in a temperate hardwood forest. Soil Biology and Biochemistry 138:107600. [Google Scholar]
- Montgomery, H. J. , Monreal C. M., Young J. C., and Seifert K. A.. 2000. Determination of soil fungal biomass from soil ergosterol analyses. Soil Biology & Biochemistry 32:1207–1217. [Google Scholar]
- Moorhead, D. L. , and Reynolds J. F.. 1992. Modeling the contributions of decomposer fungi in nutrient cycling. Pages 691–714 in Dighton J., White J., and Oudemans P., editors. The fungal community: its organization and role in the ecosystem. Marcel Dekker, Boston, Massachusetts, USA. [Google Scholar]
- Nguyen, N. H. , Song Z. W., Bates S. T., Branco S., Tedersoo L., Menke J., Schilling J. S., and Kennedy P. G.. 2016. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology 20:241–248. [Google Scholar]
- Nylund, J. E. , and Wallander H.. 1992. Ergosterol analysis as a means of quantifying mycorrhizal biomass. Methods in Microbiology 24:77–88. [Google Scholar]
- Olsson, P. A. , Larsson L., Bago B., Wallander H., and van Aarle I. M.. 2003. Ergosterol and fatty acids for biomass estimation of mycorrhizal fungi. New Phytologist 159:7–10. [DOI] [PubMed] [Google Scholar]
- Osborne, O. G. , De‐Kayne R., Bidartondo M. I., Hutton I., Baker W. J., Turnbull C. G. N., and Savolainen V.. 2018. Arbuscular mycorrhizal fungi promote coexistence and niche divergence of sympatric palm species on a remote oceanic island. New Phytologist 217:1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y. D., et al. 2011. A large and persistent carbon sink in the world's forests. Science 333:988–993. [DOI] [PubMed] [Google Scholar]
- Parrent, J. L. , and Vilgalys R.. 2007. Biomass and compositional responses of ectomycorrhizal fungal hyphae to elevated CO2 and nitrogen fertilization. New Phytologist 176:164–174. [DOI] [PubMed] [Google Scholar]
- Phillips, R. P. , Brzostek E., and Midgley M. G.. 2013. The mycorrhizal‐associated nutrient economy: a new framework for predicting carbon‐nutrient couplings in temperate forests. New Phytologist 199:41–51. [DOI] [PubMed] [Google Scholar]
- Pritchard, S. G. , Strand A. E., McCormack M. L., Davis M. A., and Oren R.. 2008. Mycorrhizal and rhizomorph dynamics in a loblolly pine forest during 5 years of free‐air‐CO2‐enrichment. Global Change Biology 14:1252–1264. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2014. R: a language and environment for statistical computing. Version 3.1.2. R Foundation for Statistical Computing, Vienna, Austria. https://www.R‐project.org/ [Google Scholar]
- Read, D. J. 1991. Mycorrhizas in ecosystems. Experientia 47:376–391. [Google Scholar]
- Rosling, A. , Midgley M. G., Cheeke T., Urbina H., Fransson P., and Phillips R. P.. 2016. Phosphorus cycling in deciduous forest soil differs between stands dominated by ecto‐ and arbuscular mycorrhizal trees. New Phytologist 209:1184–1195. [DOI] [PubMed] [Google Scholar]
- Rousk, J. , and Bååth E.. 2007. Fungal biomass production and turnover in soil estimated using the acetate‐in‐ergosterol technique. Soil Biology and Biochemistry 39:2173–2177. [Google Scholar]
- Saiya‐Cork, K. R. , Sinsabaugh R. L., and Zak D. R.. 2002. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biology & Biochemistry 34:1309–1315. [Google Scholar]
- Salmanowicz, B. B. , and Nylund J.‐E.. 1988. High performance liquid chromatography determination of ergosterolas a measure of ectomycorrhiza infection in Scots pine. European Journal of Forest Pathology 18:291–298. [Google Scholar]
- Schäfer, H. , Dannoura M., Ataka M., and Osawa A.. 2019. Decomposition rate of extraradical hyphae of arbuscular mycorrhizal fungi decreases rapidly over time and varies by hyphal diameter and season. Soil Biology and Biochemistry 136:107533. [Google Scholar]
- See, C. R. , Luke McCormack M., Hobbie S. E., Flores‐Moreno H., Silver W. L., and Kennedy P. G.. 2019. Global patterns in fine root decomposition: climate, chemistry, mycorrhizal association and woodiness. Ecology Letters 22:946–953. [DOI] [PubMed] [Google Scholar]
- Soudzilovskaia, N. A. , P. M. van Bodegom , Terrer C., Zelfde M. V. T., McCallum I., Luke McCormack M., Fisher J. B., Brundrett M. C., N. C. de Sá , and Tedersoo L.. 2019. Global mycorrhizal plant distribution linked to terrestrial carbon stocks. Nature Communications 10:5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora, J. W., et al. 2016. A phylum‐level phylogenetic classification of zygomycete fungi based on genome‐scale data. Mycologia 108:1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon, P. L. , Ramsey C. B., Ostle N., Ineson P., and Fitter A. H.. 2003. Rapid turnover of hyphae of mycorrhizal fungi determined by AMS microanalysis of C‐14. Science 300:1138–1140. [DOI] [PubMed] [Google Scholar]
- Steidinger, B. S. , et al. 2019. Climatic controls of decomposition drive the global biogeography of forest‐tree symbioses. Nature 569:404–408. [DOI] [PubMed] [Google Scholar]
- Sun, T. , Hobbie S. E., Berg B., Zhang H., Wang Q., Wang Z., and Hättenschwiler S.. 2018. Contrasting dynamics and trait controls in first‐order root compared with leaf litter decomposition. Proceedings of the National Academy of Sciences USA 115:10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, J. M. , Allison S. D., and Treseder K. K.. 2008. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Functional Ecology 22:955–963. [Google Scholar]
- Talbot, J. M. , Bruns T. D., Smith D. P., Branco S., Glassman S. I., Erlandson S., Vilgalys R., and Peay K. G.. 2013. Independent roles of ectomycorrhizal and saprotrophic communities in soil organic matter decomposition. Soil Biology & Biochemistry 57:282–291. [Google Scholar]
- Taylor, A. F. S. , Fransson P. M., Hogberg P., Hogberg M. N., and Plamboeck A. H.. 2003. Species level patterns in C‐13 and N‐15 abundance of ectomycorrhizal and saprotrophic fungal sporocarps. New Phytologist 159:757–774. [DOI] [PubMed] [Google Scholar]
- Tedersoo, L. , and Bahram M.. 2019. Mycorrhizal types differ in ecophysiology and alter plant nutrition and soil processes. Biological Reviews 94:1857–1880. [DOI] [PubMed] [Google Scholar]
- Treseder, K. K. , Allen M. F., Ruess R. W., Pregitzer K. S., and Hendrick R. L.. 2005. Lifespans of fungal rhizomorphs under nitrogen fertilization in a pinyon‐juniper woodland. Plant and Soil 270:249–255. [Google Scholar]
- Treseder, K. K. , Masiello C. A., Lansing J. L., and Allen M. F.. 2004. Species‐specific measurements of ectomycorrhizal turnover under N‐fertilization: combining isotopic and genetic approaches. Oecologia 138:419–425. [DOI] [PubMed] [Google Scholar]
- Vargas, R. , and Allen M. F.. 2008. Dynamics of fine root, fungal rhizomorphs, and soil respiration in a mixed temperate forest: Integrating sensors and observations. Vadose Zone Journal 7:1055–1064. [Google Scholar]
- Wallander, H., et al. 2013. Evaluation of methods to estimate production, biomass and turnover of ectomycorrhizal mycelium in forests soils—A review. Soil Biology & Biochemistry 57:1034–1047. [Google Scholar]
- Wallander, H. , Johansson U., Sterkenburg E., Durling M. B., and Lindahl B. D.. 2010. Production of ectomycorrhizal mycelium peaks during canopy closure in Norway spruce forests. New Phytologist 187:1124–1134. [DOI] [PubMed] [Google Scholar]
- Wallander, H. , Nilsson L. O., Hagerberg D., and Baath E.. 2001. Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytologist 151:753–760. [DOI] [PubMed] [Google Scholar]
- White, T. J. , Bruns T., Lee S., and Taylor J.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pages 315–322 in Innis D. H. G. M. A., Sninsky J. J., and White T. J., editors. PCR protocols: a guide to methods and applications. Academic Press, San Diego, California, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Appendix S2
Metadata S1
Metadata S2
Metadata S3
Data S1
Data S2
Data S3
Data Availability Statement
Sequencing data are available in the NCBI Sequence Read Archive under accession number PRJNA674497.


